The synthesis, crystal structure and some spectroscopic details for (E)-1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-(4-chlorophenyl)prop-2-en-1-one are presented.
Keywords: crystal structure, chalcone, π–π interaction
Abstract
The synthesis, crystal structure and spectroscopic analysis of (E)-1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-(4-chlorophenyl)prop-2-en-1-one (C17H13ClO3), a substituted chalcone, are described. The overall geometry of the molecule is largely planar (r.m.s. deviation = 0.1742 Å), but slightly kinked, leading to a dihedral angle between the planes of the benzene rings at either side of the molecule of 8.31 (9)°. In the crystal, only weak interactions determine the packing motifs. These include C—H⋯O and C—H⋯Cl hydrogen bonds and π–π overlap of aromatic rings.
1. Chemical context
Chalcones exhibit a wide range of fascinating biological and pharmacological properties. Some of their beneficial attributes include anti-inflammatory, antimicrobial, antifungal, antioxidant, cytotoxic and anticancer activities (Dimmock et al., 1999 ▸). Moreover, several chalcones have demonstrated significant antimalarial properties (Troeberg et al., 2000 ▸). The efficient second-harmonic generation (SHG) conversion efficiency of some chalcones has also made them promising organic nonlinear optical (NLO) materials (Sarojini et al., 2006 ▸). The presence of phenolic groups in many chalcones has garnered attention due to their radical quenching capabilities, making these compounds and chalcone-rich plant extracts potential candidates for drug development or food preservation (Dhar et al., 1981 ▸; Di Carlo et al., 1999 ▸). The design, synthesis and evaluation of 1,4-benzodioxane-substituted chalcones as selective and reversible inhibitors of human monoamine oxidase B was reported by Kong et al. (2020 ▸). Furthermore, Shinde et al. (2020 ▸) have conducted experimental and theoretical studies on the molecular structure, FT–IR, NMR, HOMO/LUMO frontier orbital, molecular electrostatic potential (MESP) and reactivity descriptors of (E)-1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one. They have also reported the synthesis, antibacterial and computational studies of halo-chalcone hybrids derived from 1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)ethan-1-one (Shinde et al., 2021 ▸) and of two trifluorinated chalcones derived from 1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)ethan-1-one (Shinde et al., 2022a
▸), as well as investigations of (E)-4-(3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-oxoprop-1-en-1-yl)benzonitrile (Shinde et al., 2022b
▸). Additionally, Zhuang et al. (2017 ▸) presented a comprehensive review on the chalcone framework as a privileged structure in medicinal chemistry, while Elkanzi et al. (2022 ▸) have published a review on the synthesis of chalcone derivatives and their biological activities.
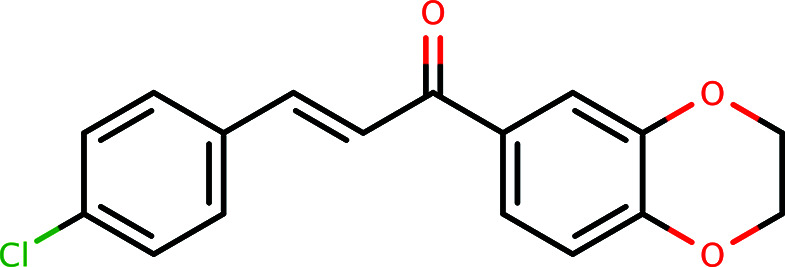
Given the general significance of chalcones, we report herein the synthesis of (E)-1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-(4-chlorophenyl)prop-2-en-1-one, C17H13ClO3 (I), along with its crystal structure and related studies.
2. Structural commentary
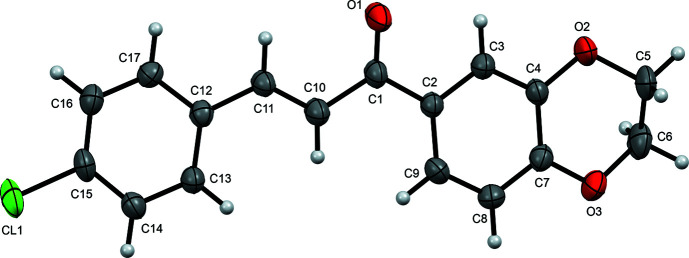
As shown in Fig. 1 ▸, the overall conformation of the molecule of I is determined by the torsion angles C3—C2—C1—C10 [169.9 (2)°], C2—C1—C10—C11 [−171.3 (3)°], C1—C10—C11—C12 [−179.0 (3)°] and C10—C11—C12—C17 [−176.7 (3)°] of the propenone moiety. In magnitude, these are all within about 10° of 180°, which makes the molecule largely flat (r.m.s. deviation for the non-H atoms is 0.1742 Å), though slightly bent, leading to a dihedral angle between the chlorophenyl and dihydrobenzodioxine rings of 8.31 (9)°. The conformation of the dihydrodioxine ring, as determined by its Cremer–Pople puckering parameters (Cremer & Pople, 1975 ▸) [Q = 0.466 (3) Å, θ = 51.2 (4)° and φ = 282.6 (4)°], is closest to half-chair (Boeyens, 1978 ▸). This puckering places atoms C5 and C6 at 0.499 (4) and −0.207 (4) Å on either side of the mean plane of the dihydrobenzodioxine ring. All bond lengths and angles lie within the typical ranges found in organic structures.
Figure 1.
The molecular structure of the title compound, showing the atom labelling and displacement ellipsoids drawn at the 50% probability level.
3. Supramolecular features
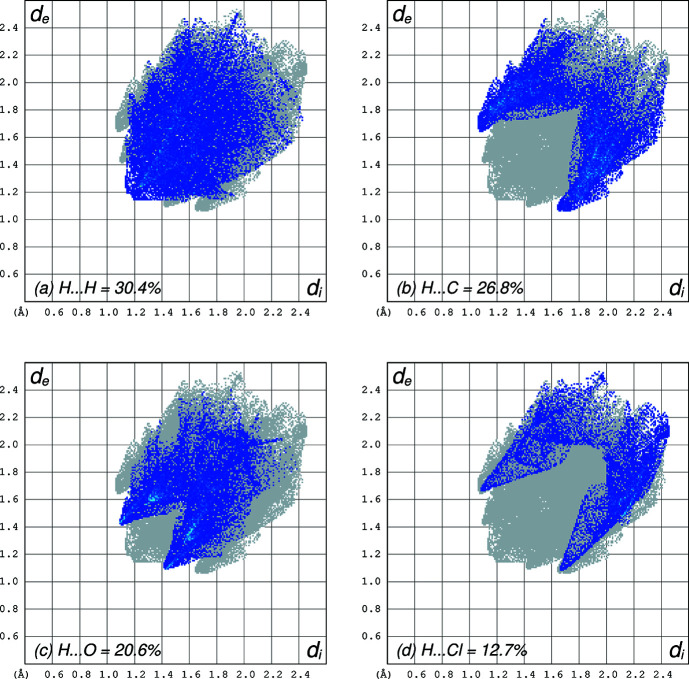
The crystal packing in I is governed solely by weak interactions, as the only possible hydrogen-bond donors are of type C—H. The absence of any sharp spikes protruding to the lower left in the Hirshfeld surface fingerprint plots (Fig. 2 ▸) calculated using CrystalExplorer (Spackman et al., 2021 ▸) clearly marks the absence of any very close contacts. Indeed, only two such contacts are flagged as intermolecular hydrogen-bond-type interactions by the default SHELXL HTAB command. These are C5—H5A⋯O3i and C5—H5B⋯Clii [symmetry codes: (i) x −
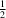 , −y +
, −y +
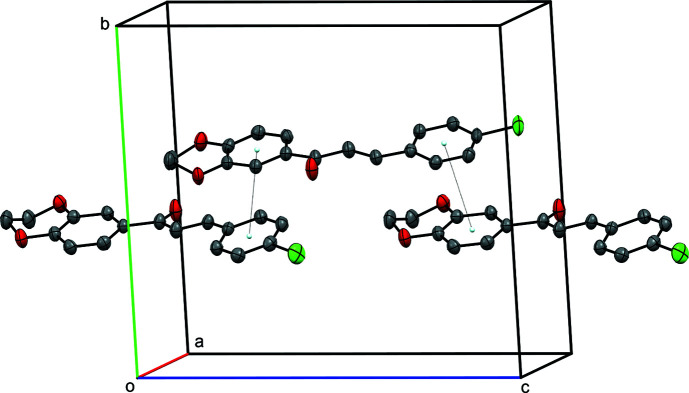
 , z; (ii) x − 1, y, z − 1], shown in Table 1 ▸. The benzene rings of the 4-chlorophenyl and dihydrobenzodioxine groups, however, exhibit π–π overlap, which link the molecules into chains that extend parallel to [001], as shown in Fig. 3 ▸. The centroid–centroid distances [3.747 (18) Å] and the dihedral angle between the participating rings [6.12 (11)°] imply that these are rather weak interactions.
, z; (ii) x − 1, y, z − 1], shown in Table 1 ▸. The benzene rings of the 4-chlorophenyl and dihydrobenzodioxine groups, however, exhibit π–π overlap, which link the molecules into chains that extend parallel to [001], as shown in Fig. 3 ▸. The centroid–centroid distances [3.747 (18) Å] and the dihedral angle between the participating rings [6.12 (11)°] imply that these are rather weak interactions.
Figure 2.
Hirshfeld surface fingerprint plots showing (a) H⋯H contacts = 30.4% coverage, (b) H⋯C = 26.8%, (c) H⋯O = 20.6% and (d) H⋯Cl = 12.7%. Reciprocal contacts are included in the coverage fractions. Other contact types were unremarkable.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5A⋯O3i | 0.99 | 2.61 | 3.595 (4) | 172 |
| C5—H5B⋯Cl1ii | 0.99 | 2.80 | 3.455 (3) | 124 |
Symmetry codes: (i)
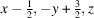 ; (ii)
; (ii)
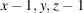 .
.
Figure 3.
A partial packing plot of I, viewed approximately down the a axis. The stacking of the aromatic rings is shown by dotted lines between ring centroids, which lead to chains of molecules that extend parallel to [001].
4. Database survey
A search of the Cambridge Structural Database (CSD, Version 5.43 with updates to November 2022; Groom et al., 2016 ▸) using a molecular fragment consisting of chalcone (C6H5C=OCH=CHC6H5) with no specific substituents returned 4725 hits. With hydrogen attached across the C=C double bond, the search gave 1200 hits. Of these entries, 155 had ‘any halogen’ at the equivalent of C15 in I, 62 of which had Cl. A search fragment without the halogen but with ‘any oxygen bound’ substituent at the equivalent of C4 and C7 produced 78 matches. Addition of the aforementioned halogen reduced this to 14 structures, seven of which were chlorides, but only five are unique. These are: BOJFIQ [(2E)-1-(1,3-benzodioxol-5-yl)-3-(4-chlorophenyl)prop-2-en-1-one; Jasinski et al., 2008 ▸], FATFIR [(E)-3-(2,4-dichlorophenyl)-1-(3,4,5-trimethoxyphenyl)prop-2-en-1-one; Wu et al., 2012 ▸], TICDIT [3-(4-chlorophenyl)-1-(3,4-dimethoxyphenyl)prop-2-en-1-one; Teh et al., 2007 ▸], VIDDIW [3-(2,4-dichlorophenyl)-1-(3,4-dimethoxyphenyl)prop-2-en-1-one; Ng et al., 2007 ▸] and XOLLOC [3-(4-chloro-3-fluorophenyl)-1-(3,4-dimethoxyphenyl)prop-2-en-1-one; Çelikesir et al., 2019 ▸]. A few other related chalcone structures include: QERYOC [(2E)-3-(1,3-benzodioxol-5-yl)-1-(4-bromophenyl)prop-2-en-1-one; Harrison et al., 2006 ▸], KUYWOR [(2E)-3-(1,3-benzodioxol-5-yl)-1-(3-bromo-2-thienyl)prop-2-en-1-one; Harrison et al., 2010 ▸], TUNTAY [(2E)-1-(1,3-benzodioxol-5-yl)-3-(2-bromophenyl)prop-2-en-1-one; Li et al., 2010 ▸], and UNUZUZ {(E)-3-(8-benzyloxy-2,3-dihydro-1,4-benzodioxin-6-yl)-1-[2-hydroxy-4,6-bis(methoxymethoxy)phenyl]prop-2-en-1-one; Zhang et al., 2011 ▸}.
5. Synthesis, crystallization, and spectroscopic data
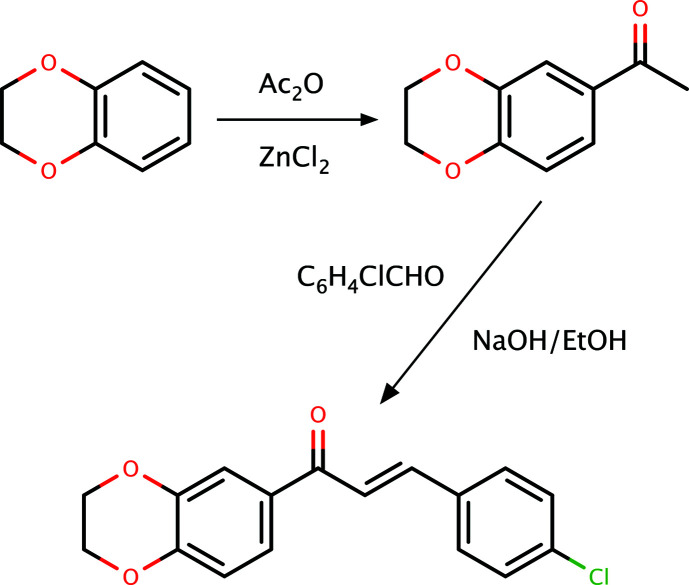
1-(2,3-Dihydrobenzo[1,4]dioxin-6-yl)ethanone was prepared by Friedel–Crafts acylation of benzo[1,4]dioxane using ether as solvent (Fig. 4 ▸). 1-(2,3-Dihydrobenzo[1,4]dioxin-6-yl)ethanone (1.78 g, 0.01 mol) and 4-chlorobenzaldehyde (1.40 g, 0.01 mol) were stirred in a 30% ethanolic NaOH and water mixture at 293 K for 4–6 h. The reaction mixture was refrigerated overnight. The precipitate that formed was filtered off and dried. X-ray-quality crystals were obtained from a solvent mixture of DMF–DMSO (dimethylformamide–dimethyl sulfoxide) (1:1 v/v) and the corresponding melting point was found to be 410–411 K.
Figure 4.
A generalized reaction scheme for the synthesis of I.
FT–IR (ν in cm−1): 3156–2934 (Ar-CH), 1655 (C=O), 1575 (C=C); 1H NMR (CDCl3, 400 MHz): δ 7.71 (d, 1H, β-CH), 7.92 (d, 2H, Ar-H), 7.62 (d, 2H, Ar-H), 7.23 (d, 1H, 2,3-dihydrobenzo[1,4]dioxine Ar-H), 7.21 (d, 1H, 2,3-dihydrobenzo[1,4]dioxine Ar-H), 7.01 (d, 1H, α-CH), 6.93 (s, 1H, 2,3-dihydrobenzo[1,4]dioxine Ar-H), 4.14 (m, 4H, 1,4-dioxane CH2); 13C NMR (CDCl3, 100 MHz): δ 196.1, 156.5, 149.8, 145.1, 133.5, 133.3, 129.0, 128.7, 122.1, 121.4, 121.3, 112.0, 106.8, 64.2.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were found in difference Fourier maps and subsequently included in the refinement using riding models, with constrained distances set to 0.95 [C(sp 2)H] and 0.99 Å (R 2CH2). U iso(H) values were set to 1.2U eq of the attached atom. The absolute structure was determined using 1035 quotients of the form [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C17H13ClO3 |
| M r | 300.72 |
| Crystal system, space group | Orthorhombic, P n a21 |
| Temperature (K) | 200 |
| a, b, c (Å) | 5.8655 (5), 14.3499 (17), 16.4803 (19) |
| V (Å3) | 1387.1 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.28 |
| Crystal size (mm) | 0.26 × 0.23 × 0.22 |
| Data collection | |
| Diffractometer | Bruker D8 Venture dual source |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.788, 0.971 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 17057, 2896, 2694 |
| R int | 0.038 |
| (sin θ/λ)max (Å−1) | 0.651 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.033, 0.083, 1.07 |
| No. of reflections | 2896 |
| No. of parameters | 190 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.21, −0.16 |
| Absolute structure | Flack x determined using 1035 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.01 (2) |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989023005613/vm2285sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023005613/vm2285Isup2.hkl
CCDC reference: 2272334
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
One of the authors (V) is grateful to the DST–PURSE Project, Vijnana Bhavana, UOM for providing research facilities. HSY thanks UGC for a BSR Faculty fellowship for three years.
supplementary crystallographic information
Crystal data
| C17H13ClO3 | Dx = 1.440 Mg m−3 |
| Mr = 300.72 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 9943 reflections |
| a = 5.8655 (5) Å | θ = 2.5–27.6° |
| b = 14.3499 (17) Å | µ = 0.28 mm−1 |
| c = 16.4803 (19) Å | T = 200 K |
| V = 1387.1 (3) Å3 | Cut block, pale yellow |
| Z = 4 | 0.26 × 0.23 × 0.22 mm |
| F(000) = 624 |
Data collection
| Bruker D8 Venture dual source diffractometer | 2896 independent reflections |
| Radiation source: microsource | 2694 reflections with I > 2σ(I) |
| Detector resolution: 7.41 pixels mm-1 | Rint = 0.038 |
| φ and ω scans | θmax = 27.6°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −7→7 |
| Tmin = 0.788, Tmax = 0.971 | k = −18→18 |
| 17057 measured reflections | l = −19→21 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | H-atom parameters constrained |
| wR(F2) = 0.083 | w = 1/[σ2(Fo2) + (0.0322P)2 + 0.4666P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 2896 reflections | Δρmax = 0.21 e Å−3 |
| 190 parameters | Δρmin = −0.16 e Å−3 |
| 1 restraint | Absolute structure: Flack x determined using 1035 quotients [(I+)-(I-)]/[(I+)+(I-)] [Parsons et al. (2013) |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.01 (2) |
Special details
| Experimental. The crystal was mounted using polyisobutene oil on the tip of a fine glass fibre, which was fastened in a copper mounting pin with electrical solder. It was placed directly into the cold gas stream of a liquid-nitrogen based cryostat (Hope, 1994; Parkin & Hope, 1998). The crystals appeared to undergo a destructive phase transition when cooled to 90K. Visual inspection of crystal integrity and diffraction quality vs temperature established a safe temperature for data collection of -73° C. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement progress was checked using Platon (Spek, 2020) and by an R-tensor (Parkin, 2000). The final model was further checked with the IUCr utility checkCIF. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.86056 (16) | 0.65710 (6) | 0.91506 (5) | 0.0577 (3) | |
| O1 | −0.0096 (3) | 0.59370 (18) | 0.48876 (12) | 0.0505 (6) | |
| O2 | 0.0030 (3) | 0.57318 (14) | 0.18914 (12) | 0.0416 (5) | |
| O3 | 0.4399 (4) | 0.64129 (16) | 0.14256 (12) | 0.0443 (5) | |
| C1 | 0.1903 (5) | 0.6112 (2) | 0.47247 (15) | 0.0338 (6) | |
| C2 | 0.2634 (4) | 0.61897 (18) | 0.38646 (15) | 0.0286 (5) | |
| C3 | 0.1080 (4) | 0.59307 (18) | 0.32641 (17) | 0.0318 (5) | |
| H3 | −0.038161 | 0.570520 | 0.341626 | 0.038* | |
| C4 | 0.1637 (4) | 0.59976 (17) | 0.24503 (16) | 0.0300 (5) | |
| C5 | 0.0414 (6) | 0.6143 (2) | 0.11107 (17) | 0.0453 (7) | |
| H5A | 0.008461 | 0.681919 | 0.113519 | 0.054* | |
| H5B | −0.062554 | 0.585853 | 0.070731 | 0.054* | |
| C6 | 0.2829 (6) | 0.5995 (2) | 0.08557 (18) | 0.0475 (8) | |
| H6A | 0.313824 | 0.531780 | 0.081738 | 0.057* | |
| H6B | 0.306758 | 0.627076 | 0.031178 | 0.057* | |
| C7 | 0.3778 (5) | 0.63300 (19) | 0.22209 (16) | 0.0320 (6) | |
| C8 | 0.5313 (5) | 0.66010 (18) | 0.28102 (18) | 0.0343 (6) | |
| H8 | 0.675979 | 0.683954 | 0.265581 | 0.041* | |
| C9 | 0.4761 (5) | 0.65280 (18) | 0.36243 (17) | 0.0329 (6) | |
| H9 | 0.584046 | 0.671004 | 0.402365 | 0.040* | |
| C10 | 0.3628 (5) | 0.62304 (19) | 0.53720 (16) | 0.0361 (6) | |
| H10 | 0.511960 | 0.643759 | 0.523877 | 0.043* | |
| C11 | 0.3090 (5) | 0.60465 (18) | 0.61436 (15) | 0.0314 (6) | |
| H11 | 0.158246 | 0.583195 | 0.624038 | 0.038* | |
| C12 | 0.4563 (4) | 0.61380 (17) | 0.68570 (15) | 0.0297 (5) | |
| C13 | 0.6738 (4) | 0.65334 (18) | 0.68259 (17) | 0.0326 (5) | |
| H13 | 0.737075 | 0.671519 | 0.631943 | 0.039* | |
| C14 | 0.7979 (5) | 0.66609 (18) | 0.75398 (18) | 0.0347 (6) | |
| H14 | 0.945533 | 0.693264 | 0.752293 | 0.042* | |
| C15 | 0.7047 (5) | 0.63898 (18) | 0.82670 (18) | 0.0355 (6) | |
| C16 | 0.4927 (5) | 0.59816 (19) | 0.83158 (18) | 0.0367 (6) | |
| H16 | 0.432553 | 0.578743 | 0.882366 | 0.044* | |
| C17 | 0.3696 (5) | 0.58612 (18) | 0.76102 (16) | 0.0330 (6) | |
| H17 | 0.222733 | 0.558409 | 0.763644 | 0.040* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0663 (5) | 0.0745 (5) | 0.0324 (4) | −0.0046 (4) | −0.0248 (4) | 0.0048 (4) |
| O1 | 0.0324 (10) | 0.0939 (18) | 0.0252 (10) | −0.0057 (11) | 0.0000 (9) | 0.0001 (10) |
| O2 | 0.0414 (11) | 0.0586 (12) | 0.0248 (9) | −0.0105 (10) | −0.0082 (9) | 0.0018 (9) |
| O3 | 0.0440 (11) | 0.0634 (14) | 0.0254 (10) | −0.0025 (10) | 0.0040 (9) | 0.0059 (9) |
| C1 | 0.0345 (14) | 0.0442 (15) | 0.0226 (13) | 0.0005 (12) | −0.0028 (11) | −0.0017 (12) |
| C2 | 0.0283 (12) | 0.0340 (13) | 0.0234 (12) | 0.0011 (10) | −0.001 (1) | 0.0006 (10) |
| C3 | 0.0312 (12) | 0.0381 (13) | 0.0261 (13) | −0.0009 (10) | −0.0011 (11) | 0.0015 (11) |
| C4 | 0.0313 (13) | 0.0327 (12) | 0.0259 (12) | −0.0001 (11) | −0.0058 (11) | 0.0007 (10) |
| C5 | 0.0526 (18) | 0.0612 (18) | 0.0221 (13) | −0.0025 (15) | −0.0101 (13) | 0.0041 (13) |
| C6 | 0.0591 (19) | 0.062 (2) | 0.0217 (13) | 0.0047 (16) | −0.0007 (14) | −0.0005 (13) |
| C7 | 0.0326 (14) | 0.0374 (13) | 0.0259 (13) | 0.0032 (11) | 0.0014 (11) | 0.004 (1) |
| C8 | 0.0288 (13) | 0.0410 (14) | 0.0330 (14) | −0.0026 (11) | 0.0014 (12) | 0.0034 (12) |
| C9 | 0.0303 (12) | 0.0380 (14) | 0.0305 (13) | −0.0033 (10) | −0.0059 (11) | −0.0012 (11) |
| C10 | 0.0326 (13) | 0.0489 (16) | 0.0268 (14) | −0.0015 (13) | −0.0020 (11) | −0.0015 (13) |
| C11 | 0.0291 (13) | 0.0403 (14) | 0.0247 (13) | −0.0004 (11) | −0.0012 (10) | −0.0031 (11) |
| C12 | 0.0316 (13) | 0.0330 (13) | 0.0246 (13) | 0.0023 (10) | 0.0005 (11) | −0.0017 (11) |
| C13 | 0.0324 (13) | 0.0396 (13) | 0.0259 (13) | −0.0005 (10) | −0.0024 (11) | 0.0010 (11) |
| C14 | 0.0321 (13) | 0.0387 (14) | 0.0332 (14) | 0.0003 (11) | −0.0069 (12) | 0.0010 (12) |
| C15 | 0.0416 (15) | 0.0393 (13) | 0.0255 (12) | 0.0060 (11) | −0.0117 (12) | 0.0000 (11) |
| C16 | 0.0430 (15) | 0.0416 (14) | 0.0256 (13) | 0.0023 (12) | −0.0005 (12) | 0.0033 (12) |
| C17 | 0.0337 (13) | 0.0362 (13) | 0.0292 (14) | −0.0005 (10) | 0.0021 (11) | 0.0013 (11) |
Geometric parameters (Å, º)
| Cl1—C15 | 1.739 (3) | C7—C8 | 1.380 (4) |
| O1—C1 | 1.229 (3) | C8—C9 | 1.384 (4) |
| O2—C4 | 1.372 (3) | C8—H8 | 0.9500 |
| O2—C5 | 1.433 (3) | C9—H9 | 0.9500 |
| O3—C7 | 1.366 (3) | C10—C11 | 1.336 (4) |
| O3—C6 | 1.446 (4) | C10—H10 | 0.9500 |
| C1—C10 | 1.480 (4) | C11—C12 | 1.465 (4) |
| C1—C2 | 1.485 (3) | C11—H11 | 0.9500 |
| C2—C3 | 1.396 (4) | C12—C13 | 1.397 (4) |
| C2—C9 | 1.396 (4) | C12—C17 | 1.399 (4) |
| C3—C4 | 1.384 (4) | C13—C14 | 1.396 (4) |
| C3—H3 | 0.9500 | C13—H13 | 0.9500 |
| C4—C7 | 1.395 (4) | C14—C15 | 1.373 (4) |
| C5—C6 | 1.493 (5) | C14—H14 | 0.9500 |
| C5—H5A | 0.9900 | C15—C16 | 1.377 (4) |
| C5—H5B | 0.9900 | C16—C17 | 1.379 (4) |
| C6—H6A | 0.9900 | C16—H16 | 0.9500 |
| C6—H6B | 0.9900 | C17—H17 | 0.9500 |
| C4—O2—C5 | 112.4 (2) | C7—C8—H8 | 119.7 |
| C7—O3—C6 | 114.7 (2) | C9—C8—H8 | 119.7 |
| O1—C1—C10 | 121.2 (2) | C8—C9—C2 | 120.7 (2) |
| O1—C1—C2 | 120.0 (2) | C8—C9—H9 | 119.7 |
| C10—C1—C2 | 118.8 (2) | C2—C9—H9 | 119.7 |
| C3—C2—C9 | 118.4 (2) | C11—C10—C1 | 120.1 (3) |
| C3—C2—C1 | 117.9 (2) | C11—C10—H10 | 119.9 |
| C9—C2—C1 | 123.7 (2) | C1—C10—H10 | 119.9 |
| C4—C3—C2 | 121.0 (2) | C10—C11—C12 | 127.4 (3) |
| C4—C3—H3 | 119.5 | C10—C11—H11 | 116.3 |
| C2—C3—H3 | 119.5 | C12—C11—H11 | 116.3 |
| O2—C4—C3 | 118.0 (2) | C13—C12—C17 | 118.7 (2) |
| O2—C4—C7 | 122.1 (3) | C13—C12—C11 | 123.0 (2) |
| C3—C4—C7 | 119.9 (2) | C17—C12—C11 | 118.2 (2) |
| O2—C5—C6 | 110.0 (3) | C14—C13—C12 | 119.9 (3) |
| O2—C5—H5A | 109.7 | C14—C13—H13 | 120.0 |
| C6—C5—H5A | 109.7 | C12—C13—H13 | 120.0 |
| O2—C5—H5B | 109.7 | C15—C14—C13 | 119.4 (3) |
| C6—C5—H5B | 109.7 | C15—C14—H14 | 120.3 |
| H5A—C5—H5B | 108.2 | C13—C14—H14 | 120.3 |
| O3—C6—C5 | 111.2 (2) | C14—C15—C16 | 122.1 (3) |
| O3—C6—H6A | 109.4 | C14—C15—Cl1 | 118.6 (2) |
| C5—C6—H6A | 109.4 | C16—C15—Cl1 | 119.3 (2) |
| O3—C6—H6B | 109.4 | C15—C16—C17 | 118.5 (3) |
| C5—C6—H6B | 109.4 | C15—C16—H16 | 120.8 |
| H6A—C6—H6B | 108.0 | C17—C16—H16 | 120.8 |
| O3—C7—C8 | 118.5 (3) | C16—C17—C12 | 121.5 (3) |
| O3—C7—C4 | 122.0 (3) | C16—C17—H17 | 119.3 |
| C8—C7—C4 | 119.5 (3) | C12—C17—H17 | 119.3 |
| C7—C8—C9 | 120.5 (2) | ||
| O1—C1—C2—C3 | −8.9 (4) | C4—C7—C8—C9 | 1.3 (4) |
| C10—C1—C2—C3 | 169.9 (2) | C7—C8—C9—C2 | −0.8 (4) |
| O1—C1—C2—C9 | 169.5 (3) | C3—C2—C9—C8 | −0.2 (4) |
| C10—C1—C2—C9 | −11.7 (4) | C1—C2—C9—C8 | −178.6 (3) |
| C9—C2—C3—C4 | 0.7 (4) | O1—C1—C10—C11 | 7.4 (4) |
| C1—C2—C3—C4 | 179.1 (2) | C2—C1—C10—C11 | −171.3 (3) |
| C5—O2—C4—C3 | 158.0 (3) | C1—C10—C11—C12 | −179.0 (2) |
| C5—O2—C4—C7 | −21.8 (4) | C10—C11—C12—C13 | 6.8 (4) |
| C2—C3—C4—O2 | −180.0 (2) | C10—C11—C12—C17 | −176.7 (3) |
| C2—C3—C4—C7 | −0.1 (4) | C17—C12—C13—C14 | −1.2 (4) |
| C4—O2—C5—C6 | 50.4 (3) | C11—C12—C13—C14 | 175.3 (2) |
| C7—O3—C6—C5 | 38.6 (4) | C12—C13—C14—C15 | 0.4 (4) |
| O2—C5—C6—O3 | −60.1 (4) | C13—C14—C15—C16 | 0.9 (4) |
| C6—O3—C7—C8 | 171.8 (3) | C13—C14—C15—Cl1 | −179.0 (2) |
| C6—O3—C7—C4 | −9.4 (4) | C14—C15—C16—C17 | −1.3 (4) |
| O2—C4—C7—O3 | 0.2 (4) | Cl1—C15—C16—C17 | 178.7 (2) |
| C3—C4—C7—O3 | −179.6 (3) | C15—C16—C17—C12 | 0.4 (4) |
| O2—C4—C7—C8 | 179.0 (2) | C13—C12—C17—C16 | 0.8 (4) |
| C3—C4—C7—C8 | −0.8 (4) | C11—C12—C17—C16 | −175.8 (2) |
| O3—C7—C8—C9 | −179.9 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5A···O3i | 0.99 | 2.61 | 3.595 (4) | 172 |
| C5—H5B···Cl1ii | 0.99 | 2.80 | 3.455 (3) | 124 |
Symmetry codes: (i) x−1/2, −y+3/2, z; (ii) x−1, y, z−1.
Funding Statement
Funding for this research was provided by: NSF (MRI CHE1625732) and the University of Kentucky (Bruker D8 Venture diffractometer) to SP.
References
- Boeyens, J. C. A. (1978). J. Cryst. Mol. Struct. 8, 317–320.
- Bruker (2016). APEX3. Bruker AXS Inc., Madison, Wisconsin, USA.
- Çelikesir, S. T., Sheshadri, S. N., Akkurt, M., Chidan Kumar, C. S. & Veeraiah, M. K. (2019). Acta Cryst. E75, 942–945. [DOI] [PMC free article] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Dhar, D. N. (1981). In The Chemistry of Chalcones and Related Compounds. New York: John Wiley.
- Di Carlo, G., Mascolo, N., Izzo, A. A. & Capasso, F. (1999). Life Sci. 65, 337–353. [DOI] [PubMed]
- Dimmock, J. R., Elias, D. W., Beazely, M. A. & Kandepu, N. M. (1999). Curr. Med. Chem. 6, 1125–1149. [PubMed]
- Elkanzi, N. A. A., Hrichi, H., Alolayan, R. A., Derafa, W., Zahou, F. M. & Bakr, R. B. (2022). ACS Omega, 7, 27769–27786. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Harrison, W. T. A., Bindya, S., Yathirajan, H. S., Sarojini, B. K. & Narayana, B. (2006). Acta Cryst. E62, o5293–o5295.
- Harrison, W. T. A., Chidan Kumar, C. S., Yathirajan, H. S., Ashalatha, B. V. & Narayana, B. (2010). Acta Cryst. E66, o2477. [DOI] [PMC free article] [PubMed]
- Jasinski, J. P., Butcher, R. J., Sreevidya, T. V., Yathirajan, H. S. & Narayana, B. (2008). Anal. Sci. X, 24, X245–X246.
- Kong, Z., Sun, D., Jiang, Y. & Hu, Y. (2020). J. Enzyme Inhib. Med. Chem. 35, 1513–1523. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Li, H., Rathore, R. S., Prakash Kamath, K., Yathirajan, H. S. & Narayana, B. (2010). Acta Cryst. E66, o1289–o1290. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Ng, S.-L., Patil, P. S., Razak, I. A., Fun, H.-K. & Dharmaprakash, S. M. (2007). Acta Cryst. E63, o1897–o1898.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sarojini, B. K., Narayana, B., Ashalatha, B. V., Indira, J. & Lobo, K. J. (2006). J. Cryst. Growth, 295, 54–59.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shinde, R. A., Adole, V. A. & Jagdale, B. S. (2022a). Polycyclic Aromat. Compd. 42, 6155–6172.
- Shinde, R. A., Adole, V. A., Jagdale, B. S. & Desale, B. S. (2021). J. Indian Chem. Soc. 98, 100051.
- Shinde, R. A., Adole, V. A., Jagdale, B. S. & Pawar, T. B. (2020). Mat. Sci. Res. Ind. 17, 54–72.
- Shinde, R. A., Adole, V. A., Shinde, R. S., Desale, B. S. & Jagdale, B. S. (2022b). Results Chem. 4, 100553.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Teh, J. B.-J., Patil, P. S., Fun, H.-K., Razak, I. A. & Dharmaprakash, S. M. (2007). Acta Cryst. E63, o1783–o1784.
- Troeberg, L., Chen, X., Flaherty, T. M., Morty, R. E., Cheng, M., Hua, H., Springer, C., McKerrow, J. H., Kenyon, G. L., Lonsdale-Eccles, J. D., Coetzer, T. H. T. & Cohen, F. E. (2000). Mol. Med. 6, 660–669. [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wu, J.-Z., Jiang, X., Zhao, C.-G., Li, X.-K. & Yang, S.-L. (2012). Z. Kristallogr. New Cryst. Struct. 227, 215–216.
- Zhang, Y., Zhang, Y.-N., Liu, M.-M., Ryu, K.-C. & Ye, D.-Y. (2011). Acta Cryst. E67, o912–o913. [DOI] [PMC free article] [PubMed]
- Zhuang, C., Zhang, W., Sheng, C., Zhang, W., Xing, C. & Miao, Z. (2017). Chem. Rev. 117, 7762–7810. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989023005613/vm2285sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023005613/vm2285Isup2.hkl
CCDC reference: 2272334
Additional supporting information: crystallographic information; 3D view; checkCIF report