Abstract
Two members of the GATA family of transcription factors, GATA-4 and GATA-6, are expressed in the developing and postnatal myocardium and are equally potent transactivators of several cardiac promoters. However, several in vitro and in vivo lines of evidence suggest distinct roles for the two factors in the heart. Since identification of the endogenous downstream targets of GATA factors would greatly help to elucidate their exact functions, we have developed an adenovirus-mediated antisense strategy to specifically inhibit GATA-4 and GATA-6 protein production in postnatal cardiomyocytes. Expression of several endogenous cardiac genes was significantly down-regulated in cells lacking GATA-4 or GATA-6, indicating that these factors are required for the maintenance of the cardiac genetic program. Interestingly, transcription of some genes like the α- and β-myosin heavy-chain (α- and β-MHC) genes was preferentially regulated by GATA-4 due, in part, to higher affinity of GATA-4 for their promoter GATA element. However, transcription of several other genes, including the atrial natriuretic factor and B-type natriuretic peptide (ANF and BNP) genes, was similarly down-regulated in cardiomyocytes lacking one or both GATA factors, suggesting that GATA-4 and GATA-6 could act through the same transcriptional pathway. Consistent with this, GATA-4 and GATA-6 were found to colocalize in postnatal cardiomyocytes and to interact functionally and physically to provide cooperative activation of the ANF and BNP promoters. The results identify for the first time bona fide in vivo targets for GATA-4 and GATA-6 in the myocardium. The data also show that GATA factors act in concert to regulate distinct subsets of genes, suggesting that combinatorial interactions among GATA factors may differentially control various cellular processes.
The vertebrate GATA transcription factors share a highly conserved domain composed of two zinc fingers (39). This domain is responsible for specific binding to a consensus WGATAR element. Based on sequence homology and tissue distribution, the vertebrate GATA family can be divided into two subgroups. The first subgroup is composed of GATA-1, -2, and -3. These three GATA factors are expressed in the hematopoietic system and are essential for normal hematopoiesis (36, 37, 39, 40, 44). The second subgroup is composed of GATA-4, -5, and -6, which are differentially expressed in the heart and gut (26). Within the heart, GATA-5 is restricted to the endocardium (22), whereas GATA-4 and -6 are expressed in the developing and postnatal myocardium (14, 18, 21, 33).
Several lines of evidence have implicated GATA-4 in diverse developmental processes including survival, differentiation, and/or migration of cardiomyocyte precursors. For example, in vitro experiments using embryonic stem cells showed that GATA-4 is essential for survival of cardioblasts and terminal cardiomyocyte differentiation (15, 16). In vivo, inactivation of the GATA-4 gene is embryo lethal at day 9.5 postcoitum due to failure of the GATA-4 null mice to develop a primitive heart tube (25, 31). Unfortunately, the requirement for GATA-4 at early stages of heart development has precluded analysis of its role in the postnatal myocardium either in vitro or in vivo. Nevertheless, two lines of evidence suggest that GATA-4 may play a critical role in postnatal cardiac transcription: GATA elements were found to be essential for activation of some cardiac promoters in adult myocardium (17, 19, 28, 30), and GATA-4 was shown to functionally and physically interact with NFAT-3, which would implicate it as a mediator of the calcineurin-dependent hypertrophic process in the myocardium (32).
Analyses of GATA-6 in the myocardium have been more limited, but the available data are consistent with a role for GATA-6 in myocardial development and gene expression. Thus, axis disruption experiments with Xenopus frogs showed that transcription of GATA-6, much like that of GATA-4 and -5, correlates with specification of cardiac progenitors and ectopic expression of either of those factors activates the α-myosin heavy-chain (α-MHC) and cardiac α-actin genes (21). Moreover, cotransfection experiments using heterologous cells showed that GATA-6 is as potent as GATA-4 in transactivating cardiac genes harboring GATA elements in their promoter, such as the cardiac troponin C (cTnC) and atrial natriuretic factor (ANF) genes (9, 33). Interestingly, Gove et al. have reported that in Xenopus embryos, GATA-6 overexpression blocks differentiation and stimulate proliferation of heart precursors (12). Recently, the inactivation of the GATA-6 gene in mice was reported to be embryo lethal at day 7.5 postcoitum, precluding analysis of its role in the heart (34). Finally, the inability of GATA-6 to compensate for the GATA-4 deficiency in GATA-4 null mice suggests that GATA-4 and GATA-6 play different roles in vivo.
The molecular basis for the differential roles of GATA-4 and -6 in the myocardium remains largely unknown but could occur via at least three nonexclusive mechanisms: differential expression of GATA-4 and GATA-6 in subsets of cardiomyocytes, differential affinity of GATA factors for GATA elements and therefore different in vivo target genes, or differential interaction of GATA-4 and -6 with cofactors. Support for this latter possibility was recently provided by Durocher et al., who showed functional and physical interaction between GATA-4, but not GATA-6, and the cardiac homeodomain protein Nkx2-5 (9, 10). However, the relative affinities of GATA-4 and -6 for their DNA-binding sites have not been determined, and whether GATA-4 and -6 localize differentially in the heart and target distinct genes is not known.
To address these issues, we have developed antibodies specific for the different cardiac GATA factors and analyzed at the cellular level the localization of GATA-4 and -6 in the myocardium. We also developed an adenovirus-mediated antisense strategy to specifically inhibit GATA-4 and GATA-6 protein production in neonatal cardiomyocytes and assess its effect on cardiac gene expression. The results indicate that several endogenous cardiac genes, including those encoding ANF, B-type natriuretic peptide (BNP), α-MHC, β-MHC, cTnI, and platelet-derived growth factor receptor β (PDGFRβ), are down-regulated in cardiomyocytes lacking either GATA-4 or GATA-6, suggesting that these genes are bona fide targets for both GATA-4 and GATA-6. Interestingly, the α- and β-MHC genes are preferential targets for GATA-4, likely due to the higher affinity of GATA-4 for their promoter GATA element. Remarkably, GATA-4 and GATA-6 colocalize in postnatal cardiomyocytes and interact functionally and physically to provide cooperative activation of the ANF and BNP promoters. These results suggest that GATA factors are involved in the maintenance of the cardiac phenotype and that expression of cardiac genes is controlled by combinatorial interactions of the different GATA proteins.
MATERIALS AND METHODS
Plasmids and adenovirus vectors.
The recombinant replication-deficient adenovirus type 5 (Ad5) expressing antisense regions directed specifically toward GATA-4 or -6 was generated by using the cloning system developed and generously provided by F. L. Graham (29). Briefly, a 358-bp EcoRI/HindIII fragment encoding the extreme N-terminal portion of rat GATA-4 and a 359-bp XbaI/StuI fragment from the 5′ untranslated region (UTR) of rat GATA-6 were subcloned into HindIII/EcoRI and EcoRV/XbaI, respectively, between left-end adenovirus sequences in Ad5 shuttle vector pΔE1sp1B/CMV/BGH, a plasmid generously provided by B. A. French (1). This plasmid was constructed by inserting the 1,276-bp BglII/PvuII fragment (containing the cytomegalovirus [CMV] immediate-early promoter, polylinker, and bovine growth hormone polyadenylation signal) from pcDNA3 (Invitrogen Corp., San Diego, Calif.) between the BglII/Klenow-blunted ClaI sites of the polylinker in Ad5 shuttle vector pΔE1sp1B (6). Each shuttle vector was cotransfected into 293 embryonic kidney cells with pJM17, which contains a circularized dl309 adenovirus genome, to generate replication-deficient viruses with substitution of the Ad5 E1 genes for the antisense GATA-4 and GATA-6 sequences (AS4 and AS6). The virus Ad5/CMV/NLS-lacZ, carrying an expression cassette in which the CMV immediate-early promoter transcribes sequences encoding the simian virus 40 (SV40) large T-antigen nuclear localization signal (NLS) fused to the Escherichia coli lacZ reporter gene, was used as control (Ctl; a generous gift from B. A. French) (11). Putative Ad5 clones were plaque purified, screened for antisense inserts, propagated, isolated, and titered according to the protocol of Graham and Prevec (13), to produce viral stocks with titers of >2 × 109 PFU/ml.
Wild-type rat ANF and BNP reporter plasmids and the wild-type rat GATA-4 expression vector (pCG-GATA-4) were described previously (14). The various deletions or mutations of the ANF promoter and GATA-4 cDNA were performed by PCR or by the Altered Sites in vitro mutagenesis system (Promega Corp., Madison, Wis.) as described by the manufacturer. The polyhistidine-tagged GATA-4 constructs used for in vitro transcription and translation were generated by insertion of the XbaI-BamHI fragment of the corresponding pCG-GATA-4 construct into the NheI-BamHI or NheI-BglII sites of pRSETA (Invitrogen Corp.). The rat GATA-6 cDNA was cloned by PCR and subcloned into the pcDNA3 expression vector. All constructs were confirmed by sequencing.
Neonatal cardiomyocyte preparation, infection, and transfection.
Primary cultures of cardiac myocytes were prepared from 4-day-old Sprague-Dawley rats as previously described (4), with minor modifications. Essentially, ventricles and atria of ∼60 to 72 hearts were digested four to five times, for 15 min each time, in Joklik’s modified Eagle’s medium (Canadian Life Technologies Inc.) containing 18 mM HEPES (pH 7.4), 0.1% collagenase (∼250 U/ml; Worthington Biochemical Corp.), and DNase I (5 μg/ml; ∼2,000 U/mg; Boehringer Mannheim Canada). The enzymatic digestion was stopped with fetal bovine serum (FBS, qualified grade; Canadian Life Technologies Inc.), and the undigested tissue was removed by filtration through nylon mesh (pore size, 100 μm). Cardiomyocytes were purified by three preplatings of 20 min each to remove residual nonmyocytes by differential adhesiveness, then plated at a density of 0.5 × 106 cells/35-mm-diameter dish (Primeria; Falcon), and cultured for 16 to 24 h in Dulbecco’s modified Eagle’s medium (DMEM; Canadian Life Technologies Inc.) containing 10% FBS. The following day, the medium was exchanged for serum-free hormonally defined medium (SFHD) as previously described (4).
Transfections were carried out by using calcium phosphate precipitation 24 h after plating. For assay of basal ANF promoter activity, cardiomyocytes were transfected with 6 μg of a wild-type or mutant ANF-luciferase reporter. At 36 h posttransfection, cells were harvested and luciferase activity was assayed with a Berthold LB 953 luminometer.
Serial dilutions of recombinant Ad5 (Ctl, AS4, or AS6) were prepared in OptiMEMI (Canadian Life Technologies Inc.). Cardiomyocytes were exposed to 200 μl of OptiMEMI containing 0.5 × 106, 2 × 106, or 8 × 106 PFU of recombinant Ad5 for 30 min at 25°C, and 2 ml of SFHD was added to each petri dish. The medium was replaced 16 h later with fresh SFHD. The cells were kept for 3 to 5 days, with the medium replaced every 24 h with fresh SFHD. Just before replacement, aliquots of the medium were taken for determination of ANF concentration.
β-Galactosidase detection.
One day after infection, cardiomyocytes were fixed in 0.5% glutaraldehyde–phosphate-buffered saline (PBS) for 10 min, washed twice for 30 min each time with PBS containing 0.02% Nonidet P-40, and stained with a mixture of 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 2 mM MgCl2, 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Boehringer Mannheim Corp.) per ml, 0.01% deoxycholic acid, and 0.02% Nonidet P-40 in PBS for 1 to 2 h at 25°C in the dark. The stained cardiomyocytes were washed with PBS, and 2 ml of 70% glycerol was added to each petri dish. The stained cells were kept at 4°C until photographed.
RNA extraction and Northern blotting.
Total RNA was isolated from cardiomyocytes by the guanidium thiocyanate-phenol-chloroform method as previously described (3). RNA was denatured with formaldehyde and formamide, size fractionated on a 1.2% agarose gel, transferred to a nylon membranes (MSI, Westborough, Mass.) by capillary blotting with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and cross-linked to the membrane with a UV Stratalinker 2400 (120 mJ; Stratagene). Blots were hybridized with random prime-labeled rat cDNA probes for GATA-4, GATA-6, ANF, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The GATA-4 and -6 cDNA fragments are the same ones used to generate the AS4 and AS6 adenoviruses. The ANF cDNA was described previously (5), and the GAPDH cDNA was generously provided by P. Jolicoeur. Blots were exposed in a PhosphorImager cassette and analyzed with ImageQuant (Molecular Dynamics).
Cell cultures and transfections.
HeLa or L cells were grown in DMEM supplemented with 10% FBS. Transfections were carried out by using calcium phosphate precipitation 24 h after plating. For overexpressions, 800,000 L cells were plated per 100-mm-diameter petri dish and transfected with 40 μg of pCG, pCG-GATA-4, pCG-GATA-4 mutant, or pCDNA3-GATA-6 expression vector. At 36 h posttransfection, cells were harvested and nuclear extracts were prepared as previously described (14). Nuclear extract protein concentrations were quantitated by the Bradford assay (Bio-Rad Laboratories, Hercules, Calif.).
For ANF promoter transactivation assays, 100,000 HeLa cells were plated per 35-mm-diameter petri dish and transfected with 3 μg of ANF-luciferase reporter plasmid and 1 μg of GATA expression vector. For synergy, 200 ng of GATA-6 and 200 ng of wild-type or mutant GATA-4 expression vector were used. The total amount of DNA was kept constant at 4 μg. At 36 h posttransfection, cells were harvested and luciferase activity was assayed.
Electrophoretic mobility shift assays (EMSAs).
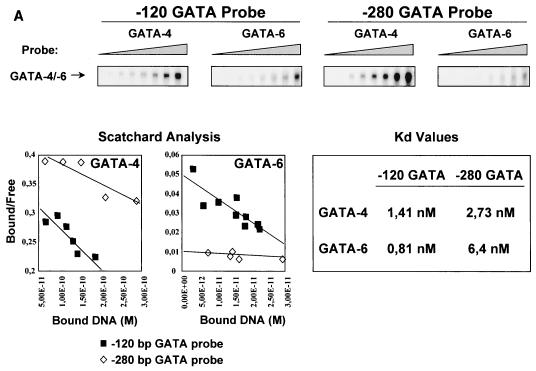
Binding reactions were performed in 20-μl reaction mixtures containing 3 μg of nuclear extracts from L cells overexpressing GATA-4 or GATA-6 in a buffer containing 12 mM HEPES (pH 7.9), 5 mM MgCl2, 60 mM KCl, 4 mM Tris-HCl (pH 7.9), 0.6 mM EDTA, 0.6 mM dithiothreitol, 0.5 mg of bovine serum albumin (BSA) per ml, 1 μg of poly(dI-dC), 12% glycerol, 20,000 cpm of radiolabeled double-stranded −120 ANF GATA probe, and increasing amounts of the appropriate unlabeled competitor for 20 min at room temperature. Reactions were then loaded on a 4% polyacrylamide gel and run at 200 V at room temperature in 0.25× Tris-borate-EDTA. The gel was then dried and exposed to a PhosphorImager (Molecular Dynamics) cassette for quantitative analysis. Relative bindings were quantitated and plotted as a function of the unlabeled competitor amount. Probes used were, from 5′ to 3′ (only the coding strand is shown), rat ANF −120 (proximal; GATCTCGCTGGACTGATAACTTTAAAAGG), rat ANF −120mut (TGACAAGCTTCGCTGGACTCCTAACTTTAAAAG), rat ANF −280 (distal; GATCTCCCAGGAAGATAACCAAGGACTCG), and rat α-MHC −265 bp (GATCCTCCTCTATCTGCCCATCA), where the WGATAR consensus motifs are underlined and the mutations are in boldface.
For DNA-binding affinity measurement, EMSAs were performed as described above but with increasing amounts of radiolabeled ANF proximal or distal GATA probe with a constant amount (3 μg) of nuclear extracts from L cells overexpressing GATA-4 or GATA-6. Scatchard analysis was then performed on the binding data by plotting the bound/free DNA ratio as a function of the bound DNA. The resulting dissociation constant (Kd) was calculated by using Microsoft Excel.
Western blots.
Three days after infection, cardiomyocytes were harvested and nuclear extracts were prepared; then 20 μg of cardiomyocyte nuclear extracts and 2 μg of GATA-4 or GATA-6-overexpressing L-cell nuclear extracts were boiled in Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a Hybond polyvinylidene difluoride membrane and immunoblotted by using the Renaissance chemiluminescence system (NEN Life Sciences, Boston, Mass.) as described by the manufacturer. Goat GATA-4 supershift antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) was used at a dilution of 1/1,000 and was revealed with an anti-goat horseradish peroxidase-conjugated antibody (Sigma, St. Louis, Mo.) at a dilution of 1/100,000. The GATA-6 antibody was made in rabbits by injection of a GATA-6-specific peptide linked to keyhole limpet hemocyanin as described elsewhere (2). The purified GATA-6 antibody was used at a dilution of 1/1,000 and was revealed with an anti-rabbit horseradish peroxidase-conjugated antibody (Sigma) at a dilution of 1/100,000.
RIA.
Immunoreactive ANF (irANF) concentration was determined in the cardiomyocyte culture medium by radioimmunoassay (RIA) as previously described (3).
Pull-down assays.
Polyhistidine-tagged GATA-4 and wild-type GATA-6 were in vitro cotranscribed and cotranslated in the presence of radiolabeled methionine according to the manufacturer’s protocol (TNT reticulocyte lysate kit; Promega Corp.). The proteins were then allowed to interact at 4°C with agitation in 400 μl of binding buffer (150 mM NaCl, 50 mM Tris-Cl [pH 7.5], 0.3% Nonidet P-40, 10 mM ZnCl2, 1 mM dithiothreitol, 0.25% BSA) as described previously (9). After 2 h, 50 μl of nickel resin (ProBond resin; Invitrogen Corp.) was added, and the reaction mixture was incubated further for 2 h at 4°C. The resin was then washed three times with binding buffer and twice with binding buffer minus BSA. Interacting proteins were resolved by SDS-PAGE (15% gel). The gel was dried and exposed to a PhosphorImager cassette.
Cross-linking.
Cardiomyocyte whole-cell extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, cocktail of protease inhibitors) by repeated aspiration through a syringe needle, followed by centrifugation to pellet cellular debris. The supernatant was then incubated on ice for increasing amounts of time in the presence of 0.02% glutaraldehyde, and the reaction was blocked by the addition of 1 M glycine (pH 7.6). To reduce nonspecific binding, the cellular extracts were preincubated at 4°C for 30 min with 1 μg of normal goat serum and 20 μl of protein A/G PLUS-Agarose (Santa Cruz Biotechnology). GATA-4/GATA-6 complexes were immunoprecipitated with 1 μl of anti-GATA-4 antibody (Santa Cruz Biotechnology) and 20 μl of protein A/G PLUS-Agarose at 4°C overnight with agitation. Subsequently, the immunoprecipitates were washed four times in RIPA buffer, resolved by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. The presence of the GATA-4/GATA-6 complexes was revealed by Western blotting using an anti-GATA-6 antibody.
Immunofluorescence.
Paraformaldehyde-fixed heart sections from neonatal mice were processed for immunofluorescence as described elsewhere (41). The GATA-4 antibody was used at a dilution of 1/500 and was revealed with a biotinylated anti-goat antibody (1/200; Vector Laboratories Inc., Burlingame, Calif.) followed by avidin-rhodamine (1/200; Vector Laboratories). The purified GATA-6 antibody was used at a dilution of 1/50 and was revealed with a fluorescein isothiocyanate-conjugated anti-rabbit antibody (1/200; Sigma).
RESULTS
In vivo target genes for GATA-4 and GATA-6 in postnatal atrial and ventricular cardiomyocytes.
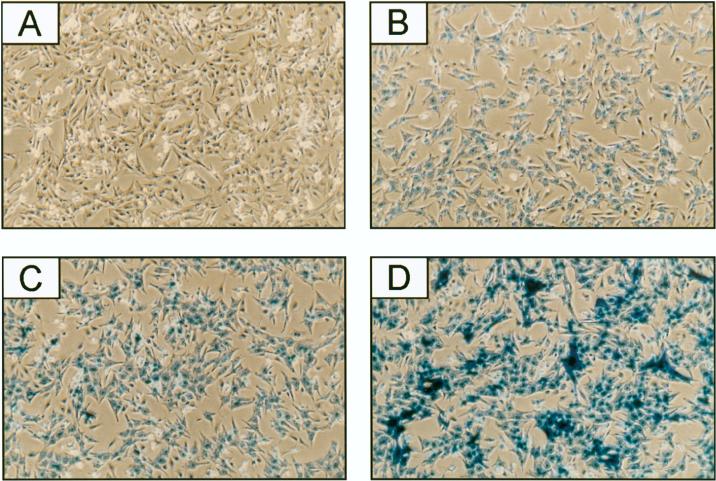
To analyze the consequence of inhibiting GATA-4 or GATA-6 expression for endogenous myocardial gene expression, we used adenovirus-mediated delivery of antisense gene regions to cardiomyocytes in primary culture. For this, we initially determined the dose of adenovirus that would efficiently infect ventricular cardiomyocytes isolated from 4-day neonatal rats, using a replication-deficient adenovirus expressing a lacZ gene fused to an SV40 NLS (Ctl adenovirus). At a multiplicity of infection (MOI) of 1, all the cardiomyocytes showed β-galactosidase activity (blue staining) in the nucleus, with staining intensity increasing in a dose-dependent manner up to an MOI of 16 (Fig. 1B to D). No staining was observed in mock-infected cardiomyocytes (Fig. 1A). Similar results were obtained with atrial neonatal cardiomyocytes (data not shown). Higher dose of the Ctl adenovirus led to cytotoxicity (data not shown). Thus, MOIs ranging from 1 to 16, which achieve efficient infection without any apparent cytotoxicity (Fig. 1), were used in subsequent experiments.
FIG. 1.
Cardiomyocytes are efficiently infected by adenovirus. Ventricular cardiomyocytes isolated from 4-day neonatal rats were mock infected (A) or infected at MOIs of 1 (B), 4 (C), and 16 (D) with the Ctl adenovirus, which expresses an NLS-lacZ gene. Twenty hours later, cells were assayed for β-galactosidase activity.
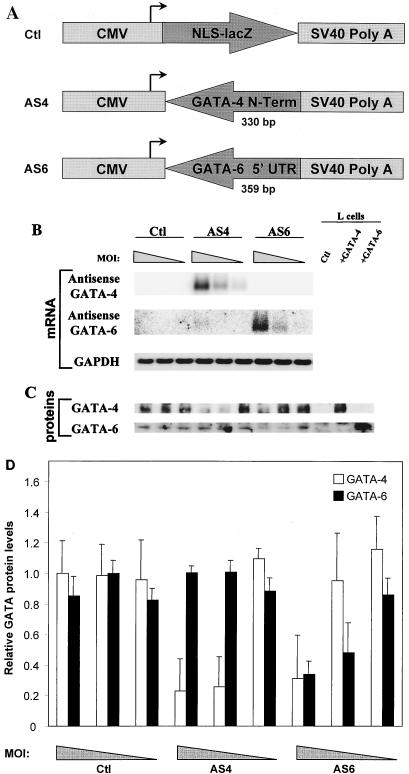
To specifically inhibit GATA-4 or GATA-6 protein production, replication-deficient adenoviruses expressing an antisense cDNA directed specifically toward GATA-4 or GATA-6 were generated. A 358-bp fragment from the extreme N-terminal portion of GATA-4 was used for the AS4 production (Fig. 2A). For AS6, a 359-bp fragment from the 5′ UTR of GATA-6 was used. These regions were chosen because of their low overall homology with other GATA factors, and as shown in Fig. 2B, the DNA fragments used to generate AS4 and AS6 show no cross-hybridization with each other’s mRNA.
FIG. 2.
Characterization of AS4 and AS6. (A) Schematic representation of the constructs used to generate the recombinant adenoviruses. NLS-lacZ, a 358-bp fragment from the extreme N-terminal portion of GATA-4, or a 359-bp fragment from the 5′ UTR of GATA-6 was cloned downstream of the CMV promoter and upstream of an SV40 poly(A) sequence in order to generate the recombinant adenoviruses. (B) Transgene expression is dose dependent. Ventricular cardiomyocytes were infected at MOIs of 16, 4, and 1 (corresponding to the progressively narrowing triangle) with Ctl, AS4, and AS6. RNA was isolated 3 days postinfection. Northern blot analysis showed that both antisense transgenes were efficiently expressed in a dose-dependent manner. (C) AS4 and AS6 are specific and efficient at decreasing GATA-4 and GATA-6 protein levels. Ventricular cardiomyocytes were infected as described above, and nuclear extracts were isolated 3 days postinfection and analyzed by Western blotting. L cells overexpressing GATA-4 or GATA-6 were used as controls for the specificity of the antibodies. (D) Quantification of the effects of AS4 and AS6 on GATA-4 and GATA-6 protein levels. The data represent the means of two independent Western blots performed as described for panel C and quantified by densitometry. At an MOI of 4, GATA-4 levels were decreased specifically by AS4 (80% reduction), while GATA-6 levels were reduced specifically by AS6 (60% reduction).
Ventricular neonatal cardiomyocytes were infected at MOIs of 1, 4, and 16 with Ctl, AS4, and AS6, and RNA was isolated 3 days postinfection. Northern blot analysis showed that both antisense transgenes were efficiently expressed in a dose-dependent manner, to a level exceeding 100-fold that of endogenous GATA-4 or -6 (Fig. 2B). AS4 and AS6 were also specific and efficient at reducing GATA-4 and GATA-6 protein levels in a dose-dependent manner, as evidenced by Western blot analysis (Fig. 2C). As shown in Fig. 2D, AS4 reduced GATA-4 levels by 80% whereas AS6 reduced GATA-6 levels by 60% at an MOI of 4. The effect of AS4 and AS6 extended also to GATA-binding activity as assessed by EMSA; a 50% decrease in GATA binding over the ANF −120 GATA probe was observed with AS4 or AS6, indicating that indeed both GATA-4 and GATA-6 contribute to the GATA-binding activity in postnatal cardiomyocytes (data not shown). Moreover, AS4 and AS6 had no effect on GATA-5 mRNA levels, suggesting that GATA-5 does not compensate for the lack of GATA-4 or GATA-6 in postnatal cardiomyocytes (data not shown).
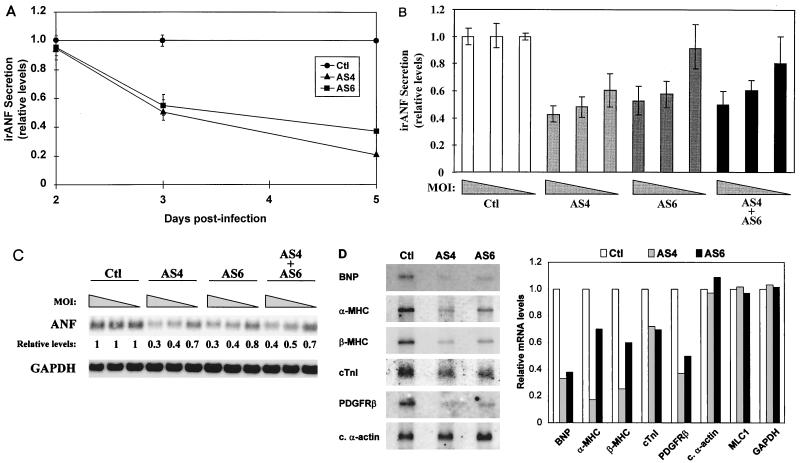
To assess the effects of reduced GATA-4 or GATA-6 activity on endogenous gene expression, we initially analyzed changes in ANF mRNA and protein levels. The ANF promoter contains two conserved GATA elements (see Fig. 4A) and was previously shown to be transactivated by GATA-4 and GATA-6 in heterologous cells (9, 14). In ventricular neonatal cardiomyocytes with reduced GATA-4 or GATA-6 activity, irANF secretion was decreased in a time-dependent manner (Fig. 3A). The effect was first observed at 3 days postinfection (50% decrease), and irANF secretion was decreased by 60 to 80% at 5 days postinfection. The decrease of secreted irANF was dose dependent (Fig. 3B). Interestingly, the reduction of both GATA-4 and GATA-6 activities (AS4 plus AS6) had the same effect on irANF secretion as reduction of GATA-4 (AS4) or GATA-6 (AS6) activity alone (Fig. 3B), suggesting that either a maximal threshold is reached or GATA-4 and GATA-6 are in the same transcriptional pathway. The effect of attenuation of GATA-4 or GATA-6 on ANF expression extended to the transcript level (Fig. 3C). Similar results were obtained with atrial cardiomyocytes (data not shown) and indicate that the ANF gene is an in vivo transcriptional target for GATA-4 and GATA-6 in postnatal cardiomyocytes.
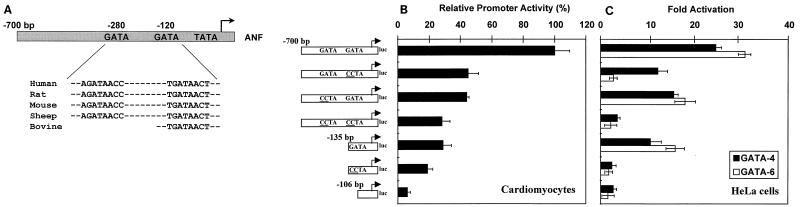
FIG. 4.
ANF is a direct transcriptional target for both GATA-4 and GATA-6. (A) Alignment of sequences of human, rat, mouse, sheep, and bovine (for which only a partial sequence is available) ANF promoters. Note that two consensus WGATAR elements, a proximal one at −120 bp and a distal one at −280 bp, are entirely conserved across species. (B) The proximal and distal GATA elements are major contributors of ANF promoter activity in neonatal cardiomyocytes. Wild-type and mutated ANF promoters fused to the luciferase (luc) reporter gene were transiently transfected into ventricular cardiomyocytes isolated from 4-day neonatal rats. Luciferase activity was assayed 36 h posttransfection. The results are expressed relative to the −700 bp ANF promoter activity. (C) GATA-4 and GATA-6 transactivate the ANF promoter. HeLa cells were cotransfected with a GATA-4 or GATA-6 expression vector and wild-type or mutated ANF promoters fused to the luciferase reporter gene. Luciferase activity was assayed 36 h posttransfection. The results are expressed as fold activation by GATA-4 or GATA-6. In all cases, the data represent the mean ± standard deviation of two or three independent experiments carried out in duplicate.
FIG. 3.
In vivo transcriptional targets for GATA-4 and GATA-6 in postnatal cardiomyocytes. (A) irANF secretion is decreased in a time-dependent manner by AS4 and AS6. Ventricular cardiomyocytes were infected at an MOI of 4 with Ctl, AS4, and AS6. Secreted irANF was assayed in the cardiomyocyte culture medium after a 24-h accumulation period at 2, 3, and 5 days postinfection. Since the effect of the antisense DNA on irANF secretion is clearly visible at 3 days postinfection, subsequent analyses were performed at this time point. (B) irANF secretion is decreased in a dose-dependent manner by AS4, AS6, and AS4 plus AS6. Ventricular cardiomyocytes were infected at MOIs of 16, 4, and 1 (corresponding to the progressively narrowing triangle) with Ctl, AS4, AS6, and AS4 plus AS6. (C) ANF mRNA levels are decreased in a dose-dependent manner by AS4, AS6, and AS4 plus AS6. Ventricular cardiomyocytes were infected as for panel B, and total RNA was analyzed by Northern blotting. The ANF mRNA levels are expressed relative to that of Ctl-infected cardiomyocytes. The GAPDH mRNA levels were unaffected. (D) Many cardiac genes are decreased by AS4 and AS6. Ventricular cardiomyocytes were infected at an MOI of 4 with Ctl, AS4, and AS6, and Northern blot analysis was performed on poly(A)+ RNA (left). Relative mRNA levels were quantified with a PhosphorImager (right). Note how α-MHC and β-MHC mRNAs were preferentially down-regulated by AS4, whereas cardiac (c.) α-actin, myosin light-chain 1 (MLC1), and GAPDH mRNA levels were not affected by AS4 or AS6.
We also verified that other cardiac genes were affected by the decrease in GATA-4 or GATA-6. BNP, α-MHC, β-MHC, cTnI, and PDGFRβ mRNA levels were down-regulated by AS4 and AS6 (Fig. 3D). Interestingly, α-MHC and β-MHC were preferentially down-regulated by AS4, suggesting that these genes are preferential targets for GATA-4. The lack of GATA factors did not affect all cardiac genes; for example, cardiac α-actin, myosin light-chain 1, and GAPDH mRNA levels were not altered by AS4 or AS6 (Fig. 3C and D and data not shown). Similar results were obtained by semiquantitative reverse transcription-PCR analysis (data not shown). Thus, GATA-4 and GATA-6 are involved in regulating specific subsets of cardiac genes in postnatal cardiomyocytes.
ANF is a direct transcriptional target for both GATA-4 and GATA-6.
To determine if ANF is a direct transcriptional target for both GATA-4 and GATA-6, a detailed analysis of the ANF promoter was performed. Comparison of the ANF promoter sequences available in the database (from human, rat, mouse, sheep, and bovine genomes) shows that two consensus WGATAR elements, a proximal one at −120 bp and a distal one at −280 bp, are entirely conserved across species (Fig. 4A), suggesting that these GATA elements may play important evolutionarily conserved functions in ANF gene regulation. Indeed, deletion or point mutation of the proximal or the distal GATA element decreased by about 50% ANF promoter activity in postnatal cardiomyocytes (Fig. 4B). A more drastic effect was observed when both elements were mutated, leaving only 30% of wild-type promoter activity. Similar results were observed for all constructs in 1-day ventricular or atrial cardiomyocytes. Thus, both proximal and distal GATA elements are major contributors of ANF promoter activity in postnatal cardiomyocytes.
Since both GATA-4 and GATA-6 are present in neonatal cardiomyocytes, we tested their relative efficiencies at transactivating the ANF promoter. The results indicate that both GATA factors are potent activators of the ANF promoter, exhibiting about 25- to 30-fold activation (Fig. 4C). However, while mutation of the proximal or distal GATA element reduced GATA-4 transactivation by approximately 50%, mutation of the proximal GATA element fully abrogated GATA-6 transactivation.
To test whether this was due to differential affinity of GATA-4 and -6 for certain GATA elements, we analyzed the binding affinities of GATA-4 and GATA-6 for both ANF GATA elements. EMSAs were performed with increasing amounts of radiolabeled probes and a constant amount of nuclear extracts from L cells overexpressing GATA-4 or GATA-6 (Fig. 5A). Scatchard analysis revealed that GATA-4 has similar relative affinities for the proximal and distal GATA elements (relative Kds of 1.41 and 2.73 nM, respectively). However, the relative affinity of GATA-6 for the distal element was 8-fold lower than that for the proximal GATA element (relative Kds of 6.4 and 0.81 nM, respectively). Similar Kds were obtained with bacterially expressed proteins (data not shown). These results show for the first time that GATA-4 and GATA-6 possess differential affinities for naturally occurring sites.
FIG. 5.
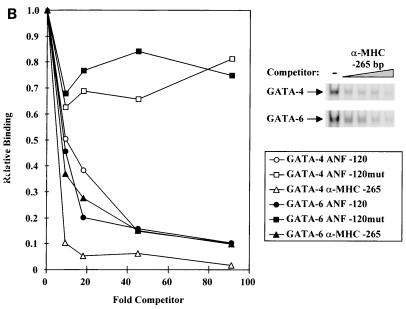
GATA-4 and GATA-6 bind GATA elements with different affinities. (A) EMSAs were performed with increasing amounts of radiolabeled probe (−120-bp GATA or −280-bp GATA) and a constant amount of nuclear extracts from L cells overexpressing GATA-4 or GATA-6. GATA binding is shown in the upper panel. Scatchard analysis were performed on the binding data, and the relative affinities (Kd values) are shown. (B) GATA-4 has higher affinity than GATA-6 for the −265-bp α-MHC GATA element. EMSAs were performed with nuclear extracts from L cells overexpressing GATA-4 or GATA-6 incubated with a radiolabeled −120-bp ANF GATA element and increasing amounts of an unlabeled competitor (−120-bp ANF GATA, −120-bp ANF GATAmut, or −265-bp α-MHC GATA; top right panel). Relative bindings were quantitated and plotted as a function of the amount of unlabeled competitor (left).
Since differential activation of the ANF promoter GATA elements by GATA-4 and -6 correlated well with the relative affinities for these sites, we tested whether the preferential regulation of α-MHC by GATA-4 was due to differential affinities of GATA factors for the α-MHC GATA element (30). As shown in Fig. 5B, the α-MHC GATA element was more efficient at competing GATA-4 than GATA-6 binding. Thus, the higher affinity of GATA-4 for this element may explain the preferential regulation of the α-MHC gene by GATA-4.
GATA-4 and GATA-6 functionally and physically interact to cooperatively activate cardiac promoters.
The fact that the reduction of both GATA-4 and GATA-6 protein levels had the same effect on ANF gene expression as the reduction of either factor alone suggests that GATA-4 and GATA-6 are members of a single functional complex and that ablation of GATA-4 or GATA-6 in that complex is sufficient to disrupt its transcriptional activity.
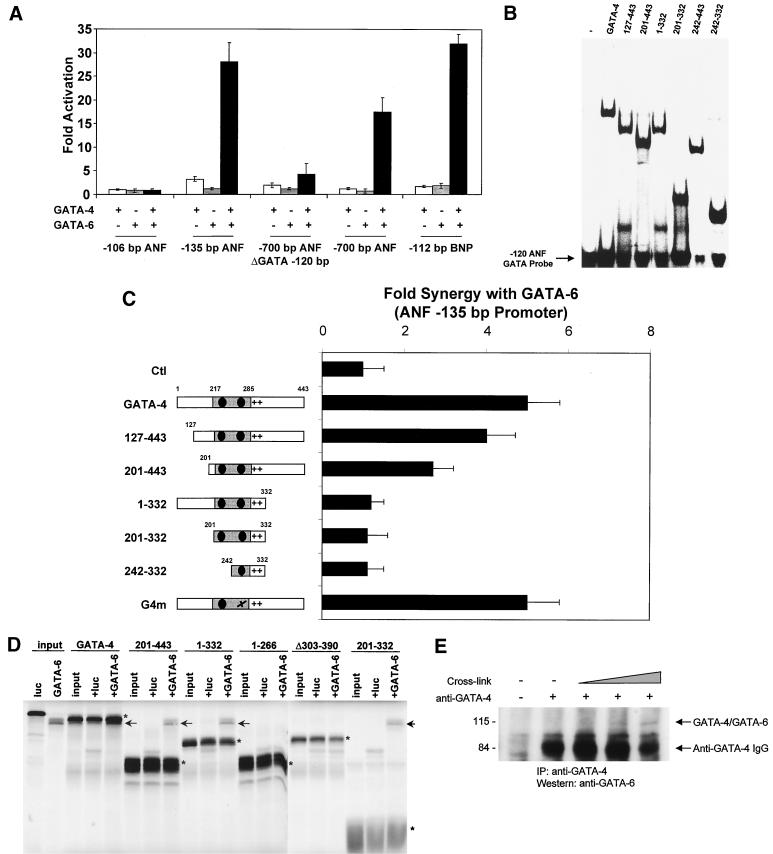
To test whether GATA-4 and GATA-6 act cooperatively, the −700-bp ANF promoter fused to luciferase was cotransfected with various doses of GATA-4 and GATA-6 expression vectors. At a low dose of expression vector where neither GATA-4 nor GATA-6 could activate transcription by itself, synergistic activation of the ANF promoter was achieved when both GATA factors were added (Fig. 6A). Interestingly, cooperative activation by GATA-4 and -6 occurred through a single GATA-binding site, as evidenced by the activation of the −135-bp ANF promoter construct. In fact, the proximal GATA element was necessary and sufficient for synergy, and the distal GATA element (−700-bp ANF ΔGATA −120 bp) could not mediate cooperative activation. Synergistic activation by GATA-4 and GATA-6 was also observed on the BNP promoter but not on a shorter ANF promoter lacking GATA elements (−106-bp ANF).
FIG. 6.
GATA-4 and GATA-6 functionally and physically interact to activate cardiac promoters. (A) GATA-4 and GATA-6 cooperatively activate cardiac promoters. ANF and BNP reporter vectors were cotransfected with 200 ng of GATA-4 and GATA-6 expression vectors in HeLa cells. Luciferase activity was assayed 36 h posttransfection. The results are expressed as fold activation by GATA-4 and/or GATA-6. (B) DNA binding by the GATA-4 mutants. EMSAs were performed on the −120-bp ANF GATA element, using various GATA-4 mutants overexpressed in L cells. The results are summarized in Fig. 7. (C) Mapping of the domain(s) required for synergy between GATA-4 and GATA-6. HeLa cells were cotransfected with GATA-6 and various GATA-4 mutant expression vectors. Luciferase activity was assayed 36 h posttransfection. The results are expressed as fold synergy of the −135-bp ANF promoter, which is defined by the activation of the −135-bp ANF promoter by both GATA-4 and GATA-6 divided by the sum of the activation by GATA-4 and GATA-6 alone. (D) The zinc fingers and the basic region of GATA-4 are sufficient for physical interaction with GATA-6. Polyhistidine-tagged GATA-4 and wild-type GATA-6 were in vitro cotranscribed and cotranslated in the presence of radiolabeled methionine. The reaction mixture was then incubated with a nickel resin in order to pull down His-tagged GATA-4 from the mixture. Interacting proteins were resolved by SDS-PAGE. Luciferase (luc) was used as a negative control for interaction. The various His-tagged GATA-4 proteins are indicated with asterisks, and the interacting GATA-6 proteins are marked by arrows. Note that due to the His tag, GATA-4 has a slightly lower electrophoretic mobility than GATA-6. (E) GATA-4 and GATA-6 interact in vivo in postnatal cardiomyocytes. Cardiomyocyte whole-cell extracts were cross-linked by using glutaraldehyde for 15, 30, and 150 min, followed by immunoprecipitation (IP) with an anti-GATA-4 antibody. The immunoprecipitates were resolved by SDS-PAGE, and the GATA-4/GATA-6 complexes were revealed by Western blotting using an anti-GATA-6 antibody. Positions of molecular weight standards (in kilodaltons) are indicated on the left. The strong signal is due to the GATA-4 antibody immunoglobulin G (IgG) heavy chains. Note that the GATA-4/GATA-6 complex migrates at about 110 kDa, which corresponds to the sum of the molecular masses of GATA-4 and GATA-6.
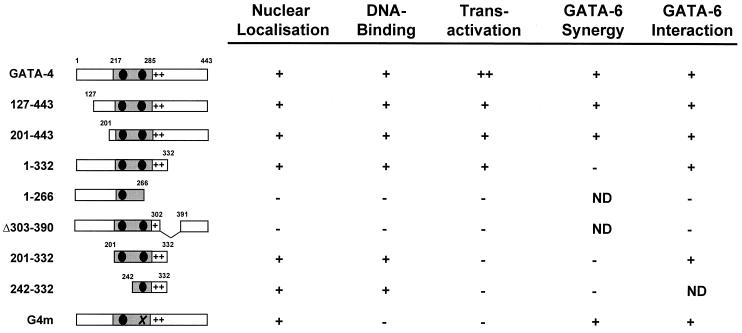
The domain(s) of GATA-4 required for synergy with GATA-6 was mapped by cotransfection in HeLa cells of GATA-6 and various GATA-4 mutant expression vectors (Fig. 6C; summarized in Fig. 7). All mutants were tested for expression and nuclear localization. The mutants used in transfection assays were expressed at similar levels, as evidenced by EMSAs and Western blot analysis (Fig. 6B and data not shown). Progressive deletion of the N-terminal (127 to 443 and 201 to 443) activation domain of GATA-4 did not drastically affect synergy with GATA-6. However, deletion of the C-terminal activation domain of GATA-4 (1 to 332) or GATA-4 mutants harboring no transcriptional activation domain (201 to 332 and 242 to 332) did not exhibit synergy with GATA-6. DNA binding by GATA-4 was not required since G4m, a GATA-4 mutant in the second zinc finger bearing no DNA-binding activity (but that still localizes to the nucleus) was able to provide synergistic activation with GATA-6. Thus, while the N-terminal activation domain of GATA-4 and GATA-4 DNA-binding ability are dispensable, the C-terminal activation domain of GATA-4 is required for synergy with GATA-6.
FIG. 7.
Summary of GATA-4 functional domains. The GATA-4 constructs used in this study are shown, along with some of their functional properties. ND, not determined.
To test whether this functional cooperation between GATA-4 and -6 involves direct interaction, polyhistidine-tagged GATA-4 and wild-type GATA-6 were in vitro cotranscribed and cotranslated in presence of radiolabeled methionine. As shown in Fig. 6D and summarized in Fig. 7, GATA-4 interacted specifically with GATA-6, and this interaction required the zinc fingers and the basic region of GATA-4. The ability of GATA-4 and GATA-6 to contact each other was further confirmed in vivo by chemical cross-linking of cardiomyocyte extracts followed by immunoprecipitation with a GATA-4 antibody and Western blotting with a GATA-6 antibody. As shown in Fig. 6E, this resulted in immunoprecipitation of a heterodimer composed of endogenous GATA-4 and GATA-6 proteins.
GATA-4 expression and GATA-6 expression colocalize in postnatal cardiomyocytes.
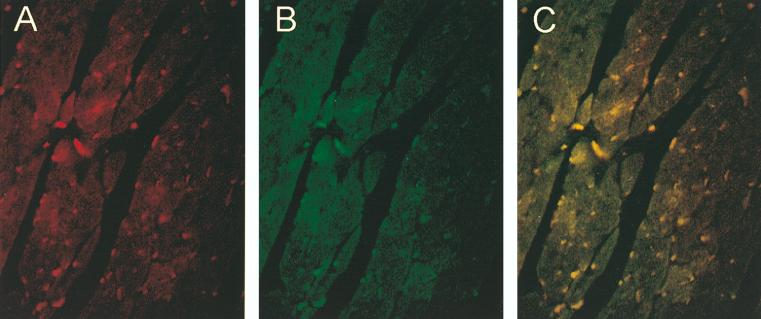
To ascertain that the immunoprecipitated GATA-4 and -6 complex reflects the ability of these proteins to associate with each other intracellularly, we verified the coexpression of GATA-4 and GATA-6 in cardiomyocytes. Immunofluorescence studies using specific anti-GATA-4 and anti-GATA-6 antibodies were performed on sections from neonatal mouse hearts and on primary rat cardiomyocyte cultures (Fig. 8 and data not shown). These studies revealed that GATA-4 and GATA-6 colocalize in most postnatal ventricular cardiomyocytes.
FIG. 8.
GATA-4 and GATA-6 colocalize in postnatal cardiomyocytes. Immunofluorescence studies were performed on heart sections from neonatal mice, using a specific anti-GATA-4 antibody (A) and a specific anti-GATA-6 antibody (B). (C) Superposition of panels A and B indicates that GATA-4 and GATA-6 colocalize in most postnatal cardiomyocytes.
DISCUSSION
Members of the GATA family of transcription factors play critical roles in diverse cellular processes. Although some members are coexpressed in specific cell types, each family member appears to fulfill essential, nonredundant functions during development. However, the mechanisms by which GATA factors control gene expression and cell fate as well as the molecular basis for their specificity remain poorly understood.
The data presented in this paper provide evidence for the existence of specific in vivo downstream targets for two members of the GATA family, GATA-4 and -6, which are coexpressed in myocardial cells. The results also show that GATA factors act in concert to regulate transcription of subsets of cellular genes, suggesting that combinatorial interactions among GATA factors may differentially control various cellular processes.
Identification of in vivo targets for GATA factors in cardiomyocytes.
Regulatory elements containing GATA-binding sites have been identified in many cardiac promoters, including BNP, α-MHC, cTnI, and cTnC, and GATA-4 was shown to transactivate several of these promoters (8, 14, 20, 28, 30, 35, 38, 42). However, several cardiac markers examined which were previously proposed to be GATA-4 targets were still expressed at high levels in GATA-4 null hearts, raising the possibility that these genes are not bona fide GATA-4 targets or that other cardiac factors, including GATA-6, whose expression is up-regulated in GATA-4 null mice, provide compensatory pathways (25, 31). Thus, determining the in vivo targets for GATA-4 and GATA-6 is crucial for understanding their role in the heart. The data obtained in this study indicate that while some cardiac genes are regulated preferentially by GATA-4, others are targets for both GATA-4 and GATA-6. This is consistent with a specialized role for each factor in heart development and the requirement for both in normal heart formation. At present, GATA-6 has been linked to cardioblast proliferation whereas GATA-4 has been associated with terminal differentiation (12, 15). In this respect, it is noteworthy that markers of later stages of cardiomyocyte differentiation—α- and β-MHC genes—appear to be preferential GATA-4 targets whereas genes expressed in precardiomyocytes prior to the beating stage, such the natriuretic peptide (ANF and BNP) and PDGFR genes (16, 24), are targeted by both GATA-4 and GATA-6. Since the adenovirus-mediated antisense strategy revealed that GATA-4 and GATA-6 are required for maintenance of the differentiated phenotype in postnatal cardiomyocytes, it could now be used to determine the role of these factors in embryonic myocytes. Finally, in light of recent reports suggesting a role for GATA-4 in mediating hypertrophic signals (17, 19), it will be interesting to use the adenovirus tools developed in this study to directly test the implication of GATA-4 or GATA-6 in cardiomyocyte hypertrophy.
Transcriptional mechanisms of GATA factors.
The data presented here show that a subset of cellular genes may be bona fide targets for more than one GATA factor, whereas others are under the control of a specific GATA factor. It may be significant that this is the first time that specific in vivo targets for cardiac GATA factors have been reported.
Differential affinity could be one of the mechanisms by which GATA factors target distinct downstream genes. In the case of the hematopoietic GATA-1, -2, and -3 proteins, in vitro binding site selection experiments have shown differences in DNA-binding specificity, although they have not yet been correlated with natural GATA elements present on hematopoietic promoters (23).
In this study, we show that GATA-4 and GATA-6 bind to the two ANF GATA elements with different relative affinities that correlate well with their ability to transactivate the ANF promoter. The results also show that the higher affinity of GATA-4 for the α-MHC GATA element correlates with the finding that the endogenous α-MHC gene is a preferential GATA-4 target. Interestingly, the ANF and α-MHC sequences that are preferential GATA-4 binding sites have an A residue at the W position of the consensus WGATAR. While ascertainment of the generality of this observation awaits additional studies, it is noteworthy that at least one other natural AGATAA site present on the BNP promoter (at −30 bp) also appears to be a preferential GATA-4-binding site (6a). The results raise the possibility of finding specific targets for GATA-4 or -6 based on differential affinities for their DNA sequences.
An important outcome of this study is the demonstration that some cardiac genes such as the ANF and BNP genes are bona fide targets for both GATA-4 and GATA-6 and that the two GATA factors form transcriptionnally active complexes over a single GATA element. Several lines of evidence supporting the existence of functional interaction between GATA-4 and GATA-6 are presented. First, the reduction of both GATA-4 and GATA-6 protein levels had the same effect on ANF expression as that of either factor alone, suggesting that GATA-4 and GATA-6 are members of a single functional complex and that the ablation of GATA-4 or GATA-6 in that complex is sufficient to disrupt its transcriptional activity. Second, when GATA-4 and GATA-6 are cotransfected in heterologous cells, they cooperatively activate ANF and BNP reporter genes. The presence of a functional complex between GATA-4 and GATA-6 is further supported by the finding that GATA-4 and GATA-6 physically interact in vitro and in vivo in postnatal cardiomyocytes. Finally, the colocalization of GATA-4 and GATA-6 in postnatal cardiomyocytes lends further credibility to the likelihood of in vivo relevance of a GATA-4 and GATA-6 interaction.
Homotypic (for GATA-1) and heterotypic interactions between hematopoietic GATA factors (GATA-1 and GATA-2 or GATA-3) via the DNA-binding domain of GATA-1 have also been reported (7, 27, 43). In the case of GATA-4 and GATA-6, physical interaction also occurs via the DNA-binding zinc finger domain; however, functional cooperativity requires the C-terminal activation domain of GATA-4, suggesting that the transcriptionally active complex includes additional cofactors that are involved in specific protein-protein interactions with one but not the other GATA member. In the case of ANF, such a cofactor may be Nkx2-5, which was shown to specifically interact with GATA-4 but not GATA-6 (9, 10). Thus, heterotypic interactions may be an intrinsic property of GATA factors, and the combinatorial interaction of different GATA factors present in various cell types with each other and with other cellular cofactors may contribute to their cell-specific mode of action. The finding that GATA-4 and GATA-6 cooperatively target the ANF and BNP genes in neonatal cardiomyocytes provides biological relevance for these heterotypic interactions.
ACKNOWLEDGMENTS
We are grateful to Gaétan Thibault for assistance with the ANF RIA, to Brent French for the Ctl adenovirus and the pΔE1sp1B/CMV/BGH shuttle vector, to Tony Antakly for production of the GATA-6 antibody, to Lynda Robitaille for technical assistance, to Lise Laroche for secretarial assistance, and to members of the Nemer laboratory for discussions and critical reading of the manuscript.
This work was supported by grants from the Canadian Medical Research Council (MRC). F.C. and O.B. are recipients of research traineeships from the Heart and Stroke Foundation of Canada, G.N. holds a GIBCO-IRCM studentship, and M.N. is an MRC Scientist.
REFERENCES
- 1.Agah R, Frenkel P A, French B A, Michael L H, Overbeek P A, Schneider M D. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Investig. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antakly T, Raquidan D, O’Donnel D, Katnick L. Regulation of glucocorticoid receptor expression. I. Use of a specific radioimmunoassay and antiserum to a synthetic peptide of the N-terminal domain. Endocrinology. 1990;126:1821–1828. doi: 10.1210/endo-126-4-1821. [DOI] [PubMed] [Google Scholar]
- 3.Ardati A, Nemer M. A nuclear pathway for α1-adrenergic receptor signaling in cardiac cells. EMBO J. 1993;12:5131–5139. doi: 10.1002/j.1460-2075.1993.tb06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argentin S, Ardati A, Tremblay S, Lihrmann I, Robitaille L, Drouin J, Nemer M. Developmental stage-specific regulation of atrial natriuretic factor gene transcription in cardiac cells. Mol Cell Biol. 1994;14:777–790. doi: 10.1128/mcb.14.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argentin S, Sun Y-L, Lihrmann I, Schmidt T J, Drouin J, Nemer M. Distal cis-acting promoter sequences mediate glucocorticoid stimulation of cardiac atrial natriuretic factor gene transcription. J Biol Chem. 1991;266:23315–23322. [PubMed] [Google Scholar]
- 6.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Charron, F., and M. Nemer. Unpublished data.
- 7.Crossley M, Merika M, Orkin S H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Lisi R, Millino C, Calabria E, Altruda F, Schiaffino S, Ausoni S. Combinatorial cis-acting elements control tissue-specific activation of the cardiac troponin I gene in vitro and in vivo. J Biol Chem. 1998;273:25371–25380. doi: 10.1074/jbc.273.39.25371. [DOI] [PubMed] [Google Scholar]
- 9.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durocher D, Nemer M. Combinatorial interactions regulating cardiac transcription. Dev Genet. 1998;22:250–262. doi: 10.1002/(SICI)1520-6408(1998)22:3<250::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.French B A, Mazur W, Ali N M, Geske R S, Finnigan J P, Rodgers G P, Roberts R, Raizner A E. Percutaneous transluminal in vivo gene transfer by recombinant adenovirus in normal porcine coronary arteries, atherosclerotic arteries, and two models of coronary restenosis. Circulation. 1994;90:2402–2413. doi: 10.1161/01.cir.90.5.2402. [DOI] [PubMed] [Google Scholar]
- 12.Gove C, Walmsley M, Nijjar S, Bertwistle D, Guille M, Partington G, Bomford A, Patient R. Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors. EMBO J. 1997;16:355–368. doi: 10.1093/emboj/16.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F L, Prevec L. Gene transfer and expression protocols. In: Murray E J, editor. Manipulation of adenovirus vectors. 1991. pp. 109–128. Methods in molecular biology. Humana Press, Clifton, N.J. [DOI] [PubMed] [Google Scholar]
- 14.Grépin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grépin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124:2387–2395. doi: 10.1242/dev.124.12.2387. [DOI] [PubMed] [Google Scholar]
- 16.Grépin C, Robitaille L, Antakly T, Nemer M. Inhibition of transcription factor GATA-4 expression blocks in vitro cardiac muscle differentiation. Mol Cell Biol. 1995;15:4095–4102. doi: 10.1128/mcb.15.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa K, Lee S J, Jobe S M, Markham B E, Kitsis R N. cis-acting sequences that mediate induction of beta-myosin heavy chain gene expression during left ventricular hypertrophy due to aortic constriction. Circulation. 1997;96:3943–3953. doi: 10.1161/01.cir.96.11.3943. [DOI] [PubMed] [Google Scholar]
- 18.Heikinheimo M, Scandrett J M, Wilson D B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 19.Herzig T C, Jobe S M, Aoki H, Molkentin J D, Cowley A W, Jr, Izumo S, Markham B E. Angiotensin II type 1a receptor gene expression in the heart: AP- 1 and GATA-4 participate in the response to pressure overload. Proc Natl Acad Sci USA. 1997;94:7543–7548. doi: 10.1073/pnas.94.14.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ip H S, Wilson D B, Heikinheimo M, Tang Z, Ting C N, Simon M C, Leiden J M, Parmacek M S. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y M, Evans T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev Biol. 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- 22.Kelley C, Blumberg H, Zon L I, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- 23.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohtz D S, Dische N R, Inagami T, Goldman B. Growth and partial differentiation of presumptive human cardiac myoblasts in culture. J Cell Biol. 1989;108:1067–1078. doi: 10.1083/jcb.108.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 26.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 27.Mackay J P, Kowalski K, Fox A H, Czolij R, King G F, Crossley M. Involvement of the N-finger in the self-association of GATA-1. J Biol Chem. 1998;273:30560–30567. doi: 10.1074/jbc.273.46.30560. [DOI] [PubMed] [Google Scholar]
- 28.McGrew M J, Bogdanova N, Hasegawa K, Hughes S H, Kitsis R N, Rosenthal N. Distinct gene expression patterns in skeletal and cardiac muscle are dependent on common regulatory sequences in the MLC1/3 locus. Mol Cell Biol. 1996;16:4524–4538. doi: 10.1128/mcb.16.8.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGrory W J, Bautista D S, Graham F L. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin J D, Kalvakolanu D V, Markham B E. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the α-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molkentin J D, Lin Q, Duncan S A, Olson E N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 32.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrisey E E, Ip H S, Lu M M, Parmacek M S. GATA-6—a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 34.Morrisey E E, Tang Z, Sigrist K, Lu M M, Jiang F, Ip H S, Parmacek M S. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy A M, Thompson W R, Peng L F, Jones L., II Regulation of the rat cardiac troponin I gene by the transcription factor GATA-4. Biochem J. 1997;322:393–401. doi: 10.1042/bj3220393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandolfi P P, Roth M E, Karis A, Leonard M W, Dzierzak E, Grosveld F G, Engel J D, Lindenbaum M H. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 37.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D’Agati V, Orkin S H, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 38.Rosoff M L, Nathanson N M. GATA factor-dependent regulation of cardiac m2 muscarinic acetylcholine gene transcription. J Biol Chem. 1998;273:9124–9129. doi: 10.1074/jbc.273.15.9124. [DOI] [PubMed] [Google Scholar]
- 39.Simon M C. Gotta have GATA. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 40.Tsai F Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 41.Viger R S, Mertineit C, Trasler J M, Nemer M. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Müllerian inhibiting substance promoter. Development. 1998;125:2665–2675. doi: 10.1242/dev.125.14.2665. [DOI] [PubMed] [Google Scholar]
- 42.Wang G F, Nikovits W, Jr, Schleinitz M, Stockdale F E. A positive GATA element and a negative vitamin D receptor-like element control atrial chamber-specific expression of a slow myosin heavy-chain gene during cardiac morphogenesis. Mol Cell Biol. 1998;18:6023–6034. doi: 10.1128/mcb.18.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H Y, Evans T. Homotypic interactions of chicken GATA-1 can mediate transcriptional activation. Mol Cell Biol. 1995;15:1353–1363. doi: 10.1128/mcb.15.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng W, Flavell R A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]