Abstract
Objectives
The number of deaths from cardiovascular disease is projected to reach 23.3 million by 2030. As a contribution to preventing this phenomenon, this paper proposed a machine learning (ML) model to predict patients with arteriosclerotic heart disease (AHD). We also interpreted the prediction model results based on the ML approach and deployed modelagnostic ML methods to identify informative features and their interpretations.
Methods
We used a hematology Electronic Health Record (EHR) with information on erythrocytes, hematocrit, hemoglobin, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, leukocytes, thrombocytes, age, and sex. To detect and predict AHD, we explored random forest (RF), XGBoost, and AdaBoost models. We examined the prediction model results based on the confusion matrix and accuracy measures. We used the Shapley Additive exPlanations (SHAP) framework to interpret the ML model and quantify the contribution of features to predictions.
Results
Our study included data from 6,837 patients, with 4,702 records from patients diagnosed with AHD and 2,135 records from patients without an AHD diagnosis. AdaBoost outperformed RF and XGBoost, achieving an accuracy of 0.78, precision of 0.82, F1-score of 0.85, and recall of 0.88. According to the SHAP summary bar plot method, hemoglobin was the most important attribute for detecting and predicting AHD patients. The SHAP local interpretability bar plot revealed that hemoglobin and mean corpuscular hemoglobin concentration had positive impacts on AHD prediction based on a single observation.
Conclusions
ML models based on real clinical data can be used to predict AHD.
Keywords: Machine Learning, Coronary Artery Disease, Hematology, Machine Learning, Supervised Machine Learning
I. Introduction
Atherosclerosis is a common condition characterized by the buildup of plaque in the arteries [1]. This accumulation can obstruct blood flow throughout the body. Consequently, when atherosclerosis affects the heart’s blood vessels, it can lead to coronary heart disease and heart attacks [1]. Cardiovascular disease (CVD) is the primary cause of death globally, accounting for approximately 17.9 million fatalities annually [2]. The Sample Registration System (SRS) 2019 report from the Ministry of Health Republic of Indonesia ranked heart disease as the second leading cause of death, following stroke. Notably, heart disease is a preventable condition [3]. Early detection and prediction, coupled with the ongoing analysis of Electronic Health Records (EHRs) by computational agents and machine learning (ML), are likely to become essential components in the management of patients with atherosclerotic heart disease (AHD) [4]. ML, a subset of artificial intelligence (AI), employs data analysis techniques to discern patterns and predict behaviors [5,6]. Predictive models developed using ML algorithms can assist in identifying patients with AHD and uncover previously unrecognized patterns of risk factors [7].
Numerous researchers have utilized a variety of ML methods to predict cardiovascular disease. One study [8] applied a random forest (RF) algorithm to predict atherosclerosis in China, using data from a retrospective study and statistical analysis. Park et al. [9] developed several ML models, including a classification and regression tree (CART) decision tree and RF, to predict the presence of coronary artery calcification. This was based on retrospective data from 3,302 Korean patients. Fan et al. [10] built ML models, specifically RF, decision tree, and eXtreme Gradient Boosting (XGBoost), to predict asymptomatic carotid atherosclerosis. This was done using EHRs from 6,553 patients in China. Ward et al. [11] employed logistic regression (LR), RF, gradient boosting (GB), and XGBoost algorithms to predict the risk of atherosclerotic CVD in a diverse patient cohort. Lastly, Terrada et al. [12] developed a medical diagnosis method to aid in predicting atherosclerosis in patients, using medical records from 835 patients.
Most previous studies have concentrated on the performance of ML models or the importance of features, with minimal focus on thoroughly understanding and explaining predictions using interpretable methods [8–13]. However, in clinical environments, models that are interpretable are generally favored over black box models [14,15]. Consequently, ML methods that are model-agnostic have been developed to identify informative features and interpret them. A model-agnostic interpretation method, such as Shapley Additive exPlanations (SHAP) framework, uses a dataset and various prediction models as inputs, applies these models to the data, and then identifies the characteristics of data features within each prediction model [16].
The objectives of this study were as follows: (1) to develop a predictive model for AHD using an ML approach and hematology EHR data, (2) to interpret the results of this predictive model using the ML approach, and (3) to construct model-agnostic ML methods for identifying informative features and interpreting them. In order to create a predictive model for AHD, we assessed the effectiveness of RF, XGBoost, and AdaBoost models, utilizing hematology EHR data. We chose to investigate RF, XGBoost, and AdaBoost as these algorithms have previously demonstrated potential in predicting CVD [8–10,12,13]. Subsequently, we evaluated the performance of each model to determine which one was superior (H1). Given that ML models are often seen as a black box, and that interpretable models are generally preferred in clinical settings, we incorporated interpretability into our ML model. This allowed us to calculate and examine the influence of features on individual and overall predictions, as well as to evaluate informative features and investigate their interpretability and characteristics. To address the hypothesis regarding how to interpret the predictive model and evaluate informative features (H2), we utilized the SHAP framework. This enabled us to further investigate their interpretability and characteristics.
Few studies have sought to answer the same questions posed in our report. We expect that our methodology will establish a foundation for future advancements by offering evidence and setting the stage for subsequent research on computational agents and ML. These tools are capable of detecting, predicting, and interpreting prediction models using EHRs.
II. Methods
1. Ethics and Data Use Agreement
We obtained EHR data for patients with heart disease from the Indonesia National Heart Center Harapan Kita EHR, under the ethical clearance number LB.02.01/VII/520/KEP014/2021. This data spans from 2016 to 2021. It was unclear when the predictors for each patient were extracted from the EHR (i.e., whether it was on the day the patient was initially diagnosed with AHD or on subsequent days).
2. Data Preprocessing
The EHR system houses both clinical and hematological test data for patients. It has stored records for 6,837 patients who have been diagnosed with heart disease by a physician using International Classification of Diseases 9th or 10th revision (ICD-9/ICD-10) codes. Subsequently, we identified the records of patients with AHD using the ICD-9/ICD-10 diagnosis code I25.1, which indicates AHD including coronary artery disease and coronary artery atheroma. We received 4,702 records from patients with AHD and 2,135 records from patients with no AHD. Patients with no AHD were those who did not have an AHD diagnosis.
The data preprocessing phase encompassed data cleaning, data integration, data transformation, and data reduction [17]. The EHR medical record table included several attributes: (1) patient information, which includes registration date, return date, patient name, medical record code, age, room code, laboratory test code, and doctor name; (2) the ICD code and its description; (3) hematology test attributes, which include erythrocytes, hematocrit, hemoglobin, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, leukocytes, thrombocyte, age, and sex. Unfortunately, we were unable to include total cholesterol, triglycerides, high-density lipoprotein (HDL), low-density lipoprotein (LDL), complete blood count with differential (CBC), and lipoprotein in our dataset as these were not recorded in our EHR. Data cleaning was the subsequent step, which involved identifying and rectifying errors or inconsistencies in the data, such as missing values and duplicates. In our dataset, we did not find any duplicate values, and we removed the row that contained missing values. After the data cleaning process, we retained 10 features as input variables. We identified eight numerical attributes: erythrocytes, hematocrit, hemoglobin, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, leukocytes, thrombocyte, and age, along with two categorical attributes—sex and diagnosis code (ICD code). Data transformation is the process of converting the data into a format suitable for analysis. For the categorical attribute of sex, we converted “female” to 0 and “male” to 1; for the diagnosis code, we converted “patient with AHD” to 1 and “patient with no AHD” to 0. We utilized the EHR diagnosis column as the data class label to identify and predict AHD patients. Table 1 displays all predictor attributes and their categorical values. All categorical values were established based on Indonesian medical laboratory testing standards for adults.
Table 1.
Predictor attributes and their baseline characteristics in the dataset (n = 6,837)
| Predictor attribute | Valuea | |
|---|---|---|
| Male | Female | |
| Sex | 5,505 | 1,332 |
| Age (yr) | 58.18 | 61.27 |
| Erythrocytes (million cells/μL) | 4.3–5.6 | 3.9–5.1 |
| Hematocrit (%) | 41–50 | 36–44 |
| Hemoglobin (g/dL) | 13–17 | 12–15 |
| MCH (pg) | 27.5–33.2 | |
| MCHC (g/dL) | 32–36 | |
| Leukocytes (103/μL) | 3.5–10.5 | |
| Thrombocyte (103/μL) | 135–317 | 157–371 |
| Diagnosis code | ||
| Number of patients with AHD | 4,702 | |
| Number of patients with no AHD | 2,135 | |
MCH: mean corpuscular hemoglobin, MCHC: mean corpuscular hemoglobin concentration, AHD: atherosclerotic heart disease.
Based on Indonesian medical laboratory testing standards for adults.
3. Machine Learning Algorithms for Detection and Prediction
The prediction experiments utilized RF, XGBoost, and Ada-Boost algorithms. RF is an ensemble of high-performing trees that are amalgamated into a single model. Notably, this algorithm surpasses the performance of the decision tree algorithm [18]. XGBoost, meanwhile, is an optimized distributed gradient boosting library, designed to be highly efficient, flexible, and portable [14]. AdaBoost is an ML approach that was originally developed as an ensemble method to enhance the performance of binary classifiers [19]. A variety of ML techniques used in heart disease prediction are outlined in Table 2 [9,15,19,20–23]. The parameter tuning for RF, XGBoost, and AdaBoost is presented in Table 3.
Table 2.
Compilation of machine learning (ML) techniques for heart disease prediction
| Study | ML technique | Dataset | Result and limitation |
|---|---|---|---|
| Almustafa [20] | NB, SGD, SVM, KNN, DT, AdaBoost | 1,025 patient records from Cleveland, Hungary, Switzerland, and Long Beach datasets. 14 attributes were used, mainly, age, sex, chest pain type, resting blood pressure, serum cholesterol, fasting blood sugar, resting electrocardiographic results, maximum heart rate achieved, exercise-induced angina, old peak, the slope of the peak exercise ST segment, number of major vessels fluoroscopy and defect along with the class attribute. | Classification algorithms for the heart disease dataset produce very promising results in terms of classification accuracy. In-depth sensitivity analysis and performance have not been performed. |
| Park et al. [9] | LRM, CART, CIT, RF | 3,302 patient records from two cohorts (Soonchunhyang University Cheonan Hospital and Kangbuk Samsung Health Study). Attributes were namely HTN (hypertension), DM (diabetes mellitus), eGFR (estimated glomerular filtration rate), BMI (body mass index), non-HDL (non-high-density lipoprotein) cholesterol, and CACS (coronary artery calcification score). | All models showed acceptable accuracies: LR (70.71%), CART (71.32%), CIT (71.32%), RF (71.02%). The cohorts used in this study had previously been enrolled in other studies, which could result in biases. |
| Su et al. [21] | RF, LR | 498 subjects were conducted in Xi’an Medical University. The risk of developing CVD can be predicted according to the individual’s age, BMI, triglycerides, and diastolic blood pressure (DBP). | The ROC-AUCs were 0.802 for random forest model and 0.843 for LR model. A retrospective study with a small number of subjects (n = 498). |
| Budholiya et al. [15] | XGboost, RF, ExtraTree classifiers | The Cleveland Heart Disease dataset obtained from the University of California, Irvine (UCI) online ML, and data mining repository. Attributes were namely, age, sex, chest pain, resting blood pressure, serum cholesterol, fasting blood sugar, resting electrocardiograph results, maximum heart rate achieved, exercise-induced angina, ST-depression, ST-slope, number of major vessels, thalassemia, num (target variable). | XGboost performed the highest prediction accuracy of 91.8%. This research has not performed interpretable methods for ML to understand and explain predictions results. |
| Cao et al. [22] | LR, BP neural network, XGBoost, RF | 553 patients in the Department of Cardiology at a tertiary hospital in Anhui Province. Clinical data sources include patients’ general data, cardiac ultrasound recording, laboratory examination results. |
The XGBoost model’s prediction value was the best. A retrospective study with a small number of subjects (n = 553). |
| Absar et al. [19] | RF, DT, Ada-Boost, KNN | The Cleveland Heart Disease dataset obtained from the University of California, Irvine (UCI) online machine learning, and data mining repository. Attributes were namely, age, sex, chest pain type, blood pressure, serum cholesterol, fasting blood sugar, resting electro-cardiographic, maximum heart rate, old peak, the slant of the peak exercise ST segment, number of major vessels, exercise-induced angina, Thalach. | AdaBoost performed the highest prediction accuracy of 100%. This research has not performed interpretable methods for ML to understand and explain predictions results. |
| Mahesh et al. [23] | NB, DT, AdaBoost | UCI Repository provided the Heart Disease dataset. Attributes were namely, age, sex, chest pain, resting blood pressure, serum cholesterol, fasting blood sugar, resting electrocardiograph, maximum heart rate achieved, exercise-induced angina, oldpeak, slope, major vessels colored by fluoroscopy, defect type. | AdaBoost-RF classifier provides 95.47% accuracy in the early detection of heart disease. This research has not performed interpretable methods for ML to understand and explain predictions results. |
NB: naïve Bayes, SGD: stochastic gradient descent, SVM: support vector machine, KNN: K-nearest neighbor, DT: decision tree, LR: logistic regression, CART: classification and regression tree, CIT: conditional inference tree, RF: random forest, BP: backpropagation
Table 3.
Parameter tuning for the algorithm
| Algorithm | Parameter tuning | Definition |
|---|---|---|
| Random forest | n_estimators =100 | The number of trees in the forest |
| Criterion = entropy | The function to measure the quality of a split | |
| max_depth = none | The maximum depth of the tree | |
| min_samples_split = 2 | The minimum number of samples required to split an internal node | |
| min_samples_leaf = 1 | The minimum number of samples required to be at a leaf node | |
| min_weight_fraction_leaf = 0.0 | The minimum weighted fraction of the sum total of weights (of all the input samples) required to be at a leaf node | |
| max_features = “sqrt” | The number of features to consider when looking for the best split | |
|
| ||
| XGBoost | loss = ‘log_loss’ | The loss function to be optimized. ‘log_loss’ refers to binomial and multinomial deviance |
| learning_rate = 0.1 | Learning rate shrinks the contribution of each tree by learning_rate | |
| n_estimators = 100 | The number of boosting stages to perform | |
| subsample = 1.0 | The fraction of samples to be used for fitting the individual base learners | |
| criterion = ‘friedman_mse’ | The function to measure the quality of a split | |
| min_samples_split = 2 | The minimum number of samples required to split an internal node | |
| min_samples_leaf = 1 | The minimum number of samples required to be at a leaf node | |
| min_weight_fraction_leaf = 0.0 | The minimum weighted fraction of the sum total of weights (of all the input samples) required to be at a leaf node | |
| max_depth = 3 | Maximum depth of the individual regression estimators | |
| min_impurity_decrease = 0.0 | A node will be split if this split induces a decrease of the impurity greater than or equal to this value | |
| Init estimator = none | An estimator object that is used to compute the initial predictions | |
| RandomState instance or None = None | Controls the random seed given to each Tree estimator at each boosting iteration | |
| max_features = none | The number of features to consider when looking for the best split. If None, then max_features=n_features | |
|
| ||
| AdaBoost | estimator object = none | The base estimator from which the boosted ensemble is built. |
| n_estimators = 50 | The maximum number of estimators at which boosting is terminated | |
| learning_rate = 1.0 | Weight applied to each classifier at each boosting iteration | |
| algorithm = ’SAMME.R’ | If ‘SAMME.R’ then use the SAMME.R real boosting algorithm | |
| RandomState instance = none | Controls the random seed given at each estimator at each boosting iteration | |
| base_estimatorobject = none | The base estimator from which the boosted ensemble is built | |
4. Performance Measures
We used a confusion matrix to evaluate the models by deriving the following metrics: true positives, true negatives, false positives, and false negatives [24]. We calculated accuracy, precision, F1-score, and recall. We also calculated the area under the receiver operating characteristic curve (AUC) value. The receiver operating characteristic (ROC) curve is a measure of the predictive quality of a classifier. The optimum position is thus in the plot’s upper left corner, where false positives equal 1 and true positives equal 0. The AUC denotes the degree or measure of separability. This shows how well the model can differentiate among classes. A higher AUC means that the model better predicts class 0 as 0 and class 1 as 1 [25].
5. Model-Agnostic Interpretation
ML models are often perceived as black boxes, accepting specific features, and producing predictions. Generally, in clinical scenarios, models that are interpretable are favored over black box models. In a computerized environment, an agnostic approach is one that can operate across various platforms [26].
SHAP is a game-theoretic approach used to interpret the output of ML models. This method ranks attributes based on their contribution to the model, and it can visually display the relationship between these attributes and the results. The absolute value of an attribute signifies its influence, while its positive or negative value indicates the attribute’s predictive power for atherosclerotic heart disease. SHAP allows for the calculation of a feature’s impact on both individual and global predictions [27]. The model’s g value is calculated using the following formula (1), where p is the number of attributes, z = [z1, z2,…, zp] is a simplification in the input, where z represents the data prediction attributes and is 1, and the unused attribute has a z value of 0. Furthermore, φi ∈ ℝ reflects each attribute’s contribution to the model [27].
| (1) |
The higher the SHAP value, the greater the positive contribution of the attributes, and vice versa [28].
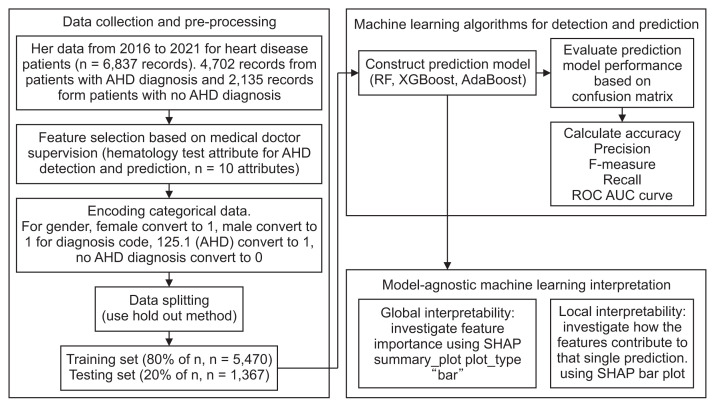
In our study, we utilized the SHAP model to generate SHAP values for our test dataset. Subsequently, we created a SHAP summary bar plot for global interpretability and a separate bar plot for local interpretability, both pertaining to the prediction model. The steps of our proposed research study are depicted in Figure 1.
Figure 1.
Steps of the proposed research study. EHR: Electronic Health Record, AHD: atherosclerotic heart disease, ROCAUC: ROC-AUC: receiver operating characteristic-area under the curve, SHAP: Shapley Additive exPlanations.
III. Results
1. Machine Learning Algorithm’s Performance for Detection and Prediction
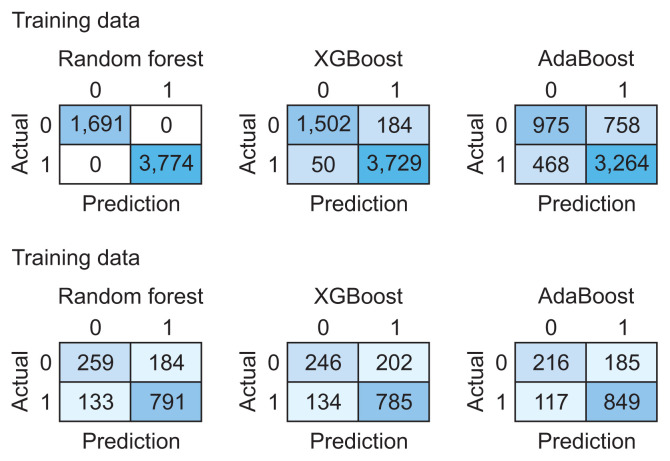
We utilized 6,837 patient records in our study. The diagnosis code was employed as the label in our model. For the ML model to be trained, the dataset needs to be divided into training and testing data [16]. We randomly split the dataset into two parts using the hold-out method, allocating 80% of the data for training (n = 5,470) and the remaining 20% for testing (n = 1,367). Figure 2 presents the confusion matrix of the training and testing data for each algorithm. Table 4 illustrates the performance of RF, XGBoost, and AdaBoost when applied to the test set. Accuracy is a measure of how many positive and negative observations were correctly classified. Precision addresses the question of what percentage of positive identifications were indeed correct. The F1-score is the harmonic mean of precision and recall, and it is not solely based on the accuracy value. Recall addresses the question of what percentage of actual positives were correctly identified [24,25]. Figure 3 showcases the ROC-AUC curve for RF, XGBoost, and AdaBoost. A ROC curve plots the true positive rate on the Y-axis and the false positive rate on the X-axis, both globally and on a per-class basis. The ideal point is located in the upper left corner of the plot, where false positives are 0 and true positives are 1. The AUC quantifies the relationship between false positives and true positives. A higher AUC indicates a better overall model [15].
Figure 2.
Confusion matrix for the training data (top) and testing data (bottom).
Table 4.
Predictive model performance for the test set
| Algorithm | Accuracy | Precision | F1-score | recall | ROC-AUC |
|---|---|---|---|---|---|
| Random forest | 0.77 | 0.81 | 0.83 | 0.86 | 0.82 |
| XGBoost | 0.75 | 0.80 | 0.82 | 0.85 | 0.80 |
| AdaBoost | 0.78 | 0.82 | 0.85 | 0.88 | 0.81 |
ROC-AUC: receiver operating characteristic-area under the curve.
Figure 3.
Receiver operating characteristic (ROC) curve and area under the curve (AUC) for (A) random forest, (B) XGBoost, and (C) AdaBoost.
2. Model-Agnostic Interpretation
Given that AdaBoost surpassed nearly all other classification and prediction metrics, we employed SHAP to elucidate the predictions of a single instance (a patient’s record), by determining the contribution of each feature to the predictions. The SHAP Python library was utilized to compute SHAP values and generate charts. We applied both global and local SHAP interpretability to demonstrate the comprehensive contribution of the feature to both global and local interpretability.
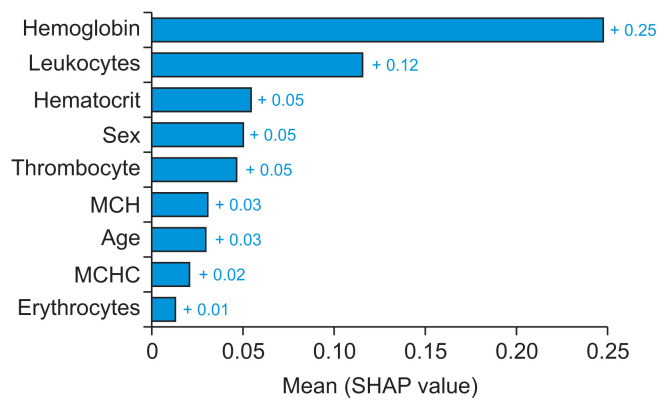
A global feature significance plot was generated by inputting a matrix of SHAP values into the bar plot function. This process assigned the global importance of each feature to correspond with the mean absolute value of that feature across all samples. The x-axis represents the average absolute SHAP value of each feature. The features are organized in descending order based on their impact on the model’s prediction. This arrangement takes into account the absolute SHAP value, meaning it is irrelevant whether the feature positively or negatively influences the prediction. Figure 4 presents a global feature importance plot for the AdaBoost algorithm, which was used to detect and predict AHD. This plot was created using a bar type in the Python library. Hemoglobin was the most significant attribute for detecting and predicting arteriosclerotic heart disease in patients, followed by leukocytes, hematocrit, sex, thrombocytes, mean corpuscular hemoglobin, age, mean corpuscular hemoglobin concentration, and erythrocytes.
Figure 4.
Global interpretability: feature importance for the AdaBoost algorithm to detect and predict atherosclerotic heart disease (as visualized by summary_plot method with plot type bar in the Python library). MCH: mean corpuscular hemoglobin, MCHC: mean corpuscular hemoglobin concentration, SHAP: Shapley Additive exPlanations.
Next, we used the plots of individual data points to evaluate the implications on a case-by-case basis. These plots illustrate the primary features that influence the prediction of a single observation, along with the magnitude of the SHAP value for each feature. The bar plot is zero-centered to emphasize the contributions of different variables. Each bar corresponds to the SHAP value of a particular feature. Positive shifts are represented by red bars, while blue bars denote negative shifts. Figure 5 provides a local interpretation of the AdaBoost algorithm’s ability to identify and predict AHD, using a bar plot from the Python library. Hemoglobin and mean corpuscular hemoglobin concentration are indicated by red bars, signifying positive shifts, while leukocyte, hematocrit, sex, thrombocyte, mean corpuscular hemoglobin, age, and erythrocyte are represented by blue bars, indicating negative shifts that affect the prediction of a single observation.
Figure 5.
Local interpretability for the AdaBoost algorithm to detect and predict atherosclerotic heart disease (as visualized by the plot method with plot type bar plot in the Python library). In data preprocessing, we converted female to 0 and male to 1. MCH: mean corpuscular hemoglobin, MCHC: mean corpuscular hemoglobin concentration SHAP: Shapley Additive exPlanations.
IV. Discussion
1. Findings
In this study, we developed a ML model to predict AHD using EHR hematology data. We evaluated three models: RF, XGboost, and AdaBoost. Our experiment demonstrated that AdaBoost outperformed the other models in nearly all classification and prediction measures (accuracy, precision, F1-score, recall), with the exception of AUC, where it ranked second, slightly below the AUC value of RF (H1). Given that AdaBoost surpassed RF and XGBoost in almost all classification and prediction measures, we employed SHAP analysis to uncover insights and patterns that were not readily discernible from the initial AdaBoost features. A global interpretability analysis using the SHAP summary_plot method with a bar plot type revealed that hemoglobin is the most critical attribute for detecting and predicting AHD patients. This was followed by leukocyte, hematocrit, sex, thrombocytes, mean corpuscular hemoglobin, age, mean corpuscular hemoglobin concentration, and erythrocytes (H2). In addition to examining the global trends in feature impact, we also used the local interpretability SHAP bar plot method to explore the contribution of individual features to individual predictions. In this context, hemoglobin and mean corpuscular hemoglobin concentration (represented by the red bar) had a positive impact, while leukocytes, hematocrit, sex, thrombocytes, mean corpuscular hemoglobin, age, and erythrocytes (represented by the blue bar) had a negative impact on the AHD prediction of a single observation (H2).
The clinical implications of our research are as follows: (1) For interpretation and prediction, we processed complex, heterogeneous data from EHR to predict patients with AHD. This enhances our understanding of the patient’s condition. (2) For comprehension, we interpreted the prediction model and evaluated informative features. We also investigated their interpretability and characteristics with the aim of explaining the predictions of an instance (patient’s record). This was achieved by calculating the contribution and impact of each attribute to the predictions. (3) For decision support, we utilized the previous steps to predict clinical outcomes. Our findings indicated that hemoglobin was the most crucial feature for detecting and predicting AHD patients, as evidenced by the SHAP value. Both hemoglobin and mean corpuscular hemoglobin concentration demonstrated positive shifts in predicting a single observation. Previous research has corroborated some of our findings, thus validating our approach to evaluating informative features of the ML prediction model and investigating their interpretability and contributions to prediction. Lee et al. [29] and Goel et al. [30] concluded that both low and high hemoglobin concentrations were associated with increased cardiovascular and all-cause mortality. This aligns with our finding that hemoglobin has a positive impact on individual predictions of AHD.
The technical implication of our research is that we uncovered trends that have seldom been investigated before. Most prior studies have concentrated on the performance of ML models or the importance of features, with little focus on fully understanding and explaining predictions using interpretable methods. In clinical settings, interpretable models are often favored over black box models [26]. Previous research has explored RF, XGBoost, and AdaBoost for heart disease prediction, yielding better results than ours. A previous study [21] utilized RF and achieved ROC-AUC of 0.802, but it involved a small sample size of 498 subjects. Another study [15] employed XGBoost and achieved a prediction accuracy of 91.8%. Furthermore, yet another study [19] used Ada-Boost and achieved a prediction accuracy of 100%. However, these studies did not employ interpretable methods for ML to understand and explain prediction results, an aspect that was addressed in our study.
Finally, our proposed ML model, along with the interpretability model, holds promise as tools for detecting and predicting AHD patients, as well as elucidating the prediction results. Our study was not designed as a prospective investigation observing disease progression over time. Instead, our proposed methodology involved a post-hoc analysis of hematological EHR data, from which we sought to extract valuable information. Utilizing this information, we constructed a prediction model for arteriosclerotic heart disease, in addition to a machine-learning agnostic model, both based on ML techniques.
2. Limitations
This preliminary study aimed to understand and explain predictions made by ML models using an interpretable method. Our research indicates that ML has significant potential to enhance clinical investigation. One of the primary challenges for ML approaches in interpreting results is the extraction of meaningful concepts and attributes from raw data and datasets. This includes building prediction models, understanding and evaluating the performance of these predictions, and interpreting the results of these predictions. At present, our study only utilized structured data from hematology EHRs and excluded information from a variety of tests used in diagnosing heart conditions. Blood tests that record characteristics such as total cholesterol, triglycerides, HDL, LDL, CBC, and lipoprotein could be considered comprehensive attributes for model prediction in future studies. These tests are used to determine the risk of CAD. In future research, we need to expand the ML model used in this study and acquire different comprehensive attributes. These attributes should contain additional information about the patient’s medical tests that may contribute to AHD. This will enhance the performance of our proposed prediction model. Furthermore, the impact of the directionality of the features that demonstrated the significance of a feature, and whether it has a positive or negative impact on prediction, should be validated with future medical research.
Acknowledgments
The research was sponsored by Research and Technology Transfer Office, Bina Nusantara University as a part of Bina Nusantara University’s International Research Grant entitled “Model Prediksi Penyakit Jantung dengan Pendekatan Data Mining Probabilistic Classifier” (Contract No. 061/VR.RTT/IV/2022, Date: April 8, 2022) and supported by the Indonesia National Heart Center Harapan Kita for data collection.
Footnotes
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
References
- 1.Vinciguerra M, Romiti S, Fattouch K, De Bellis A, Greco E. Atherosclerosis as pathogenetic substrate for Sars-Cov2 cytokine storm. J Clin Med. 2020;9(7):2095. doi: 10.3390/jcm9072095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Cardiovascular disease [Internet] Geneva, Switzerland: World Health Organization; c2023. [cited at 202. Jul 27]. Available from https://www.who.int/health-topics/cardiovasculardiseases#tab=tab_1. [Google Scholar]
- 3.Ministry of Health Republic of Indonesia . Non-communicable disease management guide. [Internet] Jakarta, Indonesia: Ministry of Health Republic of Indonesia; 2019. [cited at 2023 Jul 27]. Available from http://p2ptm.kemkes.go.id/uploads/VHcrbkVobjRzUDN3UCs4eU-J0dVBndz09/2019/03/Buku_Pedoman_Manajemen_PTM.pdf. [Google Scholar]
- 4.Capotosto L, Massoni F, De Sio S, Ricci S, Vitarelli A. Early diagnosis of cardiovascular diseases in workers: role of standard and advanced echocardiography. Biomed Res Int. 2018;2018:7354691. doi: 10.1155/2018/7354691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhammad Y, Tahir M, Hayat M, Chong KT. Early and accurate detection and diagnosis of heart disease using intelligent computational model. Sci Rep. 2020;10(1):19747. doi: 10.1038/s41598-020-76635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo CY, Wu MY, Cheng HM. The comprehensive machine learning analytics for heart failure. Int J Environ Res Public Health. 2021;18(9):4943. doi: 10.3390/ijerph18094943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karthick K, Aruna SK, Samikannu R, Kuppusamy R, Teekaraman Y, Thelkar AR. Implementation of a heart disease risk prediction model using machine learning. Comput Math Methods Med. 2022;2022:6517716. doi: 10.1155/2022/6517716. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Chen Z, Yang M, Wen Y, Jiang S, Liu W, Huang H. Prediction of atherosclerosis using machine learning based on operations research. Math Biosci Eng. 2022;19(5):4892–910. doi: 10.3934/mbe.2022229. [DOI] [PubMed] [Google Scholar]
- 9.Park S, Hong M, Lee H, Cho NJ, Lee EY, Lee WY, et al. New model for predicting the presence of coronary artery calcification. J Clin Med. 2021;10(3):457. doi: 10.3390/jcm10030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J, Chen M, Luo J, Yang S, Shi J, Yao Q, et al. The prediction of asymptomatic carotid atherosclerosis with electronic health records: a comparative study of six machine learning models. BMC Med Inform Decis Mak. 2021;21(1):115. doi: 10.1186/s12911-021-01480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward A, Sarraju A, Chung S, Li J, Harrington R, Heidenreich P, et al. Machine learning and atherosclerotic cardiovascular disease risk prediction in a multiethnic population. NPJ Digit Med. 2020;3:125. doi: 10.1038/s41746-020-00331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terrada O, Cherradi B, Raihani A, Bouattane O. A novel medical diagnosis support system for predicting patients with atherosclerosis diseases. Inf Med Unlocked. 2020;21:100483. doi: 10.1016/j.imu.2020.100483. [DOI] [Google Scholar]
- 13.Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016 Apr 13–17; San Francisco, CA. pp. 785–94. [DOI] [Google Scholar]
- 14.Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2017;2(2):204–9. doi: 10.1001/jamacardio.2016.3956. [DOI] [PubMed] [Google Scholar]
- 15.Budholiya K, Shrivastava SK, Sharma V. An optimized XGBoost based diagnostic system for effective prediction of heart disease. J King Saud Univ Comput Inf Sci. 2022;34(7):4514–23. doi: 10.1016/j.jksuci.2020.10.013. [DOI] [Google Scholar]
- 16.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30:4765–74. [Google Scholar]
- 17.Cho E, Chang TW, Hwang G. Data preprocessing combination to improve the performance of quality classification in the manufacturing process. Electronics. 2022;11(3):477. doi: 10.3390/electronics11030477. [DOI] [Google Scholar]
- 18.Johnson A, Cooper GF, Visweswaran S. A novel personalized random forest algorithm for clinical outcome prediction. Stud Health Technol Inform. 2022;290:248–52. doi: 10.3233/SHTI220072. [DOI] [PubMed] [Google Scholar]
- 19.Absar N, Das EK, Shoma SN, Khandaker MU, Miraz MH, Faruque MR, et al. The efficacy of machine-learning-supported smart system for heart disease prediction. Healthcare (Basel) 2022;10(6):1137. doi: 10.3390/healthcare10061137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almustafa KM. Prediction of heart disease and classifiers’ sensitivity analysis. BMC Bioinformatics. 2020;21(1):278. doi: 10.1186/s12859-020-03626-y. https://doi.org10.1186/s12859-020-03626-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su X, Xu Y, Tan Z, Wang X, Yang P, Su Y, et al. Prediction for cardiovascular diseases based on laboratory data: an analysis of random forest model. J Clin Lab Anal. 2020;34(9):e23421. doi: 10.1002/jcla.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao J, Zhang L, Ma L, Zhou X, Yang B, Wang W. Study on the risk of coronary heart disease in middle-aged and young people based on machine learning methods: a retrospective cohort study. PeerJ. 2022;10:e14078. doi: 10.7717/peerj.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahesh TR, Dhilip Kumar V, Vinoth Kumar V, Asghar J, Geman O, Arulkumaran G, et al. AdaBoost ensemble methods using k-fold cross validation for survivability with the early detection of heart disease. Comput Intell Neurosci. 2022;2022:9005278. doi: 10.1155/2022/9005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alelyani S. Detection and evaluation of machine learning bias. Appl Sci. 2021;11(14):6271. doi: 10.3390/app11146271. [DOI] [Google Scholar]
- 25.He S, Qu L, He X, Zhang D, Xie N. Comparative evaluation of 15-minute rapid diagnosis of ischemic heart disease by high-sensitivity quantification of cardiac biomarkers. Exp Ther Med. 2020;20(2):1702–8. doi: 10.3892/etm.2020.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S, Chen R, Wei W, Belovsky M, Lu X. Understanding heart failure patients EHR clinical features via SHAP interpretation of tree-based machine learning model predictions. AMIA Annu Symp Proc. 2022;2021:813–22. [PMC free article] [PubMed] [Google Scholar]
- 27.Futagami K, Fukazawa Y, Kapoor N, Kito T. Pairwise acquisition prediction with SHAP value interpretation. J Financ Data Sci. 2021;7:22–44. doi: 10.1016/j.jfds.2021.02.001. [DOI] [Google Scholar]
- 28.Wang K, Tian J, Zheng C, Yang H, Ren J, Liu Y, et al. Interpretable prediction of 3-year all-cause mortality in patients with heart failure caused by coronary heart disease based on machine learning and SHAP. Comput Biol Med. 2021;137:104813. doi: 10.1016/j.compbiomed.2021.104813. [DOI] [PubMed] [Google Scholar]
- 29.Lee G, Choi S, Kim K, Yun JM, Son JS, Jeong SM, et al. Association of hemoglobin concentration and its change with cardiovascular and all-cause mortality. J Am Heart Assoc. 2018;7(3):e007723. doi: 10.1161/JAHA.117.007723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goel H, Hirsch JR, Deswal A, Hassan SA. Anemia in cardiovascular disease: marker of disease severity or disease-modifying therapeutic target? Curr Atheroscler Rep. 2021;23(10):61. doi: 10.1007/s11883-021-00960-1. [DOI] [PubMed] [Google Scholar]