Abstract
Background:
The respiratory tract fungal microbiome in cystic fibrosis (CF) has been understudied despite increasing recognition of fungal pathogens in CF lung disease. We sought to better understand the fungal communities in adults with CF, and to define relationships between fungal profiles and clinical characteristics.
Methods:
We enrolled 66 adults with CF and collected expectorated sputum, spirometry, Cystic Fibrosis Questionnaire-revised, and clinical data. Fungi were molecularly profiled by sequencing of the internal transcribed spacer (ITS) region. Total fungal abundance was measured by quantitative PCR. Relative abundance and qPCR-corrected abundances were determined. Selective fungus culture identified cultivable fungi. Alpha diversity and beta diversity were measured and relationships with clinical parameters were interrogated.
Results:
Median age was 29 years and median FEV1 percent predicted 58%. Members of the Candida genus were the most frequent dominant taxa in CF sputum. Apiotrichum, Trichosporon, Saccharomyces cerevisiae, and Scedosporium were present in high relative abundance in few samples; whereas, Aspergillus species were detected at low levels. Higher FEV1% predicted and CFTR modulator use were associated with greater alpha-diversity. Chronic azithromycin use was associated with lower alpha-diversity. Patients with acute pulmonary had distinct fungal community composition compared to clinically stable subjects. Differing yeast species were mainly responsible for the community differences.
Conclusion:
The respiratory tract fungal microbiome in adults with CF is associated with lung function, pulmonary exacerbation status, macrolide use, and CFTR modulator use. Future work to better understand fungal diversity in the CF airway and its impact on lung health is necessary.
Keywords: Fungi, Microbiome, Infection, Aspergillus, Candida
1. Introduction
The early description of cystic fibrosis (CF) was marked by malnutrition and lung infections. The diagnosis and treatment of infection has transformed disease prognosis to a median predicted survival over 50 years [1]. Yet, chronic infections continue to significantly impact morbidity and mortality in CF [1,2]. Over time, the microbial epidemiology has changed, with a rise of filamentous fungi and yeast recovered by culture from the CF respiratory tract [3]. The pressures of cumulative and chronic exposure to antibiotics in the course of CF likely contributes to the observed changes in fungal detection [4].
Conventional culture-based detection methods are the primary modality to identify microorganisms in CF sputum. However, the adoption of DNA sequence-based analyses has greatly expanded our understanding of bacterial diversity in CF. Unbiased 16S rRNA gene sequencing of CF respiratory samples have yielded insight into age-specific bacterial diversity and resiliency of the bacterial community during pulmonary exacerbations [5-7]. However, mycologic profiling of the CF airway has remained limited [8-13]. Our study aimed to molecularly define the fungal composition and community structure in CF sputa and evaluate their associations with clinical characteristics and outcomes.
2. Materials and methods
2.1. Study design and participants
We conducted a prospective study of adults 18 years and older with CF followed in the adult CF center at the University of Pennsylvania from March through November 2017. We included subjects with a diagnosis of CF according to the CF Foundation diagnosis guidelines and excluded those with a history of solid organ transplantation. All eligible participants, regardless of health state, were consecutively recruited from the clinic and written informed consent was obtained. The study was approved by the University of Pennsylvania institutional review board (protocol 825979).
Demographic and clinical data were collected at the time of sampling and included comorbidities, medication use within the past three months (cystic fibrosis transmembrane conductance regulator [CFTR] modulators, chronic non-macrolide oral antibiotics, azithromycin, chronic prednisone, inhaled anti-pseudomonal antibiotics, inhaled corticosteroid, and antifungals), spirometry, clinical microbiological data in the past 24 months, and presence of pulmonary exacerbation at the time of sampling. Pulmonary exacerbation was defined as a physician diagnosis of an acute worsening of respiratory symptoms and/or lung function with or without anorexia or weight loss, fatigue, and pulmonary exam findings. Chronic use of antibiotics was defined as continuous use for at least one month during the past 3 months. Spirometry was performed according to the American Thoracic Society guidelines and FEV1% predicted reported according to the National Health and Nutrition Examination Survey III prediction equations. Respiratory-related quality-of-life scores were collected with the Cystic Fibrosis Questionnaire-revised (CFQ-R). A higher CFQ-R respiratory domain score (range 0 to 100) reflects better respiratory-related quality-of-life [14].
2.2. Sputum sample collection and analysis
Subjects spontaneously expectorated consecutive sputum samples into two sterile specimen cups during a single encounter; one for culture and one underwent DNA extraction and sequencing. Six samples of DNA-free water were collected in specimen cups at the clinic sites of sample collection to serve as environmental controls. Sputum underwent bacterial culture and semi-selective fungal culture within 8 h of collection as previously described [15]. Morphologic identification of macroscopic and microscopic features of fungal isolates was done following standard clinical microbiological procedure. Sputum for DNA extraction was placed immediately at 4 °C after collection and processed within 24 h. Dithiothreitol (DTT) was added to liquefy mucous, which was then aliquoted under sterile conditions and stored at −80 °C until sequencing. Genomic DNA was extracted from sputum in a single batch using the DNeasy PowerSoil kit (Qiagen) with an additional 10-minute heating step at 95 °C before bead beating to enhance fungal recovery. The fungal internal transcribed spacer (ITS) region was amplified using barcoded ITS1F/ITS2 primers (Table S1). The resulting library was sequences on the Illumina MiSeq instrument yielding paired end reads that were 250 bp in length. Total fungal abundance was measured by quantitative PCR (qPCR) using the FungiQuant assay, a Taqman qPCR assay targeting a 351 bp region in the fungal 18S rRNA gene [16]. Values were calculated as copy number per μL of sputum. A detailed description of the methods is outlined in the Supplement.
2.3. Bioinformatic analysis
Paired-end ITS sequence reads were analyzed using the PIPITS pipeline. Taxonomic assignments of representative ITS sequences were generated by BROCC (BLAST Read and Operational Taxonomic Unit Consensus Classifier), a validated pipeline using the nt database from NCBI. BROCC uses local alignments to classify sequences by consensus-based algorithm to accurately characterize single cell eukaryotes [17]. We determined the relative abundances of fungal taxa from ITS sequencing. To approximate absolute fungal quantities at a taxonomic level, the qPCR-corrected abundance was computed by multiplying the relative abundance of each taxon in the sample by the absolute fungal abundance as estimated by qPCR. For samples that did not have quantifiable qPCR values, we imputed the values by calculating half of the limit of detection with threshold of 40 cycles (33.4 copies per μL of sputum) as 16.7 copies. Detailed methods are included in the Supplement.
2.4. Statistical analysis
Community composition was characterized by the richness (number of different taxa) and alpha (within sample) diversity based on the Shannon index, which accounts for the number of taxa and evenness of distribution. Bray-Curtis dissimilarity was used to measure beta diversity, or the dissimilarity between fungal communities in each pair of samples. We compared the diversity measures to clinical characteristics at the time of sampling, including age, lung function (defined as FEV1 percent predicted), CFQ-R respiratory domain score, chronic inhaled antibiotic use, chronic azithromycin use, use of CFTR modulator drugs, antifungals, pulmonary exacerbation status, and culture-positive Pseudomonas aeruginosa. Least squares linear model was used to compare differences in richness and Shannon diversity. Bray-Curtis was analyzed using permutational multivariate analysis of variance (PERMANOVA) of uncorrected abundances, as implemented by the R function adonis() in the vegan package version 2.3–5. We included covariates age, sex, and FEV1 percent predicted in the PER-MANOVA model. Sensitivity analysis using beta dispersion analyses were conducted. We calculated the ratio of qPCR-corrected abundances of Candida dubliniensis and Candida albicans and determined the association between the ratio and pulmonary exacerbation state using t-test.
Statistical analyses were conducted using R version 4.0.3.
3. Results
3.1. Subject characteristics
We recruited 66 patients with CF (Table 1). Median age was 29 years and 52% were female. Pancreatic insufficiency was present in 92%. Homozygosity for the F508del mutation was present in 47% and the use of CFTR modulators (ivacaftor or lumacaftor/ivacaftor) in 36%. Median forced expiratory volume in one second percent (FEV1%) percent predicted was 58%. Thirty-five individuals (53%) were experiencing an acute pulmonary exacerbation at time of sampling.
Table 1.
Subject characteristics (n = 66).
| N (%) or median [interquartile range] |
|
|---|---|
| Demographics | |
| Age (years) | 29 [23,41] |
| Female sex | 34 (51.5) |
| White race | 62 (93.9) |
| Disease characteristics | |
| F508del homozygous | 31 (47.0) |
| Pancreatic insufficiency | 61 (92.4) |
| Body mass index (kg/m2) | 22 [19.6, 23.8] |
| FEV1% predicted | 57.5 [40,72] |
| Cystic fibrosis related diabetes | 29 (43.9) |
| ABPA | 4 (6.1) |
| CFQ-R respiratory quality-of-life* | 58.3 [38.9, 72.2] |
| Medications | |
| CFTR modulatorsǂ | 24 (36.4) |
| Ivacaftor | 6 (9.1) |
| Inhaled antibiotics | 44 (66.7) |
| Azithromycin | 41 (62.1) |
| Chronic oral antibiotics | 22 (33.3) |
| Inhaled corticosteroids | 54 (81.8) |
| Prednisone | 13 (19.7) |
| Antifungal | 7 (10.6) |
| Oral and intravenous antibiotic courses in previous 12 months | 2 [1,4] |
| IV antibiotics courses in previous 12 months | 0.5 [0,1] |
Abbreviations:FEV1 = forced expiratory volume in 1 second, ABPA=allergic bronchopulmonary aspergillosis, CFQ-R =Cystic Fibrosis Questionnaire-Revised, CFTR =cystic fibrosis transmembrane conductance regulator.
CFQ-R missing data for one subject.
includes Ivacaftor and Lumacaftor/Ivacaftor.
Table S2 describes the culture-based microbiology at the time of sampling and over the prior two years. Two subjects lacked contemporaneous fungus culture results, neither of whom had a history of fungal isolation in the prior 24 months. Fifty-seven (86.4%) participants had history of Pseudomonas aeruginosa and nine (13.6%) had a history of Aspergillus fumigatus.
3.2. Taxonomonic identification of fungi in CF sputum
A total of 9.6 million fungal ITS sequence reads were generated from 66 sputum samples. After quality filtering, 8.4 million reads were used for analysis. A total of 249 taxa were identified in the cohort, including 128 at the species level. We randomly selected four subjects and tested two replicate samples from each subject for quality-control, which confirmed that beta diversity was significantly greater between than within subjects (p = 0.01; Fig. S1a and S1b).
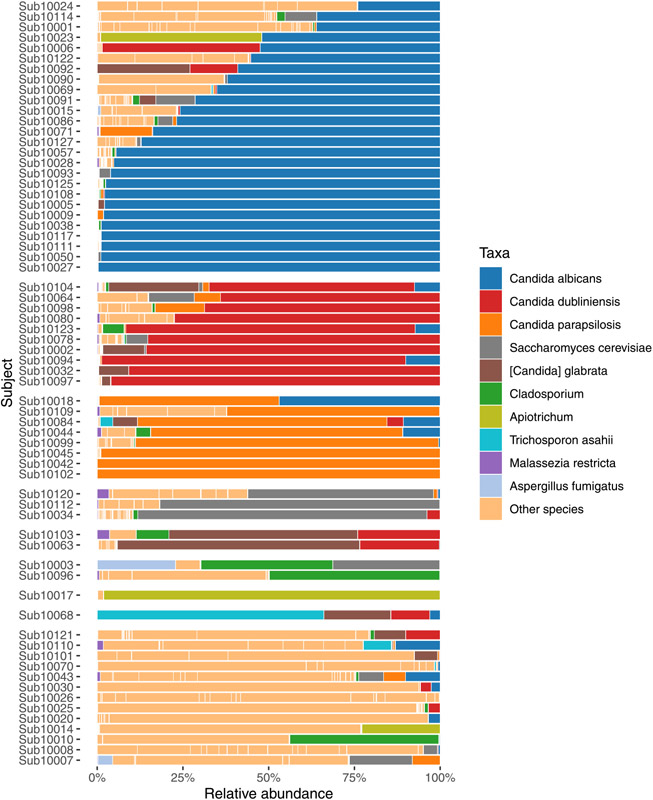
Fig. 1 shows relative abundances of fungal taxa. Candida albicans was the dominant taxon in the largest number of samples, followed by Candida dubliniensis and Candida parapsilosis. Aspergillus species was dominant taxon in one sputum sample and detected as the sub-dominant taxa in 36 additional samples (Table S4). Other fungi were present in high relative abundances in small numbers of specimens including Apiotrichum, Trichosporon, Saccharomyces cerevisiae, Cladosporium, Scedosporium, Malassezia restricta, and Diutina.
Fig. 1.
Top ten fungal taxa by relative mean abundance, grouped by the most dominant taxon. Each row represents a sputum sample from unique subject. For subjects with “Other” species listed as dominant taxon, the most abundant taxon is listed in Table S3.
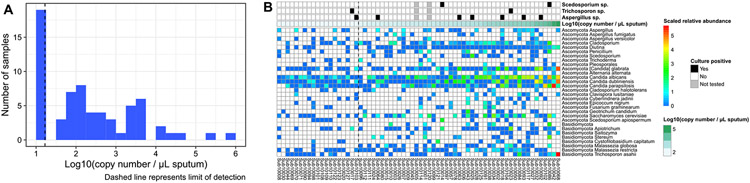
Samples underwent qPCR to measure the total fungal abundance. A total of 47 samples had fungal DNA above the limit of detection (33.43 copies per μL), most of which had low fungal load (Fig. 2A). Because the absolute abundance of fungi differed markedly among samples, which is not reflected in relative abundance measures, we calculated the qPCR-corrected fungal abundances to estimate the absolute abundance of individual fungal taxa in the ITS sequencing results (Fig. 2B; qPCR-corrected fungal abundances including background controls is shown in Fig. S2) [18]. For example, in subject 10092, the relative abundance of Aspergillus fumigatus was 3 × 10−5 and total fungal abundance measured by FungiQuant was 15,900 copy number/uL. Therefore, estimated qPCR-corrected abundance of A. fumigatus was 0.44852.
Fig. 2.
(A) Histogram of the log transformation of quantitative PCR of fungal DNA in CF samples. Dashed line represents to the limit of detection. (B) FungiQuant qPCR-corrected or absolute abundance of fungal taxa in sputum samples of 66 individual participants are shown. The corresponding culture result for Scedosporium species, Trichosporon species, and Aspergillus species are displayed in top three rows. The fourth row represents the log-transformed FungiQuant qPCR DNA count, keyed by teal-colored scale at the top right. In the samples to the left of the dashed line (representing the limit of DNA detection), total fungal abundance was imputed to 16.7 copies per μL of sputum.
Fig. 2B shows the qPCR-corrected fungal abundances in a heatmap, with the total fungal abundance for each sample. Samples to the left side of the dashed line represent the samples with fungal DNA below the limit of detection. We observed dominance of Candida species in high fungus-load specimens. To highlight correlation between sequencing and culture, we displayed the corresponding culture result for clinically relevant fungi (Aspergillus, Trichosporon, and Scedosporium species) in Fig. 2B. Of the 64 subjects that had concomitant culture evaluation, the concordance between fungal species identified on culture and the most abundant taxon identified by DNA sequencing was 64% (Table S3). However, we found 81% concordance between fungal species identified on culture and identification of fungal taxa by sequencing when including non-dominant taxa. Aspergillus species were detected in 37 samples (56%) but at mostly low levels. For the seven Aspergillus species culture-positive samples, Aspergillus genus was detected by DNA sequencing in all seven samples (Table S4). Correlation between qPCR corrected Aspergillus abundance and Aspergillus culture positivity was not observed (data not shown).
Scedosporium species were detected by sequencing in 26 (39%) individuals; but was the most abundant taxon in only two. Scedosporium apiospermum was detected by culture in subject 10086, but ITS sequencing identified S. aurantiacum in this subject. We found high Trichosporon abundance in three (4.5%) subjects and low level detection in 33 others. Trichosporon asahii was the dominant taxon in the sputum sample with the highest fungal DNA concentration, subject 10068 (Figs. 1 and 2); this subject grew Candida only on culture. Trichosporon mycotoxinivorans was cultivated in two samples (Table S3), but Apiotrichum genus was found to be abundant, while Trichosporon was not identified by ITS in those samples. Apiotrichum was found in high qPCR-corrected abundance in three (4.5%) subjects and low levels in 30 samples and eight environmental controls (Fig. S2). Overall, ITS sequencing identified the presence of clinically relevant fungi in nearly all culture-positive and several culture-negative samples.
3.3. Relationship between fungal communities in CF sputum and clinical status
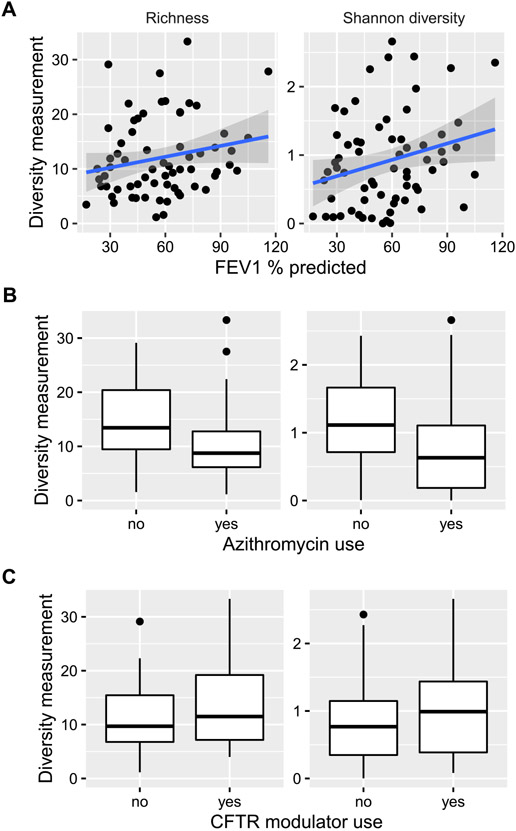
Higher FEV1% predicted was directly associated with greater fungal richness and alpha-diversity (Shannon index; p = 0.016 and 0.003, respectively; Fig. 3A). Chronic azithromycin use was significantly associated with both lower fungal richness and alpha-diversity (p = 4.77 × 10−4 and 5.16 × 10−4, respectively, Fig. 3B). CFTR modulator use was associated with higher fungal alpha-diversity, but not richness (p = 0.04 and 0.06, respectively, Fig. 3C). In contrast, there was no correlation between fungal richness or alpha-diversity and age, chronic anti-pseudomonal inhaled antibiotics, antifungal use, Pseudomonas aeruginosa colonization, or pulmonary exacerbation status (data not shown). Furthermore, fungal DNA copy number by qPCR did not correlate with clinical features (data not shown).
Fig. 3.
Fungal within-sample (alpha) diversity of CF sputum specimens correlates with lung function (3A),chronic azithromycin use (3B), and CFTR modulator use (3C) in adults with cystic fibrosis. Alpha diversity was summarized using richness and Shannon diversity index, represented on the y-axes.
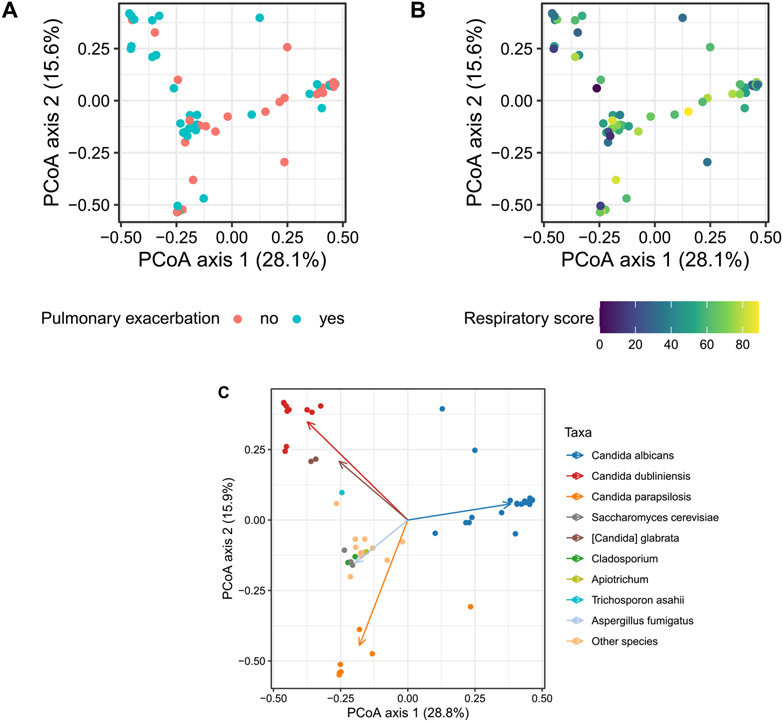
We observed a correlation between fungal community composition and pulmonary exacerbation status. CF patients experiencing an acute pulmonary exacerbation had distinct fungal profiles compared to clinically stable CF adults, adjusting for age, sex and FEV1% predicted, as measured by Bray Curtis metric (PERMANOVA R2=0.041, p = 0.01, Fig. 4A). Fungal community structure was also different in people with lower CFQ-R respiratory domain score, representing worse patient-reported respiratory health, compared to those with better respiratory health, accounting for age, sex, and FEV1% predicted (PERMANOVA R2 =0.036, p = 0.02, Fig. 4B). We followed our PERMANOVA-based analysis of beta diversity with an analysis of beta dispersion, or average within-group community distance. We observed no difference in beta dispersion according to pulmonary exacerbation (p = 0.056) but found that the community-level dispersion was slightly higher in samples with high CFQ-R score (p = 0.04, Fig. S3). Overall, differences in beta dispersion were small relative to the range of community-level distances between samples. Estimates for difference in beta dispersion were less than 20% of the IQR among within-group distances in both comparisons, suggesting that even statistically significant differences in beta dispersion were unlikely to have accounted for differences in community composition observed in our PERMANOVA comparisons.
Fig. 4.
(A) PERMANOVA reveals demonstrating microbial composition differs in CF adults with (aqua) and without (pink) pulmonary exacerbation, adjusted for age, sex, and FEV1 percent predicted, R2 = 0.041, p = 0.01. (B) Fungal composition may be associated with respiratory quality-of-life (QOL) with the yellow (light) color representing higher respiratory domain score (scale 0–100, higher respiratory domain score corresponding with better respiratory QOL), adjusted for age, sex and FEV1 percent predicted, R2 = 0.036, p = 0.02. (C) Dominant taxa groups are depicted in this clustered biplot of Bray-Curtis distance. Each dot represents unique sample color-coded by dominant taxon. Color-coded arrow (vector) represents the dominant taxon driving the differences.
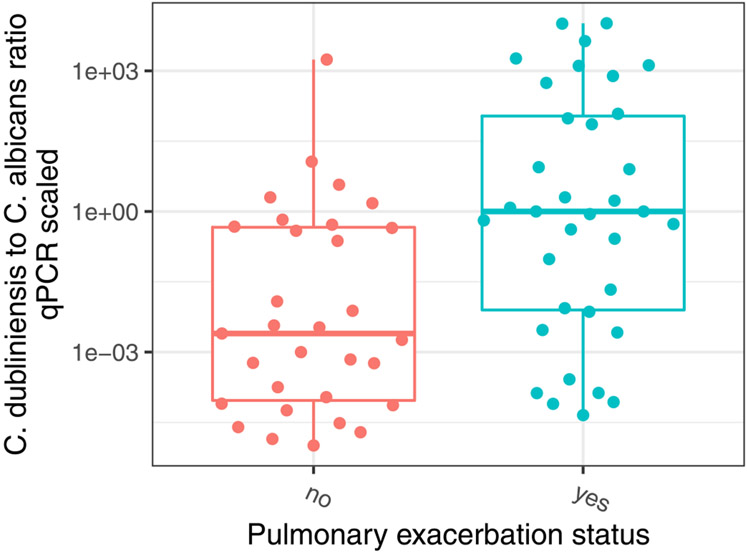
Candida albicans represented the dominant taxon in 16 (51.6%) subjects with stable clinical state (Figs. 4C and S4). In contrast, Candida dubliniensis was abundant in subjects with acute pulmonary exacerbations (Fig. S4). We therefore calculated the ratio of Candida dubliniensis and Candida albicans (C. dubliniensis/C. albicans) to assess the relationship between Candida species profile and clinical status. We observed higher C. dubliniensis/C. albicans was associated with pulmonary exacerbation state whereas a lower ratio was associated with stable state (Fig. 5, p < 0.001). Furthermore, higher C. dubliniensis/C. albicans correlated with lower CFQ-R respiratory domain score, representing worse respiratory health (data not shown).
Fig. 5.
The ratio of Candida dubliniensis to Candida albicans qPCR corrected relative abundance is associated with pulmonary exacerbation status, p<0.001. Higher C. dubliniensis/C. albicans was observed in CF patients with pulmonary exacerbation.
4. Discussion
The changing microbial epidemiology of CF airway has pro-voked investigation of the prevalence and clinical impact of filamentous fungi and yeast [3,19]. Culture-based detection has reported increasing prevalence of fungi in CF respiratory samples, which may contribute to infection and inflammation in the airways of children and adults [20,21]. By applying next-generation sequencing methods, we define the fungal microbiome in sputum and its relationship to host-specific factors and patient-reported respiratory quality-of-life in a well-characterized CF population.
Members of the Candida genus were the most prevalent and abundant fungi in CF sputum. These findings are consistent with previous descriptions of the CF fungal mycobiome [9,11,12,22]. Candida species are rarely implicated as the cause of acute lower respiratory infections and are present in the oral cavity of healthy and diseased individuals [23,24]. It is plausible that the Candida recovered in the sputum could represent contamination from passage through the mouth during sampling, but we understand that microbes in the oral compartment inform and influence the composition of the lung microbiome [25,26]. For example, Candida albicans was in the oral wash and also in stringently-collected bronchoalveolar lavage (BAL) of lung transplant recipients, some of whom had CF [10]. The pathogenicity of Candida species in the CF airways is controversial. Limited observational data have reported a potential association between C. albicans colonization and FEV1 decline and pulmonary exacerbation rates in CF, but this has yet to be reproduced [27]. Indeed, we found that fungal composition was associated with disease status and respiratory symptom burden (Fig. 4). C. dubliniensis was seen in greater abundance in sick CF patients with acute exacerbation. On the other, C. albicans was found to be more abundant in stable individuals without pulmonary exacerbation. Willger et al. investigated six CF subjects experiencing pulmonary exacerbation and similar to our study, C. albicans, C. parapsilosis, and C. dubliniensis accounted for 75–99% of the reads [11]. The mechanism for this relationship is uncertain, though inter-kingdom interactions between C. albicans and Pseudomonas aeruginosa in the airway might play a role in our findings [28].
While Aspergillus species were widely present, it was rarely the dominant taxon in sputa. For all samples culture-positive for Aspergillus, ITS also detected Aspergillus DNA; yet, we did not observe a correlation between Aspergillus DNA count and culture status. Detection of Aspergillus fumigatus in the CF airway is important for Aspergillus related lung disease. The clinical diagnostic value of Aspergillus DNA detection has yet to be understood for fungal lung disease in CF. Although a positive Aspergillus PCR cannot differentiate colonization versus infection, these more sensitive molecular assays could have a potential diagnostic role in CF patients.
We found high qPCR corrected abundances of Trichosporon asahii, a basidiomycetous yeast, in three subjects, suggesting authentic Trichosporon presence, whereas low abundance samples cannot be distinguished from background. Notably, the Trichosporon mycotoxinivorans culture-positive samples in the cohort did not identify Trichosporon by ITS sequencing; yet Apiotrichum was identified (Fig. S3). This may be explained by the recent taxonomic reassignment of Trichosporon mycotoxinivorans to Apiotrichum mycotoxinivorans and the phylogenetic similarities between the two genera [29]. Apiotrichum/Trichosporon species has been identified as a potentially pathogenic member of the CF fungal community and may contribute to lung function decline [11,30]
ITS sequencing also detected fungal taxa, such as Saccharomyces cerevisiae, Cystofilobasidium, and Malassezia, supporting previous observations [9,11-31]. While these taxa have known roles as skin and gut flora, they are of unclear significance in the CF lung. We also observed high relative abundance of Diutina in samples with three samples with high fungal load. Yet, Diutina (formerly Candida) species is a rare pathogen in human infection and has not been well-described in the CF lung [32]. When comparing culture and sequence results, discordance between cultivable agents and dominant fungal taxa did occur for a minority of samples (Table S3). Sensitivity of molecular methods may identify DNA from dead microorganisms, which may contribute to the discrepancies between culture and culture-independent diagnostics.
As CF lung disease progresses, bacterial richness and diversity are known to decrease [6], concomitant with increased absolute and relative abundance of CF pathogens, such as Pseudomonas aeruginosa. We observed lower fungal diversity in more advanced lung disease [9]. The mechanism for this is unclear. The use of chronic inhaled anti-pseudomonal antibiotics has been thought to potentially drive enrichment of fungal communities [4]. Yet, we did not observe this association. Interestingly, lower fungal alpha diversity was observed in sputa of patients on chronic azithromycin compared to those who did not use azithromycin. Chronic macrolide use has been implicated in altering the airway bacterial microbiota in non-CF bronchiectasis and asthma, though azithromycin has not been associated with changes in bacterial diversity in CF [33-35]. The immunomodulatory mechanism of azithromycin or the interaction between fungi and bacteria may be playing a role in the observed fungal diversity differences. Alternatively, individuals on chronic azithromycin in our cohort may be represent a sicker population. Adults with CF on ivacaftor or lumacaftor/ivacaftor showed trends of higher fungal richness and alpha-diversity. Greater bacterial richness and alpha diversity have been reported in CFTR modulators, but fungal diversity has not been previously described to our knowledge [36,37]. CFTR modulators are known to exhibit antimicrobial properties, but the direct mechanism of how improved CFTR function alters the microbiome remains unknown.
A novel aspect of our study is that we investigated total fungal burden and estimated abundance of each taxon adjusted for the total fungal content rather than simply relative abundances. Microbiome analyses often focus only on relative abundances, which omits potentially important information on total microbial burden and burden of each taxon. This may be particularly critical in environments like the lung, where there are very large differences in total fungal burden between subjects. In addition, fungal qPCR-corrected abundance enables better comparison between samples of interest and background, given the risk of contamination in low biomass respiratory samples, adding methodological rigor.
Our study has several limitations. Sputum has the potential to be contaminated by organisms in the oral cavity, although bacterial studies suggest that may be less problematic in CF than other states due to a higher microbial burden in the lung [22]. Nevertheless, the absence of oral samples in this study leads to potential risk. The study was cross-sectional, limiting the ability to infer causality in the observed associations, and longitudinal examination must be the next step. The risk of confounding is present, but we adjusted our models for age, sex, and lung function to address this. We used ITS1F/ITS2 primers targeting the ITS1 region, one of the several fungal ribosomal target regions, which may impact the ability to identify certain fungal taxa [38]. We did not analyze sputum bacterial communities and understanding bacterialfungal interaction will be an important topic for future investigation. While total fungal burden adds considerable information to relative abundances, the extent to which it reflects airway burden may vary among patients based on levels of sputum production. The inability to distinguish dead or alive microbes with molecular testing remains a limitation in most human microbiome studies. Finally, this cohort was studied before the introduction of elexacaftor/tezacaftor/ivacaftor (ETI), which may alter the fungal community landscape. Studies investigating the microbiological changes in CF sputa related to ETI are ongoing (NCT04038047).
In conclusion, our study contributes to the understanding of the understudied mycobiome in a CF adult population. The differences in the fungal community structure of CF sputa influenced by C. albicans and C. dubliniensis in pulmonary exacerbation states and high respiratory symptom burden requires further investigation to determine whether fungi may contribute. The associations between azithromycin, CFTR modulation, and lung function with fungal richness and diversity in CF sputa also merits closer examination. Longitudinal evaluation of the mycobiome, bacterial microbiome, and microbial interactions in a large cohort over time is necessary to elucidate these relationships further.
Supplementary Material
Acknowledgments
The authors thank the research participants who volunteered their time and information for this study. We would like to acknowledge Ariel Myatt for assistance with figure production, PennCHOP microbiome sequencing laboratory for their technical assistance of specimen analysis, and Sharon C.W. Ng, Justin Martin, and Yujia Su for subject recruitment and data collection.
Funding
The work was supported by grant funding and support from the Cystic Fibrosis Foundation (HONG16A0). Other support by the following grants: National Institutes of Health K23-HL146970 (GH), K24-HL103844 (SMK), R33-HL137063 (RGC).
Abbreviations:
- FEV1
forced expiratory volume in one second
- CFQ-R
cystic fibrosis questionnaire-revised
- DTT
dithiothreitol
- ITS
internal transcribed spacer
- QPCR
quantitative polymerase chain reaction
- OTU
operational taxonomic unit
- CFTR
cystic fibrosis transmembrane conductance regulator
- ABPA
allergic bronchopulmonary aspergillosis
- IV
intravenous
- BAL
bronchoalveolar lavage
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcf.2023.02.003.
Credit author statement
Conceptualization and design: GH, DH, SMK, RGC. Acquisition, analysis, and interpretation of the data: GH, SGD, JL, KB, LG, LMM, SMK, DJD, DH, RGC. Writing- Original draft- GH. Writing- review and Editing: GH, SGD, JL, KB, LG, LMM, SMK, DJD, DH, RGC. Funding acquisition: GH
References
- [1].2019 Annual Data Report Cystic fibrosis foundation patient registry. Bethesda, MD: Cystic Fibrosis Foundation; 2020. [Google Scholar]
- [2].Elborn JS. Cystic fibrosis. Lancet 2016;388(10059):2519–31. [DOI] [PubMed] [Google Scholar]
- [3].Sudfeld CR, Dasenbrook EC, Merz WG, Carroll KC, Boyle MP. Prevalence and risk factors for recovery of filamentous fungi in individuals with cystic fibrosis. J Cyst Fibros 2010;9(2):110–16 official journal of the European Cystic Fibrosis Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hong G, Psoter KJ, Jennings MT, Merlo CA, Boyle MP, Hadjiliadis D, et al. Risk factors for persistent Aspergillus respiratory isolation in cystic fibrosis. J Cyst Fibros 2018. official journal of the European Cystic Fibrosis Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 2015;5:10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 2010;5(6):e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carmody LA, Caverly LJ, Foster BK, Rogers MAM, Kalikin LM, Simon RH, et al. Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLoS One 2018;13(3):e0194060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mac Aogain M, Chandrasekaran R, Lim AYH, Low TB, Tan GL, Hassan T, et al. Immunological corollary of the pulmonary mycobiome in bronchiectasis: the CAMEB study. Eur Respir J 2018;52(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS One 2012;7(4):e36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med 2012;186(6):536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2014;2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cuthbertson L, Felton I, James P, Cox MJ, Bilton D, Schelenz S, et al. The fungal airway microbiome in cystic fibrosis and non-cystic fibrosis bronchiectasis. J Cyst Fibros 2021;20(2):295–302 official journal of the European Cystic Fibrosis Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Soret P, Vandenborght LE, Francis F, Coron N, Enaud R, Avalos M, et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci Rep 2020;10(1):3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Quittner AL, Modi AC, Wainwright C, Otto K, Kirihara J, Montgomery AB. Determination of the minimal clinically important difference scores for the cystic fibrosis questionnaire-revised respiratory symptom scale in two populations of patients with cystic fibrosis and chronic Pseudomonas aeruginosa airway infection. Chest 2009;135(6):1610–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hong G, Alby K, Ng SCW, Fleck V, Kubrak C, Rubenstein RC, et al. The presence of Aspergillus fumigatus is associated with worse respiratory quality of life in cystic fibrosis. J Cyst Fibros 2020;19(1):125–30 official journal of the European Cystic Fibrosis Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu CM, Kachur S, Dwan MG, Abraham AG, Aziz M, Hsueh PR, et al. Fungi-Quant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol 2012;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol 2012;13(7):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol 2014;15(10):487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Breuer O, Schultz A, Turkovic L, de Klerk N, Keil AD, Brennan S, et al. Changing prevalence of lower airway infections in young children with cystic fibrosis. Am J Respir Crit Care Med 2019;200(5):590–9. [DOI] [PubMed] [Google Scholar]
- [20].Brandt C, Roehmel J, Rickerts V, Melichar V, Niemann N, Schwarz C. Aspergillus bronchitis in patients with cystic fibrosis. Mycopathologia 2018;183(1):61–9. [DOI] [PubMed] [Google Scholar]
- [21].Breuer O, Schultz A, Garratt LW, Turkovic L, Rosenow T, Murray CP, et al. Aspergillus infections and progression of structural lung disease in children with cystic fibrosis. Am J Respir Crit Care Med 2020;201(6):688–96. [DOI] [PubMed] [Google Scholar]
- [22].Krause R, Moissl-Eichinger C, Halwachs B, Gorkiewicz G, Berg G, Valentin T, et al. Mycobiome in the lower respiratory tract - a clinical perspective. Front Microbiol 2016;7:2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Krause R, Halwachs B, Thallinger GG, Klymiuk I, Gorkiewicz G, Hoenigl M, et al. Characterisation of Candida within the mycobiome/microbiome of the lower respiratory tract of ICU patients. PLoS One 2016;11(5):e0155033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lepesqueur LSS, Tanaka MH, Lima GMG, Chiba SM, Mota AJ, Santos SF, et al. Oral prevalence and antifungal susceptibility of Candida species in cystic fibrosis patients. Arch Oral Biol 2020;116:104772. [DOI] [PubMed] [Google Scholar]
- [25].Muhlebach MS, Zorn BT, Esther CR, Hatch JE, Murray CP, Turkovic L, et al. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog 2018;14(1):e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Prevaes SM, de Steenhuijsen Piters WA, de Winter-de Groot KM, Janssens HM, Tramper-Stranders GA, Chu ML, et al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 2017;49(3). [DOI] [PubMed] [Google Scholar]
- [27].Gileles-Hillel A, Shoseyov D, Polacheck I, Korem M, Kerem E, Cohen-Cymberknoh M. Association of chronic Candida albicans respiratory infection with a more severe lung disease in patients with cystic fibrosis. Pediatr Pulmonol 2015;50(11):1082–9. [DOI] [PubMed] [Google Scholar]
- [28].Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2002;296(5576):2229–32. [DOI] [PubMed] [Google Scholar]
- [29].McTaggart LR, Copeland JK, Surendra A, Wang PW, Husain S, Coburn B, et al. Mycobiome sequencing and analysis applied to fungal community profiling of the lower respiratory tract during fungal pathogenesis. Front Microbiol 2019;10:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Esther CR Jr, Plongla R, Kerr A, Lin FC, Gilligan P. Clinical outcomes in cystic fibrosis patients with Trichosporon respiratory infection. J Cyst Fibros 2016;15(5):e45–9 official journal of the European Cystic Fibrosis Society. [DOI] [PubMed] [Google Scholar]
- [31].Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, Höfle MG. Cohort study of airway mycobiome in adult cystic fibrosis patients: differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol 2015;53(9):2900–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ming C, Huang J, Wang Y, Lv Q, Zhou B, Liu T, et al. Revision of the medically relevant species of the yeast genus Diutina. Med Mycol 2019;57(2):226–33. [DOI] [PubMed] [Google Scholar]
- [33].Acosta N, Thornton CS, Surette MG, Somayaji R, Rossi L, Rabin HR, et al. Azithromycin and the microbiota of cystic fibrosis sputum. BMC Microbiol 2021;21(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rogers GB, Bruce KD, Martin ML, Burr LD, Serisier DJ. The effect of long-term macrolide treatment on respiratory microbiota composition in non-cystic fibrosis bronchiectasis: an analysis from the randomised, double-blind, placebo-controlled BLESS trial. Lancet Respir Med 2014;2(12):988–96. [DOI] [PubMed] [Google Scholar]
- [35].Slater M, Rivett DW, Williams L, Martin M, Harrison T, Sayers I, et al. The impact of azithromycin therapy on the airway microbiota in asthma. Thorax 2014;69(7):673–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Einarsson GG, Ronan NJ, Mooney D, McGettigan C, Mullane D, NiChroinin M, et al. Extended-culture and culture-independent molecular analysis of the airway microbiota in cystic fibrosis following CFTR modulation with ivacaftor. J Cyst Fibros 2021;20(5):747–53 official journal of the European Cystic Fibrosis Society. [DOI] [PubMed] [Google Scholar]
- [37].Graeber SY, Boutin S, Wielpütz MO, Joachim C, Frey DL, Wege S, et al. Effects of lumacaftor-ivacaftor on lung clearance index, magnetic resonance imaging, and airway microbiome in Phe508del homozygous patients with cystic fibrosis. Ann Am Thorac Soc 2021;18(6):971–80. [DOI] [PubMed] [Google Scholar]
- [38].Hoggard M, Vesty A, Wong G, Montgomery JM, Fourie C, Douglas RG, et al. Characterizing the human mycobiota: a comparison of small subunit rRNA, ITS1, ITS2, and large subunit rRNA genomic targets. Front Microbiol 2018;9:2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.