Abstract
cdc25C induces mitosis by activating the cdc2-cyclin B complex. The intracellular localization of cyclin B1 is regulated in a cell cycle-specific manner, and its entry into the nucleus may be required for the initiation of mitosis. To determine the cellular localization of cdc25C, monoclonal antibodies specific for cdc25C were developed and used to demonstrate that in human cells, cdc25C is retained in the cytoplasm during interphase. A deletion analysis identified a 58-amino-acid region (amino acids 201 to 258) in cdc25C that was required for the cytoplasmic localization of cdc25C. This region contained a specific binding site for 14-3-3 proteins, and mutations in cdc25C that disrupted 14-3-3 binding also disrupted the cytoplasmic localization of cdc25C during interphase. cdc25C proteins that do not contain a binding site for 14-3-3 proteins showed a pancellular localization and an increased ability to induce premature chromosome condensation. The cytoplasmic localization of cdc25C was not altered by γ irradiation or treatment with the nuclear export inhibitor leptomycin B. These results suggest that 14-3-3 proteins may negatively regulate cdc25C function by sequestering cdc25C in the cytoplasm.
In eukaryotic cells, an active cyclin-dependent kinase complex, cdc2-cyclin B1, promotes entry into mitosis. Prior to mitosis, kinase activity is inhibited by phosphorylation of the cdc2 catalytic subunit on two residues, threonine 14 (T14) and tyrosine 15 (Y15) (reviewed in reference 46). Several kinases, including wee1 (3, 21, 48), mik1 (32), and myt1 (36, 42), phosphorylate cdc2 at residues T14 and Y15 and inhibit mitotic progression. Entry into mitosis is dependent on dephosphorylation of the T14 and Y15 residues, resulting in the formation of an active cdc2-cyclin B complex (reviewed in reference 46). Inhibition of cdc2 dephosphorylation is a target of the DNA replication and DNA damage checkpoints in both yeast and mammalian cells (reviewed in references 45 and 46).
Dephosphorylation of cdc2 and subsequent entry into mitosis are catalyzed by the dual-specificity phosphatase, cdc25C (11, 27, 33, 40, 57), which in turn is regulated by cell cycle-dependent phosphorylation events (15, 24). Hyperphosphorylation of cdc25C during mitosis is thought to stimulate its phosphatase activity (19, 22, 24, 29). Although the specific kinase that phosphorylates cdc25C during mitosis is unknown, several candidate kinases activate cdc25C in vitro. cdc25C may be phosphorylated by its own substrate, an active cdc2-cyclin B complex, to create an autoactivation loop (19, 22, 56). Other studies have reported that the mitosis-specific hyperphosphorylation of cdc25C can occur in the absence of both cdc2 and cdk2 (23), suggesting that other kinases may activate cdc25C during mitosis. A candidate for the cdc25C M-phase kinase is the product of the Xenopus Plx1 gene, a polo-like kinase. Plx1 can phosphorylate cdc25C in vitro and stimulate cdc25C phosphatase activity in vitro (28); however, it is not yet clear whether Plx1 stimulates the mitotic activation of cdc25C in vivo.
Premature activation of cdc25C is prevented by phosphorylation of specific residues in cdc25C during interphase that are distinct from the sites phosphorylated in M phase. Piwnica-Worms and colleagues have demonstrated that during interphase, the major phosphorylation site in human cdc25C is a serine residue at position 216 (S216) (47). Conversely, S216 is not phosphorylated during mitosis, suggesting that phosphorylation of this residue may contribute to the negative regulation of cdc25C activity (49, 52). Consistent with the above hypothesis, expression of a cdc25C mutant that substituted alanine for serine 216 (S216A) induced premature entry into mitosis by override of a DNA replication checkpoint and a γ-radiation-induced DNA damage checkpoint (49). Several kinases that promote the phosphorylation of the S216 residue in cdc25C have recently been identified. Piwnica-Worms and colleagues purified from HeLa cells a kinase, C-TAK1, that specifically phosphorylates residue S216 in vitro (47, 50). cdc25C can also be phosphorylated by chk1, a DNA damage checkpoint kinase that was first identified in fission yeast (9, 49, 52, 60, 61). chk1 is activated by phosphorylation in response to γ-radiation-induced DNA damage, leading to a cell cycle arrest in G2 (9, 52). Recent work with the fission yeast Schizosaccharomyces pombe has shown that cdc25 can also be phosphorylated by another kinase activated by DNA damage, cds1 (65). This pathway is replicated in other eukaryotes, as Kumagai et al. have demonstrated that cdc25C is phosphorylated and capable of responding to checkpoint control in Xenopus laevis extracts that have been depleted of chk1 (30). A human homolog of cds1, chk2, has recently been cloned and found to phosphorylate cdc25C at S216 in vitro (38). Therefore, the S216 residue in cdc25C may be a substrate for multiple kinases that specifically inhibit its activity in response to the S-phase or DNA damage checkpoints.
Phosphorylation of cdc25C at S216 by chk1 or C-TAK1 in vitro results in the generation of a binding site for the 14-3-3 family of proteins (44, 49, 50, 52). A consensus binding site for 14-3-3 was first identified by Muslin et al. as RSXpSXP (X being any amino acid and pS indicating phosphoserine) (44). Recent work by Yaffe and colleagues has led to refinement of the 14-3-3 binding consensus to R[S/Ar][+/S]pS[L/E/A/M]P as well as identification of a second consensus, RX[Ar/S][+]pS[LEAM]P (where Ar is an amino acid with an aromatic side chain and + is an amino acid with a basic side chain) (62). A peptide corresponding to the first sequence is present in residues 213 to 218 of cdc25C. 14-3-3 proteins have been shown to bind to cdc25C during interphase, at a time when S216 is phosphorylated (49). Similarly, cdc25 has been shown to form a complex with the 14-3-3ɛ and 14-3-3ζ proteins in X. laevis egg extracts (31). Given that S216 is critical for regulation of cdc25C activity and can be phosphorylated by chk1, 14-3-3 proteins may inhibit cdc25C function in response to DNA damage (49). Consistent with this model, two 14-3-3 homologues in fission yeast, rad24 and rad25, are required for the response to radiation induced DNA damage and G2 checkpoint control (1, 8). The rad24 and rad25 proteins associate with the S. pombe cdc25 protein in vitro when cdc25 is phosphorylated by either chk1 or cds1 (65). Notably, expression of 14-3-3ς can be induced by γ-radiation-induced DNA damage in human cells (18).
The activity of a number of cellular proteins is regulated by their cell cycle-dependent intracellular localization. For example, cyclin B1 is located in the cytoplasm during interphase and then enters the nucleus just prior to mitosis (51). Several reports have demonstrated that the cytoplasmic retention of cyclin B1 is maintained due to the presence of a strong nuclear export signal (NES) (13, 59, 63). Toyoshima and colleagues demonstrated that inhibition of cyclin B1 nuclear export led to an override of the DNA damage checkpoint upon treatment with caffeine (59). Similarly, Jin et al. demonstrated that a constitutively nuclear cyclin B1 (cyclin B1 was fused to the simian virus 40 [SV40] nuclear localization signal [NLS]) protein could override a DNA damage-induced checkpoint arrest in cooperation with a cdc2 mutant that could not be phosphorylated at T14 and Y15 (cdc2AF) (25). Notably, in both studies, the nuclear localization of cyclin B1 was necessary but not sufficient to override the DNA damage checkpoint and promote entry into mitosis. Similarly, Hagting et al. observed premature mitosis when an export-defective cyclin B1 mutant was coexpressed with either cdc2AF, cdc25C, or a dominantly active mutant of cdc25C, S216G (13). These results suggest that the DNA replication and DNA damage checkpoints affect mitotic progression by regulating cyclin B1 localization as well as by modulating cdc25C function.
Although cdc25C activity can be regulated by complex formation with 14-3-3 proteins, association of 14-3-3 proteins with cdc25C has not been shown to decrease cdc25C phosphatase activity in vitro (31, 49). This leads to the question of how 14-3-3 proteins may affect cdc25C function. cdc25C has been reported to contain a bipartite NLS in its N terminus (47), and at least three reports suggested that human cdc25C was a predominantly nuclear protein (10, 12, 39). However, an exogenously expressed Myc epitope-tagged human cdc25C protein was reported to be localized primarily in the cytoplasm during interphase and then translocated to the nucleus just prior to entry into M phase (16). To address this issue, a panel of monoclonal antibodies (MAbs) specific for human cdc25C was developed and used to demonstrate that the endogenous human cdc25C protein is localized in the cytoplasm during interphase in multiple cell types. The region in cdc25C required for the cytoplasmic localization was mapped to a 14-3-3 binding site in cdc25C. Thus, cytoplasmic localization of cdc25C may depend on its ability to associate with 14-3-3 proteins, suggesting that 14-3-3 proteins prevent mitotic progression by sequestering cdc25C in the cytoplasm.
MATERIALS AND METHODS
Cell strains and transfections.
The human osteosarcoma cell line U-2OS and the primary diploid human fibroblast strains MRC-5 and WI-38 were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (Cellgro) supplemented with 10% Fetal Clone-I serum (Hyclone), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were transfected by calcium phosphate precipitation as described previously (58). Briefly, 15 μg of the cdc25C plasmid was used per 100-mm-diameter dish transfected, and the total amount of DNA was adjusted to 25 μg with pBluescript (pBSK−). To perform the localization and premature chromosome condensation (PCC) assays, cells were transfected on a coverslip in a 35-mm-diameter dish; 3 μg of each cdc25C plasmid was cotransfected with either 2 μg of pBSK− or 2 μg of the hemagglutinin (HA) epitope-tagged cyclin B1 plasmid per 35-mm-diameter dish. To perform the cell cycle synchrony experiments, 15 μg of cdc25C plasmid, 8 μg of pBSK−, and 2 μg of a CD19 cDNA (64) were transfected per 100-mm-diameter dish. To perform the 14-3-3 binding assays, 15 μg of cdc25C plasmid was cotransfected with 7.5 μg of pBSK− and 2.5 μg of HA-14-3-3ɛ per 100-mm-diameter dish.
Plasmids.
The N-terminal Myc-tagged cdc25C cDNA has been described previously (16). All of the cdc25C constructs were tagged with the Myc epitope (EQKLISEEDL [5, 43]) and expressed from the SV40 promoter in plasmid pSG5-L (54). The SV40 NLS (PKKKRKVEDPAV) was fused to the C terminus of cdc25C by PCR using the primers (5′ ggg aag ctt gga tcc ATG GAG CAG AAG CTC 3′ and 5′ ggg gtc gac TCA CAC CGC CCG GAT CTT CTA CTT TCC GTT TCT TTT TGG GTG GGC TCA TGT CCT TCA CC 3′ for all oligonucleotides, coding sequences are in uppercase). The mutation of the active-site residue of the phosphatase domain, cysteine 377 to serine (C377S) (4, 11, 40), was generated by PCR using the oligonucleotides 5′ TGA GGA GAA TTC AGA GTG GAA CAC GAT 3′ and 5′ ggg tct aga gga tcc TCA TGG GCT CAT GTC CTT CAC CAG 3′ and inserted into the wild-type (WT) cdc25C constructs as a BamHI-EcoRI fragment. The deletion mutants 1–258, 1–200, and 1–150 were amplified from WT cdc25C by using the oligonucleotides 5′ ggg aga tct TCA TAA ACA TAA GCC CTT CCT 3′, 5′ ggg aga tct TCA TTT CAG GGA AAA CTC CAT 3′, and 5′ ggg aga tct TCA ATT TGC AGA TGA ACT ACA 3′, respectively, with the oligonucleotide 5′ ggg aag ctt gga tcc ATG GAG CAG AAG CTC 3′ in PCRs. The mutant 259–473 was constructed by performing a PCR with the oligonucleotides 5′ aaa aag ctt ATG GAG CAG AAG CTC ATC TCA GAG GAG GAC CTG GGA TCC AAG AAG ACA GTC TCT CTG TGT 3′ and 5′ ggg tct aga gga tcc TCA TGG GCT CAT GTC CTT CAC CAG 3′. The deletion mutants Δ 201–258 and Δ 201–258 C377S were cloned by PCR amplifying either WT cdc25C or the C377S mutant with the oligonucleotides 5′ ttt gga tcc AAG AAG ACA GTC TCT CTG TGT GAC 3′ and 5′ aaa ctc gag TTA AGA TCT TTA TCA TGG GCT CAT GTC CTT CAC CAG AAG GGC 3′ and inserted into pSG5-L as a BamHI-XhoI fragment. Subsequently, another PCR fragment generated with the oligonucleotides 5′ aaa aag ctt ATG GAG CAG AAG CTC ATC TCA GAG GAG GAC CTG 3′ and 5′ ggg gga tcc TTT CAG GGA AAA CTC CAT TAA TTC ATG TG 3′ was inserted into the above construct as a HindIII-BamHI fragment. The point mutant of the serine residue at position 216 to alanine (S216A) was generated by PCR with the oligonucleotides 5′ AAC AGG CCT AGA CTG AAG and 5′ ggg tct aga gga tcc TCA TGG GCT CAT GTC CTT CAC CAG 3′ and inserted in a three-way ligation into pSG5-L. The clones NEScdc25C and NESS216A were generated by cloning cdc25C or S216A as BamHI fragments downstream of the influenza virus HA epitope (YPYDVPDYA [6]) and the human immunodeficiency virus type (HIV-1) Rev NES (LQLPPLERLTLD [7]) in pSG5-L (55). Cyclin B1 was PCR amplified by using the primers 5′ ccc gga tcc ATG GCG CTC CGA GTC ACC AGG AAC TCG and 5′ ggg ctc gag TTA CAC CTT TGC CAC AGC CTT GGC TAA ATC 3′ and fused to the HA epitope in pcDNA3 (Invitrogen). Cyclin B1 was cloned as a BamHI-XhoI fragment downstream of the SV40 NLS and the HA epitope in pSG5-L (55) to generate HA-NLSB1. 14-3-3ɛ (62) was cloned as a BamHI-XhoI fragment into pcDNA3 downstream of the HA epitope to generate HA-14-3-3ɛ. All clones were verified by DNA sequencing.
Antibodies.
To generate specific MAbs to cdc25C, a glutathione S-transferase (GST) fusion to the first 258 amino acids of human cdc25C (GST 1–258) was constructed. The protein was purified from Escherichia coli on glutathione-Sepharose beads as described by the manufacturer (Pharmacia). The protein was eluted off the beads overnight at 4°C in 20 mM glutathione–100 mM Tris (pH 8.0)–120 mM NaCl, and dialyzed against phosphate-buffered saline (PBS). RBF/DnJ mice were immunized and fusions to NS1 myeloma cells were performed as described elsewhere (14). Four specific MAbs to cdc25C, TC14, TC15, TC19, and TC113, were identified. The antibodies were subtyped with an isotype kit (IsoStrip; Boehringer Mannheim) and belong to the immunoglobulin G1 (IgG1) subclass. The phosphorylated histone (phospho-histone) H3-specific antiserum (Upstate Biotechnology) was used at a dilution of 1:200 for immunofluorescence. Tissue culture supernatants of the mouse monoclonal hybridomas anti-HA (12CA5) and anti-Myc (9E-10) were used at a dilution of 1:50 for immunofluorescence analysis and Western blotting. Tissue culture supernatants of the hybridomas DG122 (anti-GST), TC14, TC15, and TC113 were used at a dilution of 1:50, while the TC19 supernatant was used without dilution for Western blotting of GST fusion proteins. The anti-cdc25C rabbit polyclonal antibody (C-20; Santa Cruz Biotechnology) was used at a dilution of 1:500 for Western blotting. For immunofluorescence analysis, ascites fluid was generated for TC14, TC15, TC19, and TC113 and used at a dilution of 1:50. A mixture of TC14, TC15, and TC19 ascites fluid (each at a 1:500 dilution) was used to perform Western blotting in the immunoprecipitation Western blotting (IP/Western) experiments. TC14 (1:10), TC15 (1:10), and TC113 (1:100) high-titer tissue culture supernatants were used at the dilutions indicated for direct Western blotting. The anti-Myc rabbit polyclonal antibody (A-14; Santa Cruz) was diluted 1:1,000 for immunofluorescence and 1:500 for Western blotting. Tissue culture supernatants of the cyclin B1 antibody CB169 (Upstate Biotechnology) was used at a dilution of 1:50 for immunostaining. The anti-Rb (retinoblastoma protein) MAb (MAb 245; Pharmingen) was used at a dilution of 1:500 for Western blotting, and 2 μl of antibody was used to immunoprecipitate Rb from cellular extracts. The secondary antibodies goat anti-mouse IgG-rhodamine and goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) (Boehringer Mannheim) were diluted 1:1,000 for immunofluorescence assays. The MPM2 antibody was used at a dilution of 1:1,000 for immunofluorescence.
Immunoprecipitation and Western blotting.
To determine the specificity of the anti-cdc25C MAbs, E. coli strains expressing GST fusion to full-length human cdc25A (GST-cdc25A) and human cdc25B (GST-cdc25B) or the first 258 amino acids of human cdc25C (GST-1–258) were induced to synthesize the fusion protein as described by the supplier (Pharmacia). The bacterial pellets were boiled in sodium dodecyl sulfate (SDS) buffer (58) and separated in SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes, and the immune complexes were detected as described previously (58) with an alkaline phosphate-conjugated goat anti-rabbit IgG (Boehringer Mannheim). To map the epitopes of the different antibodies, various cdc25C constructs were translated in vitro in the presence of [35S]methionine in the Promega TNT coupled transcription translation system. The in vitro translation products were incubated with the various antibodies and protein A-Sepharose in 500 μl of NET-N (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 [NP-40]) for 2 h at 4°C. The immune complexes were washed three times with NET-N, boiled in SDS buffer, separated in SDS-polyacrylamide gels, and analyzed by autoradiography.
Whole-cell extracts of U-2OS cells were prepared in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 0.1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM EDTA). Extracts were cleared by centrifugation at 12,000 × g for 15 min at 4°C. Two micrograms of EBC extract was rocked overnight at 4°C with protein A-Sepharose beads and either 200 μl of the antibody supernatants (9E-10, TC14, TC15, and TC113) or 2 μl of ascites fluid (TC19). The immune complexes were washed three times with NET-N and resolved in an SDS–7.5% polyacrylamide gel, followed by Western blotting with a mixture of TC14, TC15, and TC19 or the anti-cdc25C rabbit polyclonal antibody (Santa Cruz). Whole-cell extracts for direct Western blots were prepared by harvesting cells by trypsinization and boiling the cell pellet in 50 mM Tris (pH 8.0)–2% SDS for 10 min. The extracts were cleared by centrifugation, and 25 to 50 μg of extract was loaded onto an SDS–10% polyacrylamide gel. The antibody antigen complexes were detected by enhanced chemiluminescence as instructed by the manufacturer (Pierce).
To detect transiently transfected Myc-tagged cdc25C, 40 to 44 h posttransfection, cells were extracted in 1 ml of EBC buffer and the extracts were rocked overnight at 4°C with protein A-Sepharose beads and 100 μl of 9E-10 tissue culture supernatant. Immune complexes were washed thrice with NET-N and detected by Western blotting as described above. In the experiments where HA-14-3-3ɛ was cotransfected with cdc25C, the extracts were lysed in EBC as described above and then immunoprecipitations were performed with antibodies to either the Myc (9E-10) or HA (12CA5) epitope overnight as described above.
Cellular fractionation.
Fractionation of U-2OS cells was performed by using a modified version of a protocol described by Schreiber et al. (53). Briefly, U-2OS cells at ∼50% confluence were harvested by trypsinization. Whole-cell extracts were made with EBC. Cytoplasmic extracts were prepared by harvesting cells in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 0.1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM sodium orthovanadate) on ice for 10 min. Additional phosphatase inhibitors (10 μM cypermethrin, 200 μM dephostatin, 200 nM okadaic acid, 25 nM tautomycin) were added to buffer A when indicated. The extracts were spun for 30 s in a microcentrifuge at 4°C. The cytoplasmic extract was removed, and the nuclear pellets from the two different preparations were pooled and extracted with EBC to generate nuclear extracts. Two milligrams of each extract was immunoprecipitated with a MAb to cdc25C (TC113) or to Rb (MAb 245). The immune complexes were resolved in an SDS–7.5% polyacrylamide gel and Western blotted for the presence of either cdc25C or Rb. For direct Western blotting of the extracts, 100 μg of each fraction was separated in an SDS–10% polyacrylamide gel and then Western blotted with a mixture of the protein A-purified cdc25C MAbs.
Localization and PCC assays.
To detect endogenous cdc25C protein by immunofluorescence, cells were fixed in a solution of 4% paraformaldehyde in PBS for 20 min at room temperature (RT) and subsequently washed with PBS and permeabilized with 0.3% Triton X-100 in PBS for 10 min at RT. The fixed permeabilized cells were incubated with primary antibody, diluted 1:50 in PBS containing 3% bovine serum albumin and 0.1% NP-40, overnight at 4°C. The coverslips were washed alternately with PBS containing 0.1% NP-40 and PBS six times and then incubated with the appropriate secondary antibody in PBS containing 3% bovine serum albumin and 0.1% NP-40 for 30 minutes at RT. The cells were washed again as described above and counterstained with the DNA dye 4′,6-diamidino-2-phenylindone (DAPI) at a concentration of 5 μg/ml. Confocal images were obtained by using a Bio-Rad MRC-1024/2P instrument interfaced with a Zeiss Axiovert S100 microscope. A krypton-argon laser with emission lines at 488 and 568 nm was used for conventional excitation of fluorescein and rhodamine with bandpass emission filters at 522 and 598 nm. A Spectra-Physics Tsunami femtosecond pulsed laser tuned to 770 nm was used for the multiphoton excitation of DAPI.
To determine the localization of cdc25C in γ-irradiated cells, U-2OS cells were plated in a 100-mm-diameter dish containing a coverslip. The next day the medium was changed to medium containing 200 μM mimosine to induce a G1 arrest (26); 20 h later, the mimosine was removed, and the cells were washed twice and refed with fresh medium; 6 h after mimosine removal, cells were irradiated in a Gamma-cell 40 irradiator (MDS Nordion) at a dosage of 6 Gy. After irradiation, one set of plates was incubated with leptomycin B at a concentration of 2 ng/ml for 18 h (13, 59); 18 h postirradiation, the cells were washed with PBS, and the coverslips were removed and fixed with paraformaldehyde as described above. After fixation, the coverslips were stored in PBS and then stained with a MAb to either cdc25C (TC113) or cyclin B1 (CB169). The remainder of the cells were harvested by trypsinization, fixed in 70% ethanol, stained with propidium iodide, and processed for fluorescence-activated cell sorting (FACS) as previously described (64).
Localization of the transfected cdc25C and cyclin B1 constructs was determined by performing indirect immunofluorescence with antibodies to the HA or Myc epitope. To perform the PCC assays, asynchronously growing U-2OS cells were transfected with the various cdc25C and cyclin B1 expression plasmids. At 40 to 44 h after transfection, the cells were fixed and stained with antibodies to the appropriate epitope tags. In each experiment, at least 100 transfected cells were counted. The results presented are the averages of at least three independent experiments. To perform the cell cycle synchrony experiments, U-2OS cells were plated in a 100-mm-diameter dish containing a coverslip. The next day the medium was changed with either regular medium or medium containing 200 μM mimosine to induce a G1 arrest (26). Approximately 2 h later, the cells were transfected as described above; 6 h posttransfection, the DNA precipitate was removed, and the cells were fed twice with medium containing mimosine; 20 h after the mimosine was first added, the mimosine was removed, and the cells washed twice and fed with fresh medium. At the indicated time after mimosine removal, the cells were washed with PBS, and the coverslip was removed and fixed with paraformaldehyde as described above. After fixation, the cells on coverslips were stained with an anti-Myc rabbit polyclonal antibody (Santa Cruz) and a MAb to cyclin B1 (CB169). The remainder of the cells were harvested by trypsinization and stained with antibody to CD19 conjugated to FITC (Caltag) and then processed for FACS as previously described (64). An S-phase arrest was induced in certain transfections by the addition of 100 μM hydroxyurea (HU).
RESULTS
Generation of specific MAbs to cdc25C.
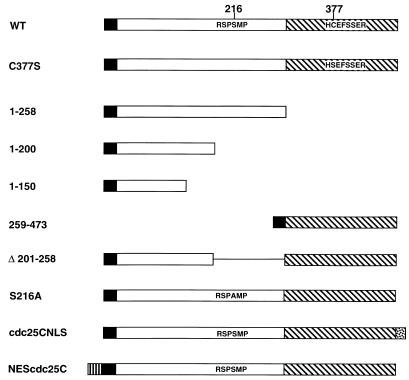
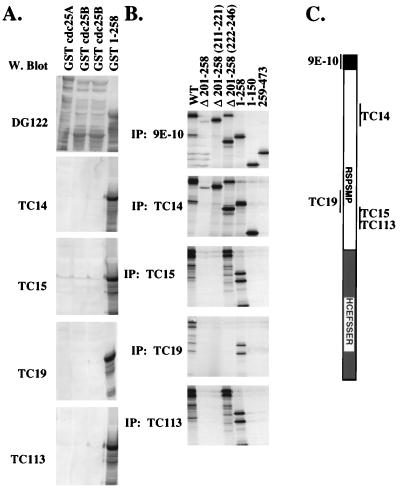
A diagram of the cdc25C constructs used in this study is shown in Fig. 1. A panel of MAbs to cdc25C, TC14, TC15, TC19, and TC113, was generated by immunizing mice with a GST fusion protein containing the first 258 residues of human cdc25C. The specificity of these antibodies for cdc25C was tested by Western blot analysis of bacterial lysates expressing GST-cdc25A (Fig. 2A, lane 1), GST-cdc25B (lanes 2 and 3), or the N-terminal GST-cdc25C fusion protein (GST 1–258; lane 4). All three fusion proteins were recognized by the anti-GST MAb DG122. In contrast, the anti-cdc25C antibodies failed to react with either GST-cdc25A or GST-cdc25B but did react with cdc25C. These results suggest that each of these antibodies could specifically detect cdc25C and not cdc25A or cdc25B.
FIG. 1.
Diagram of the human cdc25C constructs used. All cdc25C constructs contained an N-terminal Myc epitope tag (black box). A 14-3-3 binding motif of cdc25C (RSPSMP) is phosphorylated at the second serine residue (S216) in the consensus (44, 47, 49), and mutation of the serine residue to alanine abolishes binding to 14-3-3 proteins (49). The phosphatase domain (hatched box) is located in the C terminus with the active site (HCEFSSER) shown. Substitution of the cysteine residue at position 377 with serine disrupts phosphatase activity (4, 11, 40). The N-terminal box (vertical lines) represents the HA epitope and the HIV-1 Rev NES, and the C-terminal stippled box represents the SV40 large-T-antigen NLS.
FIG. 2.
Specificity of MAbs to cdc25C. (A) MAbs were raised to a GST fusion protein that contained the first 258 amino acids of human cdc25C (GST 1–258). Bacterial lysates expressing either GST-cdc25A (10 μl; lane 1), GST-cdc25B (20 and 30 μl; lanes 2 and 3, respectively), or GST 1–258 (10 μl; lane 4) were separated in an SDS–7.5% polyacrylamide gel and Western (W.) blotted with DG122, a mouse MAb specific for GST, and the four cdc25C antibodies, TC14, TC15, TC19, and TC113. (B) Indicated Myc-tagged cdc25C constructs were translated in vitro with [35S]methionine; 15 μl of in vitro translate was immunoprecipitated with the indicated antibodies, and the immunoprecipitated proteins were visualized by autoradiography. The Δ 201–258 construct contains an in-frame deletion of amino acids 201 to 258 (lane 2). Δ 201–258 (211–221) and Δ 201–258 (222–246) (lanes 3 and 4) are mutant proteins that contain insertions of the indicated amino acids into the Δ 201–258 mutant. 1–258, 1–150, and 259–473 (lanes 5 to 7) are shown in Fig. 1. (C) cdc25c diagram showing where the MAbs bind. The N-terminal myc tag is recognized by MAb 9E-10.
To map the epitopes in cdc25C recognized by the MAbs, in vitro translates of various cdc25C deletion mutants tagged with the Myc epitope (Fig. 1) were used in immunoprecipitation reactions. The anti-Myc antibody 9E-10 immunoprecipitated each of the cdc25C constructs as expected (Fig. 2B). The cdc25C MAbs could immunoprecipitate WT cdc25C as well as an N-terminal fragment expressing the first 258 residues (1–258) (Fig. 2B, lanes 1 and 5) but not a C-terminal fragment of cdc25C (259–473) that was not part of the immunizing peptide (lane 7). Of the four antibodies generated, only TC14 could immunoprecipitate 1–150 (lane 6), suggesting that it recognized an epitope contained within the first 150 amino acids of the protein. The other three MAbs failed to immunoprecipitate the Δ 201–258 or 1–150 protein (lanes 2 and 6), suggesting that they recognize epitopes between residues 201 and 258. TC15 and TC113 recognized an epitope located between amino acids 222 and 246, as determined by the observation that they could immunoprecipitate the Δ 201–258 (222–246) mutant (lane 4). TC19 recognized an epitope between amino acids 211 and 246 that was distinct from the epitope recognized by TC15 and TC113, based on its inability to immunoprecipitate either Δ 201–258 (211–221) or Δ 201–258 (222–246) (lanes 3 and 4) and its ability to immunoprecipitate both the Δ 201-210 and Δ 247–258 proteins (data not shown). Therefore the four MAbs recognize at least three different epitopes in cdc25C.
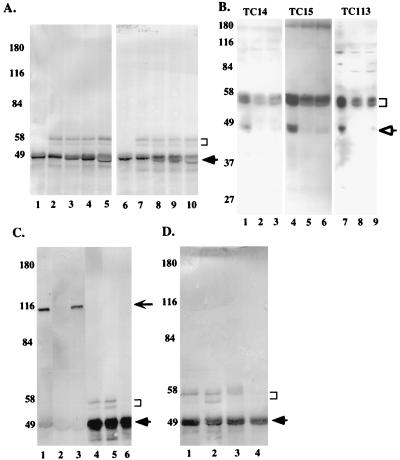
The cdc25C antibodies were tested for the ability to immunoprecipitate endogenous cdc25C in lysates prepared from U-2OS cells. All four cdc25C antibodies specifically immunoprecipitated a doublet that migrated at approximately 55 kDa in U-2OS cells upon Western blotting with either a mixture of the cdc25C MAbs or an anti-cdc25C rabbit polyclonal antiserum (C-20; Santa Cruz) raised against the C terminus of cdc25C (Fig. 3A, lanes 2 to 5 and lanes 7 to 10, respectively). A similar doublet was observed in the normal diploid human fibroblast line, MRC-5 (data not shown). The anti-Myc MAb 9E-10, which belongs to the same subclass as the anti-cdc25C antibodies, failed to immunoprecipitate the doublet (lanes 1 and 6). It has been reported previously that the slower-migrating band in this doublet is due to phosphorylation (47, 49). The immunoprecipitates were treated with lambda phosphatase, resulting in a loss of the upper band and an enhancement of the lower band, confirming that the upper band in the doublet was a phosphorylated version of the lower band (data not shown). These results suggest that all four MAbs could specifically immunoprecipitate both the phosphorylated and unphosphorylated forms of cdc25C.
FIG. 3.
cdc25C is present in the cytoplasm of asynchronously growing human cells. (A) Two-milligram aliquots of EBC extracts prepared from U-2OS cells were immunoprecipitated with cdc25C-specific antibodies TC14 (lanes 2 and 7), TC15 (lanes 3 and 8), TC113 (lanes 4 and 9), and TC19 (lanes 5 and 10) or the Myc antibody 9E-10 (lanes 1 and 6). The immune complexes resolved in SDS–7.5% polyacrylamide gels followed by Western blotting with either a mixture of TC14, TC15, and TC19 (lanes 1 to 5) or an anti-cdc25C rabbit polyclonal antibody (Santa Cruz) (lanes 6 to 10). The position of cdc25C, which migrated as a doublet, is indicated by the bracket. The arrow indicates the immunoglobulin heavy chain. The positions of the molecular weight markers are indicated in kilodaltons to the left of each gel. B. U-2OS (lanes 1, 4, and 7), MRC-5 (lanes 2, 5, and 8), and WI-38 (lanes 3, 6, and 9) cells were harvested by trypsinization, and the cell pellets were boiled in 2% SDS; 50 (lanes 1 to 6) or 25 (lanes 7 to 9) μg of protein extract was separated in an SDS–10% polyacrylamide gel, and then Western blotting was performed with the indicated antibodies. The immune complexes were detected by using the Pierce Supersignal system. TC14, TC15, and TC113 all recognized a specific doublet at ∼55 kDa in each of the different cell types. In addition to this band, they specifically recognized a band at ∼47 kDa (open arrow) in U-2OS cells. On longer exposures, this band was also detected in MRC-5 and WI-38 cells. No other specific signal was detected with these antibodies. (C) Two-milligram aliquots of whole-cell (lanes 1 and 4), cytoplasmic (lanes 2 and 5), or nuclear (lanes 3 and 6) extracts prepared from U-2OS cells were immunoprecipitated with either an anti-Rb antibody (MAb 245; Pharmingen) (lanes 1 to 3) or an anti-cdc25C antibody (TC113) (lanes 4 to 6). The immune complexes were resolved in an SDS–7.5% polyacrylamide gel and Western blotted with MAb 245 (lanes 1 to 3) or a mixture of TC14, TC15, and TC19 (lanes 4 to 6). The cdc25C doublet is indicated by the bracket. The thick arrow indicates the immunoglobulin heavy chains; the position of Rb on the gel is indicated by the thin arrow. (D) Two-milligram aliquots of whole-cell (lane 1), cytoplasmic (lanes 2 and 3), or nuclear (lane 4) extracts prepared from U-2OS cells were immunoprecipitated with the cdc25C MAb (TC113). The cytoplasmic extracts in lane 3 were prepared in the presence of a cocktail of phosphatase inhibitors (10 μM cypermethrin, 200 μM dephostatin, 200 nM okadaic acid, and 25 nM tautomycin). cdc25C is indicated by a bracket; the arrow indicates the position of the immunoglobulin heavy chain.
To further demonstrate the specificity of these antibodies, we tested their abilities to recognize cdc25C in direct Western blot analyses. Whole-cell extracts prepared from U-2OS, MRC-5, and WI-38 cells were resolved on an SDS–10% polyacrylamide gel and Western blotted with three different cdc25C MAbs (Fig. 3B). The three cdc25C MAbs recognize a doublet at ∼55 kDa in U-2OS (lanes 1, 4, and 7), MRC-5 (lanes 2, 5, and 8), and WI-38 (lanes 3, 6, and 9) cells. A band at ∼47 kDa is seen predominantly in U-2OS cells, though it can be seen in the other cell types at longer exposures. To determine if this band is related to cdc25C, we performed an immunoprecipitation with each of the MAbs followed by Western blotting with C-20. C-20 recognizes both the 55-kDa doublet and the 47-kDa band (data not shown). Similar results were obtained when the Western blotting was performed with a pool of the cdc25C MAbs (data not shown). This result suggests that the 47-kDa band is likely to be a truncated product of cdc25C and not a cross-reactive species.
To determine the intracellular localization of cdc25C, biochemical fractionation experiments were performed in asynchronously growing U-2OS cells. Whole-cell, cytoplasmic, or nuclear extracts were prepared as described in Materials and Methods. cdc25C was detected in the whole cell as well as the cytoplasmic extracts but not in the nuclear fraction (Fig. 3C lanes 4 to 6). As a control, IP/Western experiments were also performed to determine the localization of Rb, previously demonstrated to be localized to the nucleus (34, 41). As shown in Fig. 3B, Rb was present in the nuclear extracts and no Rb was detectable in the cytoplasmic fraction (lanes 1 to 3). These results suggest that the cytoplasmic extracts were not contaminated by nuclear proteins. A significant increase in amount of the faster-migrating band of the cdc25C doublet was observed when cytoplasmic fractions were prepared from U-2OS cells compared to the whole-cell extracts (Fig. 3D; compare lanes 1 and 2), suggesting that cdc25C may have been undergoing dephosphorylation while the extract was prepared. To test this hypothesis, cytoplasmic extracts were prepared in the presence of additional phosphatase inhibitors. Under these conditions, only the top band of the doublet can be seen (lane 3); cdc25C is not present in the nuclear fraction (lane 4). This latter result suggests that all of the endogenous cdc25C protein exists in the phosphorylated form.
The localization of cdc25C changes prior to mitosis in human cells.
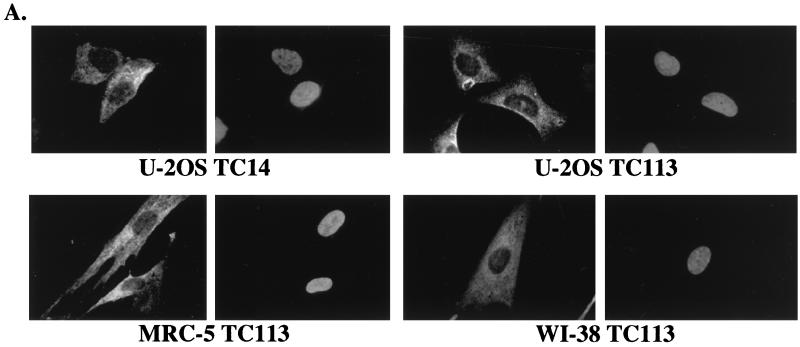
To confirm the cytoplasmic localization of endogenous cdc25C in human cells, indirect immunofluorescence analysis was performed on normal diploid human fibroblasts as well as immortal human cell lines. As shown in Fig. 4A, a specific cytoplasmic signal for cdc25C was obtained in U-2OS cells when they were immunostained with either TC14 or TC113. A similar staining pattern in U-2OS was obtained with each of the other cdc25C MAbs, TC15 and TC19 (data not shown). The normal diploid human fibroblast strains, MRC-5 and WI-38, also showed a cytoplasmic staining pattern for cdc25C with MAb TC113 (Fig. 4A) as well as with each of the other MAbs (data not shown).
FIG. 4.
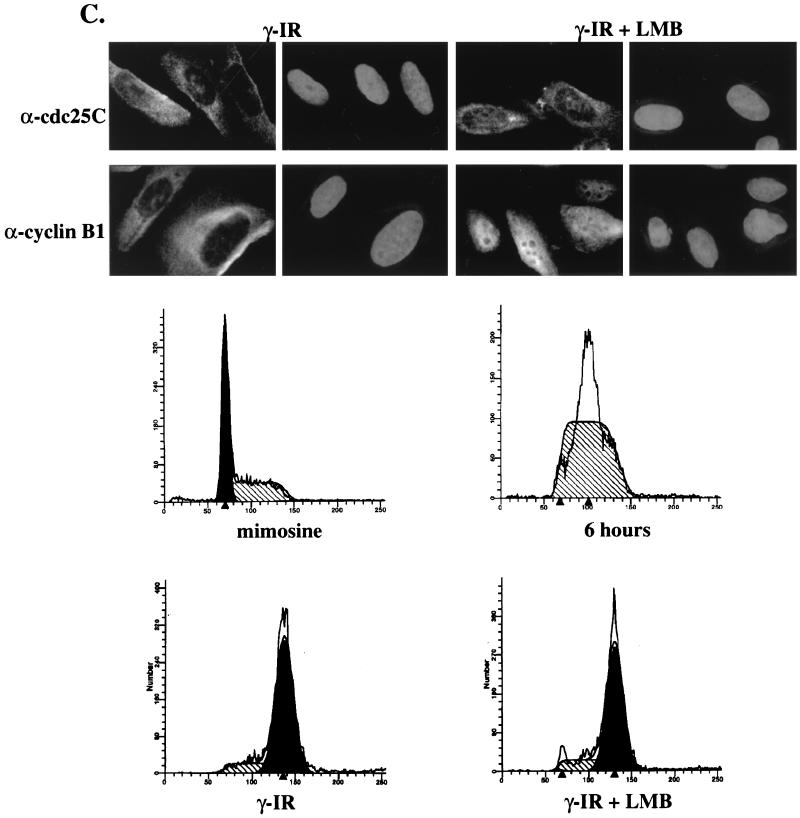
Cellular localization of cdc25C in human cells. (A) Indirect immunofluorescence with the cdc25C antibodies was performed in multiple cell types. U-2OS cells immunostained with either TC14 (left) or TC113 (right) are shown in the top panels. The primary human fibroblasts, MRC-5 (bottom left) and WI-38 (bottom right), were immunostained with antibody TC113. In each pair, the left panel shows antibody staining while the right panel shows the DAPI stain of the same field (original magnification, ×100). (B) MRC-5 cells were immunostained with polyclonal sera specific for phospho-histone H3 (α pH3) (17), a MAb to cdc25C (TC113), and DAPI, as indicated, and visualized by confocal microscopy. The merge of the cdc25C and the pH3 staining is shown in the bottom left panel. A cell that stains positively for histone H3 and shows partial chromatin condensation (DAPI) is indicated by the thick arrow. A cell that does not stain with the phospho-histone H3 antibody in which the chromatin is not condensed is indicated by the thin arrow (original magnification, ×63). (C) U-2OS cells were treated with mimosine to induce a G1 arrest. Six hours after mimosine release, cells were γ irradiated (γ-IR) with a dose of 6 Gy and then incubated in either the presence or absence of the crm1 inhibitor leptomycin B (LMB) for an additional 18 h. Cells were immunostained with either an anti-cdc25C antibody (TC113) or an anti-cyclin B1 antibody (CB169). In each pair, the left panel shows immunofluorescence while the right panel shows the DAPI stain of the same field (original magnification, ×100). The cell cycle profiles of the cells in mimosine (mimosine), cells 6 h after release from mimosine (6 hours), and the γ-irradiated cells (γ-IR) and irradiated cells treated with leptomycin B (γ-IR + LMB) are shown.
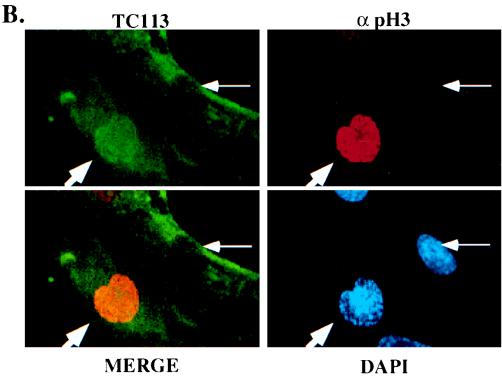
To determine whether the cellular localization of cdc25C was altered over the cell cycle, MRC-5 cells were stained with a MAb to cdc25C (TC113) and a polyclonal serum specific for phospho-histone H3 followed by confocal microscopy. Histone H3 is not phosphorylated in G1 and S phases. Phosphorylation of histone H3 is initiated during G2, and the intensity and pattern of the signal phospho-histone H3 signal changes as cells progress from G2 to M phase, with a steady accumulation of the phosphorylated form of the protein until anaphase (17). Cells that show no phospho-histone H3 in the nucleus are in G1 or S phase of the cell cycle, while cells that show a punctate pattern of phospho-histone H3 in the nucleus are in early G2 phase (17). Cells that did not stain for phospho-histone H3 showed a cytoplasmic staining pattern for cdc25C; in contrast, cells that stain positively with the antibodies to phospho-histone H3 contain cdc25C both in the nucleus and the cytoplasm, as evidenced by the yellow staining in the merged image (Fig. 4B). The timing of the change in cellular localization just prior to mitosis is similar to that reported for cyclin B1 (13, 51, 59) and suggests that, like cyclin B1, cdc25C enters the nucleus just prior to mitosis in human cells.
The human cyclin B1 protein enters the nucleus just prior to mitosis due to the inactivation of a NES in cyclin B1 (13, 59). Notably, DNA damage prevents inactivation of the NES, thus forcing cyclin B1 to be retained in the cytoplasm (13, 51, 59). To determine whether DNA damage affects the localization of cdc25C, U-2OS cells were initially synchronized in G1 phase with mimosine (Fig. 4C). Six hours after mimosine was removed, the cells had entered S phase (Fig. 4C). cdc25C and cyclin B1 could both be detected in the cytoplasm by immunostaining during S phase, while only cdc25C was expressed in cells arrested in G1 with mimosine (data not shown). At this point, the cells were γ irradiated and then incubated in the absence or presence of leptomycin B for 18 h. The γ-irradiated U-2OS cells arrested almost exclusively in the G2 phase of the cell cycle (Fig. 4C). Immunostaining of γ-irradiated U-2OS cells with a MAb to cdc25C (TC113) or cyclin B1 (CB169) demonstrated that both proteins remained in the cytoplasm (Fig. 4C). The leptomycin B-treated cells also showed a G2 arrest similar to that for the cells that were γ irradiated but not treated with leptomycin B (Fig. 4C). The localization of cdc25C was not altered when the cells were treated with leptomycin B (Fig. 4C). In contrast, in cells treated with leptomycin B, the localization of cyclin B1 was dramatically altered, with a marked increase in the nuclear signal being observed (Fig. 4C), in agreement with results published by other groups (13, 59). These results suggest that although both cyclin B1 and cdc25C are maintained in the cytoplasm in γ-irradiated cells, the cytoplasmic localization and subsequent nuclear transport of these two proteins are regulated by different mechanisms in human cells.
A 58-amino-acid domain in cdc25C controls its cellular localization.
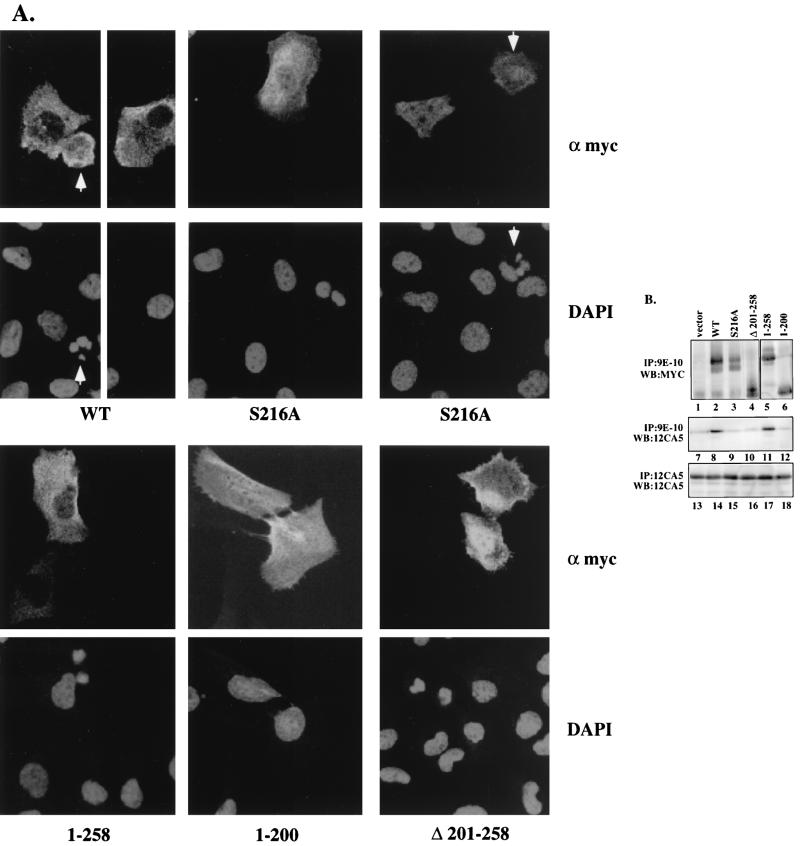
To determine the regions of cdc25C that participate in its normal cytoplasmic localization during interphase, various Myc epitope-tagged cdc25C deletion constructs were transiently expressed in U-2OS cells. The cells were immunostained with the anti-Myc MAb 9E-10 and imaged by confocal microscopy. Wild-type cdc25C was found to be localized in the cytoplasm (Fig. 5A), similar to results for the endogenous protein and to previously published results (10, 16). A full-length cdc25C protein with an inactive phosphatase domain, C377S (4, 11, 40), as well as an N-terminal cdc25C construct containing residues 1 to 258, lacking the catalytic domain, also localized to the cytoplasm (Fig. 5A and data not shown), suggesting that the phosphatase activity of cdc25C was not required for its cytoplasmic retention. Further deletion into the N terminus (construct 1–200) resulted in a pancellular distribution, with the protein being present in both the cytoplasmic and nuclear compartments (Fig. 5A). An N-terminal deletion mutant, 259–473, that contained the entire catalytic domain was also pancellular (data not shown). These results suggest that residues 201 to 258 contributed to the cytoplasmic localization of cdc25C.
FIG. 5.
Cytoplasmic localization of cdc25C is dependent on a 14-3-3 binding motif. (A) U-2OS cells were transiently transfected with the indicated cdc25C constructs and visualized by confocal microscopy with the anti-Myc antibody 9E-10 (α myc) and DAPI staining. The arrow highlights cells showing condensed fractured chromatin indicative of PCC (original magnification, ×63). (B) Extracts of U-2OS cells cotransfected with plasmids expressing an HA-tagged 14-3-3ɛ cDNA and the indicated Myc-tagged cdc25C constructs were immunoprecipitated with an anti-Myc antibody (9E-10) or an anti-HA antibody (12CA5). The immune complexes were washed and then resolved in either an SDS–7.5% polyacrylamide gel (lanes 1 to 4, top panel) or an SDS–10% polyacrylamide gel (lanes 5 and 6, top panel; lanes 7 to 18, middle and bottom panels). The cdc25C constructs were detected by Western blotting (WB) with a polyclonal Myc antibody (Santa Cruz), and the HA 14-3-3ɛ was detected by Western blotting with the anti-HA MAb (12CA5).
To determine if residues 201 to 258 were necessary for the cytoplasmic localization of cdc25C, a mutant with an in-frame deletion of residues 201 to 258 (Δ 201–258) was constructed. As shown in Fig. 5A, Δ 201–258 showed a pancellular localization similar to that observed for 1–200. A consensus binding site for 14-3-3 proteins is contained in residues 201 to 258 (213–218). Phosphorylation of the serine residue at position 216 is required for binding to 14-3-3 proteins, and mutation of S216 to alanine has been shown to prevent phosphorylation at this site, resulting in a loss of 14-3-3 binding (49). To determine whether S216 was required for cytoplasmic retention of cdc25C, we generated a substitution mutant that altered the serine residue at position 216 to alanine (S216A). As shown in Fig. 5A, S216A showed a pancellular localization similar to that of the mutant Δ 201–258, suggesting that S216 may be required for retention of cdc25C in the cytoplasm. A mutant that substituted an aspartic acid for serine, S216D, also showed a pancellular localization (data not shown). Mutation of another serine residue in the same region, S214, did not alter the cytoplasmic localization of cdc25C (data not shown).
The ability of these cdc25C mutant proteins to form a complex with 14-3-3 proteins was tested. It has been reported that the Xenopus cdc25 protein forms a complex with 14-3-3ɛ (31). To determine if human cdc25C associated with 14-3-3ɛ, wild-type cdc25C or the different mutants were transfected into U-2OS cells with an expression construct for a HA epitope-tagged 14-3-3ɛ cDNA (HA-14-3-3ɛ). Extracts from the transfected cells were prepared, and immunoprecipitations were performed with antibodies to either the HA or Myc epitope. As shown in Fig. 5B, each of the cdc25C proteins (Fig. 5B, lanes 1 to 6) and the HA-14-3-3ɛ protein (lanes 13 to 18) was expressed at equivalent levels in each transfection. Both WT cdc25C and the 1–258 protein formed a complex with 14-3-3ɛ in vivo, as demonstrated by their ability to coprecipitate the HA-14-3-3ɛ protein (lanes 8 and 11). In contrast, S216A, Δ 201–258, and 1–200 were unable to form a complex with the HA-14-3-3ɛ protein above background levels (lanes 9, 10, and 12) (49). In cells transfected with the vector and HA-14-3-3ɛ, an immunoprecipitation performed with the Myc antibody did not result in the coprecipitation of the HA-14-3-3ɛ protein (lane 7). These results demonstrate that the proteins that show a pancellular distribution by immunostaining, such as S216A, Δ 201–258, and 1–200 also failed to form a complex with 14-3-3ɛ, while the proteins that show a cytoplasmic staining pattern, such as wild-type cdc25C and 1–258, could bind to 14-3-3ɛ.
Pancellular localization of cdc25C results in an increase in its ability to induce PCC.
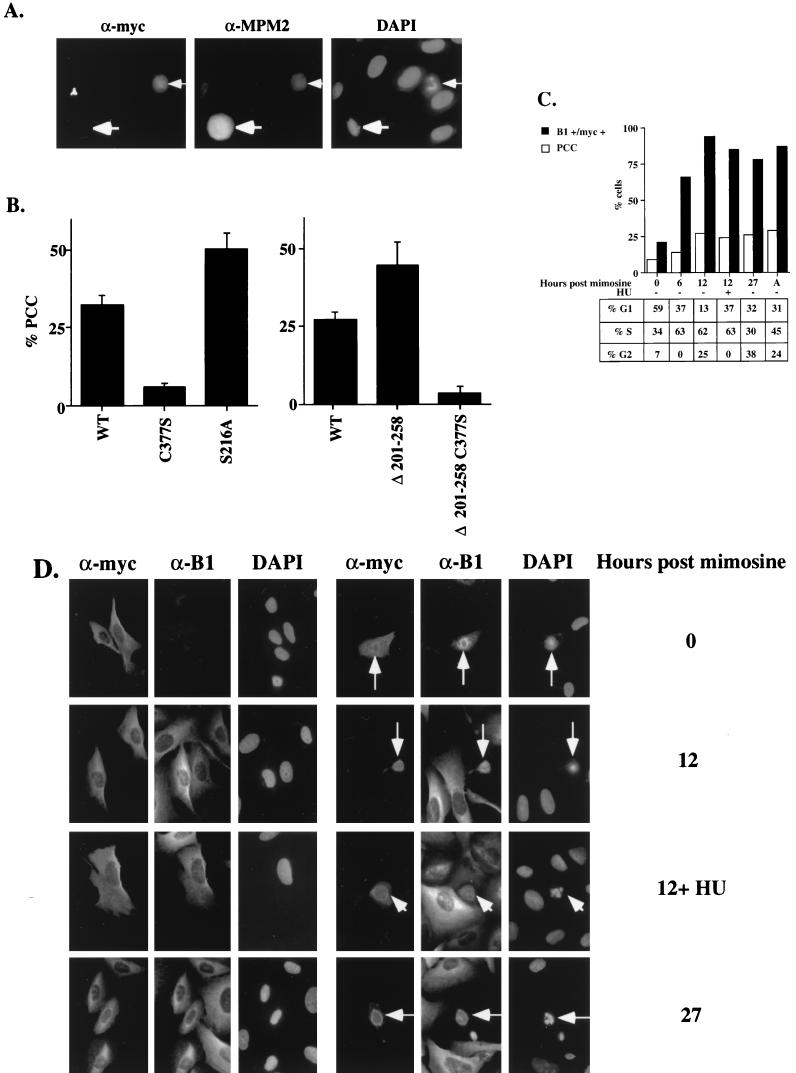
To determine whether the alteration of cdc25C localization affected its function, each of these cdc25C mutants was tested for its ability to induce PCC in U-2OS cells. A minimum amount of cdc25C expression plasmid was used in all experiments to yield equivalent levels of expression detectable by both Western blotting and immunofluorescence analysis (Fig. 5). Some of the cells expressing Myc-tagged cdc25C (Fig. 5A) or cdc25C and an HA-tagged cyclin B1 (Fig. 6A) contained condensed fractured chromatin by DAPI staining as previously reported (13, 16, 49). Cells that contain condensed fractured chromatin by DAPI staining also stained positively with the mitosis-specific antibody MPM2 (2) (Fig. 6A). The morphology of the condensed chromatin in cells undergoing PCC was readily distinguishable from a normal mitosis (Fig. 6A).
FIG. 6.
Effect of localization on cdc25C function. (A) U-2OS cells transfected with Myc-tagged cdc25C and cyclin B1 were immunostained with antibodies to the Myc epitope (α-myc) the mitosis-specific antibody MPM2 (α-MPM2) and DAPI. A cell immunostained with both the Myc and MPM2 antibodies showed condensed fractured chromatin when stained with DAPI (thin arrow). An untransfected cell undergoing mitosis is stained by the MPM2 antibody (thick arrow). (B) U-2OS cells transfected with the indicated Myc-tagged cdc25C constructs were immunostained with the anti-Myc antibody (9E-10) and DAPI. More than a 100 Myc-positive, cdc25C-expressing cells were counted, and the percentage of cells containing condensed fragmented chromatin was determined in three independent experiments. (C) U-2OS cells were treated with mimosine to induce a G1 arrest. Cells were transfected with cdc25C and a CD19 cDNA in the presence of mimosine. Six hours after transfection, the DNA was removed and the cells were refed twice with mimosine-containing medium; 20 h after mimosine was first added to the cells, cells were refed with fresh medium; cells were then harvested at the indicated time points for FACS analysis or were immunostained for Myc-tagged cdc25C and the endogenous cyclin B1 as described in the text and with DAPI to determine PCC. Six hours after mimosine was removed, 100 μM HU was added to one set of plates for 6 h. Open bars indicate the percentage of cells undergoing PCC; filled bars indicate the percentage of transfected cells staining positively for cyclin B1. A, asynchronous. The percentages of CD19-positive cells in G1, S, and G2 phases at the indicated timepoints are tabulated at the bottom. (D) Cells from the experiment described above were stained with a polyclonal antibody to the Myc epitope (α-myc) a MAb to cyclin B1 (α-B1), or DAPI. Cells arrested in mimosine (0) show little staining for cyclin B1. After mimosine release, the percentage of cells staining for cyclin B1 increases until all cells stain for cyclin B1 (12 and 12+HU). All cells undergoing PCC (thin arrow) show staining for cyclin B1 (original magnification, ×60).
To determine whether the transient expression of cdc25C was more likely to induce PCC effectively during S and G2 phases when cells expressed cyclin B1, a cell cycle synchrony experiment was performed. U-2OS cells were transfected with a wild-type cdc25C plasmid and an expression plasmid for the cell surface marker CD19 to monitor the cell cycle profile of the transfected cells. The cell cycle profile was monitored by staining cells with anti-CD19 antibodies cross-linked to FITC and propidium iodide followed by FACS analysis. During transfection, cells were initially arrested in G1 phase by mimosine treatment (26) and then released from the G1 block by removing mimosine and monitored over a period of 27 h. A transfection in which the cells were not treated with any drugs was also performed (asynchronous). Cyclin B1 expression was monitored by immunostaining with a MAb to cyclin B1 (CB169), and the transfected cells were identified by immunostaining with an anti-Myc antiserum (Santa Cruz). All Myc-tagged cdc25C-expressing cells that contained condensed fractured chromatin also stained positively for endogenous cyclin B1 (Fig. 6D). Cells in mimosine were mostly in G1 phase of the cell cycle (Fig. 6C, 0) and showed low levels of PCC (9%) and cyclin B1 (22%) staining (Fig. 6C and D). Cells maintained in mimosine for the duration of the experiment remained arrested in G1 phase. Neither the levels of PCC nor the number of cells staining positively for cyclin B1 increased significantly in those transfections (data not shown). After release from mimosine, cells progressed through the cell cycle and entered S phase (6 h, >60% S phase) and then G2 (12 h) (Fig. 6C), with a concomitant increase in the number of cells undergoing PCC (14 and 27% at 6 and 12 h, respectively) and staining for cyclin B1 (66 and 94%, respectively) (Fig. 6C and D). HU was added to one set of transfected plates after release from mimosine to induce an S-phase arrest. As can be seen in the FACS analysis, HU was effective at preventing cells from entering G2 between 6 and 12 h after mimosine removal. However, HU did not slow the increase in percentage of cells expressing cyclin B1, as the number of cells staining for cyclin B1 in cells treated with HU was similar to the number staining positively in the absence of HU at the same time point (Fig. 6C and D; compare 12 to 12+ HU). Furthermore, the percentage of cells showing condensed fractured chromatin increased in the presence of HU (Fig. 6C). The percentage of cells undergoing PCC at 27 h after release from mimosine was similar to that observed in the untreated asynchronously growing cells not treated with either mimosine or HU (asynchronous). These results suggest that the appearance of condensed fractured chromatin reflected an abnormal mitotic event that is dependent on the presence of cyclin B1 and transfected cdc25C. Furthermore, this most likely represents a premature mitotic event, given that transient expression of cdc25C could induce a similar phenotype in the presence of HU.
PCC assays were performed to compare the activity of the wild-type cdc25C construct to those of the cdc25C mutants. These assays were performed in asynchronously growing cells, and levels of PCC were determined at a time similar to the asynchronous time point in the cell cycle synchrony experiment. Approximately 30% of the cells that expressed the wild-type Myc epitope-tagged cdc25C construct showed PCC (Fig. 6B). A cdc25C mutant with an inactive phosphatase domain, C377S, was unable to induce PCC above background levels (Fig. 6B, left panel), consistent with the notion that cdc25C phosphatase activity was required for entry into mitosis. Similarly, the 1–258 and 1–200 mutants (lacking the C-terminal catalytic domain) were unable to induce PCC at levels above background (data not shown). Consistent with previous results (49), the 14-3-3 binding-defective mutant S216A induced PCC at levels greater than WT (Fig. 6B, left panel). Similarly, Δ201–258, which also does not bind 14-3-3ɛ, was able to induce PCC in about 50% of the transfected cells (Fig. 6A, right panel). The specificity of this effect could be demonstrated by the inability of the corresponding active site mutant, Δ201–258 C377S, to induce PCC at significant levels. In cells that did not undergo PCC, these proteins demonstrated a pancellular localization (Fig. 5A). Furthermore, when WT, S216A, or Δ201–258 was cotransfected into cells with an expression construct for cyclin B1, more than 90% of the transfected cells showed evidence of PCC (Fig. 7B and C and data not shown). However, the active-site mutants C377S and Δ201–258 C377S were unable to cooperate with cyclin B1 to induce PCC, with the levels of PCC remaining at background levels (data not shown). These data are consistent with the data from the cell cycle synchrony experiment that suggest that the induction of PCC by cdc25C requires the presence of cyclin B1. They also provide support for the hypothesis that an intact 14-3-3 binding motif contributes to regulation of cdc25C activity.
FIG. 7.
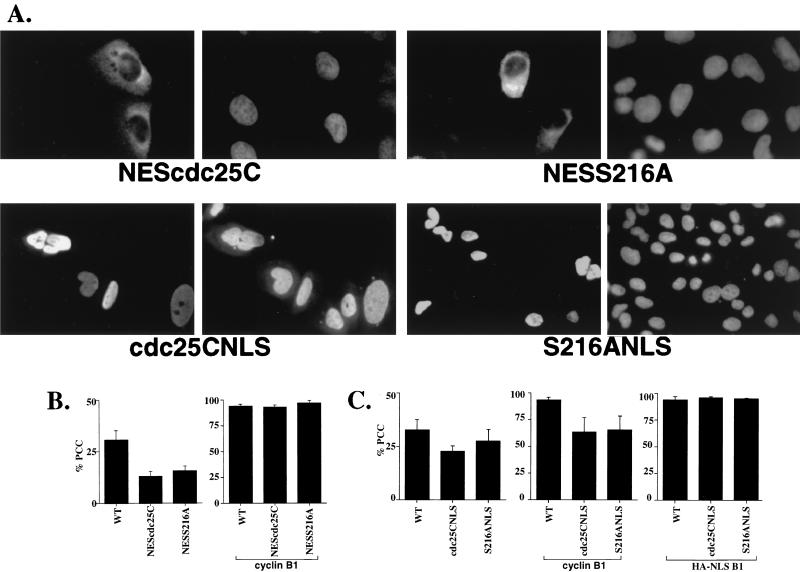
Effect of a heterologous NES or NLS on cdc25C localization and function. (A) U-2OS cells were transfected with constructs expressing either the Myc-tagged NLS or HA-tagged NES fusion of cdc25C. The cells were fixed and visualized with indirect immunofluorescence with antibody to the HA or Myc epitope (left panel of each pair) and DAPI (right panel of each pair). The original magnification on all the panels is ×60 except for the S216ANLS panel (×40). (B) WT cdc25C and the NES fusions were transfected into U-2OS cells either alone (left) or in the presence of ectopically expressed cyclin B1 (right). (C) WT and the NLS fusions were transfected into U-2OS cells alone (left), cotransfected with cyclin B1 (middle), or cotransfected with an NLS-tagged version of cyclin B1 (right).
Targeting cdc25C expression to either the cytoplasm or the nucleus results in a decrease in its ability to induce PCC.
The results described above suggest that the cytoplasmic retention of cdc25C by 14-3-3 proteins may prevent premature mitosis. Furthermore, they suggest the possibility that the entry of cdc25C into the nucleus is required to initiate mitosis. A prediction of this model would be that a cdc25C protein that cannot accumulate in the nucleus would not induce PCC as efficiently as the WT protein, while a cdc25C protein that is nuclear in interphase cells would induce PCC at levels greater than the WT protein. To test this hypothesis, cdc25C was fused to either the HIV-1 Rev NES (7) or the SV40 NLS. As shown in Fig. 7A, the NEScdc25C fusion localized to the cytoplasm in interphase cells as expected. Significantly, the NESS216A fusion, containing a substitution mutation in the 14-3-3 binding domain, was also excluded from the nucleus (Fig. 7A), a phenotype distinct from that observed for the S216A mutant. This result suggests that the heterologous NES can induce the cytoplasmic localization of cdc25C independent of its potential to bind to 14-3-3 proteins. Conversely, the cdc25CNLS fusion localized exclusively to the nucleus in interphase cells, as did the corresponding 14-3-3 binding site mutant, S216ANLS (Fig. 7A), suggesting that the SV40 NLS could act dominantly over the cdc25C cytoplasmic retention signal. Each of these cdc25C constructs was expressed at equivalent levels as determined by IP/Western analysis (data not shown).
The cdc25C constructs with heterologous localization signals were tested for the ability to induce PCC. As shown in the left panel of Fig. 7B, the NEScdc25C and NESS216A fusions induced PCC at similar levels despite the loss of the 14-3-3 binding site in NESS216A (Fig. 6B, left panel). Notably, these NES fusions did not induce PCC as well as WT. To test whether the NEScdc25C fusions could cooperate with cyclin B1 to induce PCC, they were transfected into U-2OS cells with an expression vector for an HA epitope-tagged cyclin B1. Upon coexpression of WT cdc25C and cyclin B1 in U-2OS cells, more than 90% of the transfected cells showed evidence of PCC (Fig. 7B, right panel). This effect requires cdc25C phosphatase activity, as when cyclin B1 was cotransfected with a catalytically inactive cdc25C mutant, C377S, the levels of PCC were at background levels (data not shown). Similarly, when either the NEScdc25C or NESS216A fusion was cotransfected with cyclin B1, more than 90% of the transfected cells showed PCC (Fig. 7B, right panel). These results suggest that the NES fusion proteins had no intrinsic phosphatase defect and were fully capable of inducing PCC in the presence of sufficient cyclin B1. To test if the NEScdc25C fusions could cooperate with a nuclear version of cyclin B1, they were cotransfected with a nuclear version of cyclin B1, HA-NLSB1 (B1 was fused to the T-antigen NLS), into U-2OS cells. HA-NLSB1 was predominately a nuclear protein, although there was a significant signal observed in the cytoplasm as well (data not shown). Notably, when the HA-NLSB1 construct was coexpressed with the NEScdc25C fusions, more than 90% of the cells expressing both proteins underwent PCC (data not shown). This high degree of cooperativity may be due to the presence of readily detectable amounts of the HA-NLSB1 protein in the cytoplasm of U-2OS cells (data not shown).
The ability of the nuclear versions of cdc25C to induce PCC was also compared to that of WT cdc25C. Both the cdc25CNLS and S216ANLS fusion proteins were not as effective as the WT cdc25C protein at inducing PCC in U-2OS cells (Fig. 7C, left panel). Furthermore, coexpression of these constructs with cyclin B1 resulted in only 70% of the cells showing PCC(Fig. 7C, middle panel). In cells not showing PCC, cdc25CNLS and S216ANLS were nuclear whereas the transfected cyclin B1 remained cytoplasmic (data not shown), suggesting that the NLS fusion proteins did not alter the localization of cyclin B1 and that the reduced level of PCC exhibited by these nuclear cdc25C proteins may be due to limiting amounts of cyclin B1 in the nucleus (59). To test whether an NLS-tagged version of cyclin B1 could cooperate with the NLS fusion proteins to induce PCC, cdc25CNLS and S216ANLS were cotransfected with HA-NLSB1 into U-2OS cells. When transfected alone, HA-NLSB1 did not induce PCC above background levels (data not shown). However, when HA-NLSB1 was cotransfected with either cdc25CNLS or S216ANLS, more than 90% of the transfected cells showed evidence of PCC (Fig. 7B, right panel). The specificity of this effect was demonstrated by the failure of the inactive mutant C377SNLS to induce PCC in cooperation with HA-NLSB1 (data not shown). This result suggests that the nuclear versions of cdc25C were not compromised in the ability to function as phosphatases but failed to induce PCC at WT levels due to limiting amounts of cyclin B1 in the nucleus due to the rapid nuclear export of cyclin B1 (13, 51, 59).
DISCUSSION
The results presented herein suggest that the ability of 14-3-3 proteins to regulate cdc25C function is mediated, at least in part, by promoting the retention of cdc25C in the cytoplasmic compartment during interphase. Cytoplasmic retention of cdc25C by 14-3-3 required an intact 14-3-3 binding motif. cdc25C mutants that do not contain a 14-3-3 binding motif, either by deletion or mutation of the S216 residue, showed a pancellular distribution when transiently expressed in U-2OS cells, in contrast to the predominantly cytoplasmic appearance of a WT construct. It was observed that transient expression of cdc25C with a disrupted 14-3-3 binding site led to a significantly increased tendency to induce PCC compared to WT cdc25C expressed under similar conditions. The same 14-3-3 binding site in cdc25C has been reported to represent an important regulatory motif affecting the activity of cdc25C during the DNA replication and under DNA damage checkpoint (49). Therefore, 14-3-3 proteins may serve to regulate cdc25C activity by restricting its cellular localization to the cytoplasm under various conditions.
Cytoplasmic localization of human cdc25C.
There has been some controversy in the field regarding the cellular localization of cdc25C. Previously, three studies reported that cdc25C was a nuclear protein in human cells (10, 12, 39). However, each of these studies used a polyclonal serum that had the potential to recognize both cdc25A and cdc25B, in addition to cdc25C. For example, Girard et al. used a polyclonal rabbit antiserum raised against a peptide derived from the catalytic domain of starfish cdc25 (12). Every residue within this peptide was perfectly conserved in the catalytic domains of all three human cdc25 proteins. Therefore, this antiserum may have recognized cdc25A and cdc25B in addition to cdc25C. Two other groups generated polyclonal antibodies by immunizing rabbits with bacterially produced, full-length cdc25C protein followed by affinity purification with the immunogen (10, 39). The purified antisera may have recognized highly conserved elements within cdc25A, cdc25B, and cdc25C. Notably, cdc25A was reported to be a nuclear protein with a predicted molecular mass (58 kDa) similar to that of cdc25C (53 kDa) (20). In contrast to the cdc25C antiserum, the cdc25A antibody generated in this latter study was raised against an N-terminal peptide in cdc25A not present in cdc25C and therefore was unlikely to recognize cdc25C (20).
In this report, four different MAbs were generated to the N-terminal 258 residues of cdc25C, a region not highly conserved with either cdc25A or cdc25B. These MAbs specifically recognized at least three distinct epitopes within cdc25C and could not detect cdc25A or cdc25B by Western blotting. Immunostaining of a number of primary human cell strains and immortal cell lines with each of these antibodies revealed a specific cytoplasmic signal. The cytoplasmic localization of cdc25C was confirmed by the appearance of a specific Western blot signal of the appropriate molecular weight in the cytoplasmic but not the nuclear extracts prepared from U-2OS cells. The cytoplasmic localization of the endogenous cdc25C protein was consistent with earlier reports for transiently expressed, epitope-tagged cdc25C protein. These observations were extended in this report to demonstrate that the cytoplasmic localization of transiently expressed cdc25C was dependent on an intact 14-3-3 binding motif.
Effect of 14-3-3 on cdc25C function and subcellular localization.
S216 has been identified as the major phosphorylation site in cdc25C during interphase (47, 49), suggesting that its phosphorylation may contribute to the negative regulation of cdc25C activity during interphase. It was subsequently demonstrated that phosphorylation of S216 results in the generation of a binding site for 14-3-3 proteins that may serve to inhibit cdc25C function (44, 49, 62). However, the precise manner in which 14-3-3 proteins inhibit cdc25C function was not known. The results presented here suggest that an intact 14-3-3 binding site in cdc25C may be required for the cytoplasmic retention of cdc25C during interphase. Binding to 14-3-3ɛ was disrupted by substitution of residue 216 with alanine (Fig. 5B). Loss of the 14-3-3 binding site in the transiently expressed construct, S216A or Δ 201–258, resulted in pancellular localization of cdc25C (Fig. 5A) and an increased ability to induce PCC compared to WT cdc25C (Fig. 6B). These results suggest that at least one mechanism by which 14-3-3 proteins regulate cdc25C function is by inducing its cytoplasmic localization.
At least seven different 14-3-3 proteins have been identified in mammalian cells (62). This leads to the question of which 14-3-3 protein modulates cdc25C function in response to the DNA replication or DNA damage checkpoints. 14-3-3ɛ and 14-3-3ζ have been shown to complex with cdc25 in Xenopus egg extracts (31). Another human homologue, 14-3-3ς, may have a specific role in a G2/M checkpoint, given that its expression was stimulated by DNA damage induced by γ irradiation in a p53-dependent manner (18). Furthermore, 14-3-3ς overexpression induced a G2 arrest, leading to the speculation that at least part of this effect may be dependent on its interaction with cdc25C. It is possible that cdc25C is regulated by any one of several 14-3-3 proteins in response to distinct DNA replication and damage checkpoints. In this report, 14-3-3ɛ was shown to bind specifically to cdc25C and required an intact S216 residue. Whether other 14-3-3 proteins can bind specifically to cdc25C remains to be tested.
Phosphorylation of the S216 residue may also occur in response to a variety of situations. Three kinases (chk1, chk2, and C-TAK1) have been shown to be capable of specifically phosphorylating the S216 residue in cdc25C, resulting in the generation of a consensus binding site for 14-3-3 proteins (38, 49, 50, 52). Notably, C-TAK1 was found to be cytoplasmic in human cells and could potentially promote the phosphorylation of cdc25C in the cytoplasm, resulting in association with 14-3-3 proteins and cytoplasmic retention (50). In contrast, chk1 and chk2 have been reported to be nuclear proteins (38, 52). chk1 and chk2 may phosphorylate the S216 residue when cdc25C enters the nucleus. Whether this phosphorylation of S216 can promote the nuclear export of cdc25C is not known. It has been recently reported that S. pombe cdc25 may be exported from the nucleus in complex with rad24, a 14-3-3 homolog. This export is abolished in a strain lacking chk1 (37). Therefore, in response to DNA damage, chk1 may promote the nuclear export of a cdc25 protein that has entered the nucleus prematurely.
The ability of 14-3-3 proteins to sequester important regulatory proteins was previously demonstrated for the apoptosis inducer BAD (66). Phosphorylation at serine residues within two 14-3-3 binding consensus sites in BAD promotes the formation of a complex with 14-3-3 proteins, leading to its retention in the cytoplasm. Upon dephosphorylation of these residues, BAD becomes capable of forming a complex with BCL-XL within the mitochondrial membrane fraction. Association with BAD inhibits the antiapoptotic activity of BCL-XL, resulting in apoptosis and cell death (66). Therefore, it appears that 14-3-3 proteins may serve an important role in cell survival and cell division by sequestering regulatory proteins in the cytoplasm.
Cellular localization and PCC.
In the PCC assays described here, WT cdc25C was capable of inducing fractured condensed chromatin in 25 to 30% of the transfected cells. This percentage was much higher than that reported previously (13, 49). The difference in frequency of PCC observed in these assays compared to previous reports (13, 49) may be attributable to the differences in cell type (U-2OS cells versus HeLa) or methods used for expression (transient transfection in this study versus inducible stable cell lines or microinjection of cDNA in others). Furthermore, the cell cycle synchrony experiments shown here suggest that PCC was initiated only in the presence of cyclin B1. In asynchronously growing U-2OS cells, more than 60% of the cells were in S and G2 phases of the cell cycle, with a similar percentage containing cyclin B1, as determined by immunostaining (Fig. 6). Therefore, the high numbers observed in the PCC assay may reflect the highly proliferative nature of these cells. Consistent with this finding was the observation that cotransfection of cdc25C and cyclin B1 resulted in nearly all of the cells undergoing PCC (Fig. 7) (13, 16). The higher numbers of PCC observed in this assay system permitted the analysis of cdc25C mutant proteins with lower activity than the WT construct.
The timely translocation of cdc25C from the cytoplasm to the nucleus may be an important component of the entry into mitosis. The increased tendency of the 14-3-3 binding site mutant S216A to induce PCC may indicate that cdc25C should be excluded from the nucleus until the appropriate time in the cell cycle. The requirement for nuclear entry for initiation of mitosis by cdc25C was supported by the reduced tendency of the cdc25C constructs containing the heterologous NES. Paradoxically, a cdc25C construct containing a heterologous NLS was not more active than the WT construct. Since the cytoplasmic localization of cyclin B1 in interphase cells was unaltered in the presence of NLS-tagged cdc25C, it may be argued that cdc25CNLS was unable to activate a cytoplasmic cdc2-cyclin B complex. Consistent with this possibility cdc25C-NLS was as active as WT cdc25C when coexpressed with an NLS-tagged variant of cyclin B1. The reduction in activity of the NLS and NES constructs may imply that there may be a dynamic shuttling of cdc25C into various cellular compartments during the cell cycle that was altered by the presence of the heterologous NES or NLS tag.
Cyclin B1 is maintained in the cytoplasm in interphase cells due to the presence of an N-terminal NES (13, 59, 63). Several recent reports have suggested that the entry of cyclin B1 into the nucleus is necessary but not sufficient for M-phase progression (13, 25, 59). The cytoplasmic localization signal in cdc25C shows no apparent homology to the NES of cyclin B1 (13, 51, 59), suggesting that the intracellular localization of cdc25C and cyclin B1 may be regulated by different mechanisms. This is further supported by the observation that treatment of U-2OS cells with leptomycin B led to the nuclear accumulation of cyclin B1 (13, 59, 63), but the cytoplasmic localization of cdc25C was not altered, and there was no indication of an increased tendency for cells to undergo PCC or advance past G2 phase and enter mitosis. These results are consistent with the hypothesis that cdc25C may not be excluded from the nucleus due to the presence of an NES but maybe retained in the cytoplasm by complex formation with 14-3-3 proteins in U-2OS cells. However, it has been reported that the rad24 gene product of S. pombe, a 14-3-3 homologue, promotes the nuclear export of cdc25 in a crm1-dependent manner (37). It is possible that the association of cdc25C with 14-3-3 proteins results in the nuclear export of cdc25C via a mechanism that is not sensitive to leptomycin B in U-2OS cells. The differential regulation of the intracellular localization of cdc25C and cyclin B1 by the checkpoint apparatus in human cells may provide an independent regulation of the entry into mitosis whereby, under certain conditions, cdc25C would be restricted to the cytoplasm and be unable to activate a cdc2-cyclin B complex that had already translocated to the nucleus. It has been shown in Xenopus that phosphorylation of consensus cdc2 sites in cyclin B1 are required to inhibit its nuclear export (35, 63). Given that cdc25C also undergoes hyperphosphorylation during mitosis, it is possible that the cytoplasmic retention of cdc25C is disrupted by phosphorylation of consensus cdc2 sites. Thus, activation of cdc25C in the cytoplasm could promote the nuclear transport of cyclin B1-cdc2 complexes as well as its own transport, resulting in further activation of cyclin B-cdc2 complexes in the nucleus.
We propose the following model for cdc25C regulation. cdc25C is retained in the cytoplasm by association with 14-3-3 proteins during interphase. This effect is dependent on an intact residue S216 that is most likely constitutively phosphorylated by any one of several kinases during interphase. In late G2, S216 is dephosphorylated, resulting in the loss of 14-3-3 binding and free diffusion or active transport of cdc25C into the nucleus. Independently, but perhaps at a similar time, the NES of cyclin B1 is inactivated, leading to accumulation of cyclin B1-cdc2 complexes in the nucleus (13, 35, 51, 59, 63). Subsequently, the activity of the cdc25C phosphatase is stimulated by hyperphosphorylation, resulting in the activation of nuclear cdc2-cyclin B complexes. This model does not exclude the possibility that the regulation of cdc25C by 14-3-3 proteins occurs through other mechanisms in addition to regulating the subcellular localization of cdc25C.
ACKNOWLEDGMENTS
We thank R. Scully for plasmid pSG5-L, Y. Sanchez, S. Elledge, P. Saha, and A. Datta for GST-cdc25A and GST-cdc25B, Michael Yaffe for GST 14-3-3ɛ, and W. R. Sellers for the HA-NLS and HA-NES constructs. We thank Peter Marks and William Connors at the Brigham and Womens Hospital confocal facility for help with generating the confocal images and Steven R. Grossman and Michael Rokas for their help with antibody purification. We thank D. M. Livingston, P. Adams, J. Ayté, H. Chao, S. Patankar, and H. Stubdal for critically reading the manuscript. We also thank the other members of the DeCaprio laboratory for their help and encouragement.
S.N.D. was supported by a fellowship from the Leukemia Society of America. J.A.D. is a Scholar of the Leukemia Society of America. This work was supported in part by Public Health Service grants CA-63113 and CA-50661.
REFERENCES
- 1.Al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis F M, Tsao T Y, Fowler S K, Rao P N. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Den Haese G J, Walworth N, Carr A M, Gould K L. The wee1 protein kinase regulates T14 phosphorylation of fission yeast cdc2. Mol Biol Cell. 1995;6:371–385. doi: 10.1091/mbc.6.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunphy W G, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 5.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field J, Nikawa J I, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;90:1051–1060. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 8.Ford J C, Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Carr A M. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994;265:533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- 9.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 10.Gabrielli B G, DeSouza C P C, Tonks I D, Clark J M, Hayward N K, Ellem K A O. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J Cell Sci. 1996;109:1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- 11.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 12.Girard F, Strausfeld U, Cavadore J-C, Russell P, Fernandez A, Lamb N J C. cdc25 is a nuclear protein expressed constitutively throughout the cell cycle in nontransformed mammalian cells. J Cell Biol. 1992;118:785–794. doi: 10.1083/jcb.118.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 15.Hartley R S, Rempel R E, Maller J L. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 16.Heald R, McLoughlin M, McKeon F. Human Wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 17.Hendzel M J, Wei Y, Mancini M A, Hooser A V, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann I, Clarke P R, Marcote M J, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25C by cdc2-cyclin B and its involvement in the self amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi M, Nagat A, Jinno S, Suto K, Okayama H. Wee1+ like gene in human cells. Nature. 1991;353:80–83. doi: 10.1038/353080a0. [DOI] [PubMed] [Google Scholar]
- 22.Izumi T, Maller J L. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell. 1993;4:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi T, Maller J L. Phosphorylation and activation of the Xenopus cdc25 phosphatase in the absence of cdc2 and cdk2 kinase activity. Mol Biol Cell. 1995;6:215–226. doi: 10.1091/mbc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi T, Walker D H, Maller J L. Periodic changes in phosphorylation of the Xenopus cdc25 phosphatase regulate its activity. Mol Biol Cell. 1992;3:927–939. doi: 10.1091/mbc.3.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin P, Hardy S, Morgan D O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krek W, DeCaprio J A. Cell synchronization. Methods Enzymol. 1995;254:114–124. doi: 10.1016/0076-6879(95)54009-1. [DOI] [PubMed] [Google Scholar]
- 27.Kumagai A, Dunphy W G. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell. 1991;64:903–914. doi: 10.1016/0092-8674(91)90315-p. [DOI] [PubMed] [Google Scholar]
- 28.Kumagai A, Dunphy W G. Purification and molecular cloning of Plx1, a cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 29.Kumagai A, Dunphy W G. Regulation of the cdc25 protein during the cell cycle in xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai A, Guo Z, Emami K H, Wang S X, Dunphy W G. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai A, Yakowec P S, Dunphy W G. 14-3-3 proteins act as negative regulators of the mitotic inducer cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M S, Enoch T, Piwnica-Worms H. mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J Biol Chem. 1994;269:30530–30537. [PubMed] [Google Scholar]
- 33.Lee M S, Ogg S, Xu M, Parker L L, Donoghue D J, Maller J L, Piwnica-Worms H. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol Biol Cell. 1992;3:73–84. doi: 10.1091/mbc.3.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee W-H, Shew J-Y, Hong F D, Sery T W, Donoso L A, Young L-J, Bookstein R, Lee E Y-H P. The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature. 1987;329:642–645. doi: 10.1038/329642a0. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Meyer A N, Donoghue D J. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Stanton J J, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 38.Matsuoka S, Huang M, Elledge S J. Linkage of ATM to cell cycle regulation by the chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 39.Millar J B A, Blevitt J, Gerace L, Sadhu K, Featherstone C, Russell P. p55CDC25 is a nuclear protein required for the initiation of mitosis in human cells. Proc Natl Acad Sci USA. 1991;88:10500–10504. doi: 10.1073/pnas.88.23.10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millar J B A, McGowan C H, Lenaers G, Jones R, Russell P. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 1991;10:4301–4309. doi: 10.1002/j.1460-2075.1991.tb05008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittnacht S, Weinberg R A. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 42.Mueller P A, Coleman T R, Kumagai A, Dunphy W G. Myt1: a membrane-associated inhibitory kinase that phosphorylates cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 43.Munro S, Pelham H R B. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functions of Drosophila hsp70. EMBO J. 1984;3:3087–3093. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 45.Nurse P. Checkpoint pathways come of age. Cell. 1998;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 46.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 47.Ogg S, Gabrielli B, Piwnica-Worms H. Purification of a serine kinase that associates with and phosphorylates human cdc25C on serine 216. J Biol Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 48.Parker L L, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human wee1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 49.Peng C-Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 50.Peng C Y, Graves P R, Ogg S, Thoma R S, Byrnes M J, Wu Z, Stephenson M T, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 51.Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to cdk regulation through cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 53.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ’mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully, R., and D. M. Livingston. 1997. Unpublished data.
- 55.Sellers, W. R. 1998. Unpublished data.
- 56.Strausfeld U, Fernandez A, Capony J-P, Girard F, Lautredou N, Derancourt J, Labbe J-C, Lamb N J C. Activation of p34cdc2 protein kinase by microinjection of human cdc25C into mammalian cells. Requirement for prior phosphorylation of cdc25C by p34cdc2 on sites phosphorylated at mitosis. J Biol Chem. 1994;269:5989–6000. [PubMed] [Google Scholar]
- 57.Strausfeld U, Labbe J C, Fesquet D, Cavadore J C, Picard A, Sadhu K, Russell P, Doree M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human cdc25 protein. Nature. 1991;351:242–245. doi: 10.1038/351242a0. [DOI] [PubMed] [Google Scholar]
- 58.Stubdal H, Zalvide J, DeCaprio J A. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 61.Walworth N C, Bernards R. Rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 62.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1998;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Bardes E S G, Moore J D, Brennan J, Powers M, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor, CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 66.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]