Abstract
Chronic kidney disease (CKD) refers to a range of various pathophysiological processes correlated with abnormal renal function and a progressive loss in GFR. Just as dysbiosis and altered pathology of the gut are accompanied with hypertension, which is a significant CKD risk factor. Gut dysbiosis in CKD patients is associated with an elevated levels of uremic toxins, which in turn increases the CKD progression.
According to research results, the gut-kidney axis has a role in the formation of kidney stones, also in IgAN. A number of researchers have categorized the gut microbiota as enterotypes, and others, skeptical of theory of enterotypes, have suggested biomarkers to describe taxa that related to lifestyle, nutrition, and disease status. Metabolome-microbiome studies have been used to investigate the interactions of host-gut microbiota in terms of the involvement of metabolites in these interactions and are yielded promising results. The correlation between gut microbiota and CKD requires further multi-omic researches. Also, with regard to systems biology, studies on the communication network of proteins and transporters such as SLC and ABC, can help us achieve a deeper understanding of the gut-liver-kidney axis communication and can thus provide promising new horizons in the treatment of CKD patients. Probiotic-based treatment is an approach to reduce uremic poisoning, which is accomplished by swallowing microbes those can catalyze URS in the gut. If further comprehensive studies are carried out, we will know about the probiotics impact in slowing the renal failure progression and reducing inflammatory markers.
Keywords: Chronic kidney disease, Renal failure, Glomerular filtration rate, Gut microbiota
1. Background
The kidney is a complicated organ which is responsible for maintenance of various aspects of homeostasis in the human body. The association of marked and closely connected molecular functions through various cell types outlines the factors which contributes in reduction or eliminating renal failure [1].
CKD is a stage that kidney function reaches less than 50% of its normal capacity. CKD is the 16th main agent of reduced life years on the world, defined as a persistent abnormal state in kidney (GFR<60 mL/min/1.73m2 or albuminuria ≥30 mg per 24 h) for more than 3 months [2]. CKD is depicted by progressive failure of physiological and biochemical function which is due to catabolites accumulation, confused fluid-electrolyte, hyperkalemia, hyperphosphatemia, disturbed acid-base balance, metabolic acidosis, hormonal disorders and hyperparathyroidism [3]. Due to kidney function failure, CKD patients accumulates of toxic molecules called uremic toxins [4]. Uremic toxins accumulation, may be one of the important factors of rapid progression of CKD particularly in ESRD patients that are undergoing dialysis [5].
ESRD refers to the time when kidneys are not able to function more than 10–15% of their normal ability. Unfortunately at this stage, kidney transplantation or dialysis, is necessary for the save the patient's life [6,7]. Renal transplantation (RT) is generally considered as the last resort for therapy of ESRD which is more effective and affordable than dialysis [8].
CKD is associated with a distinct elevation of mortality risk and exacerbation of the disease, which often requires hospitalization. Apart from cardiovascular causative factors, nearly 50% of deaths in CKD patients could be related to non-cardiovascular factors, including infections, which are now increasingly recognized as one of these factors, especially when a number of these infectious events are preventable [9].
The main role of the kidneys is to filter and secrete the end products of metabolism and excess electrolytes. Persistent renal failure, CKD and its inability to survive is called ESRD [10]. ESRD is associated with a large symptom burden [7]. CKD consists of five stages. The stage of kidney disease is based on the presence of kidney damage and the GFR reduction. GFR is an estimation of renal function, as CKD progresses, the GFR will decrease [11]. ESRD is one of the most frequent critical diseases. The number of ESRD patients that need dialysis or renal transplantation is increasing every year in developed and developing countries and causes a heavy socioeconomic impact on these countries [12].
According to the simulation model, the ESRD burden on the US population will increase through 2030 because of demographic, clinical and lifestyle changes in the community and also due to advances in renal replacement therapy (RRT) [13].
Results of a previous study in Kazakhstan from 2014 to 2018 showed an elevation in the incidence and prevalence of ESRD patients undergoing dialysis. However, the mortality rate in these patients reduced during this period, which indicates better access to health facilities [14].
In many countries, the most common RRTs is hemodialysis followed by kidney transplantation and peritoneal dialysis, respectively [15].
Recent data also indicate that the global prevalence of CKD is 8–16% [2,16] and is constantly increasing, which imposes a heavy economic burden in the field of health care because although the disease progresses from CKD to ESRD in only 1% of cases, ESRD is associated with elevated morbidity and mortality, and usual comorbidities include CVD, diabetes, mineral disorders, bone diseases and anemia. These comorbidities, along with infections, are major causes of hospital admissions for patients with ESRD.
Considering the subject that ESRD patients need dialysis or renal transplants, it causes ESRD to be one of the most costly chronic diseases. In fact, ESRD therapies put paid to up to 5% of the total annual health care budget of countries. To prevent the CKD progression and to decrease ESRD effect on patients and health care systems, early detection of individuals with renal dysfunction is essential [16].
Also it should be noted that there is a significantly higher prevalence of CKD earlier stages, with detrimental outcomes, consist of kidney function failure, CVD and premature death, thus a global effort would be required to improve outcomes at the earlier stages of CKD [17].
According to importance of CKD and its complications, this review intends to overview of the impact of gut microbiota dysbiosis in CKD, which can be considered as a non-traditional or unusual risk factor and tries to evaluate for multidimensional aspects related to this critical subject.
2. Materials and methods
For this review, by using keywords include CKD, ESRD, gut dysbiosis, enterotypes, IgAN, nephrolithiasis, probiotics, systems biology, the articles from databases such as PubMed, Scopus, Springer, and websites such as Nature, were searched and then based on the aim of review analyzed and extracted.
3. Physiology of the gastrointestinal tract
The parts of intestine as follows small intestine or small bowel (consists of duodenum, jejunum, and ileum) and the large intestine, these parts from compositional and structural aspects are fundamentally different. Goblet cells, for example, are abundant in the proximal colon, while Peyer plaques are situated mainly in the small intestine [18,19]. Besides, the mucin layers in the small intestine are thinner than the colon, and microvilli are more abundant.
The aforementioned diversity is correlated with multi-dimensional functions that are essential in microbiota interactions. Gut is really a mucosal surface that is continuously lay open to foreign antigens and micro-organisms, and also a vast groups of immunologically active cells and structures protect it. A population of innate lymphocytes is located within the gut epithelium and its related lamina propria, which can be stimulated to cytokines production [20].
Epithelial cells act as separator of the host and the environment, and have apical surface facilities such as microvilli, cilia, intercellular junctions, mucus secretion for their duty against microbiota [21].
About 70% of immune cells, are situated in the gastrointestinal tract: these immune cells preserve a balance between activity and immune tolerance against gut microbiota [20]. Also in the body, gut is the second nervous organ, which facilitates the link of gut with the brain [22].
The intricate vascular bed enables the gut to effectively absorb nutrients and water and maintain an oxygenation gradient along its [23].
Gut is one of the first important organs in the face of environmental factors such as food, pathogens and toxins and their interactions with immune system, endocrine and neurological factors and has an essential effect on the physiological responses of the host. The epithelial barrier, physically separates intestinal mucus from submucosa, which is continuously affected and changed by factors consists of nutrients, toxins, drugs and pathogens. Tight junction proteins protect or plug the intestinal epithelial layer and prevent the movement of gut pathogens through the epithelial barrier. The immune cells which located in the lymph glands assess the gut environment conditions and preserve the gut homeostasis [24].
Gut microbiota grow in an environment that is stable and nutrient–rich, but also these organisms serve useful utilities to the host consist of energy conserve of complex and indigestible carbohydrates, micronutrient vitamin and production, competitive exclusion of pathogens, significantly having the role in immune improvement [21].
4. Gut as an inflammation source in CKD
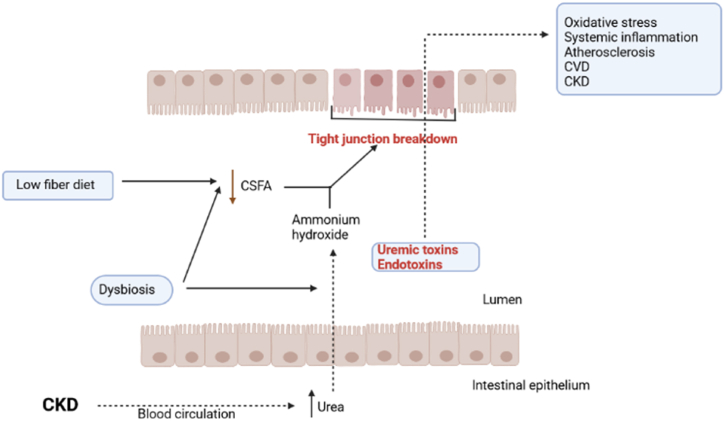
Chronic inflammation is present all over the gastrointestinal tract (from the beginning or esophagus to end or colon) of dialysis patients [25,26]. Animal studies have shown the destruction of narrow epithelial junctions of the colon by bacterial urease activity and localized inflammation through cytokine production. While, it is important that mammals cannot breakdown urea [27]. Dietary restriction in CKD patients (the low phosphorus and low potassium) results in insufficient intake of vegetable fiber and symbiotic bacteria, as well as urea influx from the circulation, alters the gut microbiota and promotes the growth of a microbial population that produces uremic toxins such as p- Cresyl and indoxyl-sulfates. Urea is broken down into ammonia by the urease of the bacteria those produces this enzyme, urea is also broken down into ammonium hydroxide (NH4OH), which in turn degrades tight junction proteins [25]. For example, a study in the intestinal tissue of rats with CKD, showed remarkable reduction of the claudin, occludin, and zona-occludens (the key molecules of epithelial tight junction) [26]. In the meantime, the altered microbiome changes the symbiotic bacteria harmony and competes with colonocytes for SCFA as an energy source. This can alter the integrity of the colonocytes and damage the mucosal barrier by influxing inflammatory leukocytes and producing cytokines (Fig. 1). Inflammation of the gut intensifies the destruction of the epithelial barrier by stimulating the endocytosis of tight junction proteins [25]. Decreased Nrf 2 expression (which is a transcription factor that adjusts the cellular defense against oxidative and toxic damages via the expression of the genes have the role in oxidative stress response and the detoxification of drugs [28]), can contribute to further destruction of the tight junction proteins. Eventually, translocation of bacterial derivatives, endotoxin and uremic toxins occures from the intestinal lumen into the blood circulation and these events leads to systemic inflammation. Progression of kidney disease results in more increase of serum urea concentrations, more increasing changes in gut microbiome and making a vicious circuit [25] (Fig. 2).
Fig. 1.
Blood urea levels raise and urea influx to gut lumen in CKD patients. Dysbiosis of gut microbiota increases some of urease containing bacteria and urease activity increases ammonium hydroxide. On the otherhand the low fiber diet of these patients decreases SCFAs production by gut bacteria which are the colonocytes nutrients. The SCFAs decrease together with ammonium hydroxide increase, damage to intestinal epithelium barrier (tight junctions) and then uremic toxins and bacterial derivatives such as endotoxins penerate from damaged epithelium and entered to blood circulation and cause to inflammatory and oxidative processes such as CVD and CKD. SCFA: Short chain fatty acid; CVD: Cardiovascular disease; CKD: Chronic kidney disease.
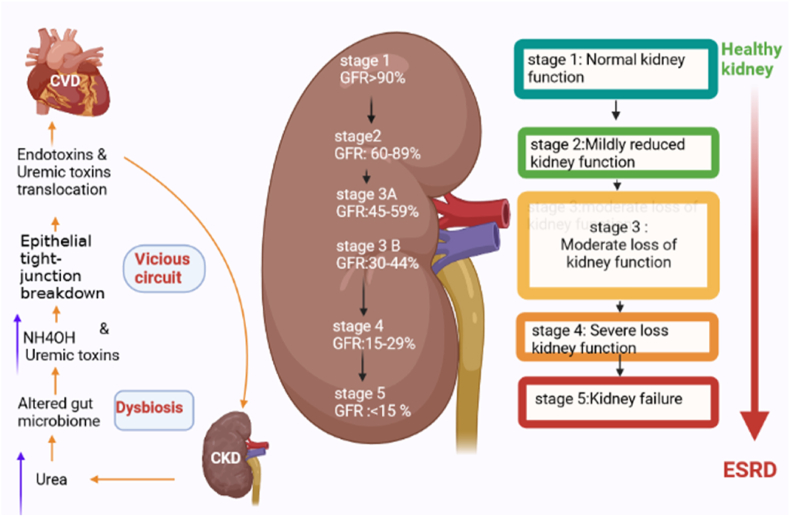
Fig. 2.
The relationship of gut dysbiosis and chronic kidney disease stages. Left: Vicious circuit that theoretically impact in health status of kidney. Kidney failure cause an increase in blood urea levels, then urea influxes to gut lumen and alters gut microbiota (dysbiosis), this condition increases uremic toxins and NH4OH and these toxins damage to gut epithelial tight junction which increases permeability of gut barrier and then uremic toxins and bacterial derivatives such as endotoxins translocate to blood circulation and damage to organs such as heart and kidney. Right: The stages of chronic kidney disease according to GFR. CKD: Chronic kidney disease; CVD: Cardiovascular disease; GFR: Glomerular filtration rate; ESRD: End stage renal disease.
In spite of severity of the consequences of uremia and despite the vast surveys achieved about this subject, the molecular and cellular mechanisms involved in syndrome development, mainly remain unclear, the major reason is the enormous and constantly expanded list of uremic toxins and solutes. More than 100 various solutes raise during uremia condition, but, the precise source for most of them is still controversial [29].
5. Gut microbiota
Intestinal microbiota is related to human health. Microbial colonization occurs with the maturation of the immune system and is associated in the physiology and regulation of the intestine. The gut harbors trillions of microbes. Primary colonization of microorganisms generally occurs transvertically at birth, then constantly evolves into a stable and adult-like state during the first 3–5 years of life [30]. But the evidence that the fetal microbiota is affected by maternal microbiota, has challenged this theory [31], which is discussed below.
Ecological and functional characteristics of gut microbiome in healthy individuals, including infants, children and adults need further investigation because knowing about functions of gut microbial populations plays a significant role in recognizing their interaction with the host [30]. The microbial community nature and its alterations in response to conditions such as dietary fibers and antibiotics consumption is dependent on the host genetic status [32].
According to the predominant bacterial phylotype, two major enterotypes could be considered for adults gut microbiota. Both of these enterotypes are highly accompanied with long-term nutrition [33]. The enterotype 1 is predominantly consists of Bacteroides, which mainly metabolize proteins, while enterotype 2 contains mainly Prevotella which is a saccharolytic bacteria. Not surprisingly, microbial metabolic profiles (e.g., SCFAs and bile acids) are mainly related to enterotypes [34]. According to D. Aguirre study, the human gut pan-microbiome provides a predominant compositional phylogenetic core, consist of separated units of varying depth existent in all studied persons, and this study suggested a novel conceptual framework which has notable possibility for progression the comprehension of microbial ecosystems, especially host-related microbiome [35].
The dynamic development of gut microbiota during early life initiates with the colonization of obligate anaerobes (mostly Proteobacteria) and follows by the anaerobic bacteria colonization (usually Lactobacillus and Bifidobacterium). Eventually, bacterial diversity (generally different species in the Bacteroides phylum) matches with energy supply [30].
Immune system of infants is immature that cannot competently defend against pathogens, which may results in severe and life threatening infections. Breastfeeding together with maternal interactions provide status, which are required for the optimum development of the immune system. The maternal milk lactose, greatly enhances the Lactobacillus growth and forms the gut microbiota in childhood. The following solid foods feeding, significantly causes the gut microbiota recomposition, which in turn suggests that environmental factors play a vital role in determining its composition [24].
Vaginal delivery and breastfeeding are very significant for the growth and health of the infant and interventions such as cesarean section, use of antibiotics, formula feeding alter the infant's microbiome and can be considered as major factors that shape a new microbiome landscape in the history of human life [31].
Creating a complete intestinal-blood barrier, identified by entire physiological and immunological protection, protects the intestinal and absorption functions and limits the pathogens and toxic metabolites entering into the bloodstream. This process mostly depends on a balanced intestinal microbiota [24]. In adults, gut microbial metabolic pathways are relatively constant, even though, as indicated earlier, environmental factors, mainly diet, deeply affect the intestinal microbiota. Age-associated alterations in the intestinal microbiota, which have also been recognized in the older individuals, are identified by a decline in diversity, a reduction in two saccharolytic intestinal bacteria, an enhancement of proteolytic bacteria, an increase in certain Proteobacteria, and a decrease in Bifidobacterium count. The levels of plasma markers that are involved in increasing intestinal permeability increase in healthy older individuals, indicating a functional intestinal disruption with age. Besides, probiotic supplements have been offered to have healthy lifespan-promoting outcomes, including: repressing low-grade chronic inflammation and prolonging the life of mice, which in turn indicates the significance of intestinal microbiota in maintaining overall health [24]. It is necessary to mention, according to a comprehensive study, there is no important difference between healthy young and aged people in gut permeability, and the functional competence of the gut barrier is maintained in aging period [36].
The gut virome exhibits more differences between individuals and compared with gut bacterial microbiota, it is less affected by changes of environment. Metagenomic studies indicate that each individual's viral population is unique [24,37]. The human gut virome contains a assembly of highly variable sequences that serve as a reservoir for viral evolution to adapt to the new environment [24].
In type 1 diabetic patients, alterations in the gut virome appear to outweigh autoimmunity, suggesting that the virome may play a role in causing the disease. In this regard, researchers have identified contigs of eukaryotic viruses and bacteriophage that are accompanied with the existence or lack of autoimmunity. Viruses have long been considered as possible causes of the onset of autoimmune diseases [38].
The gut fungal populations, do not appear to cause disease directly however may develop dysbiosis, which can be associated with systemic inflammation. Fungi have conventionally been examined with culture-based techniques, and only newly, developments in sequencing methods have provided opportunities to reveal the complexities of fungal populations colonized at mammalian mucosal surfaces. The lower gastrointestinal tract compared with upper tract, has richer and more diverse fungal population [39]. It is necessary to mention, Gialin Hu et al. observed mycobiome dysbiosis in CKD patients, and they reported that gut mycobiome dysbiosis is associated with immunological disorders. They notified that mycobiome-based therapeutic strategies might be effective [40].
The composition and metabolic activity of the gut microbiome, has a deep effect on the regulation of mucosal immunity [20].
The colon has the greatest bacterial population in the body (more than 1011 organisms gram/wet weight), and the most of these bacteria are anaerobic, of which approximately 35% belongs to Bacteroides species, which is the most predominant anaerobic bacterium in the gut. As a commensal bacterium, Bacteroides plays a good role, but plays a bad role in human disease [41]. Bacteroides fragilis is an instance to exhibit the phenotypic heterogeneity within a commensal species [42].
As mentioned, Bacteroides has a bad role in human disease. Bacteroides has important virulence factors. Various factors are responsible for the pathogenesis and persistence of Bacteroides fragilis at sites of infection.
The virulence factors include capsular polysaccharides, withstand against antimicrobial agents, existence during the extended oxidative stress conditions, quorum sensing, iron acquisition and secretion systems [43].
Bacteroides species are non-spore-forming anaerobic, resistant to bile, gram-negative rods.
Bacteroides can be transmitted from mother to infant during vaginal birth, so, they become part of the human microbial flora in the early stages of life [41,42]. However, study on vertical transmission essentially operates species-level taxonomic assignment, whilst resolution at strain-level is needed to decisively determination of transmission modes [42].
Hannah M refers to the term commensal as a very inappropriate term to describe the relationship between Bacteroides and the human host because the term commensal is used when one side benefits from the relationship and the other side does not. But the author suggests the mutualism term, which is a more appropriate description. Carbohydrates, by fermenting Bacteroides and other intestinal bacteria, create a store of short chain acids that are absorbed via the colon and then used as an energy source (and provide an important part of the host's diurnal energy needs). The gut flora thus provides a rich source of food for the host. Researches revealed that germ-free rats that lack intestinal flora compared with normal rats require 30% more calories to preserve body mass.
Other gut organisms, which lack the series of sugar-metabolizing enzymes (while Bacteroides have these enzymes), can take advantage from the presence of Bacteroides to use the sugars (generated by glycosyl hydrolase). Therefore, these organisms will be able to use sugars in other ways. Bifidobacterium langum, for example, has better systems for bringing in simple sugars than Bacteroides thetaiotaomicron, whereas Bacteroides thetaiotaomicron can degrade a wide variety of glycoside bonds and thus prepares food and then uses Bifidobacterium langum [41].
The Bifidobacterium family are gram-positive, anaerobic, immobile and non-spore-forming bacteria those belong to the Actinobacteria class. The genus Bifidobacterium has health –promoting benefits. These organisms dominates the gut of healthy breast-fed infants while in adulthood these organisms levels are lower but relatively constant [44,45].
Generally, Bacteroidetes and Firmicutes are the two important phyla in the adult intestine. The species of Clostridium genus are assorted within the Firmicutes phyla [46].
Clostridium bacteria are rod-shaped, gram-positive anaerobes that produce spores. These bacteria are found in soil, water, animals and humans intestinal tract.
Initially, these bacteria were classified according to the mentioned physiological and morphological characteristics of the genus Clostridium, but further studies have demonstrated that the heterogeneity between Clostridium species became more and more obvious.
Years ago, researchers proposed a taxonomic classification criterion on the power of phylogenetic analysis of the 16srRNA gene sequence. The genus Clostridium is classified into 19 clusters and this novel criterion recommended a number of asporulate bacteria such as Ruminococcus torques and Roseburia cecicola. Clostridium cluster I include of most prior members of Clostridium, which represented by Clostridium butyricum.
In humans and animals, Clostridium species are one of the most abundant bacterial clusters and are mostly consist of Clostridium cluster IV and XIVa. Clostridium IV is called the Clostridium leptum group, which has four members include Clostridium leptum, Clostridium sporosphaeroides, Clostridium cellulosi, and Faecalibacterium prausnitzii. Clostridium cluster XIVa, which also known as Clostridium coccoides group, include 21 species. In addition to the Clostridium spp; the asporulate bacteria include Acetitomaculum ruminis, Roseburia cecicola, Ruminococcus torques, Coprococcus eutactus, Eubacterium cellulosolvens, Streptococcus hansenii and Peptostreptococcus productus are also placed in Clostridium cluster XIVa. Clostridium species are able to metabolize large amounts of nutrients that the host is unable to digest and are able to produce large amounts of SCFAs, which play a significant role in intestinal homeostasis [47], these compounds (acetate, butyrate and propionate) are the by-product of complex and non-absorbable carbohydrates fermentation [47]. Clostridium species are generally predominant in the colon, especially in the ascending mucosal folds and have commensal life with Bacteroidaceae, Lactobacillaceae and Enterococcaceae colonized in the colon [48].
Clostridioides difficile (already was called Clostridium difficile)is an important causative agent disease and economic burden throughout the world [49]. The new name shows the taxonomic differences between this species and other members of the Clostridium genus [50]. This organism is a saprophyte anaerobe bacterium with a common environmental spreading in food, soil, feces and intestine of animals and humans. But this opportunistic bacterium, under certain conditions becomes one of the most common pathogens that induces several enteric disease [51].
In obese people, gut dysbiosis seems to be accompanied by an increase in Firmicutes phylum, Clostridium genus and species of Clostridium coccoides, Clostridium histolyticum, lactobacillus reuteri, Eubacterium rectale, Akkermansia muciniphila and Staphylococcus aureus [52].
The genus Lactobacillus survives in the gastrointestinal tract of animals and humans and includes non-spore-forming and bacilliform gram-positive bacteria, which includes more than 90 species and subspecies. Lactobacillus species belong to the Firmicutes phylum, which are considered as lactic acid bacteria. Lactobacillus species are one of the most common probiotic strains used to prevent chronic inflammatory disease and increase health status [53](Table 1).
Table 1.
The prominent bacterial genera of human gut microbiota [54].
| Bacteroides |
|---|
| Bifidobacterium |
| Clostridum |
| Enterobacteria |
| Enterococcus |
| Escherichia |
| Eubacterium |
| Klebsiella |
| Lactobacillus |
| Peptococcus |
| Peptostreptococcus |
| Staphylococcus |
| Streptococcus |
| Methanobrevibacter (Archaea) |
Understanding the compatibilities of symbiotic microbiota versus pathogens that permit them to remain or be eliminated in the intestinal mucosa, is a significant step in understanding the factors that play a role in preventing or enhancing the mobilization of immune responses to microbial epitopes [55].
6. Association of intestinal microbiota with chronic kidney disease
Gut microbiota appear to be an important factor in mediating the prevalence of chronic kidney disease. Just as dysbiosis and altered pathology of the gut are correlated with hypertension which is a key factor contributing to the CKD progression, it is not surprising that changes in the intestinal microbiota composition is correlated with CKD. Compared with healthy individuals, there is a decline in cultivable anaerobic bacteria in the feces of stage 3–4 CKD patients, but in spite of this subject, an increase in cultivable aerobic bacteria has reported in the feces of CKD patients who had not yet undergone dialysis compared with controls [24].
Some researchers found strong reasons around the gut-metabolite-kidney axis effect on CKD severity, as well as the beneficial effects of certain gut microorganisms as a potential marker of disease differentiation. They stated that these specific microbes are potential biomarkers for the initial diagnosis and prognosis of this disease, which is a global problem, and could be used as a candidate for therapeutic purposes for gut-metabolite-kidney interventions.
However, they stated that further and vast researchers are needed to measure the effects of these microbes on reducing nephrotoxins and improving renal outcomes in CKD patients [56].
CKD is associated with congestion and edema of intestinal wall, slow colonic transmission, metabolic acidosis, common consumption of antibiotics, reduced intake of iron and dietary fiber, which in turn affects intestinal tight junction and increases intestinal permeability and causes the transfer of bacterial metabolic products through the intestinal barrier. This is followed by an immune response, the occurrence of which suggests that systemic inflammation contributes to degenerative renal disease. In addition, increased gastrointestinal urea secretion causes dysbiosis of the gut microbiota increases the formation of toxic ammonia. Moreover, urea supplementation to drinking water is associated with alterations in the gut bacterial microbiota [57]. Also, in recent years, the SCFAs production of gut microbiota and modulation of inflammatory response and consequent amelioration of renal damage, has been on the focus of interest [58].
7. The evidence of gut microbiota alterations in CKD patients
In a meta-analysis, Stanford J et al. reported that 20 microbial taxa in the two groups of patients with kidney disease were more different from those in healthy controls. For example, in people with kidney disease, the abundance of Proteobacteria, Enterobacteriaceae, Streptococcaceae and Streptococcus increased relatively, and the abundance of Firmicutes, Prevotellaceae, Prevotella and Prevotella 9 decreased. The researchers found that the gut microbiota profile of CKD people differed markedly from controls [59].
Vaziri et al. in their studies on DNA extracted from 24 fecal samples of ESRD patients and 12 healthy individuals found that the frequency of these samples significantly differed in ESRD patients compared to healthy controls (approximately 190 operational taxonomic units (OTUs)). These researchers believed that uremia profoundly alters the gut microbiome composition. In the mentioned study, OTUs belonging to Brachybacterium, Catenibacterium, Enterobacteriaceae, Moraxellaceae, Halomonadaceae, Pseudomonadaceae, Nesterenkonia, Polyangiaceae and Thiothrix families were significantly increased in ESRD patients. Prevotellaceae and Lactobacillaceae (both of which are considered normal colon microbiota) were also significantly decreased [60].
The aerobic bacteria quantity, such as Enterococci and Enterobacteriaceae, in ESRD patients is higher than healthy controls. Gut microbiota dysbiosis in CKD patients is associated with an increase in the uremic toxins levels, which in turn increases the CKD progression [57].
Gut microbiota dysbiosis and CKD association is really a bidirectional communication. CKD itself can results in changes of the normal gut microbiota. The antibiotics and the special nutritious diet consumption by CKD patients may enhance the gut dysbiosis possibility [61].
Gut microbiota imbalance occurred in CKD patients both quantitatively and qualitatively and is often associated with an increased in frequency of Lachnospiracea, Enterobacteriaceae and certain Ruminococcaceae and a decrease in some of Bacteroidaceae, Prevotellaceae, and especially Lactobacillus and Bifidobacterium species [57]. The most often alterations of gut microbiome in CKD patients which has been issued are the decreased quantities of Lactobacillaceae and Bifidobacteriaceae and increased quantities of Enterobacteriaceae [62]. According to a study in Chinese population, in ESRD patients, the absolute count of feacal microbiota was declined and Prevotella was predominated in healthy controls, while the number of Bacteroides was increased in ESRD patients. The butyrate-producing bacteria consist of Clostridium, Faecalibacterium, Roseburia, Prevotella and Coprococcus were decreased in ESRD patients. Generaly, Universal bacteria, Escherichia coli, Bacteroides fragilis group, Bifidobacterium, Clostridium coccoides group (Clostridium cluster XIVa), Faecalibacterium prausnitzii, Enterococcus spp., Roseburia and Prevotella were declined in ESRD [47]. In a striking discovery, by FengXia Li et al. reported that a significant decline in abundance of Akkermansia in CKD patients compared with healthy group [63]. A study on Taiwanese CKD patients showed that dialysis patients had important differences in composition of intestinal microbiota compared with controls. The researchers detected many bacterial taxa related to specific demographics. The study represented that Bacteroides is the most abundant detected genus in CKD patients [64].
Jialin Hu et al. showed that gut mycobiome dysbiosis observes in CKD patients also this dysbiosis is accompanied by the immunological disorder [40]. YE Guirong et al. study in recipients of renal transplant, showed that alterations in the gut microbiota function and composition are markedly associated with the clinical status of these patients [8].
8. The evidence of no changes of gut bacteria in CKD patients
However, based on a study about Clostridium species diversity, despite of crucial role of gut microbiota, the researchers reported no association between studied bacteria and kidney failure. This study, revealed that abundance of Clostrdium spp. Have not differed between CKD/ESRD patients and normal controls [46]. Also according to another study about Lactobacilli species diversity in CKD patients, analyzing the abundance mean of the strains, showed no important association between disease and control groups. Nevertheless, Lactobacillus spp. Have more diversity in patients group compare to control group [53]. Also, based on another study, Bifidobacteriaceae family, are not altered in CKD/ESRD patients. This study showed that Bifidobacterium adolescentis and Bifidobacterium longum subsp. longum, were the most abundant species in both disease and control groups and Bifidobacterium animalis subsp. Lactis has the least abundance in disease group [65].
9. Intestinal microbiota as an origin of uremic toxins
Uremic retention solutes are the molecules which in the conditions that renal excretory function is damaged, accumulate in the blood. Some of these solutes at high concentrations are toxic and are commonly known “uremic toxins” [29]. It has been suggested that uremic toxins promote by damaging renal tubular cells, and overload of these compounds accelerates the glomerular sclerosis, tubulointerstitial damage and eventually renal failure [66].
The European Uremic Toxin Work Group (EUTox) reports that according to physiochemical characteristics and behavior during dialysis, uremic toxins could be divided to three groups.
-
Ⅰ)
Low molecular weight and water-soluble uremic toxins (with molecular mass less than 500 Da), such as urea, creatinine, TMAO.
-
Ⅱ)
Middle molecule uremic toxins (peptides with molecular mass more than 500 Da), such as beta 2-microglobulin.
-
Ⅲ)
Protein-bound uremic toxins (PBUTs), which are poorly dialyzed and arising from aromatic amino acids such as phenols and indoles [67].
The gut microbiota is a potential origin of Kidney-induced uremic toxins consist of pCS, IS, indole-3-acetic acid, trimethylamine N-oxide and phenylacetylglutamine which are associated with CVD, mortality in CKD patients, and other end organ-related toxicities [57].
Various organic anion transporters, mediates uremic toxins through cells and organs [68]. It is important that membrane transporters removement of uremic toxins through renal tubular secretion are essential and has been proposed that the expression of these transporters probably correlated to the toxicity of these compounds those retain in the body during CKD progression [67].
9.1. p-Cresyl sulfate
P-Cresyl sulfate (pCS) is a prototype protein-binding uremic toxin that has been shown to have many biochemical, toxic, and biological effects [69]. Really, p-Cresol, defined as precursor of p-Cresyl sulfate as the main metabolite, and p-Cresyl glucuronide, which has markedly lower concentration in humans [70]. Elevated levels of pCS is correlated with deteriorating outcomes in CKD patients [69], and pCS is considered toxic, due to its biochemical impact and high circulated levels [71]. When intestinal bacteria metabolize aromatic amino acids such as phenylalanine and tyrosine, they lead to the final phenolic compounds, such as pCS that is derived from the gut.
According to in vitro studies, it seems that the gut bacteria that produce phenolic compounds, are mostly relevant to the families Bacteroidaceae, Clostridiaceae, Bifidobacteriaceae, Lactobacillaceae, Enterobacteriaceae, Enterococcacea, Staphylococcaceae, Eubacteriaceae, Lachnospiraceae, Fusobacteriaceae, Porphyromonadaceae, Ruminococcaceae and Veillonellaceae [69]. The gut anaerobic bacteria produce p-Cresol/p-Cresyl sulfate, p-Cresol is conjugated either by gut bacteria or liver [71,72]. Because pCS is difficult to eliminate by dialysis, intestinal microbiota can be used as a target to reduce the pCS-related toxicity even in the early stages of CKD, with the aim of reducing disease progression and reducing cardiovascular burden in the future [69].
9.2. Indoxyl sulfate
Indoxyl sulfate (IS), is a typical uremic toxin, has notable significance in nephrotoxicity and CKD progression [73]. Indoxyl sulfate is an invasive uremic toxin that is considerably retained in CKD patients blood and its levels in these patients can elevate even 50- fold compared to healthy people [74].
Indoxyl sulfate and endol-3-acetic acid are derived from the tryptophan metabolism. Tryptophan is metabolized into indole by bacterial tryptophanase (such as E. coli), and the indole after absorption from intestine, is sulfated in the liver and changed into indoxyl sulfate. IS normally excreted in the urine. Considering its high affinity to bind albumin, indoxyl sulfate is not effectively excreted by conventional hemodialysis methods [57]. The adverse effects of pCS on renal tubular cells are similar to those of IS [75].
9.3. TMAO
Trimethylamine N-oxide is a circulating metabolite that is involved in atherosclerosis and CVD onset [76]. Choline and carnitine are compounds which present in the dietary food and metabolized by intestinal bacteria to produce Trimethylamine (TMA), which is the precursor of TMAO, then TMA, oxidized and altered to TMAO by the liver flavin-containing monooxygenases (FMOs) [57,77]. Actually there are two main sources for circulating TMAO levels, first, from diet, second from the gut microbiota [76]. Unlike indoxyl sulfate and pCS, which are difficult to be excreted through dialysis since they bind proteins, TMAO is easily removed by dialysis [57].
9.4. PAGIn
Phenylacetylglutamine (PAGIn) is another product of gut microbiota made from phenylalanine fermentation. Phenylalanine is metabolized by microbes to phenylacetic acid, which is conjugated to glutamine and converted to methylglutamine. Like TMAO, phenylacetylglutamine can be dialyzed [57,78]. PAGIn may increases the risk of thrombosis and CVD by stimulating of platelets [78].
9.5. Ammonia and urea
Ammonia is the end-product of mammalian protein catabolism and in higher concentrations is toxic to cells and thus via ornithine-urea cycle is converted to urea. The urease of gut bacteria cleaves and converts urea to ammonia and carbon dioxide. Also, ammonia could be produced by fermentation of amino acids such as glutamine, threonine, serine and glycine into the colon. Ammonia and ammonium hydroxide cause injury to intestinal epithelial barrier [27].
10. Kynurenine pathway in CKD
The KP is the primary metabolic pathway for tryptophan catabolism in the liver, and is responsible for degradation about 95% of dietary tryptophan [79]. Many studies have demonstrated that tryptophan metabolism through the kynurenine pathway is potently elevated during CKD. Accumulation of KP metabolites during the CKD, may bring about oxidative cell injury, which stimulate inflammatory processes [80].
The KP metabolites, have immunomodulatory activity, also other biologic functions, including antioxidative, hypotensive and IhR agonist effects. Considering the intricacies of kidney disease, and multi-organ association in CKD-related problems the description of KP metabolites impact in biological functions, may be related to conditions and therefore researchers must be careful in interpretation of this subject [79].
11. The role of gut-kidney-heart axis as an important and multidirectional link in CKD
Due to the sharp increase of scientists' attentions in the gut microbiota field, our insight about pathophysiological interactions between the gastrointestinal tract and various organs has increased dramatically. Evidence of a link between the gut microbiota, CKD and CVD creates a new mechanistic view of the gut-kidney-heart axis and emphasizes the significance of gut as a possible cause of CKD-associated problems [81].
According to emerging evidence, gut epithelial barrier is changed both structurally and functionally in CKD patients, which leading to increasing intestinal epithelium permeability [82].
Among toxic gut–derived molecules detectable in systemic blood circulation, bacterial endotoxin and gut metabolites (such as pCS and TMAO), have been studied due to their atherogenic and immunostimulatory effects [81].
Endotoxin is a phospholipid that forms the outer membrane of most gram-negative bacteria, and it constantly produced in the intestine and is carried away to the capillaries via a TLR4-dependent mechanism [83]. The bacterial endotoxin lipopolysaccharide (LPS) is the typical inducer to create numerous inflammatory disease models, particularly multiple organ damage [84], among which, cardiovascular disease is the main cause of CKD mortality [81].
Plasma endotoxin concentration of healthy people is low (1–200 pg/mL) and is caught by the liver and mononuclear phagocytes and finally removed. Endotoxin stimulates a group of host responses by binding to CD14. Endotoxin stimulates immune cells, especially endothelial cells and macrophages, to activate them and secrete various effector molecules that trigger the inflammatory response. New evidence suggests that subclinical levels of endotoxins are likely a probable inducer of inflammation in CKD patients. The role of bacteria in atherosclerosis development has been recognized for more than two decades. Newly, however, researchers zoom in endotoxin as a bacterial product and its role in causing atherosclerosis. Endotoxin is an important factor in the inception and progression of atherosclerosis, which plays a role in mediating endothelial cell damage, increasing monocyte recruitment, conversion of macrophages to foamy cells, and procoagulant activity. Besides, vascular smooth muscle cells respond strongly even to low levels of endotoxin [83].
The novel studies, have also proposed similar biologic properties (immunostimulatory and atherogenic properties) of bacterial DNA fragments in blood flow of CKD patient's, even in lack of obvious infection [81].
12. The role of gut microbiota in renal stone forming and nephrolithiasis
Nephrolithiasis is a chronic disease that is highly prevalent in Caucasian and Arab ethnicity [85].
In developing countries, nephrolithiasis can be fatal due to poor medical facilities in rural areas or lack of financial capacity for treatment.
The chemical composition of most kidney stones are calcium oxalate and calcium phosphate and are mainly associated with their metabolism in body. Both of these compounds enter our bodies through the variety of nutrients [86].
Hypercalciuria and hyperoxaluria are the most common metabolic conditions observed in calcium nephrolithiasis [85].
Oxalate is usually entered directly from food sources (exogenous source) or indirectly to be created as final metabolite in the liver (endogenous source). Dietary precursors such as glyoxylate, glycine, hydroxyproline, and ascorbic acid are metabolized to oxalate to produce NADH in the liver. Oxalate is a toxic compound that is largely found in various plants as well as in humans who consume large amounts of oxalate-containing plant foods (vegetables, fruits, grains, nuts) [86]. The role of gut microbiota in the developing of renal stone disease has not been well determined [87].
Lack of Oxalobacter formigenes can cause enteric hyperoxaluria, however the colonization rate of Oxalobacter formigenes differes in healthy people and people with urinary stones. In addition, the presence of Faecalibacterium, Enterobacter, and Dorea in kidney stone formers can reduce the probable formation of calcium oxalate stones. Faecalibacterium can decrease oxidative stress and inflammation accompanied with stone formation [88]. Culturomic analyzes revealed that the number of Oxalobacter formigenes was notably lower in the fecal sample of kidney stone formers than in control subjects. The bio-degradation of oxalate in the gut decreases its blood circulation rate, so this is considered a negative risk factor for kidney disorders. Oxalobacter formigenes, as the normal gut flora, metabolizes oxalate in the urinary tract, through two enzymes: oxalyl-CoA decarboxylase and formyl–CoA transferase [86]. The presence of oxalate-degrading bacteria in gut can restrict oxalate absorption and decrease oxalate excretion [85]. The human genome lacks genes that encode oxalate degrading enzymes. Some researchers believe that therapeutic application of Oxalobacter formigenes may ultimately provide the best approach to prevent the progression of hyperoxaluria by potassium sulfate supplementation [86], nevertheless, several prior studies could not identify an important role for Oxalobacter formigenes in nephrolithiasis, due to impracticality of Oxalobacter formigenes isolation from stool samples [87].
Findings from shotgun metagenomic studies indicate distinct microbiome composition in idiopathic kidney stone formers, suggesting that the Gut-kidney axis regulates renal stone formation [85]. In a study, the incidental and prevalent renal stone groups, exhibited an alteration in composition of gut microbiota compared with healthy controls, which could lead to nephrolithiasis. Overall, along with the identified oxalate degradation pathways, further investigations in other functional pathways is required [87].
13. Gut-kidney axis in IgA nephropathy
IgA is the prevalent mucosal secretions and is the prominent mark of the mesangial deposits in IgA nephropathy [89]. In other words, IgA nephropathy is characterized by deposition of IgA Antibodies in the mesangium. IgAN is considered the commonest primary glomerulopathy [89,90].
Secretory IgA, plays a significant role in the mucosal surfaces and is associated in intestinal homeostasis maintaining by neutralization of pathogens and toxins, modulation of gut microbiota composition, inflammatory responses downregulation and keeping from inappropriate immune responses against microbial and dietary antigens [91].
Current studies show that the gut has significant role in creation of majority of IgA, also gut utilizes IgA for keeping mucosal colonization [92].
While older theories have focused on the role of upper respiratory infections, however, new data propose that older theories need to be revised and attention should be paid to the composite role of genetic conditions, gut dysbiosis and diet in gut mucosal immunity dysregulation and consequently development and progression of IgA nephropathy [89].
IgAN is an autoimmune disorder characterize by abnormal synthesis and glycosylation of IgA1, which causes an imbalanced elevation of circulating galactose deficient IgA1. This abnormal Ab can attach with specific autoantibodies and shape immune complexes, ultimately deposit in glomeruli and induce the onset of pathologic events in mesangium and matrix [93]. Recent information indicates the role of GALT in IgA nephropathy development. The new attentions and interests based on clinical findings in GALT hyper-reactivity in IgAN disease have emerged [89].
Gut microbiota have the complex interactions with host immune system and a significant influence in regulation of the GALT, for example promotion of Treg differentiation [94]. The role of gut microbiota in IgA nephropathy patients has recently been investigated and researchers have reported differences between patients and healthy groups, also between patients with the progressive and the non-progressive form of IgAN; that is, the number of Bifidobacteria and Streptococci decreased and increased, respectively [89]. De Angelis and co workers in 2014 showed a decrease in Clostridium, Lactobacillus, Bifidobacterium, Enterococcus genera. The F/B ratio also Firmicutes and Proteobacteria phylum, were elevated in the IgAN group [92]. Another study about Henoch Schonlein Purpura in children showed that a reduction in diversity and richness in the patients group also an important negative association of serum IgA levels with Bifidobacterium genus, and a significant changes in the intestinal microbiota of HSP patients can be seen [95]. Another study found no differences in the gut microbial abundance and diversity between healthy controls and IgAN patients in Malaysia [92]. The other study, presented an extensive investigation of the gut microbiota composition in IgAN and MN patients, and represented the significant difference of gut microbiota in IgAN and MN patients compared to healthy controls [90].
At the molecular level, TLRs, such as TLR4, which binds to lipopolysaccharide, and TLR2, which binds to lipoteichoic acid, are much expressed in mucosal epithelial cells and identify the microbial molecular patterns.
An altered intestinal barrier function facilitates increased lipopolysaccharide absorption and circulation and patients with IgA nephropathy are characterized by increased gut permeability, which may facilitate elevated LPS circulation [89]. At a study was shown that LPS activates TLR4 in cultured peripheral B lymphocytes, then chaperone Cosmc to be methylated, and this process is crucial for the activity of galactosyltransferase enzyme, so that reduction of Cosmc gene expression in mentioned lymphocytes is the cause of glycosylation aberration in IgAN patients [96].
14. Microbial enterotypes and their reality
Humans are different not only because of their genetic content but also because of their gut microbiome. The gut microbiome of humans composed of at least 1800 genera and almost 15,000–36000 bacterial species, and a total bacterial count of 1013 to 1014 [97].
The gut microbiota varies greatly from person to person on a space-time scale, and this is a barrier to microbiome-based medical procedures. Enterotype theory was first proposed in 2011 to solve this problem by stratification of gut microbiota.
Arumugam et al. integrated 22 newly sequenced fecal metagenomes related to 4 countries with prior available datasets, then they concluded that these samples can be divided into three separate clusters under the influence of discriminative microbial genera, which consist of enterotype 1 (mostly includes Bacteroides), enterotype 2 (mainly includes Prevotella) and enterotype 3 (predominately includes Ruminococcus) [97,98]. Enterotypes proposed as a prognostic tool to evaluation the association of lifestyle, nourishment and disease [99]. However, today there are doubts about the being and strength of enterotypes. According to Mingue Chong et al. opinion, due to high complexity and diversity of human gut microbiota, it seems realistic for researchers to divide the gut microbiota into smaller groups. Nevertheless, existing studies are neither able to prove nor disprove the enterotype theory [97].
Ivan Bulygin and co-workers believe that the number of enterotypes proposed in the literature, range from two to four. They reanalyzed the human gut microbiome with non-linear dimensionality reduction methods and based on concluded that do not exist the clear and stable clusters that could display enterotypes [99].
According to Mingue Chong opinion, after gaining more experience on a larger scale to validate and advance enterotype theory, this theory, using a single enterotype method, this theory will made easier to understand the classification of human gut microbiota. Such a stratification will help us to achieve a better understanding the links between gut microbiota and diseases to make easier precision medicine according to gut microbiota [97].
Anastassia et al. found the term enterotype misleading because the theory indicates both an underlying consistency of community taxa and a distinct separation of human gut specimens sets, non of which is approved by the vast and larger studies. The researchers suggested using the biomarker as a better description for the taxa, related to nutrition, lifestyle and disease state [100]. For example, at a study, Prevotella-to-Bacteroides ratio (P/B ratio), was suggested as a predictive biomarker in weight management randomized to take arabinoxylan oligosaccharides [101].
Some researchers studied about the possibility of enterotypes description to use in human gut virome and their association with human disease. The experiments of these researchers showed that most individuals could be categorized into viral enterotypes [102].
15. Systems biology and kidney disease
With regard to diseases such as most renal diseases with an intricate pathophysiology and molecular complexity, we need to comprehend the physiology at the cellular networks and molecular level under genome-wide scale to identify and predict the disease course and key events to develop drugs for diseases therapy. Systems biology can be used to understand how changes in cell composition and interactions lead to the disease pathophysiology. With the advent of microarrays, NGS techniques, mass spectrometry and more sensitive and precise methods for identifying molecular species and genome-wide scale interactions, and bioinformatics tools, it is theoretically possible to have an overview of cellular functions of kidney from a molecular view, the relationship of phenotypes to molecular networks that conduct pathophysiological changes [103,104]. In fact, the systems biology approach has the advantage to use the recent advances in computational methods, machine learning approaches and omics technologies for integrating different types of information such as cellular, molecular and clinical parameters to elucidate the interactions of multiple genes, proteins, and molecular mechanisms that drive the discrete steps. The systems biology gives us the methods and tools needed to draw a holistic map and a unified image of the kidney in a health and disease states [1,105] and integrating omics information to stablish a dynamic model of molecular changes in chronic kidney disease for patients diagnosis, biomarker and drug exploration [104,106].
16. DME gene network and SLC and ABC transporters in Remote Sensing and signaling in the gut-liver-kidney axis
The ABC transporter superfamily is one of the most considerable and widely expressed superfamilies recognized. These transporters present in all living organisms and is associated in almost all cellular, biological and physiological systems and the vast majority of members of the ABC superfamily have a key role in the active transport of many substrates across biological membranes [107]. Organic anion carriers, OAT1 (SLC22A6) and OAT3 (SLC22A8), have similar specificity to drugs, but it is unclear whether this includes metabolites of endogenous substrates. In a study, Bush et al. reported that OAT3 is involved in the transport of intestinal microbiota products, key metabolites, and signaling molecules, including molecules that flow via the gut-liver-kidney axis. The main affected pathways are those engaged in nutrients, amino acids (including tryptophan derivatives, which are uremic toxins), flavonoids, bile acids and lipids metabolism. Also OAT3 has a significant role in the liver-derived phase II metabolites elimination [108].
Remote Sensing and Signaling hypothesis tries to describe how multi-specific, oligo-specific and mono-specific transporters and enzymes associated with regulatory proteins, work together to achieve optimal levels of many small molecules in various body fluids and tissues [68]. This hypothesis focuses about a network of ADME genes those essentially consist of SLC and ABC transporters, DMEs (drug metabolizing enzymes), and regulator genes, for inter-organ crosstalk through signaling molecules, metabolites, antioxidants, and also intestinal microbial products and uremic toxins.
Multispecific drug transporters, including approximately 50–100 solute carriers (SLC) and ATP-binding cassette transporters (ABC) are expressed in all epithelial cells also many non-epithelial cells of all body tissues. Also, these are a subdivision of more than 400 SLC and ABC transporters those have a range of substrate specificity; mono-specificity, oligo-specificity and multi-specificity.
These genes, together with phase Ⅰ and phase Ⅱ drug-metabolizing enzymes, have a critical role in the absorption, distribution, metabolism, and elimination (ADME) of small molecule drugs and toxins [109].
In this regard, the gut-liver-kidney axis refers to the absorption of a metabolite such as a food or a gut microbiome product via intestine and then its metabolism in the liver and finally its excretion from the kidney [108].
Based on our developing understanding about mechanistic basis of indoxylsulfate, this uremic toxin is a typical example of the necessity to further surveys beyond the toxins and study about specific signaling routes and metabolic pathways [68].
OAT1 and OAT3 interact with several uremic solutes and toxins in-vitro, and according to substantial in vivo information, OATs use many endogenous metabolites that contain a number of uremic toxins and retention substances. These transporters are placed in renal proximal tubules, they are two of the most significant transporters engaged in the absorption and secretion of drugs, nutrients, endogenous metabolites (such as uremic toxins) and exogenous toxins [110]. These membrane transporters, have protein structure and mediate the influx or efflux of molecules such as uremic toxins, these toxins can prompt signaling pathways at arrival in to the cell and then modulate cellular response, which under uremic status contribute to pathologic process of CKD [67].
Viewing uremic toxins via the multiscale systems biology aspect of the Remote Sensing and Signaling Theory recommends a lot variety of therapeutic ways to moderate the toxin effect of small protein-bound and hardly dialyzable uremic toxins [68].
17. Microbiome –metabolome
One of the microbiome interact ways with its host, is through the production of various metabolites [111]. For example, microbiota of a healthy colon, produces a large amounts of metabolites that are necessary to maintaining of the colon environment homeostasis [112]. Really, these metabolites are related to some diseases such as obesity, insulin resistance, CVD, diabetic kidney disease and colon cancer [[112], [113], [114], [115]]. The untargeted metabolomics and 16 S r RNA gene sequencing on mice and rats have been done to determination of gut microbiota and CKD correlations [116]. Mouse researches have represented that the intestinal metabolome is closely connected to the intestinal microbiome [117]. Lifestyle and nutritional factors can affect the gut microbiota composition. According to some studies, there is a link between two dominant bacterial phyla, Bacteroides and Firmicutes, with obesity in mice and humans, respectively [118]. Also researchers by db/db mouse studies, showed that intestinal microbiome dysbiosis can affect plasma metabolism and then affect renal function [119]. The therapeutic effect of magnesium lithospermate B on mouce kidney injury [115] and rhubarb enema on renal fibrosis improvement in CKD rats [120] also α-ketoacid protective mechanism on adenine-induced CKD rats have been studied [121]. Also researchers showed the correlation of PS with albuminuria in diabetic mice [122]. The results of molecular analysis have represented that the human intestinal microbiota composition is host-specific. It should be noted that antibiotic treatment not only targets pathogenic bacteria but also affects commensal bacteria for a long time. As a result, the ecological balance between humans and microbiota could be disturbed by antibiotics [123]. The integrative shotgun sequencies and metabolomics analyses have been used for the study of gut microbiota role in antibody-mediated renal allograft rejection [124].
18. Metagenomics and metabolomics approach to investigate host-gut microbiota interactions
The aim of metabolomics is to study small molecular metabolites and their biochemical functions, and these studies are promising to survey about the interactions between gut microbiota with the host. Metabolomics identifies the metabolites associations and interactions with gut microbiota and this technique focuses on determining the functional conditions of microbial-host communications in biological samples such as urine, feces, blood and tissues [112]. Metabolomics as an important method has arised as a systemic approach which can study about changes in low-molecular- weight endogenous metabolites during disease, toxic exposure and genetic variations which ultimately help us to understanding the gut microbiota function [57].
Most studies on the gut microbiome are performed with the aim of detecting disease-related metabolites or disregulated metabolic pathways [125]. Metabolomic surveys in gut microbiota have enhanced attention to generation of novel biomarkers which potentially would be useful in the nutritional and personalized therapies [126]. Metabolomics, prepares the biological information which cannot be attained by classical approaches of “omics” include transcriptomics, genomics and proteomics [127].
Mass spectrometry (MS)-based metabolomic techniques are robust techniques for measuring key capabilities involved in gut and host microbial interactions [125].
The emergence of developed next-generation sequencing techniques, including metagenomics and 16s r RNA sequencing, by their solitary advantages made probable the analysis of much larger amounts of gut microbiome [57]. It should be noted that genomic studies are about a specific organism genetic content, while metagenomics consider the genetic content of whole communities of organisms [128].
Metagenomics sequencing helps us determine the function of these sequences by random sequencing of all extracted DNA, while 16s r RNA analysis is more beneficial in finding out what types of bacteria are present in the sample [57]. Metagenomics helps us to find new genes and proteins or even the whole genomes of non-culturable organisms with higher accuracy and in less time compared with traditional methods [129].
Functional shotgun metagenomics sequencing depends largely on our information about the manner of coding enzymatic or other functions through gene sequences, and metabolic databases such as KEGG and MetaCyc, which are important sources in this area [57]. KEGG includes mainly more compounds than MetaCyc, whilst MetaCyc includes mostly more reactions and pathways compared with KEGG. Nevertheless, the number of reactions occurring in pathways, are quite similar [130].
16s r RNA characterization has not quantitative functional annotation, but fecal metabolome enables us to provide a functional read-out of microbial functions. Fecal metabolic profiling seems as a novel facility for discovering connections between microbiome composition, host phenotypes and complex trait genetics [131].
Due to the advantages of NMR include high reproducibility, the quantification feasibility, a high grade structural information, the global detection capacity for every organic compound, and its compatibility in the biological samples analysis, this technique very early arise as a choice method for clinics metabolomics works [132].
Proton NMR Spectroscopy and Mass Spectrometry are important analytical techniques for metabolomic studies [57].
In many studies, multiple MS-based platforms have been used to promot the coverage of metabolite detection. In most microbiota and metabolomic studies, MS, along with separation techniques such as liquid chromatography, gas chromatography and capillary electrophoresis, have provided a higher ability to perform qualitative and quantitative analysis. The TOFMS, (Time-Of-Flight Mass Spectrometry) method is a powerful method for the universal detection of metabolites. The Tandem Mass Spectrometry technique is useful for studying the target metabolome [125].
Metabolomics has been extensively used to improve the diagnosis and prognosis of various diseases, the discovery of biomarkers, the pharmaceutical development, and drug efficacy/toxicity evaluation, also widely used to study a variety of kidney diseases [57]. Lipidomics, is a novel methodology, and is a branch of metabolomics which encompass systematic and thorough survey of lipids and lipid-derived metabolites in health and disease. With attention to recent advances in MS-based techniques and rapid progression in chromatography, specially UPLC-MS along with bioinformatics, our knowledge about the role of derivative metabolites of lipids in kidney disease have developed [133].
Mi Y et al. used Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) analytical technique to ascertain of uremic toxins in ESRD patients. In their study, 26 uremic toxins in ESRD patients and normal subjects before and after hemodialysis were determined [134]. The LC-MS/MS has been vastly undergone development in clinical laboratories during recent decades, however, this technique has several limitations [135].
However, the use of metabolomics in samples of gut microbiome-induced kidney disease is limited. It is essential to perform such studies to understand the association between intestinal microbiota and renal disease.
Overall, given our infancy information about the gut microbiome and metabolome data, there is a need to perform multi-omics research in order to achieve a better comprehension of the mechanisms and phenotypes in connections between the gut microbiota and kidney disease [57].
Advances in diagnostic methods have broadened our understanding of the complicated mechanistic linkages between microbiome and diseases.
There are several methods for classify and measuring the diversity and composition of gut microbiota. Some methods use 16s r RNA sequencing, which gives us a good insight into the gut microbiota diversity and qualitative and quantitative information about existing microbes. FISH, DNA microarrays and NGS of the 16s r RNA gene are the examples of this technique.
Another method which previously mentioned is metagenomics or shotgun sequencing, this technique randomly sequences all DNA extracted in the sample. Considering its ability to detect microbial taxa at the species and strain level, this technique has a higher taxonomic resolution. In addition, this method provides information about the microbial community function [59].
19. Probiotics and kidney disease
In recent years, probiotics have been recognized as agents that help maintain the physiological balance of gut microbiota. Probiotic supplements can lower pCS levels and increase IL-6 levels, thereby protecting the gut epithelial barrier of patients. But, the limitations of these studies included the limited number of researches as well as the small sample size [136]. The potential role of certain probiotic strains has even been suggested as a novel adjuvant strategy for neurological disorders because gut microbiota have a significant role in bilateral gut and nervous system interactions [137].
Probiotics are defined as naturally or genetically modified microorganisms that express particular exogenous enzymes that enable them to live in gastric and bile acids. They increase concentration of colon symbionts and have positive health effects [138].
In other words, probiotics are living microorganisms that are able to live in the gut and restore the gut flora balance, and have useful effects on individual's health [3,138]. Their possible beneficial effects are as follows.
-
1.
Strengthening the intestinal barrier by enhancing mucus and epithelial tight junction integrity [138]. Probiotics can increase the secretion of mucin glycoproteins by goblet cells which causes thickening mucus layer or epithelial cells [139].
-
2.
Antimicrobial activity by lowering the local pH, production of AMPs and defensins, which restrain the pathobionts such as urease-producing bacteria [138]. Probiotics are able to promote the differentiation of B lymphocytes and therefore to stimulate the production of antibodies especially secretory IgA [140], which interferes with adhesive cell receptors on the pathogen's cell membrane and provides an additional protection from the luminal microbiota [138,[140], [141], [142]].
-
3.
Anti-inflammatory effects and immune tolerance improvement.
-
4.
Competition for nutrients and bile acid metabolism [138], also probiotics can compete with commensals and enteric pathogens for adhesion sites and mucus layers, and competitively excluding pathogens at the binding sites [58,138].
The reduction in pathobionts, limits the production of gut-derived uremic toxins.
In general, the fact that the levels of circulating lipopolysaccharides and microbiome-derived URS (indoxyl sulfate, p-Cresyl sulfate and TMAO) increased in CKD stages suggests an association between gut barrier and renal dysfunction [138]. The translocation of these uremic toxins from the leaky gut into the bloodstream further promotes systemic inflammation, detrimental cardiovascular outcomes and CKD progression [25].
Probiotic therapies offer promising opportunities considering their low cost and harmless nature. These therapies apparently are the gold standard for prophylaxis and treatment particularly subsequent to the failing in several antibiotic therapies [3,143]. Numerous experimental and clinical studies have shown that probiotic treatment is an interesting approach to reducing uremic toxicity by ingesting microbes that can catalyze URSs. Also, according to data, probiotics are able to retard the renal failure progression and decrease inflammatory markers such as plasma IS, pCS levels [3,138]. As a result, oral supplementation of selected microorganisms (in a probiotic formula), by extracting UTs in CKD patients, may exert a nephroprotective effect [144]. Also, the supplementation of SCFA, either directly or through the regulation of intestinal microbiota via SCFA-producing bacteria, could have a positive effect on the CKD control [58].
The important strains which used for the management of CKD patients, are belonging to the Lactobacillus and Bifidobacterium genera [3]. Also, Akkermansia has been suggested as a promising potential therapeutic strategy to reduce the systemic inflammation in CKD patients [63].
Novel probiotics such as Clostridium species have been discussed; but there are still some significant challenges in medication of them. For example, in some Clostridium botulinum and Clostridium butyricum type E strains, the recombination and insertion of botulinum neurotoxin complex genes were revealed, also in other gut commensal bacteria, toxins plasmids of Clostridium perfringens were detected. Also, Clostridium species must be firmly detected via safety evaluation of probiotic strains [48].
Recent studies have expressed that the intestinal microbiome could be a not discovered storage of likely useful microbes [145]. It is especially noteworthy that some researchers suggest a novel therapeutic program using the probiotic yeast Saccharomyces boulardii to modulating of intestinal microbiome composition [146].
There has been reduced desire to use probiotics due to concerns about how precisely to take advantage and mediate their beneficial effects these various organisms and also concerns about the possibility of increasing inflammation due to subsequent urea hydrolysis [138]. Overall, the researches on human cases, are still rare and therefore, obtain the approval from regulatory agencies rarely happens [145]. Also, owing to being alive the probiotics, there are many biological and biopharmaceutical restrictions in their clinical application [147].
It should be noted that the purpose of using probiotics, regardless of improving the intestinal biochemical environment, can lead to the failure of relevant studies. Therefore, further fundamental and clinical studies should be performed on the dysbiosis effect in the CKD development and its related complications [138].
20. Conclusion
Gut microbiota play an important role in a number of diseases, including CKD and nephrolithiasis. Increasing evidence suggests a bilateral link between gut microbiota in patients with a variety of renal diseases. Gut inflammation and damage to the intestinal epithelial barrier, translocation of uremic toxins such as indoxyl sulfate, p-Cresyl sulfate, TMAO are accelerated, which in turn cause oxidative stress damage to organs such as kidney, cardiovascular and endocrine system. Without doubt, investigation of gut-kidney axis consists one of the most cutting-edge areas of research on our health which has promising potential implications for better quality of our health and life.
Treatment strategies for CKD patients based on gut microbiota have already been investigated, but it is unclear whether CKD is accompanied with particular microbial profiles and metabolomes. These profiles can be beneficial biomarkers for diagnosis and severity assessment, as well as potential treatment goals.
New evidence for the association of gut microbiota with CKD, and CVD provides a mechanistic vision to the gut-kidney-heart axis, which emphasizes the significance of gut as a potential risk factor for CKD-related side-effects and complications.
Remote sensing and signaling network theory that was proposed by some researchers discusses about the existence of an ADME-gene-centered network consist of SLC and ABC transporters, DMEs and regulator genes in association with inter-organ communication through signaling molecules, metabolites, gut microbial products and uremic toxins. The network of SLC and ABC transporters and DME genes, along with protein-protein interactions, proposes how multi-specificity merges with oligo-specificity and mono-specificity to adjust the many endogenous small molecules homeostasis.
Transporters of uremic toxins (indoxyl sulfate, p-Cresyl sulfate, urate, creatinine, kynurenine) in the proximal tubule include two drug transporters belonging to the organic anion transporters of the OAT family (OAT1, OAT3). These transporters do a regulating role in the metabolism of uremic toxins; therefore, further studies on these transporters can indicate new horizons regarding the microbiota role and their metabolites in related with the gut-liver-kidney axis.
Metagenomic and metabolomic studies have been performed in kidney diseases to evaluate the function of gut microbiome-derived low-molecular weight endogenous metabolites. However, 16s r RNA sequencing has been used to evaluate the structure of the gut microbiome. Although metagenomic sequencing provides a more knowledge about existent genes, the function of most of these genes remains unknown. KEGG and MetaCyc databases are used to evaluate the function of genes. To achieve a better understanding of microbial metabolism in renal disease, advanced multi-omic integration techniques must be developed.
Probiotic therapies offer good opportunities for the prevention and treatment of diseases such as kidney disease considering their low cost and harmless nature, but because there are concerns about their side effects, further relevant larger scale studies should be conducted.
Finally, the novel topics based on recent improvements in a multidimensional view especially in multi-omics field and from a broad perspective, systems biology, have shed light on the microbial impact in health and disease such as chronic kidney disease. In other words, considering the limited and detailed nature of current studies, a holistic view of systems biology will help to increase our understanding of chronic kidney disease and its prevention, management and treatment methods. So further and vast surveys for more steps to understanding the intricate communications which cause these illnesses must be done.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
No data was used for the research described in the article.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Schaub J.A., et al. Systems biology and kidney disease. Clin. J. Am. Soc. Nephrol. 2020;15(5):695–703. doi: 10.2215/CJN.09990819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T.K., Knicely D.H., Grams M.E. Chronic kidney disease diagnosis and management: a review. JAMA. 2019;322(13):1294–1304. doi: 10.1001/jama.2019.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagundes R.A.B., et al. Probiotics in the treatment of chronic kidney disease: a systematic review. J Bras Nefrol. 2018;40(3):278–286. doi: 10.1590/2175-8239-JBN-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemczyk L., Malyszko J. Renal replacement modality affects uremic toxins and oxidative stress. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/6651367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnani S., Atti M. Uremic toxins and blood purification: a review of current evidence and future perspectives. Toxins. 2021;13(4) doi: 10.3390/toxins13040246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Your Kidney & How They Work. 2002. [Google Scholar]
- 7.O'Connor N.R., Corcoran A.M. End-stage renal disease: symptom management and advance care planning. Am. Fam. Physician. 2012;85(7):705–710. [PubMed] [Google Scholar]
- 8.Guirong Y.E., et al. [Gut microbiota in renal transplant recipients, patients with chronic kidney disease and healthy subjects] Nan Fang Yi Ke Da Xue Xue Bao. 2018;38(12):1401–1408. doi: 10.12122/j.issn.1673-4254.2018.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C.-H., et al. Infection in advanced chronic kidney disease and subsequent adverse outcomes after dialysis initiation: a nationwide cohort study. Sci. Rep. 2020;10(1):2938. doi: 10.1038/s41598-020-59794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eleftheria Tzanakaki V.B.A.S., Stylianou Kostas, Rovithis Michael, Zidianakis Zacharias. Causes and complications of chronic kidney disease in patients on dialysis Health. Sci. J. 2014;8(3) [Google Scholar]
- 11.About Chronic Kidney Disease . 2013. Guide For Patients.WWW.KIDNEY.ORG 2014; Available from: [Google Scholar]
- 12.Beladi-Mousavi S.S., et al. Long-term survival of patients with end-stage renal disease on maintenance hemodialysis: a multicenter study in Iran. Iran J Kidney Dis. 2012;6(6):452–456. [PubMed] [Google Scholar]
- 13.McCullough K.P., et al. Projecting ESRD incidence and prevalence in the United States through 2030. J. Am. Soc. Nephrol. 2019;30(1):127–135. doi: 10.1681/ASN.2018050531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaipov A., et al. Epidemiology of dialysis-treated end-stage renal disease patients in Kazakhstan: data from nationwide large-scale registry 2014-2018. BMC Nephrol. 2020;21(1):407. doi: 10.1186/s12882-020-02047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khazaei M., et al. Epidemiological characteristics and causes of end-stage renal disease in hemodialysis patients. International Journal of Epidemiologic Research. 2020;7(2):53–57. [Google Scholar]
- 16.Pedrini L.A., et al. Clinical outcomes of hemodialysis patients in a public-private partnership care framework in Italy: a retrospective cohort study. BMC Nephrol. 2019;20(1):35. doi: 10.1186/s12882-019-1224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levey A.S., et al. Definition and classification of chronic kidney disease: a position statement from Kidney Disease: improving Global Outcomes (KDIGO) Kidney Int. 2005;67(6):2089–2100. doi: 10.1111/j.1523-1755.2005.00365.x. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T.L., et al. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atuma C., et al. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280(5):G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 20.McDermott A.J., Huffnagle G.B. The microbiome and regulation of mucosal immunity. Immunology. 2014;142(1):24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neish A.S. Mucosal immunity and the microbiome. Ann Am Thorac Soc. 2014;11(Suppl 1):S28–S32. doi: 10.1513/AnnalsATS.201306-161MG. Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furness J.B., et al. The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv. Exp. Med. Biol. 2014;817:39–71. doi: 10.1007/978-1-4939-0897-4_3. [DOI] [PubMed] [Google Scholar]
- 23.Zheng L., Kelly C.J., Colgan S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 2015;309 6:C350–C360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang T., et al. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 2018;14(7):442–456. doi: 10.1038/s41581-018-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau W.L., Kalantar-Zadeh K., Vaziri N.D. The gut as a source of inflammation in chronic kidney disease. Nephron. 2015;130(2):92–98. doi: 10.1159/000381990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau W.L., Vaziri N.D. The leaky gut and altered microbiome in chronic kidney disease. J. Ren. Nutr. 2017;27(6):458–461. doi: 10.1053/j.jrn.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Ramezani A., et al. Role of the gut microbiome in uremia: a potential therapeutic target. Am. J. Kidney Dis. 2016;67(3):483–498. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He F., Ru X., Wen T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020;21(13):4777. doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popkov V.A., et al. Gut microbiota as a source of uremic toxins. Int. J. Mol. Sci. 2022;23(1) doi: 10.3390/ijms23010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez J.M., et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26 doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller N.T., et al. The infant microbiome development: mom matters. Trends Mol. Med. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujisaka S., et al. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep. 2018;22(11):3072–3086. doi: 10.1016/j.celrep.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu G.D., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou J., et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013;98(1):111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre de Cárcer D. The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-32221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilms E., et al. Intestinal barrier function is maintained with aging – a comprehensive study in healthy subjects and irritable bowel syndrome patients. Sci. Rep. 2020;10(1):475. doi: 10.1038/s41598-019-57106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minot S., et al. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 2013;110(30):12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao G., et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. USA. 2017;114(30):E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iliev I.D., Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 2017;17(10):635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J., et al. Gut mycobiome in patients with chronic kidney disease was altered and associated with immunological profiles. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.843695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wexler H.M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrow H.C., Batachari L.E., Chu H. Strain diversity in the microbiome: lessons from Bacteroides fragilis. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yekani M., et al. To resist and persist: important factors in the pathogenesis of Bacteroides fragilis. Microb. Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104506. [DOI] [PubMed] [Google Scholar]
- 44.He B.L., et al. Bifidobacterium spp. as functional foods: a review of current status, challenges, and strategies. Crit. Rev. Food Sci. Nutr. 2022:1–18. doi: 10.1080/10408398.2022.2054934. [DOI] [PubMed] [Google Scholar]
- 45.Arboleya S., et al. Gut Bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khiabani S.A., et al. Clostridium species diversity in gut microbiota of patients with renal failure. Microb. Pathog. 2022;169 doi: 10.1016/j.micpath.2022.105667. [DOI] [PubMed] [Google Scholar]
- 47.Jiang S., et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci. Rep. 2017;7(1):2870. doi: 10.1038/s41598-017-02989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo P., et al. Clostridium species as probiotics: potentials and challenges. J. Anim. Sci. Biotechnol. 2020;11(1):24. doi: 10.1186/s40104-019-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnizlein M.K., Young V.B. Capturing the environment of the Clostridioides difficile infection cycle. Nat. Rev. Gastroenterol. Hepatol. 2022;19(8):508. doi: 10.1038/s41575-022-00610-0. [DOI] [PubMed] [Google Scholar]
- 50.Czepiel J., et al. Clostridium difficile infection: review. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38(7):1211–1221. doi: 10.1007/s10096-019-03539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forti K., et al. Molecular characterization of Clostridium perfringens strains isolated in Italy. Toxins. 2020;12(10):650. doi: 10.3390/toxins12100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomes A.C., Hoffmann C., Mota J.F. The human gut microbiota: metabolism and perspective in obesity. Gut Microb. 2018;9(4):308–325. doi: 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanifi G., et al. Lactobacilli species diversity in gut microbiota of renal failure patients. J. King Saud Univ. Sci. 2020;32(4):2365–2369. [Google Scholar]
- 54.Clark M.M.J.M.D.S.D. Brock biology of microorganisms. 13th Edition. Global Edition. Pearson Higher Edocation; 2011. [Google Scholar]
- 55.Sonnenburg J.L., Angenent L.T., Gordon J.I. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004;5(6):569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 56.Wu I.W., et al. Gut microbiota as diagnostic tools for mirroring disease progression and circulating nephrotoxin levels in chronic kidney disease: discovery and validation study. Int. J. Biol. Sci. 2020;16(3):420–434. doi: 10.7150/ijbs.37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y.-Y., et al. Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease. J. Transl. Med. 2019;17(1):5. doi: 10.1186/s12967-018-1756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magliocca G., et al. Short-chain fatty acids in chronic kidney disease: focus on inflammation and oxidative stress regulation. Int. J. Mol. Sci. 2022;23(10) doi: 10.3390/ijms23105354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanford J., et al. The gut microbiota profile of adults with kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol. 2020;21(1):215. doi: 10.1186/s12882-020-01805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaziri N.D., et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83(2):308–315. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 61.Feng Z., et al. Association between gut dysbiosis and chronic kidney disease: a narrative review of the literature. J. Int. Med. Res. 2021;49(10) doi: 10.1177/03000605211053276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampaio-Maia B., et al. The role of the gut microbiome on chronic kidney disease. Adv. Appl. Microbiol. 2016;96:65–94. doi: 10.1016/bs.aambs.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Li F., et al. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front. Cell. Infect. Microbiol. 2019;9:206. doi: 10.3389/fcimb.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shivani S., et al. Uremic toxin-producing Bacteroides species prevail in the gut microbiota of Taiwanese CKD patients: an analysis using the new taiwan microbiome baseline. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.726256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanifi G.R., et al. Bifidobacteriaceae family diversity in gut microbiota of patients with renal failure. Arch Razi Inst. 2021;76(3):521–528. doi: 10.22092/ari.2020.352271.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lisowska-Myjak B. Uremic toxins and their effects on multiple organ systems. Nephron Clin. Pract. 2014;128(3–4):303–311. doi: 10.1159/000369817. [DOI] [PubMed] [Google Scholar]
- 67.Cunha R.S.d., et al. The interplay between uremic toxins and albumin, membrane transporters and drug interaction. Toxins. 2022;14(3):177. doi: 10.3390/toxins14030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lowenstein J., Nigam S.K. Uremic toxins in organ crosstalk. Front. Med. 2021;8 doi: 10.3389/fmed.2021.592602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gryp T., et al. p-Cresyl Sulfate. Toxins. 2017;9(2) doi: 10.3390/toxins9020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koppe L., et al. p-Cresyl glucuronide is a major metabolite of p-cresol in mouse: in contrast to p-cresyl sulphate, p-cresyl glucuronide fails to promote insulin resistance. Nephrol. Dial. Transplant. 2017;32(12):2000–2009. doi: 10.1093/ndt/gfx089. [DOI] [PubMed] [Google Scholar]
- 71.Maciel R.A., et al. p-cresol but not p-cresyl sulfate stimulate MCP-1 production via NF-κB p65 in human vascular smooth muscle cells. J Bras Nefrol. 2016;38(2):153–160. doi: 10.5935/0101-2800.20160024. [DOI] [PubMed] [Google Scholar]
- 72.Clayton T.A., et al. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U. S. A. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan X., et al. Indoxyl sulfate, a valuable biomarker in chronic kidney disease and dialysis. Hemodial. Int. 2017;21(2):161–167. doi: 10.1111/hdi.12483. [DOI] [PubMed] [Google Scholar]
- 74.Kamiński T.W., et al. Indoxyl sulfate - the uremic toxin linking hemostatic system disturbances with the prevalence of cardiovascular disease in patients with chronic kidney disease. BMC Nephrol. 2017;18(1):35. doi: 10.1186/s12882-017-0457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edamatsu T., et al. Classification of five uremic solutes according to their effects on renal tubular cells. Internet J. Nephrol. 2014 doi: 10.1155/2014/512178. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manor O., et al. A multi-omic association study of trimethylamine N-oxide. Cell Rep. 2018;24(4):935–946. doi: 10.1016/j.celrep.2018.06.096. [DOI] [PubMed] [Google Scholar]
- 77.Lakshmi G., et al. Gut microbiota derived trimethylamine N-oxide (TMAO) detection through molecularly imprinted polymer based sensor. Sci. Rep. 2021;11(1):1338. doi: 10.1038/s41598-020-80122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu F., et al. Gut-derived metabolite phenylacetylglutamine and white matter hyperintensities in patients with acute ischemic stroke. Front. Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.675158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wee H.N., et al. The kynurenine pathway in acute kidney injury and chronic kidney disease. Am. J. Nephrol. 2021;52(10–11):771–787. doi: 10.1159/000519811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mor A., Kalaska B., Pawlak D. Kynurenine pathway in chronic kidney disease: what's old, what's new, and what's next? Int. J. Tryptophan Res. 2020;13 doi: 10.1177/1178646920954882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sumida K., Kovesdy C.P. The gut-kidney-heart axis in chronic kidney disease. Phys. Int. 2019;106(3):195–206. doi: 10.1556/2060.106.2019.19. [DOI] [PubMed] [Google Scholar]
- 82.Meijers B., et al. Intestinal barrier function in chronic kidney disease. Toxins. 2018;10(7) doi: 10.3390/toxins10070298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramezani A., Raj D.S. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 2014;25(4):657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao H., et al. Changes of lipopolysaccharide-induced acute kidney and liver injuries in rats based on metabolomics analysis. J. Inflamm. Res. 2021;14:1807–1825. doi: 10.2147/JIR.S306789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ticinesi A., et al. Understanding the gut-kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut. 2018;67(12):2097–2106. doi: 10.1136/gutjnl-2017-315734. [DOI] [PubMed] [Google Scholar]
- 86.Sadaf H., Raza S.I., Hassan S.W. Role of gut microbiota against calcium oxalate. Microb. Pathog. 2017;109:287–291. doi: 10.1016/j.micpath.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 87.Kim H.-N., et al. Gut microbiota and the prevalence and incidence of renal stones. Sci. Rep. 2022;12(1):3732. doi: 10.1038/s41598-022-07796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jung H., Lee J.Y. Impact of the human microbiome on nephrolithiasis. Urogenital Tract Infection. 2021;16:25–31. [Google Scholar]
- 89.Coppo R. The gut-kidney axis in IgA nephropathy: role of microbiota and diet on genetic predisposition. Pediatr. Nephrol. 2018;33(1):53–61. doi: 10.1007/s00467-017-3652-1. [DOI] [PubMed] [Google Scholar]
- 90.Dong R., et al. A comparative study of the gut microbiota associated with immunoglobulin a nephropathy and membranous nephropathy. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.557368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pietrzak B., et al. Secretory IgA in intestinal mucosal secretions as an adaptive barrier against microbial cells. Int. J. Mol. Sci. 2020;21(23) doi: 10.3390/ijms21239254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sugurmar A.N.K., et al. Gut microbiota in immunoglobulin A nephropathy: a Malaysian perspective. BMC Nephrol. 2021;22(1):145. doi: 10.1186/s12882-021-02315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gutiérrez E., et al. A personalized update on IgA nephropathy: a new vision and new future challenges. Nephron. 2020;144(11):555–571. doi: 10.1159/000509997. [DOI] [PubMed] [Google Scholar]
- 94.Shreiner A.B., Kao J.Y., Young V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015;31(1):69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X., et al. Gut microbiota dysbiosis is associated with Henoch-Schönlein Purpura in children. Int. Immunopharm. 2018;58:1–8. doi: 10.1016/j.intimp.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 96.Qin W., et al. External suppression causes the low expression of the Cosmc gene in IgA nephropathy. Nephrol. Dial. Transplant. 2008;23(5):1608–1614. doi: 10.1093/ndt/gfm781. [DOI] [PubMed] [Google Scholar]
- 97.Cheng M., Ning K. Stereotypes about enterotype: the old and new ideas. Dev. Reprod. Biol. 2019;17(1):4–12. doi: 10.1016/j.gpb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arumugam M., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bulygin I., et al. 2021. Absence of Enterotypes in the Human Gut Microbiomes Reanalyzed with Non-linear Dimensionality Reduction Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gorvitovskaia A., Holmes S.P., Huse S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4:15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Christensen L., et al. Microbial enterotypes beyond genus level: Bacteroides species as a predictive biomarker for weight change upon controlled intervention with arabinoxylan oligosaccharides in overweight subjects. Gut Microb. 2020;12(1) doi: 10.1080/19490976.2020.1847627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song L., Zhang L., Fang X. Characterizing enterotypes in human metagenomics: a viral perspective. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.740990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He J.C., et al. Systems biology of kidney diseases. Kidney Int. 2012;81(1):22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cisek K., et al. The application of multi-omics and systems biology to identify therapeutic targets in chronic kidney disease. Nephrol. Dial. Transplant. 2016;31(12):2003–2011. doi: 10.1093/ndt/gfv364. [DOI] [PubMed] [Google Scholar]
- 105.Abedi M., et al. Systems biology and machine learning approaches identify drug targets in diabetic nephropathy. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-02282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miao H., et al. 2022. Membranous Nephropathy: Systems Biology-Based Novel Mechanism and Traditional Chinese Medicine Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mercado-Lubo R., McCormick B.A. The interaction of gut microbes with host ABC transporters. Gut Microb. 2010;1(5):301–306. doi: 10.4161/gmic.1.5.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bush K.T., et al. The drug transporter OAT3 (SLC22A8) and endogenous metabolite communication via the gut-liver-kidney axis. J. Biol. Chem. 2017;292(38):15789–15803. doi: 10.1074/jbc.M117.796516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosenthal S.B., Bush K.T., Nigam S.K. A network of SLC and ABC transporter and DME genes involved in Remote sensing and signaling in the gut-liver-kidney Axis. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-47798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu W., Bush K.T., Nigam S.K. Key role for the organic anion transporters, OAT1 and OAT3, in the in vivo handling of uremic toxins and solutes. Sci. Rep. 2017;7(1):4939. doi: 10.1038/s41598-017-04949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Misheva M., Ilott N.E., McCullagh J.S.O. Recent advances and future directions in microbiome metabolomics. Current Opinion in Endocrine and Metabolic Research. 2021;20 [Google Scholar]
- 112.Yuan C., et al. Mucosal microbiota and metabolome along the intestinal tract reveal a location-specific relationship. mSystems. 2020;5(3) doi: 10.1128/mSystems.00055-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yuan C., et al. Mucosal microbiota and metabolome along the intestinal tract reveal a location-specific relationship. mSystems. 2020;5(3) doi: 10.1128/mSystems.00055-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Q., et al. The role of gut microbiota and microbiota-related serum metabolites in the progression of diabetic kidney disease. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.757508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao J., et al. Magnesium lithospermate B improves the gut microbiome and bile acid metabolic profiles in a mouse model of diabetic nephropathy. Acta Pharmacol. Sin. 2019;40(4):507–513. doi: 10.1038/s41401-018-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang B., et al. Characteristics of serum metabolites and gut microbiota in diabetic kidney disease. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.872988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee-Sarwar K.A., et al. Metabolome–microbiome crosstalk and human disease. Metabolites. 2020;10(5):181. doi: 10.3390/metabo10050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee-Sarwar K.A., et al. Metabolome-microbiome crosstalk and human disease. Metabolites. 2020;10(5) doi: 10.3390/metabo10050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu C., et al. Interaction between plasma metabolomics and intestinal microbiome in db/db mouse, an animal model for study of type 2 diabetes and diabetic kidney disease. Metabolites. 2022;12(9) doi: 10.3390/metabo12090775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ji C., et al. Rhubarb enema decreases circulating trimethylamine N-oxide level and improves renal fibrosis accompanied with gut microbiota change in chronic kidney disease rats. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.780924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mo Y., et al. Microbiome-metabolomics analysis reveals the protection mechanism of α-ketoacid on adenine-induced chronic kidney disease in rats. Front. Pharmacol. 2021:1129. doi: 10.3389/fphar.2021.657827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kikuchi K., et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat. Commun. 2019;10(1):1835. doi: 10.1038/s41467-019-09735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jernberg C., et al. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology (Read.) 2010;156(Pt 11):3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 124.Li X., et al. Integrative metagenomic and metabolomic analyses reveal the role of gut microbiota in antibody-mediated renal allograft rejection. J. Transl. Med. 2022;20(1):614. doi: 10.1186/s12967-022-03825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen M.X., et al. Metabolome analysis for investigating host-gut microbiota interactions. J. Formos. Med. Assoc. 2019;118(Suppl 1):S10–s22. doi: 10.1016/j.jfma.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 126.Vernocchi P., Del Chierico F., Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kapoor R., Ladak S., Gomase V. MALDI-TOF based Metabolomic approach. Int. J. Genet. 2009;1 [Google Scholar]
- 128.Aguiar-Pulido V., et al. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis:supplementary issue: bioinformatics methods and applications for big metagenomics data. Evol. Bioinf. Online. 2016;12s1 doi: 10.4137/EBO.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Escobar-Zepeda A., Vera-Ponce de León A., Sanchez-Flores A. The road to metagenomics: from microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2015;6:348. doi: 10.3389/fgene.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Altman T., et al. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinf. 2013;14:112. doi: 10.1186/1471-2105-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zierer J., et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018;50(6):790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Letertre M.P.M., Giraudeau P., de Tullio P. Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: current challenges and perspectives. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.698337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao Y.Y., Vaziri N.D., Lin R.C. Lipidomics: new insight into kidney disease. Adv. Clin. Chem. 2015;68:153–175. doi: 10.1016/bs.acc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 134.Ma Y.-r., et al. An LC-MS/MS analytical method for the determination of uremic toxins in patients with end-stage renal disease. J. Pharmaceut. Biomed. Anal. 2020;191 doi: 10.1016/j.jpba.2020.113551. [DOI] [PubMed] [Google Scholar]
- 135.Grebe S.K., Singh R.J. LC-MS/MS in the clinical laboratory - where to from here? Clin. Biochem. Rev. 2011;32(1):5–31. [PMC free article] [PubMed] [Google Scholar]
- 136.Jia L., et al. Efficacy of probiotics supplementation on chronic kidney disease: a systematic review and meta-analysis. Kidney Blood Press. Res. 2018;43(5):1623–1635. doi: 10.1159/000494677. [DOI] [PubMed] [Google Scholar]
- 137.Carabotti M., et al. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- 138.Koppe L., Mafra D., Fouque D. Probiotics and chronic kidney disease. Kidney Int. 2015;88(5):958–966. doi: 10.1038/ki.2015.255. [DOI] [PubMed] [Google Scholar]
- 139.van Zyl W.F., Deane S.M., Dicks L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microb. 2020;12(1) doi: 10.1080/19490976.2020.1831339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yan F., Polk D.B. Probiotics and immune health. Curr. Opin. Gastroenterol. 2011;27(6):496–501. doi: 10.1097/MOG.0b013e32834baa4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaur I.P., Chopra K., Saini A. Probiotics: potential pharmaceutical applications. Eur. J. Pharmaceut. Sci. 2002;15(1):1–9. doi: 10.1016/s0928-0987(01)00209-3. [DOI] [PubMed] [Google Scholar]
- 142.Parvez S., et al. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006;100(6):1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 143.Stavropoulou E., et al. Focus on the gut-kidney Axis in health and disease. Front. Med. 2020;7 doi: 10.3389/fmed.2020.620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lu P.H., et al. The therapeutic strategies for uremic toxins control in chronic kidney disease. Toxins. 2021;13(8) doi: 10.3390/toxins13080573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Filippis F., Esposito A., Ercolini D. Outlook on next-generation probiotics from the human gut. Cell. Mol. Life Sci. 2022;79(2):76. doi: 10.1007/s00018-021-04080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Villar-García J., et al. Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: a double-blind, randomised, placebo-controlled trial. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0173802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baral K.C., et al. Advancements in the pharmaceutical applications of probiotics: dosage forms and formulation technology. Int. J. Nanomed. 2021;16:7535–7556. doi: 10.2147/IJN.S337427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.