Summary
Polypharmacology aids in the identification of multiple protein targets involved in disease pathology and selecting appropriate therapeutic compounds interacting with protein targets. Here, we present a protocol to identify the targets involved in obesity-linked diabetes and suitable phytocompounds to bind with the identified target. We describe steps to install and use softwares for identifying several protein targets by linking multiple diseases. This protocol allows the use of therapeutic compounds of both phytochemical and synthetic origins.
For complete details on the use and execution of this protocol, please refer to Martiz et al.,1 and Maradesha et al.2
Subject areas: Bioinformatics, High Throughput Screening, Protein Biochemistry, Structural Biology
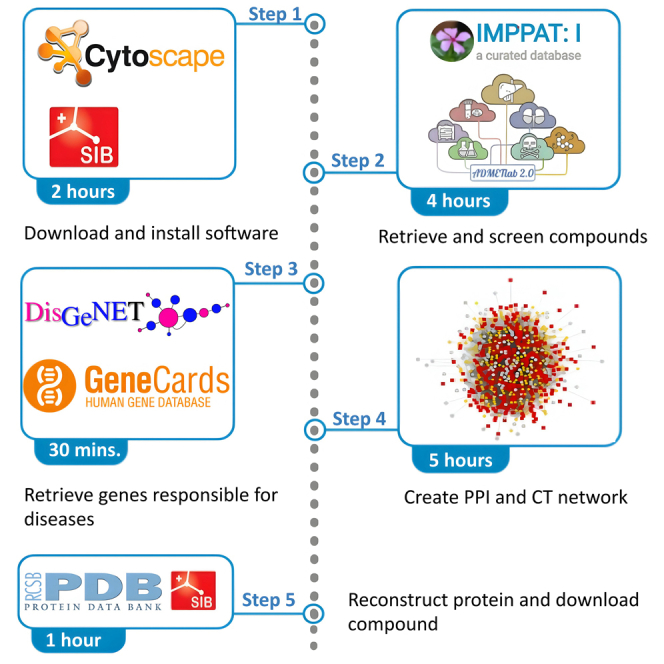
Graphical abstract
Highlights
-
•
Protocol to identify multiple protein targets belonging to different diseases
-
•
Steps to build a network of pathogenic proteins and selecting the best targets
-
•
Construction of a protein-compound network to identify the best therapeutic compounds
-
•
Exploration of multiple protein targets and their suitable binding compounds
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Polypharmacology aids in the identification of multiple protein targets involved in disease pathology and selecting appropriate therapeutic compounds interacting with protein targets. Here, we present a protocol to identify the targets involved in obesity-linked diabetes and suitable phytocompounds to bind with the identified target. We describe steps for installing and using software, using these tools to identify several protein targets by linking multiple diseases. This protocol allows the use of therapeutic compounds of both phytochemical and synthetic origins.
Before you begin
In this protocol, we detail the process of obtaining a high-confidence target protein and a suitable therapeutic compound to bind to the identified protein target. We choose 2 lifestyle-based disorders that are closely linked – obesity and diabetes, treatment by the phytochemicals of the Camellia sinensis plant.
-
1.
Initially, obtain the phytochemicals from IMPPAT website, download the structures in 3D SDF format and subject to ADMET analysis using ADMETlab 2.0 software.
-
2.
Further, the identification of protein targets of the phytochemicals using SWISS target prediction, enlisting genes relating to disease using DisGeNET database.
-
3.
Subsequently, construction of CTD network using Cytoscape 3.9.1 displaying intersecting genes and PPI network construction using STRING database.
-
4.
Further, import PPI network from STRING database, visualize, and identify final target gene of interest using Cytoscape.
-
5.
Identify final protein target, retrieve from the RCSB PDB database, Model verification using Swiss PDB Viewer, retrieval of phytocompounds from PubChem.
However, one can choose multiple diseases/disorders and can perform target identification. Regardless of their origin, synthesized, semi-synthesized, and natural compounds can also be used to find the best therapeutic compound using this protocol. The purpose of this protocol is to identify common target/s related to multiple health aberrations, which can be bound with the best suitable chemical compounds to interact. This is a complete in silico protocol and needs to be done using a laptop/PC with good configurations.
Hardware and software applications
This in silico pipeline is designed to be run on Windows OS. Basic requirements: Authentic Windows OS 7, 8, 10, 11 with 4 GB of RAM and 512 GB of ROM. Advanced requirements: Windows 64-bit 10 or 11 with 16 GB of RAM and 1 TB of ROM. Cytoscape version 3.9.1 is essential, Download Swiss PDB viewer using the website mentioned in the key resources table. Use ADMETlab 2.0 web tool for ADMET. For gene data retrieving and construction of PPI network, use DisGeNET, Gene Cards and STRING web tools respectively. To get the compound structure in SMILES format, use PubChem database. To construct Venn diagrams Bioinformatics & Evolutionary Genomics use Venny web tools.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| Cytoscape | Cytoscape Team | https://cytoscape.org/development_team.html |
| SWISS PDB Viewer | SIB Swiss Institute of Bioinformatics | https://spdbv.unil.ch/refs.html |
| Other | ||
| DisGenNET | Integrative Biomedical Informatics Group | https://www.disgenet.org/ |
| GeneCards | GeneCards Suit Project | https://www.genecards.org |
| ADMETlab 2.0 | CBDD Team, Central South University, China | https://admetmesh.scbdd.com/ |
| PubChem | National Center for Biotechnology Information (NCBI) | https://pubchem.ncbi.nlm.nih.gov/ |
| Venny | Juan Carlos Oliveros, Spanish National Biotechnology Centre (CNB-CSIC) | https://bioinfogp.cnb.csic.es/tools/venny/ |
| Windows 11 64-bit, 11th Gen Intel(R) Core (TM) i7-1165G7 @ 2.80GHz 2.80 GHz, 16 GB, RAM, 512 GB ROM. | – | – |
| IMPPAT | Institute of Mathematical Sciences, Chennai | https://cb.imsc.res.in/imppat/ |
| Swiss Target Prediction | SIB Swiss Institute of Bioinformatics | http://www.swisstargetprediction.ch/ |
Step-by-step method details
Part 1. Download and install Cytoscape and Swiss PDB viewer softwares
Timing: 2 h
This section accomplishes the procedure for downloading and installing both Cytoscape and Swiss PDB viewer softwares.
Cytoscape is an open-source software platform for visualizing molecular interaction networks and biological pathways and fusing these networks with annotations, gene expression profiles, and other state data.
-
1.
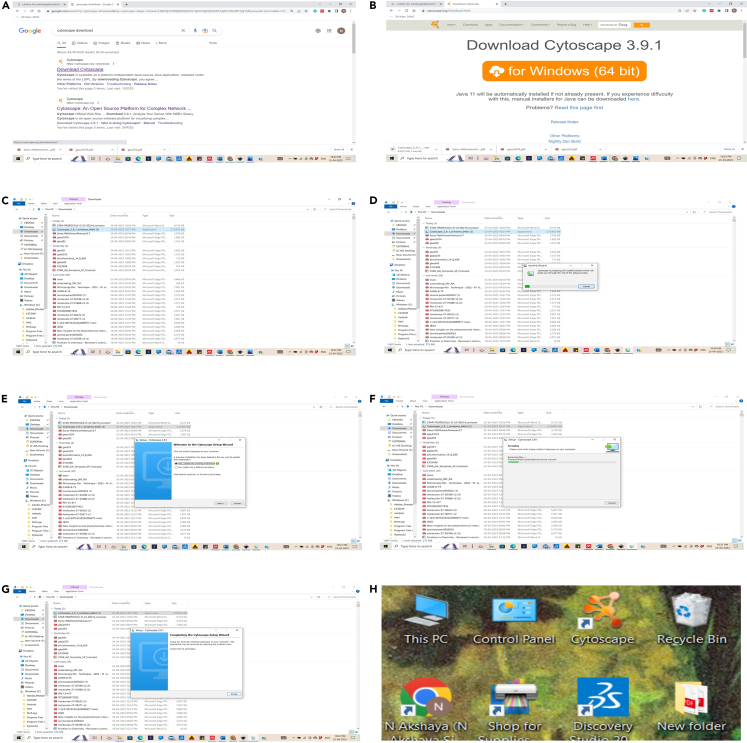
Download it for free using this link (https://cytoscape.org/download.html).
-
2.
Cytoscape is a Java application verified to run on the Linux, Windows, and Mac OS X platforms. Although not officially supported, other UNIX platforms such as Solaris or FreeBSD may run Cytoscape if Java version 8 is available for the platform.
Note: Cytoscape requires Java 11. We recommend running the latest version of Cytoscape (v 3.9.1) with a 64-bit Java Runtime Environment (JRE) such as OpenJDK 11.
-
3.Install Cytoscape.
-
a.Downloading and Installing.There are several options for downloading and installing Cytoscape. Refer to the download page at the http://cytoscape.orgwebsite for all options.
-
i.Automatic installation packages exist for Windows, Mac OS X, and Linux platforms – best for most users.
-
ii.User can install Cytoscape from a compressed archive distribution.
-
iii.User can build Cytoscape from the source code. You can check out the latest and greatest software from our Git repository (https://github.com/cytoscape/cytoscape). Figure 1 explains the steps involved in downloading Cytoscape.
-
i.
-
a.
-
4.
A fundamental set of features for data integration, analysis, and visualization are offered by the Cytoscape core distribution.
-
5.
Applications with additional functionalities are accessible (formerly called Plugins).
-
6.
These include apps for network and molecular profiling analysis, new layouts, extra file format support, scripting, and database connectivity.
-
7.
Anybody can create by utilizing the Cytoscape open API based on JavaTM technology, and community development of applications are encouraged.
-
8.
Download most of the applications for free from the Cytoscape App Store.
-
9.
Swiss-PDB Viewer (also known as Deep View) is a program that offers a user-friendly interface that permits the simultaneous analysis of many proteins (https://spdbv.unil.ch/disclaim.html).
-
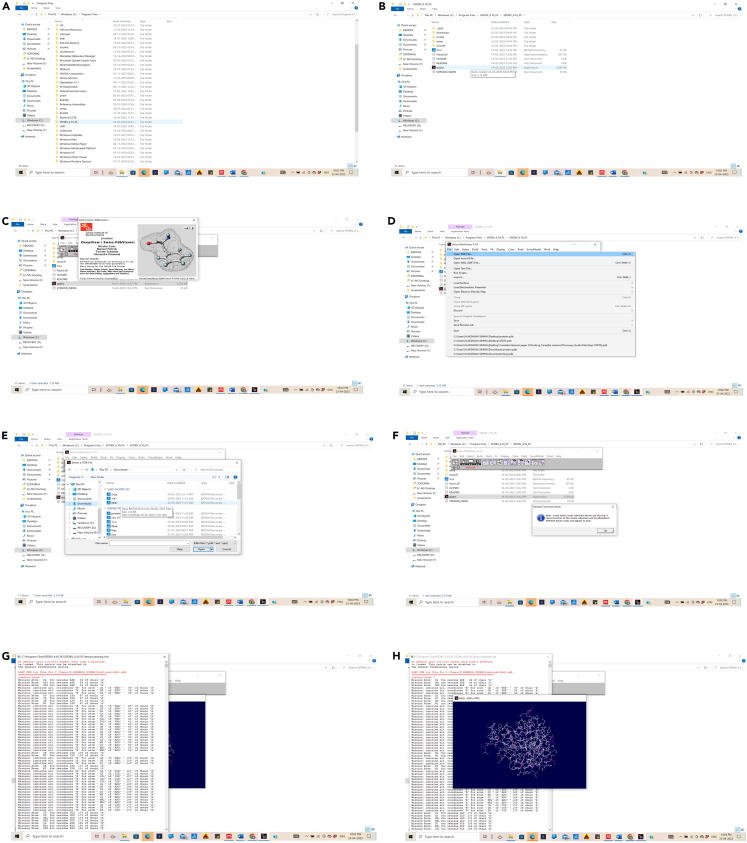
10.Download deepview from http://www.expasy.org/spdbv/ or any of the mirror sites mentioned.
-
a.Download & install Swiss-PdbViewer. Distribution of deepview either as a self-extractable archive (.exe) or as a zip archive (.zip): · (.exe): Double click the file. By default, a directory called spdbv will be created in your C: drive.
-
b.Move this directory where the user require on hard drive. Be sure to maintain the directory content.
-
c.To launch Deep View, double click the application icon (). · (.zip): Expand the file using WinZip. In this case, be sure to configure WinZip to keep the directory hierarchy. Figure 2 explains steps to install SWISS PDB database.
-
a.
-
11.
Compare the proteins' active sites or other important components and structural alignments can be inferred by superimposing them.
-
12.
The simple graphic and menu interface makes it simple to find amino acid changes, H- bonds, angles, and distances between atoms.
-
13.
Swiss-PDB Viewer offers several tools to build into the density and can read electron density maps as well.
-
14.
Furthermore, integration of different modelling tools, and alteration of residues. In order to create beautiful ray-traced quality photos, create POV-Ray sceneries from the present perspective.
Figure 1.
Explanation of the installation procedure of Cytoscape 3.9.1
Figure 2.
Explanation of the protein structure visualization using SWISSPDB viewer
Part 2. Enlisting and downloading of phytochemicals from IMPPAT website in 3D SDF format and virtual screening using ADMET properties
Timing: 4 h
This section accomplishes the complete retrieval of IMPPAT phytochemicals and their ADMET evaluation.1,2
-
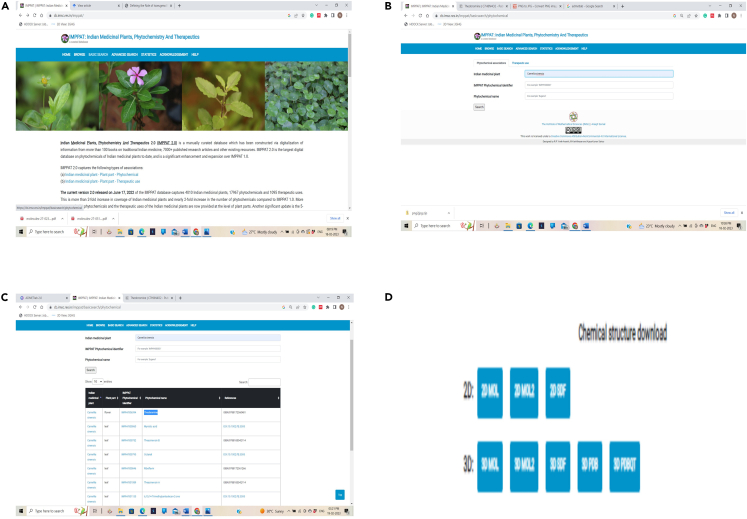
15.Open IMPPAT database (https://cb.imsc.res.in/imppat/)3 in browser.
-
a.Click on BASIC RESEARCH option.
-
b.Under the PHYTOCHEMICAL ASSOCIATIONS option, enter name of the plant i.e., Camellia sinensis and click on SEARCH option.
-
c.In the list, select the phytochemical and click on it. Out of all the different formats available, click on 3D SDF and download the file.
-
d.Save all the phytochemicals in a separate folder by assigning a suitable name. Figure 3 explains the downloading protocol of phytochemicals from IMPPAT.
-
a.
-
16.
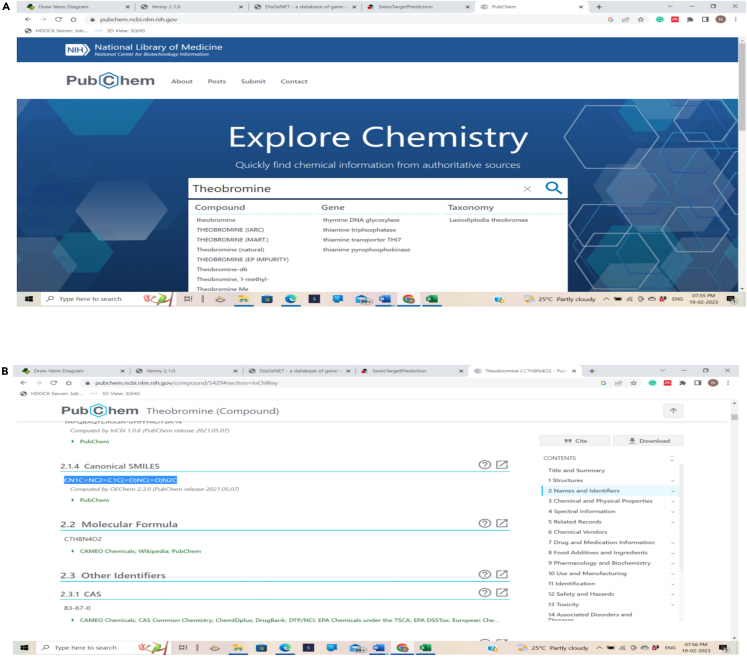
Copy the name of the phytochemical, i.e., Theobromine and paste in PubChem search bar.
-
17.
Open the details of the phytochemical, select the CANONICAL SMILES, copy, and paste in ADMET lab evaluation https://admetmesh.scbdd.com/service/evaluation/index window and click on submit option.4
-
18.
Figure 4 explains obtaining the compound structure in SMILES format using PubChem, whereas Figure 5 explains finding the bioavailability of phytochemicals using ADMETlab 2.0.1,2
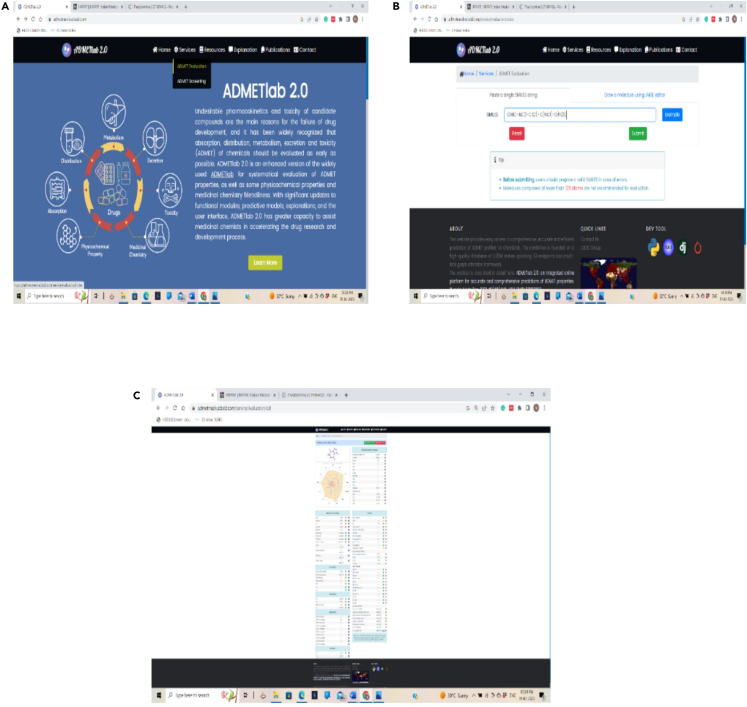
Note: Analyze the phytochemical for the following criterion – oral bioavailability (OB ≥ 30%), blood–brain barrier (BBB), Drug half-life (HL < 3 h), Lipinski’s RO5 (rule of five), Intestinal epithelial permeability Caco-2 cells, drug-induced liver injury (DILI), Clearness (CL > 15 mL/min/kg), molecular weight (MW 100–600), hydrogen bond donors (HbD 0–7), hydrogen bond acceptors (HbA 0–12), topological polar surface area (TPSA), and pan assay interference compounds (PAINS) in alerts.
Figure 3.
Retrieval of phytochemical compounds in SDF format using IMPPAT database
(A–D) (A) IMPPAT database Web page, (B) Search results of compounds, and (C) List of Compounds and (D) Compound available in 3D SDF format.
Figure 4.
Obtaining compounds in SMILES format using PubChem database
(A and B) (A) Pasting the compound name in PubChem, (B) Copying the SMILES format from the PubChem database.
Figure 5.
Virtual screening of compounds based on their ADMET properties
(A–C) (A) ADMETlab 2.0 evaluation, (B) Pasting of SMILES format, and (C) Results with ADMET properties of a compound.
Part 3. Identification of protein targets using SWISS target prediction, enlisting genes relating to disease using DisGeNET database
Timing: 30 min
This section accomplishes prediction of protein targets of the IMPPAT phytochemicals, retrieval of genes related to disease.1,2
-
19.
Prepare 3 network tables of a) Botanical; b) Target and c) Disease genes using Microsoft Excel.
-
20.
Prepare disease network table by downloading the both the disease genes from DisGeNET database (https://www.disgenet.org/)5 and Target network table using SwissTargetPrediction (http://www.swisstargetprediction.ch/).6
Figure 6 explains the preparation of disease network by retrieving all the disease genes. Figure 7 explains the prediction of targets of the phytochemicals.
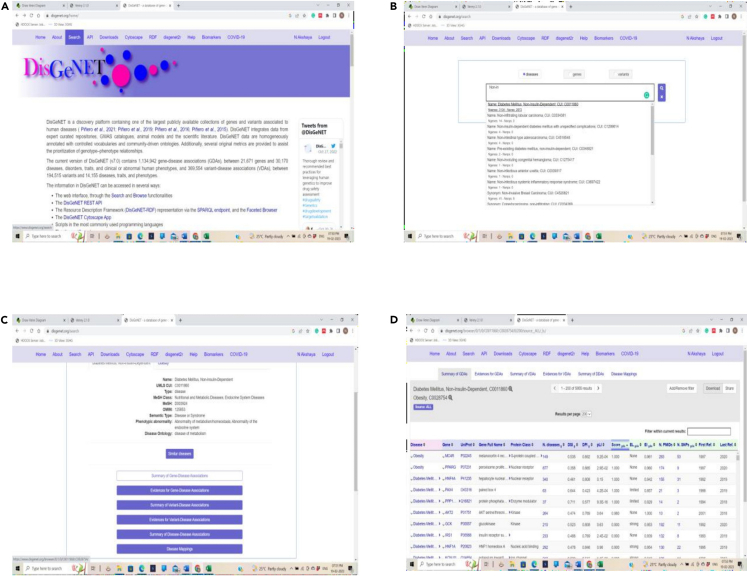
Note: You can also use Gene Cards database (https://www.genecards.org/)7 instead of DisGeNET. However, usage of both databases is also recommended.
-
21.Open https://bioinfogp.cnb.csic.es/tools/venny/ an online tool for creation of Venn diagram.
-
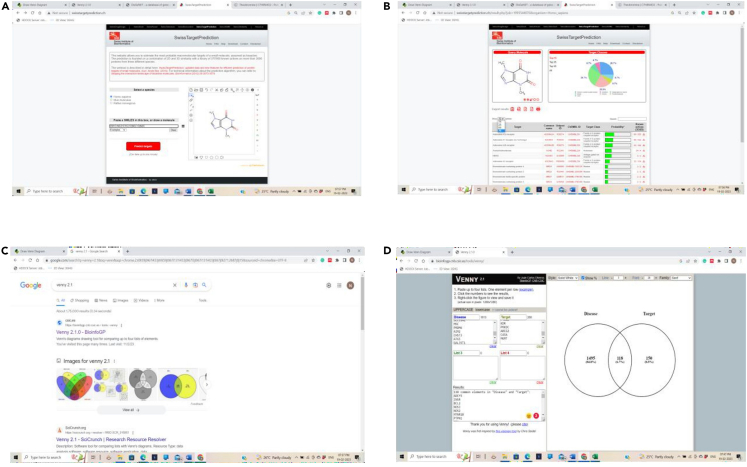
a.First add T2DM (Type 2 Diabetes Mellitus) genes in list 1 and then add obesity genes in list 2.Note: Automatically the web tool creates the Venn diagram.
-
b.Click on the intersecting genes which separately gives the list of intersecting genes between two diseases.
-
c.Then create second Venn diagram
- i.
-
a.
Figure 6.
Obtaining genes from DisGeNET database
(A–D) (A) DisGeNET search bar, (B) DisGeNET showing list of disease, (C) Selection of a specific disease, and (D) Obtaining all the genes for a disease.
Figure 7.
Prediction of target for the selected compounds
(A–D) (A) SWISS Target prediction, (B) SWISS target prediction predicting the targets of the phytochemicals, (C) Venny web-tool, and (D) Identification and enlisting of the intersecting targets using Venny web tool.
Part 4. Construction of CTD network using cytoscape 3.9.1 displaying intersecting genes and PPI network construction using STRING database
Timing: 4 h
This section accomplishes the construction of CTD network and PPI network using Cytoscape and STRING database respectively.1,2
-
22.
Open Cytoscape 3.9.1 or 3.8 (https://cytoscape.org/)8 and above version using the browser.
-
23.
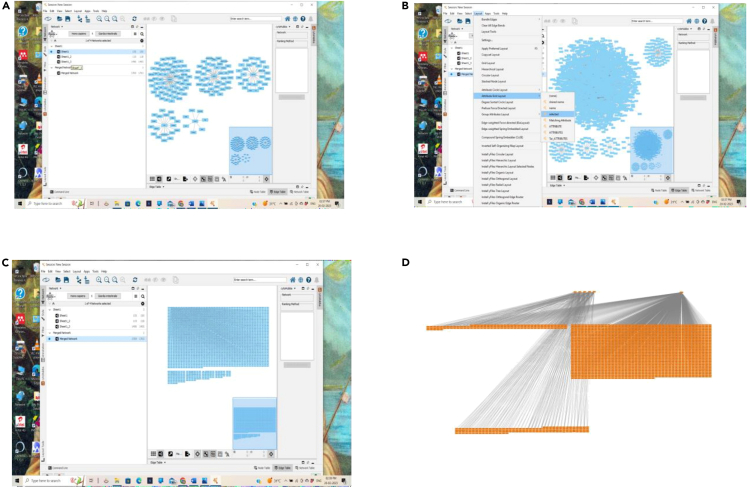
Import the network tables – Targets of phytochemicals (150), disease intersecting genes (1495) and final intersecting targets (118). Further, merge the network with union, which automatically creates the big network.
-
24.
By using filters, find out 118 intersecting genes, set the layout and separate the network, further find out the 150 targets of phytochemicals, set the layout and separate the network and remaining network with the 1495 disease target network. Figure 8 explains the steps of CTD network construction using Cytoscape.
Note: The output of step 6 is known as CTD network.
-
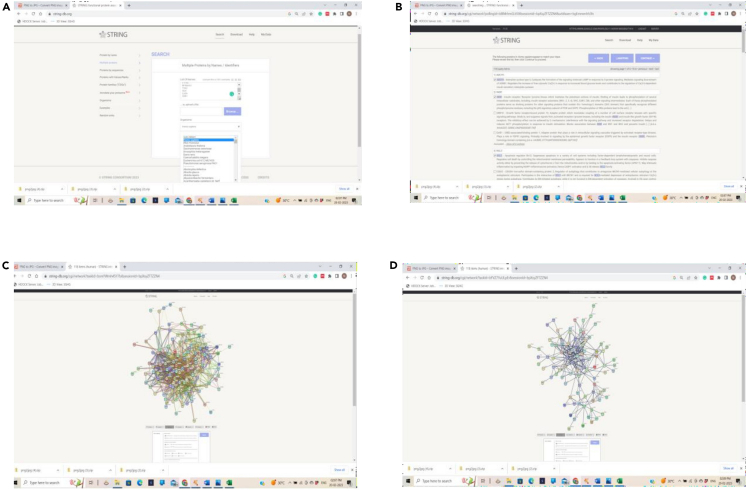
25.Open STRING database (https://string-db.org/).9
-
a.Upload intersecting targets, select multiple protein filter with organism as “Homo sapiens” and select SUBMIT option.
-
b.Further, Click on CONTINUE option, under settings apply different filters as full STRING network, confidence with highest confidence level 0.900, interactive network and hide disconnected nodes in the network for the first PPI network and click on UPDATE option.
-
a.
Figure 8.
Construction of CTD network using Cytoscape
(A–D) (A) Protein targets of compounds, (B) Specific protein targets related with disease, (C) Merged network of protein targets, intersecting targets, and disease targets, and (D) Final CTD network emerged as a common network from all the networks mentioned in Figure 7C.
Part 5. Importing of PPI network from STRING database, visualization, and identification of final target gene of interest using cytoscape
Timing: 1 h
This section accomplishes the identification of final target gene of interest from PPI network.1,2
-
26.
Open Cytoscape of 3.8 (https://cytoscape.org/) and above version using the browser for second time.
-
27.
Select the option SEND NETWORK TO CYTOSCAPE which is below the PPI network.
-
28.
Now visualize the STRING network (PPI network), analyze the network, and note down the result.
-
29.
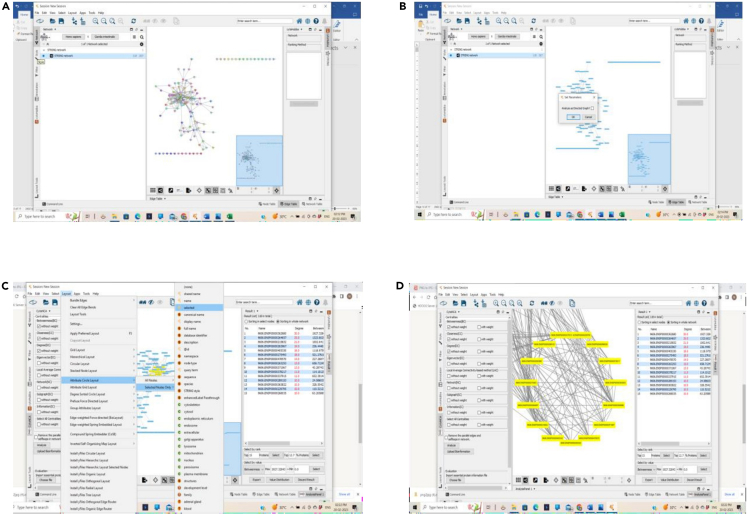
Open CytoNCA from applications menu of the Cytoscape and analyze the top 15 targets of the STRING network by selecting 3 topological characteristics Betweenness, Closeness and Degree and save the images of the above 3 topological characteristics.
Note: Repeat the step 4 for other 2 topological characteristics i.e., closeness and degree.
-
30.
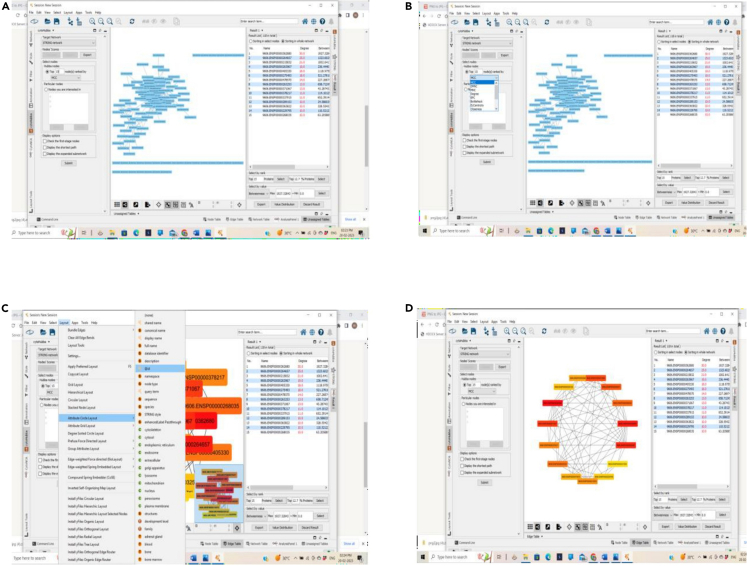
Now open Cytohubba and generate subnetworks by analyzing top 15 nodes using various algorithms such as MCC, MNC, EPC, DMNC, Eccentricity, Closeness, bottleneck and betweenness.
Note: Repeat step 5 for the generation of subnetworks using other algorithms after MCC.
-
31.
Identify the final target /targets by analyzing the 3 clusters and all the subnetworks.1,2 Figure 9 explains the steps of creating a PPI network using the STRING database and importing it to Cytoscape.
-
32.
Analysis of the constructed PPI network using CytoNCA and CytoHubba has been given in Figures 10 and 11, respectively.
Note: Step 4 in the graphical abstract corresponds to Part 4 and Part 5 of the manuscript.
Figure 9.
Construction of PPI network using STRING database
(A) Uploading of intersecting (disease-compound) gene targets to the STRING, (B) Processing of the genes to the STRING, (C) Crude network constructed, and (D) Actual PPI network constructed by applying filters.
Figure 10.
Analysis of the constructed PPI network using CytoNCA
(A–D) (A) Importing a constructed network from STRING to Cytoscape, (B) Analysis of constructed network, (C) Identification of network by CytoNCA, and (D) Extraction of clusters from CytoNCA.
Figure 11.
Analysis of the constructed PPI network using CytoHubba
(A–D) (A) Initiation of analysis using CytoHubba, (B) Stages of Analysis – top 15 modes, (C) Identification of clusters using various algorithms of CytoHubba, and (D) core key subnetwork of top 15 nodes analyzed by CytoHubba – Color intensity indicates the degree of final targets.
Part 6. Identification and retrieval of the final protein target from the RCSB PDB database, model verification using Swiss PDB viewer, retrieval of phytocompounds from PubChem
Timing:1h
This section accomplishes the identification and retrieval of final protein target (structural biology of protein target) and phytochemicals. The RCSB PDB database also provides insights into the target protein biochemistry.10,11
-
33.
Search the protein name and identify the correct protein using RCSB PDB database.
-
34.
Download the protein structure from RCSB PDB database in PDB format.
-
35.
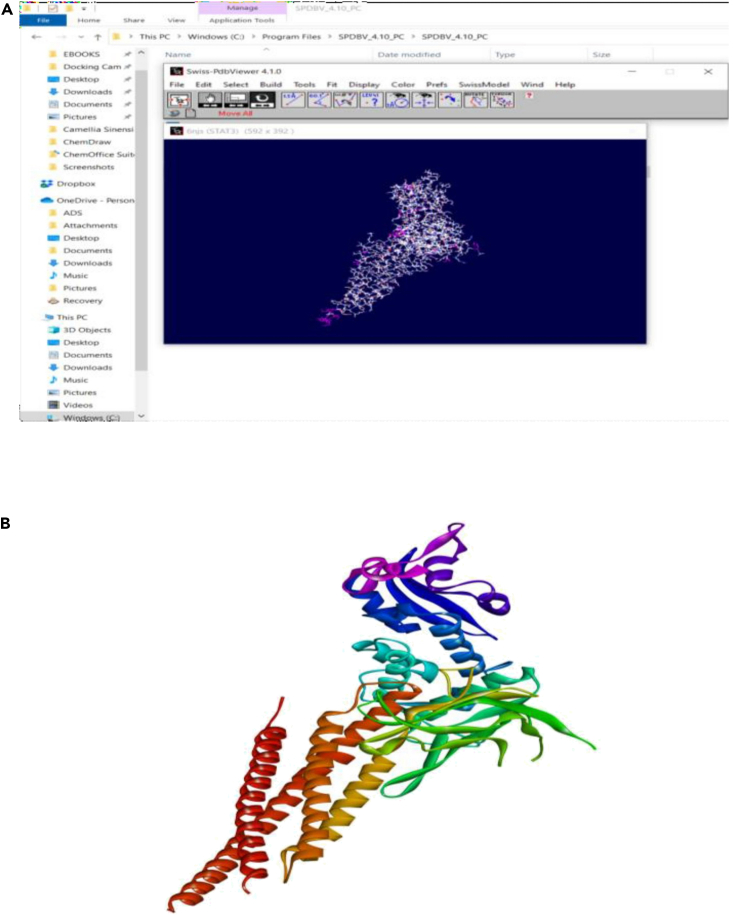
To add missing amino acid residues, open the downloaded PDB in Swiss PDB viewer software.
-
36.
Once the model is completely built, save it in the PDB format. The protein is now ready for further in silico studies like molecular docking. Figure 12 explains the procedure to identify and retrieve the final protein target.
-
37.
Similarly obtain the top phytochemicals with highest bioavailability using PubChem database in 3D SDF format. These phytochemicals act as ligands for further in silico studies. Figure 13 explains the procedure to identify and retrieve the phytocompounds from PubChem.
-
38.
The entire protocol has been summarized in the given Methods item (Methods S1: Workflow diagram of the entire protocol to identify multiple protein targets and therapeutic compounds using an in silico polypharmacological approach, relating to the entire protocol from Part 1 to Part 6).
Figure 12.
Identification and reconstruction of correct protein model
(A and B) (A) Reconstruction of the protein model using SWISS PDB Viewer – the missing amino acids appear in pink, (B) Completely built protein model ready for further analyses (PDB ID: 6NJS in this study).
Figure 13.
Selection of best compounds in ADMET evaluation that are suitable for interact with the protein model 6NJS
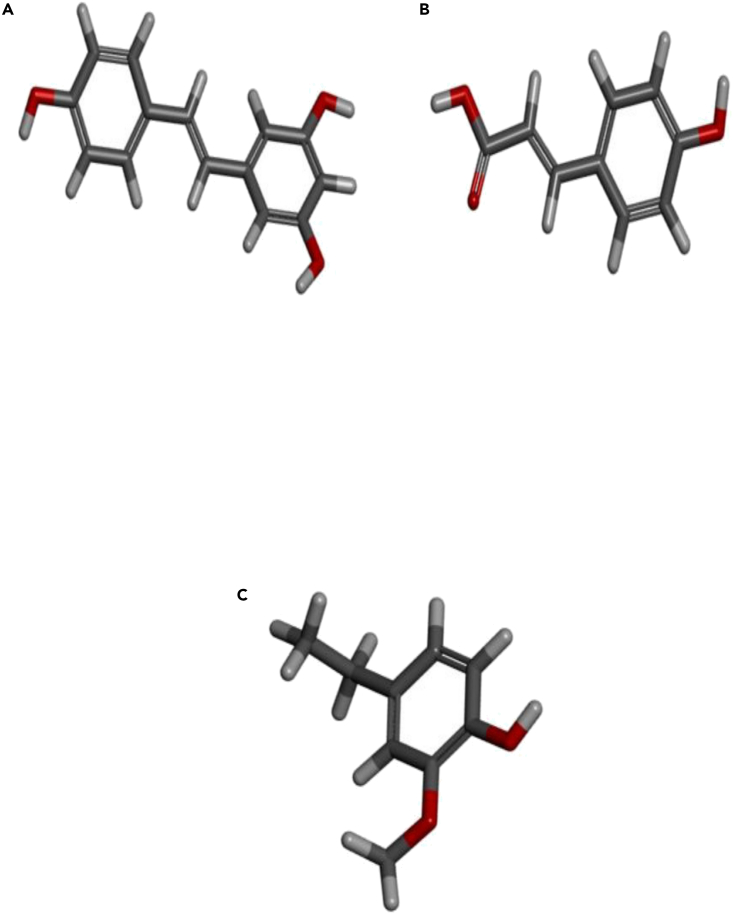
(A–C) (A) Resveratrol, (B) 4-hydroxy cinnamic acid, and (C) 4-ethyl-2-methoxy phenol.
Expected outcomes
This protocol helps in the identification of multiple targets related with more than one disease or disorders. It also provides the best therapeutic targets to bind with the target protein/s. One can consider taking more than 2 or 3 protein targets which are common with the selected number of diseases or disorders. Also, more than 1 therapeutic compound can be selected for further analyses like molecular docking and molecular dynamics simulation. Here, we have selected only 1 protein target STAT3 (PDB ID: 6NJS) which is common for diabetes mellitus and obesity. We have selected 3 compounds based on their bioavailability rank - Resveratrol, 4-hydroxy cinnamic acid, and 4-ethyl-2-methoxy phenol. These proteins and compounds could be subjected to in vitro and in vivo studies if in silico results are acceptable. In the end, we can conclude that this polypharmacology-based protocol helps largely in the discovery of novel protein targets as well as their best therapeutic compounds.
Limitations
The procedure followed in the protocol needs significant amount of time yet will yield promising targets related to multiple health aberrations. However, there are chances of miscalculations/errors during retrieval of genes, obtaining common genes using Venn diagrams, merging 2 or more networks, etc. Careful and observant handling of resources and following the protocol could get the best results possible. Networks can be generated only if the network table is created properly. Proper way of creating network table is by using appropriate spacings and font size.
Troubleshooting
Problem 1
Improper Network generation (related to step 1 of Part 3).
Potential solution
-
•
To generate the proper network, initially the network table should be prepared appropriately. Proper way of creating network table is by using appropriate spacings and font size.
Problem 2
Exporting of PPI network from STRING tool to Cytoscape (related to step 2 of part 4).
Potential solution
-
•
Despite of using updated version of Cytoscape, STRING and high-speed internet, during exporting of network from STRING to Cytoscape, an error message occurs as “confirm whether you are using updated version of Cytoscape 3.8 + and above. This can be resolved by exiting from STRING and Cytoscape softwares and repeating the steps of generating the STRING PPI network and exporting procedure and closing the other tabs and programs running on the system.
Problem 3
Troubleshooting with respect to Cytoscape tool (step 3 and 4 of Part 1).
Potential solutions to various issues
Installation issues
-
•
If you encounter problems during the installation process, make sure that your system meets the minimum requirements for running Cytoscape. You can also try downloading the latest version of the software from the Cytoscape website and reinstalling it.
Network loading issues
-
•
If you are having trouble loading a network into Cytoscape, make sure that the file format is supported by the software. Cytoscape supports many different file formats, including GraphML, XGMML, SIF, and CSV. You can also try opening the file in a text editor to check for any errors or formatting issues.
Performance issues
-
•
If Cytoscape is running slowly or freezing, try closing any other programs that may be using up system resources. You can also try adjusting the settings in the Cytoscape preferences menu to optimize performance.
Java issues
-
•
Cytoscape requires Java to run, so if you are encountering errors related to Java, make sure that you have the latest version installed and that your system meets the Java requirements for Cytoscape.
Network layout issues
-
•
If you are having trouble with the layout of your network, try using different layout algorithms or adjusting the settings for the current layout algorithm. You can also try manually moving nodes and edges to improve the visualization
Problem 4
Due to the scarcity of databases with open access, target identification often relies on one or a small number of databases. This occasionally produces unfinished results. Additionally, there can be undiscovered new targets that are a component of the bioactive' mechanism of action. Multiple databases should be taken into consideration for target identification to address this network disparity.
Potential solution
-
•
This issue could also be resolved by integrating databases that perform related tasks. Furthermore, the network predictions will be supported by experimental confirmation of the target molecules utilizing investigations of protein-protein interactions or gene expression.8
Problem 5
Troubleshooting with respect to ADMET Lab 2.0 web tool (related to step 3 of Part 2).
The primary problem occurs with respect to ADMET lab tool is sometimes it consumes more time to open.
Potential solution
-
•
Resolved by closing the web tool and re-opening or reloading the page until it opens and make sure to close unwanted tabs and applications.
The secondary problem is displaying incomplete results after ADMET evaluation.
Potential solution
-
•
Resolved by re-loading the webtool and submitting the SMILES once again.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, [Ramith Ramu] (ramithramu@jssuni.edu.in).
Materials availability
This study did not generate any unique reagents/materials since this is a complete in silico-based protocol. All the required resources are mentioned in key resources table (KRT).
Data and code availability
Data generated using the procedures presented in this paper are published in Martiz et al. (2022),1 and Maradesha et al. (2023).2
Acknowledgments
The authors are thankful to JSS AHER for their kind support and necessary facilities. S.M.P. thanks JSS AHER, Mysore, India for awarding the student research fellowship (JSSAHER/REG/RES/JSSURF/29(2)/2020-21) dated 06.03.2023.
Author contributions
Conceptualization, R.R.; data analysis and method development, A.S.N., S.M.P.; original draft preparation and writing, A.S.N., A.C., S.M.P.; supervision and editing, S.A.M., R.R. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102440.
Contributor Information
A.M. Satish, Email: drsatisham@gmail.com.
Ramith Ramu, Email: ramith.gowda@gmail.com.
Supplemental information
References
- 1.Martiz R.M., Patil S.M., Abdulaziz M., Babalghith A., Al-Areefi M., Al-Ghorbani M., Mallappa Kumar J., Prasad A., Mysore Nagalingaswamy N.P., Ramu R. Defining the Role of Isoeugenol from Ocimum tenuiflorum against Diabetes Mellitus-Linked Alzheimer’s Disease through Network Pharmacology and Computational Methods. Molecules. 2022;27:2398. doi: 10.3390/molecules27082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maradesha T., Martiz R.M., Patil S.M., Prasad A., Babakr A.T., Silina E., Stupin V., Achar R.R., Ramu R. Integrated network pharmacology and molecular modeling approach for the discovery of novel potential MAPK3 inhibitors from whole green jackfruit flour targeting obesity-linked diabetes mellitus. PLoS One. 2023;18 doi: 10.1371/journal.pone.0280847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanraj K., Karthikeyan B.S., Vivek-Ananth R.P., Chand R.P.B., Aparna S.R., Mangalapandi P., Samal A. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics. Sci. Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., García-García J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42:W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 8.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 11.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated using the procedures presented in this paper are published in Martiz et al. (2022),1 and Maradesha et al. (2023).2