Abstract
We have screened for proteins that interact with v-SNAREs of the late secretory pathway in the yeast Saccharomyces cerevisiae. A novel protein, designated Vsm1, binds tightly to the Snc2 v-SNARE in the two-hybrid system and can be coimmunoprecipitated with Snc1 or Snc2 from solubilized yeast cell extracts. Disruption of the VSM1 gene results in an increase of proteins secreted into the medium but does not affect the processing or secretion of invertase. In contrast, VSM1 overexpression in cells which bear a temperature-sensitive mutation in the Sec9 t-SNARE (sec9-4 cells) results in the accumulation of non-invertase-containing low-density secretory vesicles, inhibits cell growth and the secretion of proteins into the medium, and blocks rescue of the temperature-sensitive phenotype by SNC1 overexpression. Yet, VSM1 overexpression does not affect yeast bearing a sec9-7 allele which, in contrast to sec9-4, encodes a t-SNARE protein capable of forming a stable SNARE complex in vitro at restrictive temperatures. On the basis of these results, we propose that Vsm1 is a novel v-SNARE-interacting protein that appears to act as negative regulator of constitutive exocytosis. Moreover, this regulation appears specific to one of two parallel exocytic paths which are operant in yeast cells.
SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein [SNAP] receptor) proteins comprise distinct families of membrane-associated receptors that are components of the vesicle docking and fusion machinery in eukaryotes (62; reviewed in references 35 and 54). Evidence from evolutionarily divergent systems, e.g., from yeast to mammals, suggests that the role of SNAREs in protein transport is highly conserved (reviewed in references 7 and 21). SNAREs act throughout the secretory pathway to confer the trafficking of cargo-containing carrier vesicles and may also participate in compartmental organization. SNAREs found on vesicular compartments (v-SNAREs) are proposed to interact with specific cognate receptors (t-SNAREs) present on target compartments to form a prefusion SNARE complex. Thus, v-SNARE–t-SNARE (v-t SNARE) assembly confers vesicle docking; one of the first events leading to membrane fusion. Moreover, recent work has suggested that together v- and t-SNAREs may provide the minimal requirements necessary for conferring both membrane association and bilayer fusion, using a liposome-based in vitro assay (70). However, there is still some controversy surrounding the role of SNAREs in the fusion event itself (13, 65).
To confer specificity to protein transport, SNARE assembly leading to membrane fusion is expected to occur only on appropriate target membranes. Yet, the v- and t-SNAREs involved in Golgi and post-Golgi trafficking events passage through the early secretory pathway and are likely to reside together on identical endoplasmic reticulum (ER)- and Golgi-derived transport vesicles. Therefore, some mechanism(s) should prevent nonproductive SNARE partnering from occurring between opposing membranes early in the pathway. Likewise, it is reasonable to assume that other cellular mechanisms regulate specific v-t SNARE interactions in order to confer both temporal and spatial regulation of membrane docking and fusion, perhaps at the level of the target compartments themselves. Thus, certain constraints may prevent nonproductive SNARE partnering early in the pathway and, perhaps, ready SNAREs for assembly upon reaching their appropriate target membranes. Moreover, additional steps to remove these constraints may be prerequisites for docking and fusion to proceed in vivo. These steps should include dissociation of preformed SNARE complexes, as proposed earlier by Ungermann et al. (64), as well as the removal of other inhibitory constraints placed on the individual SNARE elements. Such constraints may include negative-acting SNARE regulatory proteins, which we have designated SNARE-masters (reviewed in reference 30). By definition, these regulators are expected to associate directly with specific v- or t-SNAREs and act to downregulate trafficking functions by modulating their entry into SNARE complexes. Thus, dissociation of SNARE regulators from SNAREs, or their inactivation, is expected to precede complex assembly and membrane fusion.
A conserved family of SNARE regulators which function in constitutive and regulated secretory systems is that of yeast Sec1 and its homologs found in higher organisms (1, 23, 26, 34, 47, 55). SEC1 was identified in the original sec mutant screen (45) and encodes a soluble protein that interacts with members of the Sso/syntaxin family of t-SNAREs (2, 23, 47, 48). Studies on mammalian Sec1 proteins have shown that they are not components of the SNARE complex and act to restrict SNARE partnering, probably by preventing t-SNAREs from assembling into binary complexes (24, 34, 48). In addition, studies in vivo have shown that the overexpression of rop1, a Drosophila homolog of Sec1, inhibits neurotransmitter release at the neuromuscular junction in larvae (60). Thus, Sec1-like molecules may act as potential SNARE-masters for the Sso/syntaxin family members by virtue of their ability to inhibit t-SNARE functioning.
A possible regulator of v-SNAREs in regulated exocytosis is synaptophysin, a membrane protein from synaptic vesicles (37, 71) which forms complexes with members of the synaptobrevin/VAMP (vesicle-associated membrane protein) family (10, 19, 69). Interestingly, these complexes were shown not to contain any of the known t-SNARE partners (19, 22, 69), suggesting that the synaptophysin-synaptobrevin interaction may prevent synaptobrevin from undergoing assembly into the ternary complex. The mechanism by which these proteins disassociate to allow for formation of the fusion complex remains unknown.
A large class of potential SNARE regulators consists of the Rab GTPases, which appear to confer SNARE complex assembly. Rabs have been suggested to regulate the specificity in membrane trafficking steps, due to their distinct subcellular localizations and interactions with SNARE components (reviewed in reference 46). Rabs have been implicated at the level of SNARE activation, and in the case of ER-Golgi transport in yeast, the Ypt1 GTPase has been proposed to bind directly to the Sed5 t-SNARE and to displace the Sec1 homolog, Sly1 (42). Genetic studies performed with yeast have demonstrated that overexpression of v- and t-SNAREs can suppress the temperature sensitivity of mutant Rab proteins that are specific to the transport step on which the SNAREs function (9, 16, 40). Thus, Rab proteins not only may regulate the fidelity of docking and fusion but could serve to dissociate proteins that act as negative regulators for SNARE complex formation (30). More recently, Rabs have been suggested to act in the tethering of vesicles to their acceptor compartments, prior to SNARE assembly (12, 65).
Another class of SNARE regulators includes the NSF/Sec18 and SNAP/Sec17 protein families which function upon membrane transport at various stages of the secretory pathway. These proteins were originally proposed to mediate both the disassembly of ternary SNARE complexes and ATP-dependent membrane fusion but more recently have been proposed to prime the docking step in SNARE assembly (64, 74). Such proteins appear to act as general SNARE regulators and, thus, disassemble preformed v-t SNARE complexes present in the same membrane (64) and, potentially, regulate either the association or dissociation of other, more specific, SNARE regulators. In the latter example, a novel SNARE regulator that participates in homotypic vacuolar fusion was identified. LMA1 (72, 73) is a heterodimer consisting of thioredoxin and the IB2 protease B inhibitor, which cooperates with (and may be initially bound to) Sec18 to release Sec17 and to stabilize the activated t-SNAREs (64, 74). Moreover, LMA1, like Sec17/Sec18, may act on different levels of the secretory pathway in yeast (5).
As no structural homologs of the synaptophysin family are encoded by the yeast genome, it is unclear whether negative regulators (aside, perhaps, from Sec1) act upon constitutive exocytosis in lower eukaryotes. To identify potential SNARE-masters for SNAREs which function in exocytosis, we used the two-hybrid system to screen for proteins that interact with the Snc v-SNAREs. The yeast Snc1 and Snc2 proteins (28, 49) are archetypal exocytic v-SNAREs which bear high structural homology to members of the synaptobrevin/VAMP/cellubrevin family of vesicle-associated membrane proteins (6, 44, 63) and are engaged in similar functions (14, 29, 49; reviewed in references 7 and 21). Like their brethren, Snc proteins localize to secretory vesicles (49) and interact physically with t-SNAREs from the plasma membrane (e.g., Sec9, Sso1, and Sso2) to form a ternary SNARE complex in vitro (9, 53). This was confirmed in vivo by genetic studies which showed that cells bearing disruptions in both SNC genes (a conditional lethal effect) and possessing a temperature-sensitive (ts) allele of those late-acting SEC genes which are involved in membrane fusion (e.g., sec17-1, sec9-4, and sso2-1) are inviable (14, 17, 29). Structure-function analyses have shown that the region of these SNAREs required to mediate exocytosis and cell viability localizes to the α-helical portion of the cytoplasmic domain (29). This region is conserved evolutionarily and is required for VAMP to form SNARE complexes in vitro (11, 36) and in permeabilized cells (50).
Here, we describe the identification of a protein that acts as a potential SNARE-master for the Snc v-SNAREs. The Vsm1 (v-SNARE-master 1) protein was identified by using the yeast two-hybrid system and was found to coimmunoprecipitate preferentially with the Snc2 v-SNARE. Deletion of VSM1 in yeast results in the enhanced secretion of proteins into the medium, and overexpression of the VSM1 gene was found to (i) inhibit the secretion and growth of cells bearing a mutant Sec9 t-SNARE, (ii) block rescue of the sec9-4 mutant allele by overexpression of the Snc1 v-SNARE, and (iii) result in the accumulation of low-density secretory vesicles (LDSVs) at the bud tips. On the basis of these results, we propose that Vsm1 may function as a negative regulator of exocytosis in yeast.
MATERIALS AND METHODS
Media, DNA, and genetic manipulations.
DNA endonucleases and modification enzymes were used as recommended by the suppliers. Molecular cloning techniques were performed as described by Sambrook et al. (56). DNA sequencing was performed by the dideoxynucleotide chain termination method (57). PCRs and subcloning of PCR products were carried out as described previously (27).
Saccharomyces cerevisiae strains were grown in standard growth medium containing either 2% glucose or 3.5% galactose as a carbon source. Synthetic complete and dropout media were similar to those described by Rose et al. (52). Standard methods were used for the introduction of DNA into yeast, for preparation of genomic DNA, and for tetrad dissection (52).
The yeast two-hybrid screen was performed as described by Durfee et al. (18), using Y153 cells. Quantitative assays for β-galactosidase activity were performed as described elsewhere (75).
Yeast strains.
Yeast strains used in this study are listed in Table 1. A VSM1 disruption strain in a wild-type background was created by transforming the SphI-SalI fragment containing the vsm1::URA3 disruption into diploid yeast strain W303. A disruption of one of the VSM1 loci was verified by Southern analysis; this diploid (VL1) was sporulated, and the resulting tetrads were dissected to yield haploid vsm1 cells (VL2 and VL3). VL2 and VL3 cells were then crossed to a variety of early and late sec mutants, end4 yeast, and myo2 yeast to give diploid strains which were sporulated and dissected to yield haploid yeast bearing both mutations (Table 1).
TABLE 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| W303-ab | Mata/α can1 his3 leu2 lys2 trp1 ura3 ade2 | J. Hirsch |
| W303-1a | Mata can1 his3 leu2 lys2 trp1 ura3 ade2 | J. Hirsch |
| JG8 | Mata can1 his3 leu2 trp2 snc1::URA3 snc2::ADE8 | J. Gerst |
| sf750-14Dα | Matα ura3-52 leu2-3,112 his4-580 sec1-1 | R. Schekman |
| NY770 | Mata ura3-52 leu2-3,112 sec2-41 | P. Novick |
| NY774 | Matα ura3-52 leu2-3,112 sec4-8 | P. Novick |
| NY776 | Matα ura3-52 leu2-3,112 sec5-24 | P. Novick |
| NY778 | Matα ura3-52 leu2-3,112 sec6-4 | P. Novick |
| RSY979 | Matα ura3-52 sec7-5 | R. Schekman |
| NY780 | Matα ura3-52 leu2-3,112 sec8-9 | P. Novick |
| NY782 | Mata ura3-52 leu2-3,112 sec9-4 | P. Novick |
| BY70 | Mata ura3-52 leu2-3,112 sec9-7 | P. Brennwald |
| NY784 | Matα ura3-52 leu2-3,112 sec10-2 | P. Novick |
| NY786 | Mata ura3-52 leu2-3,112 sec15-2 | P. Novick |
| NY1217 | Mata ura3-52 leu2-3,112 sec18-1 | P. Novick |
| RSY324 | Mata ura3-52 sec22-2 | R. Schekman |
| RH268-1C | Mata can1 his4 leu2 trp1 ura3 bar1-1 end4-1 | H. Riezman |
| NY1002 | Mata ura3-52 leu2-3,112 myo2-66 | P. Novick |
| H458 | Mata ura3-52 leu2-3,112 sso1::HIS3 sso2-1 | H. Ronne |
| WCG4-1-1a | Mata ura3-52 leu2-3,112 his3 pre1-1 | D. Wolf |
| BJ1991 | Matα ura3-52 leu2-3,112 trp1 gal2 prb1-1 pep4-3 | ATCCa |
| BJ5462 | Matα ura3-52 leu2Δ1 trp1 pep4::HIS3 prb1Δ1 | ATCC |
| VL1 | Mata/α his3 leu2 trp1 ade2 vsm1::URA3 | This work |
| VL2 | Matα his3 leu2 trp1 ade2 vsm1::URA3 | This work |
| VL3 | Mata his3 leu2 trp1 ade2 vsm1::URA3 | This work |
| VL3-1 | Mata his3 leu2 trp1 ade2 vsm1::URA3 sec1-1 | This work |
| VL3-4 | Mata his3 leu2 trp1 ade2 vsm1::URA3 sec4-8 | This work |
| VL3-5 | Mata his3 leu2 trp1 ade2 vsm1::URA3 sec5-24 | This work |
| VL2-9-4 | Mata his3 leu2 trp1 ade2 vsm1::URA3 sec9-4 | This work |
| VL2-9-7 | Mata leu2 trp1 ade2 vsm1::URA3 sec9-7 | This work |
| VL2-10 | Mata his3 leu2 trp1 ade2 vsm1::URA3 sec10-2 | This work |
| VL2-15 | Matα his3 leu2 vsm1::URA3 sec15-24 | This work |
| VL2-18 | Mata leu2 trp1 ade2 vsm1::URA3 sec18-1 | This work |
| VL2-22 | Mata leu2 trp1 ade2 vsm1::URA3 sec22-2 | This work |
| VL2-E4 | Mata his3 leu2 trp1 ade2 vsm1::URA3 end4-1 | This work |
| VL2-M2 | Mata his3 leu2 trp1 ade2 vsm1::URA3 myo2-66 | This work |
| VL2-S2 | Mata leu2 trp1 ade2 vsm1::URA3 sso1::HIS3 sso2-1 | This work |
ATCC, American Type Culture Collection.
Antibodies, immunoprecipitation, and immunoblot analysis.
A polyclonal antiserum to Vsm1 was created by expressing recombinant Vsm1 in bacteria and injecting the affinity-purified protein into rabbits. A bacterial expression construct, pHISVSM1, was created so as to allow for the expression of a His6-tagged Vsm1 protein (molecular mass ≅ 64 kDa) in Escherichia coli TOP10 (see Fig. 3). Bacterial extracts were purified over Ni2+ chelation resin (Ni2+-nitrilotriacetic acid; Qiagen) to yield protein that was used for injection into rabbits. Polyclonal antisera 3609 and 3610 were obtained in this fashion and were used to detect both native and tagged Vsm1 in cell extracts.
FIG. 3.
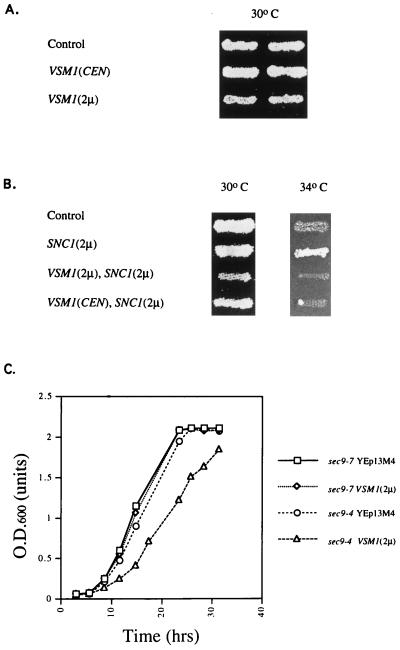
Overexpression of VSM1 specifically inhibits the growth of sec9-4 cells and blocks rescue by SNC1 overexpression. (A) sec9-4 cells were transformed with plasmids expressing VSM1 in single copy [VSM1(CEN)] and multicopy [VSM1(2μm)] or with a control plasmid (Control). Cells were patched onto selective medium prior to being replica plated onto fresh plates and allowed to grow for 2 days at 30°C. (B) sec9-4 cells bearing plasmids expressing VSM1 in single copy [VSM1(CEN)] and multicopy [VSM1(2μm)] or a control plasmid were transformed with a plasmid expressing SNC1 under the control of a constitutive promoter [SNC1(2μm)]. Control cells bearing an empty vector were transformed either with the SNC1 plasmid [SNC1(2μm)] or with a second empty vector (Control). Cells were patched onto selective medium prior to being replica plated onto fresh plates and allowed to grow for 2 days at 30 and 34°C. (C) sec9-4 and sec9-7 yeast strains were transformed either with a control plasmid (YEp13M4) or with a plasmid expressing VSM1 in multicopy [VSM1(2μm)]. Cells were grown to saturation under selective conditions, seeded at a density of 0.02 OD600 units/ml in fresh medium, and allowed to grow till saturation at 26°C. OD600 was measured at different times.
Affinity-purified anti-hemagglutinin epitope (HA) mouse monoclonal antibody 12CA5 as well as anti-HA antibody from mouse ascites fluid (gift of M. Wigler) were used in these experiments. Other affinity-purified antisera included mouse monoclonal anti-Dpm1 (Molecular Probes) and polyclonal anti-Sso (gift of P. Brennwald). Other polyclonal antisera included anti-Emp47, anti-Wbp1, anti-Mnn1 (gifts of S. Emr), anti-Gas1, anti-Sec22 (gifts of R. Schekman), anti-Snc (46), and anti-Sso (gift of S. Keränen).
Protein expression in log-phase-grown yeast cells was verified by immunoblot analysis using chemiluminescence as described elsewhere (14) or by quantitative Western analysis using 125I-labeled protein A (1 μCi/blot; ICN) as described previously (27). Quantitation of the latter was performed with a Fuji phosphorimager. Immunoprecipitation was also performed essentially as described previously (14, 15) except that EDTA was added to a final concentration of 2 mM and 1.0% Nonidet P-40 was used instead of Triton X-100 in the coimmunoprecipitation experiments.
Plasmids.
Vectors included YEp13M4, a 2μm plasmid bearing the LEU2 marker; pAD4Δ, a similar plasmid which bears the ADH1 constitutive promoter; pAD54, a plasmid identical to pAD4Δ but containing an oligonucleotide encoding a peptide derived from HA; and pAD6, a similar plasmid which bears an oligonucleotide encoding a peptide derived from the Myc protein. Directional subcloning into pAD54 or pAD6 allows for the in-frame fusion between the sequence encoding the epitope and the coding region of the subcloned gene of interest. Centromeric vectors included pRS315, which bears a LEU2 marker, and pSE358, which bears a TRP1 marker.
Other plasmids included pADH-SNC1, which contains genomic SNC1 cloned into pAD4Δ (28); pADH-HASNC1, which contains genomic SNC1 cloned into pAD54; and pTGAL-SNC1, a plasmid which expresses SNC1 under the control of the GAL10 promoter in pSE358 (49).
Plasmids created for the two-hybrid assay included pTA-SNC2Δ and pAS-SNC2Δ. pTA-SNC2Δ bears a fragment encoding SNC24-93 cloned into the pT7blue cloning vector (Novagen). This fragment was generated by PCR using the JG300 (5′-ACGATGTCGGCCATGGTGCCATACGAT-3′) and JG301 (5′-AACAACTAAGAAGGATCCCTATCTCATTTTTAG-3′) oligonucleotides, which bear NcoI and BamHI sites (underlined), respectively. The mutant SNC2 gene bears a termination signal proximal to the encoded transmembrane domain and results in the expression of truncated gene product. This was cloned into the BamHI-NcoI sites of the pAS1 (18) two-hybrid vector to generate an in-frame gene fusion with the region encoding the DNA binding domain of Gal4. Expression in S. cerevisiae Y153 cells resulted in the expression of fusion protein with an apparent molecular mass of 32 kDa (data not shown).
VSM1-containing plasmids included pACT-VSM1, which contains VSM1 cloned into the XhoI site of pACT1 (18) and which was isolated in the two-hybrid screen; pTA-VSM1, which contains VSM1 generated by PCR using the VL1 (5′-CGCAAATATAGTCGACGATGGATTTA-3′) and VL2 (5′-TATCAGTGGGGAGCTCATTCGAA-3′) oligonucleotides, which bear SalI and SacI sites (underlined), respectively; pADH-HAVSM1, which contains VSM1 cloned into the SalI and SacI sites of pAD54; pLADH-HAVSM1, which contains a BamHI fragment from pADH-HAVSM1 that contains both the ADH1 promoter and VSM1 gene, cloned into the BamHI site of pRS315; and pHISVSM1, which contains VSM1 generated by PCR using the VL1 and VL7 (5′-TATCAGTGGTACCTCATTGGGAA-3′) oligonucleotides, which bear SalI and KpnI sites, respectively, and was cloned into the XhoI and KpnI sites of pTRCHISB.
Other plasmids created for this study included pADH-mycSNC2, which contains a SalI-SacI fragment of SNC2 cloned into the SalI and SacI sites of pAD6; pUADH-mycSNC2, which carries a BamHI fragment from pADH-mycSNC2 that contains both the ADH1 promoter and SNC2 gene, cloned into the BamHI site of YCp50; pTADH-HASNC1, which carries a BamHI fragment from pADH-HASNC1 that contains both the ADH1 promoter and SNC1 gene, cloned into the BamHI site of pSE358; and pTADH-HASNC2, which carries a BamHI fragment from pADH-HASNC2 that contains both the ADH1 promoter and SCN2 gene, cloned into the BamHI site of pSE358. A disruption plasmid for VSM1, pVSM1U, was created by first cloning a 2.7-kb genomic fragment containing VSM1, amplified from yeast genomic DNA by using the VL8 (5′-GAGCAACCTCTTGAGGTCGAGA-3′) and VL9 (5′-ATTCAATGCAGCAAGATTGTCA-3′) oligonucleotides, into the pGEM cloning vector (Promega) to give pVSM1. pVSM1 was then digested with XbaI, blunt ended, and religated to create a frameshift mutation at bp 158 of VSM1, which terminates translation at bp 208, to give pVSM1FS. Next, pVSM1FS was digested with ClaI, which cuts at bp 548 and 854, to excise a fragment, and the URA3 selectable marker was inserted into this site of VSM1 by blunt-end ligation to create pVSM1U. The vsm1::URA3 fragment used to disrupt VSM1 in yeast cells was released from pVSM1U by digestion with the SphI and SalI restriction endonucleases.
Cellular fractionation and vesicle preparations.
Cellular fractionation of yeast cells was performed by standard techniques. Briefly, cells (25 OD600 [optical density at 600 nm] units) were harvested during log phase and lysed using glass beads in 300 μl of lysis buffer containing 25 mM potassium phosphate (pH 7.0), 100 mM NaCl, 2 mM EDTA, and the following protease inhibitors: aprotinin (1 μg/ml), leupeptin (2 μg/ml), pepstatin (1 μg/ml), soybean trypsin inhibitor (10 μg/ml), and phenylmethylsulfonyl fluoride (1 mM). After lysis, extracts were spun at 600 × g for 4 min to remove intact cells and cell wall debris to yield the total cell lysate; the latter was then centrifuged at 10,000 × g for 10 min to yield the S10 supernatant and P10 pellet fractions. The S10 was next centrifuged at 100,000 × g for 1 h to yield the S100 supernatant and P100 pellet fractions. Protein determination was performed by using the micro-bicinchoninic acid technique (Pierce). For experiments involving the treatment of membranes with either high salt (0.5 to 1.5 M NaCl) or low pH (200 mM glycine [pH 2.5]), aliquots of the S10 fraction were treated with the above reagents for 15 min on ice prior to centrifugation at 100,000 × g for 1 h. Quantitation of proteins present in the various fractions was performed by using specific antisera and quantitative Western analysis (described above).
Vesicle preparations and density gradient centrifugation were performed as described by Harsay and Bretscher (33). sec6 or sec9 cells were either maintained continually at 26°C or temperature shifted to 37°C for 2 h (to induce vesicle accumulation) prior to harvesting. Prior to the temperature shift, cells were incubated for 1.5 h either in low (0.05%)-glucose YPD medium, to induce invertase expression, or in phosphate-depleted YPD medium, to induce acid phosphatase expression. After centrifugation at 100,000 × g for 19 h, 15 to 30% Nycodenz gradients were prepared and collected, using a Buchler Autodensi-Flow IIc gradient builder. For enzymatic assays and Western analyses, fractions from the gradient (either 350 or 700 μl) were aliquoted and frozen at −70°C until use. For uranyl acetate staining (described below), aliquots from different fractions were fixed and processed directly. Samples were taken to determine protein concentration, using the Bradford protein-dye binding assay (Bio-Rad), and density.
Enzymatic assays.
Acid phosphatase and exoglucanase were assayed by the methods of Van Rijn et al. (66) and Santos et al. (59), respectively, as described by Harsay and Bretscher (33). Activity is expressed in arbitrary units based on absorbance measured at 415 nm. ATPase activity was assayed by the method of Bowman and Slayman (8), with some modifications of its application described by Harsay and Bretscher (33). First, substrate concentration was modified to 2 mM ATP, as higher concentrations result in an elevated background. Second, blanks were measured to determine the rate of endogenous ATP hydrolysis and were subtracted from values obtained with samples from the gradient. Inorganic phosphate was assayed by the method of Ames (3). Optical density was measured after 10 min, and activity was expressed in arbitrary units, based on absorption at 820 nm. Invertase activity was assayed by the method of Goldstein and Lampen (31) and is expressed either in arbitrary units based on absorption at 540 nm or in units, where 1 U = 1 μmol of glucose released/min/100 mg of dry cells.
Immunofluorescence and electron microscopy.
For immunofluorescence, cells containing the appropriate plasmids were grown to log phase at 25°C and fixed by the direct addition of 1 M KPO4 (pH 6.5) and 37% formaldehyde to reach final concentrations of 0.1 M KPO4 and 3.7% formaldehyde. After 30 min, 5 to 10 OD600 units of cells was harvested by centrifugation for 5 min at 2,000 rpm and resuspended in 5 ml of a solution containing 0.1 M KPO4 (pH 6.5) and 3.7% formaldehyde. Cells were incubated at room temperature for 1.5 h with gentle rocking, washed once with 5 ml of 0.1 M KPO4 (pH 7.5), and stored overnight at 4°C in 5 ml of a solution containing 0.1 M KPO4 (pH 7.5) and 1.2 M sorbitol. Cells were harvested by centrifugation, and spheroplasts were prepared by incubating half of the cells for 30 min at 30°C in 1 ml of a solution containing 0.1 M KPO4 (pH 7.5), 1.2 M sorbitol, 0.25 mg of zymolase per ml, and 0.5% β-mercaptoethanol. Spheroplasts were pelleted at 2,000 rpm for 2 min and washed twice with a 1 ml of a solution of 0.1 M HEPES (pH 7.4) and 1.0 M sorbitol (HS buffer). Spheroplasts were then permeabilized with HS buffer containing 0.5% sodium dodecyl sulfate (SDS) at room temperature for 5 min. The cells were washed three times with 1 ml of HS buffer and resuspended in the same. About 20 μl of fixed spheroplasts was placed onto coverslips precoated with 0.1% polylysine for 20 min. Coverslips were washed with phosphate-buffered saline containing 1 mg of bovine serum albumin per ml and 0.02% Tween 20 (PBT), incubated for 1.5 h with primary antibody, and washed extensively with PBT. Next, secondary antibody (fluorescein isothiocyanate [FITC]-conjugated anti-mouse or anti-rabbit) along with normal goat serum (1:100 dilution) was added for 1.5 h, followed by extensive washing with PBT. For visualization of HA-tagged protein, preabsorbed affinity-purified anti-HA monoclonal antibody (1:1,000 dilution) was used. For the visualization of Dpm1, a commercial affinity-purified mouse monoclonal anti-Dpm1 antibody was used (3 μg/ml). For visualization of Sso, an affinity-purified polyclonal anti-Sso antibody (1:500) was used. For labeling with secondary antibody, FITC-labeled goat anti-mouse or anti-rabbit antibody (Jackson Laboratories) at a dilution of 1:200 was used. After extensive washing with PBT, coverslips were labeled with propidium iodide (1 μg/ml) in PBT for 20 min. Next, coverslips were washed extensively with PBT, aspirated, and, after addition of mounting medium, placed onto slides. Proteins were visualized by confocal microscopy.
The fixation, thin sectioning, and electron microscopy of yeast were performed as described previously (75). Uranyl acetate staining of membranes from density gradients was performed by first diluting two or three fractions of a given region of enzymatic activity with gradient buffer (33) lacking Nycodenz, followed by the addition of glutaraldehyde to a final concentration of 3%. After 2 to 4 h with shaking, the membranes were pelleted at 100,000 × g and resuspended in 20 μl of buffer. Next, grids bearing collodion support film were incubated with the membranes for 1 min, followed by staining with 1% uranyl acetate (1 min). Membranes were visualized by electron microscopy.
Metabolic labeling studies.
Pulse-chase studies using [35S]methionine (Amersham) were performed as described previously (15). For medium protein secretion experiments, cells were labeled for 30 min and chased with medium containing excess methionine and cysteine for 30 min. Next, cells were centrifuged at 10,000 × g for 2 min, following which the medium was transferred to a fresh tube and the proteins were precipitated by using Strataclean resin (Stratagene). Precipitated proteins were washed and counted in a scintillation counter. For autoradiography, precipitates were resolved on SDS-polyacrylamide gels, which were then fixed, incubated with a fluorescence enhancer, dried, and exposed to X-ray film.
Nucleotide sequence accession number.
The GenBank accession number for VSM1 is AF034895.
RESULTS
Isolation of a v-SNARE-interacting protein.
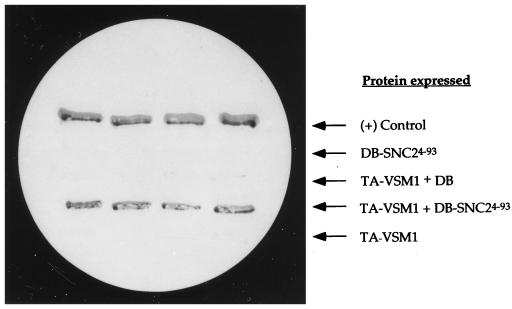
In an effort to isolate proteins which interact directly with v-SNAREs of the late secretory pathway in yeast, we used the two-hybrid system (18) with a C-terminally truncated form of Snc2 (Snc24-93) as the bait. The truncated protein lacks the membrane-spanning domain of the v-SNARE, which is essential for conferring exocytosis in vivo but is not likely to be essential for forming complexes with the t-SNAREs (29, 53). In screening a yeast cDNA library fused to the transactivation domain of Gal4, we isolated one clone out of 400,000 colonies screened which conferred robust β-galactosidase activity and growth in the presence of 3-aminotriazole in a plasmid-dependent fashion (Fig. 1 and data not shown). 3-Aminotriazole is a metabolic inhibitor of the HIS3 gene product which serves as a selection marker in the two-hybrid screen (18). The cDNA insert contained within the plasmid was cloned and was found to encode an open reading frame (ORF) from the yeast genome (YER143w; GenBank accession no. U18917 and AF034895). The protein encoded by this ORF consists of 428 amino acids and has a calculated molecular mass of about 48 kDa (Fig. 2). The translated sequence has no potential membrane-spanning domains and, despite bearing four possible α-helical segments, has little propensity for forming coiled coils, as determined by using secondary structure algorithms (data not shown). YER143w has high homology to a smaller ORF in the fission yeast Schizosaccharomyces pombe, which encodes a putative 36-kDa protein (accession no. Q10256). The translated proteins were found to be 41% identical, suggesting that they are probable homologs. In addition, sequences with significant homology to portions of YER143w are found in Caenorhabditis elegans (accession no. U50068) and Leishmania (accession no. AE001274). These sequence blocks are 40 and 49% identical over 174 and 155 amino acid residues, respectively.
FIG. 1.
The gene product of YER143w (Vsm1) interacts with Snc24-93 in the yeast two-hybrid system. Yeast Y153 cells transformed with various plasmids were tested for β-galactosidase on nitrocellulose filters. Yeast expressing the Gal4 DNA binding domain alone (DB) or fused to Snc24-93 (DB-SNC24-93) were transformed with a plasmid that expresses the transactivating domain of Gal4 (TA) fused to the YER143w (VSM1) gene product (TA-VSM1), or with a control plasmid, and were patched onto selective medium. After 2 days, the cells were replica plated onto nitrocellulose filters, freeze-fractured in liquid nitrogen, and assayed visually for β-galactosidase activity (see Materials and Methods). A positive (+) control consisted of yeast expressing the Gal4 DB fused to the retinoblastoma gene product Rb and the Gal4 TA fused to the PP1α phosphatase.
FIG. 2.
The amino acid sequence of Vsm1 (YER143w). Numbers correspond to amino acid residues.
Overproduction of Vsm1 inhibits the growth of cells bearing a mutant Sec9 t-SNARE and blocks rescue by Snc1.
Since YER143w encodes a potential partner for the Snc v-SNAREs, we tested for its ability, when overexpressed from multicopy plasmids, to suppress growth defects seen in ts mutants of the yeast secretory pathway. Interestingly, overexpression of YER143w was found to inhibit the growth of sec9-4 cells at temperatures normally permissive for growth (≤30°C) (Fig. 3) and led to an accumulation of secretory vesicles within the cells (Fig. 4b). In contrast, overproduction was not found to have effects in any of the other early or late sec mutants tested (sec1, sec2, sec4, sec5, sec6, sec7, sec8, sec10, sec15, sec18, and sec22) as well as in snc null cells (49) or in cells bearing a ts mutation in the gene encoding the Sso2 t-SNARE (sso1Δ sso2-1) (data not shown). Similarly, overexpression of this ORF had no effect on the growth of cells bearing mutations in other genes which have been shown to regulate protein trafficking along different stages of the secretory pathway, including the end4, myo2, vps4, vps5, vps33, vps45, and pep12 mutants (data not shown).
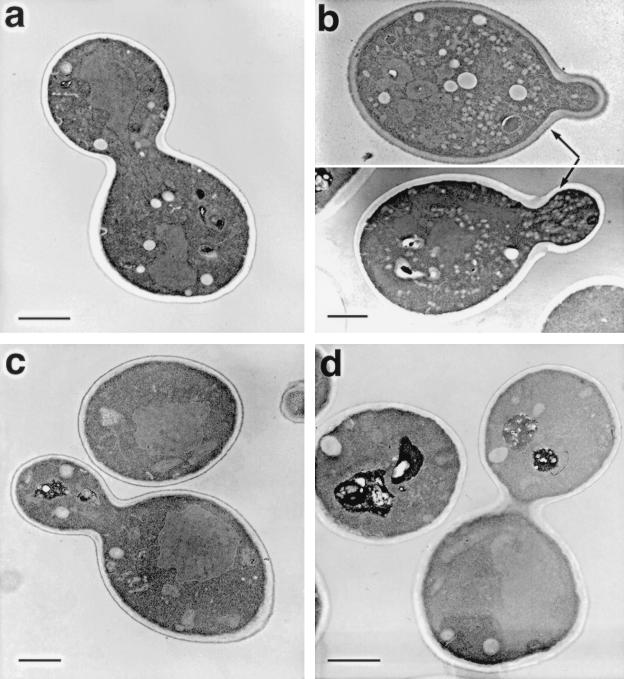
FIG. 4.
Overexpression of VSM1 in sec9-4 cells results in the accumulation of secretory vesicles at the bud tip. sec9-4 cells bearing a control plasmid (A) or a multicopy plasmid which expresses VSM1 (B) were grown to log phase and fixed for thin sectioning and electron microscopy. Thin sections of wild-type cells (W303-1a) (C) and vsm1Δ cells (VL2) (D) are also shown for comparison. Bars, 1 μm.
We have previously shown that SNC1 overexpression suppresses ts defects in cells bearing mutations in the genes encoding the Sec9 and Sso2 t-SNAREs (sec9-4 and sso2-1, respectively) (14, 29). We have suggested that overexpression of the Snc v-SNARE stabilizes the mutant t-SNARE at nonpermissive temperatures, leading to formation of a functional ternary complex and, thus, heightened viability (14, 29). Since the protein encoded by YER143w binds to Snc2 and inhibits the growth of sec9-4 cells at permissive temperatures, we determined whether its overproduction also blocked the ability of Snc1 to suppress the ts defects of sec9-4 cells. Overproduction of YER143w in sec9-4 cells, from either single-copy or multicopy plasmids, was found to effectively block the suppression conferred by Snc1 (Fig. 3B). Similar results were obtained when SNC2 was used instead of SNC1 to suppress the sec9-4 allele (data not shown). Thus, this protein appears to inhibit the functioning of the Snc v-SNAREs, perhaps as a result of its direct association therewith.
The Sec9-4 mutant protein was shown in in vitro binding experiments to be deficient in its ability to enter into the ternary SNARE complex after being shifted to nonpermissive temperatures (53). In contrast, another mutant Sec9 protein, Sec9-7, was found to enter into the ternary complex but to be unable to confer secretion (53). When YER143w was overexpressed in sec9-7 cells, we found no defects in the growth of such cells on liquid medium at permissive temperatures (Fig. 3C). In contrast, its overexpression in sec9-4 cells resulted in a substantial reduction in the growth rate, leading to a 15 to 20% increase in the length of the division time (Fig. 3C). Moreover, when sec9-4 cells overexpressing the ORF were examined by thin sectioning and electron microscopy, we found that such cells accumulated large numbers of 100-nm vesicles primarily in the bud tips (Fig. 4b). Specifically, we found that sec9-4 cells overproducing YER143w accumulated 7.1 vesicles/μm2 in cross-sectioned cells (n = 65 cells), while control sec9-4 cells (Fig. 4a) accumulated <0.5 vesicle/μm2 in cross-sectioned cells (n = 80 cells) at temperatures permissive for growth. Vesicle accumulation and localization appeared independent of whether cells were at early or late stages of budding. Thus, overproduction of this novel protein appears to inhibit the docking and fusion of secretory vesicles, resulting in their accumulation and a decrease in the growth rate of the cells.
On the basis of these experiments, we have renamed this gene VSM1 to signify its encoding of a putative v-SNARE-master from yeast. While this work was in progress, this gene was also identified as being adjacent to the MAG1 gene and was given the name DDI1, due to its induction upon treatment with DNA-damaging agents (41). We maintained usage of the name Vsm1, as it more closely approximates the biological function of this protein.
Analysis of the disruption of VSM1.
Disruption of the VSM1 gene was performed by homologous recombination. A URA3 selectable marker was cloned into the VSM1 locus carried on a genomic fragment of DNA and was transformed into a diploid wild-type strain. After sporulation and tetrad dissection, all spores were found to have undergone germination, and analysis of the resulting meiotic segregants showed no obvious defects in vsm1::URA3 cells. Cells bearing a disruption in VSM1 were found to grow normally, were not ts, and did not result in the accumulation of secretory vesicles or other membrane, as revealed by electron microscopy (data not shown and Fig. 4d). In addition, no defects in either the kinetics or extent of the intracellular processing of invertase, a secreted enzyme, were detected in cells lacking VSM1 (data not shown). Likewise, no defects in the uptake and internalization of an endocytic marker, FM4-64, were detected (data not shown).
To demonstrate possible genetic interactions between the disruption of VSM1 and known ts mutants of the secretory pathway, we crossed haploid vsm1::URA3 cells with other haploid cells bearing the appropriate ts alleles or gene disruptions. We examined possible synthetic interactions between vsm1Δ and mutant alleles of SEC1, SEC2, SEC4, SEC5, SEC6, SEC7, SEC9 (e.g., sec9-4 and sec9-7), SEC10, SEC15, SEC18, SEC22, SSO2 (sso1Δ sso2-1), END4, and MYO2. Diploid cells were sporulated, and the progeny was analyzed by tetrad dissection and analysis. We were unable to uncover any synthetic interactions between the vsm1::URA3 disruption and these other mutant genes. Combined mutations did not lead to any changes in cell growth and did not lead to either an enhancement of or reduction in temperature sensitivity (data not shown).
However, wild-type cells or sec cells bearing deletions in VSM1 examined for the secretion of proteins into the medium by 35S labeling and precipitation of the secreted proteins (25) were found to secrete significantly higher levels of protein, based upon the scintillation counts (Table 2). On average, vsm1Δ cells were found to secrete about 20 to 30% more protein. In contrast, no significant increase in the overall rate of protein biosynthesis, as measured by total incorporation of [35S]methionine into the cells, was observed (data not shown). Gel electrophoresis and autoradiography of proteins precipitated from the culture medium of labeled vsm1Δ cells revealed the same 9 to 10 proteins typically secreted from wild-type yeast (17, 25) (data not shown). As expected, all medium proteins were present at levels higher than those found in precipitates from wild-type cells, while no variation in the relative amounts of individual proteins was found (data not shown). Thus, the effect of VSM1 disruption on the secretion of proteins into the medium appears nonspecific. Correspondingly, cells overexpressing VSM1 were found to secrete 12% less protein overall into the medium (Table 2).
TABLE 2.
Secretion of proteins into the medium
| Strain | Δ (% secreted) |
|---|---|
| sec9-4 | 0 |
| sec9-4 VSM1(2μm) | −11.6 ± 0.1 |
| sec9-4 VSM1Δ | +33.0 ± 8.6 |
| WTa | 0 |
| WT VSM1Δ | +26.9 ± 1.6 |
WT, wild type.
Because vsm1Δ cells secrete more protein into the medium, we next examined their ability to secrete invertase, a standard marker for secretion competence. Surprisingly, despite repeated analyses, we detected no change in the synthesis or secretion of invertase in either wild-type or sec cells lacking the VSM1 gene (Table 3 and data not shown). Moreover, overexpression of VSM1, which results in the accumulation of secretory vesicles, did not result in an intracellular accumulation of invertase. This finding suggests that Vsm1 may exert its effect on a non-invertase-containing class of secretory vesicles.
TABLE 3.
Invertase secretiona
| Strain | Invertase (%)
|
|||
|---|---|---|---|---|
| 26°C
|
37°C
|
|||
| NS | S | NS | S | |
| sec9-4 | 19.0 ± 2.4 | 81.0 ± 2.4 | 83.9 ± 1.7 | 16.0 ± 1.7 |
| sec9-4 VSM1(2μm) | 14.0 ± 1.5 | 85.8 ± 1.4 | 84.4 ± 2.3 | 15.6 ± 2.4 |
| sec9-4 | 19.3 ± 5.6 | 80.2 ± 5.4 | 88.7 ± 1.7 | 11.3 ± 1.7 |
| sec9-4 VSM1Δ | 20.7 ± 3.6 | 79.3 ± 3.6 | 88.3 ± 1.7 | 11.7 ± 1.7 |
| WT | 4.3 ± 0.7 | 94.2 ± 0.9 | 4.9 ± 0.4 | 95.1 ± 0.4 |
| WT VSM1Δ | 10.1 ± 0.2 | 89.9 ± 0.2 | 4.1 ± 0.6 | 95.8 ± 0.6 |
NS, nonsecreted; S, secreted; WT, wild type.
Vsm1 is membrane associated and may localize to the plasma membrane.
We examined the expression and localization of Vsm1 in yeast cells by Western analysis and confocal microscopy. Tools created to perform these experiments included an epitope-tagged form of Vsm1, which contains the HA epitope located at the amino terminus of the protein, as well as a polyclonal antiserum generated against bacterially expressed His6-tagged Vsm1 (see Materials and Methods).
Detection of the native and HA-tagged forms of Vsm1 was performed in cell extracts derived from both wild-type and vsm1Δ yeast strains. We found that native Vsm1 runs as a protein doublet of 52/54 kDa, while the HA-tagged form runs as a doublet of 54/56 kDa (Fig. 5A). The native doublet was absent from vsm1Δ yeast, indicating that both protein bands arise from expression of the intact VSM1 gene. In contrast, recombinant bacterially expressed His6-Vsm1 runs as a single band corresponding to ∼64 kDa, due to the presence of additional residues encoded by the bacterial expression vector (see Materials and Methods). Pulse-chase experiments using [35S]methionine indicate that both VSM1-derived proteins are generated in roughly equal amounts that were both present at time zero of the chase (data not shown). The nature of this phenomenon and possible processing of the Vsm1 protein will be described separately.
FIG. 5.
Vsm1 is a membrane-associated protein that localizes to the plasma membrane. (A) Vsm1 exists as a protein doublet in yeast extracts. Cell lysates were prepared from wild-type yeast (lane 1), vsm1Δ yeast expressing HA-tagged Vsm1 from single-copy (CEN) (lane 2) or multicopy (2μm) (lanes 3 and 4) plasmids, and vsm1Δ yeast lacking plasmids (lane 5) and were electrophoresed on SDS-polyacrylamide gels. In lanes 1 to 3 and 5, 50 μg of protein was added; 25 μg was added in lane 4. In addition, lane 6 contained 1 μg of affinity-purified recombinant His6-tagged Vsm1. Detection was performed with a polyclonal anti-Vsm1 antiserum (1:5,000). (B) Vsm1 associates with membranes and is enriched in the P100 fraction. sec6-4 cells maintained at 26°C or temperature shifted (2 h) to 37°C were fractionated as described in Materials and Methods. Aliquots of the various fractions were electrophoresed on SDS–10% polyacrylamide gels and blotted onto nitrocellulose membranes. Quantitative Western analysis was performed with specific antibodies (Ab) (anti-Gas1 [1:10,000], anti-Vsm1 [1:5,000], anti-Sso [1:5,000], and anti-Snc [1:1,000]) and 125I-labeled protein A (1 μCi/blot). In the left panel, 50-μg protein aliquots from the different fractions obtained from cells maintained at 26°C were electrophoresed; 25-μg aliquots of protein were used in the experiment shown in the middle panel. In the right panel, aliquots of the S10 fraction were treated with 1.5 M NaCl–200 mM glycine buffer (pH 2.5) or with additional lysis buffer (lacking salt or glycine) prior to centrifugation at 100,000 × g to yield the supernatant (sup.) and pellet fractions. Samples (15 μg) were electrophoresed, blotted, and detected for protein as described above. (C) Levels of Vsm1 are elevated in cells lacking vacuolar hydrolase, but not proteosome, activities. Total cell lysates (TCL) were prepared from sec6-4 yeast and strains which bear mutations in the protein degradative pathways (e.g., pre1-1 cells, which are deficient in the 20S proteosome activity, and pep4-3 prb1-1 and pep4-3 prb1Δ cells, which are deficient in the vacuolar hydrolase activity). Aliquots of total cell lysates (50 μg) were electrophoresed, blotted, and probed with anti-Vsm1 antibody (1:5,000). Detection was performed quantitatively, using 125I-labeled protein A (1 μCi). (D) Vsm1 localizes to the plasma membrane. Localization of Vsm1 was performed in wild-type cells by indirect immunofluorescence and confocal microscopy. a, cells expressing HA-tagged Vsm1 (expressed from a single-copy plasmid), detected with an affinity-purified anti-HA antibody (1:1,000) and FITC-labeled second antibody; b, staining of Dpm1 with an affinity-purified anti-Dpm1 antibody (3 μg/ml) and FITC-labeled second antibody; c, staining of HA-tagged Snc1 (expressed from a single-copy plasmid) with an affinity-purified anti-HA antibody (1:1,000) and FITC-labeled second antibody; d, staining of Sso protein with an affinity-purified anti-Sso antibody (1:100) and an FITC-labeled second antibody. Staining of the cell nucleus was performed with propidium iodide (1 μg/ml) and visualized via the rhodamine channel.
To verify whether Vsm1 is a soluble protein or localizes to the membrane fraction, we prepared cell extracts from sec6-4 yeast either maintained at permissive temperatures or shifted to 37°C for 1 h to induce vesicle accumulation. The latter is used to obtain a 100,000 × g membrane (P100) fraction that while containing ER, Golgi, and plasma membrane markers (references 15 and 31 and Fig. 6E) is enriched in secretory vesicles. Cell fractionation experiments revealed that Vsm1 is present in the 10,000 × g and 100,000 × g soluble (S10 and S100) and pellet (P10 and P100) fractions (Fig. 5B and C; Table 4). In a representative experiment (Table 4; Fig. 5B, left panel), roughly 20% of cellular Vsm1 was found in the P10 fractions (at either 26 or 37°C), whereas 40 to 50% or more of the Gas1 and Sso plasma membrane proteins are present in this fraction, which constitutes about one-quarter of the total cellular protein. The bulk of Vsm1 (nearly 80%) remained in the S10 fraction and, following centrifugation at 100,000 × g, was evenly split between the S100 and P100 fractions. This contrasts with Gas1 and Sso, levels of which are about four- to fivefold higher in the P100 fraction. Quantitatively, both Vsm1 and Gas1 were enriched twofold or more in the P100 fraction obtained from temperature-shifted cells in comparison to nonshifted cells (Table 4). In another experiment, distribution to the S100 and P100 fractions of Vsm1 was monitored in parallel to the Snc and Sso membrane proteins, which serve as vesicle and plasma membrane markers (Fig. 5B, middle panel). It should be noted that Snc serves as a vesicle marker in the membrane preparation obtained from temperature-shifted cells. We found that the percentage of Sso and Snc that distributes between the S100 and P100 fractions was basically similar to that for Vsm1 (Fig. 5C, middle panel). Densitometric analysis of this representative experiment revealed that about 70% of Vsm1, 80% of Snc1,2, and nearly 90% of Sso1,2 were present in the P100 fraction at either permissive or restrictive temperatures. As also shown above, all three proteins (Snc1,2, Vsm1, and Sso1,2) were enriched approximately two- to threefold in the P100 obtained from cells shifted to restrictive conditions relative to the amount obtained from cells maintained at permissive conditions. We suppose that the small amounts of membrane proteins present in the S100 in these experiments likely resulted from incomplete pelleting at 100,000 × g. Finally, we performed initial studies to determine the basis for association of Vsm1 with the P100 membrane fraction (Fig. 5B, right panel). Aliquots of the S10 fraction were exposed to high ionic strength (0.5 to 1.5 M NaCl) or low pH (200 mM glycine [pH 2.5]) prior to centrifugation at 100,000 × g. We found that treatment with either could substantially, but not completely, reduce the amount of Vsm1 found in the P100 fraction. In the experiment shown in Fig. 5B (right panel), we found that fourfold more Vsm1 was present in the S100 when treated with high salt compared to untreated (no added salt) samples (51% versus 12%). Moreover, 15-fold more Vsm1 was present in the S100 when exposed to low pH compared to untreated (no glycine buffer) samples (29% versus 2%). It should be noted that some Gas1 could also be removed by high-salt treatment but was less than that for Vsm1. Sso protein, on the other hand, could not be removed by treatment with high salt, and over 98% was present in the P100 fraction. On the basis of these experiments, we suggest that Vsm1 is a membrane-associated protein that is enriched in the P100 fraction. However, we note that some Vsm1 resides in the cytosolic fraction during cell fractionation.
FIG. 6.
Vsm1 does not localize to secretory vesicles. (A) Two types of secretory vesicles accumulate in sec6-4 and sec9-4 cells. sec6-4 cells expressing HA-VSM1 from a multicopy plasmid and MYC-SNC2 from a single-copy plasmid were grown to log phase, shifted to low-phosphate-containing medium, and then either maintained at permissive conditions (26°C) or shifted to 37°C to induce vesicle accumulation. sec9-4 cells expressing HA-VSM1 and MYC-SNC2 from single-copy plasmids were grown to log phase, shifted to low-glucose-containing medium, and then either maintained at permissive conditions or shifted to 37°C to induce vesicle accumulation. Secretory vesicles from both strains were resolved by differential centrifugation and separation on 15 to 30% Nycodenz gradients. Aliquots of the fractions obtained by density gradient centrifugation were analyzed for density, protein concentration, and the following enzymatic activities: H+-ATPase, acid phosphatase (Acid Pho.), invertase, and exoglucanase (see Materials and Methods). Enzyme activities are expressed in arbitrary units based on absorbance; acid phosphatase and exoglucanase were measured at 415 nm, ATPase was measured at 820 nm, and invertase was measured at 540 nm. (B and C) HA-Vsm1 does not localize to the secretory vesicle fraction. Aliquots of fractions from the density gradients were electrophoresed, blotted, probed with anti-HA antibody (1:5,000), and detected by chemiluminescence. Samples of total cell lysates (50 μg) from sec6-4 (B) and sec9-4 (C) cells shifted to 37°C or maintained at 26°C were run along with 40-μl aliquots from the gradients (TCL). The solid arrow indicates the position of the low-density peak of vesicles (present at 37°C), while the hatched arrow indicates the high-density peak (present at 37°C). (D) Vsm1 does not localize to the LDSV population that accumulates in snc vbm cells. Aliquots (40 μl) of fractions from the density gradients shown in Fig. 6 of reference 17 were electrophoresed, blotted, probed with anti-Vsm1 antibody (1:5,000), and detected by chemiluminescence. Samples of total cell lysates (50 μg) from snc vbm1 and snc vbm2 cells were run in parallel (TCL). The solid arrow indicates the position of the single peak of vesicles. (E) Western analysis of ER, Golgi, and post-Golgi markers in the Nycodenz gradient fractions of sec9-4 cells. Aliquots (40 μl) of the fractions were electrophoresed, blotted, and probed with the following antisera: anti-Wbp1 (1:6,000), anti-Mnn1 (1:1,500), anti-Sec22 (1:2,500), anti-Emp47 (1:3,000), anti-Snc (1:500), anti-Sec4 (1:1,000), anti-Sso (1:5,000), anti-Gas1 (1:5,000), and anti-Hsp150 (1:1,000). Detection was performed by chemiluminescence. Molecular masses are indicated on the left.
TABLE 4.
Cellular fractionationa
| Fraction | % of total
|
|||
|---|---|---|---|---|
| S10 | P10 | S100 | P100 | |
| 37°C | ||||
| Total protein | 70 | 26 | 34 | 12 |
| Vsm1 | 77 | 23 | 28 | 31 |
| Gas1 | 55 | 45 | 9 | 38 |
| Sso | 52 | 48 | 7 | 24 |
| 26°C | ||||
| Total protein | 73 | 21 | 33.5 | 6 |
| Vsm1 | 83 | 17 | 16 | 16 |
| Gas1 | 67 | 33 | 8 | 16 |
| Sso | 49 | 51 | 4 | 22 |
sec6-4 cells maintained at 26°C or shifted to 37°C for 1.5 h were lysed and fractionated as described in Materials and Methods. Total protein per fraction was determined by micro-bicinchoninic acid assay, and the relative amounts of Vsm1, Gas1, and Sso proteins were determined by quantitative Western analysis as described in Materials and Methods. The amounts of individual proteins were determined by autoradiography and densitometry. The percentage of each protein present in the various fractions was determined relative to the total density value for any given protein. The latter was calculated from the sum of values obtained from the S10 and P10 fractions. This combined value was basically identical to that determined for the total cell lysate.
We also note that both the higher- and lower-molecular-weight forms of Vsm1 associate with the P100 fraction in roughly similar amounts, indicating that its association with membranes is probably independent of this phenomenon. In addition, we found that the intracellular levels of Vsm1 are significantly higher in cells bearing mutations in vacuolar hydrolases (e.g., pep4 prb1 and pep4 prb1Δ) (Fig. 5C). The latter finding suggests that Vsm1 degradation in vivo may be dependent on these hydrolases, although we cannot preclude the possibility that reduced proteolytic activity in the lysates contributed to this result.
We next examined the localization of Vsm1 in wild-type yeast by immunofluorescence microscopy. Yeast cells expressing HA-tagged Vsm1 from single-copy plasmids were permeabilized and incubated with an affinity-purified monoclonal HA antibody and then with a FITC-labeled second antibody (Fig. 5D, panel a). The cell periphery was strongly labeled by using the anti-HA antibodies and did not correspond to the labeling of the nucleus, visualized by using propidium iodide (Fig. 5D, panel a). Moreover, the localization of Vsm1 was identical to that described previously for Sec9 (9) and for Sso protein, shown here for affinity-purified anti-Sso antibodies (Fig. 5C, panel d). This contrasts with labeling of the ER, revealed by anti-Dpm1 staining (Fig. 5C, panel b), or with labeling of the Golgi, revealed by anti-Mnn1 staining (17). Labeling of an HA-tagged Snc1 protein with affinity-purified anti-HA antibody (Fig. 5C, panel c) showed that Snc localizes to areas peripheral to the nucleus, possibly ER-Golgi, and also to the plasma membrane. Earlier studies, using thin-section microscopy and immunogold labeling, showed that Snc proteins label the plasma membrane of wild-type cells, along with some poorly defined areas in the cell, probably ER-Golgi (49). Vesicle localization of Snc protein is apparent only when cells accumulate secretory vesicles (15, 49). Together, these results (Fig. 5) suggest that Vsm1 is primarily membrane associated and may localize to the plasma membrane in wild-type cells.
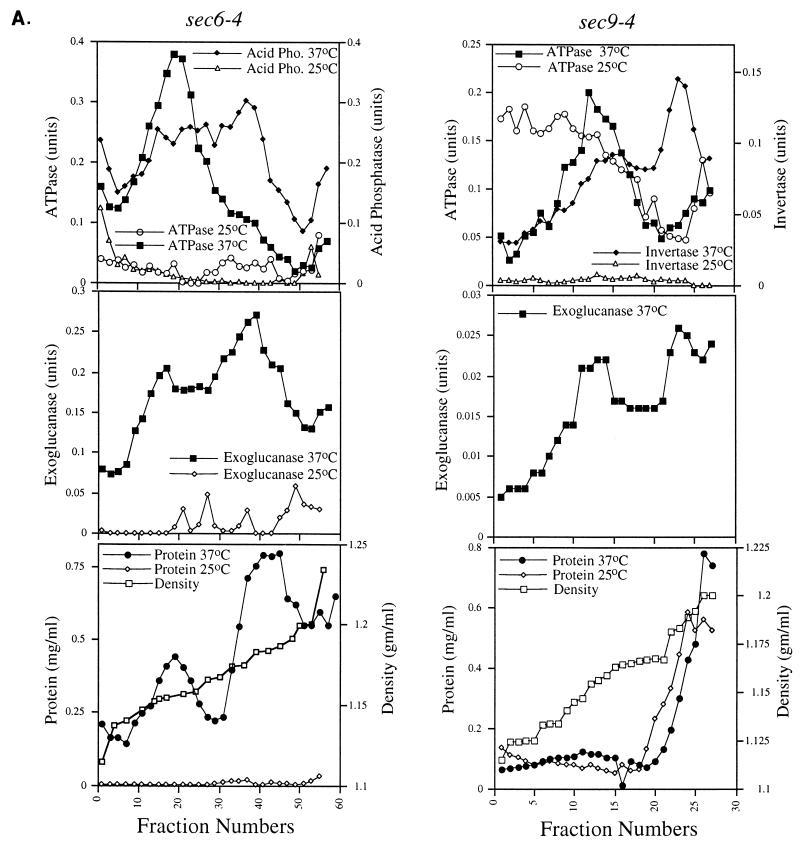
Vsm1 does not localize to secretory vesicles.
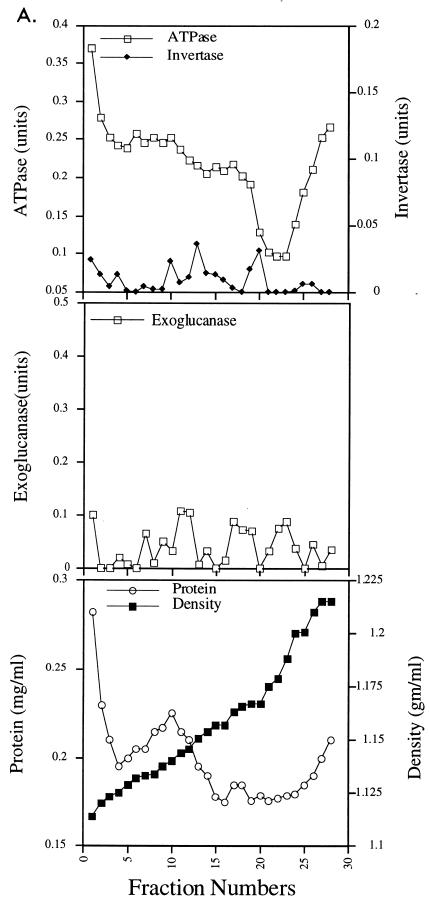
To determine whether Vsm1 also localizes to secretory vesicles, we determined whether the protein is present on vesicles that accumulate in late-acting sec mutants shifted to nonpermissive temperatures. Fractions enriched in the two different secretory vesicle populations, LDSVs and high-density secretory vesicles (HDSVs), which differ in both density and protein cargo content (17, 33), were separated by differential Nycodenz gradient centrifugation (Fig. 6A). In these experiments, we used both the sec6-4 and sec9-4 late-acting sec mutants to identify the LDSV and HDSV populations. We found that HDSVs fractionated at a density of 1.18 g/ml of Nycodenz and contain the soluble-secreted enzymes acid phosphatase, invertase, and exoglucanase. In contrast, LDSVs fractionated at a density of 1.156 g/ml of Nycodenz and are known to contain the Pma1 plasma membrane H+-ATPase activity. These values correspond well with results described by Harsay and Bretscher (33) and later by us (17). At permissive conditions, no significant amounts of enzymatic activity are found at these densities, indicating that vesicles do not accumulate (Fig. 6A, upper left and middle left panels; references 17 and 33). However, we did note that the membrane preparations from sec9-4 cells expressing VSM1 from a single-copy plasmid had elevated levels of ATPase activity in the early fractions of the gradient (see below).
Since Vsm1 is present in the 100,000 × g pellet (Fig. 5B and C), it was necessary to determine in definitive fashion whether Vsm1 colocalizes with secretory vesicles. To do so, we performed Western analysis on fractions obtained from the gradients that were electrophoresed on SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. We found that neither HA-tagged nor native Vsm1 protein colocalized significantly with the LDSV or HDSV population but rather eluted after the HDSV population (Fig. 6B). The HDSV fractions nominally contain secreted proteins and proteins associated with the plasma membrane, and not ER or Golgi markers (Fig. 6E; references 17 and 33). Specifically, they contain the Sso and Snc SNAREs and the secreted proteins, Hsp150 and Gas1 (Fig. 6E). Importantly, Vsm1 associated with the high-density fractions from sec6 (Fig. 6B) or sec9 (Fig. 6C) cells which were not temperature shifted and are not likely to accumulate secretory vesicles. Similar results were obtained for membrane preparations from snc null cells, which also accumulate the LDSV and HDSV populations (reference 17 and data not shown). In snc vbm1 or snc vbm2 cells, which accumulate only one population of vesicles at low density (17), Vsm1 was also found to elute before or after the vesicle peak (Fig. 6D). These results (Fig. 6) suggest that Vsm1 does not localize with the secretory vesicles that accumulate in the various late-acting sec and snc mutants. Moreover, they imply that Vsm1 associates with membranes in a manner which is largely independent of Snc protein.
Interestingly, we note that the Sec4 GTPase did not elute with the HDSV fractions but was clearly detectable early in the gradient and on the LDSVs. This finding suggests that Sec4 may not be a stable component of the HDSV population, in contrast to the exocytic v- and t-SNAREs.
LDSVs accumulate in sec9-4 cells which overexpress Vsm1.
Several results indicated that Vsm1 might act upon a specific class of vesicles. As shown in Fig. 3 and 4, sec9-4 cells overexpressing VSM1 grow slowly and accumulate vesicles in the buds of dividing cells under normally permissive conditions (26°C). Yet, invertase secretion from these cells was shown to be unaffected (Table 3). Moreover, elevated levels of ATPase activity were found in the fractions of lower density obtained from sec9-4 cells that overexpress VSM1 from a single-copy plasmid (Fig. 6A, upper right panel). This result was not observed in sec6-4 cells which overexpress VSM1.
To determine the type of vesicle that accumulates in sec9-4 cells overexpressing VSM1, we performed density gradient separation of 100,000 × g membrane preparations obtained from cells grown at permissive temperatures. Similar to results shown for sec9-4 cells in Fig. 6A, we found that sec9-4 cells overexpressing VSM1 from multicopy plasmids had even more elevated levels of H+-ATPase activity in the lower-density fractions of the gradient (Fig. 7A).  While eluting in the initial fractions of the gradient, the bulk of the H+-ATPase activity corresponded to the LDSV population, both on the basis of density as well as by visualization of the membrane that elutes in these fractions. Uranyl acetate staining and electron microscopy of membrane derived from fractions corresponding to the LDSV peak show the presence of 90- to 120-nm vesicles (fractions 14 and 15 [Fig. 7C]), which are similar to those shown previously to accumulate in sec6-4 or snc cells (17, 33). These vesicles were also found in the initial fractions of the gradient (fractions 4 and 5 [Fig. 7B]), along with the 200- to 300-nm microsomes and other membranes typically detected in these fractions (reference 17 and Fig. 7B). In contrast, the presence of an HDSV population was not detected in these gradients, as neither invertase nor exoglucanase activities were found in the high-density fractions (Fig. 7A) and no significant staining of the vesicle membrane was observed at the corresponding density (data not shown). Thus, the effects of VSM1 overproduction in sec9-4 yeast are likely to be due to the accumulation of the LDSV population of vesicles alone.
While eluting in the initial fractions of the gradient, the bulk of the H+-ATPase activity corresponded to the LDSV population, both on the basis of density as well as by visualization of the membrane that elutes in these fractions. Uranyl acetate staining and electron microscopy of membrane derived from fractions corresponding to the LDSV peak show the presence of 90- to 120-nm vesicles (fractions 14 and 15 [Fig. 7C]), which are similar to those shown previously to accumulate in sec6-4 or snc cells (17, 33). These vesicles were also found in the initial fractions of the gradient (fractions 4 and 5 [Fig. 7B]), along with the 200- to 300-nm microsomes and other membranes typically detected in these fractions (reference 17 and Fig. 7B). In contrast, the presence of an HDSV population was not detected in these gradients, as neither invertase nor exoglucanase activities were found in the high-density fractions (Fig. 7A) and no significant staining of the vesicle membrane was observed at the corresponding density (data not shown). Thus, the effects of VSM1 overproduction in sec9-4 yeast are likely to be due to the accumulation of the LDSV population of vesicles alone.
FIG. 7.
LDSVs accumulate in sec9-4 cells overexpressing VSM1. (A) sec9-4 cells expressing HA-VSM1 from a multicopy plasmid were grown to log phase, shifted to low-glucose-containing medium, and then processed to yield secretory vesicles as described in Materials and Methods. Vesicle-containing membrane fractions were resolved by differential centrifugation and separation on 15 to 30% Nycodenz gradients. Aliquots of the fractions obtained by density gradient centrifugation were analyzed for density, protein concentration, and the following enzymatic activities: H+-ATPase, invertase, and exoglucanase. Enzyme activities are expressed in arbitrary units based on absorbance; exoglucanase was measured at 415 nm, ATPase was measured at 820 nm, and invertase was measured at 540 nm. (B and C) Uranyl acetate-stained membranes from the early (B) and later (C) fractions of the gradient (fractions 4 and 5 and fractions 14 and 15, respectively). Bars represent 100 nm.
Vsm1 coprecipitates with Snc1 and Snc2 from detergent-solubilized cell extracts.
To verify the results obtained from the two-hybrid assay, we determined whether Vsm1 coimmunoprecipitates with Snc protein. We immunoprecipitated an epitope-tagged form of Snc2 (HA-Snc2) which was expressed from a single-copy plasmid. Analysis of immunoprecipitates probed with anti-Vsm1 antibody showed that the Vsm1 doublet can be coprecipitated with Snc2 protein (Fig. 8A and data not shown). This interaction appears specific and could be blocked when HA peptide was added in excess to the immunoprecipitation reaction. Thus, the protein-protein interaction identified previously between Vsm1 and Snc2 appears genuine. In contrast, in the representative experiment shown we were unable to demonstrate a similar interaction between Vsm1 and HA-tagged Snc1 when HA-Snc1 was expressed via a single-copy plasmid. This finding suggested either that Vsm1 binding is specific to Snc2 or that the affinity of the Snc1-Vsm1 interaction is substantially lower.
FIG. 8.
Vsm1 coimmunoprecipitates with Snc1 and Snc2. Cell lysates prepared from wild-type yeast expressing HA-tagged Snc1 or Snc2 from either single-copy (A) or multicopy (B) expression plasmids were subjected to immunoprecipitation (IP) with anti-HA antibody (ab). Duplicate samples were immunoprecipitated with excess HA peptide (pep) (75 μg). Immunoprecipitates were electrophoresed, blotted, and probed with either anti-Vsm1 (1:5,000) or anti-HA (1:5,000) antibody. Detection was performed by chemiluminescence (A) and by 125I-protein A labeling and autoradiography (B). For panel A, cell lysates were prepared from sec18-1 cells expressing HA-Snc2 that were either shifted to 37°C or maintained at 26°C (room temperature [RT]). Immunoprecipitation and detection were performed in a similar manner. Molecular masses are indicated on the left.
To verify whether the latter is true, we overproduced either HA-Snc1 or HA-Snc2 via multicopy plasmids in wild-type cells and immunoprecipitated the protein with anti-HA antibodies. This overexpression corresponds to roughly 10-fold that of the native level of expression (15). Quantitative immunoblot analysis revealed that Vsm1 could coprecipitate with HA-Snc1 (Fig. 8B), although the amount precipitated was at least 15-fold less than that which could coprecipitate with a corresponding amount of HA-Snc2, based on densitometric analysis of the precipitate and supernatant fractions (n = 2 experiments). This suggests that the affinity of Vsm1 for Snc1 may be substantially less than that for Snc2. Quantitative analysis of the immunoblots also revealed that over 60% of cellular Vsm1 could be precipitated with HA-Snc2 upon overexpression. At native levels of expression, we estimate that only about 20% of cellular Vsm1 can coprecipitate with Snc protein (data not shown).
The yeast NSF homolog Sec18 has been suggested to act at a level either preceding SNARE complex assembly (43) or occurring after complex formation, leading to disassembly (61). To determine whether the association of Vsm1 to Snc2 is affected by the inactivation of Sec18, we performed immunoprecipitation experiments in sec18-1 cells either maintained at permissive temperatures or shifted to restrictive conditions for 15 min. In these experiments, we were unable to detect any significant difference in the amounts of Vsm1 bound to HA-Snc2 under the different conditions (Fig. 8A). This makes it unlikely that the Snc2-Vsm1 interaction is directly affected by the loss of Sec18 function. In parallel immunoprecipitation experiments, we also determined that Vsm1 could not coprecipitate with the Sso proteins (data not shown).
DISCUSSION
In screening for interacting partners for the Snc v-SNAREs, we isolated a gene, designated VSM1, which encodes a novel SNARE-binding protein that may act at the level of secretory vesicle docking and fusion. The protein encoded by VSM1 binds tightly to the Snc2 v-SNARE and less so to Snc1, as evidenced by direct immunoprecipitation experiments (Fig. 8). This finding suggests that there may be a preference in the specificity of action of this SNARE-binding protein. Specificity in other SNARE-binding proteins has been described for the mammalian Sec1 protein, which interacts solely with syntaxins and not with either VAMP or SNAP-25 (24, 34, 48), as well as for synaptophysin, which interacts solely with synaptobrevin/VAMP (19, 69).
Genetic studies reveal important clues regarding the function of Vsm1 in yeast. First, overexpression of VSM1 in cells bearing a ts sec9-4 allele results in an inhibition of cell growth (Fig. 3) and an accumulation of vesicles (Fig. 4) at permissive temperatures and, at nonpermissive temperatures, blocks rescue conferred by overproduction of either Snc v-SNARE (Fig. 3 and data not shown). This phenotype is allele specific, as VSM1 overexpression does not inhibit the growth of cells bearing a sec9-7 allele (Fig. 3C), which encodes a mutant Sec9 t-SNARE that is capable of forming a ternary complex in vitro but does not confer secretory functions in vivo at restrictive temperatures (53). Second, we found that VSM1 overexpression specifically results in the accumulation of LDSVs which possess H+-ATPase activity (Fig. 7A) and a modest decrease in protein secretion into the medium (Table 2). In contrast, its deletion results in an increase in medium protein secretion (Table 3). Third, neither overexpression nor deletion of VSM1 affects the synthesis, processing, and secretion of invertase (Table 2 and data not shown). Together, these results imply that the inhibitory effect of Vsm1 overproduction may be conferred at the level of SNARE assembly, resulting in a partial block in the docking and fusion of the LDSVs. The results suggest that Vsm1 acts in a restrictive capacity prior to SNARE complex formation, although this remains to be formally proven. However, we cannot rule out an additional role for Vsm1 after membrane fusion has occurred.
Earlier results suggested that the two Snc v-SNAREs may fulfill slightly different functions although, individually, each confers normal cellular viability and the secretion of invertase and fully couples vesicle transport in the absence of the other isoform (29, 49). Previously, we had found that only SNC1 overexpression could suppress ts defects exhibited by the sec9-4 and sso2-1 alleles, whereas Snc2 either could not or did so weakly (14, 29). The basis for this finding has remained unclear, although it could reflect alternative modes of SNARE regulation, perhaps by specific SNARE binding proteins. This possibility is partially supported by the experiments performed here, which demonstrate a weaker interaction between Vsm1 and Snc1 than with Snc2 (Fig. 8). Since these v-SNAREs differ significantly only in the first 30 amino acids, the so-called variable region that is not essential for exocytic function (29), we suggest that it may be important for Vsm1 binding. The ability of Vsm1 to bind Snc proteins is expected to block productive interactions with the t-SNARE partners and, thus, prevent rescue of the ts defects in t-SNARE mutants (29), as shown here (Fig. 3B and data not shown). Yet, the deletion of VSM1 in sec9-4 cells did not render them less sensitive to temperature (data not shown). Thus, it appears unlikely that Vsm1 is the sole regulator of the Snc v-SNAREs. Deletion of VSM1 does not result in any deleterious phenotype or yield synthetic lethal interactions with known components of the secretory pathway, which indicates that Vsm1 does not fulfill an essential role in mediating cellular secretion. Alternatively, it may share redundant functions with another protein that could act more specifically upon Snc1. As no other structural homolog of Vsm1 is encoded by the yeast genome, such a regulator will have to be identified by using other techniques.
The results shown in this study suggest that Vsm1 plays a distinct role in the trafficking of one of two classes of secretory vesicles found in yeast. Vsm1 overproduction selectively results in the accumulation of the low-density class of vesicle, which indicates that LDSV docking and fusion may be more strongly influenced by the levels of this protein. What distinguishes between these two classes and renders LDSVs more sensitive to Vsm1 regulation is unclear. The results of this study suggest that the Vsm1-regulated class of vesicles, the LDSVs, are directly responsible for the delivery of medium proteins (e.g., HSP150) to the cell surface, in contrast with the HDSV class, which delivers soluble secreted periplasmic enzymes such as acid phosphatase, invertase, and exoglucanase to the surface (17, 33). Vsm1 could also play a role in the trafficking of other vesicle types in yeast (32, 38, 58), but this has not yet been determined.
Immunofluorescence (Fig. 5D) and membrane fractionation (Fig. 5B and C; Fig. 6) studies support the idea that Vsm1 is associated with the plasma membrane and not directly with secretory vesicles. This observation contrasts with our initial prediction that Vsm1 acts throughout the secretory pathway to restrict Snc v-SNARE function and suggests that their association may occur solely at the level of the plasma membrane. Thus, Vsm1 is likely to be a late-acting factor in the docking and fusion steps, in addition to its predicted inhibitory role. Alternatively, it could play a role in the retrieval of Snc proteins from the plasma membrane, although there is no pressing reason to believe that there is a deficiency in Snc protein available during LDSV biogenesis. Its precise physiological role remains, therefore, unclear, and in vitro binding studies may shed light on the nature and timing of the Vsm1-Snc interactions. Quantitative analysis of the immunoprecipitation experiments revealed that only 20% of cellular Vsm1 can be precipitated with Snc protein at native levels of expression. Vsm1 is mostly membrane associated, even in cells lacking Snc protein (Fig. 6D and data not shown), which suggests that other sites of anchorage are operant at the level of the membrane.
Here, we have cloned and initially characterized a novel SNARE binding protein that may modulate constitutive secretion from yeast. If Vsm1 is indeed a negative regulator of constitutive exocytosis, there may a temporal and spatial need to regulate the trafficking of the different vesicle populations, presumably in coordinating bud growth and septation once establishment of a secretion landmark and rearrangement of the cortical actin cytoskeleton have occurred. Interestingly, we note that VSM1 was identified independently as a gene adjacent to MAG1 and, like MAG1, is induced upon DNA damage and in the presence of cycloheximide (41). This finding suggests that under conditions in which growth arrest is paramount (i.e., during environmental stress), genes like VSM1 may be up-regulated in order to block cellular secretion and growth. Whether Vsm1 has a role in gene regulation, as suggested by Liu et al. (41), we cannot say, but such a role would seem less likely in the context of the results shown here.
ACKNOWLEDGMENTS
We are grateful to Tamara Doering, Scott Emr, Erin Gaynor, Todd Graham, Peter Novick, Patrick Brennwald, Howard Riezmann, Randy Schekman, and Michael Wigler for the generous gifts of antibodies and yeast strains. A special acknowledgment goes to Peter Novick, who coined the term SNARE-master. Thanks go to Vera Shinder for electron microscopy and Shmuel Pietrovsky for database analyses.
This work was supported by grants to J.E.G. from the Ebner Family Foundation for Biomedical Research; Forchheimer Center for Molecular Genetics; and Minerva Foundation, Munich, Germany. J.E.G. is holder of the Henry Kaplan Chair in Cancer Research.
REFERENCES
- 1.Aalto M K, Ruohonen L, Hosono K, Keranen S. Cloning and sequencing of the yeast Saccharomyces cerevisiae SEC1 gene localized on chromosome IV. Yeast. 1992;8:587–588. doi: 10.1002/yea.320080710. [DOI] [PubMed] [Google Scholar]
- 2.Aalto M K, Ronne H, Keränen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames B N. Assay of inorganic phosphate, total phosphate, and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 4.Araki S, Tamori Y, Kawanishi M, Shinoda H, Masugi J, Mori H, Niki T, Okazawa H, Kubota T, Kasuga M. Inhibition of the binding of SNAP-23 to syntaxin 4 by Munc18c. Biochem Biophys Res Commun. 1997;234:257–262. doi: 10.1006/bbrc.1997.6560. [DOI] [PubMed] [Google Scholar]
- 5.Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert M, Maycox P R, Navone F, DeCamilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicle of rat brain. EMBO J. 1989;8:379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett M K, Scheller R H. The molecular machinery for secretion is conserved from yeast to neurons. Proc Natl Acad Sci USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman B J, Slayman C W. The effects of vanadate on the plasma membrane ATPase of Neurospora crassa. J Biol Chem. 1979;254:2928–2934. [PubMed] [Google Scholar]
- 9.Brennwald P, Kerns B, Champion K, Keränen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be an effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 10.Calakos N, Scheller R H. Vesicle-associated membrane protein and synaptophysin are associated on the synaptic vesicle. J Biol Chem. 1994;269:24534–24537. [PubMed] [Google Scholar]
- 11.Calakos N, Bennett M K, Peterson K E, Scheller R H. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- 12.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNAREs. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coorsen J R, Blank P S, Tahara M, Zimmerberg J. Biochemical and functional studies of cortical vesicle fusion: the SNARE complex and Ca2+ sensitivity. J Cell Biol. 1998;143:1845–1857. doi: 10.1083/jcb.143.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couve A, Gerst J E. Yeast Snc proteins complex with Sec9: functional interactions between putative SNARE proteins. J Biol Chem. 1994;269:23391–23394. [PubMed] [Google Scholar]
- 15.Couve A, Protopopov V, Gerst J E. Yeast synaptobrevin homologs are modified post-translationally by palmitate addition. Proc Natl Acad Sci USA. 1995;92:5987–5991. doi: 10.1073/pnas.92.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dascher C, Ossig R, Gallwitz D, Schmitt H D. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David D, Sundarababu S, Gerst J E. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee T, Becherer K, Chen P-L, Yeh S H, Yang Y, Kilburn A E, Kee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 19.Edelmann L, Hanson P I, Chapman E R, Jahn R. Synaptobrevin binding to synaptophysin: a potential mechanism for controlling the exocytotic fusion machine. EMBO J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elferink L A, Trimble W S, Scheller R H. Two vesicle-associated membrane protein genes are differentially expressed in the rat central nervous system. J Biol Chem. 1989;264:11061–11064. [PubMed] [Google Scholar]
- 21.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 22.Galli T, McPherson P S, DeCamilli P. The V0 sector of the V-ATPase, synaptobrevin, and synaptophysin are associated on synaptic vesicles in a Triton X-100-resistant, freeze-thawing sensitive, complex. J Biol Chem. 1996;271:2193–2198. doi: 10.1074/jbc.271.4.2193. [DOI] [PubMed] [Google Scholar]
- 23.Garcia E P, Gatti E, Butler M, Burton J, DeCamilli P. A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci USA. 1994;91:2003–2007. doi: 10.1073/pnas.91.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia E P, McPherson P S, Chilcote T J, Takei K, DeCamilli P. rbSec1A and B colocalize with syntaxin 1 and SNAP-25 throughout the axon, but are not in a stable complex with syntaxin. J Cell Biol. 1995;129:105–120. doi: 10.1083/jcb.129.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaynor E C, Emr S D. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gengyo-Ando K, Kamiya Y, Yamakawa A, Kodaira K, Nishiwaki K, Miwa J, Hori I, Hosono R. The C. elegans unc-18 gene encodes a protein expressed in motor neurons. Neuron. 1993;11:703–711. doi: 10.1016/0896-6273(93)90080-b. [DOI] [PubMed] [Google Scholar]
- 27.Gerst J E, Ferguson K, Vojtek A, Wigler M, Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol Cell Biol. 1991;11:1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerst J E, Rodgers L, Riggs M, Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc Natl Acad Sci USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerst J E. Conserved α-helical segments on yeast homologs of the synaptobrevin/VAMP family of v-SNAREs mediate exocytic function. J Biol Chem. 1997;272:16591–16598. doi: 10.1074/jbc.272.26.16591. [DOI] [PubMed] [Google Scholar]
- 30.Gerst, J. E. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell. Mol. Life Sci., in press. [DOI] [PMC free article] [PubMed]
- 31.Goldstein A, Lampen J O. β-d-Fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- 32.Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hata Y, Slaughter C A, Sudhof T C. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 35.Hay J C, Scheller R H. SNAREs and NSF in targeted membrane fusion. Curr Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof T C, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn R, Schiebler W, Ouimet C, Greengard P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci USA. 1985;83:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston G C, Prendergast J A, Singer R A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for the vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katagiri H, Terasaki J, Murata T, Ishihara H, Ogihara T, Inukai K, Fukushima Y, Anai M, Kikuchi M, Miyazaki J. A novel isoform of syntaxin-binding protein homologous to yeast Sec1 expressed ubiquitously in mammalian cells. J Biol Chem. 1995;270:4963–4966. doi: 10.1074/jbc.270.10.4963. [DOI] [PubMed] [Google Scholar]
- 40.Lian J P, Ferro-Novick S. Bos1p, an integral membrane protein of endoplasmic reticulum to Golgi transport vesicles is required for fusion competence. Cell. 1993;73:504–511. doi: 10.1016/0092-8674(93)90253-m. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Dai H, Xiao W. UAS(MAG1), a yeast cis-acting element that regulates the expression of MAG1, is located within the protein coding region of DDI1. Mol Gen Genet. 1997;255:533–542. doi: 10.1007/s004380050526. [DOI] [PubMed] [Google Scholar]
- 42.Lupashin V V, Waters M G. SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- 43.Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion in yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 44.McMahon H T, Ushkaryov Y A, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Sudhof T C. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- 45.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 46.Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 47.Pevsner J, Hsu S C, Scheller R H. n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci USA. 1994;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pevsner J, Hsu S C, Braun J E, Calakos N, Ting A E, Bennett M K, Scheller R H. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- 49.Protopopov V, Govindan B, Novick P, Gerst J E. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- 50.Regazzi R, Sadoul K, Meda P, Kelly R B, Halban P A, Wollheim C B. Mutational analysis of VAMP domains implicated in Ca2+-induced insulin exocytosis. EMBO J. 1996;15:6951–6959. [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson J S, Klionsky D J, Banta L M, Emr S D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 53.Rossi G, Salminen A, Rice L, Brünger A T, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C-terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16623. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- 54.Rothman J E, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 55.Salzberg A, Cohen N, Halachmi N, Kimchie Z, Lev Z. The Drosophila Ras2 and Rop gene pair: a dual homology with a yeast Ras-like gene and a suppressor of its loss-of-function phenotype. Development. 1993;117:1309–1319. doi: 10.1242/dev.117.4.1309. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos T, del Rey F, Conde J, Villanueva J R, Nombela C. Saccharomyces cerevisiae mutant defective in exo-1,3-β-glucanase production. J Bacteriol. 1979;139:333–338. doi: 10.1128/jb.139.2.333-338.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulze K L, Littleton J T, Salzberg A, Halachmi N, Stern M, Lev Z, Bellen H J. rop, a Drosophila homolog of yeast Sec1 and vertebrate n-Sec1/Munc-18 proteins, is a negative regulator of neurotransmitter release in vivo. Neuron. 1994;13:1099–1108. doi: 10.1016/0896-6273(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 61.Søgaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Söllner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 62.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 63.Trimble W S, Cowan D M, Scheller R H. VAMP-1: a synaptic vesicle-associated integral membrane protein. Proc Natl Acad Sci USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ungermann C, Nichols B J, Pelham H R B, Wickner W. A v-t SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140:61–70. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ungermann C, Sato K, Wickner W. Defining functions of trans-SNARE pairs. Nature. 1998;396:543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- 66.Van Rijn H J M, Boer P, Steyn-Parvé E P. Biosynthesis of acid phosphatase of baker’s yeast. Factors influencing its production by protoplasts and characterization of the secreted enzyme. Biochim Biophys Acta. 1972;268:431–441. doi: 10.1016/0005-2744(72)90339-7. [DOI] [PubMed] [Google Scholar]
- 67.Walworth N C, Novick P J. Purification and characterization of constitutive secretory vesicles from yeast. J Cell Biol. 1987;105:163–174. doi: 10.1083/jcb.105.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walworth N C, Brennwald P, Kabcenell A K, Garrett M, Novick P. Hydrolysis of GTP by Sec4 plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Washbourne P, Schiavo G, Montecucco C. Vesicle-associated membrane protein-2 (synaptobrevin-2) forms a complex with synaptophysin. Biochem J. 1995;305:721–724. doi: 10.1042/bj3050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Sollner T H, Rothman J E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 71.Wiedenmann B, Francke W W. Identification and localization of synaptophysin, an integral membrane glycoprotein of Mr 38,000 characteristic of presynaptic vesicles. Cell. 1985;41:1017–1028. doi: 10.1016/s0092-8674(85)80082-9. [DOI] [PubMed] [Google Scholar]
- 72.Xu Z, Wickner W. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J Cell Biol. 1996;132:787–794. doi: 10.1083/jcb.132.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Z, Mayer A, Muller E, Wickner W. A heterodimer of thioredoxin and I(B)2 cooperates with Sec18p (NSF) to promote yeast vacuole inheritance. J Cell Biol. 1997;136:299–306. doi: 10.1083/jcb.136.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Z, Sato K, Wickner W. LMA1 binds to vacuoles at Sec18p (NSF), transfers upon ATP hydrolysis to a t-SNARE (Vam3p) complex, and is released during fusion. Cell. 1998;93:1125–1134. doi: 10.1016/s0092-8674(00)81457-9. [DOI] [PubMed] [Google Scholar]
- 75.Zelicof A, Protopopov V, David D, Lin X-Y, Lustgarten V, Gerst J E. Two separate functions are encoded by the carboxyl-terminal domains of the yeast cyclase-associated protein and its mammalian homologs: dimerization and actin-binding. J Biol Chem. 1996;271:18243–18252. doi: 10.1074/jbc.271.30.18243. [DOI] [PubMed] [Google Scholar]