Abstract
Background:
Regulator of calcineurin 1.4 (RCAN1.4) is a functionally downregulated metastasis progression suppressor (MPS) in thyroid cancer; however, the mechanisms for RCAN1.4 loss in thyroid cancer have not yet been reported. The RCAN1.4 promoter and gene contain several cytosine-guanine (CG)-rich regions, some of which are reported to be hypermethylated in nonthyroid tissues. We, therefore, hypothesized that RCAN1.4 downregulation in thyroid cancer was in part due to hypermethylation.
Methods:
Studies were performed in 5 thyroid cancer cell lines (TPC1, FTC133, BCPAP, C643, and 8505C) with different genetic drivers, and in 18 paired normal and thyroid cancer human thyroid cancer tissues. Basal RCAN1.4 messenger RNA (mRNA) and protein levels were assessed in all of the cell lines. Cell lines with lowest RCAN1.4 expression levels were treated with the DNA methyl transferase inhibitor, decitabine. Normal/tumor tissue pairs were analyzed for methylation of three CG-rich regions both by capture of methylated DNA by MBD2 protein and by methylation-specific polymerase chain reaction (MSPCR).
Results:
In all assessed cell lines, RCAN1.4 mRNA and protein levels increased after decitabine treatment. In silico analysis of the RCAN1.4 gene identified 3 CG-rich regions as possible methylation targets: 1 in the proximal promoter and 2 in intron 1. Hypermethylation of the intron 1 CG-rich regions was identified by both the capture method and MSPCR. In contrast, hypermethylation of the CG-rich region of the proximal promoter was not identified. Gene expression confirmed that hypermethylation in thyroid cancer samples in intron 1 of RCAN1.4 was associated with lower levels of RCAN1.4 mRNA. Finally, the cancer samples demonstrated increased NFE2L3 expression, a downstream marker of functional RCAN1.4 loss.
Conclusions:
The MPS gene, RCAN1.4, is downregulated in thyroid cancer cells and human thyroid cancer in part by hypermethylation of CG-rich regions in intron 1.
Keywords: NFE2L3, metastatic dormancy, gene regulation
Introduction
Thyroid cancer is the most common malignancy of a classical endocrine organ, accounting for an estimated 43,800 new cases and 2230 deaths in 2022 in the United States.1 The progressive increase in frequency of thyroid cancer diagnosis has stabilized in recent years with adherence to new guidelines.2,3 However, the number of individuals who die from thyroid cancer has not reduced, mostly due to progressive disease and metastasis.2 It is well recognized that many patients with thyroid cancer distant metastasis have nonprogressive disease for decades, and that clinically silent distant metastases are common on autopsy.4–6
Thus, thyroid cancer is an important model to study “gatekeepers” of progression, including gain of secondary drivers and loss of metastasis progression suppressor (MPS) genes, which might serve as predictive biomarkers of disease-specific mortality and/or as new therapeutic targets.4,7
The multiple steps involved in cancer metastasis have been extensively studied and modeled.8,9 For most tumor types, a subset of cancer cells undergo an epithelial-to-mesenchymal transition, enabling local invasion and motility.10–12 Individual or clusters of cancer cells invade, intravasate into the vasculature, circulate, and a subpopulation extravasates and survives in the metastatic environment (disseminated tumor cells [DTCs]).13,14 The process also involves release of factors from the primary tumor that support development of a premetastatic niche.15–17
Surviving metastatic DTCs can immediately proliferate leading to cancer progression or enter dormancy in which they survive in a quiescent state.18 These “dormant” cells are relatively therapeutically resistant, thus, when they escape from dormancy, they are difficult to treat.19 Understanding the factors that lead cancers to emerge from dormancy represents a key translational need to properly target therapies at the time of cancer progression. In metastatic thyroid cancer, despite the tendency for prolonged dormancy, new driver gene mutations unique to newly invasive primary cancers or growing metastases are relatively uncommon.20,21
This feature has enabled the use of primary tumor genomics to predict response to targeted therapies,20,22 but also defines the need for a deeper understanding of factors leading to cancer progression to improve outcomes since current therapies are not curative. Due to this relative paucity of new driver mutations in progressive thyroid cancer, we posited that loss of MPSs may be important in thyroid cancer progression.4,19 MPS genes encode proteins that when lost facilitate primary cancer growth, invasion, metastasis, and progression, but not transformation separating them from tumor suppressors.7,23
We initially identified regulator of calcineurin 1.4 (RCAN1.4) as a potential MPS in an objective screen for downstream pathways of the KiSS1/GPR54 MPS pathway.24,25 Subsequent studies demonstrated its MPS function in vitro and in vivo and confirmed reduced levels in advanced thyroid cancer samples and metastatic lesions.24,26 RCAN1.4 is one of two expressed isoforms of the RCAN1 gene (RCAN1.1 and RCAN1.4) expressed with unique promoters from the same gene that originally was identified as the Down's syndrome critical region 1 (DSCR1) gene on chromosome 21.27–30
RCAN1.4 was identified as the primary inducible RCAN1 transcript. RCAN1.4 functions as a negative regulator of calcineurin/NFAT signaling, thereby reducing cell proliferation, migration, and apoptosis.29,31–34 Interestingly, individuals with Down's syndrome who express 3 copies of chromosome 21 have very low incidence of solid tumors.35 This reduced cancer incidence can be partially attributed to the three copies of RCAN1 in mouse model systems.33,36–38
Using transcriptomics, we identified that RCAN1.4 loss also results in an increase in NFE2L3 levels, a member of the Cap N’ Collar family of basic leucine zipper transcription factors, and that this gene is functionally required for RCAN1.4 loss-mediated cancer progression.26 An antiprogression role for RCAN1.4 subsequently has been confirmed for renal cell carcinoma,39 hepatocellular cancer,32 osteosarcoma,31 pancreatic cancer,40 and breast cancer.36 RCAN1.4 also was reported to be a functional target of the prometastatic mIR-619-5p that suppresses RCAN1.4 levels, leading to angiogenesis and metastasis nonsmall cell lung cancer.41
Hypermethylation of the RCAN1.4 proximal promoter or intron 1 has been reported in liver fibrosis and hepatocellular carcinoma cells, respectively,32,42 but has not been studied in the thyroid cancer. This study aims to test the hypothesis that RCAN1.4 levels are reduced in thyroid cancer in part due to hypermethylation. Our results demonstrate that RCAN1.4 levels are reduced in thyroid cancer by methylation of cytosine-guanine (CG)-rich regions of intron 1 in association with increased expression of NFE2L3.
Materials and Methods
Cell lines, cell culture, in vitro drug reagent
Five thyroid cancer cell lines were used with mutational information detailed by Landa et al.,43 8505C (anaplastic thyroid cancer [ATC]; BRAFV600E), TPC1 (papillary thyroid cancer [PTC]; RET/PTC1), BCPAP (PTC; BRAFV600E), FTC-133 (follicular thyroid cancer, PTEN null), and C643 (ATC; HRAS). All cell lines were independently DNA fingerprinted for identification. 8505C, BCPAP, and C643 cells were cultured in RPMI with TPC1 and FTC-133 cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37°C. RPMI, DMEM, and FBS were obtained from ThermoFisher Scientific (118775119, Gibco; 11960069, Gibco; 26140079, Gibco, respectively).
Cells were split approximately every 3–4 days using 0.05% trypsin–EDTA (25300120; Gibco). For experiments, cells were split, washed, and placed in culture conditions as already mentioned except with 1% FBS for 24 hours. The DNA methyltransferase inhibitor decitabine (S1200; Selleckchem) diluted in 0.2% dimethyl sulfoxide (DMSO), or diluent alone, was added at the shown concentrations.
Western blot
Cells were plated on 10 cm dishes and allowed to incubate for 24 hours. Medium was changed to 1% FBS and allowed to incubate for 24 hours. Plates were treated with decitabine at 0, 5, and 10 μM concentrations for 72 hours; cells were isolated in phosphate-buffered saline (PBS; 10010-023; Gibco); and lysed with MPER buffer (78501; ThermoFisher). Protein concentrations were measured and quantified by Pierce BCA Protein Assay Kit (23225; ThermoFisher).
Protein was combined with NuPAGE LDS sample buffer (4 × NP0007; ThermoFisher), DTT, and ddH2O, boiled for 5 minutes, and 20 μg protein per well for Western blot. Samples were run, transferred, and blocked with 5% bovine serum albumin (BSA) for 1 hour. Membranes were incubated in primary antibody overnight at 4°C, washed 3 times with PBS, and incubated with secondary antibody at room temperature for 1 hour. After washing, blots were imaged using the Odyssey CLx LI-COR Imager and ImageStudio software (LI-COR). Primary antibodies were the following: anti-DSCR1 (RCAN; 1:1000, D6694-200uL; Millipore) and β-Actin (1:5000, sc-8432; Santa Cruz).
Human samples, DNA extraction, and sequencing
Eighteen papillary thyroid tumor and opposite lobe histologically normal tissues from consecutive unselected patients were collected after obtaining informed consent as part of an Ohio State University IRB-approved protocol (2006C0047) in accordance with the Declaration of Helsinki (revised 2013). Deidentified clinical/pathological information is listed in Supplementary Table S2. DNA was isolated from the paired thyroid tissue samples and five cell lines using phenol–chloroform extraction. RNase A (12091021; Invitrogen) was applied during the extraction procedure. Methods for BRAF and TERT promoter sequencing are included in Supplementary Methods.
DNA methylation analysis
Capture of methylated DNA by MBD protein
DNA fragmentation was obtained by restriction digestion using MseI; efficiency was confirmed by agarose gel electrophoresis. Three DNA fragment positions are: chr21:34527031–34527182, chr21:34525269–34525646, and chr21:34524894–34525268 (hg38). Methylated DNA was isolated using CpG MethylQuest DNA Isolation Kit (Cat. No. 17-10035; Millipore) according to the manufacturer's protocol. In brief, 400 ng MseI-digested DNA fragments were incubated with CpG MethylQuest beads. The beads were washed three times and eluted to isolate the methylated DNA.
Primers were designed to amplify the CG-rich regions in RCAN1.4 proximal promoter and the two CG-rich regions of intron 1 individually. Quantitative polymerase chain reaction (qPCR) was performed with methylated DNA using Fast SYBR Green Master mix (ThermoFisher), and the percentage of methylation then was calculated as the amount over input DNA. Primers are given in Supplementary Table S1.
Qualitative methylation-specific polymerase chain reaction
DNA was converted by using EpiTect Bisulfite Kit (Cat. No. 59104; Qiagen) as per the manufacturer's protocol. Methylated and unmethylated DNA-specific primers for intron 1 are from Jin et al.32 and are listed in Supplementary Table S1. Forty cycles of polymerase chain reaction (PCR) were performed and results were analyzed qualitatively. Methylation-specific PCR (MSPCR) was not possible in the proximal promoter region, and there were not suitable primers that could amplify the two intron 1 regions separately. Thus, this method was used for intron 1 methylation encompassing both regions.
RNA preparation, messenger RNA analysis, quantitative real-time PCR
Thyroid tissue total RNA was extracted by using TRIzol (15596018; Invitrogen). One microgram RNA was treated with DNA-free DNA Removal Kit (AM1906; ThermoFisher) and reverse transcribed to complementary DNA (cDNA; High-Capacity cDNA Reverse Transcription Kit; 4368814, Applied Biosystems). qPCR was performed with Fast SYBR Green Master mix for RCAN1.4 and TaqMan Fast Universal PCR Master Mix (4352042; ThermoFisher) for NFE2L3. GAPDH was used as an internal control to calculate the relative expression levels for gene RCAN1.4 and NFE2L3. PCR primers are shown in the Supplementary Table S1.
Data analyses
All cell line experiments were repeated at least three times. For real-time PCR (RT-PCR) and methylation experiments, technical replicates were performed for each independent experiment. One-way analysis of variance was used to determine the significant differences (p < 0.05) between groups in protein and messenger RNA (mRNA) data. After stabilizing methylation proportions by the arcsine square root transformation, nonpaired or paired Student t-tests were applied appropriately to compare groups.
Quantitative real-time PCR (qRT-PCR) relative expression data were log2 transformed and tumor versus normal comparison analyses were performed by applying paired Student t-tests. Western blots were quantified using ImageJ (1.53a; National Institute of Health). Statistical analysis and graph generation were done using Prism GraphPad (9.2.0) and R software.
Results
Decitabine upregulates RCAN1.4 mRNA and protein levels in thyroid cancer cells
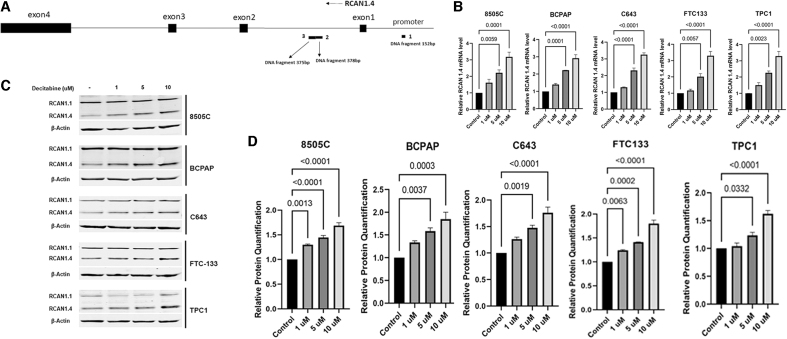
After initial time course studies, the 5 thyroid cancer cell lines were treated with decitabine for 48 hours and RNA was extracted. Then, 1, 5, and 10 μM doses were used along with a nontreatment control. qRT-PCR was performed. Results from 3 independent experiments show RCAN1.4 RNA levels increase with increasing drug concentration, with significant increases after treatment with 5 and 10 μM decitabine for all cell lines (Fig. 1B).
FIG. 1.
Decitabine increases protein and mRNA levels. (A) Diagram of RCAN1.4 showing the three CG-rich regions in the promoter and intron 1 with the sizes of the DNA fragments. (B) Cells were treated with increasing doses of decitabine (0, 1, 5, and 10 μM) for 48 hours, mRNA levels were measured; RCAN1.4 levels increased after treatment with 5 or 10 μM decitabine. (C, D) Cells were treated with increasing doses of decitabine (0, 1, 5, and 10 μM) for 72 hours. RCAN1.4 protein levels increased in all cell lines with 5 or 10 μM dosing. β-actin was used as loading control, n = 3 for all experiments. Statistical comparisons were performed using one-way ANOVA; p < 0.05 is significant. ANOVA, analysis of variance; mRNA, messenger RNA; RCAN1.4, regulator of calcineurin 1.4.
To assess effect of demethylation on RCAN1.4 protein levels, after initial time course studies, thyroid cancer cells were treated with decitabine (0, 1, 5, and 10 μM) for 72 hours. RCAN1.4 protein levels significantly increased in all cell lines after decitabine treatment in three independent experiments (Fig. 1C, D). RCAN1.1 protein levels did not show an increase, suggesting that the regulatory regions for RCAN1.1 gene expression that are unique from RCAN1.430 are not hypermethylated. β-actin was used as loading control for all experiments.
Hypermethylation was detected at RCAN1.4 intron 1 with thyroid cancer cell lines and PTC tissues
Previous studies reported hypermethylation in the 5′ regulatory region and intron 1 of RCAN1.4.32 To determine whether RCAN1.4 downregulation in thyroid cancer tissues is associated with DNA methylation at these sites, methylation analysis at the three potential methylation sites in RCAN1.4 promoter and intron 1 was performed (Fig. 1A).
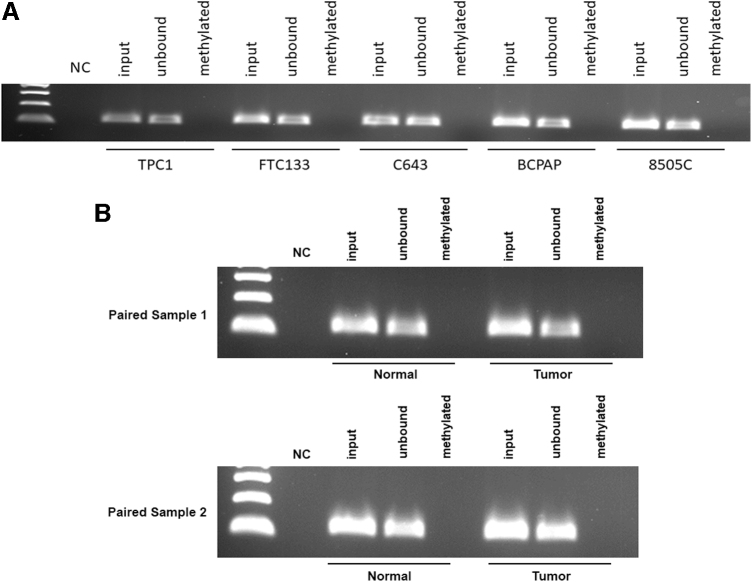
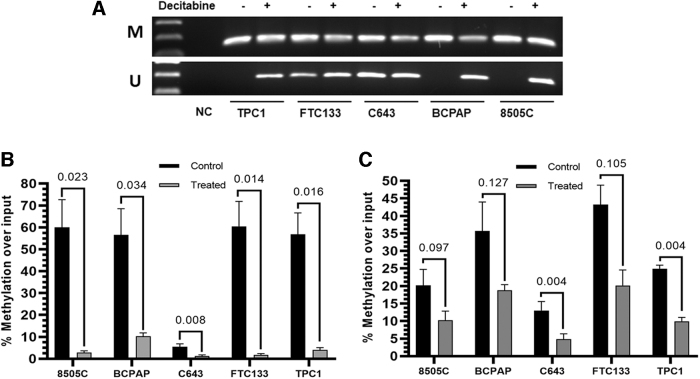
Using the quantitative capture assay, we did not detect methylation in the CG-rich region proximal to the RCAN1.4 promoter region in the five thyroid cancer lines (Fig. 2A, B). MSPCR was not possible in the promoter region as there were not suitable sequences for primers. To assess methylation in intron 1, qualitative methylation-specific PCR identified methylation in the five cell lines. Subsequent studies demonstrated that the intron 1 methylation was partially reversed with decitabine (Fig. 3A).
FIG. 2.
(A) Promoter region shows no hypermethylation in all five cell lines. (A) DNA was isolated from five cell lines and quantitative capture of methylated DNA by MBD2 protein bead precipitation and PCR was performed, n = 3. (B) DNA from 18 paired normal and tumor thyroid samples were analyzed by quantitative capture of methylated DNA by MBD2 protein bead precipitation. Two samples shown in Figure 2, all samples are shown in Supplementary Figure S1. NC, negative control; PCR, polymerase chain reaction.
FIG. 3.
- Decitabine decreases RCAN1.4 intron 1 methylation in thyroid cancer cell lines (A) Cells were treated with decitabine (10 μM) for 72 hours and DNA isolated. Qualitative methylation-specific PCR for both regions of intron 1 was performed and demonstrated relatively less methylation after decitabine treatment for most samples. (B, C) Quantitative capture of methylated DNA by MBD2 protein bead precipitation and PCR was performed for region 2 (B) and region 3 (C). Significant reductions of methylation were identified for region 2 in all cell lines and in 2 of 5 cell lines for region 3 (n = 3). Student t-tests were used to compare results in (B, C), p < 0.05 is considered significant. M, methylated; U, unmethylated.
This method was not able to be performed separately for the two adjacent GC-rich regions in intron 1. To confirm and extend the data quantitatively for each region, subsequent studies using the quantitative capture assay confirmed hypermethylation in both regions of intron 1. They also demonstrated quantitative reversal with decitabine treatment more consistently in region 2 versus region 3 (Fig. 3B).
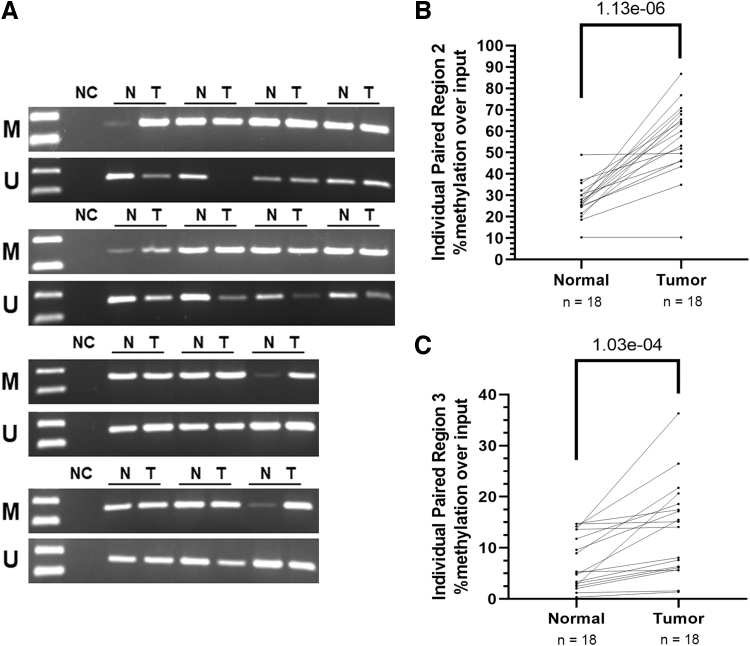
To validate the findings in human thyroid cancer tissues, we performed DNA methylation analysis with 18 paired PTC normal and tumor tissue samples. As for the cell lines, there was no evidence of methylation of the proximal promoter region (Supplementary Figs. S1 and S2). Qualitative analysis of intron 1 by MSPCR in 14 of the sample pairs suggested a relative increase in methylation in most of the samples (Fig. 4A). Four of the samples did not have adequate DNA from complete pairs for both methods and we prioritized the quantitative region-specific capture assay.
FIG. 4.
RCAN1.4 intron 1 is hypermethylated in tumor PTC vs. paired normal tissue samples. Paired thyroid cancer normal and tumor samples were analyzed for RCAN1.4 methylation of intron 1. (A) Qualitative methylation-specific PCR was performed to assess methylation of intron 1 (both regions) in 14 PTC normal and tumor paired samples with adequate DNA with results suggesting relatively higher methylated: unmethylated DNA in tumor samples. (B) Quantitative capture of methylated DNA by MBD2 at RCAN1.4 intron 1 for all 18 pairs confirmed higher levels of methylation in both regions 2 and 3 in thyroid cancer samples. Paired Student t-test was used for statistical analysis; p < 0.05 is significant. N, normal; PTC, papillary thyroid cancer; T, tumor.
Using this assay, higher methylation in both intron 1 regions in tumor versus normal tissue was identified (region 2: p = 1.13e-06, region 3: 1.03e-04) (Fig. 4B). Region 2 effects were more consistent than region 3 effects (higher in 16/18 tumors vs. normal), a result that is similar to the thyroid cancer cell lines.
RCAN1.4 expression is lower and NFE2L3 expression is higher in PTC samples
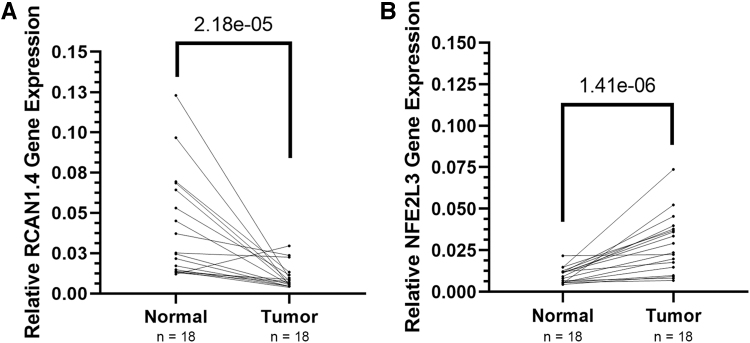
To determine whether the human tumor samples had lower levels of RCAN1.4 versus normal tissue we performed qRT-PCR and identified lower levels in the tumor samples versus paired normal samples (Fig. 5, p = 2.18e-05). As a measure of biological effects of RCAN1.4 loss in the paired thyroid cancer samples, we analyzed gene expression for NFE2L3 to assess for the predicted inverse results based on our prior functional studies.26 Results in the paired samples show the predicted higher levels of NFE2L3 in tumor versus the normal samples (Fig. 5, p = 1.41e-06).
FIG. 5.
RCAN1.4 gene expression is downregulated while NFE2L3 gene expression is upregulated in PTC tissues. RCAN1.4 and NFE2L3 gene expression levels in 18 paired papillary thyroid cancer normal and tumor samples were assessed by qRT-PCR. (A) Reduced levels of RCAN1.4 mRNA were found in 16 of 18 samples (p = 2.18e-05). (B) Increased levels of NFE2L3 were identified in 17 of 18 samples (p = 1.41e-06). Graphs represent relative gene expression compared with normal tumor samples, with each normal sample paired with the corresponding tumor sample. Data were log2 transformed and analyses were performed using paired Student t-tests; p < 0.05 is significant. qRT-PCR, quantitative real-time PCR.
Discussion
Differentiated thyroid cancer typically is characterized by relatively slow growth rates and prolonged stability in both primary and distant metastatic sites. However, local invasion and progression of distant metastases are the most common causes of thyroid cancer morbidity and mortality. Thus, understanding the molecular causes of progression of thyroid cancer represents an important opportunity to identify new biomarkers and therapeutic targets (reviewed in Summers et al.,19 Ringel,44 and Ganesh and Massagué45). We previously identified and characterized RCAN1.4 as a bona fide functional MPS in thyroid cancer that is downregulated in some primary and most metastatic lesions, a finding that has been confirmed in a number of other malignancies.24–26,31,32,36,39,40
Although RCAN1.4 is known to regulate calcineurin signaling, and does so in thyroid cancer cells,24,30 we identified a new functional downstream target, NFE2L3 (Nrf3), for RCAN1.4 loss in cancer cells.26 NFE2L3 is a member of the Cap N’ Collar family of transcription factors known to be important in promoting cancer development and progression.46–50 In addition, association studies confirmed a progressive increase in NFE2L3 levels in more aggressive thyroid cancers and metastatic lesions.26 Although these data defined a new RCAN1.4/NFE2L3 regulatory pathway for cancer progression, the causes of the reduction of RCAN1.4 levels in thyroid cancers had not been addressed.
In this study, we hypothesized that the reduced levels of RCAN1.4 in thyroid cancer would be, at least in part, due to hypermethylation of regulatory sequences. This hypothesis is built on an initial analysis of the RCAN1.4 promoter and introns for CG-rich regions and published data demonstrating regulation of RCAN1.4 in the proximal promoter or in intron 1 in hepatic stellate cells or hepatocellular carcinoma cells in vitro.32,42 Our data demonstrate that the proximal promoter is not hypermethylated either in tested thyroid cancer cell lines or in human thyroid cancers. In contrast, hypermethylation was identified in in intron 1 in the two predicted regions, most convincingly in region 2 (Fig. 1A).
The intron 1 hypermethylation was reversed with decitabine in concert with the predicted increase in RCAN1.4 levels. Importantly, human thyroid cancer samples (mostly PTC) showed similar RCAN1.4 methylation and gene expression patterns. Functional activation of the pathway is suggested by the increased expression of NFE2L3 in association with the RCAN1.4 loss.26
Epigenetic methylation is important in regulating expression of genes important for tumor progression.51,52 For instance, clustered and single circulating tumor cells express differential methylation patterns in accordance with upregulated proliferation and stem cell genes.53 Experiments are ongoing to identify the upstream regulators of the hypermethylated intron 1 regions. Intriguing recent data identified a superenhancer in intron 1 of RCAN1.4 in breast cancer.36 It is possible that hypermethylation reduces access to the superenhanced region, thereby downregulating RCAN1.4 expression after cellular stress.
This hypothesis requires further evaluation of superenhancer activity in the intron 1 sites in the thyroid cell context. Finally, RCAN1.4 has been reported to be phosphorylated by several kinases in some cellular contexts.54–57 It is possible that these modifications, and others, regulate activity and/or stability of the protein in thyroid cells. Thus, hypermethylation may not account for all downregulation of RCAN1.4 activity.
We identified the hypermethylated regions in thyroid cancer cell lines and validated the findings in human thyroid cancers. These results provide a basis for future studies assessing a potential role for RCAN1.4 methylation as a predictive or prognostic biomarker in thyroid cancer. Although MPS gene reductions can enable primary cancer progression as well as metastases, it is possible that the quantitative level of RCAN1.4 intron 1 hypermethylation may increase in metastatic lesions versus paired primary tumor tissue.
This may be suggested by the progressive increase in NFE2L3 in metastatic lesions we identified in our prior studies.26 Further studies using paired samples from patients with metastatic lesions are planned to assess this possibility. It is of interest that all of the human tumor samples in this study harbored BRAFV600E mutations. While the cell lines with hypermethylation were not specific for this mutation, analysis of larger and different histologies will be needed to determine whether this association is related to BRAFV600E.
In conclusion, we demonstrated for the first time that intron 1 of RCAN1.4 is hypermethylated at CG-rich regions in thyroid cancer cells. We also report that a methyltransferase inhibitor reverses the methylation and increases RCAN1.4 mRNA and protein. Our results also demonstrate hypermethylation of this same region in human PTC in association with reduced expression of RCAN1.4. The reduced RCAN1.4 is inversely correlated with NFE2L3 expression, a pattern consistent with functional loss of RCAN1.4 that also is consistent with prior association data.26 These results demonstrate that reduction of the RCAN1.4 MPS gene in thyroid cancer is in part due to hypermethylation of intron 1, thereby providing a regulatory mechanism for loss of this key regulator of thyroid cancer progression.
Supplementary Material
Authors' Contributions
T.K. contributed to methodology, investigation, analysis, and writing; N.R. was involved in methodology, investigation, analysis, and writing; W.L. carried out methodology, investigation, analysis, and writing; S.L. took charge of analysis and writing; M.D.R. was in charge of conceptualization, methodology, investigation, analysis, and resources.
Author Disclosure Statement
The authors all declare that there are no conflicts of interest.
Funding Information
This study was supported by grants R01CA240302 and R01CA227847 to M.D.R. and in part by the OSUCCC Core Grant P30CA0166058. T.K., N.R., W.L., and S.L. were supported by the mentioned grants to M.D.R.
Supplementary Material
References
- 1. Surveillance, Epidemiology, and End Results Program (SEER). SEER Cancer Stat Facts: Thyroid Cancer. Bethesda, MD; 2022. Available from: https://seer.cancer.gov/statfacts/html/thyro.html [Last accessed: December 1, 2022].
- 2. American Cancer Society (ACS). Thyroid Cancer Survival Rates, by Type and Stage. 2022. Available from: https://www.cancer.org/cancer/thyroid-cancer/detection-diagnosis-staging/survival-rates.html [Last accessed: December 1, 2022].
- 3. Pereira M, Williams VL, Johnson JH, et al. Thyroid Cancer Incidence Trends in the United States: Association with Changes in Professional Guideline Recommendations. Thyroid 2020;30(8):9., doi: 10.1089/thy.2019.0415 [DOI] [PubMed] [Google Scholar]
- 4. Rajan N, Khanal T, Ringel MD. Progression and dormancy in metastatic thyroid cancer: Concepts and clinical implications. Endocrine 2020;70(1):12; doi: 10.1007/s12020-020-02453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robenshtok E, Neeman B, Reches L, et al. Adverse histological features of differentiated thyroid cancer are commonly found in autopsy studies: Implications for treatment guidelines. Thyroid 2022;32(1):37–45; doi: 10.1089/thy.2021.0268 [DOI] [PubMed] [Google Scholar]
- 6. Hugen N, Sloot YJE, Netea-Maier RT, et al. Divergent metastatic patterns between subtypes of thyroid carcinoma results from the nationwide Dutch pathology registry. J Clin Endocrinol Metab 2020;105(3):e299–e306; doi: 10.1210/clinem/dgz078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hurst DR, Welch DR. Metastasis suppressor genes: At the interface between the environment and tumor cell growth. Int Rev Cell Mol Biol 2011;286:107–180; doi: 10.1016/B978-0-12-385859-7.00003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011;147(2):18; doi: 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res 2019;79(12):17; doi: 10.1158/0008-5472.CAN-19-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bakir B, Chiarella AM, Pitarresi JR, et al. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol 2020;30(10):13; doi: 10.1016/j.tcb.2020.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jolly MK, Ware KE, Gilja S, et al. EMT and MET: Necessary or permissive for metastasis? Mol Oncol 2017;11(7):20; doi: 10.1002/1878-0261.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suhail Y, Cain MP, Vanaja K, et al. Systems biology of cancer metastasis. Cell Syst 2019;9(2):19; doi: 10.1016/j.cels.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeeshan R, Mutahir Z. Cancer metastasis—Tricks of the trade. Bosnian J Basic Med Sci 2017;17(3):11; doi: 10.17305/bjbms.2017.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Majidpoor J, Mortezaee K. Steps in metastasis: An updated review. Med Oncol 2020;38(3):17; doi: 10.1007/s12032-020-01447-w [DOI] [PubMed] [Google Scholar]
- 15. Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther 2021;221:107753; doi: 10.1016/j.pharmthera.2020.107753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell 2016;30(5):14; doi: 10.1016/j.ccell.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 17. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res 2019;79(18):10; doi: 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neophytou CM, Kyriakou T-C, Papageorgis P. Mechanisms of metastatic tumor dormancy and implications for cancer therapy. Int J Mol Sci 2019;20(24):21; doi: 10.3390/ijms20246158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Summers MA, McDonald MM, Croucher PI. Cancer cell dormancy in metastasis. Cold Spring Harb Perspect Med 2020;10(4):9; doi: 10.1101/cshperspect.a037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shonka DC Jr., Ho A, Chintakuntlawar AV, et al. American Head and Neck Society Endocrine Surgery Section and International Thyroid Oncology Group consensus statement on mutational testing in thyroid cancer: Defining advanced thyroid cancer and its targeted treatment. Head Neck 2022;44(6):1277–1300; doi: 10.1002/hed.27025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Justiniano SE, McElroy JP, Yu L, et al. Genetic variants in thyroid cancer distant metastases. Endocr Relat Cancer 2016;23(10):L33–L36; doi: 10.1530/ERC-16-0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krane JF, Cibas ES, Endo M, et al. The Afirma Xpression Atlas for thyroid nodules and thyroid cancer metastases: Insights to inform clinical decision-making from a fine-needle aspiration sample. Cancer Cytopathol 2020;128(7):452–459; doi: 10.1002/cncy.22300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Horak CE, Lee JH, Marshall J-C, et al. The role of metastasis suppressor genes in metastatic dormancy. J Pathol Microbiol Immunol 2008;116(7–8):21; doi: 10.1111/j.1600-0463.2008.01027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Espinosa AV, Shinohara M, Porchia LM, et al. Regulator of calcineurin 1 modulates cancer cell migration in vitro. Clin Exp Metastasis 2009;26(6):517–526; doi: 10.1007/s10585-009-9251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stathatos N, Bourdeau I, Espinosa AV, et al. KiSS-1/G protein-coupled receptor 54 metastasis suppressor pathway increases myocyte-enriched calcineurin interacting protein 1 expression and chronically inhibits calcineurin activity. J Clin Endocrinol Metab 2005;90(9):5432–5440; doi: 10.1210/jc.2005-0963 [DOI] [PubMed] [Google Scholar]
- 26. Wang C, Saji M, Justiniano SE, et al. RCAN1-4 is a thyroid cancer growth and metastasis suppressor. JCI Insight 2017;2(5):16; doi: 10.1172/jci.insight.90651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lao M, Zhang X, Ma T, et al. Regulator of calcineurin 1 gene isoform 4 in pancreatic ductal adenocarcinoma regulates the progression of tumor cells. Oncogene 2021;40(17):3136–3151; doi: 10.1038/s41388-021-01763-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuentes JJ, Pritchard MA, Planas AM, et al. A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum Mol Genet 1995;4(10):1935–1944; doi: 10.1093/hmg/4.10.1935 [DOI] [PubMed] [Google Scholar]
- 29. Oh M, Dey A, Gerard RD, et al. The CCAAT/enhancer binding protein beta (C/EBPbeta) cooperates with NFAT to control expression of the calcineurin regulatory protein RCAN1–RCAN4. J Biol Chem 2010;285(22):16623–16631; doi: 10.1074/jbc.M109.098236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davies KJ, Ermak G, Rothermel BA, et al. Renaming the DSCR1/Adapt78 gene family as RCAN: Regulators of calcineurin. FASEB J 2007;21(12):3023–3028; doi: 10.1096/fj.06-7246com [DOI] [PubMed] [Google Scholar]
- 31. Huang B, Jiang Z, Wu S, et al. RCAN1.4 suppresses the osteosarcoma growth and metastasis via interfering with the calcineurin/NFAT signaling pathway. J Bone Oncol 2021;30:100383; doi: 10.1016/j.jbo.2021.100383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin H, Wang C, Jin G, et al. Regulator of calcineurin 1 gene isoform 4, down-regulated in hepatocellular carcinoma, prevents proliferation, migration, and invasive activity of cancer cells and metastasis of orthotopic tumors by inhibiting nuclear translocation of NFAT1. Gastroenterology 2017;153(3):799..e33–811.e33; doi: 10.1053/j.gastro.2017.05.045 [DOI] [PubMed] [Google Scholar]
- 33. Minami T, Yano K, Miura M, et al. The Down syndrome critical region gene 1 short variant promoters direct vascular bed-specific gene expression during inflammation in mice. J Clin Invest 2009;119(8):2257–2270; doi: 10.1172/JCI35738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iizuka M, Abe M, Shiiba K, et al. Down syndrome candidate region 1, a downstream target of VEGF, participates in endothelial cell migration and angiogenesis. J Vasc Res 2004;41(4):334–344; doi: 10.1159/000079832 [DOI] [PubMed] [Google Scholar]
- 35. Hasle H, Friedman JM, Olsen JH, et al. Low risk of solid tumors in persons with Down syndrome. Genet Med 2016;18(11):1151–1157; doi: 10.1038/gim.2016.23 [DOI] [PubMed] [Google Scholar]
- 36. Deng R, Huang J-H, Wang Y, et al. Disruption of super-enhancer-driven tumor suppressor gene RCAN1.4 expression promotes the malignancy of breast carcinoma. Mol Cancer 2020;19(122):19; doi: 10.1186/s12943-020-01236-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baek KH, Zaslavsky A, Lynch RC, et al. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 2009;459(7250):1126–1130; doi: 10.1038/nature08062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryeom S, Baek KH, Zaslavsky A. Down's syndrome: Protection against cancer and the therapeutic potential of DSCR1. Future Oncol 2009;5(8):1185–1188; doi: 10.2217/fon.09.88 [DOI] [PubMed] [Google Scholar]
- 39. Song Z, Cao Q, Ruan H, et al. RCAN1.4 acts as a suppressor of cancer progression and sunitinib resistance in clear cell renal cell carcinoma. Exp Cell Res 2018;372(2):11; doi: 10.1016/j.yexcr.2018.09.017 [DOI] [PubMed] [Google Scholar]
- 40. Lao M, Zhang X, Yang H, et al. RCAN1-mediated calcineurin inhibition as a target for cancer therapy. Mol Med 2022;28(1):69; doi: 10.1186/s10020-022-00492-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim DH, Park S, Kim H, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett 2020;475:2–13; doi: 10.1016/j.canlet.2020.01.023 [DOI] [PubMed] [Google Scholar]
- 42. Pan XY, You HM, Wang L, et al. Methylation of RCAN1.4 mediated by DNMT1 and DNMT3b enhances hepatic stellate cell activation and liver fibrogenesis through Calcineurin/NFAT3 signaling. Theranostics 2019;9(15):4308–4323; doi: 10.7150/thno.32710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Landa I, Pozdeyev N, Korch C, et al. Comprehensive genetic characterization of human thyroid cancer cell lines: A validated panel for preclinical studies. Clin Cancer Res 2019;25(10):11; doi: 10.1158/1078-0432.CCR-18-2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ringel MD. New horizons: Emerging therapies and targets in thyroid cancer. J Clin Endocrinol Metab 2021;106(1):e382–e388; doi: 10.1210/clinem/dgaa687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ganesh K, Massagué J. Targeting metastatic cancer. Nat Med 2021;27(1):34–44; doi: 10.1038/s41591-020-01195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kobayashi A, Ito E, Toki T, et al. Molecular cloning and functional characterization of a new Cap'n’ collar family transcription factor Nrf3. J Biol Chem 1999;274(10):6443–6452; doi: 10.1074/jbc.274.10.6443 [DOI] [PubMed] [Google Scholar]
- 47. Chevillard G, Blank V. NFE2L3 (NRF3): The Cinderella of the Cap'n'Collar transcription factors. Cell Mol Life Sci 2011;68(20):3337–3348; doi: 10.1007/s00018-011-0747-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chowdhury A, Katoh H, Hatanaka A, et al. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci Rep 2017;7(1):12494; doi: 10.1038/s41598-017-12675-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bury M, Le Calve B, Lessard F, et al. NFE2L3 controls colon cancer cell growth through regulation of DUX4, a CDK1 inhibitor. Cell Rep 2019;29(6):1469..e9–1481.e9; doi: 10.1016/j.celrep.2019.09.087 [DOI] [PubMed] [Google Scholar]
- 50. Kobayashi A. Roles of NRF3 in the hallmarks of cancer: Proteasomal inactivation of tumor suppressors. Cancers (Basel) 2020;12(9):2681; doi: 10.3390/cancers12092681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kulis M, Esteller M.. 2—DNA Methylation and Cancer. In: Advances in Genetics. (Herceg Z, Ushijima T. eds.) Academic Press, Elsevier: Oxford, UK; 2010; p. 396. [DOI] [PubMed] [Google Scholar]
- 52. Yu B, Yu X, Xiong J, et al. Methylation modification, alternative splicing, and noncoding RNA play a role in cancer metastasis through epigenetic regulation. Biomed Res Int 2021;2021:4061525; doi: 10.1155/2021/4061525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 2019;176(1–2):15; doi: 10.1016/j.cell.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Q, Busby JC, Molkentin JD. Interaction between TAK1-TAB1-TAB2 and RCAN1-calcineurin defines a signalling nodal control point. Nat Cell Biol 2009;11(2):154–161; doi: 10.1038/ncb1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Song WJ, Song EA, Choi SH, et al. Dyrk1A-mediated phosphorylation of RCAN1 promotes the formation of insoluble RCAN1 aggregates. Neurosci Lett 2013;554:135–140; doi: 10.1016/j.neulet.2013.08.066 [DOI] [PubMed] [Google Scholar]
- 56. Kim SS, Lee EH, Lee K, et al. PKA regulates calcineurin function through the phosphorylation of RCAN1: Identification of a novel phosphorylation site. Biochem Biophys Res Commun 2015;459(4):604–609; doi: 10.1016/j.bbrc.2015.02.155 [DOI] [PubMed] [Google Scholar]
- 57. Han KA, Yoo L, Sung JY, et al. Leucine-rich repeat kinase 2 (LRRK2) stimulates IL-1beta-mediated inflammatory signaling through phosphorylation of RCAN1. Front Cell Neurosci 2017;11:125; doi: 10.3389/fncel.2017.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.