Abstract
In response to activation of the Wnt signaling pathway, β-catenin accumulates in the nucleus, where it cooperates with LEF/TCF (for lymphoid enhancer factor and T-cell factor) transcription factors to activate gene expression. The mechanisms by which β-catenin undergoes this shift in location and participates in activation of gene transcription are unknown. We demonstrate here that β-catenin can be imported into the nucleus independently of LEF/TCF binding, and it may also be exported from nuclei. We have introduced a small deletion within β-catenin (Δ19) that disrupts binding to LEF-1, E-cadherin, and APC but not axin. This Δ19 β-catenin mutant localizes to the nucleus because it may not be efficiently sequestered in the cytoplasm. The nuclear localization of Δ19 definitively demonstrates that the mechanisms by which β-catenin localizes in the nucleus are completely independent of LEF/TCF factors. β-Catenin and LEF-1 complexes can activate reporter gene expression in a transformed T-lymphocyte cell line (Jurkat) but not in normal T lymphocytes, even though both factors are nuclear. Thus, localization of both factors to the nucleus is not sufficient for activation of gene expression. Excess β-catenin can squelch reporter gene activation by LEF-1–β-catenin complexes but not activation by the transcription factor VP16. Taken together, these data suggest that a third component is necessary for gene activation and that this third component may vary with cell type.
Wnt signals initiate at the plasma membrane of receptive cells when the secreted ligand Wnt binds to frizzled, a seven-transmembrane receptor on the plasma membrane (9, 10, 15, 43). For Wnt-1, this binding event activates a multistep cascade that directs nuclear transport of a cytoplasmic protein named β-catenin. Wnt-1 directs nuclear localization of β-catenin in part by inhibiting the activity of the serine/threonine kinase GSK-3β via the action of a cytoplasmic phosphoprotein named dishevelled (14, 35, 61, 68). Most models portray GSK-3β, along with another protein called APC (for adenomatous polyposis coli), as promoting β-catenin degradation via the ubiquitin-proteasome pathway (1, 52, 57). Under these circumstances, β-catenin is stable only at the plasma membrane in cell adherens junctions as an adapter protein between the cytoplasmic tail of E-cadherin receptors and α-catenin, a cytoskeleton binding protein. A Wnt-dependent increase in cytosolic β-catenin is soon followed by the appearance of β-catenin in the nucleus, where it cooperates with LEF/TCF (for lymphoid enhancer factor and T-cell factor) proteins to alter transcription of gene targets (29, 44, 49, 53). In addition to these parts of the cascade, additional steps are involved because proteins such as axin/conductin/axil and GBP have been identified recently as regulatory components of this pathway (6, 32, 34, 69, 70, 72). The final steps of this cascade, nuclear accumulation and gene activation by β-catenin, are not well characterized.
β-Catenin is known to interact with a number of proteins, and the list of these interactions is growing (conductin/axin/axil, APC, α-catenin, E-cadherin, presenilin, LEF/TCF proteins) (10, 71). These proteins are localized to different subcellular compartments, such as the plasma membrane (E-cadherin, α-catenin), the endoplasmic reticulum and Golgi (presenilin and derivative fragments), the cytoplasm (APC, conductin/axin/axil), and the nucleus (LEF/TCF transcription factors). Although interaction domains for each individual protein have not been precisely delimited, it appears that β-catenin interacts with these proteins through overlapping portions of its armadillo repeat array (reference 10 and references therein). The repeating unit of this array, the armadillo repeat (referred to here as the arm repeat) is so named because of its sequence similarity to a reiterated, degenerate 37- to 43-amino-acid (aa) motif in the Drosophila melanogaster ortholog armadillo (41, 50). Multiple arm repeats occur in tandem arrays from as few as 4 to as many as 13 repeats, and together they function as a protein interaction domain. The crystal structure for the 12 arm repeats of β-catenin was solved recently (28). Structural analysis has revealed that arm repeats are alpha-helical and pack against one another to form an elongated superhelix of alpha-helices. The groove created by the twisted superhelical structure is highly charged and proposed to be the interacting surface for β-catenin targets. The structure of a second armadillo repeat protein, yeast importin α/karyopherin α/Srp1, was recently determined, and while the amino acid sequence of its 9- or 10-arm repeat array is only 17% similar to β-catenin, the repeats fold into a similar twisted structure with a groove (13). Importin α, and a second arm repeat protein, importin β, function together as receptors for nuclear localization signals (NLSs) in proteins (26, 47). NLSs are highly basic, whereas β-catenin targets are enriched in acidic residues. Thus, even though arm repeat arrays are found in a wide variety of proteins and the amino acid sequences of their targets vary, there are unifying structural similarities in the way these proteins bind to their targets.
A major unanswered question is how β-catenin localizes to different subcellular compartments. For example, since β-catenin does not have an obvious NLS for importin α/β receptors, it was proposed that β-catenin must enter the nucleus by binding to NLS-containing LEF/TCF transcription factors in the cytoplasm (29, 33, 43, 44). An alternative model, in which β-catenin can access the nucleus independently of LEF/TCF binding (17), has been proposed. This model was derived from a study of nuclear localization of recombinant β-catenin in digitonin-permeabilized cells where β-catenin entered the nucleus on its own, in the absence of exogenously added extract (17). The inference from this study was that β-catenin transport was independent of importin/karyopherins and also of any LEF/TCF protein.
In the nucleus, β-catenin is known to interact with at least one class of transcription factors, the LEF/TCF family of proteins (36, 44, 62, 64, 66). LEF/TCF factors are sequence-specific DNA binding proteins that function as context-dependent transcription factors, unable to work on their own but requiring the cooperative effort of adjacent DNA binding transcription proteins or non-DNA binding cofactors (11, 21). LEF/TCF proteins have a single HMG (high-mobility-group) DNA binding domain that includes an HMG box, which functions as a DNA binding-bending motif, and a B box, a very basic cluster of 8 or 9 aa with dual functions in DNA binding and nuclear localization (20, 23, 40, 54). β-Catenin binds to a region within the first 56 aa of all known LEF/TCF proteins (7, 29, 44). The highly conserved β-catenin binding domain in LEF/TCF proteins is separate and independent from the HMG DNA binding domain, which is located in the middle or near the C terminus. A model has been proposed whereby the tethering of β-catenin to promoters or enhancers by LEF/TCF factors enables a transcription activation domain in β-catenin, perhaps in cooperation with LEF/TCF sequences, to regulate transcription of Wnt-responsive genes (27, 63). Reporter plasmids containing only multimerized LEF/TCF binding sites are activated by this complex in a context-independent manner that apparently does not require additional factors binding to flanking regulatory elements (27, 29, 36, 44, 45, 53, 59, 63). However, whether there is a requirement for additional factors that do not bind DNA is not known. We have constructed a deletion mutant that does not bind LEF/TCF proteins and have used this mutant protein to explore the mechanisms by which β-catenin accumulates in nuclei and activates gene expression. We show that β-catenin localizes to nuclei independently of its interaction with LEF/TCF. We also show that in the nucleus β-catenin cooperates with LEF-1 and perhaps at least one other factor to activate reporter gene expression.
MATERIALS AND METHODS
Cell culture, DNA transfections, luciferase and β-galactosidase assays.
Cos-1 cells were cultured in high-glucose (4.5-g/l) Dulbecco’s modified Eagle’s medium (Gibco). 293 cells were cultured in Eagle’s minimum essential medium with nonessential amino acids (Irvine Scientific). Jurkat cells were cultured in RPMI 1640 (Irvine Scientific). All media contained 10% fetal bovine serum (Irvine Scientific), 100 U of penicillin per ml, 100 μg of streptomycin per ml, 250 ng of amphotericin B (Gibco) per ml, and 50 μM β-mercaptoethanol. Cell lines were grown in a humidified atmosphere containing 5% CO2 at 37°C. Cos-1, 293, and Jurkat cells were transiently transfected by electroporation. Normal human peripheral blood mononuclear cells (PBMCs) were prepared and transfected as previously described (30). For reporter gene expression, 1 μg of a reporter gene plasmid containing five tandemly repeated Gal4 recognition elements upstream of a luciferase reporter gene (generous gift of P. Carlsson, Göteborg University, Göteborg, Sweden) was cotransfected with various amounts of G4LEF-1, β-catenin, and control expression plasmids as indicated in the figure legends. One microgram of the reporter plasmid Top TK (generous gift of H. Clevers, University of Utrecht) was cotransfected with 10 μg each of wild-type β-catenin and wild-type LEF-1 expression plasmids for reporter gene expression in PBMCs. Cells were harvested at 15 to 24 h posttransfection, and cell lysates were analyzed for luciferase activity as described previously (42). To normalize between transfections, 0.5 μg of LacZ expression plasmid was included in each sample and β-galactosidase activity was determined with the Galacto-Light/Plus kit (Tropix, Inc.).
Plasmid construction.
c-Myc epitope-tagged Xenopus full-length β-catenin from pCS2 (generous gift of B. Gumbiner, Memorial Sloan-Kettering Cancer Center, New York, N.Y.) was cloned into pBluescript at the BamHI and HincII sites. An internal deletion of aa 346 to 364 of β-catenin (Δ19 β-catenin) was constructed by digesting β-catenin in pBluescript with AflII and HindIII. Blunt ends were generated with Klenow enzyme, and the ends were religated. Δ19 β-catenin was then transferred to pCS2 in the same orientation as β-catenin in pCS2. An N-terminal deletion of the first 148 aa of Δ19 β-catenin (ΔNΔ19 β-catenin) was constructed by digesting MT10 β-catenin in pCS2 (aa 149 to 525) (generous gift of B. Gumbiner) with XhoI and SnaBI and replacing this fragment with a fragment from Δ19 β-catenin digested at the identical sites. All mutations were sequenced to ensure that no second-site mutations were introduced during the cloning procedure. Construction of G4LEF, G4LEF (aa 80 to 256), FL LEF, G4VP16, and ΔNLEF (aa 67 to 399) is mentioned elsewhere (11).
GST fusion protein purification.
Glutathione S-transferase (GST) and full-length GST–hLEF-1 were purified as described previously (54). GST–importin-β (generous gift of S. A. Adam, Northwestern University) was purified as above in the presence of 0.1 mM ZnSO4.
In vitro binding and immunoprecipitation assays.
Radiolabeled full-length β-catenin, Δ19 β-catenin, the C-terminal tail of E-cadherin (generous gift of P. Polakis, Onyx Pharmaceuticals), full-length pendulin/Rch1/importin-α2, and an N-terminal deletion of pendulin (aa 62 to 529 [ΔN importin-α2]) (55) were prepared with a coupled in vitro transcription/translation system (Promega) and [35S]methionine (DuPont-NEN). The in vitro binding assay was performed as previously described (55). For the coimmunoprecipitation assay, 293 cells were transiently transfected by electroporation with 10 μg of the plasmids described in the figure legends. At 24 h after transfection, cells were harvested and washed once in 1× phosphate-buffered saline (PBS). Each cell pellet was resuspended in ice-cold IP buffer, which contains 20 mM Tris (pH 8.0), 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 50 mM NaF, 1 mM NaVO4, 20 μg of soybean trypsin inhibitor per ml, 10 μg of aprotinin per ml, 4 μg of leupeptin per ml, and 1 μg of pepstatin A per ml. Lysates were microcentrifuged at 14,000 rpm for 10 min at 4°C. Two microliters of an anti-c-Myc monoclonal antibody (MAb) (9E10) (Santa Cruz Biotechnology) was added to each cleared lysate and incubated on a rotating platform for 1 h at 4°C. Forty microliters of a 50% slurry of protein G-Sepharose beads (Pharmacia) diluted in IP buffer was added to each lysate and incubated for 1 h at 4°C. The precipitates were centrifuged for 5 min at 2,000 rpm, washed three times with 1 ml of IP buffer for each wash, and boiled in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer. The samples were analyzed by Western blotting with protein A-purified hLEF-1 polyclonal antiserum and a secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham) and detection by enhanced chemiluminescence (ECL) (Amersham). In samples with additional hLEF-1 recombinant protein, 5 μg (100 pmol) of recombinant, purified full-length LEF-1 protein (46a) was added and the samples were incubated for 30 min on a rotating platform at 4°C prior to incubation with anti-c-Myc antibody. For normalizing levels of β-catenin, the immunoblot was stripped and reblotted with anti-c-Myc MAb, followed by incubation with horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham) and detection with the ECL kit.
The in vitro coimmunoprecipitation assay represented by Fig. 2C was performed as described previously (46) except as follows. Five microliters of 35S-labeled in vitro-translated wild-type or Δ19 β-catenin was incubated with 1.5 μg of purified recombinant APC2 (aa 1034 to 2130), APC3 (aa 2130 to 2844), or axin (aa 320 to 530) (generous gifts from P. Polakis, Onyx Pharmaceuticals) or no protein, and the volume of the mixture was brought to 30 μl with buffer B (20 mM Tris [pH 8.0], 150 mM NaCl, 0.5% Nonidet P-40) plus 0.2% bovine serum albumin (BSA). Two micrograms of Glu-Glu antibody (gift of P. Polakis) and 10 μl of protein G-Sepharose beads were added to immunoprecipitate Glu-Glu-tagged APC2, APC3, and axin.
FIG. 2.
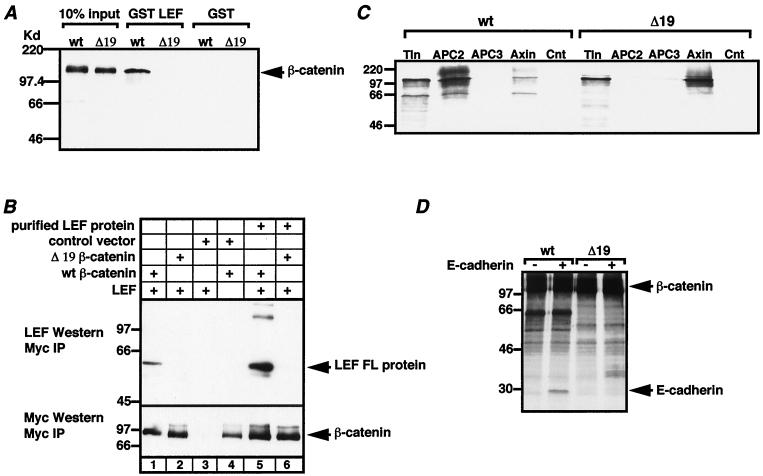
Deletion of 19 aa within the arm repeat array of β-catenin disrupts interaction with LEF-1, APC, and E-cadherin but not axin. (A) Wild-type (wt) β-catenin and a 19-aa deletion mutant of β-catenin (Δ19) were produced by a coupled transcription/translation system (Promega) in the presence of [35S]methionine. These proteins were incubated with recombinant GST–LEF-1 or GST alone in a GST pull-down binding assay (see Materials and Methods). The first two lanes indicate the amount of β-catenin protein added (10% input). Glutathione beads are able to cosediment wild-type β-catenin with GST–LEF-1 but not GST alone. In contrast, Δ19 β-catenin is unable to bind to GST–LEF-1. (B) Whole-cell extracts were prepared from 293 cells that had been transiently transfected with the indicated expression plasmids (see Materials and Methods). Anti-Myc MAb was used to coimmunoprecipitate β-catenin-associated proteins. A Western blot of the immunoprecipitates was probed with LEF-1 antisera. LEF-1 can be detected in coimmunoprecipitates with wild-type (lane 1) but not Δ19 β-catenin (lane 2). Purified recombinant LEF-1 protein (5 μg) added to cell extract prior to immunoprecipitation (lanes 5 and 6) is able to interact only with wild-type β-catenin and not even weakly with Δ19 β-catenin. In lanes 3 and 4, a control eukaryotic expression plasmid was cotransfected with either LEF-1 (lane 3) or wild-type β-catenin (lane 4) to show that LEF-1 coimmunoprecipitates specifically by association with β-catenin and not nonspecifically with the anti-Myc MAb. The same Western blot was stripped and reprobed with anti-Myc MAb to determine that the protein levels of wild-type and Δ19 β-catenin were equivalent. (C) In vitro-translated wild-type or Δ19 β-catenin was incubated with two purified APC fragments (APC2 and APC3), an axin fragment (Axin), or no protein (Cnt). Anti-Glu-Glu antibody and protein G-Sepharose beads were added in a coimmunoprecipitation assay. Ten percent of the amount of β-catenin protein added is shown (Tln). APC2 and axin are coimmunoprecipitated with wild-type β-catenin but only axin is coimmunoprecipitated with Δ19 β-catenin. (D) In vitro-translated wild-type or Δ19 β-catenin and the C-terminal tail of E-cadherin were incubated with anti-Myc MAb and protein G-Sepharose beads in a coimmunoprecipitation assay. E-cadherin can be detected in coimmunoprecipitates with wild-type but not Δ19 β-catenin. The number of additional bands is due to partial translation products of β-catenin which are coimmunoprecipitated with anti-Myc MAb.
For the in vitro coimmunoprecipitation assay represented by Fig. 2D, 35S-labeled in vitro-translated C-terminal tail of E-cadherin was incubated with either radiolabeled full-length β-catenin or Δ19 β-catenin in TBST (200 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.2% Tween 20) with 0.2% BSA in a 100-μl total volume for 30 min at room temperature on a rotating platform. One microliter of anti-c-Myc MAb and 20 μl of a 50% slurry of protein G-Sepharose beads (Pharmacia) washed in TBST–0.2% BSA were added and incubated for 1 h at room temperature on a rotating platform. The precipitates were centrifuged for 5 min at 2,000 rpm, washed three times with 1 ml of TBST for each wash, and boiled in 15 μl of 2× SDS sample buffer. The samples were analyzed by SDS-polyacrylamide gel electrophoresis, and the gel was treated with En3Hance (DuPont-NEN) and exposed to film overnight.
Fluorescence microscopy.
Full-length β-catenin in pCS2, MT10 in pCS2 (generous gifts of B. Gumbiner, Memorial Sloan-Kettering Cancer Center, New York, N.Y.), and Δ19 β-catenin in pCS2 were transiently transfected into Cos-1 and Jurkat cells, followed by immunofluorescence, as described previously (54), with anti-c-Myc MAb and fluorescein isothiocyanate (FITC)-conjugated or Texas red-conjugated anti-mouse (Amersham) antibodies. Jurkat cells were washed in 1× PBS and diluted to ∼1 × 106 cells/ml, and 100 μl of cells were cytospun onto poly-l-lysine-coated slides prior to fixation. For detection of endogenous β-catenin, anti-β-catenin MAb (Transduction Laboratories) was used. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) prior to being mounted slides with mounting media. Normal human PBMCs were prepared and transfected as previously described (30). Four hours after transfection, PBMCs were treated with media containing 25 ng of phorbol-12-myristate-13-acetate (PMA) per ml and 1 μg of ionomycin per ml or with media containing an identical amount of ethyl alcohol carrier control (untreated) for 3 h at 37°C. For export of β-catenin, 10 μg of actinomycin D per ml and 100 μg of cycloheximide per ml were added to PBMCs treated with PMA and ionomycin and cells were incubated for 3 h at 37°C. Cells were washed with PBS containing identical amounts of PMA and ionomycin for treated cells, PBS alone for untreated cells, and PMA, ionomycin, actinomycin D, and cycloheximide for export conditions and then resuspended to ∼1 × 106 cells/ml. One hundred microliters of this cell preparation was cytospun onto poly-l-lysine-coated slides. Cells were fixed in 3.7% formaldehyde–1× PBS, and immunofluorescence was performed as described above. Slides were examined as described previously (55).
RESULTS
An internal deletion of arm repeat 6 destroys LEF/TCF binding in vitro and in vivo.
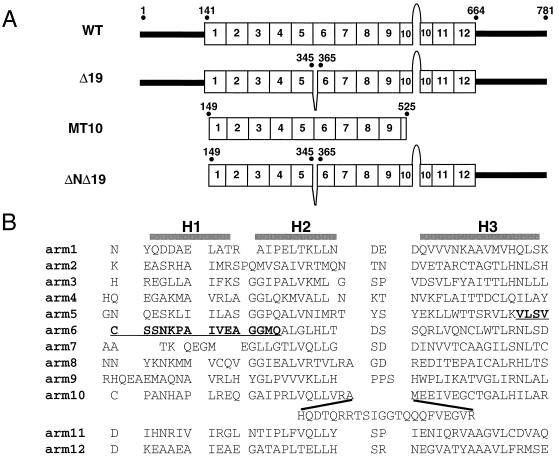
In determining which region of the β-catenin arm repeat array recognizes LEF/TCF proteins, we found that a deletion of part of the sixth arm repeat could destroy binding. Figure 1A depicts the location and scale of the deletion mutation (Δ19). Based on the arrangement of alpha-helices in the arm repeat array, the 19-aa deletion removes the final four residues of helix 3 of the fifth arm repeat and 15 aa (helix 1 and 2) of the sixth arm repeat (Fig. 1B). This region is well conserved between all β-catenin orthologs as well as catenin-related proteins such as plakoglobin. Mutant Δ19 was tested for binding to human LEF-1 in a GST pull-down experiment. Wild-type β-catenin and Δ19 were labeled with [35S]methionine by in vitro transcription/translation procedures and incubated with either GST–LEF-1 or GST recombinant fusion protein. Figure 2A shows that while wild-type β-catenin binds specifically to GST–LEF-1, the Δ19 β-catenin protein does not bind. Neither the wild type nor Δ19 binds nonspecifically to the GST control fusion protein. The loss of LEF-1 binding was examined in vivo in transfected 293 embryonic kidney cells. A eukaryotic expression vector producing Myc-tagged wild-type or Δ19 β-catenin protein was cotransfected with an expression vector producing wild-type LEF-1. Extracts from these transfected cells were prepared and anti-Myc MAb was added to coimmunoprecipitate β-catenin and interacting proteins. Using LEF-1 polyclonal antisera, a Western analysis was performed to determine if LEF-1 protein was present in these coimmunoprecipitates. Figure 2B shows that LEF-1 interacts with wild-type β-catenin in vivo but not with Δ19 β-catenin (compare lanes 1 and 2). Five micrograms (∼1 × 10−10 mol) of recombinant, full-length LEF-1 protein was added to cell extracts to determine whether a weak interaction between LEF-1 and Δ19 could be detected, but even an excess amount of LEF-1 protein was unable to reveal any binding to Δ19 (lanes 5 and 6). This total lack of an interaction is not due to differential stability of the Δ19 mutant in 293 cells, because reprobing of the same Western blot with anti-Myc antibody showed that the expression levels of wild-type and Δ19 β-catenin proteins are nearly the same (compare lanes 1 and 2 and lanes 5 and 6). Western analysis of total cell lysates from transiently transfected cells also showed that the expression levels of wild-type and Δ19 β-catenin vary by less than twofold (data not shown).
FIG. 1.
Amino acid sequence and structure of the arm repeat array of wild-type β-catenin and deletion mutants. (A) Schematic of wild-type β-catenin, a 19-aa deletion mutant of β-catenin (Δ19); MT10 β-catenin, which contains only the first nine arm repeats and no flanking sequences; and ΔNΔ19, which is identical to Δ19 but contains a deletion of the first 148 aa. (B) Amino acid alignment of the arm repeats of β-catenin. According to the recently solved structure for β-catenin, each arm repeat consists of three alpha-helices, which are denoted by H1, H2, and H3 (28). The sequence deleted in the Δ19 β-catenin mutant is in boldface type and underlined.
Mutant Δ19 binds to axin but is defective for binding to APC and the cytoplasmic tail of E-cadherin.
Large deletions of the arm repeat region have been shown to destroy binding to APC, axin, and E-cadherin (18, 31, 32, 48). We tested whether the much smaller Δ19 deletion might also disrupt interactions with these three proteins (Fig. 2C and D). Wild-type β-catenin and Δ19 mutant proteins were in vitro translated and tested for binding to two Glu-Glu-tagged protein fragments of APC (APC2 and APC3) and one Glu-Glu-tagged protein fragment of axin (Fig. 2C). APC2 (aa 1034 to 2130) and axin (aa 320 to 530) each contain a β-catenin binding domain, while APC3 (aa 2130 to 2844) does not and serves as a negative control (58). Anti-Glu-Glu antibody and protein G-Sepharose beads were added to immunoprecipitate Glu-Glu-tagged protein. Wild-type β-catenin coimmunoprecipitated with APC2 and axin but not with APC3 or protein G-Sepharose beads alone. Δ19 mutant protein coimmunoprecipitated with axin at levels that were significantly greater than that observed with wild-type β-catenin. In contrast, Δ19 did not bind to APC2. Next, wild-type β-catenin or Δ19 mutant protein was incubated with the cytoplasmic tail of E-cadherin (Fig. 2D). Anti-Myc antibody and protein G-Sepharose beads were added to immunoprecipitate Myc-tagged β-catenin protein. E-cadherin coimmunoprecipitated with wild-type β-catenin but not with Δ19. Thus, sequences within or near the sixth arm repeat are required for binding to three of the four proteins tested. These sequences could be directly involved in interactions with E-cadherin, APC, and LEF/TCF. Alternatively, because tandem arm repeats are interdependent for correct folding, removal of these sequences may have caused structural distortions in neighboring regions of the arm repeat array. However, this region must not be grossly misfolded because removal of half of the sixth arm repeat does not disrupt axin recognition of arm repeats 3 through 7 (6, 32).
Nuclear localization of β-catenin is LEF/TCF independent.
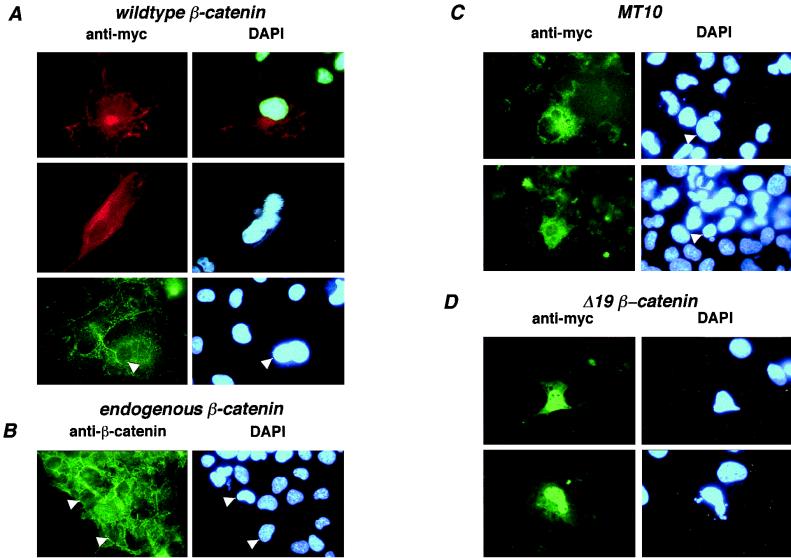
Since Δ19 is unable to bind to LEF/TCF proteins, this mutant allowed us to test the model that β-catenin can move into the nucleus independent of LEF/TCF binding in vivo. Immunofluorescence detection of transiently expressed β-catenin protein in Cos-1 cells was carried out to determine patterns of subcellular localization. Overexpressed β-catenin was detected with an anti-Myc MAb. Localization of transiently expressed protein was similar to that of endogenous β-catenin (Fig. 3A and B); the protein localized to plasma membrane and cytoplasmic sites. A mutant β-catenin protein (MT10), missing the flanking NH2- and COOH-terminal regions as well as arm repeats 10 through 12, was also observed to localize to membranes and cytoplasm (Fig. 1A and 3C) (16). In marked contrast, most of the Δ19 protein was present in the nucleus (Fig. 3D). This surprising pattern of nuclear localization directly shows that β-catenin can move into the nucleus independently of LEF/TCF binding. The mechanism by which β-catenin gains entry to the nucleus is not understood. Nuclear localization of recombinant β-catenin in digitonin-permeabilized cells does not require cytoplasmic extracts which contain importin α/β NLS receptor complexes (2, 25). While these data suggest that β-catenin is capable of entering nuclei independently of importins, a direct test for recognition of β-catenin by an NLS receptor complex has not been reported.
FIG. 3.
Δ19 β-catenin localizes constitutively to the nucleus. Plasmids encoding Myc epitope-tagged wild-type β-catenin (A), MT10 (C), and Δ19 (D) were transiently transfected into Cos-1 cells. Subcellular localization was determined 48 h later by immunofluorescence with anti-Myc MAb to detect transfected β-catenin expression plasmids or anti-β-catenin MAb to detect endogenous β-catenin. In some cases, FITC antimouse or Texas red antimouse secondary antibody was used. (A to C) Wild-type β-catenin, endogenous β-catenin, and MT10 localize to various sites in the cytoplasm and the plasma membrane. Background levels are higher for MT10, since immunofluorescence of MT10 was weak and exposure times were longer than for the other β-catenin proteins. (D) In contrast to wild-type and MT10 β-catenin, Δ19 β-catenin localizes predominantly to the nucleus. DAPI staining of DNA indicates the locations of nuclei.
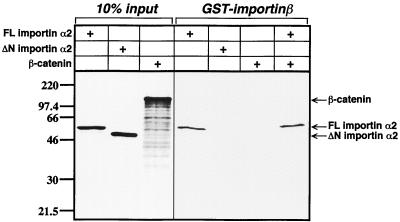
To test for a direct interaction between β-catenin and the importin α/β NLS receptor, β-catenin and importin α2 (also known as pendulin/Rch1) were translated in vitro and incubated with recombinant GST-importin β (Fig. 4). An N-terminal deletion mutant of importin α2 defective for GST-importin β binding was also generated by in vitro translation and used as a control for binding specificity in the assay. Full-length importin α2 but not the N-terminal deletion (ΔN importin α2) bound specifically to GST-importin β, as expected. β-Catenin did not bind to importin α2 or GST-importin β. There was no interaction even if all three proteins were incubated together. A test for binding of β-catenin alone with importin α2 was also negative (data not shown). In vitro-translated β-catenin is capable of binding to LEF-1 protein, indicating that the in vitro-translated protein product is functional in other well-known binding activities (Fig. 2A). These data suggest that β-catenin does not interact with classic NLS receptor complexes such as importin α2-importin β. It is possible that β-catenin could interact with another importin α or β subtype not tested here. However, a lack of an interaction is consistent with a model that β-catenin enters the nucleus via a mechanism independent of the NLS receptor pathway (17).
FIG. 4.
β-Catenin does not interact with importin α2 or importin β. A GST pull-down experiment with 35S-labeled, in vitro-translated wild-type β-catenin, importin α2, and recombinant GST-importin β was performed. An N-terminal deletion mutant of importin α2 that removes the importin β interaction domain (ΔN importin α2) was included as a negative control for nonspecific binding. The first three lanes indicate the amount of importin α2 and β-catenin proteins added (10% input). While full-length (FL) importin α2 readily interacts with GST-importin β, β-catenin does not, either in the presence or absence of importin α2.
Localization of β-catenin in normal peripheral blood lymphocytes and Jurkat T lymphocytes.
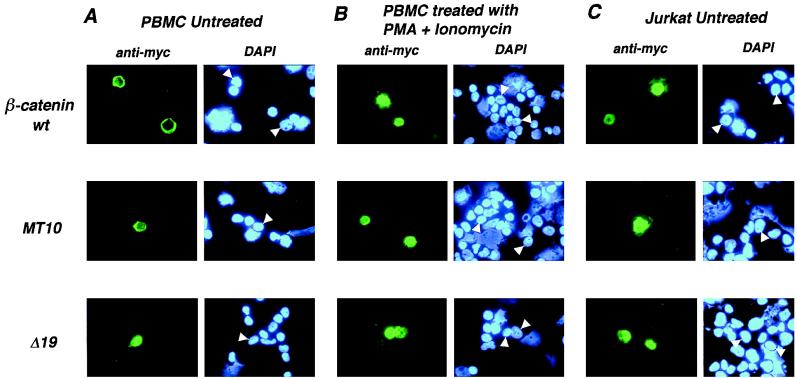
Many studies that have examined β-catenin activation of gene transcription have used the B-lymphocyte cell line IIA1.6 (36, 37, 44, 63). Likewise, T-lymphocyte cell lines have often been used to study gene activation by LEF-1 and TCF-1 (11, 21, 22, 62, 64). In contrast to transcription activation, most studies of β-catenin localization have used adherent cell lines or normal cells in whole animal model systems. Therefore, we wished to determine the subcellular localization of β-catenin in normal, untransformed T lymphocytes as well as a T-lymphocyte cell line. Preparations of PBMCs from normal human blood can be transiently transfected with plasmid DNA (30). The overwhelming majority of cells that are transfected in this procedure are T lymphocytes (>98%), and we used this method to assess the localization of β-catenin in nontransformed T cells. Both wild-type β-catenin and MT10 localized to the plasma membrane or cytoplasm, while Δ19 was localized to the nucleus (Fig. 5A) (lymphocytes have very little cytoplasm compared to nucleus, making it difficult to distinguish between plasma membrane and cytoplasm). The localization patterns of wild-type and mutant β-catenins in these cells were similar to those in Cos-1 cells (Fig. 3).
FIG. 5.
Localization of β-catenin in normal human peripheral blood lymphocytes and Jurkat T lymphocytes. (A and B) PBMCs were transfected with the indicated expression constructs. Four hours after transfection, cell preparations were treated with PMA and ionomycin or treated with ethanol as a mock control (untreated) for 3 h. (C) Jurkat cells were transiently transfected with the indicated β-catenin constructs, and subcellular localization was determined 24 h later. Transiently expressed β-catenin proteins were detected with anti-Myc MAb and FITC-antimouse secondary antibody. DAPI staining of DNA indicates the locations of nuclei. (A) In untreated cells, both the wild type and MT10 are cytoplasmic while Δ19 is nuclear. (B) When cells are treated with PMA and ionomycin, both wild-type β-catenin and MT10 localize to the nucleus. (C) Wild-type, MT10, and Δ19 β-catenin all localize to the nucleus in Jurkat cells.
Activation of T lymphocytes through T-cell receptor stimulation or by PMA-ionomycin treatment can mimic some of the effects of activating the Wnt signal transduction pathway. For example, GSK3-β, the serine/threonine kinase that negatively regulates β-catenin, is inhibited upon T-cell stimulation (67). This causes at least one transcription factor (NF-AT) to accumulate in the nuclear compartment, in part because GSK-3β-dependent nuclear export of NF-AT is inhibited (4, 5). We tested whether activation of transfected human T lymphocytes by PMA-ionomycin treatment would cause a shift in the localization pattern of transiently expressed β-catenin. Transiently transfected PBMC preparations were treated with PMA and ionomycin for 3 h prior to fixation; within this time, there was a quantitative shift in localization of both wild-type β-catenin and MT10 protein from the plasma membrane or cytoplasm to the nucleus in all transfected cells (Fig. 5B). The nuclear localization of Δ19 remained unchanged.
Jurkat T cells are transformed T lymphocytes; these cells behave similarly to activated T lymphocytes in that they synthesize and secrete the cytokine interleukin 2, a marker of T-cell activation (60). Transient expression of wild-type, MT10, and Δ19 β-catenin protein in these cells exhibited a pattern of subcellular localization nearly identical to that of activated normal T cells (Fig. 5C). All three forms were constitutively nuclear; there was no dramatic difference between wild-type and Δ19 β-catenin as there was in the Cos-1 cells. These results again confirmed that nuclear localization of β-catenin is independent of LEF/TCF binding (Δ19 is nuclear) and that a nuclear localization domain or determinant is located within the first nine arm repeats of the protein (MT10 is nuclear). However, these results also suggested that the ability of β-catenin to accumulate in the nucleus differs between cell types.
The PMA-ionomycin-directed shift in subcellular localization of β-catenin in normal T cells suggested either that the ability of β-catenin to access its nuclear transport pathway is regulated or that the transport pathway used by β-catenin is itself under tight restriction. Another possibility is that β-catenin might be capable of both nuclear import and export. In a manner similar to that observed for NF-AT, PMA-ionomycin treatment may cause nuclear accumulation of β-catenin by shifting the kinetic balance to favor import over export. If this were true, the constitutive nuclear accumulation of Δ19 might be due to an inability of the Δ19 mutant protein to be exported from the nucleus.
Export of β-catenin from nuclei in normal human peripheral blood lymphocytes.
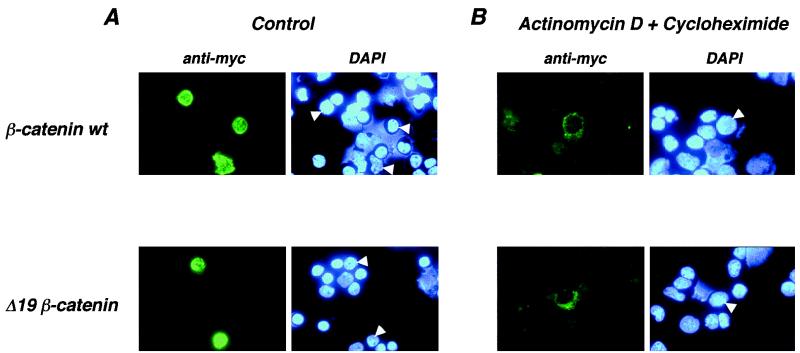
We tested whether wild-type β-catenin could undergo nuclear export, and we also tested whether Δ19 exhibited any differences with respect to this activity. A common method to test for nuclear export of proteins is to block nuclear import with actinomycin D treatment (a transcription inhibitor) and to block protein synthesis with cycloheximide such that no newly synthesized protein accumulates in the cytoplasm (51). Therefore, any formerly nucleus-localized protein that accumulates in the cytoplasm in these treated cells must have undergone active export from the nucleus. PBMC preparations were transiently transfected with either wild-type or Δ19 β-catenin expression plasmid and then treated with PMA-ionomycin to direct all β-catenin protein to the nucleus (Fig. 6A). Actinomycin D and cycloheximide were added to half of the cells to block import and de novo protein synthesis (Fig. 6B). After 3 h, cells were processed for immunofluorescence. Both wild-type and Δ19 β-catenin staining was observed only in the cytoplasm and not the nucleus, suggesting that both were actively exported from the nucleus during actinomycin D-cycloheximide treatment. Overall fluorescence was weaker in cells treated with these reagents because protein synthesis was completely inhibited for 3 h, and exported β-catenin may have been subject to degradation in the cytoplasm. A less likely explanation is that a low level of cytoplasmic β-catenin remained stable throughout the 3-h incubation with cycloheximide, and nuclear β-catenin was preferentially degraded. Taken together, these data show that the differing localization patterns between wild-type β-catenin (cytoplasm) and Δ19 (nucleus) in unstimulated T lymphocytes are not due to differences in import or export.
FIG. 6.
Wild-type and Δ19 β-catenins are exported from the nucleus in normal human peripheral blood lymphocytes. PBMCs were transfected with the indicated expression constructs. (A) Six hours prior to harvest, cell preparations were treated with PMA and ionomycin (Control). (B) Three hours prior to harvest, cell preparations were treated with actinomycin D and cycloheximide. Transiently expressed β-catenin was detected with anti-Myc MAb and FITC–anti-mouse secondary antibody. DAPI staining of DNA indicates the locations of nuclei. In cells treated with PMA and ionomycin, both wild-type and Δ19 β-catenins are localized to the nucleus. Subsequent treatment with actinomycin D and cycloheximide inhibits nuclear import and protein synthesis. With these treatments, wild-type and Δ19 β-catenins are exported to the cytoplasm.
β-Catenin- and LEF-1-dependent activation of transcription varies in different cell types.
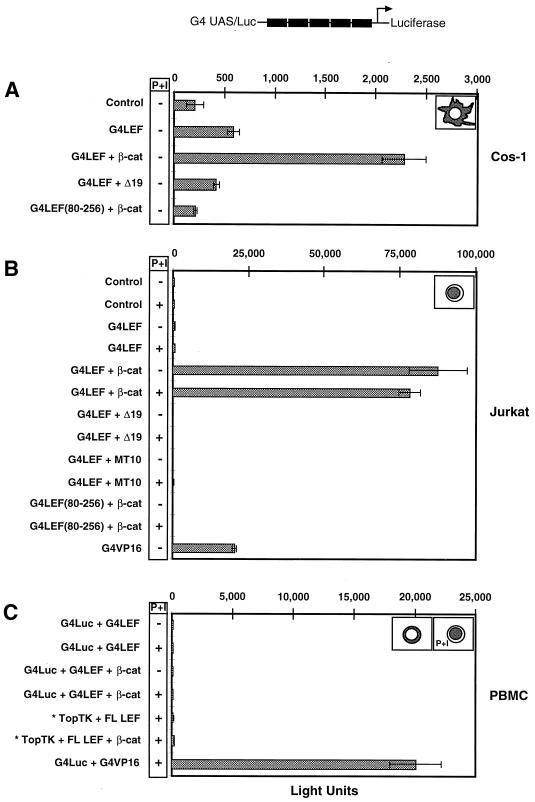
In addition to the dramatic effects that the Δ19 mutation has on localization, we also determined the effect of the mutation on reporter gene activation in the nucleus. We examined the transactivation potential of LEF-1 and β-catenin in Cos-1 cells, Jurkat T cells, and normal human T lymphocytes, all of which are different in some aspect of β-catenin localization (Fig. 7). In order to examine the ability of only the transiently expressed proteins to work together, a LEF-1 fusion protein, in which the first 256 aa of LEF-1 were fused to the yeast Gal4 DNA binding domain, was used in place of full-length LEF-1 protein (G4LEF) (Fig. 7). The reporter plasmid contained five Gal4 upstream activating sites (UAS) placed upstream of a minimal promoter driving luciferase coding sequences (G4 UAS/Luc). Transfection of the G4LEF expression plasmid with the G4 UAS/Luc reporter plasmid did not produce significant luciferase expression in Cos-1 cells (Fig. 7A). However, if a β-catenin expression plasmid was included, a fourfold increase in reporter gene expression was observed. Cotransfection of Δ19 with G4LEF yielded only basal levels of expression, confirming that, in vivo, the Δ19 mutant does not interact with G4LEF even though both proteins are concentrated in the nucleus (see Fig. 3 for β-catenin localization). The modest fourfold activation is due to a bona fide interaction between LEF-1 and β-catenin, because an N-terminal deletion of LEF-1 missing the β-catenin binding domain does not activate reporter gene expression [G4LEF(80-256) + β-cat; Fig. 7A]. In Cos-1 cells, overexpression of G4LEF does not cause a change in the subcellular distribution of β-catenin protein, similar to what is observed in NIH 3T3 cells (27, 55a). Instead, β-catenin remains mostly localized to the plasma membrane and cytoplasm, thereby accounting for the modest levels of activation observed in these cells.
FIG. 7.
A direct interaction between β-catenin and LEF-1 is necessary but not sufficient to activate gene expression. Wild-type, Δ19, or MT10 β-catenin expression plasmids were cotransfected with the Gal4 DNA binding domain (DBD) fused to LEF-1 aa 1 to 256 (G4LEF) and a reporter plasmid containing five tandemly repeated Gal4 UAS directing luciferase reporter gene (G4 UAS/Luc) expression in Cos-1 and Jurkat cells and PBMCs. A summary of the β-catenin localization in each of these cell lines is shown in the upper right corner of each panel. (A) Five micrograms of β-catenin, 5 μg of G4LEF expression plasmids, and 1 μg of reporter plasmid were transfected into Cos-1 cells. (B) Seven hundred fifty nanograms of β-catenin and 50 ng of G4LEF expression plasmids or 750 ng of G4VP16 expression plasmid and 1 μg of reporter plasmid were transfected into Jurkat cells. In the absence of β-catenin, the range of luciferase activity varied from 574 to 825. (C) The indicated expression plasmids (10 μg) were cotransfected with 10 μg of reporter plasmid into PBMCs. P + I, treatment of cell preparations with PMA plus ionomycin for 3 h prior to harvest of cells; ∗, use of the Top TK reporter plasmid in place of the G4 UAS/Luc reporter plasmid. A plasmid (0.5 μg) containing the lacZ gene under the control of the CMV promoter was cotransfected as an internal control for transfection efficiency. G4LEF (aa 80 to 256), which does not bind to β-catenin, was used as a negative control (A and B). An empty eukaryotic expression plasmid was used to equalize the amount of DNA used in each transfection. Cells were harvested at 15 to 24 h posttransfection, and cell lysates were analyzed for luciferase and β-galactosidase activity. Wild-type β-catenin and LEF-1 transactivated the reporter gene construct in Cos-1 and Jurkat cells, while neither Δ19 nor MT10 cooperated with LEF-1 to activate gene expression. In addition, the level of transcription activation of β-catenin–LEF-1 complexes in Jurkat cells was higher than that observed with G4VP16 (750 ng). β-Catenin and G4LEF-1 or full-length LEF-1 (FL LEF-1) did not transactivate reporter gene expression in PBMCs, although G4VP16 (10 μg) was able to transactivate the reporter gene in PBMCs. Error bars were calculated by using the average of duplicate points (A and C) or triplicate points (B). Values shown are from one of three replicate experiments.
In Jurkat T lymphocytes, transiently expressed β-catenin protein is localized to the nucleus (Fig. 5C). Cotransfection of G4LEF and β-catenin expression plasmids in these cells stimulated a large increase in reporter gene expression (150-fold) (G4LEF + βcat [Fig. 7B]), an even greater activation than what can be achieved by the strong activating fusion protein Gal4-VP16 (G4VP16) (35-fold). As observed for Cos-1 cells, cotransfection of Δ19 β-catenin did not activate gene expression at all, nor did MT10 or the G4LEF mutant missing the β-catenin binding domain, even though all of these proteins are localized to the nucleus. Treatment of Jurkat cells with PMA-ionomycin did not change the transactivation potential of LEF-1 and β-catenin. Either β-catenin has a strong costimulatory effect in Jurkat T cells due to the overall strength of its nuclear localization or another transcription cofactor or basal factor is abundant in these cells.
We tested whether strong activation could be observed in normal human T lymphocytes (Fig. 7C). Surprisingly, β-catenin and G4LEF were unable to activate gene expression, even in the presence of PMA-ionomycin treatment (which causes a quantitative shift of β-catenin into the nucleus [see Fig. 5B for localization patterns]). In contrast, an equivalent amount of G4VP16 was able to produce a 115-fold increase in reporter gene expression. Thus, activation of reporter gene expression by the VP16 transcription activation domain is possible in normal T cells, but β-catenin and LEF-1 are not able to elicit detectable amounts of luciferase gene expression. This dramatic difference between the activities of β-catenin and LEF-1 in normal versus transformed lymphocytes is not due to differential localization or to dramatic differences in protein stability, as our immunofluorescence data indicate. To rule out that G4LEF was the cause of this differential activation, we tested whether full-length β-catenin and full-length, wild-type LEF-1 could activate gene expression with the Top TK reporter plasmid (44). This plasmid contains five multimerized LEF/TCF binding sites and has been used extensively to examine LEF-1–β-catenin reporter gene activation in many different cell types (27, 29, 36, 44, 45, 59, 63). Wild-type LEF-1 and wild-type β-catenin were unable to activate Top TK reporter gene expression. In addition, the Top TK reporter plasmid was tested in Jurkat cells. High levels of wild-type LEF-1 and wild-type β-catenin were able to activate reporter gene expression, which is similar to the results in Fig. 7B with the G4 UAS/Luc reporter plasmid (data not shown). Thus the G4 UAS/Luc reporter plasmid functions in a manner similar to the wild-type LEF/TCF binding site reporter plasmid. One possibility for the different activities of β-catenin and LEF-1 is that another cofactor is required to activate gene expression. This cofactor might be abundant in Jurkat cells but absent or limiting in normal human T lymphocytes. Such a cofactor may not be an essential basal RNA polymerase II factor, as G4VP16 fusion protein is capable of activating reporter gene expression in either cell type.
Δ19 can squelch β-catenin-activated gene expression but not VP16-activated gene expression.
To test whether another nuclear component is required for β-catenin–LEF-1 complexes to activate reporter gene expression, a squelching experiment was designed. It has been proposed that β-catenin contains transcription activation domains in the C terminus and the N terminus of the protein (27, 63). Although these regions do not have sequence similarity or amino acid compositions similar to known transcription activation domains, they can activate gene expression when fused to heterologous DNA binding domains such as LEF-1 or Gal4 (27, 59, 63). If either domain of β-catenin is interacting with a third required transcription factor, then overexpression of this domain should sequester the third component from wild-type β-catenin–LEF-1 complexes and inhibit activation of gene transcription. This method of inhibition is referred to as squelching.
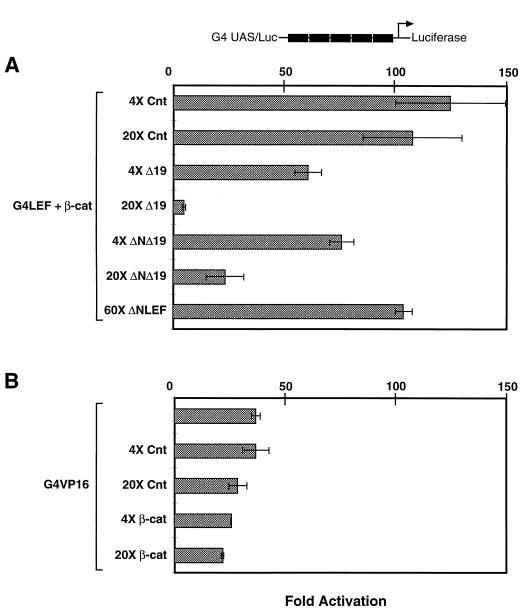
To carry out the experiment, overexpression of the Δ19 β-catenin mutant was used to test for squelching. Since Δ19 is completely negative for binding to LEF/TCF proteins in vitro and in vivo (Fig. 2 and 7), no squelching should occur due to simple sequestration of LEF-1 protein. Secondly, Δ19 protein localizes constitutively to the nucleus of all cells where squelching of transcription factor components should occur. Jurkat cells were used because the level of activation is very high. In the experiment represented by Fig. 8A, G4LEF and β-catenin activated luciferase reporter gene expression 125-fold. Excess plasmid containing the strong cytomegalovirus (CMV) promoter was used to control for inhibitory effects due to competition for basal polymerase II components. As this balancer plasmid was increased from a 4-fold to a 20-fold excess, a 15% drop in G4LEF/β-catenin-dependent luciferase activity was observed (Fig. 8A). Thus, large amounts of competitive CMV promoter-containing plasmid may account for a minor inhibitory effect. There was a 2-fold drop in luciferase activity when a 4-fold excess of Δ19 plasmid was included, and a 20-fold excess of Δ19 plasmid caused a 22-fold drop in activation. This suggests that overexpression of β-catenin can indeed squelch activation of reporter gene expression by sequestering a required component. The Δ19 mutant β-catenin was further modified to remove the hydrophilic N terminus (ΔNΔ19) (Fig. 1). Fourfold and 20-fold excess ΔNΔ19 plasmid also exhibited squelching, albeit to a slightly lesser extent. Finally, a 60-fold excess of an expression plasmid encoding ΔNLEF, an N-terminal deletion mutant of wild-type LEF-1 missing the β-catenin binding domain, was unable to squelch reporter gene expression, suggesting that the squelching activity is specific to a region of β-catenin. To determine whether Δ19 β-catenin was squelching by inhibiting a general step in polymerase II transcription, beyond the point of initiation, 4-fold and 20-fold excess β-catenin was tested against G4VP16 activation of the same G4 UAS/Luc reporter plasmid (Fig. 8B). Addition of 20-fold excess balancer plasmid elicited a 20% decrease in G4VP16 activation from 35-fold activation to 29-fold activation. Addition of a 4-fold excess of β-catenin plasmid caused a 32% decrease in G4VP16 activation, and a 20-fold excess caused a 31% decrease, compared to matched controls. This amount of inhibition is minor compared to the striking effect on G4LEF and β-catenin shown in Fig. 8A. Furthermore, the level of inhibition does not change when the excess of β-catenin plasmid is increased from 4-fold to 20-fold but stays constant at approximately 30%, suggesting that this minor inhibitory effect is due to reasons other than specific squelching. We conclude that β-catenin can squelch reporter gene expression because a domain within the arm repeat array or the C terminus is interacting with a third component required by β-catenin–LEF-1 complexes to activate gene transcription.
FIG. 8.
Excess β-catenin squelches transactivation of β-catenin and LEF-1 but not transactivation by VP16. (A) Wild-type β-catenin expression plasmid (750 ng) was cotransfected with 50 ng of G4LEF (aa 1 to 256), 1 μg of G4 UAS/Luc reporter plasmid, and the following plasmids used as competitors: 4- and 20-fold (3 and 15 μg, respectively) excess amounts of Δ19 β-catenin, control plasmid (empty eukaryotic expression plasmid), and ΔNΔ19 β-catenin (see Fig. 1) and a 60-fold (3 μg) excess amount of ΔNLEF (hLEF-1 aa 67 to 399). Excess amounts of Δ19 and ΔNΔ19 β-catenin, but not control plasmid or ΔNLEF, were able to squelch wild-type β-catenin–LEF-1 reporter gene activation. (B) G4VP16 (750 ng) was cotransfected with 1 μg of G4 UAS/Luc reporter plasmid and the following plasmids used as competitors: 4- and 20-fold (3 and 15 μg, respectively) excess amounts of control plasmid or a wild-type β-catenin construct. Wild-type β-catenin does not squelch G4VP16 transactivation. A plasmid (0.5 μg) containing the lacZ gene under the control of the CMV promoter was cotransfected as an internal control for transfection efficiency. Error bars were calculated by using the average of duplicate points. Values shown are from one of three replicate experiments. Fold activation was calculated by comparing levels of luciferase activity to the G4 UAS/Luc reporter alone.
DISCUSSION
We have examined nuclear localization of β-catenin and demonstrate directly that nuclear import is independent of LEF/TCF binding; we also show data to suggest that β-catenin can be exported from nuclei. Below, we discuss a model in which subcellular localization of β-catenin is shaped by competing cytoplasmic and nuclear sequestration and the relative rates of nuclear import and nuclear export. β-Catenin–LEF-1 complexes do not activate reporter gene expression in normal T lymphocytes even though both factors are abundantly and predominantly in the nucleus. Thus, gene activation by β-catenin does not always correlate with nuclear localization. This fact, coupled with our observation that overexpression of β-catenin can squelch activation by LEF-1–β-catenin complexes but cannot squelch activation by the herpesvirus VP16 transcription activator, leads us to propose that an additional transcription component is required for β-catenin to activate gene expression.
Three models could explain how a small deletion of 19 aa in arm repeat 6 of β-catenin causes a dramatic change in subcellular localization. First, the 19-aa deletion could have unmasked or generated a strong, cryptic NLS. Second, if β-catenin is a cytoplasmic-nuclear shuttling protein with export capabilities, the 19-aa deletion might prevent export such that newly imported Δ19 β-catenin protein remains trapped in the nuclear compartment. Third, β-catenin might be sequestered in the cytoplasm and plasma membrane, and the 19-aa deletion could disrupt one or more of these interactions.
The first model is unlikely because no obvious clustered or bipartite NLS sequence is evident around the deleted region. Furthermore, larger deletions that remove other portions of the arm repeat array also create nucleus-localized protein (48). While the exact region specifying nuclear transport has not been defined, we show that nuclear import can be carried out by arm repeats 1 through 9 (MT10) (Fig. 5 and 6). Others have also observed MT10 to be localized to the nucleus (16). The ability of this region to direct nuclear transport is remarkably resilient, even when subjected to large deletions. The subcellular localization of a series of armadillo deletion mutants was determined in Drosophila embryos (48). Deletion of arm repeat 5, 8, or 3 through 6 did not destroy nuclear localization; indeed, no deletion mutation has yet been reported to impair nuclear localization. Instead, deletion of repeats 3 through 6 created a constitutively nucleus-localized armadillo protein, much like the constitutively localized Δ19 mutant described here (48). Either nuclear import is directed by a discrete signal motif that has yet to be specifically deleted or it is specified by any one of a number of redundant determinants present in the arm repeat array.
We have presented data to suggest that wild-type β-catenin is capable of nuclear export (Fig. 6). While this lends support to the second model, in which Δ19 is localized to the nucleus because it has lost the ability to be exported, we show that there is no difference between wild-type β-catenin and Δ19 localization (Fig. 6). Therefore, the second model is insufficient to explain the contrasting patterns of localization. Even so, regulated shifts in subcellular localization of β-catenin could involve a modulation of the relative rates of import and export. Indeed, we observe that localization of β-catenin in normal T lymphocytes can be shifted from cytoplasm to nucleus with PMA-ionomycin treatment (Fig. 5). Either this T-cell-activating signal modulates relative rates of import and export, as it does for NF-AT, or it somehow causes release of β-catenin from anchoring sites in the cytoplasm or plasma membrane. Future studies of β-catenin localization should include a consideration of the ability of this protein to shuttle in and out of the nucleus.
Our data are most consistent with the third model, in which Δ19 localizes to the nucleus because it cannot be efficiently sequestered in the cytoplasm and plasma membrane. We have shown that Δ19 is completely defective for LEF/TCF binding, but we have also shown that it no longer interacts with the cytoplasmic tail of E-cadherin or the cytoplasmic tumor suppressor protein APC (Fig. 2). Thus, Δ19 may not be easily anchored in the cytoplasm, and this might be at least part of the reason for the predominant nuclear localization of this mutant. In light of the dramatic localization of Δ19 protein to the nucleus, it is surprising that it binds well to axin. Obviously, this ability to interact with axin does not prevent nuclear localization, but more work is necessary to determine why this is the case. In their study of armadillo deletion mutants in Drosophila embryos, Orsulic and Peifer found that the strongest correlation for nuclear accumulation of armadillo mutants was a loss of E-cadherin binding; loss of interactions with other cytoplasmic proteins discovered more recently might produce the same effect (48). Overexpressed E-cadherin protein can prevent nuclear localization of β-catenin, and it has been suggested that cadherins act as regulators of β-catenin signaling by depleting the soluble cytoplasmic pool (16). Likewise, cytoplasmic extracts inhibit nuclear accumulation of β-catenin in digitonin-permeabilized cells, again suggesting that an anchoring activity in the cytoplasmic extract prevents β-catenin from transporting through nuclear pores. Another explanation for the localization pattern of Δ19 would be that this protein is much more stable because it has lost the ability to interact with conductin/axin/axil and APC. Indeed, an increase in the stability of β-catenin is one of the major effects of the Wnt signaling cascade, and this increase correlates with an appearance of the protein in the nucleus. However, there are no apparent differences between the expression levels of wild-type β-catenin and Δ19 (Fig. 2 and data not shown). This is surprising given that Δ19 no longer binds to APC. Δ19 β-catenin is able to bind to axin as well as, if not better than, wild-type β-catenin. Perhaps axin is still capable of mediating some form of degradation. It is curious that Δ19 can localize to nuclei even though it can bind to axin; this would appear to be inconsistent with a cytoplasmic anchoring model. Perhaps axin/conductin/axil does not play a strong role in sequestration compared to E-cadherin. Immunolocalization studies of exogenously expressed conductin show that the protein localizes throughout the cell (6). Another possibility is that axin relocalizes with Δ19 to the nucleus. Taken together, our data support a model in which varying patterns of subcellular localization are due in part to sequestration by β-catenin binding proteins.
It is important to point out that since β-catenin may be exported, subcellular localization might also be influenced by sequestration of β-catenin in the nucleus and competing rates of import and export. Early reports of an interaction between β-catenin and LEF/TCF factors showed that transiently expressed β-catenin was cytoplasmic unless coexpressed with a LEF/TCF protein (7, 29, 44). In those situations, coexpressed LEF/TCF proteins may have been preventing cytoplasmic anchoring by competing with cytoplasmic or plasma membrane proteins for binding to the same or overlapping portion of the arm repeat array. But it is also plausible that the high levels of transiently expressed LEF/TCF protein may have trapped β-catenin in the nucleus and prevented export. A nuclear entrapment or sequestering of β-catenin by LEF/TCF proteins may explain why we observe that transiently expressed β-catenin is predominantly nuclear in cells that express high levels of endogenous LEF-1 and TCF-1 (Jurkat) but is cytoplasm or membrane bound in cells that express little or no LEF/TCF protein (PBMCs and Cos-1 cells, respectively). Consistent with this idea is the fact that wild-type β-catenin cannot be exported from Jurkat nuclei (55a). In other cell lines, such as NIH 3T3 and Cos-1 cells, overexpression of LEF-1 is not sufficient to direct endogenous β-catenin to the nucleus (27, 53a). Interestingly, Wnt-1 treatment of NIH 3T3 cells is also insufficient to cause nuclear accumulation, even though it stimulates a large increase in total levels of β-catenin protein. Instead, it is necessary to coexpress LEF-1 and Wnt-1 in NIH 3T3 cells to drive endogenous β-catenin into the nucleus and keep it there (27).
Once β-catenin accumulates in nuclei, it complexes with LEF/TCF proteins to regulate gene expression. β-Catenin and LEF/TCF proteins have been able to elicit increases in reporter gene expression in different cell lines including Neuro 2A, NIH 3T3, IIA1.6 lymphocytes, and others. These data and others, including the ability of the C-terminal hydrophilic region of armadillo to activate reporter gene expression in yeast, have led to a straightforward model in which recruitment of β-catenin by a LEF/TCF protein to the promoter template may be all that is required to activate gene transcription. We tested whether Cos-1 cells, Jurkat cells, and PBMCs could support β-catenin-dependent activation of reporter gene expression and whether the level of reporter gene activation correlated with the level of nuclear localization in different cells, and it did not (Fig. 7). We observed that activation of gene expression by this complex varied greatly between cell types. LEF-1 and β-catenin activated reporter gene expression poorly in Cos-1 cells. Wild-type β-catenin does not localize efficiently to the nucleus in these cells, and coexpression of LEF-1 does not force a shift of β-catenin to the nucleus (53a). Similar observations have been made with NIH 3T3 cells; LEF-1 overexpression is insufficient to relocalize β-catenin (27). Thus, the low level of activation in Cos-1 cells could simply correlate with the poor ability of β-catenin to transport to nuclei. This is clearly not the case for normal peripheral lymphocytes: LEF-1 and β-catenin could not activate transcription in PBMCs even when PMA and ionomycin were added to direct β-catenin to the nucleus (Fig. 7C). These cells are certainly capable of supporting reporter gene activation, as they have been used extensively to study regulation of interleukin 2 promoter activity (30), and we observe a 115-fold activation of the reporter by G4VP16 (Fig. 7C). Perhaps an inhibitory factor is abundant in normal T lymphocytes, or a third required component or modification of β-catenin or LEF-1 protein is limiting in these cells.
We tested these notions by overexpressing β-catenin and LEF-1 mutants that could not engage in complex formation to see whether these mutants could act negatively by squelching a required cofactor (Fig. 8A). The Δ19 mutant was able to effectively squelch reporter gene activation when provided at 20-fold excess over wild-type β-catenin, as was a related mutant missing the N-terminal region (Fig. 8A). A 60-fold excess of LEF-1 missing the β-catenin binding domain did not squelch. These data suggest that a region in the arm repeat array or the C terminus of the protein is able to specifically sequester a required positive cofactor for activation, although they do not rule out the possibility that an additional inhibitory factor is present in normal T lymphocytes. The sequestered factor is not likely to be involved in general steps of transcription downstream of RNA polymerase II initiation, because β-catenin does not squelch G4VP16 activation. The transcription activation domain of VP16 is highly acidic and known to affect both initiation and processivity of RNA polymerase II transcription through multiple contacts with basal factors and TATA binding protein-associated factors (8, 24, 39). The Δ19 squelching activity must not be interfering with these contacts or downstream events.
The C-terminal domain of β-catenin has been proposed to be a transcription activation domain (48, 63). However, most of the C-terminal region is not nearly as well conserved between catenin orthologs and plakoglobin (which can also activate reporter gene expression) as the arm repeats, making it difficult to identify a specific conserved region as a candidate transcription activation domain. Perhaps the C-terminal domains adopt similar functional structures despite their modest level of sequence similarity, or they each activate transcription through different sequences. For the squelching results presented here, it is equally likely that a region other than the divergent C terminus is responsible and that a region of the arm repeat array, not perturbed by the Δ19 mutation, is competing for a required transcription component.
Pontin52, a nuclear protein that binds to Tata binding protein, has also been shown to bind to arm repeats 2 through 5 of β-catenin (3). Pontin52 can be coimmunoprecipitated within a larger complex containing β-catenin and LEF-1 and is proposed to provide a bridging function between LEF-1–β-catenin complexes and basal transcription machinery. If this is true, Pontin52 could presumably be a component that is squelched by excess Δ19 protein. However, we show that MT10, a β-catenin deletion mutant containing arm repeats 1 through 9, does not activate reporter gene expression even though it should be fully capable of binding Pontin52 (Fig. 7B). Therefore, an additional protein or complex is likely to be involved. Clearly, more work is necessary to test these possibilities.
Three additional nuclear proteins, Groucho, CREB binding protein (CBP), and Teashirt, have recently been shown to interact with either LEF-1 (Groucho and CBP) or β-catenin/armadillo (Teashirt) (12, 19, 38, 56, 65). Groucho, a transcriptional repressor, has been shown to bind to TCF-1 near the HMG DNA binding domain (12, 38, 56). CBP, a coactivator-acetyltransferase, binds to the HMG DNA binding domain of LEF-1 and is proposed to antagonize β-catenin binding (65). For our experiments, we have used a G4LEF (aa 1 to 256) expression plasmid that does not contain the Groucho and CBP binding regions. Thus the lack of activation obtained in normal T lymphocytes is not a result of Groucho repression or antagonistic actions by CBP. Teashirt, a Zn-finger Drosophila protein expressed in a trunk-specific pattern, requires a direct interaction with armadillo/β-catenin to regulate the expression of some Wingless gene targets but not others (19). Teashirt may be a tissue-specifically expressed DNA binding protein that recruits β-catenin to a subset of gene targets independent of LEF-1, or it may be a coactivating component of a larger complex involving all three proteins. Whether a mammalian ortholog of Teashirt is expressed in Jurkat cells or normal T lymphocytes is not known, but it is unlikely. It is more likely that there are other β-catenin binding proteins in addition to LEF/TCFs, Pontin52, and Teashirt that are involved in transcription activation. Future studies should be focused on identifying these additional factors.
ACKNOWLEDGMENTS
We thank Chris Hughes, Javier Mestas, and other members of the Hughes laboratory for invaluable help with transfection of PBMC preparations and generous sharing of time and reagents. We are grateful to Barry Gumbiner for sharing full-length and MT10 β-catenin constructs and to Steve Adam for the GST-importin β plasmid. Many thanks go to Harry Mangalam, Karine Hovanes, Chris Hughes, Bert Semler, Larry Marsh, and Adeela Syed for critical reading of the manuscript.
This work was supported by grant CA 62069 from NIH and in part by grant RPG-97-156-CSM from the American Cancer Society and by the Chao Cancer Center at UCI. M.L.W. is a member of the Developmental Biology Center and the Cancer Research Institute at UCI.
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Sterne-Marr R E, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci USA. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beals C, Clipstone N, Ho S, Crabtree G. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 5.Beals C, Sheridan C, Turck C, Gardner P, Crabtree G. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 6.Behrens J, Jerchow B-A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhn M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 7.Behrens J, von Kries J P, Kuh M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 8.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J, Moon R. Wnt signaling: why is everything so negative? Curr Opinion Cell Biol. 1998;10:182–187. doi: 10.1016/s0955-0674(98)80140-3. [DOI] [PubMed] [Google Scholar]
- 10.Cadigan K, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1998;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 11.Carlsson P, Waterman M, Jones K. The hLEF/TCF-1α HMG protein contains a context-dependent transcriptional activation domain that induces the TCRα enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 12.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 13.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 14.Cook D, Fry M, Hughes K, Sumathipala R, Woodgett J, Dale T. Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 15.Dale T C. Signal transduction by the Wnt family of ligands. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagotto F, Funayama N, Gluck U, Gumbiner B M. Binding to cadherins antagonizes the signaling activity of β-catenin during axis formation in Xenopus. J Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagotto F, Gluck U, Gumbiner B. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 18.Funayama N, Fagotto F, McCrea P, Gumbiner B M. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallet A, Erkner A, Charroux B, Fasano L, Kerridge S. Trunk-specific modulation of wingless signalling in Drosophila by teashirt binding to armadillo. Curr Biol. 1998;8:893–902. doi: 10.1016/s0960-9822(07)00369-7. [DOI] [PubMed] [Google Scholar]
- 20.Giese K, Cox J, Grosscheldl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–196. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 21.Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 23.Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci USA. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich D, Kostak S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 26.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 27.Hsu S-C, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber A H, Nelson W J, Weis W I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 29.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 30.Hughes C C, Pober J S. Transcriptional regulation of the interleukin-2 gene in normal human peripheral blood T cells. Convergence of costimulatory signals and differences from transformed T cells. J Biol Chem. 1996;271:5369–5377. doi: 10.1074/jbc.271.10.5369. [DOI] [PubMed] [Google Scholar]
- 31.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinzler K, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 34.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 35.Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- 36.Korinek V, Barker N, Morin P, van Wichen D, de Weger R, Kinzler K, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 37.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 40.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 41.Malik H, Eickbush T, Goldfarb D. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangalam H J, Albert V R, Ingraham H A, Kapiloff M, Wilson L, Nelson C, Elsholtz H, Rosenfeld M G. A pituitary POU domain protein, Pit-1, activates both growth hormone and prolactin promoters transcriptionally. Genes Dev. 1989;3:946–958. doi: 10.1101/gad.3.7.946. [DOI] [PubMed] [Google Scholar]
- 43.Miller J, Moon R. Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 44.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTCF-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 45.Morin P, Sparks A, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 46.Munemitsu S, Albert I, Rubinfeld B, Polakis P. Deletion of an amino-terminal sequence stabilizes β-catenin in vivo and promotes hyperphosphorylation of the adenomatous polyposis coli tumor suppressor protein. Mol Cell Biol. 1996;16:4088–4094. doi: 10.1128/mcb.16.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Munguia, J., and M. L. Waterman. Unpublished results.
- 47.Nigg E. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 48.Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peifer M, Berg S, Reynolds A B. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 51.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 52.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 53.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a beta-catenin-LEF-1 complex by Wnt-1 and transforming mutants of beta-catenin. Oncogene. 1998;15:2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 53a.Prieve, M. G. Unpublished results.
- 54.Prieve M, Guttridge K L, Munguia J E, Waterman M. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-NLS receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- 55.Prieve M G, Guttridge K L, Munguia J, Waterman M L. Differential importin α recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol Cell Biol. 1998;18:4819–4832. doi: 10.1128/mcb.18.8.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Prieve, M. G., and M. L. Waterman. Unpublished results.
- 56.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 57.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3β to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 58.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. The APC protein and E-cadherin form similar but independent complexes with alpha-catenin, beta-catenin, and plakoglobin. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 59.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze’ev A. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swain S L, Croft M, Dubey C, Haynes L, Rogers P, Zhang X, Bradley L M. From naive to memory T cells. Immunol Rev. 1996;150:143–167. doi: 10.1111/j.1600-065x.1996.tb00700.x. [DOI] [PubMed] [Google Scholar]
- 61.Theisen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh J. Dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development. 1994;120:347–360. doi: 10.1242/dev.120.2.347. [DOI] [PubMed] [Google Scholar]
- 62.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 63.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 64.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 66.Waterman M L, Fischer W H, Jones K A. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 67.Welsh G I, Miyamoto S, Price N T, Safer B, Proud C G. T-cell activation leads to rapid stimulation of translation initiation factor eIF2B and inactivation of glycogen synthase kinase-3. J Biol Chem. 1996;271:11410–11413. doi: 10.1074/jbc.271.19.11410. [DOI] [PubMed] [Google Scholar]
- 68.Woodgett J. Regulation and functions of the glycogen synthase kinase-3 subfamily. Semin Cancer Biol. 1994;5:269–275. [PubMed] [Google Scholar]
- 69.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Axil, a member of the Axin family, interacts with both glycogen synthase kinase 3β and β-catenin and inhibits axis formation of Xenopus embryos. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yost C, Gist III H F, Pierce S B, Ferkey D M, Chen M M, Kimelman D. GBP, and inhibitor of GSK-3, is implicated in Xenopus development and oncogenesis. Cell. 1998;93:1031–1041. doi: 10.1016/s0092-8674(00)81208-8. [DOI] [PubMed] [Google Scholar]
- 71.Yu G, Chen F, Levesque G, Nishimura M, Zhang D-M, Levesque L, Rogaeva E, Xu D, Lian Y, Duthei M, St. George-Hyslop P, Fraser P E. The presenilin 1 protein is a component of a high molecular weight intracellular complex that contains β-catenin. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- 72.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek T J, Perry W L, Lee J J, Tilghman S M, Gumbiner B M, Costantini F. The mouse fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]