Abstract
Ure2p of Saccharomyces cerevisiae normally functions in blocking utilization of a poor nitrogen source when a good nitrogen source is available. The non-Mendelian genetic element [URE3] is a prion (infectious protein) form of Ure2p, so that overexpression of Ure2p induces the de novo appearance of infectious [URE3]. Earlier studies defined a prion domain comprising Ure2p residues 1 to 64 and a nitrogen regulation domain included in residues 66 to 354. We find that deletion of individual runs of asparagine within the prion domain reduce prion-inducing activity. Although residues 1 to 64 are sufficient for prion induction, the fragment from residues 1 to 80 is a more efficient inducer of [URE3]. In-frame deletion of a region around residue 224 does not affect nitrogen regulation but does eliminate prion induction by the remainder of Ure2p. Larger deletions removing the region around residue 224 and more of the C-terminal part of Ure2p restore prion-inducing ability. A fragment of Ure2p lacking the original prion domain does not induce [URE3], but surprisingly, further deletion of residues 151 to 157 and 348 to 354 leaves a fragment that can do so. The region from 66 to 80 and the region around residue 224 are both necessary for this second prion-inducing activity. Thus, each of two nonoverlapping parts of Ure2p is sufficient to induce the appearance of the [URE3] prion.
We discovered that the non-Mendelian genetic elements [URE3] and [PSI] were infectious protein forms (prions) of the Saccharomyces cerevisiae Ure2p and Sup35p, respectively, based on their genetic properties (47). We proposed three genetic criteria to identify prions: (i) reversible curability, (ii) induction of prion formation de novo by overexpression of the normal protein, and (iii) a unique phenotypic relationship between the presence of a prion and mutation in the chromosomal gene encoding the protein (47). We noted that [URE3] and [PSI], two non-Mendelian genetic elements of S. cerevisiae, both satisfy all three of these genetic criteria as prion forms of Ure2p and Sup35p, respectively (47). Biochemical and further genetic data have supported this proposal (6, 12, 21, 25, 30, 31, 34–36). For example, fragments of Ure2p in extracts of [URE3] cells are more resistant to treatment with proteinase K than are similar extracts of wild-type cells (31). Recently, these same genetic criteria have been used to identify another prion, the [Het-s] non-Mendelian genetic element of the filamentous fungus Podospora anserina (10).
The discovery that [URE3] and [PSI] were prions showed that proteins can, like nucleic acids, be informational molecules. Moreover, their primary sequence is not the only form in which they carry this information. Although [URE3] and [PSI], like scrapie, are diseases, [Het-s] appears to be a prion carrying normal information for Podospora (10; reviewed in reference 48). It is possible that mammals may also have prions responsible for inherited traits, or that this same or similar mechanisms operate in certain cases of information transfer within an organism.
The concept of an infectious protein (a prion) originates with studies of the mammalian transmissible spongiform encephalopathies, such as scrapie and Creutzfeldt-Jakob disease (1, 22; reviewed in references 7, 17, and 38). PrP involvement in scrapie was first shown by genetic (13) and biochemical (2) evidence, the two brought together by the identification of the mouse Sinc (for scrapie incubation) gene as the structural gene for PrP (5, 8, 33). PrP is necessary for scrapie (4) but has not yet been shown to be sufficient. Specifically, none of the three genetic criteria that we proposed for a prion (see above and reference 47) have yet been shown to be satisfied by PrP and scrapie.
To further substantiate [URE3] as a prion, we showed that it is overexpression of the URE2-encoded protein, and not its transcript or the gene itself in high copy number, that induces the de novo formation of [URE3] (30). Moreover, [URE3] was really arising de novo, not as a mutant form of a previously present nucleic acid replicon (30, 47). Finally, the presence of [URE3] can be demonstrated in cells that are not derepressed for nitrogen metabolism (30), showing that the [URE3] state is not a stable regulatory state such as has been shown for some other inheritable traits (32).
Ure2p is a 354-amino-acid protein (9) that can be divided into two domains, an amino-terminal prion domain of residues 1 to 64 and a carboxyl-terminal nitrogen regulation domain of residues 66 to 354 (30, 31). The N-terminal prion domain, when transiently overexpressed in a normal cell, induces the de novo appearance of [URE3] at frequencies 100 times higher than the rate for similarly overexpressed intact Ure2p and several thousand-fold higher than the background rate (31). This prion domain can propagate [URE3] in the complete absence of the C-terminal domain (30). The C-terminal domain can carry out the nitrogen regulation function of Ure2p in the complete absence of the prion domain, although the regulation is leaky unless this nitrogen regulation domain is overexpressed (9, 31). In a strain with the prion [URE3], the nitrogen regulation function of molecules carrying a prion domain is inactivated, but those lacking this domain remain active (30, 31).
The prion domain of Ure2p (31) is quite rich in asparagine (40% of residues) including several runs (9). The prion domain of Sup35p (12, 44) is likewise rich in asparagine and glutamine (24, 28, 49), and these residues are important for [PSI] (11). However, neither PrP (8, 33) nor the Het-s protein (46) has such regions. It has been suggested that the expansion of glutamine repeats in several triplet-repeat diseases is responsible for protein aggregation and the pathology of these diseases (37, 41). It is thus possible that the asparagines of Ure2p mediate the prion properties of this protein.
Recent data suggest a biochemical basis for [URE3]. Fusion proteins of Ure2p and green fluorescent protein are aggregated specifically in [URE3] cells but not in [ure-o] or cured cells (15). Synthetic Ure2p1-65 (the prion domain peptide) spontaneously forms amyloid filaments in vitro (43). These filaments are essentially all β-sheet, and they give green birefringence when stained with Congo red. Ure2p1-65 also induces filament formation by native purified Ure2p, and the latter filaments can seed further filament formation by Ure2p. All of these filaments have the properties of amyloid (43). These results suggest that [URE3] is due to amyloid formation by Ure2p.
The experiments reported here concern the dissection of the molecular determinants of prion inducibility by Ure2p. We have identified a second region of the molecule that is necessary for Ure2p to induce de novo formation of [URE3] but which is not necessary for the previously determined prion domain (residues 1 to 64) to do so. We find that prion induction is possible in the absence of the 1–64 region and requires two other regions. We also explore the role of the runs of asparagine in the prion domain and the interactions among other sites.
MATERIALS AND METHODS
Yeast strains and methods.
Diploid strain 3469 (MATα/MATa kar1/kar1 ura2/ura2 leu2/leu2 +/his− trp1/+ [ure-o]) and haploid strain 3385 (MATa kar1 ura2 leu2 his− [ure-o]) were used to assess induction of [URE3], and strain 3718 (MATα his3 leu2 ura2 ure2::URA3) was used to determine complementation of ure2Δ. SD and SGal are minimal media with dextrose and galactose, respectively, as carbon sources (40). Transformation was done by the lithium method (23).
Plasmid constructions.
Plasmid p646 (YEp351G-URE2) is a LEU2 2 μm DNA shuttle plasmid with URE2 under the GAL1 promoter (47). To construct p773, p774, p776, p778, and p646SD, each bearing one deletion in the URE2 gene, pGAL1::URE2 (p646) was used as template in the Kunkel method (Bio-Rad Mutagene kit) with the mutagenic oligonucleotides D18-27, D44-50, D56-62, D66-80, and DSphI (Table 1), respectively. Constructs p682, p664, p683, p680, pe2+3, and p725 are detailed in previous publications (30, 31, 47) and in Table 1. The constructs p725SK, p718N, pD44L, pD55M, and pD66I, with two deletions in URE2, were obtained by using p725 as the template with the mutagenic primers DSphI, D18-27, D44-50, D56-62, and D66-80 (Table 1), respectively. pD80-1, pD89-4, and p725D80-1 were made by PCR using p646 or p725 (for p725D80-1) as the template and the 5′ primer D3-80 or D3-89 (Table 1) with the 3′ primer LSacII (5′TGCCTTGATGACCGCGGGTCTTCT3′). The fragments obtained were cut with BamHI and SacII and subcloned into p646 cut with BamHI and SacII and thus lack the part of URE2 3′ to the SacII site within the URE2 open reading frame.
TABLE 1.
Plasmid constructions
| Plasmid | Ure2p residues expresseda | Description or reference |
|---|---|---|
| p554 | None | YEp351G vector |
| p646 | 1–354 | Ure2p in YEp351G with f1 origin of replication (47) |
| pKT35-1 | 1–44stop | Kim Taylor |
| p680 (=EagI) | 1–64*b | Minimal prion domain (31) |
| 728 | 1–64stop | 31 |
| pe2+3 | 1–80*c | 30 |
| pX7D | 1–80stop | PCR on p774 with oligosf Bam2 (5′CCTCCTGGATCCCAAATGATG3′) and X470 (5′CCACCACTCGAGTTTAGGTATTCTTGATATT3′) |
| pA2 | 1–89*d | PCR on p646 with oligos Bam2 and N448 (5′CCACCACTCGAGCTGTTGTTGTTGTCG3′) |
| pX4C | 1–89stop | PCR on p646 with oligos Bam2 and X448 (5′CCACCACTCGAGTTTACTGTTGTTGTTGTCG3′) |
| p679STO11 | 1–153stop | |
| p682 (=D1-3) | 1–2, 66–354 | N-regulation domain (31) |
| pD89h1 | 1–2, 90–354 | 3-89Δ HindIII fragment from pD89-5 into HindIII-cut p646 |
| p683 (=D4-1) | 1–2, 125–354 | 47 |
| p664 (=S1) | 1–347*e | 31 |
| Internal deletions | ||
| p773 | 1–17, 28–354 | 18-27Δ oligo D18-27 (5′ACTTTGATCGGTGGTTGTACGGAGCGCATTGGAG3′) |
| p774 | 1–43, 51–354 | 44-50Δ oligo D44-50 (5′GTTATTACTACTGCTTACACCTGTTGAAAA3′) |
| p776 | 1–55, 63–354 | 56-62Δ oligo D56-62 (5′GGCTACCATTGCGGCCATTACTACTGCTATTG3′) |
| p778 | 1–65, 81–354 | 66-80Δ oligo D66-80 (5′GTCGATGTTGTTCTAAGCGGCCGCTGTTATTG3′) |
| pML | 1–43, 45–354 | 44Δ oligo ML (5′ACTACTGCTATTGTTATTATTATTATTTACACCTGTTGA3′) |
| pMLS3 | 1–43, 45–64*b | pML cut with NotI-SphI and religated |
| p725 | 1–150, 159–354 | ApaIΔ (31) |
| p646SD | 1–220, 228–354 | SphIΔ oligo DSph (5′GAAATGTAAAGCTTGTCCCCTTTGGAAGAACAACC3′) |
| Multiple deletions | ||
| p4B, p4C | 1–2, 66–150, 159–354 | Prion domain Δ, ApaIΔ; oligos LSacII and D3-65 (5′CTTCTTGGATCCCAAATGATGAATGGTAGCCAA3′) |
| p725SK | 1–150, 159–220, 228–354 | ApaIΔ SphIΔ |
| p682Sc6 | 1–2, 66–347*e | Prion domainΔ, SacIIΔ |
| pD80-1 | 1–2, 81–347*e | 3-80Δ oligo D3-80 (5′CCTCCTGGATCCCAAATGATGTTAGAACAACATCGACAACAA3′) |
| pCS7 | 1–2, 66–150, 158–347*e | Prion domainΔ, ApaIΔ, SacIIΔ |
| pB2 | 1–43, 51–89*d | 45-54Δ from prion domain |
| K18STO4 | 1–43, 51–64stop | |
| p4X7-7 | 1–43, 51–80stop | PCR on p774 with oligos Bam2 and X470 (5′CCACCACTCGAGTTTAGGTATTCTTGATATT3′) |
| p4X4-5 | 1–43, 51–89stop | PCR on p774 with oligos Bam2 and X448 (5′CCACCACTCGAGTTTACTGTTGTTGTTGTCG3′) |
| pP409STO1 | 1–43, 51–153stop | |
| p718N | 1–17, 28–150, 159–354 | 18-27Δ, ApaIΔ |
| pD44L | 1–43, 51–150, 159–354 | 44-50Δ, ApaIΔ |
| pD55M | 1–55, 63–150, 159–354 | 56-62Δ, ApaIΔ |
| pD66I | 1–65, 81–150, 159–354 | 66-80Δ, ApaIΔ |
| p725D80 | 1–2, 81–150, 159–347*e | lg prion domainΔ, ApaIΔ |
| p725D80+5 | 1–2, 81–150, 159–354 | 3-80Δ, ApaIΔ |
| pSKX | 1–2, 66–150, 159–220, 228–347*e | 3-65Δ, ApaIΔ, SphIΔ, SacIIΔ |
| p725Hd+5 | 1–2, 81–150, 158–354 | 3-80Δ, ApaIΔ |
| pD89-4 | 1–2, 90–347*e | Oligo D3-89 (5′CAACTTGGATCCCAAATGATGGCATTTTCGGATATGATGCAC3′) |
Asterisks mark fusion peptides with out-of-frame sequences.
Residue 64 of Ure2p is followed by PSLALAVVLQRRDWENPGVTQLNRLAAHPPFASWRNSEEARTDRPSQQLRSLNGEWRLMRYFLLTHLCGISHRIDPVRRter encoded by the vector YEp351G.
Residue 79 of Ure2p is followed by I LRTTSTTTTG I FGYESRGVFQNYKIFSRTTTGG IYPFLSQVCAter encoded by the −1 frame of URE2.
Residue 89 of Ure2p is followed by LDLQACKLGTGRRFTTS encoded by the vector YEp351G.
Residue 347 of Ure2p is followed by EKESNRKQRHPCL encoded by a region 3′ of URE2.
Oligos, oligonucleotides.
Constructs P409 and NSK18 were obtained by cutting p774 with ApaI and SphI or with NotI and SphI, respectively, followed by filling with Klenow DNA polymerase I and ligation. Plasmids P409, NSK18, and 679 were cut with HindIII, and oligonucleotide SPOH (5′AGCTGCATGCTTAATTAATTAAGCATGC3′) was inserted to add a stop codon producing P409STO1, K18STO4, and 679STO11.
The constructs pA2 and pB2 were obtained by PCR with the 5′ primer Bam2 and the 3′ primer N448 on the templates p646 and p774, respectively. The product was cut with BamHI and XhoI and ligated to YEp351G cut with BamHI and SalI. The construct p4B was made by PCR using p725 as the template with primers D3-65 and LSacII. pSKX was made by PCR using p725SK as the template with primers D3-65 and LSacII. In each case, the product was cut with BamHI and SacII and ligated with p646 cut with the same enzymes.
Construct pML was obtained by the Kunkel method with the mutagenic primer ML and p646 as the template. Construct pMLS3 was obtained by cutting pM44L with SphI and NotI, followed by Klenow filling and ligation.
p725D80+5 was made from 725D80-1 by cutting with HindIII and religation to the large HindIII fragment from p646. Constructs pCS7 and 682Sc6 were obtained by cutting p4B and p682 with SacII and religation. Constructs p4X4-5 and pX4C were made by PCR using p774 and p646, respectively, as templates with primers X448 and Bam2. Constructs p4X7-7 and pX7D were made by PCR using p774 and p646, respectively as templates with primers X470 and Bam2. These PCR fragments were cut by BamHI and XhoI and ligated to 554 cut with BamHI and SalI.
All constructs were checked by sequencing, and at least three isolates of each construction were transformed into yeast and assayed for the ability to induce de novo generation of [URE3].
Western blot analysis.
Transformants of strain 3469 were grown on SGal plates with uracil for 3 days, then inoculated into liquid medium composed of 50% YPAD and 50% SGal, and grown for 4 h. Cells were then washed and transferred to liquid SGal medium with uracil for 12 h. Proteins were extracted as described elsewhere (29) but without boiling and in the presence of 100 μg of phenylmethylsulfonyl fluoride per ml. The same amount of protein from each extract, as determined by the Bio-Rad protein assay, was heated for 5 min at 96°C and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% gel). Transfer of the proteins to a polyvinylidene difluoride (Millipore) membrane was done in 10% methanol–25 mM Tris–192 mM glycine buffer (pH 8.3) for 1 h at 300 mA. The membrane was dried and incubated 2 h in blocking buffer (TTBS; 0.05% Tween 20, 10 mM Tris borate [pH 7.4] 150 mM NaCl) with 5% dried milk. After 4 h of incubation with the primary antibody, the membrane was washed with TTBS and incubated for 1 h with the secondary antibody, a mouse anti-rabbit antibody conjugated to alkaline phosphatase (Promega Western Blue). Blots were developed as described previously (39). Primary antibodies were polyclonal rabbit antisera U2 (47), K1, K2, and K3 (42), made to full-length Ure2p, and C3, made to Ure2p66-354 (42).
Characterization of [URE3] phenotype: induction, curing, and propagation assays.
The haploid strain 3385 and the diploid strain 3469 were transformed with YEp351URE2 carrying different deletions in URE2. Ten transformants selected on Leu-deficient medium were mixed in water and grown on an SGal plate with uracil for 3 days. Several dilutions of these cells were made and spread on SD containing 100 μg of USA. USA+ colonies, indicating the development of the [URE3] state, were counted after 5 days. Each average number shown in the tables was calculated from at least three repeats of each experiment. The data in the figures are for the diploid strain 3469. Similar results were obtained with strain 3385, but the induction frequencies were generally higher.
Samples of USA+ colonies were assayed for the ability to propagate [URE3] and to be cured by guanidine HCl. The ability to again form single colonies on USA medium, indicating propagation of the trait, but inability to grow on USA medium after growth on YPAD supplemented with 5 mM guanidine HCl, indicating curing of the trait, was taken as evidence that the tested strain carried [URE3].
Complementation assay.
Strain 3718 (ure2Δ) was transformed with YEp351G-URE2 carrying different deletions in URE2. The transformants selected on Leu-deficient medium were streaked on SGal with uracil and histidine. After 3 days, colonies were replica plated to SD with USA and histidine. Failure to grow on SD with USA and histidine indicates complementation of the nitrogen regulation function of URE2.
RESULTS
Asparagine-rich subregions are necessary for the minimal prion domain.
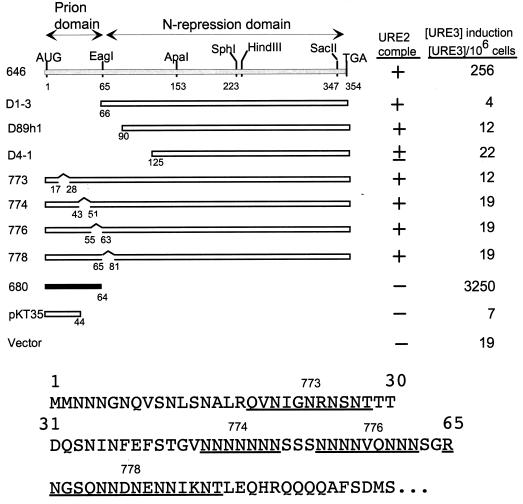
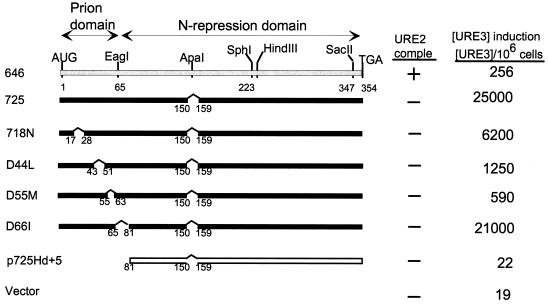
The minimal prion domain was within residues 1 to 64, based on the ability of this fragment to efficiently induce [URE3] when overexpressed and the inability of a C-terminal fragment lacking residues 1 to 65 to induce [URE3] (31) (Fig. 1). We have made smaller deletions within this prion domain and tested the ability of the overexpressed remainder to induce [URE3] in a strain with a normal copy of URE2 (Fig. 1). Each of three deletions of asparagine-rich regions within the prion domain (773, 774, and 776) resulted in loss of most of the prion-inducing activity of the remainder of the molecule. Deletion of the asparagine-rich residues 66 to 80 also reduces prion-inducing activity, a first indication that this region can be involved in prion induction (Fig. 1, 778).
FIG. 1.
Asparagine runs are important for prion-inducing activity of Ure2p. A diploid strain (3469) was transformed with each plasmid and grown on galactose for 3 days to overproduce the indicated part of Ure2p; cells were plated on SD with ureidosuccinate in place of uracil, selective for [URE3] cells. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. The N-terminal part of the Ure2p sequence is shown below, with the regions deleted in p773, p774, p776, and p778 underlined. comple, complementation.
Residues 66 to 80 enhance the prion-inducing activity of residues 1 to 64.
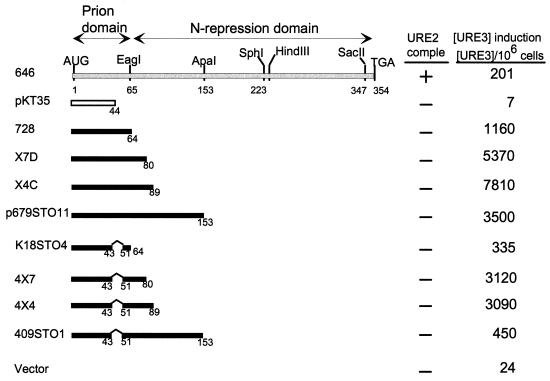
Although overexpression of residues 1 to 64 induces [URE3] formation at rates 100-fold higher than rates for the intact protein and comparable to those for C-terminal deletions, we find that including through residues 80 or 89 increases prion induction a further 10-fold (Fig. 2). Further extension results in a consistent severalfold decrease in inducing activity (Fig. 2, 672STO11). These results suggest that residues 66 to 80 can cooperate with residues 1 to 64 in the prion generation process, consistent with our finding (Fig. 1) that deletion of residues 66 to 80 reduces prion-inducing activity in the otherwise intact protein. Our results also suggest that there is a region downstream of residue 89 that modestly hinders the induction process.
FIG. 2.
Residues 66 to 80 enhance prion induction. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
Deletion of residues 45 to 54 from the 1–64 prion domain or the extended prion domains from 1 to 89 or 1 to 153 again lowered the prion-inducing activity. However, unlike the effect on the intact protein, the deletion of residues 45 to 54 did not eliminate prion-inducing activity of these smaller fragments (Fig. 2).
A region outside the 1–89 region is necessary for prion induction by intact Ure2p.
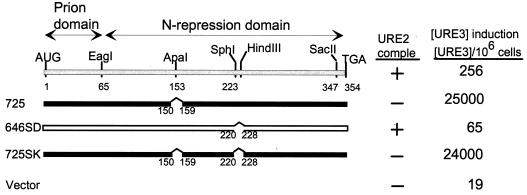
In-frame deletion of residues 151 to 158 or deletion of seven residues from the C-terminal extremity (348 to 354) results in a 100-fold increase in [URE3]-inducing activity and loss of ability to complement ure2Δ for nitrogen regulation (31) (Fig. 3, 725; Fig. 4, S1).
FIG. 3.
Ure2p residues 221 to 227 are needed for optimal [URE3] induction by intact Ure2p. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
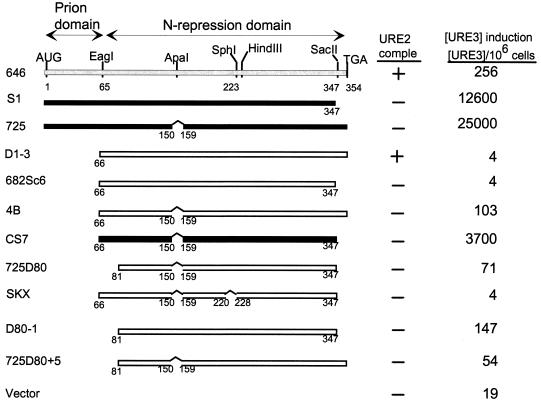
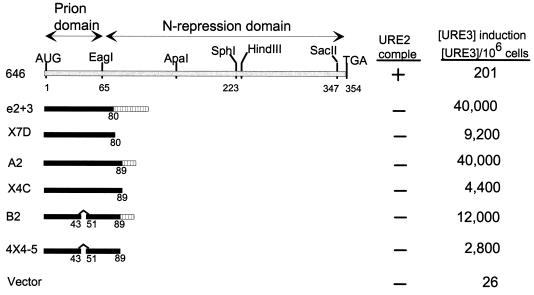
FIG. 4.
A fragment of Ure2p lacking the 1–64 prion domain induces [URE3]. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
In contrast, deletion of residues 221 to 227 results in loss of ability to induce [URE3] and retention of ability to complement ure2Δ (Fig. 3, 646SD). This region is not necessary for prion induction by the 1–64 prion domain or by the longer 1–89 prion domain, as both fragments lack the 221–227 region. Combining the 221–227 deletion with the 151–158 deletion shows that the latter is epistatic: the doubly deleted molecule has the properties of the single 151–158 deletion (Fig. 3, 725SK).
A second prion domain.
Although the N-terminal 64 residues are sufficient to induce [URE3] at high frequency, deletion of either the 151–158 region or the C-terminal seven residues of Ure2p dramatically enhance prion induction by the remainder of the molecule (Fig. 4). Moreover, we have presented evidence above that the 221–227 region is necessary for prion induction by the whole Ure2p and that residues 66 to 80 enhance the activity of the N-terminal 64 residues. Starting with a fragment lacking the 1–65 prion domain (Fig. 4, D1-3), deletion of either the 151–158 region (p4B) or the C-terminal 7 residues (p682Sc6) did not give a fragment showing substantial [URE3] induction. However, deletion of both of these small regions resulted in substantial prion induction, even though it completely lacks the 1–65 segment (Fig. 4, pCS7).
Our two leading candidates for the parts of the molecule important in this second prion domain of Ure2p were the 221–227 and 66–80 regions, since each is necessary for prion induction by the full-length Ure2p (Fig. 1 and 3). Deletion of the 66–80 segment from pCS7 dramatically reduces prion induction (compare pCS7 and p725D80 in Fig. 4). Likewise, deleting the 221–227 region eliminates prion-inducing activity (compare pCS7 and pSKX in Fig. 4). These results suggest that both the 221–227 region and the 66–80 segment have roles in the second prion domain. Other constructs (not shown) support our interpretation that the second prion domain is active only after deletion of both the 151–157 and 348–354 regions and disappears after further deletion of either the 66–80 or 221–227 region.
Asparagine-rich regions are less critical in derepressed Ure2p fragments.
Deletion of any of four asparagine-rich segments within the 1–80 region resulted in nearly complete elimination of prion-inducing activity of the otherwise intact molecule (Fig. 1). We tested the effect of deleting the same asparagine-rich segments on the very high activity seen with Ure2p fragments lacking the 151–158 region (Fig. 5). Although deletion of residues 56 to 62 decreased [URE3] induction in one strain, it did not impair induction in another (not shown). In smaller fragments, a decrease in prion induction by deletion of residues 44 to 50 is also seen (Fig. 2). Overall, the results indicate that the individual asparagine-rich regions are less critical for prion induction by the fragments derepressed for prion induction than they are in the intact molecule. Nonetheless, elimination of all of these regions by the 3–80 deletion does eliminate all known prion-inducing activity (Fig. 4, 725Hd+5).
FIG. 5.
Asparagine runs are important but not essential for prion induction by derepressed Ure2p fragments. Note that C3, B2, and A2 all have identical C-terminal tails. The C-terminal tail of 679 differs from those of P215 and P408. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
C-terminal tails affect prion-inducing activity.
The effect of the presence of a C-terminal tail consisting of vector sequences or out-of-frame URE2 sequences was tested in comparison with constructs with a termination codon at the indicated site (Fig. 6). In each case, the presence of the tail increased prion-inducing activity 4- to 10-fold compared to the tail-less construct.
FIG. 6.
C-terminal tails can affect prion induction efficiency. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
Steady-state levels of Ure2p fragments.
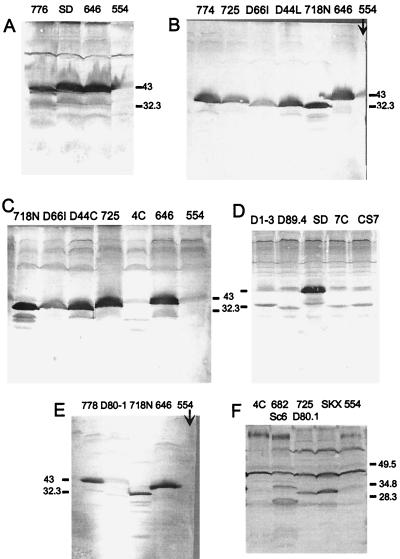
The data presented above concerning the roles of various segments in the induction of [URE3] do not distinguish effects on protein function from effects on protein stability. It is particularly important to determine whether the deletion mutants that lack prion-inducing activity are present at levels comparable to those of the normal protein. However, since fragments of Ure2p are the best inducers of [URE3] appearance, protein instability does not necessarily explain failure to induce [URE3].
We examined the steady-state levels of a number of the constructs used here by Western blot analysis of extracts of cells grown on galactose. The antibody used in our earlier studies (31) reacts mostly with the N-terminal region (16). We also used other polyclonal antibodies made to the full-length protein or the C-terminal nitrogen regulation domain (42). Although the 1–80 fragment gave the highest induction of [URE3] observed, it was not detected by our antibody specific for the N-terminal domain (data not shown). Constructs derived from the intact Ure2p by deletion of the 44–50 region, residues 56 to 62, or residues 66 to 80 each gave poor induction of [URE3] but showed levels of protein comparable to those of the intact protein (Fig. 7, constructs 774, 776, and 778). Likewise, deletion of the 221–227 region resulted in absence of prion induction by the remainder of Ure2p, but this fragment was present at levels similar to or greater than those of the intact protein (Fig. 7A, construct SD).
FIG. 7.
Steady-state levels of mutant Ure2 proteins measured by Western blotting. Construct names are as in Table 1 and Fig. 1 to 6. 554 is the vector alone, and 646 expresses full-length Ure2p. Anti-Ure2p antibodies used were U2 and affinity-purified K1, K2, K3, and C3 (A), affinity-purified U2 (B), U2, K2, and C3 (C and D), U2 (E), and affinity-purified U2, K1, K2, K3, and C3 (F). SD is 646SD. Positions of selected size markers are shown in kilodaltons. Equal amounts of total protein were analyzed.
Deletion of residues 3 to 65 or 3 to 89 did appear to decrease the steady-state levels of Ure2p, but this appearance may be partly due to some specificity of the polyclonal antibodies for the N-terminal region. Among constructs lacking residues 1 to 65, CS7, which was active in [URE3] induction, had the same apparently low steady-state level as D1-3, D89.4, and 7C, all of which were inactive (Fig. 7D). Constructs 4C and D80-1 showed particularly low levels, suggesting that they were unstable.
Deletion of the 151–158 region did not result in higher levels of protein, although the prion-inducing activity was dramatically increased (Fig. 7B). Further deletion of the asparagine-rich 18–27, 44–50, or 66–80 segment did not substantially alter steady-state levels (Fig. 7B and C).
In conclusion, the prion-inducing activity is not consistently correlated with the steady-state levels of the various fragments of Ure2p. This suggests that for most constructs, it is the tendency of these fragments to undergo the prion change and their ability to transmit that change to normal molecules, rather than their relative stability, that are being measured in these experiments. On the contrary, it remains possible that it is specific breakdown products of the full-length Ure2p, rather than the intact molecule, that actually induce [URE3].
Residue 44 is not critical for [URE3] induction.
Deletion of one base in codon 44 to change the reading frame of URE2 eliminates prion induction without reducing the amount of mRNA (30). We show here that there is no unique function of the base deleted in this experiment, since deletion of the entire codon (encoding Asn), which maintains the reading frame, does not lower the prion-inducing activity of the remainder of Ure2p. The vector gave 22 [URE3] colonies per 106 cells, while full-length Ure2p produced 303 colonies and two mutants lacking only codon 44 produced 350 and 398 colonies. Similarly, the 1–64 segment lacking codon 44 gave 1,360 colonies, while the 1–64 segment with codon 44 intact gave 1,304 colonies in the same experiment.
DISCUSSION
The N-terminal 64 amino acid residues of Ure2p are necessary for prion-inducing activity by the whole protein and are sufficient to carry out this function at higher efficiency than the intact Ure2p (31) (Fig. 1). Furthermore, the C-terminal part of Ure2p, lacking residues 3 to 65, is not inactivated on introduction of [URE3] (30). This work has given a picture of two protein domains interacting with each other, but each able to carry out its function in the absence of the other. The N-terminal domain can propagate [URE3] in the absence of the C-terminal part and the C-terminal part can regulate nitrogen metabolism in the absence of the N-terminal part (30, 31). However, the C-terminal part appears less efficient in performing the nitrogen regulatory function, and the N terminus is more efficient at inducing [URE3] in the absence of the other part, indicating that each domain stabilizes the other.
The second prion domain.
We find that there are parts of the protein lacking residues 3 to 65 which are capable of prion induction. The first clue was that deletion of the 66–80 or the 221–227 region reduced the prion-inducing activity of the intact protein dramatically. In addition, the prion-inducing activity of residues 1 to 80 was actually 10-fold higher than that of the 1–64 fragment. This suggested that either of these regions could have a positive role in initiation or propagation of the prion state.
Deletion of the 151–158 region or the C-terminal seven residues dramatically increases the prion-inducing ability of Ure2p (31). We find that together, these two deletions make prion-inducing activity reappear in a construct lacking residues 3 to 65. Removing residues 66 to 80 from the doubly deleted construct eliminates prion-inducing activity. This finding suggests that the asparagine-glutamine-rich 66–80 region has a role in this second prion-inducing activity. Deletion of the 221–227 region has a similar effect. Note that the 221–227 region has no asparagine or glutamine residues.
We show here that the lack of prion-inducing activity by some constructs is not in general due to degradation of the protein. While this distinction is critical if one is considering, for example, an enzymatic or other function that requires an essentially intact protein, it is not so clear for prion induction. Because small fragments of Ure2p can induce [URE3] 100 times more efficiently than the intact protein, instability may be an advantage rather than a disadvantage.
What does it mean that two nonoverlapping regions of Ure2p can each induce [URE3] at high efficiency? No natural enzyme can be cut in two and have each half carry out the very same reaction. We can guess that the [URE3] induction by these two different parts of Ure2p must be in some way different reactions. Different strains of scrapie are well documented with different forms of PrP (3), and genetically distinct forms of [PSI] have also been observed (12). Perhaps the two different parts of Ure2p induce different forms of [URE3]. If [URE3] consists of a self-propagating amyloid formed of Ure2p (15, 43), it is not difficult to imagine how different parts of Ure2p could initiate aggregation, perhaps by formation of aggregates with different symmetry properties or different conformations of Ure2p, or with the aggregation due to interactions of different parts of the Ure2p molecule.
One essential part of the second prion domain is residues 66 to 80, adjacent to the originally defined first prion domain. One could view residues 1 to 80 as a single prion domain, arbitrarily divided in the earlier studies by an EagI site. The interesting point, however, is that Ure2p could be so divided and still leave activity in each of the nonoverlapping parts. Moreover, the 66–80 segment is not sufficient for activity.
Asparagine-rich regions and prion induction.
The 1–64 region first found to induce the [URE3] prion change is rich in asparagine residues, including several runs of asparagine (31). The prion-inducing domain of Sup35p is likewise rich in glutamine and asparagine residues (44), and these residues are indeed important for [PSI] (11). Our studies show that three asparagine-rich regions in the N-terminal part of Ure2p are important in prion induction. Further, the asparagine-rich residues 66 to 80 are essential for the second prion-inducing fragment. Moreover, the asparagine-rich regions each contribute to the frequency of prion induction.
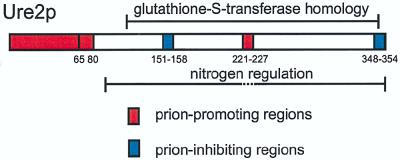
Ure2p has modest similarity to glutathione S-transferases from about residue 110 to near the C terminus (9). A Schizosaccharomyces pombe protein with 53% identity throughout its length to Ure2p lacks the N-terminal 110 residues of the S. cerevisiae protein (50). Thus, the S. pombe protein appears to have the nitrogen regulation domain but to lack the prion domains of Ure2p (Fig. 8).
FIG. 8.
Prion-promoting and -inhibiting domains of Ure2p compared to the glutathione S-transferase homology region and the nitrogen regulation domain.
Deletion of residues 3 to 80 of Ure2p leaves a fragment that is apparently unable to induce [URE3] formation even with the 151–158 and C-terminal deletions. However, it is clear that the prion induction process is more complicated than simply the presence of runs of asparagine. Deletion of the 221–227 region does not change the asparagine content of Ure2p and yet it eliminates the prion-inducing activity of the remainder of the molecule. Other deletions in the C-terminal domain also do not change the asparagine content of Ure2p but dramatically increase the prion-inducing activity of Ure2p and make it less dependent on single runs of asparagine residues. Perutz and colleagues have suggested that expanded runs of glutamine in certain inherited neurodegenerative diseases can produce aggregation of proteins mediated by β-sheet formation (37, 41). It is possible that this is a component of the mechanism of [URE3] propagation, but understanding of prion induction will require a detailed analysis of the structure of normal and prion forms and studies of their interaction with each other.
How do deletions in the C-terminal domain enhance prion induction?
Deletion of residues 151 to 158 or of seven residues at the extreme C terminus enhances induction of [URE3] about 100-fold (Fig. 4). Each of these deletions results in inactivation of the nitrogen regulation function of Ure2p, but it is unlikely that this inactivation is the cause of the enhanced prion induction. The relationships of nitrogen regulation with prion induction and propagation have been explored previously and found to be unconnected (30). The C-terminal mutations might enhance [URE3] induction by disrupting interaction of Ure2p with another protein factor, such as Gln3p, its main target of regulation. This question has not yet been adequately explored.
We suggest that the C-terminal domain both carries out the nitrogen regulation function and stabilizes the N-terminal prion domains in their normal form, perhaps by interacting with these domains. Indeed, there is recent two-hybrid evidence for interaction of N-terminal and C-terminal domains (18). Presumably, these mutations disrupt both functions of the C-terminal domain.
Parallels with scrapie, [PSI], and [Het-s].
None of the other prion proteins has been examined in sufficient detail to determine whether nonoverlapping domains can induce the prion form. Mutations distributed throughout the length of the Prnp gene encoding PrP result in dominant inherited human spongiform encephalopathy (38). Many deletions in Ure2p dramatically increase the frequency with which [URE3] arises but are outside either of the prion domains. We suggest that these mutations eliminate stabilizing interactions with the prion domains. Likewise, many Creutzfeldt-Jakob disease mutations may produce a PrP molecule in which internal interactions are replaced by intermolecular (aggregative) interactions, favoring prion development.
Sup35p has been divided into an N-terminal prion domain, a middle region whose function is not apparent, and a C-terminal domain essential for translation termination and for growth (12, 14, 44, 45). The essential character of the C-terminal domain complicates the type of studies reported here, but it has been shown that, as for Ure2p, mutations in the C-terminal domain of Sup35p result in a dramatic increase in the frequency with which the N-terminal domain induces the prion change (27). The Podospora het-s protein has not yet been examined in detail.
Comparison with mammalian amyloidoses.
Recent evidence suggests that [URE3] is due to amyloid formation by Ure2p. Ure2p is aggregated in cells carrying [URE3] but is evenly distributed in cells not carrying the prion (15). Native Ure2p, purified from normal yeast, is a stable, soluble dimer (42). Residues 1 to 65 of Ure2p spontaneously form amyloid in vitro and initiate amyloid formation by the native Ure2p (43). This parallels the 100-fold-higher efficiency of in vivo prion induction by overproduced Ure2p1-65 than by the same amount of the intact protein. The fact that fragments of Ure2p are far more efficient than the intact protein at inducing de novo [URE3] suggests that proteolytic fragments of Ure2p may be involved in its spontaneous generation even when only the full-length gene is present. Alzheimer’s disease plaques contain the Aβ peptide, a 40- or 42-residue fragment derived by proteolysis from a large precursor protein (19, 26). It is possible that only proteolytic fragments can become part of these plaques. In vitro amyloid formation by the monoclonal light chains produced by some multiple myelomas occurs only after partial proteolysis (20). However, it has been pointed out that these amyloids have been in the tissues for months or years, and so proteolytic digestion may occur after amyloid formation rather than before. There are little data for mammals comparable to those provided by our yeast prion (amyloid) induction experiments. Our finding that fragments have far higher prion (amyloid)-inducing activity than the intact protein provides evidence that fragments may play a dominant role in the initiation of amyloid formation.
REFERENCES
- 1.Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 2.Bolton D C, McKinley M P, Prusiner S B. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 3.Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 4.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 5.Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Linkagae of prion protein and scrapie incubation time genes. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 6.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 7.Chesebro B. BSE and prions: uncertainties about the agent. Science. 1998;279:42–43. doi: 10.1126/science.279.5347.42. [DOI] [PubMed] [Google Scholar]
- 8.Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith J M, Garon C, Hasse A. Identification of scrapie prion protein-specific mRNA in scrapie-infected brain. Nature. 1985;315:331–333. doi: 10.1038/315331a0. [DOI] [PubMed] [Google Scholar]
- 9.Coschigano P W, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePace A H, Santoso A, Hillner P, Weissman J S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 12.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson A G, Meikle V M H, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie in mice. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 14.Doel S M, McCready S J, Nierras C R, Cox B S. The dominant PNM2-mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edskes H K, Gray V T, Wickner R B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edskes, H. K., and K. L. Taylor. Unpublished data.
- 17.Farquhar C F, Somerville R A, Bruce M E. Straining the prion hypothesis. Nature. 1998;391:345–346. doi: 10.1038/34818. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Bellot E, Guillemet E, Baudin-Baillieu A, Gaumer S, Komar A A, Cullin C. Characterization of the interaction domains of Ure2p, a prion-like protein of yeast. Biochem J. 1999;338:403–407. [PMC free article] [PubMed] [Google Scholar]
- 19.Glenner G G. Amyloid deposits and amyloidosis: the beta-fibrillosa. N Engl J Med. 1980;302:1333–1343. doi: 10.1056/NEJM198006123022403. [DOI] [PubMed] [Google Scholar]
- 20.Glenner G G, Ein D, Eanes E D, Bladen H A, Terry W, Page D L. Creation of “amyloid” fibrils from Bence Jones proteins in vitro. Science. 1971;174:712–714. doi: 10.1126/science.174.4010.712. [DOI] [PubMed] [Google Scholar]
- 21.Glover J R, Kowal A S, Shirmer E C, Patino M M, Liu J-J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 22.Griffith J S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 23.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikuchi Y, Shimatake H, Kikuchi A. A yeast gene required for the G1 to S transition encodes a protein containing an A kinase target site and GTPase domain. EMBO J. 1988;7:1175–1182. doi: 10.1002/j.1460-2075.1988.tb02928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King C-Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisilevsky R, Fraser P E. Aβ amyloidogenesis: unique, or variation on a systemic theme? Crit Rev Biochem Mol Biol. 1997;32:361–404. doi: 10.3109/10409239709082674. [DOI] [PubMed] [Google Scholar]
- 27.Kochneva-Pervukhova N V, Poznyakovski A I, Smirnov V N, Ter-Avanesyan M D. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae. Curr Genet. 1998;34:146–151. doi: 10.1007/s002940050379. [DOI] [PubMed] [Google Scholar]
- 28.Kushnirov V V, TerAvanesyan M D, Telckov M V, Surguchov A P, Smirnov V N, Inge-Vechtomov S G. Nucleotide sequence of the SUP2(SUP35) gene of Saccharomyces cerevisiae. Gene. 1988;66:45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 30.Masison D C, Maddelein M-L, Wickner R B. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masison D C, Wickner R B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 32.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B, Aebersold R, Barry R A, Tempst P, Templow D B, Hood L E, Prusiner S B, Weissmann C. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 34.Patino M M, Liu J-J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 35.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 36.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 37.Perutz M F, Johnson T, Suzuki M, Finch J T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prusiner S B. Prions. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Raven Publishers; 1996. pp. 2901–2950. [Google Scholar]
- 39.Ribas J C, Fujimura T, Wickner R B. Essential RNA binding and packaging domains of the Gag-Pol fusion protein of the L-A double-stranded RNA virus of Saccharomyces cerevisiae. J Biol Chem. 1994;269:28420–28428. [PubMed] [Google Scholar]
- 40.Sherman F. Getting started with yeast. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press; 1991. pp. 3–21. [Google Scholar]
- 41.Stott K, Blackburn J M, Butler P J G, Perutz M. Incorporation of glutamine repeats makes protein oligomerize: implications for neurodegenerative diseases. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, K., and R. B. Wickner. Unpublished data.
- 43.Taylor K L, Cheng N, Williams R W, Stevens A C, Wickner R B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 44.TerAvanesyan A, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.TerAvanesyan M D, Kushnirov V V, Dagkesamanskaya A R, Didichenko S A, Chernoff Y O, Inge-Vechtomov S G, Smirnov V N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 46.Turcq B, Deleu C, Denayrolles M, Begueret J. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet. 1991;288:265–269. doi: 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 47.Wickner R B. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 48.Wickner R B. A new prion controls fungal cell fusion incompatibility. Proc Natl Acad Sci USA. 1997;94:10012–10014. doi: 10.1073/pnas.94.19.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson P G, Culbertson M R. SUF12 suppressor protein of yeast: a fusion protein related to the EF-1 family of elongation factors. J Mol Biol. 1988;199:559–573. doi: 10.1016/0022-2836(88)90301-4. [DOI] [PubMed] [Google Scholar]
- 50.Wood, V., M. A. Rajandream, B. G. Barrell, and M. Rieger. 1998. Direct submission to Genbank, accession no. 3136036; EMBL accession no. AL023590.