Atypical Teratoid/Rhabdoid Tumors (AT/RT) are malignant pediatric tumors of the Central Nervous System (CNS) and are molecularly characterized by a biallelic alteration of the SMARCB1 (95%) or SMARCA4 (5%) genes [1]. Transcriptional and DNA-methylation analyses can be used to classify these tumors into three distinct subgroups: SHH (overexpressing genes implicated in neurogenesis and NOTCH signaling, such as ASCL1, with additional sub-entities recently reported), TYR (overexpressing melanosomal marker genes such as TYR, MITF, and OTX2), and MYC (overexpressing MYC and HOXC clusters). These subgroups seem to be associated with distinct genetic and clinical features [1–3] but their significance in terms of prognosis is still unclear given the discrepant results reported in cohorts of patients treated with different therapies [1]. Thus, the prognosis and theranostic significance of these molecular subgroups still needs to be performed, which may be facilitated in near future prospective trials, and by using routine subgroup-specific biomarkers.
Our work studied a case series of 51 pediatric AT/RT (all SMARCB1-deficient) to evaluate the sensitivity/specificity of various combinations of immunohistochemical (IHC) markers to predict the molecular subgrouping. Using previously reported results and in-house datasets [3, 4], we retained Tyrosinase (Clone OCA1/812, 1:400, Abcam, Cambridge, United Kingdom), OTX2 (Clone 1H12C4B5, 1:600; Thermo Fisher, Rockford, USA), MITF (Clone D5, 1:100; Dako, Glostrup, Denmark), MYC (Clone Y69, 1:100, Abcam, Cambridge, United Kingdom), ASCL1 (polyclonal, 1:50; Sigma-Aldrich, Saint-Louis, USA), and SOX11 (Clone MRQ58, 1:100, Sigma-Aldrich, Saint-Louis, USA) as a potentially discriminating panel of markers. The DNA methylation analysis was performed with the Illumina EPIC 850k array, using the v12.5 of the Heidelberg classifier (https://www.molecularneuropathology.org/mnp/). Consistently, 42/51 were assigned to a subgroup of AT/RT with a calibrated score (≥ 0.9) (10 MYC, 24 SHH, and 8 TYR). The nine remaining cases were classified as AT/RT-TYR (n = 4), AT/RT-MYC (n = 4), and control tissue (n = 1) but with a low calibrated score and were thus excluded from subsequent analyses. Their overall distribution within subgroups is depicted by the specificity of t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis (Supplementary Material 1: Fig. S1). Altogether, 42 well-defined AT/RT were considered for further comparison with immunohistochemical subtyping.
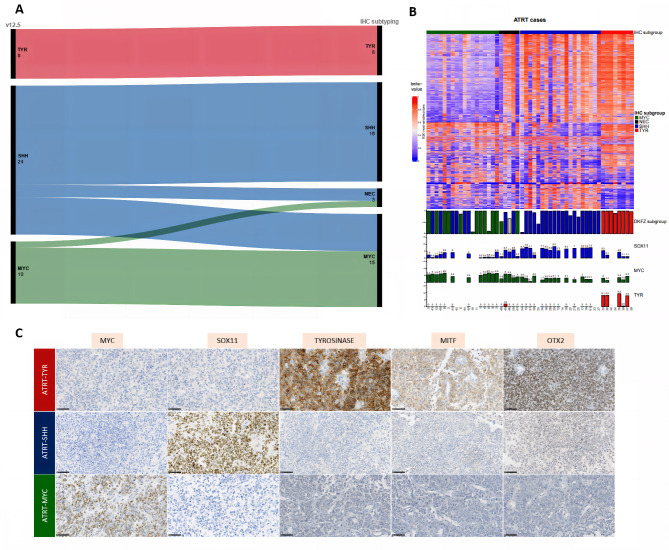
First, because of its low sensitivity (54% of correct subtyping), ASCL1 was excluded from our analyses. Next, nineteen different combinations of five different antibodies were tested to evaluate the accuracy of AT/RT subgrouping using the IHC markers we selected (Supplementary Material 2: Fig. S2). Panels 5 (SOX11-MYC-Tyrosinase), 15 (MYC-Tyrosinase), 16 (MYC-OTX2-MITF), 17 (MYC-OTX2-Tyrosinase), and 18 (MYC-MITF-Tyrosinase) presented the highest accuracy, in perfect concordance with the DNA-methylation profiling for 34/42 (81%) cases) (Supplementary Material 2: Fig. S2A and Supplementary Material 3: Table S1). Particularly, there was perfect concordance between IHC and DNA-methylation profiling for the TYR subgroup (100%). However, in 6/42 (14.3%) cases, IHC and DNA-methylation lead to discrepant conclusions (Fig. 1A-B), with the cases being labeled as belonging to the MYC subgroup by IHC and SHH by DNA-methylation (Supplementary Material 2: Fig. S2B and Table S1). Of note, RNA-sequencing analysis was available for five of these cases, which, consistently with IHC, all clustered with the AT/RT-MYC tumors (Fig. 1C). Finally, IHC failed to assign a subgroup in 2/42 cases (4.7%). Among the multi-marker panels, the best results were obtained with the combination of SOX11-MYC-Tyrosinase (panel 5), with a sensitivity and specificity to diagnose each methylation-defined subtype of: 71% and 100% (for SHH), 90% and 81% (for MYC), and 100% and 100% (for TYR). However, because a subset of AT/RT-MYC may express SOX11, IHC MYC has to be negative in the face of SOX11 positivity to suggest an AT/RT-SHH.
Fig. 1.
Comparison of molecular sub-typing by DNA-methylation analysis and immunohistochemistry. A: Alluvial diagram showing the assignment to subgroups by IHC subtyping using the panel 5 (left) and DNA-methylation profiling using the version v12.5 of the classifier. Cases with a calibrated score ≤ 0.9 were included in the “no match” group. B: Heatmap of DNA methylation beta-value using the top 5000 most variable probes. Samples are grouped according to the AT/RT subgroup predicted by our IHC method (top annotation). Unsupervised hierarchical clustering was applied to probes using Euclidean metric and Ward linkage. Sample subgroups identified using the DKFZ brain tumor classifier are plotted in the first layer of bottom annotation with the color indicating the AT/RT subgroup (red: TYR, blue: SHH and green: MYC). The height of the bar corresponds to the classification score (from 0 to 1). The three other layers of bottom annotation indicate respectively the expression level of SOX11, MYC and TYR genes in log2(TPM + 1) whenever RNA-seq data were available. C: A representative case with immunohistochemical findings for all subgroups (magnification x400). Black scale bars represent 50 μm
Comparison of molecular sub-typing by DNA-methylation analysis and immunohistochemistry
IHC: immunohistochemistry; NEC: Not Elsewhere Classified (the tumor was not classified in a subgroup)
This case series is the first to demonstrate the interest for the use of an antibody panel when sub-typing AT/RT. Our results confirm previously published outcomes showing that Tyrosinase expression is correlated to the AT/RT-TYR subgroup but may be encountered in a subset of SHH [5, 6]. OTX2 immunostaining is routinely used by neuropathologists in medulloblastoma subgrouping and represents a good candidate in the IHC panel for AT/RT subtyping [7]. We evidenced for the first time that SOX11 may constitute a good surrogate for the SHH subgroup whereas the sensitivity/specificity of ASCL1 was lowly informative in our series as reported elsewhere [3]. We also evidenced that a subset of AT/RT SHH - defined by methylation profiling - do express proteins that would be expected to relate them to another group based on RNA-expression data. The consistency between transcript and protein expressions suggests that the discrepancies between expression and methylation may reveal an actual diversity within the SHH group predicted by the current version of the classifier, with some AT/RT-SHH harboring markers predicted as being expressed in the MYC subgroup. Remarkably, the concordance between epigenetic and IHC subtyping is perfect for infratentorial AT/RT SHH (10/10 with a concordant sub-typing), but much less so for supratentorial AT/RT SHH (5/11 for cases with available data). This difference may have a potential biological significance and needs to be further explored.
To conclude, an immunostaining panel that includes MYC, SOX11 and Tyrosinase should be included in future clinical trials to study their clinicopathologic relevance and potential for use as surrogate markers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Fig. S1. t-distributed stochastic neighbor embedding (t-SNE) analysis of the DNA methylation profiles of the 51 investigated tumors alongside selected reference samples of the DKFZ classifier (v12.5). Reference DNA methylation classes: AT/RT, MYC (Atypical teratoid/rhabdoid tumor, MYC-subtype); AT/RT, SHH (Atypical teratoid/rhabdoid tumor, SHH-subtype), AT/RT, TYR (Atypical teratoid/rhabdoid tumor, TYR-subtype), CNS NB, FOXR2 (CNS neuroblastoma, FOXR2-activated), CNS tumor, BCOR ITD (CNS tumor with BCOR internal tandem duplication); CTRL, HEMI (Control tissue, cerebral hemisphere); ETMR, C19MC (Embryonal tumor with multilayered rosettes, C19MC-altered); ETMR, DICER1 (Embryonal tumor with multilayered rosettes, DICER1-altered); MB, non-WNT/ non-SHH (Medulloblastoma, non-WNT, non-SHH); MB, SHH (Medulloblastoma, SHH-activated); MB, WNT (Medulloblastoma, WNT-activated). Cohort cases are designated by their number
Supplementary Material 2: Fig. S2. Additional immunohistochemical results.A: Comparison of results for molecular subtyping using nineteen different immunohistochemical panels (x: panels; y: number of cases). NEC: Not Elsewhere Classified. *designate the panels with the highest accuracy for subtyping. B: Discrepant cases with immunohistochemical findings (magnification x400). Black scale bars represent 50 μm
Supplementary Material 3: Detailed data of the cohort and synthesis
Acknowledgements
We are thankful for the laboratory technicians at GHU Paris Neuro Sainte-Anne Hospital for their assistance, and the Integragen platform for their technical assistance with DNA-methylation analyses.
Author contributions
ATE, JMP, FB and MA participated in conception, design, collection and assembly of data. ATE, AME and PV conducted the neuropathological examinations. JMP, MA, PS, FS, AVD an OD conducted the molecular somatic and germline analyses. ATE drafted the manuscript. All authors reviewed the manuscript and approved the final version.
Funding
No funding.
Data Availability
Proteomic datasets were deposited to the Proteomics Identifications Database (PRIDE) with ac-cession number PXD016832.
Declarations
Ethics Approval
This study was approved by the local ethical committees from GHU Paris Psychiatry and Neurosciences, Sainte-Anne Hospital, of Necker Enfants Malades Hospital. Informed consent was obtained specifically from eachpatient/family for the constitutional genetic study.
Conflict of interest
The authors declare that they have no conflict of interest directly related to the topic of this arti-cle
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torchia J, Golbourn B, Feng S, Ho KC, Sin-Chan P, Vasiljevic A, et al. Integrated (epi)-Genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell. 2016;30:891–908. doi: 10.1016/j.ccell.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, et al. Atypical Teratoid/Rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29:379–393. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Ho B, Johann PD, Grabovska Y, De Dieu Andrianteranagna MJ, Yao F, Frühwald M, et al. Molecular subgrouping of atypical teratoid/rhabdoid tumors-a reinvestigation and current consensus. Neuro-Oncol. 2020;22:613–624. doi: 10.1093/neuonc/noz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leruste A, Tosello J, Ramos RN, Tauziède-Espariat A, Brohard S, Han Z-Y, et al. Clonally expanded T cells reveal immunogenicity of Rhabdoid Tumors. Cancer Cell. 2019;36:597–612e8. doi: 10.1016/j.ccell.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Hasselblatt M, Thomas C, Nemes K, Monoranu C-M, Riemenschneider MJ, Koch A, et al. Tyrosinase immunohistochemistry can be employed for the diagnosis of atypical teratoid/rhabdoid tumours of the tyrosinase subgroup (ATRT-TYR) Neuropathol Appl Neurobiol. 2020;46:186–189. doi: 10.1111/nan.12560. [DOI] [PubMed] [Google Scholar]
- 6.Zin F, Cotter JA, Haberler C, Dottermusch M, Neumann J, Schüller U, et al. Histopathological patterns in atypical teratoid/rhabdoid tumors are related to molecular subgroup. Brain Pathol Zurich Switz. 2021;31:e12967. doi: 10.1111/bpa.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tauziède-Espariat A, Huybrechts S, Indersie E, Dufour C, Puget S, Chivet A, et al. Diagnostic accuracy of a reduced immunohistochemical panel in Medulloblastoma Molecular Subtyping, correlated to DNA-methylation analysis. Am J Surg Pathol. 2021;45:558–566. doi: 10.1097/PAS.0000000000001640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Fig. S1. t-distributed stochastic neighbor embedding (t-SNE) analysis of the DNA methylation profiles of the 51 investigated tumors alongside selected reference samples of the DKFZ classifier (v12.5). Reference DNA methylation classes: AT/RT, MYC (Atypical teratoid/rhabdoid tumor, MYC-subtype); AT/RT, SHH (Atypical teratoid/rhabdoid tumor, SHH-subtype), AT/RT, TYR (Atypical teratoid/rhabdoid tumor, TYR-subtype), CNS NB, FOXR2 (CNS neuroblastoma, FOXR2-activated), CNS tumor, BCOR ITD (CNS tumor with BCOR internal tandem duplication); CTRL, HEMI (Control tissue, cerebral hemisphere); ETMR, C19MC (Embryonal tumor with multilayered rosettes, C19MC-altered); ETMR, DICER1 (Embryonal tumor with multilayered rosettes, DICER1-altered); MB, non-WNT/ non-SHH (Medulloblastoma, non-WNT, non-SHH); MB, SHH (Medulloblastoma, SHH-activated); MB, WNT (Medulloblastoma, WNT-activated). Cohort cases are designated by their number
Supplementary Material 2: Fig. S2. Additional immunohistochemical results.A: Comparison of results for molecular subtyping using nineteen different immunohistochemical panels (x: panels; y: number of cases). NEC: Not Elsewhere Classified. *designate the panels with the highest accuracy for subtyping. B: Discrepant cases with immunohistochemical findings (magnification x400). Black scale bars represent 50 μm
Supplementary Material 3: Detailed data of the cohort and synthesis
Data Availability Statement
Proteomic datasets were deposited to the Proteomics Identifications Database (PRIDE) with ac-cession number PXD016832.