Abstract
The chaperone function of the mammalian 70-kDa heat shock proteins Hsc70 and Hsp70 is modulated by physical interactions with four previously identified chaperone cofactors: Hsp40, BAG-1, the Hsc70-interacting protein Hip, and the Hsc70-Hsp90-organizing protein Hop. Hip and Hop interact with Hsc70 via a tetratricopeptide repeat domain. In a search for additional tetratricopeptide repeat-containing proteins, we have identified a novel 35-kDa cytoplasmic protein, carboxyl terminus of Hsc70-interacting protein (CHIP). CHIP is highly expressed in adult striated muscle in vivo and is expressed broadly in vitro in tissue culture. Hsc70 and Hsp70 were identified as potential interaction partners for this protein in a yeast two-hybrid screen. In vitro binding assays demonstrated direct interactions between CHIP and both Hsc70 and Hsp70, and complexes containing CHIP and Hsc70 were identified in immunoprecipitates of human skeletal muscle cells in vivo. Using glutathione S-transferase fusions, we found that CHIP interacted with the carboxy-terminal residues 540 to 650 of Hsc70, whereas Hsc70 interacted with the amino-terminal residues 1 to 197 (containing the tetratricopeptide domain and an adjacent charged domain) of CHIP. Recombinant CHIP inhibited Hsp40-stimulated ATPase activity of Hsc70 and Hsp70, suggesting that CHIP blocks the forward reaction of the Hsc70-Hsp70 substrate-binding cycle. Consistent with this observation, both luciferase refolding and substrate binding in the presence of Hsp40 and Hsp70 were inhibited by CHIP. Taken together, these results indicate that CHIP decreases net ATPase activity and reduces chaperone efficiency, and they implicate CHIP in the negative regulation of the forward reaction of the Hsc70-Hsp70 substrate-binding cycle.
Multiprotein complexes, the product of protein-protein interactions, participate in varied essential cellular processes, including cell signaling, gene regulation, protein assembly and degradation, and mechanical events such as sarcomere shortening. Conserved structural motifs have evolved to facilitate the interaction of specific proteins in the assembly of these complexes. The tetratricopeptide repeat (TPR) domain is one such protein-protein interaction motif that was originally identified by sequence comparisons among yeast proteins (14, 33). The TPR domain consists of a 34-amino-acid motif with a loose consensus and is present, usually as multiple tandem repeats, in proteins with many cellular functions, including mitosis, transcription, protein transport, and development (reviewed in references 12 and 22). Structural analysis of the TPR domain demonstrates that it forms two α-helical regions separated by a turn, such that apposed bulky and small side chains form a “knob and hole” structure (7, 14). The hydrophobic surface of this structure mediates protein-protein interactions between TPR- and non-TPR-containing proteins.
A number of TPR-containing proteins participate in interactions with the major members of the heat shock protein family, Hsp70, Hsc70, and Hsp90, and are necessary for the appropriate regulation of protein folding and transport. The TPR domains of protein phosphatase 5, cyclophilin 40 (CyP-40), and FKBP52 mediate binding of these proteins to Hsp90 so that they may assist in trafficking of nuclear hormone receptors (20). A different set of proteins with TPR domains interacts with Hsc70 and Hsp70. Hsc70-interacting protein (Hip; also known as p48) binds to the ATPase domain of Hsc70, stabilizes the ADP-bound conformation, and increases the affinity for substrate proteins (17). Hsc70-Hsp90-organizing protein (Hop; also known as p60 or Sti1) serves as a coupling factor that facilitates the cooperation between Hsc70 and Hsp90, although it does not directly assist in chaperoning functions (31, 34). In contrast to Hip, Hop interacts with the carboxy-terminal domain of Hsc70 (8).
In addition to Hip, at least two other proteins regulate the reaction cycle of mammalian Hsc70 and Hsp70. Hsp40 stimulates the ATPase activity of Hsc70 and thus promotes the conversion of ATP-bound, low-substrate-affinity Hsc70 to ADP-bound, high-substrate-affinity Hsc70 (reviewed in reference 15). The reverse reaction, which involves exchange of ATP for ADP and loss of substrate affinity, was recently shown to be facilitated by the antiapoptotic protein BAG-1 (16, 35, 39). Whereas Hip inhibits this reverse reaction and stabilizes the ADP-bound conformation, no cellular inhibitors of the forward reaction of the Hsc70 cycle have been identified.
Because TPR-containing proteins in general play diverse and important roles in cellular function, we assayed for the presence of novel TPR-containing proteins in the human heart. In this report, we present the cloning and characterization of CHIP (carboxyl terminus of Hsc70-interacting protein). CHIP is highly conserved across species and is expressed preferentially in adult striated muscle and brain. The amino-terminal region contains three tandem TPR domains that are similar to such domains in other chaperone accessory molecules. Using the yeast two-hybrid system, we have identified CHIP as an interaction partner with Hsc70 and Hsp70. Biochemical assays demonstrate that recombinant CHIP negatively regulates ATPase and chaperone activities of heat shock proteins, making CHIP an interaction partner with this class of proteins and a candidate negative regulator of the Hsc70-Hsp70 substrate-binding cycle.
MATERIALS AND METHODS
cDNA cloning of human, mouse, and Drosophila CHIP.
A fragment corresponding to nucleotides 721 to 1150 of the human CyP-40 cDNA (21) was radiolabeled with [α-32P]dCTP and used to screen a phage library of human heart cDNA in the vector λgt11 (Clontech) as described elsewhere (29). Phage colonies that hybridized preferentially under low-stringency (42°C) but not high-stringency (55°C) final washes were analyzed. Of 12 colonies characterized, 8 contained human CyP-40 cDNA sequences and 4 encoded a sequence with no homology to known genes. This sequence was analyzed using BLAST 2.0 and the GenBank databases, and three human expressed sequence tag (EST) sequences were found to be identical to the cDNA sequence obtained by phage screening: clones 548268, 177869, and 647520. Clones 548268 and 177869 contained polyadenylated sequence at the 3′ end. Plasmids containing these EST sequences were obtained from Genome Systems, Inc. The 5′ end of the cDNA was defined by 5′ rapid amplification of cDNA ends using human heart mRNA and primers designed on the basis of EST sequences. Products of these reactions, as well as plasmids containing EST fragments, were sequenced as described previously (29), and a single contiguous sequence was assembled. Homologous mouse and Drosophila melanogaster cDNAs were identified in a similar manner, based on EST clones 525111 and 846365 (for mouse) and LD 16049 (for Drosophila). Sequence comparisons were made by using GeneWorks 2.5.1 software (IntelliGenetics, Inc.).
Cell culture and mRNA isolation.
Saos-2 human osteosarcoma cells, HeLa human epidermoid carcinoma cells, HepG2 human hepatoma cells, human fibroblasts, U937 human histiocytic lymphoma cells, RD human embryonal rhabdomyosarcoma cells, IM-9 human lymphoblastoid cells, HCN-1A human neuronal cells, and COS-7 monkey kidney-derived cells were obtained from the American Type Culture Collection. Primary-culture human skeletal muscle and aortic smooth muscle cells were obtained from Clonetics Corp. Primary-culture human umbilical vein endothelial cells, also obtained from Clonetics, were grown in endothelial growth medium containing 2% fetal calf serum (Clonetics). All other cell types were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum as described elsewhere (29). Total RNA from cells in culture was prepared by guanidinium isothiocyanate extraction and centrifugation through cesium chloride (29).
Northern blot analysis.
RNA blots were hybridized as described elsewhere (29). Total RNA (10 μg) from cells in culture was fractionated on a 1.3% formaldehyde-agarose gel and transferred to a nitrocellulose filter. Multiple-tissue Northern blots were purchased from Clontech. Full-length human CHIP (hCHIP) and β-actin cDNAs were labeled with 32P by random priming and used to hybridize filters. Filters were autoradiographed for 16 h on Kodak XAR film at −80°C.
Intracellular localization.
Full-length hCHIP cDNA was inserted into the green fluorescent protein (GFP) fusion vector pEGFP-C3 (Clontech) to create plasmid pEGFP-CHIP. Plasmid pEGFP-GKLF (400-483), expressing a fragment of the gut-enriched Krüppel-like factor (GKLF) as a fusion with GFP, was a gift from Janiel M. Shields (The Johns Hopkins University School of Medicine) and served as a control for nuclear localization. Plasmid pEGFP-C3, expressing GFP alone, served as a control for nonlocalized expression. Plasmids (2 μg) were transfected into COS-7 cells by the Lipofectin method (Life Technologies) as described elsewhere (38). After 48 h, cells were examined for GFP expression by direct epifluorescence using a Nikon Diaphot 300 microscope.
In vitro transcription-translation and recombinant protein production.
35S-labeled in vitro-transcribed and -translated proteins were produced by using a T7- or T3-coupled rabbit reticulocyte lysate system (Promega) according to the manufacturer’s instructions. To determine the molecular mass at which native hCHIP migrates, clone 526948, which contains the entire open reading frame of the hCHIP cDNA in pBluescript SK, was used. For binding assays, the human open reading frame was inserted into vector pCITE-4a (Novagen) as a 43-kDa S-tagged fusion to optimize in vitro translation.
The open reading frame of hCHIP was inserted in frame into vector pET-30a (Novagen) as a His-tagged fusion to create plasmid pET-hCHIP. Glutathione S-transferase (GST) fusions were created by inserting the entire open reading frame or appropriate restriction fragments in frame in vector pGEX KG (Pharmacia) to create plasmids pGST-hCHIP (1-303), pGST-hCHIP (1-142), pGST-hCHIP (1-197), pGST-hCHIP (143-303), and pGST-hCHIP (198-303). GST fusion plasmids pGEXHsc70-(1-540), pGEXHsc70-(373-540), pGEXHsc70-(373-650), and pGEXHsc70-(540-650), containing the indicated fragments of Hsc70, were a kind gift from Ernst Ungewickell (Washington University School of Medicine) and have been previously described (37). Plasmid pGEX-2T (Pharmacia), expressing GST alone, served as a control. Recombinant proteins were expressed in Escherichia coli BL21. The cells were grown for 4 h at 37°C, then 1 mM isopropyl-1-thio-β-d-galactopyranoside was added, and the cells were grown 5 h at 37°C. Bacteria were lysed by sonication. GST fusion proteins were purified by affinity chromatography on glutathione-Sepharose 4B (Pharmacia) according to the manufacturer’s instructions. His-tagged recombinant proteins were purified by Ni2+ chelation chromatography. Recombinant BAG-1 produced in a baculovirus expression system was generously provided by Jörg Höhfeld (Max Planck Institute, Munich, Germany). Protein concentrations of recombinant proteins were determined by a modified Lowry procedure (DC protein assay; Bio-Rad) and were confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel fractionation of samples followed by Coomassie blue staining.
Yeast two-hybrid system.
A yeast two-hybrid screen (13) was performed with the MATCHMAKER two-hybrid system (Clontech). Full-length hCHIP was produced as a fusion with the GAL4 DNA-binding domain in plasmid pAS2-1 and used as a bait. A human heart cDNA library in pACT2, which expresses fusions with the GAL4 activation domain, was screened for potential prey in the yeast strain CG-1945. A total of 2.5 × 106 transformants were screened. The screen is dependent on interactions between the bait and prey to complement His auxotrophy. Of 15 His+/Trp+/Leu+ colonies obtained, 10 were also LacZ+. pACT2-based plasmids from these colonies were isolated from yeast, transformed into bacteria, and candidate cDNA inserts were sequenced.
Binding assays.
To test for interactions between Hsc70 domains and in vitro-translated hCHIP, binding assays were performed with 3 μg of pGEXHsc70 deletion mutants bound to glutathione-Sepharose 4B beads and 3 μl of in vitro translation mixture containing 35S-labeled hCHIP. Binding assays were performed in binding buffer (50 mM NaCl with 1 mg of bovine serum albumin per ml) with rocking for 30 min at 4°C. Beads were pelleted at 1,500 × g for 5 min and washed four times with 0.1% Nonidet P-40 in phosphate-buffered saline. The final pellet was resuspended in 2× Laemmli reducing buffer. Duplicate SDS–12% polyacrylamide gels were run; one gel was stained by Coomassie blue and dried in cellophane, while the other was dried onto 3MM chromatography paper for autoradiography. Additional in vitro binding assays were performed with 1 μg of purified or recombinant protein (Hsc70, Hsp70, or Hsp40) and 15 μg of GST fusion protein (pGEXHsc70 or pGST-hCHIP) bound to glutathione-Sepharose 4B beads in binding buffer. After binding, washing and electrophoresis as described above, gels were transferred to nitrocellulose. Bound proteins were detected by immunoblotting. Bovine Hsc70, human Hsp70, and human Hsp40 were obtained from Stressgen Biotechnologies Corp.
Immunoblotting.
To ensure protein transfer, blots were stained with fast green (Sigma), destained with Milli-Q water, and blocked in 5% dry milk–TBST (20 mM Tris, 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature. Blots were washed three times in TBST and incubated with the appropriate protein-specific primary antibody for 2 h in TBST. Following three washes in TBST, blots were incubated for 1 h with the appropriate horseradish peroxidase-conjugated anti-immunoglobulin G (IgG) secondary antibody in TBST. Immunoreactive protein bands were detected by chemiluminescence. Mouse anti-human Hsp70-Hsc70 monoclonal antibodies (clone N27F3-4) and rabbit anti-human Hsp40 polyclonal antibodies were from Stressgen Biotechnologies. All horseradish peroxidase-conjugated anti-IgG secondary antibodies were from Sigma.
Antisera.
Polyclonal antisera were generated in rabbits against recombinant hCHIP, coupled to keyhole limpet hemagglutinin by standard procedures. Preimmune serum from inoculated rabbits served as a control. The antiserum (hCHIP-3067) was shown to be specific for hCHIP in whole-cell lysates by Western blotting with competing protein (1a).
Immunoprecipitation.
To test for in vivo binding partners, confluent 150-mm-diameter dishes of human skeletal muscle cells were lysed in 0.75 ml of radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1 mM EGTA, 0.25% sodium deoxycholate, 1% Nonidet P-40, 1 mM sodium orthovanadate, protease inhibitors). After a 10-min incubation on ice, cells were scraped and centrifuged at 13,000 × g for 10 min to remove cellular debris. The protein concentration of the whole-cell lysate was determined, and equal amounts of lysate were incubated with the appropriate antibodies (rabbit anti-hCHIP antiserum, rabbit preimmune serum, mouse anti-Hsp70/Hsc70-agarose conjugate, or a mouse IgG-agarose conjugate specific for nitric oxide synthase as a negative control) overnight at 4°C. The anti-hCHIP and preimmune serum complexes were incubated with protein A-Sepharose beads for 2 h at 4°C, and all complexes were pelleted at 3,000 × g for 3 min. The beads were washed three times with radioimmunoprecipitation assay buffer and once with phosphate-buffered saline and resuspended in 2× Laemmli reducing buffer. The samples were separated on 8% polyacrylamide gels by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and immunoblotted as described above for the presence of Hsp70-Hsc70 and CHIP. The mouse anti-Hsp70-Hsc70-agarose conjugate (W27) and the nitric oxide synthase-agarose conjugate were purchased from Santa Cruz Biotechnology. Mouse anti-human Hsp70-Hsc70 monoclonal antibodies (clone 3a3) used for immunoblotting in these experiments were purchased from Affinity Bioreagents.
ATPase activity.
ATPase assays were carried out as described by Boice and Hightower (4). Hsc70, Hsp70, Hsp90, and Hsp40 proteins were obtained from Stressgen Biotechnologies. Briefly, duplicate 25-μl reaction mixtures contained 1.25 μg of heat shock protein (Hsc70, Hsp70, Hsp40, and/or Hsp90), 50 μM ATP, and 1 μCi of [γ-32P]ATP (3,000 Ci/mmol; NEN/Dupont) in the presence or absence of 1 μg of recombinant hCHIP in buffer C (20 mM HEPES, 10 mM (NH4)2SO4, 25 mM KCl, 2 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM dithiothreitol [DTT]). One series of experiments was performed with 10 μg of hCHIP; no difference in ATPase activity was observed compared with experiments performed with 1 μg (not shown). At the beginning of the reaction time and after 20 min, 8 μl was removed from each reaction tube and placed into 0.25 ml of acidified, activated charcoal (50 mM HCl, 5 mM H3PO4, and 7% [wt/vol] activated charcoal), which binds the free nucleotide. These tubes were centrifuged at 4,000 rpm for 20 min at 20°C. Duplicate 100-μl aliquots of the supernatant (containing 32P-labeled inorganic phosphate) were counted in a Beckman LS 3801 scintillation counter. The ATPase activity at each time point was calculated by subtracting the final counts per minute from the initial background counts per minute. Three independent experiments were performed as described, and at least six measurements of ATPase activity were determined for each experimental condition. Data were normalized to the ATPase activity of Hsc70 alone.
Nucleotide binding assay.
Measurement of nucleotide species bound to Hsc70 was determined by previously described methods (23, 26). Briefly, 1 μg of Hsc70 was incubated with 1 μg of recombinant CHIP, Hsp40, and/or BAG-1 in 25 μl of buffer C containing 1 μCi of [α-32P]ATP (3,000 Ci/mmol). Under these conditions, ATP is rapidly hydrolyzed (within 1 min), consistent with previous reports (26), and ADP dissociation can be measured. After 20 min, nucleotides bound to Hsc70 were separated from the free nucleotides by size exclusion chromatography using a G-50 spin column. Aliquots (2 μl) were analyzed by thin-layer chromatography on Selecto cellulose-polyethyleneimine sheets, using 1 M formic acid–1 M LiCl. Unlabeled ATP and ADP were run simultaneously as standards and identified by UV light. Sheets were dried and exposed to film, and binding of ATP and ADP was determined by densitometry.
Rhodanese aggregation assay.
The aggregation of denatured rhodanese was measured by a modification of the protocol described by Minami et al. (26). Bovine liver rhodanese (30 μM; Sigma) was denatured in 6 M guanidine-HCl–30 mM morpholinepropanesulfonic acid (MOPS)-KOH (pH 7.2)–2 mM DTT at room temperature for 30 min. The denatured rhodanese was diluted to a final concentration of 1.2 μM in buffer E (10 mM MOPS-KOH [pH 7.2], 50 mM KCl, 3 mM MgCl2, 2 mM ATP, 2 mM DTT) in the presence or absence of 4 μM Hsp70, 2.5 μM Hsp40, and/or 2.5 μM recombinant hCHIP. The aggregation of rhodanese was measured by the absorbance of light at 340 nm. The measured optical densities were normalized (to account for the addition of different proteins to the wells) to the zero reading for each individual well and the increase in absorbance plotted as a percentage of the total increase for rhodanese alone. Each condition was repeated for a total of eight replicates. Absorbance curves were drawn using Microsoft Excel 98 to a polynomial of the fourth order.
Denatured luciferase binding assay.
Binding of denatured luciferase to Hsc70 was performed with modifications of a previously described method (26). Hsc70 (0.57 μM) was incubated in the presence or absence of CHIP (1.1 μM) and/or Hsp40 (1.1 μM) in 50 μl of buffer C with 2 mM ATP for 5 min at 25°C. Luciferase, which was denatured as described for the luciferase folding assay, was added to a concentration of 0.16 μM, and the reaction mixture was incubated again for 15 min. Proteins interacting with Hsc70 were immunoprecipitated with the W27 antibody and washed as described above. The samples were separated by SDS-PAGE and blotted with an antiluciferase antibody (Rockland Immunochemicals).
Luciferase folding assay.
Refolding of thermally denatured firefly luciferase was performed with modifications of the procedure described by Lu and Cyr (24). Luciferase (14.8 mg/ml; Promega) was diluted to 129 nM in buffered solution (25 mM HEPES, pH 7.4, 50 mM KCl, 5 mM MgCl2) and denatured by heat at 42°C for 20 min. During this time, reaction tubes containing 0.2 μM Hsp70 and/or 0.4 μM Hsp40 in the absence or presence of 0.4 μM recombinant hCHIP in 50 μl in the same buffered solution with 1 mM ATP were prepared on ice. Refolding assays were initiated by adding 2 μl of denatured luciferase to these reaction tubes at 30°C. At various time points from 0 to 120 min, 2-μl aliquots were removed from the reaction tubes and mixed with 60 μl of luciferase assay reagent (Promega) to measure luciferase activity on a Packard TopCount Microplate scintillation counter. Luciferase activity for each reaction was normalized to luciferase in refolding buffer alone. Experiments were repeated for a total of at least 12 measurements of refolding activity for each experimental condition. Refolding curves were drawn using Microsoft Excel to a polynomial of the second order.
Nucleotide sequence accession numbers.
Nucleotide sequences for Drosophila, human, and mouse CHIP have been deposited in GenBank and assigned accession no. AF129084, AF129085, and AF129086, respectively.
RESULTS
Cloning of a novel TPR-containing protein from human heart.
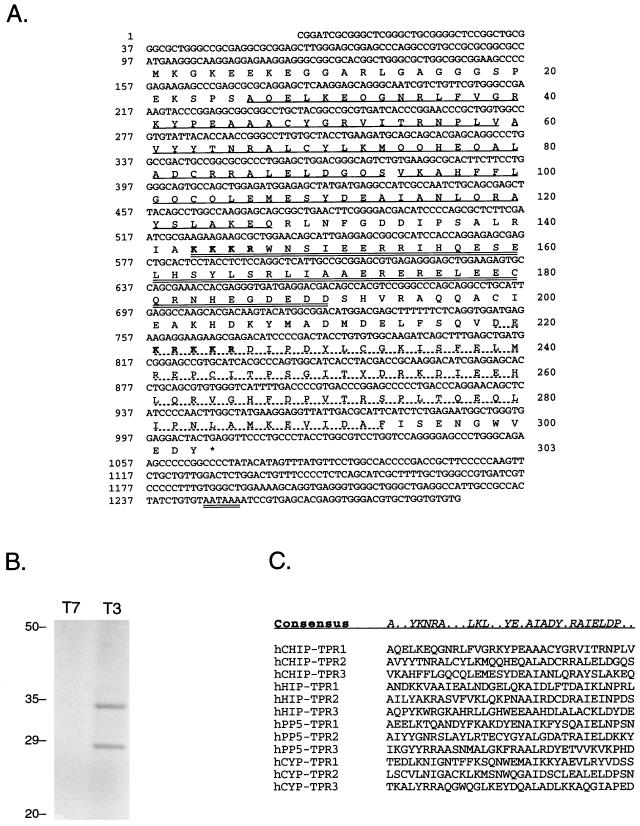
We screened a human heart cDNA library at low stringency with the cDNA fragment of human CyP-40 corresponding to its three carboxy-terminal TPR domains (21). Of 12 positive colonies, 8 contained human CyP-40 sequences and 4 encoded a novel partial cDNA sequence. The full-length cDNA of the novel sequence is 1,286 nucleotides in length and contains a single open reading frame encoding a deduced amino acid sequence of 303 residues (Fig. 1A), thus predicting a translated protein of 34.8 kDa. In vitro transcription and translation using this cDNA as a template produced a specific protein product of the expected size (Fig. 1B), validating the predicted open reading frame. The primary amino acid sequence contains three 34-amino-acid TPR domains at its amino terminus, a central domain rich in charged residues, and two potential nuclear localization signals. The amino terminus shares similarity with many TPR-containing proteins (Fig. 1C), most particularly those that interact with members of the heat shock protein family: Hip, protein phosphatase 5, and CyP-40. The carboxy-terminal region contains no defining structural motifs, although amino acids 218 to 293 show approximately 50% similarity with three proteasome-linked proteins, UFD2, NOSA, and the p40 subunit of the 26S proteasome (9, 19, 30).
FIG. 1.
The hCHIP cDNA. (A) The complete nucleotide (upper line) and deduced amino acid (lower line) sequences of hCHIP. Residues comprising the TPR domains are singly underlined, a region rich in highly charged residues is doubly underlined, and a sequence similar to ubiquitin-proteasome-related proteins is dash underlined. Potential nuclear localization signals are in boldface. (B) In vitro transcription and translation of the hCHIP cDNA in the sense (T3) and antisense (T7) directions demonstrates a major band migrating with the 35-kDa molecular weight marker (position indicated on the left). The faster-migrating species likely represents a degradation product and was noted with in vitro but not in vivo protein expression. (C) Comparison of the TPR motifs of hCHIP with those of human Hip, protein phosphatase 5 (PP5), and CyP-40 (CYP) defines a consensus sequence for this class of TPR domains.
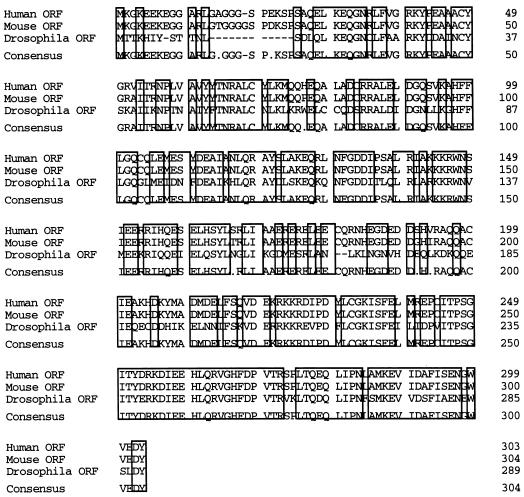
We cloned the mouse and Drosophila homologues of this protein, which we have termed CHIP, to determine its conservation across species and define potential functional domains. Compared with the human amino acid sequence, we found 97% identity and 98% similarity with the mouse sequence and 53% identity and 60% similarity with the Drosophila sequence (Fig. 2). Remarkably, the most highly conserved region is comprised of the 94 residues of the carboxy terminus, which have 87% similarity among these species. Whereas the amino-terminal residues place CHIP in the general category of TPR-containing proteins, the unique, highly conserved carboxyl terminus suggests a cellular function different from that of other members of this class.
FIG. 2.
Comparison of the human, mouse, and Drosophila CHIP amino acid sequences. The deduced amino acid sequences derived from the human, mouse, and Drosophila open reading frames (ORFs) were aligned with GeneWorks 2.5.1. Similar or identical residues are boxed.
Tissue distribution of CHIP.
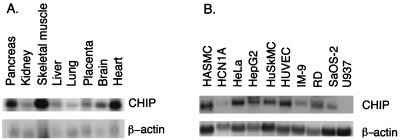
As a first step in the characterization of CHIP, we examined the expression of its mRNA in human tissues and cell lines by Northern blot analysis. Hybridization with the full-length hCHIP cDNA resulted in a single band approximately 1.3 kb in size (Fig. 3A). CHIP was most highly expressed in human striated muscle (skeletal muscle and heart), with somewhat lesser expression in pancreas and brain and relatively little message present in lung, liver, placenta, and kidney. In tissue culture, CHIP mRNA was easily detected in most human cell lines and primary culture cells tested (Fig. 3B). The major exceptions were cells of hematopoietic origin (IM-9 lymphoblastoid and U937 monocytic cells) and HCN-1A undifferentiated neuronal cells.
FIG. 3.
Tissue distribution of hCHIP. (A) Northern analysis was performed with a hCHIP cDNA probe on polyadenylated RNA from human tissues. A single hCHIP mRNA transcript of approximately 1.3 kb is visible. Filters were hybridized with β-actin to visualize RNA loading. (B) Northern analysis was performed as described above with 10 μg of total RNA from the cell lines indicated. HASMC, human aortic smooth muscle cells; HuSkMC, human skeletal muscle cells; HUVEC, human umbilical vein endothelial cells.
Intracellular localization of CHIP.
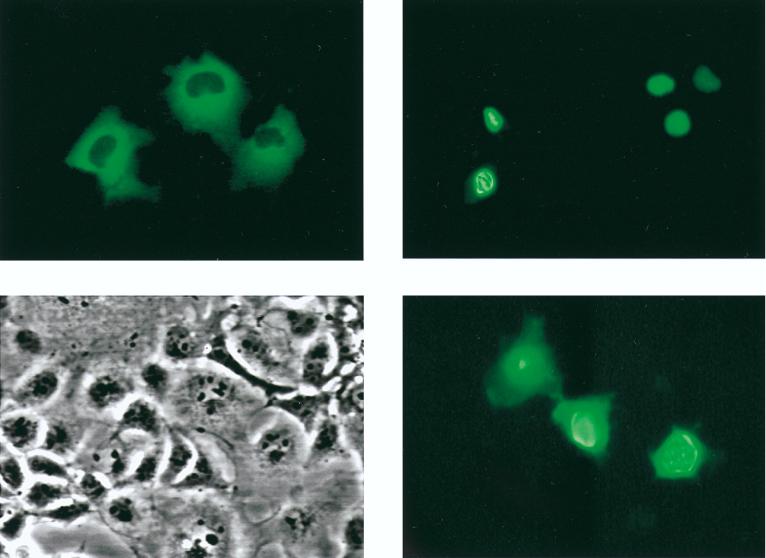
We expressed fusions of full-length hCHIP with GFP by transient transfection in COS-7 cells to localize expression of CHIP within the cell. As controls, we also expressed GFP alone and GFP fused to a nuclear localizing fragment of GKLF (32), using the same method. The GKLF fragment localized GFP to the nucleus (Fig. 4, upper right), and GFP alone exhibited diffuse expression throughout the cell (lower right). In contrast, hCHIP localized GFP strictly to the cytoplasm in a diffuse pattern (upper left). Thus, in spite of having two potential nuclear localization signals, the CHIP-GFP fusion protein localizes to the cytoplasm. Based on these experiments, however, we cannot exclude the possibility that native CHIP is transported out of the cytoplasm in response to an appropriate stimulus.
FIG. 4.
Cellular localization of hCHIP. COS-7 cells were transiently transfected with GFP fusion plasmids, and localization was assessed by direct epifluorescence 48 h after transfection. Upper left, expression of GFP-hCHIP fusions detected by epifluorescence with cytoplasmic localization; lower left, the same field analyzed by light microscopy; upper right, expression of GFP-GKLF fusion demonstrating nuclear expression; lower right, GFP alone demonstrating both nuclear and cytoplasmic localization. Original magnification, ×100.
CHIP interacts with Hsp70 and Hsc70 in the yeast two-hybrid system.
We used the yeast two-hybrid system (13) to identify proteins that interact with hCHIP. A bait vector was constructed to express full-length hCHIP fused to the GAL4 DNA-binding domain. We screened a human heart cDNA expression library with this construct and identified 10 positive (His+ LacZ+) clones from 2.5 × 106 transformants. Of these, five encoded fragments of Hsc70 and three encoded fragments of Hsp70 in frame with the GAL4 activation domain. All clones identified contained the carboxyl terminus of either Hsp70 or Hsc70; the shortest clone encoded 225 amino acids of the carboxy-terminal domain of Hsp70. CHIP did not interact with p53, the simian virus 40 large T antigen, or GAL4 alone in this assay, nor did Hsc70 or Hsp70 interact with lamin C or GAL4 alone, demonstrating the specificity of these in vivo interactions.
CHIP interacts with Hsc70 and Hsp70 in vitro.
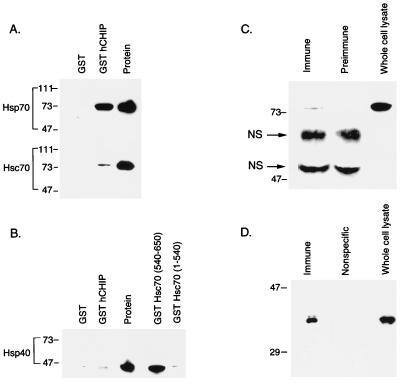
We performed in vitro binding assays using a GST-hCHIP fusion protein to confirm the interactions between CHIP and Hsc70 detected in the yeast two-hybrid screen and to exclude the possibility that these interactions were indirect, requiring additional proteins. Hsc70 and Hsp70 both bound to GST-hCHIP but not to GST alone (Fig. 5A). In contrast, Hsp40 did not interact with hCHIP in this assay (Fig. 5B), although it did interact specifically with the carboxy-terminal residues 540 to 650 of Hsc70 (a test of the binding activity of this Hsp40 preparation), as has been shown previously (8). It should be noted that these binding assays were performed in the absence of ATP. Neither ATP nor ADP influenced the affinity of hCHIP for Hsc70 in these assays (not shown). These experiments demonstrate that CHIP interacts specifically with Hsc70 and Hsp70 in in vitro binding assays in an ATP-independent fashion.
FIG. 5.
CHIP interacts with both Hsp70 and Hsc70 in vitro and in vivo. (A) Binding assays were performed with 1 μg of Hsp70 or Hsc70 plus 15 μg of GST or GST-hCHIP bound to glutathione-Sepharose 4B beads. Western blots were probed with an anti-Hsp70-Hsc70 monoclonal antibody. The protein lane contains 1 μg of the specific protein indicated at the left. Sizes are indicated in kilodaltons. (B) Binding assays were performed with 1 μg of Hsp40 plus 15 μg of GST, GST-hCHIP, GST-Hsc70(540-650), or GST-Hsc70(1-540). Western blots were probed with an anti-Hsp40 polyclonal antibody. The protein lane contains 1 μg of Hsp40. (C) Human skeletal muscle whole cell lysates were immunoprecipitated with rabbit immune serum to hCHIP or rabbit preimmune serum and analyzed by Western blotting with an anti-Hsp70-Hsc70 monoclonal antibody. Human skeletal muscle whole-cell lysates (50 μg) were loaded as a control. The two bands between 47 and 73 kDa are nonspecific (NS); the lower band is the heavy chain of IgG. (D) Human skeletal muscle whole-cell lysates were immunoprecipitated with an anti-Hsp70-Hsc70 monoclonal antibody or a monoclonal antibody recognizing nitric oxide synthase (Nonspecific) and analyzed by Western blot with rabbit anti-hCHIP serum. Whole-cell lysates (50 μg) were loaded as a control. The specific 35-kDa band seen in whole-cell lysates was detected only in immunoprecipitates obtained with the anti-Hsp70-Hsc70 antibody.
CHIP interacts with Hsc70 in vivo.
We performed coimmunoprecipitation experiments to demonstrate that the interactions between CHIP and Hsc70-Hsp70 identified in the in vitro binding assays and in the yeast two-hybrid system also occur under physiologic conditions in mammalian cells. Immunoprecipitates of human skeletal muscle cellular lysates using a polyclonal anti-hCHIP antibody contained, in addition to hCHIP itself (not shown), a single, specific 73-kDa band which was identified by an anti-human Hsp70-Hsc70 monoclonal antibody. This band is identical in size to the band identified by this antibody in whole-cell lysates (Fig. 5C) and comigrates with purified Hsc70 (not shown). Similarly, immunoprecipitates of skeletal muscle lysates obtained by using an anti-Hsp70-Hsc70-agarose conjugate contained a 35-kDa band recognized by the anti-hCHIP antiserum and identical in size to hCHIP (Fig. 5D). In contrast, the specific 35-kDa band was not detected after immunoprecipitation with a monoclonal mouse IgG recognizing nitric oxide synthase. hCHIP was also not detected after immunoprecipitation with rabbit preimmune serum or other nonspecific antibodies (not shown). Given the concordance of these data with those from the yeast two-hybrid system, it is most likely that these experiments indicate the formation of stable complexes between hCHIP and Hsc70-Hsp70 under physiologic conditions in mammalian cells, although the possibility that complexes were formed after lysis cannot be excluded.
CHIP interacts with the carboxy-terminal domain of Hsc70 in vitro.
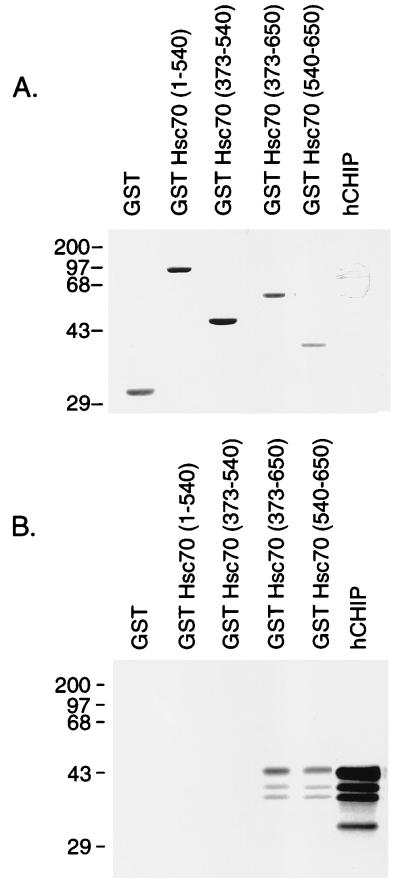
To characterize the Hsc70 domains necessary for interaction with CHIP, we tested binding between 35S-labeled, in vitro-translated hCHIP and a series of GST-Hsc70 amino- and carboxy-terminal deletion mutants (37). These mutants were designed to delineate the binding activity of the nucleotide-binding domain (amino acids 1 to 138), the substrate-binding domain (amino acids 383 to 543), and the carboxy-terminal domain (amino acids 540 to 650) of Hsc70. Affinity purification of the GST fusions by glutathione-Sepharose chromatography was efficient and nearly equivalent after the binding reactions, demonstrating appropriate folding of the fusion proteins (Fig. 6A). hCHIP bound to the carboxy-terminal Hsc70 fragments containing amino acids 373 to 650 and 540 to 650 but not to fragments lacking amino acids 540 to 650 or to GST itself (Fig. 6B). Consistent with our observations in the yeast two-hybrid system, the carboxy-terminal domain seems to be both necessary and sufficient for hCHIP interaction with Hsc70, whereas the ATPase and substrate-binding domains of Hsc70 are expendable. This specific interaction provides the basis for defining this protein as CHIP, carboxyl terminus of Hsc70-interacting protein.
FIG. 6.
CHIP interacts with the carboxy-terminal domain of Hsc70 in vitro. Binding assays were performed with 3 μl of in vitro translation mixture containing 35S-labeled hCHIP with 3 μg of GST-Hsc70 deletion mutants. (A) Coomassie blue staining of the SDS-polyacrylamide gel demonstrates equivalent loading and appropriate folding of the fusion proteins. In vitro-translated 35S-labeled hCHIP is not visualized by Coomassie blue staining. Sizes are indicated in kilodaltons. (B) An autoradiogram of a duplicate gel shows that binding of the 35S-labeled hCHIP occurs to the carboxy-terminal GST-Hsc70 fusion proteins containing amino acids 373 to 650 and 540 to 650 of Hsc70 but not to those containing amino acids 1 to 540 or 373 to 540. An aliquot of 35S-labeled hCHIP was loaded into the last lane; lower-molecular weight species represent degradation of the in vitro-translated protein.
The TPR domain of CHIP is necessary but not sufficient for binding with heat shock proteins.
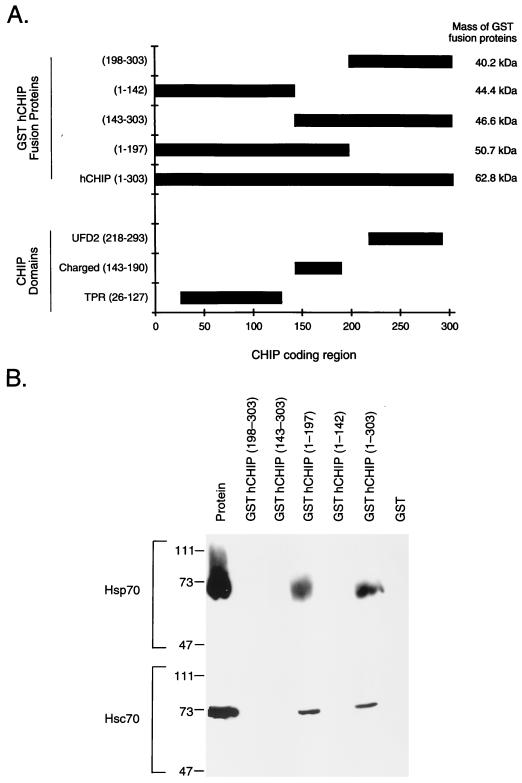
We created a series of hCHIP fragments fused to GST to determine the domains of CHIP that are required for protein-protein interactions with heat shock proteins. The constructs generated, in relationship to hCHIP domains, are shown in Fig. 7A. Binding of these GST fusions with Hsp70 and Hsc70 was tested by in vitro binding assays. Whereas full-length hCHIP interacted with Hsc70 in these experiments, fusions containing amino acids 143 to 303 alone did not (Fig. 7B), demonstrating the importance of the amino terminus of CHIP in these interactions. Amino acids 1 to 142, which contain the three TPR domains, were not sufficient for this interaction either; instead, residues 143 to 197 were also required for binding with Hsc70 to occur. As expected, similar binding requirements were found for interactions with Hsp70. Although both Hip and Hop interact with Hsc70 by means of TPR domains (8), these experiments establish that both the TPR domain and an adjacent charged domain are required for interactions between CHIP and heat shock proteins.
FIG. 7.
The TPR domain of CHIP is necessary but not sufficient for binding with Hsp70 and Hsc70. (A) Diagram depicting the location of the TPR, charged, and UFD2-like domains within the CHIP coding region. The diagram also shows the constructs of the GST-hCHIP fusion proteins that contain one or more of these domains. (B) Binding assays were performed with 1 μg of Hsp70 or Hsc70 and 15 μg of GST or GST-hCHIP fusion proteins bound to glutathione-Sepharose 4B beads. Western blots were probed with an anti-Hsp70-Hsc70 monoclonal antibody. The protein lane contains 1 μg of the specific protein indicated at the left. Sizes are indicated in kilodaltons.
Effects of recombinant hCHIP on ATPase and chaperone activities of heat shock proteins.
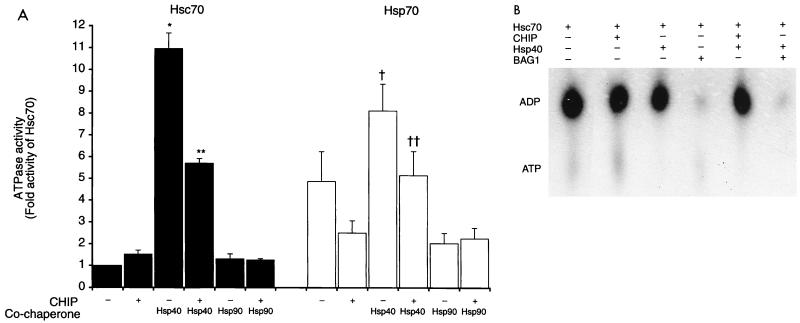
Hsp70 and Hsc70 contain an amino-terminal ATP-binding domain. The intrinsic ATPase activity of these proteins is facilitated by cofactors such as Hip and Hsp40 (17, 23). This cycle is necessary for peptide binding and folding, the so-called chaperone activity of heat shock proteins (reviewed in reference 28). Given the interactions of CHIP with Hsc70 and Hsp70, we examined whether hCHIP had an effect on their ATPase and chaperone activities, in the presence or absence of Hsp40. Recombinant hCHIP expressed in bacteria was used in these studies, given the precedence for retention of ATPase and folding activity when other proteins have been expressed in this system (17, 26, 27). We confirmed that recombinant hCHIP retained binding activity to Hsc70, to ensure that it is correctly folded and processed in bacteria (not shown). The rate of ATPase activity of Hsc70 was significantly (11-fold) increased by the addition of Hsp40 (P < 0.05 [Fig. 8A]), consistent with previous observations (11, 17). hCHIP had little effect on basal Hsc70 ATPase activity but significantly blunted the augmentation of Hsc70 activity by Hsp40 (P < 0.05). In similar experiments performed with Hsp70, the basal ATPase activity of Hsp70 was higher and the enhanced induction by Hsp40 was lower in comparison with Hsc70 (Fig. 8A), which likely reflects some Hsp40-like activity contaminating the preparation of Hsp70. In spite of this, CHIP blocked the ATPase activity of Hsp70 and Hsp40 to a degree similar to that observed for Hsc70 and Hsp40 (P < 0.05).
FIG. 8.
Effects of CHIP on ATPase activities and nucleotide binding of heat shock proteins. (A) The ATPase activities of Hsc70 (black bars) and Hsp70 (white bars) were measured over 20 min in the presence or absence of CHIP, Hsp40, and/or Hsp90 as indicated. Data were normalized to the ATPase activity of Hsc70 alone. ∗ (or †) indicates a significant difference (P < 0.05) between the ATPase activity of Hsc70 alone (or Hsp70 alone) and Hsc70 plus Hsp40 (or Hsp70 plus Hsp40). ∗∗ (or ††) indicates a significant difference (P < 0.05) between the ATPase activity of Hsc70 plus Hsp40 (or Hsp70 plus Hsp40) and Hsc70 plus Hsp40 plus CHIP (or Hsp70 plus Hsp40 plus CHIP). Each condition was repeated for a total of six replicates. (B) The nucleotide species bound to Hsc70 in the absence or presence of CHIP, Hsp40, and/or BAG-1 was determined by thin-layer chromatography. A representative autoradiograph is shown.
The effects of CHIP on ATPase activity could be due to interference with ATP binding, inhibition of ATP hydrolysis, or modulation of nucleotide release. We performed thin-layer chromatography to determine whether CHIP affected the nucleotide-bound state of Hsc70 under steady-state conditions. Under these conditions, ATP is limiting and is rapidly and nearly completely hydrolyzed to ADP. Consistent with previous reports (16), BAG-1 potently stimulated release of bound ADP from Hsc70 (Fig. 8B). By comparison, hCHIP did not elicit nucleotide release from Hsc70, whether or not Hsp40 was present, suggesting that its regulation of Hsc70 function is not dependent on modulation of nucleotide release.
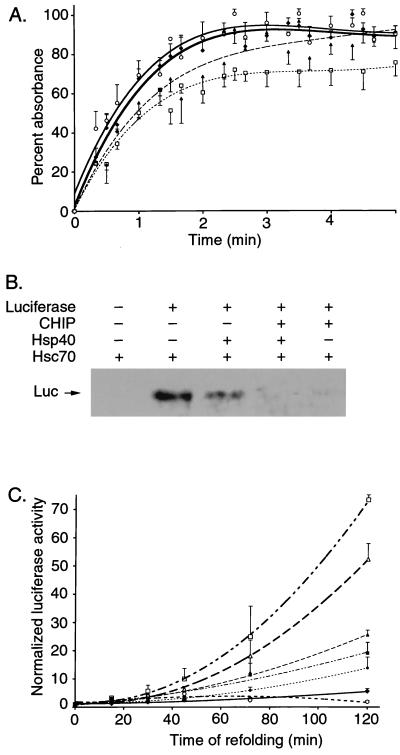
We measured aggregation of denatured rhodanese by spectrophotometry to assess the effect of hCHIP on Hsp70 substrate binding. Under the conditions used, rhodanese alone maximally aggregated after 2.5 min (Fig. 9A). hCHIP alone had no effect on rhodanese aggregation. Addition of Hsp70 and Hsp40 reduced rhodanese aggregation by 30%, indicative of binding to denatured rhodanese. Addition of hCHIP markedly reduced the ability of Hsp70 and Hsp40 to inhibit rhodanese aggregation, and aggregation of denatured rhodanese in the presence of hCHIP, Hsp70, and Hsp40 at 5 min was no different from that of rhodanese alone. To confirm these results, we measured the ability of Hsc70 to interact with denatured luciferase by coimmunoprecipitation with an anti-Hsc70 antibody. Denatured luciferase was present in immunoprecipitates when incubated with Hsc70 or Hsc70 plus Hsp40 (Fig. 9B). However, recovery of luciferase was significantly retarded when hCHIP was incubated with either Hsc70 or Hsc70 plus Hsp40. Taken together, these data indicate that hCHIP blocks the ability of the Hsp70-Hsp40 complex to bind denatured luciferase or prevent aggregation of denatured rhodanese, presumably by interfering with binding of substrates to Hsc70-Hsp70.
FIG. 9.
Effects of CHIP on chaperone functions of heat shock proteins. (A) The aggregation of rhodanese was measured at 340 nm over 5 min in the absence (⧫) or presence of CHIP (○), Hsp70-Hsp40 (□), or CHIP-Hsp70-Hsp40 (▴). The measured optical densities were normalized to the zero reading for each individual well, and the increase in absorbance was plotted as a percentage of the total increase of rhodanese alone. Each condition was repeated for a total of eight replicates, and points represent the mean ± standard error of the mean. (B) Binding assays were performed with denatured luciferase (Luc) and Hsc70 in the presence or absence of hCHIP and/or Hsp40. Binding of denatured luciferase was measured by coimmunoprecipitation using an anti-Hsc70 antibody, followed by blotting with an antiluciferase antibody. A representative experiment is shown. (C) Luciferase activity was measured as an indication of refolding after thermal denaturation. The refolding reactions were performed with Hsc70 (▵), Hsp40 (○), or both (□) in the absence (open symbols) and presence (closed symbols) of CHIP (⧫, CHIP alone). Luciferase activity was measured at various intervals between 0 and 120 min, and the activity for each reaction was normalized to luciferase in refolding buffer alone. Each condition was repeated for a total of 12 replicates, and points represent the mean ± standard error of the mean.
We used the well-characterized luciferase refolding assay as a measure of Hsp70 chaperone function. CHIP alone had little effect on the rate of luciferase refolding, when corrected for that which occurred in buffer alone (Fig. 9C). Hsp70 markedly enhanced the rate of refolding, an effect which was potentiated by Hsp40. In contrast, hCHIP decreased the folding activity of Hsp70 or Hsp70-Hsp40 by 65 or 64%, respectively. The extent to which hCHIP inhibited refolding was similar regardless of whether hCHIP protein was present in molar excess or molar equivalency with other proteins in the assay (not shown), indicating that specific interactions with Hsp70, rather than nonspecific competition for substrates, is the more likely explanation for these results. These experiments indicate that CHIP functions at least in part as a negative regulator of Hsp70- and Hsc70-mediated chaperone functions. This occurs, most likely, by inhibiting the forward reaction of the substrate-binding cycle, resulting in lower substrate affinity of Hsc70 and Hsp70.
DISCUSSION
TPR domain-containing proteins participate in a variety of cellular functions, and whereas many of these proteins do not interact with heat shock proteins, a specific subset does. As demonstrated here, CHIP is a previously undescribed cytoplasmic protein that can complex with Hsc70 and Hsp70. CHIP is distinguished by the degree to which its amino acid sequence is conserved across species, from Drosophila to humans. As expected, the three TPR domains are highly conserved (70% similarity across all three species); even more remarkable is the similarity across species in the carboxy-terminal region (87% similarity from amino acids 210 to 303 of the human sequence). The CHIP carboxy-terminal domain is not similar in sequence to that of any other protein known to interact with heat shock proteins, such as cyclophilins or molecular chaperones, making it likely that the cellular function of CHIP is unique among heat shock protein-interacting proteins.
Examination of the tissue distribution of CHIP mRNA reveals notable differences in its expression. In the adult human, expression is highest in striated muscle (skeletal muscle and heart), less in the pancreas and brain, and negligible in other tissues. Common features of the highly expressing organs include a large proportion of terminally differentiated, nonproliferating cells and relatively high levels of metabolic activity. CHIP is also expressed in a developmentally and spatially regulated fashion in the mouse embryo, particularly in the developing nervous system and during the course of cardiac and skeletal myogenesis (28a). The functional significance of the tissue-restricted expression pattern of CHIP in vivo is not clear at present, although it is possible that CHIP is associated in some way with developmental processes in the nervous system and muscle. An appreciation for the general role of Hsc70-related proteins in the control of developmental processes (for example, regulation of WT1 activity) is emerging (1, 25).
The presence of multiple TPR domains in CHIP suggested to us that CHIP participates in interactions with other proteins. We used the yeast two-hybrid system to define potential CHIP binding partners in vivo, and we found both Hsc70 and Hsp70 to be the most frequently identified interaction partners. There is concern that interactions with Hsp70 or Hsc70 in this assay may reflect a nonspecific interaction with bait proteins that misfold in yeast; interactions of this type would represent Hsc70-Hsp70 chaperone activities rather than functional protein-protein interactions. However, four lines of evidence suggest that this is not the case: (i) these interactions are also detected in vitro with bacterially expressed and in vitro-translated CHIP; (ii) the interaction domain of CHIP contains TPR motifs similar to those of other members of heat shock protein-associated complexes; (iii) CHIP interacts with the carboxy-terminal domain of Hsp70 and Hsc70 in vitro and in vivo, rather than with their substrate-binding domains, which are known to interact with misfolded proteins (5); and (iv) CHIP-Hsc70 complexes can be detected in vivo by immunoprecipitation. Based on the interactions observed in vitro and in vivo, we consider CHIP to be a bona fide interaction partner with Hsp70 and Hsc70.
A number of other proteins interact with Hsp70 and Hsc70 to regulate their cellular functions. Hip and BAG-1 interact with the ATPase domain to stabilize ADP binding and stimulate its release, respectively (17, 35, 39), and auxilin binds the ATPase domain to assist in uncoating clathrin-coated vesicles (36, 37). In addition to mediating homo-oligomeric interactions of Hsc70 and Hsp70 (2), the 10-kDa carboxy-terminal domain (amino acids 537 to 646 of Hsc70) provides noncompetitive and probably independent binding sites for Hop and Hsp40 (8). Hop serves as an organizing factor mediating the interactions between Hsp70 and Hsp90 (6, 34), whereas Hsp40 is a cochaperone that enhances ATPase activity and stable associations with unfolded peptides (11, 26). We now demonstrate that the carboxy-terminal domain is both necessary and sufficient for interactions between CHIP and Hsp70-Hsc70, making CHIP the third member of the group of known carboxyl terminus-binding proteins. We do not yet know whether CHIP interacts with either the Hsp40 or Hop binding site or whether there is a distinct binding site for CHIP in the carboxy-terminal domain. Both Hop and CHIP are TPR domain-containing proteins, which suggests that they may interact with Hsc70 and Hsp70 through common mechanisms; however, it is equally plausible that CHIP interacts in some way with the Hsp40 binding site, since the biochemical properties of CHIP could be explained by interference of Hsp40 binding with Hsc70.
It is notable that CHIP, which binds to the carboxyl termini of Hsc70 and Hsp70, shows strong structural similarities with TPR domain-containing proteins (Hip, Cyp-40, and protein phosphatase 5) that interact with Hsp90 or with the ATPase domains of Hsc70 and Hsp70 (Fig. 1C). This finding suggests that TPR motifs of proteins that interact with members of the heat shock protein family form a structurally related subset within the larger family of TPR-containing proteins. If this is indeed the case, what then determines the specificities of interactions with heat shock proteins? Although the answer to this question is likely to be determined by structural comparisons, it is worth noting that the TPR domains of CHIP alone are necessary but not sufficient for interactions with Hsc70 and Hsp70 (Fig. 7) and that an adjacent charged domain is also required. Since a positively charged domain adjacent to the TPR domain of Hip is similarly required for interactions with the ATPase domain of Hsc70 (18), it is possible that the binding specificities for this class of TPR-containing proteins are determined by sequences adjacent to the TPR domains.
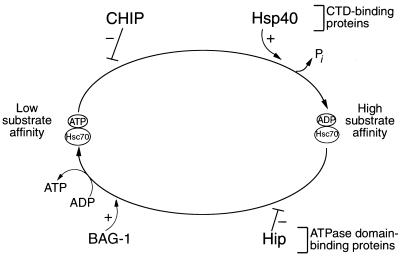
Previously, three proteins have been shown to regulate the Hsc70-Hsp70 substrate-binding cycle. Hsp40 enhances the forward reaction of this cycle by increasing Hsc70 ATPase activity and thus promoting the formation of the ADP-bound, high-substrate-affinity Hsc70 conformation (26). Hip stabilizes ADP-bound Hsc70 (17); therefore, both Hip and Hsp40 enhance Hsc70-mediated refolding. The antiapoptotic protein BAG-1 has been shown recently to facilitate the reverse reaction by promoting the exchange of ATP for ADP and release of substrate (16); BAG-1 therefore opposes the actions of Hip and attenuates Hsc70 chaperone activity. Whereas Hip might be considered to antagonize the reverse reaction of the Hsc70 cycle, a negative regulator of the forward reaction has not been described. Based on its ability to decrease the ATPase activity of Hsp70 and Hsp40, CHIP is the first described candidate protein to inhibit the forward reaction cycle. Consistent with this role, CHIP inhibits luciferase refolding and substrate binding that occurs in the presence of Hsp70 and Hsp40, although CHIP has no effect by itself in either of these assays. Based on these results, a model can be proposed whereby CHIP and BAG-1 act on the forward and reverse Hsp70-Hsc70 reaction cycles, respectively, to maintain the low substrate affinity conformation; in contrast, Hip and Hsp40 coordinately regulate the two cycles to favor the ADP-bound state (Fig. 10). Potential molecular mechanisms for the effects of CHIP on the reaction cycle include prevention of ATP binding or inhibition of ATP hydrolysis; future studies will be needed to address these possibilities.
FIG. 10.
Model of the eukaryotic reaction cycle in the presence of CHIP, Hsp40, Hip, and BAG-1. The forward reaction, in which ATP is hydrolyzed to ADP and inorganic phosphate (Pi) is released, is enhanced by Hsp40. The biochemical data suggest that CHIP blocks this forward reaction. Hip stabilizes the ADP-bound, high-substrate-affinity conformation of Hsc70 and thus enhances chaperone activity. Conversely, BAG-1 accelerates nucleotide exchange, promoting substrate release and the formation of the low-substrate-affinity, ATP-bound conformation of Hsc70. In this model, both BAG-1 and CHIP would favor the low-affinity Hsc70 conformation, whereas Hip and Hsp40 would favor the high-affinity conformation. CTD, carboxy-terminal domain.
At present, it is unclear whether the effects of CHIP on the Hsp70-Hsc70 substrate-binding cycle are mediated solely through the TPR and charged domains (perhaps by competing with or otherwise altering the affinity of Hsp40 for binding to Hsc70) or whether the highly conserved carboxy-terminal domain also regulates Hsc70-related functions in some way. The three proteins sharing similarity with this domain—UFD2, NOSA, and the p40 subunit of the 26S proteasome—have functions linked to the ubiquitin-proteasome pathway (9, 19, 30). Although a function has not yet been ascribed to the similar region in any of these proteins, it is tempting to speculate that CHIP may serve in some way to mediate interactions between the chaperoning and ubiquitin-proteasome systems, as Hsc70 and Hsp70 are required for ubiquitin-dependent degradation of some, but not all, protein substrates (3, 10). The amino acid sequence similarities between proteasome-associated proteins and CHIP, a regulator of the Hsc70-Hsp70 substrate-binding cycle, emphasize the need to define better the relationship between chaperoning and proteolysis.
ACKNOWLEDGMENTS
This work was supported by the Sealy and Smith Foundation and by grants HL03658 and AG15234 to Cam Patterson.
We thank Gordon Huggins for assistance with protein expression and binding assays, Mansoor Sajid for assistance with immunoblotting, Shiloe Souza for technical assistance, and Joann Aaron for editorial assistance. The following scientists generously provided reagents: Jörg Höhfeld (BAG-1 protein), Janiel Shields (GFP-GKLF plasmids), and Ernst Ungewickell (GST-Hsc70 plasmids). We are grateful to Marschall S. Runge for his continued support and encouragement.
REFERENCES
- 1.Angelier N, Moreau N, Rodriquez-Martin M L, Penrad-Mobayed M, Prudhomme C. Does the chaperone heat shock protein Hsp70 play a role in the control of developmental processes? Int J Dev Biol. 1996;40:521–529. [PubMed] [Google Scholar]
- 1a.Ballinger, C. A., and C. Patterson. Unpublished observations.
- 2.Benaroudj N, Fouchaq B, Ladjimi M M. The COOH-terminal peptide binding domain is essential for self-association of the molecular chaperone Hsc70. J Biol Chem. 1997;272:8744–8751. doi: 10.1074/jbc.272.13.8744. [DOI] [PubMed] [Google Scholar]
- 3.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 4.Boice J A, Hightower L E. A mutational study of the peptide-binding domain of Hsc70 guided by secondary structure prediction. J Biol Chem. 1997;272:24825–24831. doi: 10.1074/jbc.272.40.24825. [DOI] [PubMed] [Google Scholar]
- 5.Chappell T G, Konforti B B, Schmid S L, Rothman J E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 6.Chen S, Prapapanich V, Rimerman R A, Honore B, Smith D F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins Hsp90 and Hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 7.Das A K, Cohen P T W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demand J, Lüders J, Höhfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubiel W, Ferrell K, Dumdey R, Standera S, Prehn S, Rechsteiner M. Molecular cloning and expression of subunit 12: a non-MCP and non-ATPase subunit of the 26 S protease. FEBS Lett. 1995;363:97–100. doi: 10.1016/0014-5793(95)00288-k. [DOI] [PubMed] [Google Scholar]
- 10.Fisher E A, Zhou M, Mitchell D M, Wu X, Omura S, Wang H, Goldberg A L, Ginsberg H N. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- 11.Frydman J, Nimmesgern E, Ohtsuka K, Hartl F-U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 12.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 13.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 14.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 15.Höhfeld J. Regulation of the heat shock cognate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol Chem. 1998;379:269–274. [PubMed] [Google Scholar]
- 16.Höhfeld J, Jentsch S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höhfeld J, Minami Y, Hartl F-U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 18.Irmer H, Höhfeld J. Characterization of functional domains of the eukaryotic co-chaperone Hip. J Biol Chem. 1997;272:2230–2235. doi: 10.1074/jbc.272.4.2230. [DOI] [PubMed] [Google Scholar]
- 19.Johnson E S, Ma P C M, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 20.Kay J E. Structure-function relationships in the FK506-binding protein (FKBP) family of peptidylprolyl cis-trans isomerases. Biochem J. 1996;314:361–385. [PMC free article] [PubMed] [Google Scholar]
- 21.Kieffer L J, Seng T W, Li W, Osterman D G, Handschumacher R E, Bayney R M. Cyclophilin-40, a protein with homology to the P59 component of the steroid receptor complex: cloning of the cDNA and further characterization. J Biol Chem. 1993;268:12303–12310. [PubMed] [Google Scholar]
- 22.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 23.Liberak K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Z, Cyr D M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 25.Maheswaran S, Englert C, Zheng G, Lee S B, Wong J, Harkin D P, Bean J, Ezzell R, Garvin A J, McCluskey R T, DeCaprio J A, Haber D A. Inhibition of cellular proliferation by the Wilms tumor suppressor WT1 requires association with the inducible chaperone Hsp70. Genes Dev. 1998;12:1108–1120. doi: 10.1101/gad.12.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami Y, Höhfeld J, Ohtsuka K, Hartl F U. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 27.Naylor D J, Stines A P, Hoogenraad N J, Høj P B. Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the co-chaperones of specific Hsp70 members. J Biol Chem. 1998;273:21169–21177. doi: 10.1074/jbc.273.33.21169. [DOI] [PubMed] [Google Scholar]
- 28.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 28a.Patterson, C. Unpublished observations.
- 29.Patterson C, Perrella M A, Hsieh C-M, Yoshizumi M, Lee M-E, Haber E. Cloning and functional analysis of the promoter for KDR/flk-1, a receptor for vascular endothelial growth factor. J Biol Chem. 1995;270:23111–23118. doi: 10.1074/jbc.270.39.23111. [DOI] [PubMed] [Google Scholar]
- 30.Pukatzki S, Tordilla N, Franke J, Kessin R H. A novel component involved in ubiquitinization is required for development of Dictyostelium discoideum. J Biol Chem. 1998;273:24131–24138. doi: 10.1074/jbc.273.37.24131. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher R J, Hurst R, Sullivan W P, McMahon N J, Toft D O, Matts R L. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 32.Shields J M, Yang V W. Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J Biol Chem. 1997;29:18504–18507. doi: 10.1074/jbc.272.29.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 34.Smith D F, Sullivan W P, Marion T N, Zaitsu K, Madden B, McCormick D J, Toft D O. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol. 1993;13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takayama S, Bimstone D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ungewickell E, Ungewickell H, Holstein S E, Lindner R, Prasad K, Barouch W, Martin B, Greene L E, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 37.Ungewickell E, Ungewickell H, Holstein S E H. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J Biol Chem. 1997;272:19594–19600. doi: 10.1074/jbc.272.31.19594. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Ruef J, Rao G, Patterson C, Runge M S. Differential transcriptional regulation of the human thrombin receptor gene by the Sp family of transcription factors in human endothelial cells. Biochem J. 1998;330:1469–1474. doi: 10.1042/bj3301469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeiner M, Gebauer M, Gehring U. Mammalian protein RAP46: an interaction partner and modulator of 70 kDa heat shock proteins. EMBO J. 1997;16:5483–5490. doi: 10.1093/emboj/16.18.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]