Abstract
Societies in East Asia have utilized domesticated cattle for over 5000 years, but the genetic history of cattle in East Asia remains understudied. Genome-wide analyses of 23 ancient Mongolian cattle reveal that East Asian aurochs and ancient East Asian taurine cattle are closely related, but neither are closely related to any modern East Asian breeds. We observe binary variation in aurochs diet throughout the early Neolithic, and genomic evidence shows millennia of sustained male-dominated introgression. We identify a unique connection between ancient Mongolian aurochs and the European Hereford breed. These results point to the likelihood of human management of aurochs in Northeast Asia prior to and during the initial adoption of taurine cattle pastoralism.
One-Sentence Summary:
Ancient interbreeding of East Asian aurochs and cattle suggests management, but leaves no signature in modern eastern breeds.
Paleogenomic studies of cattle domestication largely focus on their origins in Western Eurasia, despite knowledge of multiple indigenous domestications of bovines such as yaks in the east. Cattle pastoralism has a long history in East Asia, but many questions remain about the origins of East Asian domestic cattle, including the possibility of indigenous domestication from local extinct wild aurochs (Bos primigenius). Current genetic evidence shows that modern cattle lineages largely derive from two aurochs domestication episodes: one in the Near East by 10,000 years ago that gave rise to taurine cattle (Bos taurus), and one in South Asia by 8000 years ago that gave rise to indicine/zebu cattle (Bos indicus) (1–5). Both genetic and archaeological evidence suggest the possibility of additional domestication events, most notably in North Africa (6–9), but the ability to conclusively recognize introgression over indigenous domestication remains largely unresolved. Regardless, wild introgression is acknowledged as critical to the development of modern lineages (10–15).
In East Asia today, taurine cattle breeds are more common in the north and indicine cattle breeds are more common in the south. There are many taurine-indicine hybrid breeds, and admixture between domestic cattle and other indigenous East Asian bovines such as yak, gayal/mithun, and banteng is common (15–18), with taurine-yak hybrids especially common in Central Mongolia. The north/south division between taurine and indicine cattle use appears to have deep roots related to the initial adoption of domestic cattle in East Asia. Taurine cattle were introduced to Northeast Asia at least 5000 years ago. Their bones are found at Afanasievo sites in Mongolia by about 5000 years ago (19), northeastern China by about 5300 years ago (20), and the Yellow River Valley by about 4500 years ago (21). Indicine cattle have played a minor role in Northeast Asia and are less well-understood, being present in southern China by the late Warring States period (ca. 500 BCE) (22). Here we explore the unique genetic history of aurochs and taurine cattle in Northeast Asia.
There is growing archaeological evidence that people in Northeast Asia hunted and possibly managed wild aurochs for thousands of years prior to the adoption of domestic taurine cattle. On the Mongolian steppe, sedentary societies during the mid-Holocene relied heavily on aurochs, with increasingly sedentary groups continuing to specialize in big-game hunting (23, 24). In northeastern China and the Yellow River Valley, several late Neolithic and early Bronze Age sites contain both aurochs and taurine cattle remains (25, 26). For example, at Houtaumuga in Jilin, archaeologists have uncovered pits full of aurochs bones. At this same site, geneticists have also identified the earliest East Asian cattle bone belonging to a taurine mtDNA haplogroup directly radiocarbon dated to 5500–5300 cal. BP (20). These finds indicate that there would have been opportunities for admixture between wild and domestic herds as soon as taurine cattle were first introduced to East Asia. However, only one East Asian aurochs mitogenome has been previously published (27) and there have not yet been any genome-wide ancient DNA studies of East Asian aurochs. Many questions remain, including how are East Asian aurochs populations related to East Asian domestic cattle? Were aurochs domesticated in East Asia? And did aurochs and domestic cattle interbreed when taurine cattle were first adopted in East Asia?
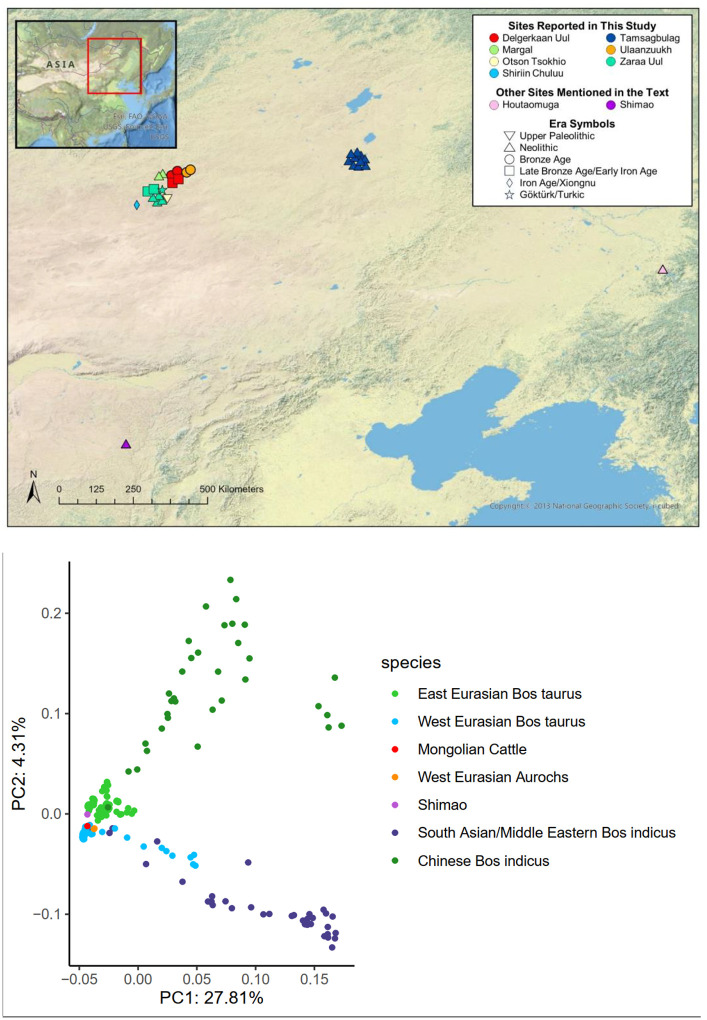
We sequenced low coverage genomes of 23 cattle excavated from seven archaeological sites in eastern Mongolia dated to between 30,000 years ago and 2,000 years ago (Figure 1A; Table S1–S2). The 30,000-year-old Paleolithic individual from Otson Tsokhio (28) represents the oldest paleogenomic data from an East Asian aurochs published to date. Early Neolithic samples from Tamsagbulag, Zaraa Uul, and Margal were directly radiocarbon dated to between 8,000–5,500 years ago and pre-date the introduction of taurine cattle. Bronze Age and later samples from Zaraa Uul (including the highest coverage sample B17), Delgerkhaan Uul, Shiriin Chuluu, and Ulaanzuukh date to after 4000 years ago. The genomes are low coverage (ranging from 0.01–0.94x), but they are still informative for investigating the deep history of cattle in Northeast Asia. We compared the ancient Mongolian samples with a reference panel of over 300 ancient and modern cattle genomes from around the world, including ancient taurine cattle from the 4000-year-old site of Shimao in the Ordos region of China (Table S3) (5, 17, 29). Shimao was one of the first sites in the Yellow River Valley region to use domesticated cattle in large numbers (30, 31). We also compared mitochondrial d-loop regions with previously published samples from China including the 6000-year-old individuals from Houtaomuga (20).
Fig. 1. Geographical distributions and principal component analysis of ancient Mongolian cattle samples.
(A) map showing the locations of samples according to time period. (B) PCA showing highest coverage ancient samples from Zaraa Uul (B17, early Bronze Age, labeled “Mongolian Cattle”) and Shimao (Shimao05, early Bronze Age) projected onto modern cattle individuals. High coverage ancient cattle individuals from Verdugo et al. (2019) are also included in the projection.
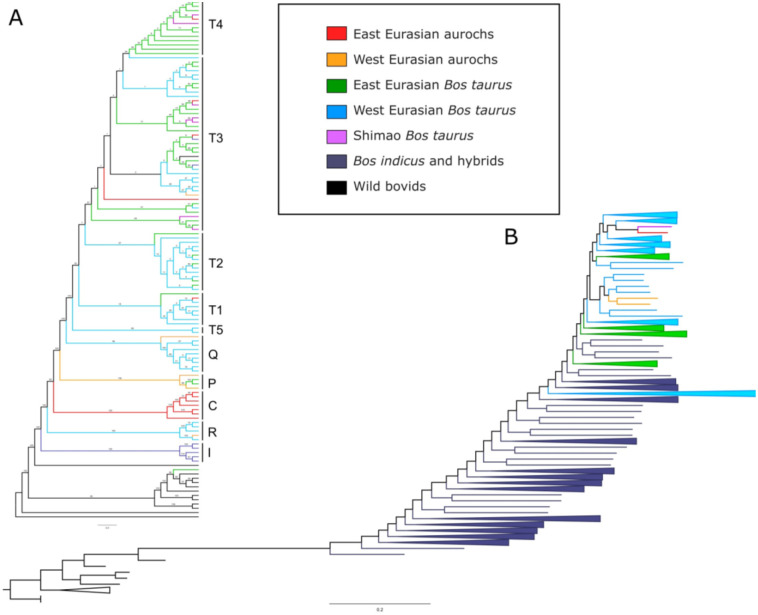
Mongolian aurochs show a close relationship to other sequenced aurochs and taurine cattle, and are distinct from indicine cattle (Figure 1B). The ancient Mongolian cattle form a single clade, suggesting that all sampled individuals represent a continuous population (Figure 2B). Neolithic Mongolian aurochs are part of the East Asian mitochondrial haplogroup C that has been previously identified exclusively in East Asian aurochs dating from between 10,000 to 4000 years ago (20, 27, 32) (Figure 2A). Interestingly, the Mongolian aurochs group together separately from African, European, and the Middle Eastern aurochs, but are still part of the clade that includes all sequenced aurochs and taurine cattle. Indicine cattle represent an outgroup. This suggests that aurochs across Northern Africa and Eurasia represent one large, panmictic population. Another possibility is that the Mongolian aurochs are part of the taurine cattle clade because they genetically contributed to early domesticated cattle in a significant way, although this hypothesis is difficult to test with low-coverage data.
Fig. 2. Phylogenetic relationships for cattle.
(A) Maximum likelihood tree of whole mitochondrial genome sequences of modern and ancient cattle and bovids. (B) Maximum likelihood tree of genome-wide SNPs for all high-coverage bovids and ancient and modern cattle. Individuals are color-coded by geographic region and species. Hybrid cattle with any amount of indicine admixture are labeled as ‘Bos indicus and hybrids.’
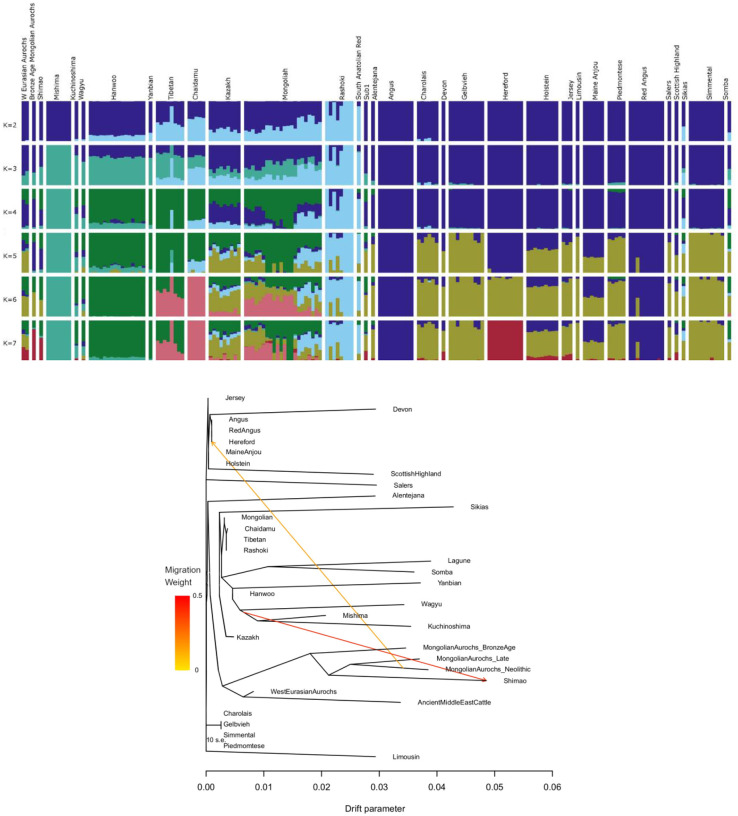
The ancient Mongolian cattle are most closely related to 4000-year-old individuals from Shimao. Previous examination of modern Eurasian cattle breeds suggests three structured geographic groups: West Eurasian, Central Eurasian, and East Asian (17). The East Asian component of ancestry is found today primarily in Japanese cattle such as Mishima and was proposed to derive from gene flow between East Asian aurochs and domesticated taurine cattle. However, we do not observe any of this ancestry in ancient Mongolian aurochs, suggesting that another wild bovine population may have contributed to East Asian cattle (Figure 3A).
Fig. 3. Admixture between Mongolian cattle and other populations.
(A) Admixture plot showing population structure in taurine cattle and aurochs. At K=7 Mongolian aurochs, ancient Chinese cattle from Shimao, and Hereford have a shared ancestry component. (B) Treemix plot showing 2 migration edges among taurine cattle populations. Here, B01 is labeled as MongolianAurochs_Neolithic, B17 is MongolianAurochs_BronzeAge, and B28 is MongolianAurochs_Late.
Additionally, most modern cattle breeds seem to show very little to no ancestry from ancient Mongolian cattle. The one exception is Hereford, a British breed of beef cattle. Previous work has shown a connection between Hereford and cattle from Shimao (17), which are genetically similar to the ancient Mongolian cattle analyzed here. Our analyses show evidence for gene flow from Neolithic Mongolian aurochs into Hereford (Figure 3B). Herefords originated as traction and meat cattle from the long-isolated region of Herefordshire, England, where the cattle were heavily influenced by stock from the north of Wales but otherwise isolated until after 1700 (33). Perhaps the connection with ancient Mongolian cattle results from close genetic similarity between Mongolian aurochs and those of the British Isles, or perhaps both European and Mongolian aurochs contributed to early ancient taurine lineages, but their genetic legacy has since been swamped in other breeds. The connection might also be explained by more recent gene flow from a modern Asian cattle breed with Mongolian aurochs ancestry, but we find this hypothesis unlikely for two reasons. First, Mongolian aurochs ancestry is found almost exclusively in European breeds of cattle while modern cattle breeds from Mongolia and other parts of East Asia have little to no Mongolian aurochs ancestry (Figure 3A). Second, the gene flow from Mongolian aurochs seems to come specifically from Neolithic individuals, which pre-date the introduction of taurine cattle to East Asia (Figure 3B). Additional sequencing of ancient cattle from across Eurasia would be needed to clarify this connection.
Our analysis also reveals one additional admixture event from Japanese cattle breeds into Shimao (Figure 3B). The gene flow from East Asian cattle lineages into Shimao is consistent with previous work that shows a connection between ancient Shimao cattle and modern northeast Asian breeds (17). Cattle from Shimao form a clade with the ancient Mongolian cattle and also have taurine mitochondrial haplogroups, suggesting hybridization between East Asian aurochs and taurine cattle. All individuals predating the arrival of taurine cattle are from mitochondrial haplogroup C. While haplogroup P, which is found in European aurochs, has been identified in modern Japanese cattle (34), we did not observe haplogroup P in the Neolithic Mongolian aurochs. Most later individuals are mitochondrial haplogroup T3, which is most common in both East Asia and western Eurasia. One individual (B20) is haplogroup T1, which is common today only in southern Europe and Africa (35). One individual (B12) from the sixth century is Haplogroup T4, but still bears significant evidence of Mongolian aurochs ancestry. The T4 haplogroup is distinct to East Asian and believed to have diverged locally from earlier arrivals of T3 or admixture with East Asian aurochs (10, 36). We do not think that the introduction of taurine cattle represents population replacement because local Bronze Age cattle are still very similar to Neolithic aurochs and genetically distinct from ancient western Eurasian cattle (5). These preliminary findings suggest that early gene flow was primarily between male aurochs and female taurine cattle. The lack of modern cattle with aurochs DNA suggests that these aurochs lineages were replaced over time, likely due to interbreeding with other Eurasian cattle breeds. Aurochs persisted in China into the Zhou Dynasty (1046-256 BCE) and probably much later (37). More geographically and temporally extensive genomic analyses of ancient East Asian cattle would provide additional insights into how East Asian cattle populations changed through time.
Stable isotope analysis was carried out on 7 of the samples, of which 4 Neolithic individuals (B06, B13, B15, B23) had elevated δ13C and δ15N ratios locally consistent with foddering (38) (Table S1). Elevated δ13C and δ15N is observed to be characteristic of Late Neolithic aurochs at the Honghe site in Northeast China (26). C4 plants are not common in these predominantly C3 landscapes and our findings point to human intervention in diet. To test the possibility of aurochs management, we looked at measures of heterozygosity (Figure S8) in order to determine whether Mongolian individuals show changes in genetic diversity through time. A reduction in genetic diversity might be evidence for bottlenecks during management or domestication. Although heterozygosity may be slightly lower in the Bronze Age individuals compared to Neolithic individuals, we also see that genetic diversity remains high in some later samples such as the Xiongnu individual from Shiriin Chuluu. We do not observe a consistent trend in heterozygosity that shows direct evidence for aurochs management, nor is there an overlap between individuals that may have been foddered and individuals with taurine mitochondrial haplotypes. Individuals that may have been foddered are all early Neolithic (Oasis 2, 8500–5000 cal BP) aurochs and tend to group more closely in our phylogenetic tree regardless of age or geographic region (Figure S9). The results point to variation in aurochs diet where some pre-taurine individuals were being grazed on or foddered with C4 plants while others were uncontrolled grazers. Deeper coverage genomic and additional isotope analyses are needed to confirm whether elevated δ13C and δ15N is connected to distinct lineages and how that may or may not have related to later interbreeding. Our current results suggest that diets of the first taurine herds were similar to ones associated with open grazing practices followed by modern local herders. More broadly, our results show the persistence of aurochs ancestry through time, even in taurine cattle from the Xiongnu and Turkic periods, which suggests widespread intentional interbreeding. This goes beyond what we might expect from opportunistic mating with wild bulls during the early stages of introduction or periods of stress (12). The isotopic results support the possibility of indigenous aurochs management in Mongolia by ~7800 cal BP, with genetic evidence for interbreeding between aurochs and taurine herds by ~2900 cal BP.
Management of herd animals in East Asia is often viewed as an external process where fully domesticated cattle, caprines, and horses were introduced to eastern Siberia, Mongolia, and China around the same time that the first pastoralist groups arrived from western Eurasia (19, 39–42). East Asia has not traditionally been recognized as a center for cattle domestication. Our results instead suggest that humans were likely already managing aurochs before the arrival of taurine cattle to Northeast Asia around 5000 years ago, after which they began intentionally interbreeding the two populations. The earliest taurine cattle introduced to East Asia are known to have been used in dairying (43), which highlights the importance of female taurine lineages. Male based introgression in cattle has been previously posited for Eurasia and Africa (9, 12, 17), but the unreliability of Y-chromosome data and a lack of genome-wide studies for ancient samples has obfuscated the evidence. Previous work has underscored the importance of introgression in environmental adaptation during adoption of domesticates (5, 12, 15). Based on the dominance of domesticated taurine mtDNA, the importance of dairying in this region, and the sustained nature of interbreeding in our sample, we hypothesize that in Mongolia this process is more accurately characterized as the introgression of female dairy cattle into indigenous herds in order to facilitate milk production. Attempts to improve fecundity and offspring survivorship might also have played a role. A greater emphasis on controlling female lineages for milk production would also explain the Eurasian expansion of haplogroup T3 ~7.5kyBP (12) after the emergence of dairying in the Near East (44).
Zooarchaeological and paleogenetic research increasingly shows that humans have managed animals in a variety of ways that do not always match the expectations of traditional models of animal domestication (45–48). Domestic animal populations often originate through multilocal processes, and admixture between wild and domestic populations was common for many taxa (5, 49–51). Our results show that these insights apply to cattle domestication in East Asia as well. Archaeological research focused on the presence and use of aurochs in and around habitation sites, as well as their individual life histories is ongoing and this work can add to our understanding of hunter-gatherer interactions with aurochs in Mongolia and also broader questions of aurochs management across East Asia.
The expansion of cattle pastoralism in East Asia intensified after about 4000 years ago. This coincided with broader changes in subsistence, ritual practices, and landscape use. Archaeological finds of dairy products (52) and identification of milk residues in pottery and human dental calculus suggests that milk was important to Bronze Age and later pastoralist peoples across the mountain, steppe, and desert regions of East Asia (43, 53, 54). Cattle bones were heavily utilized for oracle bone divination in ancient China by 4000 years ago (55), and cattle were also used in large numbers for subsistence, bone artifact production, and ritual sacrifice at early urban centers by 3000 years ago (56). Increasing reliance on cattle coincides with a decline in wild fauna such as deer, suggesting changes in human landscape use away from the mosaic habitats that facilitated interactions between humans and wild fauna (57). We do not know when during these transitions wild aurochs went extinct, but our results suggest that we should not ignore the possibility that people actively managed aurochs both before and long after the introduction of taurine cattle. The history of domesticated cattle in East Asia from initial domestication to the present remains enigmatic and further study of the archaeological and genetic history of ancient populations will be critical for understanding aurochs management and how they contributed to early cattle husbandry and modern breeds.
Supplementary Material
Acknowledgments:
We thank the Gobi-Steppe Neolithic Project, Mongolian National University of Education, Mongolian Academy of Sciences, Mongolian National University, University of Illinois, Chicago sequencing and research informatics cores, Rush University Medical Center Genomics and Microbiome Core Facility, Trent University, and the Wesleyan high powered computing cluster. Special thanks to Sohini Ramachandran and the Ramachandran lab, Emelia Huerta-Sanchez and the Huerta-Sanchez lab, and the Brown Center for Computational Molecular Biology for their support and insights during the research for this project. Part of this research was conducted using computational resources and services at the Center for Computation and Visualization, Brown University. We thank Guy Bennevat Haninovich for assistance compiling modern reference genomes, Nina Hirai for assistance producing the map in Figure 1, and Henk Meij for assistance with the Wesleyan HPCC, Canyang Ye for assistance researching bovine use in the Chinese-language literature, and Adiyasuren Molor for help with Mongolian-English translation.
Funding:
Social Sciences and Humanities Research Council of Canada (435-2020-0701) 20 We thank Wesleyan University for computer time supported by the NSF under grant number CNS-0619508 and CNS-0959856.
KW and DP were supported by NIH grant no. R35GM128946 and DP is also a trainee supported under the Brown University Predoctoral Training Program in Biological Data Science NIH T32 GM128596. 25
Bioinformatics analysis in the project described was performed by the UIC Research Informatics Core, supported in part by NCATS through Grant UL1TR002003.
Fieldwork conducted at Shiriin Chuluu by BD and AC was funded by grants from American Center for Mongolian Studies, the American Philosophical Society, and from Yale University funding from the Council on East Asian Studies, Yale Institute for Biospheric 30 Studies, and the Augusta Hazard Fund (Yale University).
Fieldwork conducted at Delgerkhaan Uul was funded by The Wenner Gren Foundation and U.S. National Endowment for the Humanities (RZ-249831-16).
Fieldwork conducted at Zaraa Uul by DO, LJ, BD was funded by SSHRC 430-2016-00173. Fieldwork conducted at Tamsagbulag was funded in 2018 by a National Geographic Society 35 Standard Grant (188R-18) and in 2021 by The Wenner Gren Foundation (9050).
Funding Statement
Social Sciences and Humanities Research Council of Canada (435-2020-0701) 20 We thank Wesleyan University for computer time supported by the NSF under grant number CNS-0619508 and CNS-0959856.
KW and DP were supported by NIH grant no. R35GM128946 and DP is also a trainee supported under the Brown University Predoctoral Training Program in Biological Data Science NIH T32 GM128596. 25
Bioinformatics analysis in the project described was performed by the UIC Research Informatics Core, supported in part by NCATS through Grant UL1TR002003.
Fieldwork conducted at Shiriin Chuluu by BD and AC was funded by grants from American Center for Mongolian Studies, the American Philosophical Society, and from Yale University funding from the Council on East Asian Studies, Yale Institute for Biospheric 30 Studies, and the Augusta Hazard Fund (Yale University).
Fieldwork conducted at Delgerkhaan Uul was funded by The Wenner Gren Foundation and U.S. National Endowment for the Humanities (RZ-249831-16).
Fieldwork conducted at Zaraa Uul by DO, LJ, BD was funded by SSHRC 430-2016-00173. Fieldwork conducted at Tamsagbulag was funded in 2018 by a National Geographic Society 35 Standard Grant (188R-18) and in 2021 by The Wenner Gren Foundation (9050).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All raw reads from the project are available on the NCBI SRA archive at [links will be added at time of publication]; code is available on Github [links will be added at time of publication]; MtDNA alignments are available at [links will be added at time of publication].
References and Notes
- 1.Chen S. et al. , Zebu Cattle Are an Exclusive Legacy of the South Asia Neolithic. Molecular Biology and Evolution 27, 1–6 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Grigson C., in Recent Advances in Indo-Pacific Prehistory, Misra V. N., Bellwood P., Eds. (Oxford and IBH, New Delhi, 1985), pp. 425–428. [Google Scholar]
- 3.Loftus R. T. et al. , Mitochondrial genetic variation in European, African and Indian cattle populations. Animal Genetics 25, 265–271 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Troy C. S. et al. , Genetic evidence for Near-Eastern origins of European cattle. Nature 410, 1088–1091 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Verdugo M. P. et al. , Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. Science 365, 173–176 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Brass M., Early North African Cattle Domestication and Its Ecological Setting: A Reassessment. Journal of World Prehistory 31, 81–115 (2018). [Google Scholar]
- 7.Linseele V. et al. , New Archaeozoological Data from the Fayum “Neolithic” with a Critical Assessment of the Evidence for Early Stock Keeping in Egypt. PLOS ONE 9, e108517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock F., Gifford-Gonzalez D., Genetics and African Cattle Domestication. African Archaeological Review 30, 51–72 (2013). [Google Scholar]
- 9.Ginja C. et al. , Iron age genomic data from Althiburos--Tunisia renew the debate on the origins of African taurine cattle. iScience 26, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achilli A. et al. , Mitochondrial genomes of extinct aurochs survive in domestic cattle. Current Biology 18, R157–R158 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Achilli A. et al. , The Multifaceted Origin of Taurine Cattle Reflected by the Mitochondrial Genome. PLOS ONE 4, e5753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubric-Curik V. et al. , Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evolutionary Applications 15, 663–678 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker J. E. et al. , Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle. PLOS Genetics 10, e1004254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Pardal L. et al. , Multiple paternal origins of domestic cattle revealed by Y-specific interspersed multilocus microsatellites. Heredity 105, 511–519 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Wu D.-D. et al. , Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nature Ecology & Evolution 2, 1139–1145 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Cai D. et al. , The origins of Chinese domestic cattle as revealed by ancient DNA analysis. Journal of Archaeological Science 41, 423–434 (2014). [Google Scholar]
- 17.Chen N. et al. , Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nature Communications 9, 2337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinding M.-H. S. et al. , Kouprey (Bos sauveli) genomes unveil polytomic origin of wild Asian Bos. iScience 24, 103226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honeychurch W. et al. , The earliest herders of East Asia: Examining Afanasievo entry to Central Mongolia. Archaeological Research in Asia 26, 100264 (2021). [Google Scholar]
- 20.Cai D. et al. , Ancient DNA reveals evidence of abundant aurochs (Bos primigenius) in Neolithic Northeast China. Journal of Archaeological Science 98, 72–80 (2018). [Google Scholar]
- 21.Brunson K. et al. , Zooarchaeology, ancient mtDNA, and radiocarbon dating provide new evidence for the emergence of domestic cattle and caprines in the Tao River Valley of Gansu Province, northwest China. Journal of Archaeological Science: Reports 31, 102262 (2020). [Google Scholar]
- 22.Brunson K., Lander B., Schneider M., in Cattle and People: Interdisciplinary Approaches to an Ancient Relationship, Wright E., Ginja C., Eds. (Lockwood Press, Atlanta, Georgia, 2022), pp. 277–297. [Google Scholar]
- 23.Janz L., Rosen A. M., Bukhchuluun D., Odsuren D., Zaraa Uul: An archaeological record of Pleistocene-Holocene palaeoecology in the Gobi Desert. PLOS ONE 16, e0249848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C., Janz L., Bukhchuluun D., Odsuren D., Neolithic pathways in East Asia: early sedentism on the Mongolian Plateau. Antiquity 95, 45–64 (2021). [Google Scholar]
- 25.Brunson K. et al. , New insights into the origins of oracle bone divination: Ancient DNA from Late Neolithic Chinese bovines. Journal of Archaeological Science 74, 35–44 (2016). [Google Scholar]
- 26.Liang Q. et al. , Subsistence strategies in the Late Neolithic and Bronze Age in Nenjiang River Basin: A zooarchaeological and stable isotope analysis of faunal remains at Honghe site, Northeast China. International Journal of Osteoarchaeology 33, 271–284 (2023). [Google Scholar]
- 27.Zhang H. et al. , Morphological and genetic evidence for early Holocene cattle management in northeastern China. Nature Communications 4, 2755 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Odsuren D., Janz L., Fox W., Bukhchuluun D., Otson Tsokhio and Zuun Shovkh: the Initial Upper Palaeolithic in Eastern Mongolia. Journal of Paleolithic Archaeology 6, 10 (2023). [Google Scholar]
- 29.Chen N. et al. , Ancient genomes reveal tropical bovid species in the Tibetan Plateau contributed to the prevalence of hunting game until the late Neolithic. Proceedings of the National Academy of Sciences 117, 28150–28159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu S., Yang M., Sun Z., & Shao J., Research on faunal remains from the Shimao site in Shenmu, Shaanxi Province (based on 2012–2013 excavations) 2012-2013 年度陕西神木石峁遗 址出土动物遗存研究. Archaeology and Cultural Relics 考古与文物 4, 109–121 (2016). [Google Scholar]
- 31.Owlett T. E., Hu S., Sun Z., Shao J., Food between the country and the city: The politics of food production at Shimao and Zhaimaoliang in the Ordos Region, northern China. Archaeological Research in Asia 14, 46–60 (2018). [Google Scholar]
- 32.Brunson K., He N., Dai X., Sheep, Cattle, and Specialization: New Zooarchaeological Perspectives on the Taosi Longshan. International Journal of Osteoarchaeology 26, 460–475 (2016). [Google Scholar]
- 33.Heath-Agnew E., A History of Hereford Cattle and Their Breeders. (Duckworth, London, 1983). [Google Scholar]
- 34.Noda A., Yonesaka R., Sasazaki S., Mannen H., The mtDNA haplogroup P of modern Asian cattle: A genetic legacy of Asian aurochs? PLOS ONE 13, e0190937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonfiglio S. et al. , Origin and Spread of Bos taurus: New Clues from Mitochondrial Genomes Belonging to Haplogroup T1. PLOS ONE 7, e38601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannen H. et al. , Independent mitochondrial origin and historical genetic differentiation in North Eastern Asian cattle. Molecular Phylogenetics and Evolution 32, 539–544 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Tang Z., Recognition of Historical and Geographical Distribution of Bos Primigenius in Northern China 中国北方原始牛历史地理分布的再认识. Agricultural Archaeology 农业考古 3, 7–12 (2020). [Google Scholar]
- 38.Makarewicz C., Tuross N., Foddering by Mongolian pastoralists is recorded in the stable carbon (δ13C) and nitrogen (δ15N) isotopes of caprine dentinal collagen. Journal of Archaeological Science 33, 862–870 (2006). [Google Scholar]
- 39.Jeong C. et al. , A Dynamic 6,000-Year Genetic History of Eurasia’s Eastern Steppe. Cell 183, 890–904.e829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motuzaite Matuzeviciute G. et al. , Climatic or dietary change? Stable isotope analysis of Neolithic–Bronze Age populations from the Upper Ob and Tobol River basins. The Holocene 26, 1711–1721 (2016). [Google Scholar]
- 41.Taylor W. T. T. et al. , Early Pastoral Economies and Herding Transitions in Eastern Eurasia. Scientific Reports 10, 1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsybiktarov A. D., Genesis of Nomadic Cattle Breeding in Mongolia and Southern Trans-Baikal Territory: Prerequisites and Reasons. Journal of Siberian Federal University Humanities & Social Sciences 14, 148–162 (2021). [Google Scholar]
- 43.Wilkin S. et al. , Dairy pastoralism sustained eastern Eurasian steppe populations for 5,000 years. Nature Ecology & Evolution 4, 346–355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evershed R. P. et al. , Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature 455, 528–531 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Frantz L. A. F., Bradley D. G., Larson G., Orlando L., Animal domestication in the era of ancient genomics. Nature Reviews Genetics 21, 449–460 (2020). [DOI] [PubMed] [Google Scholar]
- 46.MacHugh D. E., Larson G., Orlando L., Taming the Past: Ancient DNA and the Study of Animal Domestication. Annual Review of Animal Biosciences 5, 329–351 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Marshall F. B., Dobney K., Denham T., Capriles J. M., Evaluating the roles of directed breeding and gene flow in animal domestication. Proceedings of the National Academy of Sciences 111, 6153–6158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeder M. A., The Domestication of Animals. Journal of Anthropological Research 68, 161–190 (2012). [Google Scholar]
- 49.Daly K. G. et al. , Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science 361, 85 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Frantz L. A. F. et al. , Ancient pigs reveal a near-complete genomic turnover following their introduction to Europe. Proceedings of the National Academy of Sciences 116, 17231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlando L., in Paleogenomics: Genome-Scale Analysis of Ancient DNA, Lindqvist C., Rajora O. P., Eds. (Springer International Publishing, Cham, 2018), pp. 325–351. [Google Scholar]
- 52.Yang Y. et al. , Proteomics evidence for kefir dairy in Early Bronze Age China. Journal of Archaeological Science 45, 178–186 (2014). [Google Scholar]
- 53.Janz L., Cameron A., Bukhchuluun D., Odsuren D., Dubreuil L., Expanding frontier and building the Sphere in arid East Asia. Quaternary International 559, 150–164 (2020). [Google Scholar]
- 54.Xie M. et al. , Identification of a dairy product in the grass woven basket from Gumugou Cemetery (3800 BP, northwestern China). Quaternary International 426, 158–165 (2016). [Google Scholar]
- 55.Flad R. K., Divination and Power: A Multiregional View of the Development of Oracle Bone Divination in Early China. Current Anthropology 49, 403–437 (2008). [Google Scholar]
- 56.Campbell R. B., Li Z., He Y., Jing Y., Consumption, exchange and production at the Great Settlement Shang: bone-working at Tiesanlu, Anyang. Antiquity 85, 1279–1297 (2011). [Google Scholar]
- 57.Lander B., Brunson K., Wild Mammals of Ancient North China. Journal of Chinese History 2, 291–312 (2018). [Google Scholar]
- 58.Pleuger S. et al. , What’s in a Hearth?: Preliminary Findings from the Margal Huntergatherer Habitation in the Eastern Mongolian Gobi Desert. Asian Perspectives 62, (2023). [Google Scholar]
- 59.Tumen D., Khatanbaatar D., Erdene M., Bronze Age graves in the Delgerkhaan Mountain area of eastern Mongolia. Asian Archaeology 2, 40–49 (2014). [Google Scholar]
- 60.Wright J., Ganbaatar G., Honeychurch W., Byambatseren B., Rosen A., The earliest Bronze Age culture of the south-eastern Gobi Desert, Mongolia. Antiquity 93, 393–411 (2019). [Google Scholar]
- 61.Derevianko A. P., Dorj D., in History of civilization of Central Asia, Volume 1: The dawn of civilization, earliest times to 700 BC, Dani A. H., Masson V. M., Eds. (UNESCO Publishing, Paris, 1992), vol. 1, pp. 169–189. [Google Scholar]
- 62.Dorj D., Neolit voctochnoy Mongolii [Neolithic of Eastern Mongolia]. (MASIHA, Ulaanbaatar, 1971). [Google Scholar]
- 63.Odsuren D., Bukhchuluun D., Janz L., Preliminary results of Neolithic research conducted in eastern Mongolia. Studia Archaeologica 35, 72–96 (2015). [Google Scholar]
- 64.Beaumont W., Beverly R., Southon J., Taylor R. E., Bone preparation at the KCCAMS laboratory. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 268, 906–909 (2010). [Google Scholar]
- 65.Crann C. A. et al. , First Status Report on Radiocarbon Sample Preparation Techniques at the A.E. Lalonde AMS Laboratory (Ottawa, Canada). Radiocarbon 59, 695–704 (2017). [Google Scholar]
- 66.Reimer P. J., Brown T. A., Reimer R. W., Discussion: Reporting and Calibration of Post-Bomb 14C Data. Radiocarbon 46, 1299–1304 (2004). [Google Scholar]
- 67.Stuiver M., Polach H. A., Discussion Reporting of 14C Data. Radiocarbon 19, 355–363 (1977). [Google Scholar]
- 68.Bronk Ramsey C., Bayesian Analysis of Radiocarbon Dates. Radiocarbon 51, 337–360 (2009). [Google Scholar]
- 69.Reimer P. J. et al. , The IntCal20 Northern Hemisphere Radiocarbon Age Calibration Curve (0–55 cal kBP). Radiocarbon 62, 725–757 (2020). [Google Scholar]
- 70.Szpak P. et al. , Regional differences in bone collagen δ13C and δ15N of Pleistocene mammoths: Implications for paleoecology of the mammoth steppe. Palaeogeography, Palaeoclimatology, Palaeoecology 286, 88–96 (2010). [Google Scholar]
- 71.Dabney J., Meyer M., in Ancient DNA: Methods and Protocols, Shapiro B. et al. , Eds. (Springer; New York, New York, NY, 2019), pp. 25–29. [Google Scholar]
- 72.Rogers L. L., Kaestle F. A., Analysis of mitochondrial DNA haplogroup frequencies in the population of the slab burial mortuary culture of Mongolia (ca. 1100–300 BCE). American Journal of Biological Anthropology 177, 644–657 (2022). [Google Scholar]
- 73.Schubert M., Lindgreen S., Orlando L., AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Research Notes 9, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN]. 2013. [Google Scholar]
- 75.Li H. et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neukamm J., Peltzer A., Nieselt K., DamageProfiler: fast damage pattern calculation for ancient DNA. Bioinformatics 37, 3652–3653 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao L. et al. , Population genetics of wild Macaca fascicularis with low-coverage shotgun sequencing of museum specimens. American Journal of Physical Anthropology 173, 21–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stamatakis A., RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patterson N., Price A. L., Reich D., Population Structure and Eigenanalysis. PLOS Genetics 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for Ancient Admixture between Closely Related Populations. Molecular Biology and Evolution 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pickrell J., Pritchard J., Inference of population splits and mixtures from genome-wide allele frequency data. Nature Precedings, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tamura K., Stecher G., Kumar S., MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edwards C. J. et al. , Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proceedings of the Royal Society B: Biological Sciences 274, 1377–1385 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leigh J., Bryant D., PopART: Full-feature software for haplotype network construction. Methods Ecol Evol 6, 1110–1116 (2015). [Google Scholar]
- 87.Stothard P. et al. , A large and diverse collection of bovine genome sequences from the Canadian Cattle Genome Project. GigaScience 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heaton M. et al. , Using diverse U.S. beef cattle genomes to identify missense mutations in EPAS1, a gene associated with high-altitude pulmonary hypertension [version 1; peer review: 2 approved]. F1000Research 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whitacre L. K. et al. , Elucidating the genetic basis of an oligogenic birth defect using whole genome sequence data in a non-model organism, Bubalus bubalis. Scientific Reports 7, 39719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mei C. et al. , Whole-genome sequencing of the endangered bovine species Gayal (Bos frontalis) provides new insights into its genetic features. Scientific Reports 6, 19787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin D.-H. et al. , Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genomics 15, 240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawahara-Miki R. et al. , Whole-genome resequencing shows numerous genes with nonsynonymous SNPs in the Japanese native cattle Kuchinoshima-Ushi. BMC Genomics 12, 103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuda K. et al. , Abundant sequence divergence in the native Japanese cattle Mishima-Ushi (Bos taurus) detected using whole-genome sequencing. Genomics 102, 372–378 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Bickhart D. M. et al. , Diversity and population-genetic properties of copy number variations and multicopy genes in cattle. DNA Research 23, 253–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gautier M. et al. , Deciphering the Wisent Demographic and Adaptive Histories from Individual Whole-Genome Sequences. Molecular Biology and Evolution 33, 2801–2814 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang K. et al. , The genome sequence of the wisent (Bison bonasus). GigaScience 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qiu Q. et al. , Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nature Communications 6, 10283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park S. D. E. et al. , Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biology 16, 234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Q. et al. , Whole genome analyses revealed genomic difference between European taurine and East Asian taurine. Journal of Animal Breeding and Genetics 138, 56–68 (2021). [DOI] [PubMed] [Google Scholar]
- 100.Edwards C. J. et al. , A Complete Mitochondrial Genome Sequence from a Mesolithic Wild Aurochs (Bos primigenius). PLOS ONE 5, e9255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeyland J. et al. , Complete mitochondrial genome of wild aurochs (Bos primigenius) reconstructed from ancient DNA. Polish Journal of Veternary Sciences 16, 265–273 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Zeyland J. et al. , Tracking of wisent–bison–yak mitochondrial evolution. Journal of Applied Genetics 53, 317–322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bonfiglio S. et al. , The Enigmatic Origin of Bovine mtDNA Haplogroup R: Sporadic Interbreeding or an Independent Event of Bos primigenius Domestication in Italy? PLOS ONE 5, e15760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Horsburgh K. A. et al. , The Genetic Diversity of the Nguni Breed of African Cattle (Bos spp.): Complete Mitochondrial Genomes of Haplogroup T1. PLOS ONE 8, e71956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xia X.-T. et al. , Mitochondrial genomes from modern and ancient Turano-Mongolian cattle reveal an ancient diversity of taurine maternal lineages in East Asia. Heredity 126, 1000–1008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hiendleder S., Lewalski H., Janke A., Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy and domestication. Cytogenetic and Genome Research 120, 150–156 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Anderson S. et al. , Complete sequence of bovine mitochondrial DNA conserved features of the mammalian mitochondrial genome. Journal of Molecular Biology 156, 683–717 (1982). [DOI] [PubMed] [Google Scholar]
- 108.Mannen H., Morimoto M., Oyama K., Mukai F., Tsuji S., Identification of mitochondrial DNA substitutions related to meat quality in Japanese Black cattle1,2. Journal of Animal Science 81, 68–73 (2003). [DOI] [PubMed] [Google Scholar]
- 109.Hassanin A. et al. , Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. Comptes Rendus Biologies 335, 32–50 (2012). [DOI] [PubMed] [Google Scholar]
- 110.Na R.-S. et al. , Complete mitochondrial genome of the Yakow (Bos primigenius taurus × Bos grunniens) in China. Mitochondrial DNA Part A 27, 3826–3827 (2016). [DOI] [PubMed] [Google Scholar]
- 111.Gu Z., Zhao X., Li N., Wu C., Complete sequence of the yak (Bos grunniens) mitochondrial genome and its evolutionary relationship with other ruminants. Molecular Phylogenetics and Evolution 42, 248–255 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Marsolier-Kergoat M.-C. et al. , Hunting the Extinct Steppe Bison (Bison priscus) Mitochondrial Genome in the Trois-Frères Paleolithic Painted Cave. PLOS ONE 10, e0128267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw reads from the project are available on the NCBI SRA archive at [links will be added at time of publication]; code is available on Github [links will be added at time of publication]; MtDNA alignments are available at [links will be added at time of publication].