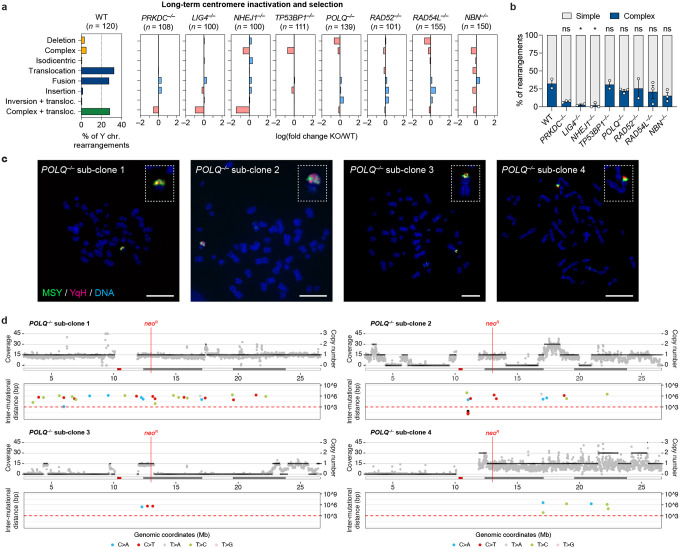
Figure 2. DSB repair pathways beyond NHEJ are dispensable for generating complex rearrangements from micronuclei.
a) Left: Distribution of Y chromosome rearrangement types as determined by metaphase FISH following continuous passage in DOX/IAA and G418 for ~30 days. Data are pooled from 2 independent experiments. Right: Plots depict the mean fold change in each rearrangement type as compared to WT cells. Sample sizes indicate the number of rearranged Y chromosomes examined; data are pooled from 2 or 3 individual KO clones per gene. See also Extended Data Figure 3c. b) Proportion of Y chromosomes exhibiting simple or complex rearrangements, as determined by metaphase FISH, following sustained centromere inactivation. Data represent the mean ± SEM of n = 2 independent experiments for WT, n = 2 KO clones for LIG4 and RAD52, and n = 3 KO clones for PRKDC, NHEJ1, TP53BP1, POLQ, RAD54L, and NBN; statistical analyses were calculated by ordinary one-way ANOVA test with multiple comparisons. ns, not significant; *P ≤ 0.05. c) Cytogenetic characterization of POLQ KO sub-clones harboring complex Y chromosome rearrangements following sustained centromere inactivation. Scale bar, 10 μm. d) Whole-genome sequencing analyses of POLQ KO sub-clones with complex Y chromosome rearrangements exhibiting oscillating DNA copy-number patterns. For each subclone, sequencing depth (grey dots), copy number information (black lines; top), and inter-mutational distances (bottom) for the mappable regions of the Y chromosome are shown.