Abstract
Directed evolution methods based on high-throughput growth selection enable efficient discovery of enzymes with improved function in vivo. High-throughput selection is particularly useful when engineering oxygenases, which are sensitive to structural perturbations and prone to uncoupled activity. In this work, we combine the principle that reactive oxygen species (ROS) produced by uncoupled oxygenase activity are detrimental to cell fitness with a redox balance-based growth selection method for oxygenase engineering that enables concurrent advancement in catalytic activity and coupling efficiency. As a proof-of-concept, we engineered P450-BM3 for degradation of acenaphthene (ACN), a recalcitrant environmental pollutant. Selection of site-saturation mutagenesis libraries in E. coli strain MX203 identified P450-BM3 variants GVQ-AL and GVQ-D222N, which have both improved coupling efficiency and catalytic activity compared to the starting variant. Computational modeling indicates that the discovered mutations cooperatively optimize binding pocket shape complementarity to ACN, and shift the protein’s conformational dynamics to favor the lid-closed, catalytically competent state. We further demonstrated that the selective pressure on coupling efficiency can be tuned by modulating cellular ROS defense mechanisms.
Keywords: growth selection, high-throughput, P450-BM3, directed evolution, coupling, oxidative stress
Graphical Abstract

Biooxygenation provides a viable alternative to traditional means of synthetic chemistry for the selective activation of C–H bonds. Members of the diverse oxygenase families such as two-component flavin hydroxylases,1,2 Cytochrome P450s,3 and Baeyer–Villiger monooxygenases (BVMOs)4 show considerable potential as industrial catalysts. Directed evolution has been widely exploited to tailor these oxygenases for desired reactions; however, their full potential is limited by the relatively low throughput of downstream selection technologies. To address this issue, advancements in designing efficient libraries with smaller theoretical sizes5 and ultrahigh-throughput screening methods utilizing microfluidic devices and fluorescence sorting6–9 have been developed. Although these processes have facilitated the successful directed evolution of a number of enzymes, there is a need for more general and accessible methods that do not require specialized reagents or expensive equipment.
Recently, we reported a growth-based, high-throughput selection platform for engineering NADPH-dependent oxygenases.10 We used this selection platform to rapidly remodel the Pseudomonas aeruginosa 4-hydroxybenzoate hydroxylase (PobA) active site to efficiently accept a non-native substrate 3,4-dihydroxybenzoic acid (3,4-DHBA). This selection platform, in E. coli strain MX203, was constructed based on the principle of cofactor redox balance (Figure 1A,B), where growth of the metabolically engineered E. coli cells is inhibited because NADPH overaccumulates and cannot be recycled back to NADP+. Rescue of cell growth requires expression of an NADPH-consuming enzyme capable of utilizing the desired substrate supplied in the culture media. The same redox balance principle has served as the basis for several other growth-based selection platforms.11–15
Figure 1.
Growth selection for coupled and uncoupled NADPH oxidation. The selection principle is founded on growth inhibition via incomplete native NADPH cycling and the toxicity of reactive oxygen byproducts. (A) In the selection strain MX203, excessive NADPH is accumulated via redirection of central metabolism through the pentose phosphate (PP) pathway and disruption of native oxidation routes, resulting in inhibited growth. (B) NADPH cycling via coupled BM3 activity restores NADP(H) balance and rescues growth in the selection strain. (C) Formation of reactive oxygen species (ROS) from uncoupled BM3 activity inhibits growth. Application of strain MX203 can be expanded to select for improved coupling efficiency.
Growth-based selection methods are high-throughput (>106 candidates per round) and use growth as a facile readout. Importantly, they directly yield enzymes that are active in vivo. However, the selection platforms currently do not account for the unique engineering requirements of oxygenases. Oxygenases execute a complex orchestration of reaction mechanisms to couple NAD(P)H consumption to substrate conversion. However, this coupling is often disrupted in engineered oxygenases, resulting in futile NAD(P)H consumption to reduce O2 in lieu of substrate conversion16,17 (Figure 1C). The frequency of the completed reaction cycle can be described by the product formed relative to the NADPH consumed and is reported as the coupling efficiency. In existing platforms, it is unclear whether the growth-based selection methods can accommodate the need for improving coupling efficiency.
Cytochrome P450s are a promiscuous class of heme-containing monooxygenases that are able to incorporate molecular oxygen with high regio- and stereoselectivity onto inert C–H bonds. Expanding the hydroxylation activity of reductase-fused P450s, both natural and synthetic, has been the goal of many engineering efforts oriented toward synthesizing pharmaceutically relevant natural products,18,19 generating metabolized pharmaceutical intermediates,20 and designing bioremediation applications.21,22 In particular, the naturally chimeric, NADPH-dependent P450 BM3 is prolific as an engineering target because it is self-sufficient, readily soluble, and possesses the highest turnover rate of any known P450.23 Extensive studies aimed at engineering BM3 to modify substrate scope beyond its native fatty acid preference have faced a reoccurring challenge to control electron coupling of the catalytic cycle.24 During the reaction cycle, the enzyme is activated by the transfer of electrons from NAD(P)H through a FAD and subsequent FMN coenzyme on the reductase domain. Following this initial transfer, electrons are mediated through a series of residues serving as electron transport pathways to the heme center of the P450 domain. Upon activation, the reactive heme is able to bind oxygen and generate an iron–oxygen complex that proceeds through a series of reductive steps. When disrupted through engineering, the precise electron transfer dynamics can be shifted out of tune with substrate binding, and the activated oxygen can decay into reactive oxygen species (ROS).
Despite their broad substrate scope, engineered BM3 variants with non-native substrates frequently demonstrate extremely low coupling between NADPH consumption and product formation.23 BM3 engineering strategies frequently target bulky active site residues close to the internal heme such as F87 for mutagenesis to smaller residues to allow substrate access to the reactive center. Although this approach is effective, these preliminary mutants do not always provide the expected promiscuity,25 and frequently display low coupling with the desired non-native substrates ranging from 1 to 14% for panels of bulky polyaromatic hydrocarbons,26 and 35% for the artemisinin precursor amorphadiene.18 Subsequent engineering typically aims to reshape the binding pocket with targeted mutations to establish improved coupling through complementary steric packing. Although matching the wild-type coupling efficiency of >80% is ideal, the coupling efficiency required will vary significantly with the intended application. BM3 variants with 50–63% coupling for amorphadiene were regarded as suitable for fermentation application, but this level of coupling for in vitro conversion significantly increases the need for feedstock material and reduces the economic efficiency of biocatalysis.
Because excessive uncoupling wastes reducing power and produces reactive oxygen species (ROS) that can inactivate the enzyme, it is essential to pursue both high activity and high coupling efficiency simultaneously in oxygenase engineering. Although redox balance-based growth selection platforms are responsive to NAD(P)H consumption, their ability to distinguish coupled versus uncoupled activity of oxygenases remains uncharacterized. In this report, NADPH-dependent growth selection strain MX20310 demonstrated responsiveness to uncoupling, likely because ROS produced during uncoupled reactions are detrimental to cell fitness. As a proof-of-concept, we simultaneously improved both the activity and the coupling efficiency of an engineered P450-BM3 variant for an unnatural substrate, acenaphthene (ACN). The discovered variant may be applied in environmental remediation to facilitate the degradation of this persistent pollutant.22,26 Importantly, computational modeling indicates that we obtained synergistic mutations reshaping the substrate binding pocket and identified mutations distal to the active site shifting the global conformational dynamics of P450-BM3. This highlights the power of high-throughput selection methods for directed evolution in discovering mutations that optimize multiple criteria in concert. Furthermore, we showed that the selection pressure on coupling efficiency can be tuned by modulating the cell’s sensitivity to ROS, through disruption of various ROS defense mechanisms.
This work augments the scope of redox balance-based growth selection methods and highlights their application for improving the coupling efficiency of oxygenase enzymes. We envision that this approach is not limited to the directed evolution of NADPH-dependent enzymes and will inform future selection methods.
RESULTS AND DISCUSSION
Growth-Based Selection Platform Is Responsive to Coupling Efficiency of Oxygenase.
An aerobic NADPH accumulation strain MX203 (Figure 1A, Table S1) has been developed by channeling glucose metabolism through the pentose phosphate pathway and eliminating native NADPH sinks.27 The selection platform utilizes the principle of cofactor redox balance, where NADPH accumulates in engineered E. coli cells to a toxic level. The cell growth is restored by expressing a heterologous oxygenase mutant with the desired activity13,27 to consume NADPH and alleviate imbalance (Figure 1B). This general selection criteria poses a limitation on engineering monooxygenases with multistep reaction mechanisms with particularly unstable intermediates. In these reactions NADPH consumption does not always warrant substrate conversion and engineered variants can tend toward incomplete reaction cycles with non-native substrates.23
Although the reliance on NADPH consumption for growth can potentially lead to selection of highly decoupled variants, these variants are likely to produce an excess of ROS products. Toxic ROS can initiate reduction of cell viability due to DNA, lipid, and protein damage.22 Whether cell growth can provide a sensitive readout for the coupling efficiency of a heterologously expressed oxygenase has yet to be determined.
To examine the potential correlation between growth and coupling efficiency, we employed a panel of P450-BM3 variants previously reported to exhibit a diverse range of coupling efficiencies in the hydroxylation of lauric acid, their native substrate (Figure 2). During coupled reactions, lauric acid is hydroxylated at the expense of NADPH. During uncoupled reactions, NADPH is still consumed, but the substrate remains unmodified while hydrogen peroxide (H2O2), superoxide (O2−), or water (H2O) are produced (Figure 2A). When three P450-BM3 variants were introduced to the previously developed NADPH-dependent growth selection strain, MX203 (Figure 1A, Table S1),10 we observed correlation between coupling efficiency and growth restoration capacities (Figure 2B): the strain harboring the wild-type (WT) enzyme (75% coupling efficiency,28 as defined by the moles of hydroxylated lauric acid formed over the moles of NADPH consumed) grew the fastest, followed by F87V (50% coupling efficiency28). The poorly coupled R47A-Y51A-F87V (6% coupling efficiency28) did not rescue growth. Notably, all three P450-BM3 enzymes have high and comparable activities in consuming NADPH (Figure 2C), which discounts the likelihood that the poor growth associated with F87V and R47A-Y51A-F87V was solely due to insufficient restoration of NADPH redox balance.
Figure 2.
Uncoupled NADPH-oxidation inhibits growth restoration in NADPH-selection strain. (A) Coupled BM3 reaction with lauric acid results in the hydroxylation of either the ω−1, ω−2, or ω−3 position of the native fatty acid substrate. Uncoupled NADPH oxidation can result in a panel of oxygen species detrimental to cell health and protein integrity. (B) In M9 minimal glucose media with the addition of 0.2 g/L lauric acid, growth restoration of the selection strain was achieved by heterologous expression of BM3 WT (green) and BM3 F87V (yellow), but not BM3 R47A-Y51A-F87V (red). The selection strain demonstrated apparent sensitivity to reported coupling efficiencies of the BM3 panel. (C) Total in vitro specific activity of three BM3 variants in the presence of lauric acid. All variants exhibited significant NADPH oxidation rates.
In order to further examine the role of oxidative stress, we sought to relieve this burden by supplementing pyruvate as an antioxidant to the same panel of variants. Pyruvate, as well as other endogenous α-keto acids, is known to nonenzymatically consume H2O2 and protect against oxidative stress.29 With the addition of 50 mg/L pyruvate, we observed that growth with the BM3 mutants was enhanced (Figure S1A). Growth with F87V achieved a similar profile as the WT BM3, while the R47A-Y51A-F87V growth was still reduced in comparison. Interestingly, with the addition of an H2O2 antioxidant to the selection media, the growth trend demonstrated by the panel of variants more closely matched the relative levels of NADPH oxidation activity in contrast to the coupling efficiency. Although pyruvate can serve as an alternative carbon source, minimal supplementation did not restore MX203 growth without expression of a heterologous NADPH-consuming enzyme (Figure S1B). These results highlight the combined effect of both NADPH activity and oxidative stress in modulating selection strain growth.
Selection strain MX203 has already been shown to be responsive to the target enzyme’s NADPH-consuming activity;10 therefore, the correlations between growth and coupling efficiency support its expanded application in selecting more coupled oxygenases.
Improving P450 BM3 for Acenaphthene Hydroxylation Using Growth-Based Selection.
As a proof-of-concept, we sought to apply strain MX203 (Figure 1A, Table S1) for the high-throughput selection of P450-BM3 variants with both higher activity and improved coupling efficiency for a non-native substrate. Unlike the wild type enzyme with native fatty acid substrates, P450-BM3 variants engineered to take non-native substrates often suffer from low coupling efficiencies.18,26,30 We chose to improve P450-BM3 variant GVQ (A74G-F87V-L188Q) toward acenaphthene (ACN), which typifies the discrepancy of high activity (previously reported as 2020 nmol min−1 nmol−1) and low coupling (5.42%).21 P450 BM3-mediated hydroxylation of ACN to 1-acenapthenol (Figure 3A) would greatly increase the solubility and biodegradability of this harmful fuel-based pollutant.31,32
Figure 3.
Identification of P450-BM3 variants with improved coupling efficiency and activity with acenaphthene in growth selection. (A) Coupled BM3 reaction with the non-native substrate acenaphthene results in the hydroxylated product 1-acenaphthenol. (B) In M9 minimal glucose media with the addition of 0.2 g/L acenaphthene (ACN), growth restoration of selection strain is achieved by heterologous expression of selected BM3 variants with improved coupling, GVQ-AL (orange) and GVQ-D222N (green). (C) GVQ-AL and GVQ-D222N displayed increased coupling efficiency in the formation of 1-acenaphthenol with NADPH compared to GVQ (teal). (D) Total in vitro NADPH oxidation rate of the three BM3 variants with ACN. Selected variants GVQ-AL and GVQ-D222N displayed increased NADPH activity over GVQ. Despite GVQ’s high total NADPH oxidation rate, uncoupled activity with acenaphthene likely inhibits growth restoration of the selection strain.
Among the diverse P450-BM3 engineering strategies, we were interested in investigating two contrasting approaches: substrate active site (SAS) engineering (by targeting residues V78 and A328) and electron transport (ET) pathway modulation (by targeting residues F393 and M490).33 Although the former is a more established approach to accommodate non-native substrates,18,22,24 the throughput of our selection platform enables rapid parallel library screening to investigate more nontraditional approaches that hold promise as more general strategies for tuning reactivity.
We constructed two libraries by simultaneously randomizing each residue pair using NNK degenerate codons. The resulting SAS and ET libraries (pLS208 and pLS219, respectively, Table S1) were individually transformed into selection strain MX203, yielding ∼8.4 × 106 and 2.3 × 106 independent transformants, respectively (see Methods in Supporting Information), which was sufficient to cover the theoretical library sizes of 202 = 400. Selection of the SAS library was performed in 10 independent liquid cultures with 2 g/L D-glucose in M9 minimal medium and 0.2 g/L acenaphthene, at 30 °C for ∼72 h. Library selection was monitored by simple OD measurements. As the cultures approached mid log phase, small volumes of selection cultures were plated on rich nonselection media to isolate potential variants. ET library selection was performed on agar plates with the same media composition, at 30 °C for ∼72 h. Library selection was monitored by observing colony growth. MX203 cells harboring P450 BM3-GVQ served as a negative control for both selections. While the starting template GVQ cannot rescue growth, the best variants obtained from each library (described below) supported robust growth of MX203 (Figure 3B).
Characterization of P450 BM3 Variants Obtained from Selection.
From the SAS library we picked one colony for each liquid culture that grew during selection. Sequencing of the 10 independent candidates revealed 6 unique residue combinations (Table S2). Repeated residue pairs found within the 10 candidates suggest that the sites chosen may be subjected to strong evolutionary pressure during selection.
We picked three candidates to further characterize because their growth restoration capability was most significantly dependent on the presence of ACN (Figure S2). Of the three variants (GVQ-AL, GVQ-SL, and GVQ-SFC, Table S1), the former two demonstrated improved coupling efficiency, with the best variant, GVQ-AL, exhibiting 3-fold higher coupling efficiency compared to GVQ toward ACN (Figure 3C).
Several variants obtained from the selection (GVQ-FF, GVQ-SF, and GVQ-AF, Table S2) demonstrated growth even without ACN (Figure S2), which suggests substrate binding site modification may have improved their activity and coupling efficiency for native metabolites present in vivo, such as fatty acids. Further biochemical characterization is needed to prove this hypothesis. Previous efforts to minimize off-target oxidation by BM3 variants in vivo have successfully employed fatty acid blocking mutations at the substrate entrance channel, R47L and Y51F.18 These residues are known to interact with the fatty acid carboxyl group and are associated with stabilizing these substrates in the binding pocket during catalysis.28
From the ET library, three colonies were chosen for further sequencing and characterization. Remarkably, wild type residues were found in all variants at the targeted sites (F393 and M490), indicating that there was strong selection pressure on these sites against any mutations. We hypothesized that although modulating these sites may improve coupling efficiency as previously reported,33 the enzyme’s overall activity might also be severely impacted given the essential roles these residues play in catalysis.
Interestingly, two of the variants obtained (GVQ-D222N and GVQ-K224N) contained spontaneous mutations outside the targeted sites which led to improved coupling efficiency. The best variant, GVQ-D222N, exhibited a 2-fold higher coupling efficiency compared to GVQ toward ACN (Figure 3C). The spontaneous mutations may have been introduced due to imperfect DNA replication fidelity during library construction in vitro or growth selection in vivo. The fact that we obtained these rare mutations during selection highlights the advantage of having a very high throughput.
The best variants obtained from both libraries, GVQ-AL and GVQ-D222N, also demonstrated enhanced total activity (as measured by NADPH consumption rate) by 1.8- and 2.5-fold, respectively, compared to the starting variant GVQ (Figure 3D). Taken together, these results suggest that the growth-based selection platform is effective in simultaneously improving oxygenase’s activity and coupling efficiency in combination. Because growth of this selection strain is not directly tied to a particular essential enzyme or the conversion of a specific product, we anticipate this platform will be broadly applicable to the selection of other NADPH-dependent oxygenases with a variety of non-native substrates.
Investigating the Mechanism of Mutations Using Computational Modeling.
We hypothesized that P450-BM3 GVQ-AL (A74G-F87V-L188Q-V78A-A328L), the best variant obtained from substrate active site (SAS) engineering library, has improved active site shape complementarity to ACN. To investigate this hypothesis, we performed computational docking with Rosetta to model the substrate binding pose (Figure 4).
Figure 4.
Docked models of the ACN binding poses. (A) In the WT, steric clash from the bulky F87 and V78 block the ACN from positioning the catalytic carbon over the heme oxygen. (B) In GVQ, the mutation F87V results in greater volume for the ligand to maneuver over the heme oxygen, but the extension of V78 prevents the ACN left surface from reaching optimal hydrophobic packing against F87V. (C) GVQ-AL displays improved binding pocket shape complementarity for ACN binding, F87V and V78A reduce steric hindrance, while A328L provides increased nonpolar surface area to face the ACN rings.
Docking of ACN in the wild type P450-BM3 results in an unproductive binding pose, with the ACN catalytic carbons positioned too far (>3.0 Å) from the heme oxygen for catalysis (Figure 5A). In P450-BM3 GVQ (A74G-F87V-L188Q), which was the starting point of engineering, the F87V mutation partially relieves steric hindrance, but the hydrophobic packing against ACN is not optimal (Figure 4B). By targeting V78 and A328 for saturation mutagenesis, the active site is further contoured to narrowly enclose ACN and limit unfavorable solvent interactions or excessive ligand mobility, while creating headspace for the ligand to readily maneuver the catalytic carbon over the heme oxygen (Figure 4C). V78A clears vertical space to accommodate ACN and allows F87V to adopt a different rotamer state compared to in the GVQ model, which creates even more volume. On the other side, A328L, with its increased bulk, packs tightly against the face of ACN. Since P450-BM3 GVQ-D222N (A74G-F87V-L188Q-D222N), the best variant obtained from electron transport (ET) pathway modulation library, has key mutations distal from the active site, we hypothesized that their beneficial effect arises from altering the enzyme conformational dynamics to more frequently sample catalytically productive states. To evaluate this hypothesis, we performed molecular dynamics (MD) simulation (Figure 5).
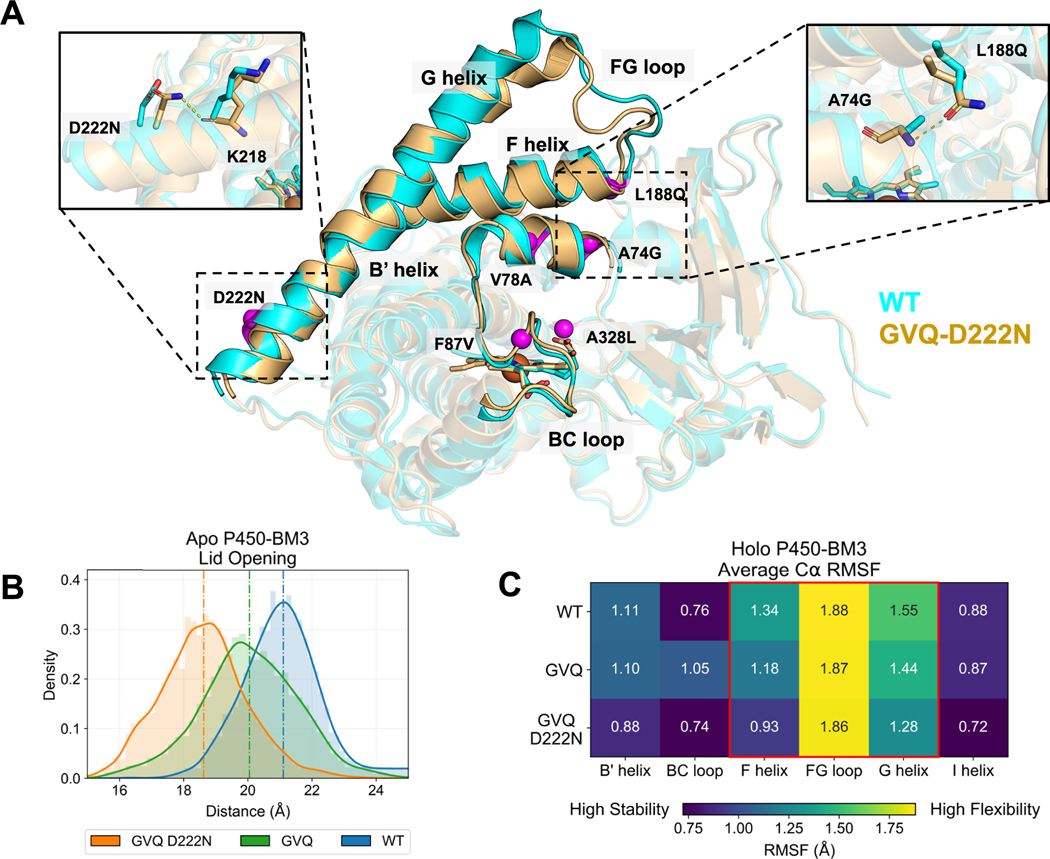
Figure 5.
Selected mutations alter P450-BM3 conformational dynamics. (A) Overlay of WT (cyan) and GVQ-D222N (orange) average structures from MD simulations of the apo structures with mutated positions highlighted as purple spheres. GVQ-D222N favors adopting the catalytically active closed conformation with the G helix lowered while the WT maintains the open conformation. L188Q forms a hydrogen bond to A74G to fasten the F helix, and D222N potentially acts as an anchor to curb lid opening motions by establishing a backbone hydrogen bond with the K218 carbonyl to stabilize the base of the G helix. (B) The lid distance characterizing the substrate channel opening is defined as the length between PRO196 and PRO45 alpha carbons. GVQ-D222N samples conformations resembling the active closed state, while GVQ experiences intermediate states, and the WT tends to stay open. (C) The WT holoprotein shows the highest mobility at the F/G-helix as measured by α carbon RMSF. GVQ has reduced flexibility at the lid regions, and GVQ-D222N is the most stable throughout.
The flexible nature of P450-BM3 is reflected by crystal structures with the F helix, F/G loop, and G helix regions (Figure 5A), which are known to act as a lid that moves during catalytic cycle, in varying positions with the “closed” lid often associated with the substrate bond, catalytically active state (Figure S3).24 We compared representative models of wild type and GVQ-D222N apo enzymes (see details in Methods) and our simulations indicate that the mutations promote lid closing (Figure 5A,B). The A74G-L188Q mutations function cooperatively, forming a novel hydrogen bond between the B′ helix and the F helix. The function of the hydrogen bond is suggested to fasten the F helix, minimizing the mobility of the lid. The D222N mutation occurs at the base of the G helix and forms a novel polar contact with the backbone carbonyl of K218, which may function to anchor the G helix and reduce lid flexibility (Figure 5A).
The effect of the mutations on P450-BM3 dynamics is first evaluated from trajectories started from the lid-open, no substrate bound states. We compare the distribution of lid distances in wild type (WT), GVQ, and GVQ-D222N (Figure 5B): WT samples the largest lid distances with an average of 21.1 Å, indicating that it favors maintaining the unproductive open state; GVQ samples more intermediate distances averaging 20.0 Å, suggesting greater disposition to closing than the WT; and GVQ-D222N favors occupying the fully closed conformation with average lid distance 18.6 Å. These results are consistent with the hypothesis that novel hydrogen bonds formed by the evolved mutations are critical to decreasing the free energy barrier of transitioning from open to closed forms and stabilizes the closed state.
We then analyzed trajectories started from the lid-closed, ACN bound state. While all enzyme variants tend to maintain the closed state, the stability of the lid varies (Figure 5C): The FG loop shows similar root mean square fluctuation (RMSF) for all samples, but the RMSF for the F helix declines from 1.34 Å in the WT, to 1.18 Å in GVQ, and finally to 0.93 Å in GVQ-D222N, while the G helix RMSF trends identically with 1.55 Å for WT, 1.44 Å for GVQ, and 1.28 Å for GVQ-D222N. The decrease in RMSF upon accumulation of the selected mutations supports the role of the novel hydrogen bonds in stabilizing the closed state and reducing excess flexibility of the lid region.
Tuning the Selection Pressure for Oxygenase Uncoupling.
The coupling efficiency (∼12%) of the P450-BM3 variants obtained is still relatively low. To further improve the coupling efficiency, strains with higher sensitivity to oxidative stress are needed to support more stringent selection. To explore this hypothesis, we disrupted genes encoding E. coli’s cellular defense mechanisms for toxic ROS including catalase (katG), alkyl hydroperoxide reductase (ahpC), and superoxide dismutase (sodA),34 and tested the growth behavior of resulting strains (Figure 6). While GVQ-AL can readily restore growth of MX203, MX203 with ΔahpC single knockout, ΔkatG single knockout, and ΔahpCΔkatG double knockout showed progressively decreased growth with this BM3 variant. Furthermore, MX203ΔsodA strain expressing GVQ-AL showed a complete lack of growth (Figure 6). Importantly, growth of MX203ΔsodA can be rescued by a well-coupled (>80%) NADPH-consuming cyclohexanone monooxygenase (CHMO)35,36 (Figure 6), indicating that the growth rate difference between the panel of knockout strains and the original MX203 selection strain was due to differing sensitivities to the oxidative stress produced by uncoupled oxygenases. Further work is needed to demonstrate the applicability of these strains and characterize the cellular response to oxidative stress in these selections. Understanding which ROS play a role in growth repression will be important for engineering the selection strain for heightened sensitivity to oxidative stress. Promising in vivo approaches to assess the intracellular state of strains expressing our BM3 variants include measuring the induction of ROS responsive genes utilizing lacZ expression driven by promoters activated by the ROS,37 and measuring the relative hydrogen peroxide produced via the uncoupled activity of the BM3 panels using a modified E. coli strain expressing a hydrogen peroxide response fluorescent reporter, HyPer strain.38
Figure 6.
Tuning selection via genetic disruption of cellular defense mechanisms for oxidative stress. Deletion of alkyl hydroperoxide reductase ahpC, catalase katG, or superoxide dismutase sodA heightened selection strain sensitivity to uncoupled activity in M9 minimal glucose media. Heterologous expression of GVQ-AL with 0.2 g/L acenaphthene (solid lines) restored growth to varying degrees in strain panel. Severe growth inhibition was observed in sodA mutant with the BM3 variant, but heterologous expression of well-coupled, NADPH-dependent cyclohexanone monooxygenase with its native substrate cyclohexanone (dashed line) restored significant growth. Strains engineered with this elevated selection pressure may enable selections with more stringent coupling criteria.
MATERIALS AND METHODS
Strain and Plasmid Construction.
Cloning was carried out with E. coli XL-1 blue cells in 2×YT medium containing 16 g/L Tryptone, 10 g/L Yeast Extract, and 5 g/L NaCl. The detailed cloning methods, including the method for BM3 library construction, are described in the Supporting Information. Construction of strain MX203 was described previously.10 Genetic disruption during the construction of the high-oxidative-stress derivatives of MX203 was performed using P1 transduction. The detailed strain construction methods are described in the Supporting Information. Strains and plasmids used are summarized in Table S1.
Growth and Selection Conditions.
MX203 and its high-oxidative-stress derivatives are regularly maintained in 2×YT medium. Selection of P450-BM3 libraries in MX203 was performed in M9 selection medium containing 1 mM MgSO4, 0.1 mM CaCl2, trace metal mix A5 with Co (H3BO3 2860 μg/L, MnCl2·4H2O 1810 μg/L, ZnSO4 7H2O 222 μg/L, Na2MoO4, 2H2O 390 μg/L, CuSO4, 5H2O 79 μg/L, Co(NO3)2·6H2O 49 μg/L), and BD Difco M9 salts (Na2HPO4 6.78 g/L, KH2PO4 3g/L, NaCl 0.5 g/L, NH4Cl 1 g/L), 2 g/L D-glucose, 0.01 g/L thiamine, 0.04 g/L FeSO4·7H2O. BM3 substrates lauric acid and acenaphthene were individually added to each culture at a concentration of 0.2 g/L prepared from a DMSO stock when appropriate. During pyruvate supplementation growth experiments, pyruvate was added to M9 selection medium at a concentration of 0.05 g/L. CHMO (on pLS201) substrate cyclohexanone was added at a concentration of 2 g/L when appropriate. For solid media, 15 g/L agar was added in addition. Concentrations for antibiotic selection were 100 mg/L for ampicillin, 50 mg/L for kanamycin, 50 mg/L for spectinomycin, and 10 mg/L for tetracycline. 0.1 mM IPTG was added for strains harboring plasmids with Plac promoter. Selection was performed at 30 °C for 60–70 h. Detailed methods on growth selection are described in the Supporting Information.
Characterization of P450-BM3 Variants.
Protein expression and purification methods are described in the Supporting Information. Total activity measurement was performed as reported previously,26 with modifications. The reaction (100 μL) contains NADPH (1 mM for lauric acid conversion or 0.4 mM for ACN conversion), 50 mM Sodium Phosphate buffer (pH 7.4), 0.3 μM FMN, protein (20 nM for lauric acid conversion or 12.5 nM for ACN conversion), 30 U/mL catalase (From Bovine Liver, Sigma-Aldrich), and 0.3 mM lauric acid or acenaphthene (Stock prepared in DMSO, final DMSO concentration in reaction is 2%). Protein was incubated with substrate and buffer (for 5 min for lauric acid conversion or 2 min for ACN conversion), before reaction was started by adding NADPH. NADPH consumption was monitored by absorbance at 340 nm (for 50 s at for lauric acid conversion or 12 s for ACN conversion 30 °C). Protein dilutions were prepared in 50 mM Sodium Phosphate Buffer (pH 7.4) immediately prior to assay mixture incubation.
The coupling efficiency was measured using previously reported methods.26 Reactions were started by the addition of 0.4 mM NADPH to the assay mixture (200 μL) containing 100 mM Tris-HCL (pH 7.4), ∼300 nM protein, 30 U/mL catalase (From Bovine Liver, Sigma-Aldrich) and 0.5 mM acenaphthene (Stock prepared in DMSO, final DMSO concentration in reaction is 2%). Protein was incubated with assay mixture for 10 min before starting the reaction by adding NADPH. Reaction was allowed to proceed for 4 h in an open microcentrifuge tube at 30 °C to allow complete consumption of NADPH. Acenaphthene and 1-acenaphthenol were extracted with 200 μL of chloroform added directly to reaction vessel. Mixture was vortexed for 1 min and subsequently centrifuged at 15 000g for 2 min. Aqueous layer absorbance was measured at 340 nm to confirm NADPH consumption and organic layer was measured using the gas chromatography (GC) method below. Coupling efficiency was calculated using product formed divided by NADPH consumed.
GC–FID Analytical Methods.
All GC analysis was performed on an Agilent 6850 (Agilent Technologies) equipped with an FID. An Agilent DB-WAXetr capillary column (30 m × 0.56 mm × 1 μm) was used for separation. The inlet and detector were held at 250 and 260 °C, respectively. The GC was operated in constant pressure mode with a pressure of 3.66 psi. Helium was used as the carrier gas. Air and hydrogen were supplied to the FID at 350 and 40 mL min−1, respectively. All gases were purchased from Airgas and 5 μL of sample was injected with a split ratio of 2:1. For analysis of acenaphthene and its hydroxylated product 1-acenaphthenol, the oven was held at 230 °C for 20 min. Acenaphthene and 1-acenaphthenol eluted at 4.4 and 17.5 min, respectively. Myristic acid was used as an internal standard.
ACN Docking.
The substrate acenaphthene (ACN) was docked into the P450-BM3 binding pocket with Rosetta.39 The crystal structure 1ZO940 of P450-BM3 with N-palmitoylmethionine (EPM) bound in the closed conformation was used as the starting template. The heme was modeled in the catalytically active Fe(IV)-oxo compound 1 state. The coordinates for EPM were removed from the structure, the ACN model was downloaded from the PubChem database,41 and ACN was initially placed in the open region above the heme oxygen. The Rosetta docking protocol consisted of mutation from the WT structure, perturbation of the ACN binding pose through random translation and rotation, and optimization of active site rotamers through Monte Carlo evaluation. A distance restraint was imposed between the heme oxygen and ACN reactive carbons (due to ligand symmetry two carbons have the potential to be hydroxylated) to focus sampling on the protein–ligand intermediate state preceding catalysis, and full flexibility for protein backbone torsions was allowed. A total of 1000 docking trials were completed for each variant, the top 100 models filtered on total Rosetta energy were further sorted based on ACN interface energy, and the model with the most favorable interface energy was selected as the reference. Protein figures were generated with PyMOL.42
Evaluation of P450-BM3 Conformational Dynamics.
MD simulations of P450-BM3 variants were completed with PMEMD43 from the AMBER 1844 package utilizing the ff14sb force field45 and 8 Å Particle Mesh Ewald real space cutoff.46 Compound 1 heme parameters were obtained from Shahrokh et al.,47 and protonation states of titratable residues were determined with the H++ web server.48 The TLEAP program was utilized to solvate the complexes with TIP3P water molecules in a truncated octahedron with 10 Å buffer and neutralizing Na+/Cl− counterions. Minimization was performed in two stages, first with 2500 steps of steepest descent and 2500 steps of conjugate gradient with non-hydrogen solute atoms restrained with a 20 kcal mol−1 Å−2 force to relieve solvent clash. The second stage minimization to remove solute steric clashes was run with the same cycle settings and restraints removed. Heating from 0 to 298 K was performed over 0.5 ns with 10 kcal mol−1 Å−2 restraints on all non-hydrogen solute atoms under NPT conditions at 1 atm pressure with Langevin thermostat and 1 fs time step. Structural artifacts from the heating step were cleared with solvent density equilibration over 5 ns with 5 kcal mol−1 Å−2 restraints on all solute atoms and an unrestrained 10 ns equilibration using 2 fs time step. Production MD trajectories were carried out for 250 ns with 2 fs time step, SHAKE restraints on hydrogens, NVT ensemble, Langevin thermostat with collision frequency 1.0 ps–1, and periodic boundary conditions.
The apo models for P450-BM3 variants were initiated from the open state with 2HPD49 as the template, amino acid substitutions were introduced with Rosetta and structures were relaxed to relieve unfavorable contacts. The holostructures for P450-BM3 variants with ACN bound started from the representative Rosetta docking models. Average structures based on α carbon coordinates were calculated for each apo trajectory by aligning all snapshots to the starting frame and averaging the α carbon positions. Direct coordinate averaging distorts bond angles and lengths, to depict a realistic model we identified the frame with the minimum α carbon RMSD to the calculated average model as an instance of the mean structure. Lid opening distances describing the positioning of the G helix were recorded as the length between PRO146 and PRO45 alpha carbons. Root mean square fluctuation (RMSF) over α carbon atoms was measured for trajectories of the holomodels and aggregated over secondary structure elements to compare flexibility.
Supplementary Material
ACKNOWLEDGMENTS
H.L. acknowledges support from University of California, Irvine, the National Science Foundation (NSF) (award no. 1847705), and the National Institutes of Health (NIH) (award no. DP2 GM137427). S.M. acknowledges support from the NSF Graduate Research Fellowship Program (grant no. DGE-1839285).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00380.
Further materials and methods, plasmids and strains used in this study (Table S1), mutations in the BM3 variants obtained from the liquid selection (Table S2), growth of BM3 panel with pyruvate supplementation (Figure S1), liquid growth of MX203 strain mediated by expression of BM3 variants obtained in selection (Figure S2), and P450-BM3 structural flexibility model (Figure S3) (PDF)
AUTHOR INFORMATION
Complete contact information is available at: https://pubs.acs.org/10.1021/acssynbio.0c00380
The authors declare no competing financial interest.
Contributor Information
Sarah Maxel, Department of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Edward King, Department of Molecular Biology and Biochemistry, University of California, Irvine, California 92697, United States.
Yulai Zhang, Department of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
Ray Luo, Department of Molecular Biology and Biochemistry, Department of Chemical and Biomolecular Engineering, Department of Biomedical Engineering, and Department of Materials Science and Engineering, University of California, Irvine, California 92697, United States.
Han Li, Department of Chemical and Biomolecular Engineering, University of California, Irvine, California 92697, United States.
REFERENCES
- (1).Lin Y, and Yan Y. (2014) Biotechnological Production of Plant-Specific Hydroxylated Phenylpropanoids. Biotechnol. Bioeng 111 (9), 1895–1899. [DOI] [PubMed] [Google Scholar]
- (2).Furuya T, and Kino K. (2014) Catalytic Activity of the Two-Component Flavin-Dependent Monooxygenase from Pseudomonas Aeruginosa toward Cinnamic Acid Derivatives. Appl. Microbiol. Biotechnol 98 (3), 1145–1154. [DOI] [PubMed] [Google Scholar]
- (3).Urlacher VB, and Eiben S. (2006) Cytochrome P450 Monooxygenases: Perspectives for Synthetic Application. Trends Biotechnol. 24 (7), 324–330. [DOI] [PubMed] [Google Scholar]
- (4).Schmidt S, Scherkus C, Muschiol J, Menyes U, Winkler T, Hummel W, Gröger H, Liese A, Herz HG, and Bornscheuer UT (2015) An Enzyme Cascade Synthesis of ε-Caprolactone and Its Oligomers. Angew. Chem., Int. Ed 54 (9), 2784–2787. [DOI] [PubMed] [Google Scholar]
- (5).Cahn JKB, Werlang CA, Baumschlager A, Brinkmann-Chen S, Mayo SL, and Arnold FH (2017) A General Tool for Engineering the NAD/NADP Cofactor Preference of Oxidoreductases. ACS Synth. Biol 6 (2), 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).de Rond T, Gao J, Zargar A, de Raad M, Cunha J, Northen TR, and Keasling JD (2019) A High-Throughput Mass Spectrometric Enzyme Activity Assay Enabling the Discovery of Cytochrome P450 Biocatalysts. Angew. Chem., Int. Ed 58 (30), 10114–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gielen F, Hours R, Emond S, Fischlechner M, Schell U, and Hollfelder F. (2016) Ultrahigh-Throughput-Directed Enzyme Evolution by Absorbance-Activated Droplet Sorting (AADS). Proc. Natl. Acad. Sci. U. S. A 113 (47), E7383–E7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Debon A, Pott M, Obexer R, Green AP, Friedrich L, Griffiths AD, and Hilvert D. (2019) Ultrahigh-Throughput Screening Enables Efficient Single-Round Oxidase Remodelling . Nat. Catal 2 (9), 740–747. [Google Scholar]
- (9).Siedler S, Schendzielorz G, Binder S, Eggeling L, Bringer S, and Bott M. (2014) SoxR as a Single-Cell Biosensor for NADPH-Consuming Enzymes in Escherichia Coli. ACS Synth. Biol 3 (1), 41–47. [DOI] [PubMed] [Google Scholar]
- (10).Maxel S, Aspacio D, King E, Zhang L, Acosta AP, and Li H. (2020) A Growth-Based, High-Throughput Selection Platform Enables Remodeling of 4-Hydroxybenzoate Hydroxylase Active Site. ACS Catal. 10 (12), 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Machado HB, Dekishima Y, Luo H, Lan EI, and Liao JC (2012) A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng 14 (5), 504–11. [DOI] [PubMed] [Google Scholar]
- (12).Liang K, and Shen CR (2017) Selection of an Endogenous 2,3-Butanediol Pathway in Escherichia Coli by Fermentative Redox Balance. Metab. Eng 39, 181–191. [DOI] [PubMed] [Google Scholar]
- (13).Zhang L, King E, Luo R, and Li H. (2018) Development of a High-Throughput, in vivo Selection Platform for NADPH-Dependent Reactions Based on Redox Balance Principles. ACS Synth. Biol 7 (7), 1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Calzadiaz-Ramirez L, Calvo-´Tusell C, Stoffel GMM, Lindner SN, Osuna S, Erb TJ, Garcia-Borras M, Bar-Even A,` and Acevedo-Rocha CG (2020) In vivo Selection for Formate Dehydrogenases with High Efficiency and Specificity toward NADP+. ACS Catal. 10, 7512–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kramer L, Le X, Rodriguez M, Wilson MA, Guo J, and Niu W. (2020) Engineering Carboxylic Acid Reductase (CAR) through A Whole-Cell Growth-Coupled NADPH Recycling Strategy. ACS Synth. Biol 9, 1632. [DOI] [PubMed] [Google Scholar]
- (16).Suzuki H, Inabe K, Shirakawa Y, Umezawa N, Kato N, and Higuchi T. (2017) Role of Thiolate Ligand in Spin State and Redox Switching in the Cytochrome P450 Catalytic Cycle. Inorg. Chem 56 (8), 4245–4248. [DOI] [PubMed] [Google Scholar]
- (17).Morlock LK, Böttcher D, and Bornscheuer UT (2018) Simultaneous Detection of NADPH Consumption and H 2 O 2 Production Using the Ampliflu™ Red Assay for Screening of P450 Activities and Uncoupling. Appl. Microbiol. Biotechnol 102 (2), 985–994. [DOI] [PubMed] [Google Scholar]
- (18).Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MCY, Baker D, and Keasling JD (2009) A Novel Semi-Biosynthetic Route for Artemisinin Production Using Engineered Substrate-Promiscuous P450BM3. ACS Chem. Biol 4 (4), 261–267. [DOI] [PubMed] [Google Scholar]
- (19).Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, and Ajikumar PK (2016) Overcoming Heterologous Protein Interdependency to Optimize P450-Mediated Taxol Precursor Synthesis in Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A 113 (12), 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Park SH, Kim DH, Dooil K, Kim DH, Jung HC, Pan JG, Taeho A, Donghak K, and Yun CH (2010) Engineering Bacterial Cytochrome P450 (P450) BM3 into a Prototype with Human P450 Enzyme Activity Using Indigo Formation. Drug Metab. Dispos 38 (5), 732–739. [DOI] [PubMed] [Google Scholar]
- (21).Li QS, Ogawa J, Schmid RD, and Shimizu S. (2001) Engineering Cytochrome P450 BM-3 for Oxidation of Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol 67 (12), 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Carmichael AB, and Wong LL (2001) Protein Engineering of Bacillus Megaterium CYP102: The Oxidation of Polycyclic Aromatic Hydrocarbons. Eur. J. Biochem 268 (10), 3117–3125. [DOI] [PubMed] [Google Scholar]
- (23).Whitehouse CJC, Bell SG, and Wong LL (2012) P450 BM3 (CYP102A1): Connecting the Dots. Chem. Soc. Rev 41 (3), 1218–1260. [DOI] [PubMed] [Google Scholar]
- (24).Whitehouse CJC, Bell SG, and Wong L-L (2012) P450BM3 (CYP102A1): Connecting the Dots. Chem. Soc. Rev 41, 1218. [DOI] [PubMed] [Google Scholar]
- (25).Rousseau O, Ebert MCCJC, Quaglia D, Fendri A, Parisien AH, Besna JN, Iyathurai S, and Pelletier JN (2020) Indigo Formation and Rapid NADPH Consumption Provide Robust Prediction of Raspberry Ketone Synthesis by Engineered Cytochrome P450 BM3. ChemCatChem 12 (3), 837–845. [Google Scholar]
- (26).Li QS, Ogawa J, Schmid RD, and Shimizu S. (2001) Engineering Cytochrome P450 BM-3 for Oxidation of Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol 67 (12), 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Maxel S, Aspacio D, King E, Zhang L, Acosta AP, and Li H. (2020) A Growth-Based, High-Throughput Selection Platform Enables Remodeling of 4-Hydroxybenzoate Hydroxylase Active Site. ACS Catal. 10, 6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cowart LA, Falck JR, and Capdevila JH (2001) Structural Determinants of Active Site Binding Affinity and Metabolism by Cytochrome P450 BM-3. Arch. Biochem. Biophys 387 (1), 117–124. [DOI] [PubMed] [Google Scholar]
- (29).Kim JG, Park SJ, Sinninghe Damsté JS, Schouten S, Rijpstra WIC, Jung MY, Kim SJ, Gwak JH, Hong H, Si OJ, Lee S, Madsen EL, and Rhee SK (2016) Hydrogen Peroxide Detoxification Is a Key Mechanism for Growth of Ammonia-Oxidizing Archaea. Proc. Natl. Acad. Sci. U. S. A 113 (28), 7888–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Whitehouse CJC, Bell SG, Tufton HG, Kenny RJP, Ogilvie LCI, and Wong LL (2008) Evolved CYP102A1 (P450BM3) Variants Oxidise a Range of Non-Natural Substrates and Offer New Selectivity Options. Chem. Commun 0 (8), 966–968. [DOI] [PubMed] [Google Scholar]
- (31).Johnsen AR, Wick LY, and Harms H. (2005) Principles of Microbial PAH-Degradation in Soil. Environ. Pollut 133 (1), 71–84. [DOI] [PubMed] [Google Scholar]
- (32).Haritash AK, and Kaushik CP (2009) Biodegradation Aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. J. Hazard. Mater 169 (1–3), 1–15. [DOI] [PubMed] [Google Scholar]
- (33).Darimont D, Weissenborn MJ, Nebel BA, and Hauer B. (2018) Modulating Proposed Electron Transfer Pathways in P450BM3 Led to Improved Activity and Coupling Efficiency. Bioelectrochemistry 119, 119–123. [DOI] [PubMed] [Google Scholar]
- (34).Park S, You X, and Imlay JA (2005) Substantial DNA Damage from Submicromolar Intracellular Hydrogen Peroxide Detected in Hpx- Mutants of Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A 102 (26), 9317–9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sheng D, Ballou DP, and Massey V. (2001) Mechanistic Studies of Cyclohexanone Monooxygenase: Chemical Properties of Intermediates Involved in Catalysis. Biochemistry 40 (37), 11156–11167. [DOI] [PubMed] [Google Scholar]
- (36).van Beek HL, Wijma HJ, Fromont L, Janssen DB, and Fraaije MW (2014) Stabilization of Cyclohexanone Monooxygenase by a Computationally Designed Disulfide Bond Spanning Only One Residue. FEBS Open Bio 4, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, and Liao JC (2012) Integrated Electromicrobial Conversion of CO2 to Higher Alcohols. Science 335, 1596. [DOI] [PubMed] [Google Scholar]
- (38).Lim JB, and Sikes HD (2015) Use of a Genetically Encoded Hydrogen Peroxide Sensor for Whole Cell Screening of Enzyme Activity. Protein Eng., Des. Sel 28 (3), 79–83. [DOI] [PubMed] [Google Scholar]
- (39).Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, and Baker D. (2011) Rosettascripts: A Scripting Language Interface to the Rosetta Macromolecular Modeling Suite. PLoS One 6 (6), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hegde A, Haines DC, Bondlela M, Chen B, Schaffer N, Tomchick DR, Machius M, Nguyen H, Chowdhary PK, Stewart L, Lopez C, and Peterson JA (2007) Interactions of Substrates at the Surface of P450s Can Greatly Enhance Substrate Potency. Biochemistry 46 (49), 14010–14017. [DOI] [PubMed] [Google Scholar]
- (41).Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, and Bolton EE (2019) PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 47 (D1), D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, 2015. [Google Scholar]
- (43).Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, and Walker RC (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 9 (9), 3878–3888. [DOI] [PubMed] [Google Scholar]
- (44).Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, and Woods RJ (2005) The Amber Biomolecular Simulation Programs. J. Comput. Chem 26 (16), 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C. (2015) Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput 11 (8), 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995) A Smooth Particle Mesh Ewald Method. J. Chem. Phys 103 (19), 8577–8593. [Google Scholar]
- (47).Bonagura CA, Bhaskar B, Shimizu H, Li H, Sundaramoorthy M, McRee DE, Goodin DB, and Poulos TL (2003) High-Resolution Crystal Structures and Spectroscopy of Native and Compound I Cytochrome c Peroxidase. Biochemistry 42 (19), 5600–5608. [DOI] [PubMed] [Google Scholar]
- (48).Anandakrishnan R, Aguilar B, and Onufriev AV (2012) H++ 3.0: Automating PK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 40 (W1), 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ravichandran K, Boddupalli S, Hasermann C, Peterson J, and Deisenhofer J (1993) Crystal Structure of Hemoprotein Domain of P450BM-3, a Prototype for Microsomal P450’s. Science (Washington, DC, U. S.) 261 (5122), 731–736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.