Abstract
The hemagglutinin (HA) protein undergoes a low-pH-induced conformational change in the acidic milieu of the endosome, resulting in fusion of viral and cellular membranes. A class of compounds that specifically interact with the HA protein of H1 and H2 subtype viruses and inhibit this conformational change was recently described (G. X. Luo et al., Virology 226:66–76, 1996, and J. Virol. 71:4062–4070, 1997). In this study, purified HA trimers (bromelain-cleaved HA [BHA]) are used to examine the properties and binding characteristics of these inhibitors. Compounds were able to inhibit the low-pH-induced change of isolated trimers, as detected by resistance to digestion with trypsin. Protection from digestion was extremely stable, as BHA-inhibitor complexes could be incubated for 24 h in low pH with almost no change in BHA structure. One inhibitor was prepared as a radiolabeled photoaffinity analog and used to probe for specific drug interactions with the HA protein. Analysis of BHA after photoaffinity analog binding and UV cross-linking revealed that the HA2 subunit of the HA was specifically radiolabeled. Cross-linking of the photoaffinity analog to BHA under neutral (native) pH conditions identified a stretch of amino acids within the α-helix of HA2 that interact with the inhibitor. Interestingly, cross-linking of the analog under acidic conditions identified a different region within the HA2 N terminus which interacts with the photoaffinity compound. These attachment sites help to delineate a potential binding pocket and suggest a model whereby the BHA is able to undergo a partial, reversible structural change in the presence of inhibitor compound.
Influenza virus contains a lipid envelope that must fuse with host cell membranes in order to initiate virus infection (42, 43, 49). The hemagglutinin (HA) protein, a trimeric glycoprotein embedded in the viral membrane, is responsible for specific binding to cell surface sialic acid-containing receptors (46) and for the fusion of the two membranes (51). Although the mechanism of viral fusion is not fully elucidated, it is known that the fusion event is preceded by a conformational change occurring in the HA trimer that is triggered by the decreasing pH encountered during endosomal passage of the virus (23, 43, 49, 50). The HA trimer is composed of three identical monomers, each containing two protein subunits (designated HA1 and HA2) attached to each other via a disulfide linkage (36, 52). These monomer subunits are formed from a single chain precursor HA (HA0) that undergoes cleavage during transport from the Golgi to the cell surface (27). Entry of the influenza virus into host cells is facilitated through receptor binding by the HA1 subunit to the sialic acid-containing receptor. The conformational change brought on by the low pH of the endosome exposes the hydrophobic amino terminus of the HA2 subunit, which is believed to be a trigger in the fusion process (8, 17, 19, 40). It is postulated that the native state of the HA is a spring-loaded coiled coil and upon acidification, the hydrophobic fusion peptide is translocated toward the target membrane (9–11). This exposed hydrophobic amino terminus is believed to mediate fusion with the cell membrane (8, 19).
Influenza virus HA can be cleaved from viral membrane surfaces with bromelain protease to create a soluble form of the protein (bromelain-cleaved HA [BHA]) (5, 52). The soluble HA remains a trimer with properties identical to those of the native membrane bound protein (44). Upon acidification, BHA undergoes a conformational change and forms rosettes caused by the aggregation of the exposed hydrophobic fusogenic domains of the HA2 subunit (14, 40). In this conformation, the BHA is susceptible to trypsin digestion, while it is resistant to this protease in its native conformation (15, 40).
We have previously reported on the identification of a class of compounds that can inhibit influenza virus fusion (29, 30). These compounds are able to inhibit the low pH induced conformational change in the HA protein of H1 and H2 subtype viruses but not of the H3 subtype virus. Of these three subtypes, precise structural information is available only for H3 HA (8, 20, 37, 38, 45, 48). Previously a model of H1 HA was constructed using H3 HA crystal structure data (52) and a potential fusion inhibitor-binding pocket was identified within HA2 based on resistant mutation analysis and inhibitor selectivity (30). In order to probe this binding model and better understand the mechanism of action of these compounds, experiments were carried out with isolated H1 BHA. Various analogs were able to protect BHA from protease digestion following acid treatment and subsequent neutralization. A radiolabeled analog which possessed a photoactivatable azide moiety was synthesized (16). Affinity labeling at a neutral or acidic pH produced very different profiles of labeled amino acids, although in each case the amino acids were in or near the proposed binding pocket in the HA2. The consequences of the differences in HA2 photoaffinity labeling patterns with regard to the mechanism of action of these fusion inhibitors are discussed below.
MATERIALS AND METHODS
Chemical synthesis of influenza virus fusion inhibitors.
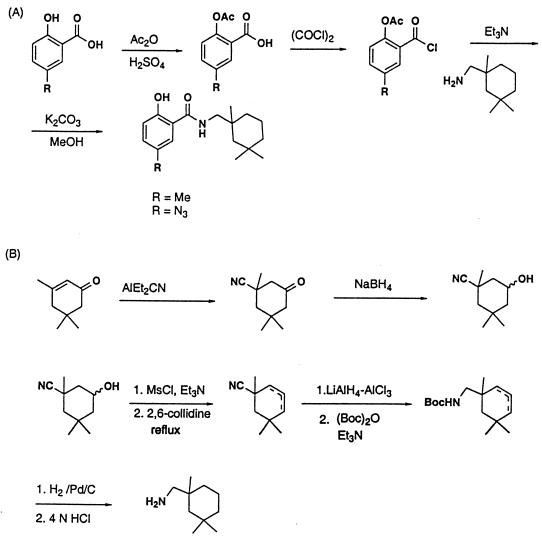
BMY-27709 has been described previously (29, 30). The structurally related BMS-199945 and BMS-201160 were synthesized by coupling acetyl 5-methylsalicylic acid chloride and acetyl 5-azido-salicylic acid chloride, respectively, with 1,3,3-trimethylcyclohexan-1-yl-methylamine in methylene chloride, followed by hydrolysis of the acetates with potassium carbonate in methanol. The acid chlorides were prepared by first treating both 5-methylsalicylic acid and 5-azido-salicylic acid with acetic anhydride and sulfuric acid and reacting the acetates with oxalyl chloride in the presence of a catalytic amount of N,N-dimethylformamide (Fig. 1A) (39).
FIG. 1.
(A) Synthesis scheme for preparation of compounds BMS-199945 and BMS-201160. (B) Synthesis scheme for preparation of the 1,3,3-trimethylcyclohexan-l-yl-methylamine intermediate used in the preparation of BMS-199945 and BMS-201160. For the preparation of [3H]BMS-201160, tritium was used. Abbreviations: Ac, acetyl; Et, ethyl; Me, methyl.
1,3,3-Trimethylcyclohexan-1-yl-methylamine was prepared from isophorone by the following sequence of steps (Fig. 1B): (i) 1,4-addition of cyanide to isophorone using aluminum diethylaluminum cyanide, followed by reduction of the ketone with sodium borohydride to give a 3-cyano-3,5,5-trimethylcyclohexanol; (ii) mesylation of the alcohol with methanesulfonyl chloride and triethylamine; (iii) elimination of methane-sulforic acid by refluxing in 2,6-collidine to afford a mixture of the olefins; (iv) reduction of the nitrile with lithium aluminum hydride-aluminum chloride and protection of the resulting amine with a t-butoxycarbonyl (Boc) group; (v) hydrogenation of the olefin followed by deprotection of the Boc group with 4 N HCl, providing the amine intermediate. For the preparation of [3H]BMS-201160 tritium was introduced during the hydrogenation of the olefins.
The tritiated analog of BMS-201160, 5-azido-2-hydroxy-N-[(1,3,3-trimethyl-4,5,6-3H-cyclohexyl)methyl] benzamide, was synthesized as described elsewhere (16). Tritiation was performed at the National Tritium Labeling Facility, Lawrence Berkeley National Laboratories, Berkeley, Calif.
Viruses and cells.
Influenza virus A/PR/8/34 (H1N1) was grown in 10-day-old embryonated chicken eggs at 37°C. Viruses were harvested 48 h after inoculation as described previously (2). Virus was collected from pooled allantoic fluid by centrifugation and was resuspended in NTE (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], and 1 mM EDTA). Virus was further purified on 30-to-60% sucrose gradients. Influenza A/WSN/33 (H1N1) virus was propagated in Madin-Darby bovine kidney (MDBK) cells grown in minimal essential medium (GIBCO/BRL, Gaithersburg, Md.) with 10% fetal bovine serum (Sigma, St. Louis, Mo.) (30). Chicken erythrocytes (RBC) were purchased from Spafas (Preston, Conn.).
Hemolysis inhibition assay.
The hemolysis inhibition assay was modified from that described previously (29, 30). One hundred microliters of influenza A/WSN/33 (H1N1) virus (∼6 μg of protein) was incubated with an equal volume of phosphate-buffered saline (PBS) containing various concentrations of inhibitor at 37°C for 1 h. Two hundred microliters of a 2.0% solution of chicken RBC in PBS were added to the reaction mixture and incubated at 37°C for 10 min. The virus-bound chicken RBC were pelleted by centrifugation at 1,600 rpm for 8 min. The RBC pellet was resuspended in 450 μl of low-pH PBS buffer (pH 5.0) containing the corresponding concentration of inhibitor and incubated at 37°C for 15 min. The reaction mixture was neutralized to pH 7.0 by the addition of 1 N NaOH. Cell debris and unlysed cells were pelleted by centrifugation at 2,000 rpm for 8 min. Three hundred μl of supernatant was transferred to a 96-well tissue culture plate (Corning Glass Works, Corning, N.Y.) for measurement of optical density at a wavelength of 540 nm by using a Multiscan MCC/340 plate reader (Titertek, Huntsville, Ala.).
HA purification.
BHA was prepared by a modification of the published procedure (5). One hundred milligrams of sucrose gradient-purified influenza A/PR/8/34 virus was pelleted at 100,000 × g for 60 min and resuspended by successive shearing through needles of decreasing gauge (18 to 27 gauge) in 10 ml of buffer (0.1 M Tris-HCl [pH 7.4], 1 mM EDTA, 50 mM 2-β-mercaptoethanol) containing bromelain (2 mg/ml; Sigma). The virus suspension was incubated at 37°C for 16 h. Virus was pelleted, and the supernatant containing the BHA and bromelain was concentrated in a Centriprep-30 concentrator (Amicon, Beverly, Mass.) to a volume of 5 ml. The NaCl concentration of the sample was adjusted to 0.5 M, and the solution was applied to a 35-by-2.8-cm Sephacryl-400 (Sigma) gel filtration column equilibrated with 0.1 M Tris-HCl (pH 7.5)–0.5 M NaCl. Elution was carried out at a flow rate of 1 ml/min. Fractions containing BHA were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and concentrated by Centriprep-30 centrifugation. Purified BHA was stored at 4°C.
Trypsin protection assay.
Native BHA is resistant to trypsin cleavage, while the conformationally changed BHA is sensitive to trypsin treatment (40). To assay the trypsin sensitivity of BHA in the presence of inhibitors, 38 μl of a solution of 6 μM influenza A/PR/8/34 (H1N1) virus BHA was incubated with various concentrations of compounds in 2 μl of dimethyl sulfoxide at 31°C for 15 min. The reaction was acidified with 0.25 M citrate (pH 4.2) to achieve a final pH of 5.0 and incubated an additional 15 min at 31°C. The solution was then neutralized with 0.25 M Tris-HCl, pH 9.0 (final pH 7.5). Four micrograms of trypsin (TPCK [tolylsulfonyl phenylalanyl chloromethyl ketone] treated; Sigma) was added and digestion was carried out for 1 h at 37°C. Digestion was terminated by the addition of 10× SDS sample buffer and heating at 95°C for 5 min. The extent of HA digestion was analyzed by SDS-PAGE on 12% Tris-Glycine ReadyGels (Bio-Rad, Hercules, Calif.). Cleaved and uncleaved peptides of BHA identified on Coomassie-stained gels were quantitated through the use of a Molecular Dynamics (Sunnyvale, Calif.) Personal Densitometer SI with ImageQuanT software.
Affinity labeling of BHA.
Thirty-eight microliters of 6 μM BHA and 2 μl of 1.5 μM [3H]BMS-201160 (∼70 Ci/mmol) in dimethyl sulfoxide were incubated at 31°C for 15 min. Some samples were acidified prior to irradiation as described in the trypsin protection assay. Reaction mixtures in open Eppendorf tubes were irradiated for 7 min from above with an 8-W Sylvania UV germicidal lamp at a distance of 5 cm at 25°C. After addition of SDS sample buffer and a 5-min, 95°C incubation, samples were electrophoresed in 4-to-20% acrylamide Tris-Glycine ReadyGels (Bio-Rad). Gels were fixed and treated with En3Hance (New England Nuclear, Boston, Mass.) according to manufacturer instructions, and radiolabeled proteins were detected by autoradiography.
Isolation of affinity-labeled peptides.
Two hundred micrograms of BHA affinity labeled with [3H]BMS-201160 was precipitated with 8 volumes of acetone or methanol and pelleted by centrifugation at 16,000 rpm. Pellets were air dried, dissolved in 50 μl of 8 M urea–0.1 M Tris-HCl (pH 8.0), reduced by dithiothreitol (DTT) treatment, and cysteine modified as described previously (12). The peptides were directly loaded onto a Vydac Protein C4 column (The Separations Group, Hesperia, Calif.) in the buffer described above. The HA1 subunit was separated from the HA2 subunit through reverse-phase high-performance liquid chromatography (HPLC) using a 0.1% triflouoroacetic acid mobile phase and eluted with an acetonitrile gradient. The acetonitrile concentration was increased at a rate of 1% per minute by using a Waters HPLC system (Millipore, Milford, Mass.). Under these conditions, the HA1 subunit eluted 7 min earlier than the more hydrophobic HA2 polyprotein. Fractions containing radiolabeled HA2 were lyophylized to dryness.
For CNBr cleavage, the dry HA2 pellet was solubilized in 50 to 100 μl of 70% trifluoroacetic acid. Solid CNBr (Pierce, Rockford, Ill.) was added at a 500-fold molar excess and allowed to react for 16 h in the dark at 25°C (1). The reaction was terminated by three cycles of lyophilization and resuspension in 100 μl H2O, and the pellet was dissolved in SDS sample buffer.
Endoproteinase Lys-C and trypsin digestion were carried out by resuspending the dry HA2 pellet in 8 M urea–40 mM NH4HCO3–10 mM DTT and incubating at 50°C for 15 min. Samples were diluted fivefold with H2O, followed by the addition of trypsin (TPCK treated; Sigma) or endoproteinase Lys-C (Sigma) at a ratio of 5 μg enzyme for each 100 μg of labeled HA2. Endoproteinase digestions were carried out at 37°C for 16 h. Samples were lyophilized and dissolved in SDS sample buffer to give a 7 M urea final concentration.
Protease-digested HA2 samples were electrophoresed in 16.5% Tris-Tricine ReadyGels (Bio-Rad) and electrophoretically blotted to polyvinylidine difluoride (PVDF) (Bio-Rad) membrane according to the manufacturer’s instructions. Following membrane transfer, peptide bands were visualized with Coomassie blue R-250. Dried PVDF membranes were autoradiographed, and stained bands corresponding to radiolabeled peptides were cut out for sequencing. Sequence analysis was performed at the peptide sequencing core facility (Hershey Medical Center, Hershey, Pa.).
RESULTS
Activity of fusion inhibitors against purified BHA.
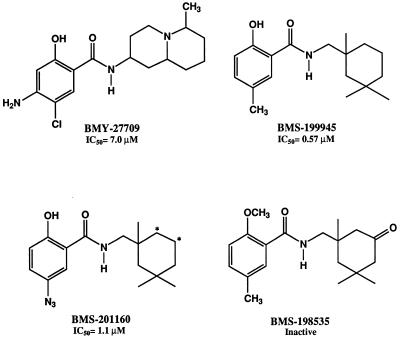
The three fusion inhibitor compounds used in this study are presented in Fig. 2. BMY-27709 was the first compound of the series identified as an inhibitor of the conformational change of the influenza virus H1 HA (29, 30). BMY-27709 inhibited replication of influenza A/WSN/33 virus in tissue culture and was also active in an RBC hemolysis assay, indicating that it blocked virus-cell fusion. BMS-199945 is a more potent analog than BMY-27709. BMS-201160 is equally active and structurally related to BMS-199945, with a photoactivatable azide group replacing the methyl group on the salicylic acid moiety. BMS-198535 is an inactive analog of BMS-199945. The 50% inhibitory concentrations (IC50s) for influenza A/WSN/33 virus-induced RBC hemolysis are 7, 0.57, and 1.1 μM for BMY-27709, BMS-199945, and BMS-201160, respectively (Fig. 2).
FIG. 2.
Structures of influenza virus fusion inhibitors used along with their respective IC50s in the influenza A/WSN/33 virus-induced hemolysis of chicken RBC. ∗, tritiation site for [3H]BMS-201160.
Bromelain-cleaved and purified HA possesses properties similar to those of native HA present on viral membranes, existing as a trimer and retaining receptor binding capability (44). In addition, exposure to low pH causes a gross conformational change in the molecule that exposes the hydrophobic amino terminus of the HA2 subunit, causing rosettes to form (40). This conformational change can be monitored through sensitivity to trypsin treatment. Whereas the neutral-pH form of native HA or purified BHA is resistant to trypsin treatment, the low-pH form becomes susceptible, leading to digestion of the BHA (40).
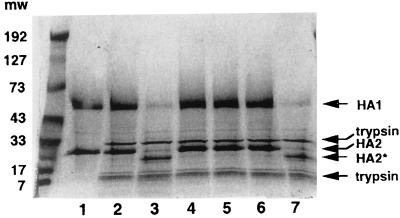
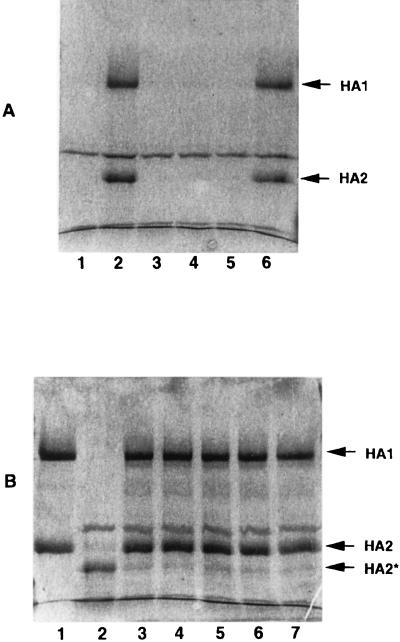
The three compounds were incubated with purified BHA isolated from influenza A/PR/8/34 virus. After acidification to pH 5.0, the solution was neutralized and treated with trypsin for 1 h as described above. Figure 3 shows the results of this experiment. The purified BHA (lane 1) is resistant to trypsin before acid treatment (lane 2), but after low-pH exposure, it becomes susceptible and is digested (lane 3). In this case, the HA1 subunit of the A/PR/8/34 virus BHA is ∼75% digested and cannot be detected as smaller proteolytic fragments. This differs somewhat from the reported behavior of H3, where the HA1 subunit is digested, but another lighter staining peptide can be detected migrating at a lower molecular weight (MW) (40). We also observe that the HA2 subunit is clipped so that it migrates at a lower MW, similar to that shown with H3 BHA (9). Prior incubation of 6 μM BHA with 100 μM compound is able to completely protect the BHA from subsequent trypsin digestion (Fig. 3, lanes 4 to 6). None of the compounds tested had any inherent trypsin inhibiting activity (data not shown). An analog with no activity in the virus-induced RBC hemolysis assay (BMS-198535) does not provide BHA protection from trypsin digestion (lane 7). Therefore, these compounds are able to inhibit the conformational change of the purified BHA.
FIG. 3.
SDS-PAGE of trypsin protection assay examining the ability of fusion inhibitors to protect BHA from tryptic digestion. BHA (6 μM) was incubated with 100 μM compound for 15 min at 31°C in pH 7.5 buffer prior to acidification to pH 5.0. Samples were neutralized and treated with trypsin for 1 h and then subjected to SDS-PAGE on 12% acrylamide gels and stained with Coomassie blue. Lane 1, 6 μM BHA alone; lane 2, BHA plus trypsin without acid; lane 3, Acidified BHA plus trypsin; lanes 4 to 7, BHA pretreated with 100 μM BMS-199945, BMS-201160, BMY-27709, and an inactive analog (BMS-198535), respectively, prior to acidification and trypsin digestion. Arrows indicate HA1, HA2, cleaved HA2*, and trypsin proteins. MWs are shown in thousands.
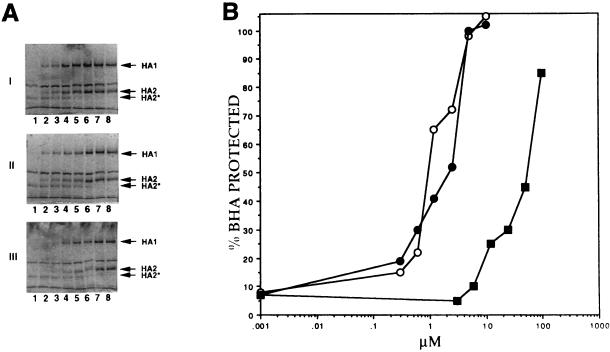
Antiviral 50% effective concentration measurements in tissue culture growth or virus-induced RBC hemolysis assays provide functional data for the inhibitors, but these assays cannot be used to estimate the stoichiometry of compound needed to inhibit each HA trimer. The use of purified BHA should allow for a more precise examination of the stoichiometry necessary for blocking the conformational change. BHA (6 μM) purified from A/PR/8/34 virus was incubated with various concentrations of compound, and following acid treatment, trypsin protection experiments were performed. The digested BHA products were examined by SDS-PAGE (Fig. 4A), and the intact and digested HA fragments were quantitated by densitometry (Fig. 4B). BMY-27709 was the least active, exhibiting 50% protection at a concentration of 55 μM. BMS-199945 and BMS-201160 exhibited higher potency, with IC50 values of approximately 1 and 2 μM compound in the trypsin protection assay, respectively. Since we previously reported a 10-fold increase in BMS-27709 IC50s in hemolysis inhibition with A/PR/8/34 rather than A/WSN/33 virus (29), these values are comparable to those achieved in the virus-induced RBC hemolysis assay.
FIG. 4.
Quantitative trypsin protection assay. (A) SDS-PAGE of BHA trypsin protection assays performed in the presence of various concentrations of the three inhibitors: I, BMS-201160; II, BMS-199945; and III, BMY-27709. For panels I and II, concentrations tested are 0, 0.15, 0.3, 0.6, 1.2, 2.5, 5, and 10 μM for lanes 1 through 7, respectively, and those for panel III are 0, 1.5, 3, 6, 12, 25, 50, and 100 μM for lanes 1 through 7, respectively. Lane 8 in all panels contains nonacidified BHA treated with trypsin in the absence of compound. (B) Densitometric scan of protected HA1 bands from stained SDS gels expressed as percent of trypsin-treated BHA without acid treatment (lane 8). Symbols: open circles, BMS-199945; solid circles, BMS-201160; solid squares, BMY-27709.
Reversibility of fusion inhibitors at neutral and acidic pH and stability of inhibitor-BHA complex at acidic pH.
BMY-27709 was previously shown to be a reversible inhibitor in virus-induced RBC hemolysis inhibition assays using intact influenza virus (29). Due to the nature of the experiments, reversibility using RBC could only be achieved at physiologic pH. Therefore, trypsin protection experiments were performed on BHA to determine if these inhibitors are also reversible at acidic pH (Fig. 5A). BMS-201160 (20 μM) was incubated with 6 μM BHA at neutral pH. As determined by the experiments described in Fig. 4B, there is enough compound to completely protect BHA from trypsin digestion following acidification and neutralization (lanes 1 and 2). Unprotected and BMS-201160-inhibited BHA mixtures were either diluted 100-fold in neutral buffer for 15 min and then acidified or acidified first and then diluted 100-fold in acidic buffer prior to trypsin digestion. Samples were concentrated by 10% trichloroacetic acid (TCA) precipitation prior to SDS-PAGE (TCA does not precipitate the proteolytically cleaved BHA2 fragment). If the compound binding is readily reversible, dilutions should decrease the level of protection from subsequent trypsin treatment. This is observed at a neutral pH, where dilution reverses the BMS-201160 protection (lane 4). However, BMS-201160-treated BHA diluted in pH 5.0 buffer still exhibited significant protection against trypsin digestion (Fig. 5A, lane 6). This suggests that the acidic treatment of the BHA results in a much more stable compound-protein interaction within the trimer. This could be due to a partial conformational change that either traps the compound in the trimer or creates an environment where the compound binds with much higher affinity. This hypothesis is further supported by experiments that examine the stability of the BHA in the presence of BMS-201160 at acidic pH. A 20 μM concentration of compound was incubated with 6 μM BHA, and the mixture acidified with citrate buffer. At various times up to 24 h, a sample was extracted and neutralized. Examination of BHA through trypsin sensitivity shows that protection by BMS-201160 is almost complete even after 24 h of incubation at pH 5.0 (Fig. 5B).
FIG. 5.
(A) Reversibility of inhibitor protection of BHA, examined by the ability of 20 μM BMS-201160 to protect 6 μM BHA from trypsin digestion following 100-fold dilution at pH 7.5 or pH 5.0. Samples were neutralized prior to trypsin treatment, TCA precipitated, and analyzed by SDS-PAGE. The HA2* and certain trypsin digestion products observed in earlier figures are not recovered during TCA precipitation. Lane 1, no inhibitor added, undiluted control; lane 2, 20 μM BMS-201160 added, undiluted control; lane 3, no inhibitor added, diluted 100-fold at pH 7.5; lane 4, 20 μM BMS-201160 added, diluted 100-fold at pH 7.5; lane 5, No inhibitor added, diluted 100-fold at pH 5.0; lane 6, 20 μM BMS-201160 added, diluted 100-fold at pH 5.0. (B) Stability of inhibitor-BHA complex: analysis of the ability of 20 μM BMS-201160 to inhibit 6 μM BHA from becoming trypsin sensitive after prolonged incubation at pH 5.0. Lane 1, pH 7.5, no trypsin added; lane 2, no inhibitor added, pH 5.0, trypsin treatment following neutralization; lanes 3 through 7, 20 μM BMS-201160 incubated with BHA at pH 5.0 for 1, 2, 4, 8, and 24 h, respectively, prior to neutralization and trypsin digestion.
Covalent labeling of the HA2 subunit of HA with [3H]BMS-201160 at neutral pH.
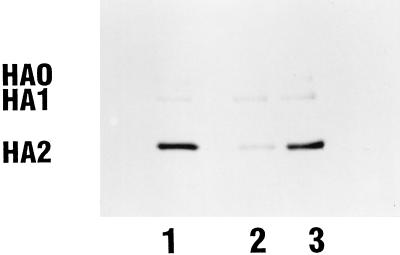
Experiments detailed above demonstrate that the photoaffinity analog BMS-201160 is able to inhibit the acid-induced conformational change associated with trypsin sensitivity. The azide group of BMS-201160 provides a photoreactive moiety through photolysis to generate the corresponding nitrene (6). This reactive moiety can react with proximate functionalities within the binding pocket of the protein. Radiolabeled [3H]BMS-201160, containing two tritium atoms attached to the cyclohexane moiety, was prepared at a specific activity of approximately 70 Ci/mmol. [3H]BMS-201160 was incubated with BHA at neutral pH and activated through irradiation with UV light, and the mixture was analyzed by SDS-PAGE (Fig. 6). A band with an MW of approximately 28,000 is the predominantly labeled protein fragment (Fig. 6, lane 1). This band corresponds to the size of the HA2 subunit of the A/PR/8/34 BHA. In addition, a minor band with an MW of 49,000, corresponding to the HA1 subunit, is also observed along with another minor band with an MW of ∼78,000, which corresponds to the uncleaved HA0 protein. Western blot analysis has confirmed these assignments (data not shown). The sample in Fig. 6, lane 2, had a 50-fold molar excess of an unlabeled competitor compound (BMS-199945) added to the reaction mixture. In this case, labeling of the HA2 subunit is significantly reduced, indicating specific labeling of the HA2 subunit. The extent of radioactivity present in the HA1 band is not appreciably altered by competitor, suggesting that the covalent attachment of compound to the HA1 subunit is nonspecific. The sample in Fig. 6, lane 3, contained an excess of an unlabeled related analog which is inactive as a fusion inhibitor and shown not to bind to HA (data not shown). There is no significant difference in radiolabeling intensity between this sample and that of [3H]BMS-201160 alone, illustrating that the covalent labeling of the HA2 subunit by BMS-201160 is specific.
FIG. 6.
Covalent labeling of the BHA2 HA subunit with [3H]BMS-201160 at neutral pH. Lane 1, BHA reacted with [3H]BMS-201160; lane 2, BHA reacted with [3H]BMS-201160 and a 50-fold molar excess of unlabeled BMS-199945; lane 3, BHA reacted with [3H]BMS-201160 and a 50-fold molar excess of a related inactive analog.
Peptide mapping and amino acid sequencing of covalently labeled HA2 subunit.
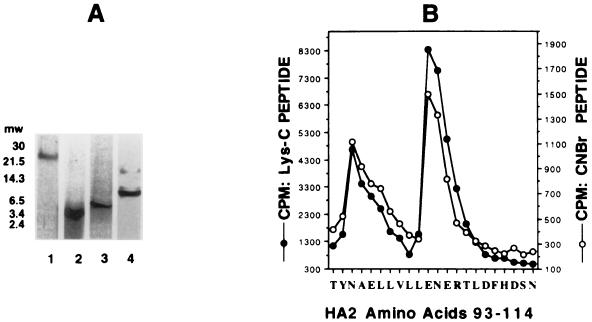
The BHA protein covalently labeled with [3H]BMS-201160 was subjected to DTT reduction and cysteine modification, and the HA2 subunit was purified by reverse-phase HPLC. After lyophilization, HA2 was digested either with trypsin or endoproteinase Lys-C or chemically cleaved by treatment with CNBr. In each case, a single radiolabeled peptide was generated. Figure 7A, lane 1, represents the HPLC-purified HA2 after covalent labeling with [3H]BMS-201160. Trypsin treatment of HA2 produces a radiolabeled peptide of ∼2,700 Da (lane 2), while Lys-C cleavage produces a radiolabeled fragment of ∼3,900 Da (lane 3). CNBr treatment of the covalently labeled HA2 produced a predominantly radiolabeled peptide of ∼8,500 Da (lane 4). Based upon the molecular weights of the labeled peptides, sequence of the HA2 subunit of A/PR/8/34 virus and recognition sites of the various cleavage treatments, tentative assignments of the radiolabeled peptides could be made. These bands correspond to overlapping protease fragments encompassing amino acids 84 to 106 (MW, 2,738) for the trypsin treatment, amino acids 84 to 116 (MW, 3,895) for the Lys-C treatment, and amino acids 78 to 149 (MW, 8,502) for the CNBr treatment. Therefore, [3H]BMS-201160 is covalently attached to residues between amino acids 84 and 106 of the HA2 subunit.
FIG. 7.
Peptide mapping and amino acid sequencing of BHA2 covalently labeled at neutral pH. (A) SDS–16% PAGE of peptides generated from purified BHA2 covalently labeled with [3H]-BMS-201160. Lane 1, uncleaved BHA2; lane 2, trypsin-digested BHA2; lane 3, Lys-C digested BHA2; lane 4, CNBr-cleaved BHA2. (B) Peptide sequence and tritium levels detected at each cycle. Results were generated for BHA2 amino acids 93 to 114.
Cleaved peptides were electroblotted onto PVDF membranes and subjected to autoradiography. Dried membranes were then stained with Coomassie blue R-250, and the labeled peptides were aligned with the stained membrane and cut out. Labeled peptides corresponding to the Lys-C and CNBr cleavages were sequenced, and each cycle was assayed for radiolabeling through scintillation counting. The results are shown in the graph in Fig. 7B. Both peptides produced identical results. The major peak of radiolabeling corresponded to a trio of residues representing Glu-103, Asn-104, and Glu-105. This region is part of the long α-helix A within the HA2 and helps comprise the pocket previously predicted to be part of the binding site for these fusion inhibitors (30). A second peak of radiolabeling was also observed at Asn-95, Ala-96, and Glu-97, a region which lies slightly above the pocket.
BHA affinity labeling with [3H]BMS-201160 under acidic conditions.
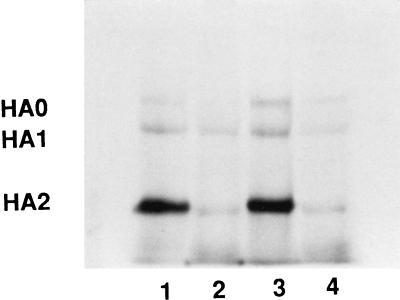
Data on the reversibility of compounds in the trypsin protection assay at neutral and acidic pH suggest that there are differences in inhibitor-protein interactions as the pH is varied. The ability of BMS-201160 to block the acid-induced conformational change (Fig. 3 and 4) indicates that the compound is bound in the trimer, allowing for cross-linking of the compound to the BHA trimer at acidic pH as well. Therefore, affinity labeling experiments at neutral or acidic pH were performed with [3H]BMS-201160 and BHA purified from A/PR/8/34 (Fig. 8). Lanes 1 and 2 were irradiated at a neutral pH as before. Lanes 3 and 4 were acidified for 10 min and then irradiated at pH 5.0. BHA2 is the predominantly labeled subunit, whether cross-linking is performed at pH 7.4 or pH 5.0 (compare lanes 1 and 3, respectively). An excess of BMS-199945 (lanes 2 and 4) competitively inhibits [3H]BMS-201160 binding at both neutral and acid pH.
FIG. 8.
SDS-PAGE comparison of BHA2 labeling by [3H]BMS-201160 under neutral and acidic conditions. Lane 1, cross-linking performed at pH 7.5 described before; lane 2, cross-linking performed in the presence of 50-fold molar excess of BMS-199945 at pH 7.5; lane 3, cross-linking performed at pH 5.0 after preincubation at pH 7.5; lane 4, cross-linking performed at pH 5.0 after preincubation at pH 7.5 in the presence of 50-fold molar excess of BMS-199945.
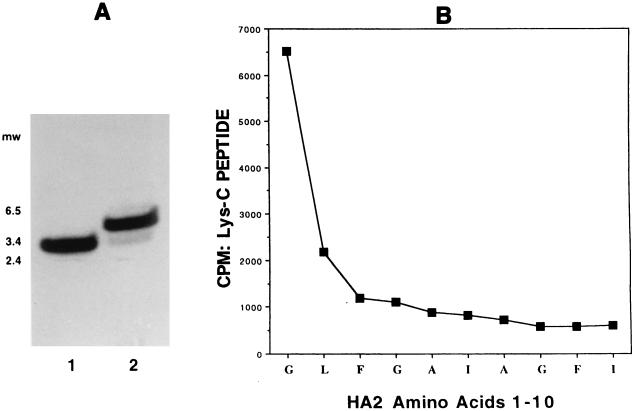
Both HA2 peptides were isolated by HPLC, subjected to endoproteinase Lys-C digestion, and analyzed by SDS-PAGE (Fig. 9A). Lys-C-digested protein which was covalently labeled at pH 5.0 (lane 2) produced a labeled HA2 peptide different from the fragment obtained when labeling was performed at pH 7.4 (lane 1). Whereas the peptide observed in lane 1 corresponds to amino acids 84 to 116 of the HA2 (MW, 3,895), the major radiolabeled peptide observed in lane 2 migrates more slowly, with an apparent MW of ∼4,200. The ∼3,900-MW peptide is also radiolabeled at a low pH, although to a much lower extent than the ∼4,200-MW peptide. The identity of this band is presumably the fragment containing amino acids 84 to 116, although it was not pursued. The only endoproteinase Lys-C-derived fragment of the HA2 subunit that is predicted to be larger than the peptide from amino acids 84 to 116 constitutes a peptide derived from residues 1 to 39 of the HA2. Indeed, when this fragment was purified and sequenced, it was identified as the amino terminus of HA2. Scintillation counting of the successive cycles is shown in Fig. 9B. Nearly the entire radioactivity is eluted with the N-terminal Gly of the hydrophobic fusion peptide, with minor amounts of radiolabel eluting with Leu-2 and trace amounts eluting thereafter. Thus, affinity labeling at a low pH results in substantially different binding patterns, strongly suggesting that alterations in compound binding or HA2 protein conformation are taking place when the pH is lowered from 7.5 to 5.0.
FIG. 9.
Affinity labeling BHA2 at pH 5.0. (A) SDS-PAGE comparison of Lys-C peptides generated after affinity labeling BHA2 with [3H]BMS-201160 under neutral and acidic conditions. Lane 1, Lys-C digest of BHA2 cross-linked at pH 7.5; lane 2: Lys-C digest of BHA2 cross-linked at pH 5.0 after preincubation at pH 7.5. (B) Peptide sequence and tritium released at each cycle. Results for BHA2 amino acids 1 to 10 are shown.
DISCUSSION
New antivirals would have significant potential for the prophylaxis and treatment of influenza virus infection (13, 31, 32). Inhibition of influenza virus receptor binding or fusion is an attractive target for drug development. Antivirals targeting the HA would combat infection prior to virus entry.
The involvement of a low-pH-induced conformational change in the HA protein of influenza virus as a prerequisite to membrane fusion has been well documented (8, 10, 11, 17, 19, 40). The commonality of a conformational change mechanism during the entry of enveloped viruses into host cells has been implied through structural similarities among the viral membrane proteins thought to be responsible for fusion (3, 21, 22, 28, 47, 48, 50). For influenza virus, it has been shown that acid-pH-induced structural rearrangement of HA is required for fusion and that the result of this conformational change is that the highly hydrophobic amino terminus of HA2 is exposed and projects outward from the virus membrane (17, 40). Interaction of this fusogenic peptide with the endosomal membrane is an integral step in the fusion process. There have been several reports of small molecules that inhibit influenza virus growth in cells through blockade of the acid-induced rearrangement of HA (4, 29, 30, 35, 41). Recently, an inhibitor was identified that induces the conformational change of HA (25). A precise understanding of the mechanism of these inhibitors could provide key insights into the steps leading to the final conformational change in the HA molecule and the subsequent fusion of the viral and cellular membranes.
The fusion inhibitor series discussed herein are active against H1 and H2 subtype HAs, subtypes for which successful crystallization conditions have not yet been developed. Therefore, biochemical and affinity labeling experiments were used to probe the actual binding mode of our inhibitor series. Since previous studies had only been carried out using whole virus, experiments were first designed to examine the effect of the conformational change inhibitors on bromelain-treated and isolated HA trimers. These trimers have been shown to react to low-pH conditions in a manner similar to HA embedded in lipid membrane (44). The conformational change exposes the hydrophobic amino terminus of the HA2 subunit, resulting in the formation of rosettes (40) and allowing for susceptibility to various protease treatments. Three compounds were examined for their ability to inhibit the low-pH conformational change in isolated A/PR/8/34 BHA trimers as measured by their ability to protect BHA from trypsin digestion. BMY-27709, BMS-199945, and BMS-201160 were all able to protect the BHA from trypsin digestion after low-pH incubation, showing that they can prevent the conformational change in the purified trimers from occurring. This strongly suggests that these compounds are able to bind to the purified trimers and function in much the same way that they act on membrane-bound protein.
The activity of these compounds against purified BHA allows for a more direct examination of potency, since known amounts of compound and trimer can be added together. Examination of various amounts of these compounds for their protection of 6 μM BHA are shown in Fig. 3. Quantitation of the amount of undigested HA1 showed that BMY-27709 protected 50% of the input BHA at a concentration of 55 μM, while BMS-199945 and BMS-201160 exhibited 50% protection at 1.0 and 2.0 μM, respectively. The values for BMS-199945 and BMS-201160 mirror the IC50s obtained in influenza virus-induced RBC hemolysis. Interestingly, the more active BMS-199945 and BMS-201160 are inhibiting at or near stoichiometric levels compared to the amount of BHA added (2 μM trimer).
Inhibition of fusion in the virus-induced RBC hemolysis (29) or inhibition of the conformational change by this class of inhibitor is readily reversible at a neutral pH (Fig. 4). However, under acidic conditions, the BHA-inhibitor complex is much more stable. The BHA-inhibitor complex can be incubated at pH 5.0 for 24 h, with a significant proportion of BHA remaining protected from trypsin treatment. Titration back to a neutral pH produces native HA and again renders the compound readily reversible (unpublished data). This suggests that changes may have occurred in the BHA molecule under acidic conditions to stabilize the binding of inhibitor. One possibility is that protonation of various amino acid residues in or around the binding site allows for additional interactions between the protein and small molecules. In this way, the compounds could be bound more tightly to the protein, retaining the HA in its native conformation at a low pH for long periods. An alternative explanation is that the BHA can undergo a partial structural change in the presence of an inhibitor. In this model, the inhibitor is thought to block the complete change from occurring but in the process becomes trapped in the BHA molecule. By this mechanism, the compound could be inhibiting the complete conformational change by acting as a wedge. This hypothetical partial conformational change would be reversible with the metastable native state.
A practical approach to determine the binding site of this inhibitor class within the HA is that of affinity labeling, especially with the lack of crystallization conditions for the H1 and H2 HAs. Affinity labeling of the HA with a compound that can prevent the acid-induced conformational change would not only identify important amino acids involved in inhibitor binding, but also indicate other residues involved in HA protein function. Hydrophobic photoaffinity labeling has previously been used to successfully determine the transmembrane domain of influenza virus HA (6) and the fusion domain of rabies and vesicular stomatitis virus glycoproteins (7, 18). We anticipated that a photoaffinity analog of one of the fusion inhibitors could be used similarly to map the portions of the hemagglutinin involved in inhibitor binding. The photoaffinity probe, BMS-201160, was made as an analog of BMS-199945, in which the azide moiety replaces the 5-methyl substituent (Fig. 2). Upon treatment with UV light, the azide photolyzes to form a reactive nitrene, which can react with proximal amino acid residues.
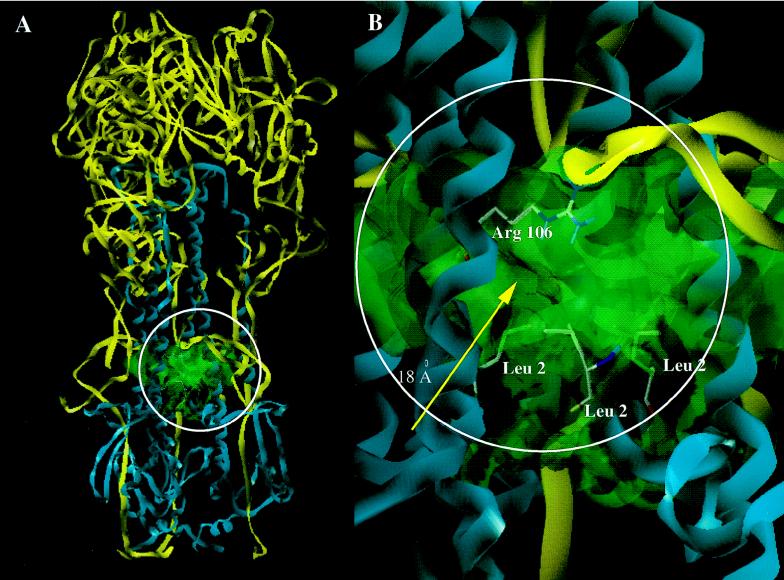
Once BMS-201160 was shown to be able to protect BHA from trypsin digestion after acidification, a tritiated form was synthesized. [3H]BMS-201160 was shown to specifically label the HA2 subunit at a neutral pH (Fig. 6). Labeling of the HA2 subunit could be competed out by the active analog BMS-199945, but not by a related inactive analog, indicating that the radiolabeling of the HA2 subunit is due to specific binding to the trimer. Affinity-labeled HA2 was HPLC purified and then fragmented by different methods. Peptide mapping analysis by SDS-PAGE, using trypsin and Lys-C endoproteinases along with CNBr chemical cleavage, identified the covalent attachment site to be within an HA2 peptide containing amino acids 84 to 106. This was confirmed by direct sequencing of isolated peptides from the Lys-C endoproteinase and CNBr treatments. Sequence cycles were counted for tritium, and the results showed that the major covalent attachment site of the affinity label was a region comprised of Glu-103, Asp-104, and Glu-105. A second close attachment site, with about 50% as much derivatization, was an area encompassing Asp-95, Ala-96, and Glu-97. The residues between these two regions (amino acids 98 through 103) comprise a hydrophobic stretch composed of amino acids LLVLL. Since nitrenes have been shown to be less reactive towards hydrophobic side chains than to more polar acidic side groups or nucleophiles (6, 26), this labeling pattern could reflect interaction of the compound with the entire stretch of amino acids but covalent labeling only on the more reactive side chains on amino acids 95 to 97 and amino acids 103 to 105. This photoaffinity labeled region corresponds to part of the pocket proposed through molecular modeling to constitute the binding site for BMY-27709 (30). The binding model suggests that a single molecule of inhibitor binds per trimer and interacts with the N-terminal portion of HA2. This model was designed to rationalize two experimental results, namely, the location of amino acid residues conferring resistance to inhibitor and the selectivity of these inhibitors for H1 and H2 subtypes of HA to the exclusion of H3. A cavity that extends 18 Å to the center of the HA structure and then bifurcates to the other entrances of the symmetric trimer was originally identified as the potential binding pocket. An additional attractive feature of this crevice is that the amino acid sequence is well conserved in all three subtypes of HA, varying in just two amino acids, 105 and 106 (24, 33). These amino acids are charged (Glu-Arg) in H1 and H2 and are neutral (Gln-His) in H3. However, these two amino acid changes are enough to account for our inhibitors having submicromolar potency against H1 and H2 HAs and yet remain relatively inactive against H3 HA. This is consistent with the hypotheses that the salicylamide moiety of the inhibitor forms a tight bidentate hydrogen bonding interaction with the guanidine of Arg-106 in H1 and H2 (Fig. 10) and that the salicylamide was unable to engage in this interaction with the smaller and much less basic imidazole of His-106 in H3. When affinity labeling experiments are performed with H1 BHA and [3H]BMS-201160 at a neutral pH, Glu-103, Asn-104, and Glu-105 comprise the preferred covalent coupling site. Based upon this model, we would have predicted covalent labeling to a region surrounding amino acid 105. Covalent labeling at amino acids 103 to 105 does support the model. The minor labeling of amino acids 93 to 95 of the HA2 subunit are slightly below the proposed binding site. However, this could be indicative of the flexibility in the binding mode at neutral pH and the readily reversible nature of the binding. However, we cannot rule out the possibility that the two peaks of labeling reflect covalent linkage to two different monomers within a single HA trimer. The area affinity labeled is on a region of α-helix A directly opposing α-helix CD (8), which is known to contain residues which are important for retaining sensitivity to this class of compounds. Resistance-inducing mutations have been mapped to amino acids 47, 50, 51, 52, and 55 of α-helix CD (30). Resistance to these compounds is proposed to result from alteration of the size of the binding pocket through ionic interactions between α-helix CD with α-helix A. Similarly, Staschke et al. (41) found mutations in both A and CD helices that are resistant to methyl-o-methyl-7-ketopodocarpate. Altering the size of the binding pocket would potentially alter the proposed inhibitor interaction with the N terminus of HA2 (30).
FIG. 10.
The proposed binding site of the fusogenic inhibitors. (A) Ribbon trace of the HA trimer highlighting the proposed binding site. HA1 is traced in yellow, while HA2 is displayed in cyan. The binding site is outlined as a van der Waals surface in green. (B) Close-up of region circled in panel A with a van der Waals surface outlining the 18Å cavity. Notice that the fusogenic peptide (shown as the sidechains of Leu-2 from each HA2 monomer) is the base of this cavity while Arg-106 lines the top.
BMS-201160 is able to inhibit the conformational change of the HA trimer at a low pH. In addition, the native structure of the BHA is preserved in the presence of compound for many hours at a low pH. This allows affinity labeling of the BHA to be performed under acidic conditions. Interestingly, under acidic conditions, BMS-201160 was found to specifically cross-link to the amino-terminal Gly-1 of the HA2 subunit, with minor amounts of tritium detected attached to Leu-2. In the binding model, the fusogenic N terminus is located at the center of the protein structure, and one inhibitor molecule is proposed to interact with all the N termini from each monomer, so while there are three potential entrances for the inhibitor, only one molecule of inhibitor could be accommodated at one time in the trimer. The complete change in the major affinity-labeled amino acid under acidic conditions suggests that some degree of structural change in the HA has occurred. Intermediates in the conformational change of HA resulting in partial exposure of the fusion peptide have previously been detected at pH 5.0 with incubation at 4°C (34). It is clear that at pH 5.0 there is a close association of the inhibitor with the N-terminus. We hypothesize that these inhibitors function by acting as a wedge, preventing the fusogenic peptide from slipping past the α-helix region where the drug may be binding. While in this intermediate HA conformation, the inhibitors are trapped within the trimer. Neutralization of the mixture is proposed to relax the BHA trimer back to its native metastable structure. At this point, the inhibitors would again be readily reversible (unpublished data). If correct, successful cocrystallization studies of these inhibitors within the H1 HA trimer at a low pH could provide a snapshot into the mechanism of events leading to the fusion active and final stable forms of the HA trimer.
ACKNOWLEDGMENTS
Work involving the tritiation of BMS-201160 was supported in part by the Biomedical Research Technology Program, National Center for Research Resources, U.S. National Institutes of Health, under grant P41 RR01237, through contract DE-AC03-76SF00098 with the U.S. Department of Energy.
We thank Anne Stanley, Peptide Sequencing Core Facility, Hershey Medical Center, for timely sequence analyses.
REFERENCES
- 1.Aitken A, Geisow M J, Findlay J B C, Holmes C, Yarwood A. Peptide preparation and characterization. Oxford, United Kingdom: IRL Press; 1989. pp. 63–65. [Google Scholar]
- 2.Barrett T, Inglis S C. Growth, purification and titration of influenza viruses. Oxford, United Kingdom: IRL Press; 1985. pp. 119–150. [Google Scholar]
- 3.Blacklow S C, Lu M, Kim P S. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 4.Bodian D L, Yamasaki R B, Buswell R L, Stearns J F, White J M, Kuntz I D. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 5.Brand C M, Skehel J J. Crystalline antigen from the influenza virus envelope. Nat New Biol. 1972;238:145–147. doi: 10.1038/newbio238145a0. [DOI] [PubMed] [Google Scholar]
- 6.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 7.Brunner J, Zugliani C, Mischler R. Fusion activity of influenza virus PR8/34 correlates with a temperature-induced conformational change within the hemagglutinin ectodomain detected by photochemical labeling. Biochemistry. 1991;30:2432–2438. doi: 10.1021/bi00223a019. [DOI] [PubMed] [Google Scholar]
- 8.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 9.Carr C M, Chaundhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr C M, Kim P S. Flu virus invasion: halfway there. Science. 1994;266:234–236. doi: 10.1126/science.7939658. [DOI] [PubMed] [Google Scholar]
- 11.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 12.Charbonneau H. Strategies for obtaining partial amino acid sequence data from small quantities (<5 nmol) of pure or partially purified protein. In: Masudaira P T, editor. A practical guide to protein and peptide purification for microsequencing. San Diego, Calif: Academic Press; 1989. pp. 15–30. [Google Scholar]
- 13.Cianci C, Krystal M. Development of antivirals against influenza. Expert Opin Investig Drugs. 1998;7:1–16. doi: 10.1517/13543784.7.2.149. [DOI] [PubMed] [Google Scholar]
- 14.Daniels R S, Douglas A R, Skehel J J, Wiley D C. Analyses of the antigenicity of influenza haemagglutinin at the pH optimum for virus-mediated membrane fusion. J Gen Virol. 1983;64:1657–1662. doi: 10.1099/0022-1317-64-8-1657. [DOI] [PubMed] [Google Scholar]
- 15.Daniels R S, Downie J C, Hay A J, Knossow M, Skehel J J, Wang M L, Wiley D C. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 16.Dischino, D. 1998. Unpublished data.
- 17.Doms R W, Helenius A H, White J M. Membrane fusion activity of the influenza virus hemagglutinin: the low pH conformational change. J Biol Chem. 1985;260:2973–2981. [PubMed] [Google Scholar]
- 18.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 19.Durrer P, Gaudin Y, Ruigrok R W H, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis virus. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M B, Sabesan S, Skehel J J, Wiley D C. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 21.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 22.Gaudin Y, Ruigrok R W H, Brunner J. Low-pH induced conformational changes in viral fusion proteins: implications for the fusion mechanism. J Gen Virol. 1995;76:1541–1556. doi: 10.1099/0022-1317-76-7-1541. [DOI] [PubMed] [Google Scholar]
- 23.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 24.Hiti A L, Davis A R, Nayak D R. Complete sequence analysis shows that the hemagglutinins of the H1 and H2 subtypes of human influenza virus are closely related. Virology. 1981;111:113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman L R, Kuntz I D, White J M. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotzyba-Hibert F, Kapfer I, Goeldner M. Recent trends in photoaffinity labeling. Angew Chem Int Ed Engl. 1995;34:1296–1312. [Google Scholar]
- 27.Lazarowitz S G, Choppin P W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 28.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 29.Luo G X, Colonno R, Krystal M. Characterization of a hemagglutinin-specific inhibitor of influenza A virus. Virology. 1996;226:66–76. doi: 10.1006/viro.1996.0628. [DOI] [PubMed] [Google Scholar]
- 30.Luo G X, Torri A, Harte W E, Danetz S, Cianci C, Tiley L, Day S, Mullaney D, Yu K L, Ouellet C, Dextraze P, Meanwell N, Colonno R, Krystal M. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol. 1997;71:4062–4070. doi: 10.1128/jvi.71.5.4062-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meanwell N A, Krystal M. Taking aim at a moving target: inhibitors of influenza virus. I. Virus adsorption, entry and uncoating. Drug Discov Today. 1996;1:316–324. [Google Scholar]
- 32.Meanwell N A, Krystal M. Taking aim at a moving target: inhibitors of influenza virus. II. Viral replication, packaging and release. Drug Discov Today. 1996;1:388–397. [Google Scholar]
- 33.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 34.Pak C C, Krumbiegel M, Blumenthal R. Intermediates in influenza virus PR/8 haemagglutinin-induced membrane fusion. J Gen Virol. 1994;75:395–399. doi: 10.1099/0022-1317-75-2-395. [DOI] [PubMed] [Google Scholar]
- 35.Plotch S J, O’Hara B, Morin J, Palant O, LaRocque J, Bloom J D, Lang S A, Jr, DiGrandi M J, Bradley M, Nilakantan R, Gluzman Y. Inhibition of influenza A virus replication by compounds interfering with the fusogenic function of the viral hemagglutinin. J Virol. 1999;73:140–151. doi: 10.1128/jvi.73.1.140-151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth M G, Gething M-J, Sambrook J. Membrane insertion and intracellular transport of influenza virus glycoproteins. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 219–267. [Google Scholar]
- 37.Ruigrok R W H, Aitken A, Calder L J, Martin S R, Skehel J J, Wharton S A, Weiss W, Wiley D C. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J Gen Virol. 1988;69:2785–2795. doi: 10.1099/0022-1317-69-11-2785. [DOI] [PubMed] [Google Scholar]
- 38.Sauter N K, Glick G D, Crowther R L, Park S-J, Eisen M B, Skehel J J, Knowles J R, Wiley D C. Crystallographic detection of a second ligand binding site in influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1989;89:324–328. doi: 10.1073/pnas.89.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz M A. A 125I-radiolabel transfer crosslinking reagent with a novel cleavable group. Anal Biochem. 1985;149:142–152. doi: 10.1016/0003-2697(85)90487-7. [DOI] [PubMed] [Google Scholar]
- 40.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staschke K A, Hatch S D, Tang J C, Hornback W J, Munroe J E, Colacino J M, Muesing M A. Inhibition of influenza virus hemagglutinin-mediated membrane fusion by a compound related to podocarpic acid. Virology. 1998;248:264–278. doi: 10.1006/viro.1998.9273. [DOI] [PubMed] [Google Scholar]
- 42.Stegmann T, Helenius A. Influenza virus fusion: from models toward a mechanism. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 89–111. [Google Scholar]
- 43.Steinhauer D A, Sauter N K, Skehel J J, Wiley D C. Receptor binding and cell entry by influenza viruses. Semin Virol. 1992;3:91–100. [Google Scholar]
- 44.Takemoto D K, Skehel J J, Wiley D C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology. 1996;217:452–458. doi: 10.1006/viro.1996.0139. [DOI] [PubMed] [Google Scholar]
- 45.Watowich S J, Skehel J J, Wiley D C. Crystal structures of influenza virus hemagglutinin in complex with high-affinity receptor analogs. Structure. 1994;2:719–731. doi: 10.1016/s0969-2126(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 46.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;332:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 47.Weissenhorn W, Calder L J, Dessen A, Laue T, Skehel J J, Wiley D C. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 49.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 50.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 51.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 52.Wilson I A, Skehel J J, Wiley D C. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]