Abstract
Domestic cattle have spread across the globe and inhabit variable and unpredictable environments. They have been exposed to a plethora of selective pressures and have adapted to a variety of local ecological and management conditions, including UV exposure, diseases, and stall-feeding systems. These selective pressures have resulted in unique and important phenotypic and genetic differences among modern cattle breeds/populations. Ongoing efforts to sequence the genomes of local and commercial cattle breeds/populations, along with the growing availability of ancient bovid DNA data, have significantly advanced our understanding of the genomic architecture, recent evolution of complex traits, common diseases, and local adaptation in cattle. Here, we review the origin and spread of domestic cattle and illustrate the environmental adaptations of local cattle breeds/populations.
Keywords: Cattle, Origin, Domestication, Migration route, Environmental adaptation, Selective pressure
Introduction
Domestic cattle are descended from the aurochs (Bos primigenius) (Ajmone-Marsan et al. 2010), which were widely distributed in Europe, Asia and northern Africa during the Holocene and went extinct in 1624 (Felius et al. 2014). Modern cattle were estimated have been domesticated ~ 10,000 years before present (BP) in Southwest Asia and ~ 8,000 years BP in South Asia (Larson et al. 2014; Pitt et al. 2019).
At present, approximately 1.5 billion cattle are kept on all inhabited continents, in a variety of climatic zones under diversified conditions (www.fao.org/faostat/en/). Domestic cattle are divided into humpless taurine cattle (Bos taurus taurus) and humped indicine/zebu cattle (Bos taurus indicus), local populations of which have undergone continuous admixture with other bovine species (Chen et al. 2018a; Chen and Lei 2021; Wu et al. 2018). Taurine cattle are largely confined to temperate and cold climates and are widely distributed in the Northern Hemisphere; some breeds are distributed in tropical Africa and America. Indicine cattle are found in southern Asia, Africa, northern Australia, the southern US, and Latin America (Utsunomiya et al. 2019; Zhang et al. 2020). Indicine cattle differ from taurine cattle in various ways: they exhibit a muscular fatty hump of variable size on their shoulders, a larger dewlap, drooping ears, and a tolerance of semiarid and tropical environments. Indicine cattle have a lower basal metabolic rate, water, and nutrient requirements. Moreover, they are generally more resistant to ticks and intestinal parasites than taurine cattle (Utsunomiya et al. 2019).
Global dispersal of cattle
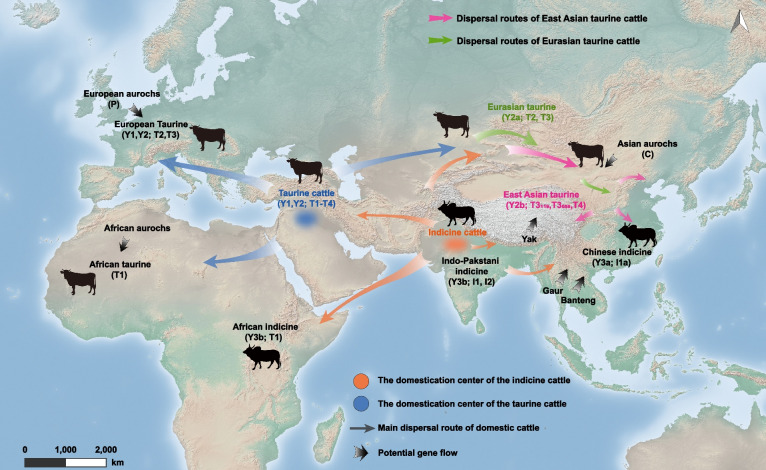
Over the past 10,000 years, cattle domestication has been followed by several major migrations, leading to their presence on all inhabited continents (Felius et al. 2014). Environmental conditions of heat or cold, high altitudes or lowlands, and arid zones or humid tropical environments have contributed to the many adaptations of cattle and pronounced genomic diversity among breeds/populations (Fig. 1).
Fig. 1.
Domestication and main migration routes of Bos taurus and Bos indicus. Paternal (Y) and maternal (taurine, T; indicine, I; and aurochs, P and C) haplogroups are shown in parentheses, separated by a semicolon
Taurine cattle were introduced from the Middle East into Africa 6,800 years ago (Felius et al. 2014) and probably influenced by local aurochs, which likely contributed to the ancestry of modern African taurine cattle (Pitt et al. 2019; Verdugo et al. 2019). The importation of male indicine cattle to Africa was initiated as early as 4,000 years ago and intensified at AD 700 (Ajmone-Marsan et al. 2010). At the end of the nineteenth century, cattle of the original taurine ancestry were largely replaced by indicine cattle, which were less vulnerable to devastating rinderpest epidemics (Felius et al. 2014). A major taurine-indicine admixture event was dated to approximately 750–1,050 years (approximately 150 generations) ago (Kim et al. 2020). This male-mediated indicine introgression into local taurine herds generated African indicine populations with a variable taurine/indicine genomic composition that are better adapted to dry climates. Over time, there was a stepwise transition from taurine to indicine diversity in Africa.
Approximately 4,200 years ago, climate change caused a male-mediated westward migration of indicine cattle from the Indus Valley to the Near East, which resulted in a change in genetic composition as cattle adapted to a dry climate (Verdugo et al. 2019). The eastward migration of taurine cattle reached the northern part of East Asia between 5,000 and 4,000 years ago, accompanied by the rapid adaptive evolution of ancestral taurine cattle to extremely low temperatures, as found in Siberia and the Qinghai-Tibetan Plateau (Felius et al. 2014). Around 3,000 years ago, indicine cattle migrated to Indochina and southern China. Following the contact of indicine cattle with early imports of taurine cattle, a north-to-south taurine-to-indicine cline was established at both the phenotypic and genomic levels in China (Gao et al. 2017). Intermediate taurine-indicine populations exhibited various combinations of taurine and indicine ancestries. The importation of indicine cattle to Southeast Asia likely began 1,500 years ago. Cattle were imported into North and South America from Europe and Africa since 1492 (Ajmone-Marsan et al. 2010; Felius et al. 2014). During their continuous dispersal in the tropical zones of Asia, Africa, and America, indicine cattle encountered southwestern and eastern Asian, African, and American taurine populations, respectively (Chen et al. 2018a; Utsunomiya et al. 2019), driving the emergence of several hybrid populations. Both southwestern Asia and central China are now characterized as typical taurine–indicine transition zones.
Molecular evidence of uniparental and autosomal markers has confirmed that taurine and indicine cattle are derived from two geographically separated and genetically differentiated aurochs progenitors from West and South Asia, respectively. Among modern cattle, there are seven major mitochondrial haplogroups (taurine T1, T2, T3, T4, and T5 as well as indicine I1 and I2) (Chen et al. 2010; Lenstra et al. 2014; Xia et al. 2019a); the rare mitochondrial haplogroups E, R, P, Q and C, supporting sporadic aurochs introgressions (Zhang et al. 2020; Xia et al. 2021; Cubric-Curik et al. 2022); five Y chromosome haplogroups (taurine Y1, Y2a, and Y2b as well as indicine Y3a and Y3b) (Xia et al. 2019b; Cao et al. 2019; Edwards et al. 2011; Pérez-Pardal et al. 2018); and at least eight major autosomal ancestral groups (Chen et al. 2018a, 2020) as follows: (1) African taurine cattle living in humid and sub-humid, tsetse fly-infested, tropical environments in West Africa (Gautier et al. 2009; Kim et al. 2017); (2) East Asian taurine cattle in Northeast Asia and the Qinghai-Tibetan Plateau, which are adapted to extremely cold and hypoxic environments, and some of them carry alleles arising from yak introgression (Chen et al. 2018a; Wu et al. 2018); (3) Eurasian taurine cattle in semiarid regions in Central Asia (Chen et al. 2021; Kantanen et al. 2009); (4) European taurine cattle inhabiting temperate climates that carry alleles arising through admixture with European aurochs and are the ancestors of most globalized industrial breeds (Achilli et al. 2008; Daetwyler et al. 2014; Park et al. 2015); (5) Indian-Pakistani indicine cattle in hot and semiarid regions (Chen et al. 2010); (6) African indicine cattle in semiarid East and Central Africa with a mixed ancestry of African taurine and South Asian indicine breeds (Bahbahani et al. 2017; Kim et al. 2017, 2020); (7) Diversified East Asian indicine cattle that inhabit hot-humid environments and carry alleles from other wild and/or domestic Asian bovine species (Chen et al. 2018a; Sinding et al. 2021); and (8) Indonesian breeds in hot-humid environments, which show a mix of indicine, banteng and/or Bali cattle ancestries (Mohamad et al. 2009; Sudrajad et al. 2020). For a more detailed and comprehensive classification of modern cattle, see Felius et al. (2011).
Adaptation to a cold environment
Cold climates are likely to affect the phenotypic characteristics and metabolism of cattle. Northern Fennoscandia and the Republic of Sakha, Russia, represent the northernmost regions inhabited by humans and are home to cattle breeds adapted to extremely cold environments (Weldenegodguad et al. 2018) (Fig. 2). For example, the Yakut cattle can be found above the Arctic Circle, and they have adapted to extremely cold winters (-50 °C). Recent genome-wide scans found that all Yakut cattle carry a breed-specific missense mutation in an evolutionarily conserved NRAP gene involved in heart function (Buggiotti et al. 2021) (Table 1). This change is shared by most hibernating mammals but absent from many mammalian species and other modern and ancient cattle breeds and bovine species. NRAP encodes the nebulin-related-anchoring protein enabling actin filament-binding activity and is abundantly expressed in striated and cardiac muscles involved in myofibrillar assembly and force transmission in the heart (Truszkowska et al. 2017). Thus, this young convergent NRAP amino acid change in Yakut cattle suggests that they may slow down their metabolism but enhance their heart function to supply blood during the winter periods (Buggiotti et al. 2021).
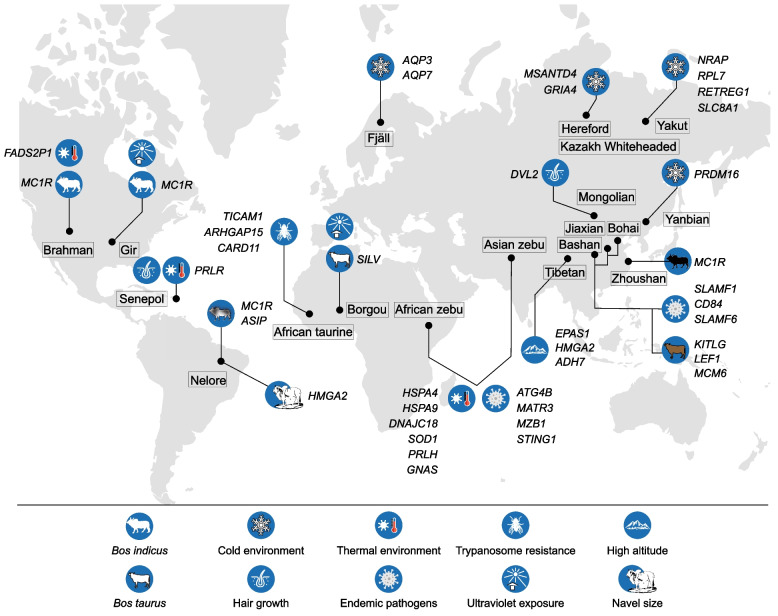
Fig. 2.
Examples of local adaptations in cattle breeds/populations, each labelled with the candidate gene or genes under selection
Table 1.
Overview of known genes under local adaptation in cattle populations
| Category | Breed/population | Method | Gene | Refs |
|---|---|---|---|---|
| Cold adaptability | Yakut cattle | RFMix, allele frequency | NRAP | (Buggiotti et al. 2021) |
| Cold adaptability | Russian native cattle | DCMS and hapFLK | RETREG1 | (Yurchenko et al. 2018) |
| Cold adaptability |
Hereford and Kazakh Whiteheaded cattle bred in Siberia |
GWAS and DCMS | MSANTD4, GRIA4 | (Igoshin et al. 2019) |
| Cold adaptability | Fjäll cattle | DCMS | AQP3, AQP7 | (Ghoreishifar et al. 2020) |
| Cold adaptability | Yanbian and Mongolian cattle | FST, Pi, Tajima's D | PRDM16 | (Yan et al. 2022) |
| Navel size | Nellore cattle | GWAS | HMGA2 | (Aguiar et al. 2018) |
| Heat adaptability | African and Asian zebus | LOTER, iHS, FST, XP-EHH, XP-CLR, Hp | HSPA4, HSPA9, DNAJC18, SOD1 | (Bahbahani et al. 2017; Kim et al. 2017, 2020) |
| Slick-hair phenotype and Thermoregulation | Senepol cattle | Association analysis | PRLR | (Littlejohn et al. 2014) |
| Thermoregulation | African zebu | XP-EHH | PRLH | (Kim et al. 2017) |
| Water reabsorption | African humped cattle | LOTER, iHS, FST | GNAS | (Kim et al. 2020) |
| Heat adaptability | Brahman | FST | FADS2P1 | (Low et al. 2020) |
| Adaptation to endemic pathogens | African taurine cattle | PBS | TICAM1, ARHGAP15, CARD11 | (Kim et al. 2020; Noyes et al. 2011) |
| Adaptation to endemic pathogens | African and Asian zebus | LOTER, iHS, FST | ATG4B, MATR3, MZB1, STING1 | (Kim et al. 2020) |
| Light coat color | Brahman, Nellore, and Gir | GWAS, di, Tajima’s D | MC1R, ASIP | (Mei et al. 2018; Trigo et al. 2021; Xu et al. 2015) |
| White to cream coat | Borgou cattle | iHS and Rsb | SILV | (Flori et al. 2014) |
| Dark brown coat | Chinese cattle | XtX statistics | KITLG, LEF1, MCM6 | (Gao et al. 2017) |
| Black coat | Zhoushan cattle | FST | MC1R | (Jiang et al. 2021) |
| Hypoxia adaptation | Tibetan cattle | FST and XP-EHH | EPAS1 | (Wu et al. 2020) |
| Short stature | Tibetan cattle | FST and XP-EHH | HMGA2, ADH7 | (Wu et al. 2020) |
| Adaptive immune responses | Jiaxian Red, Bashan, and Bohai Black cattle | FST and XP-EHH | SLAMF1, CD84, SLAMF6 | (Ma et al. 2022; Sun et al. 2021; Xia et al. 2021) |
| Hair growth | Mongolian cattle | FST and θπ ratio | DVL2 | (Mei et al. 2021) |
Previous studies revealed several candidate genes that might be related to cold acclimation, including RETREG1, RPL7, and SLC8A1, in Yakut cattle of Russian (Weldenegodguad et al. 2018; Yurchenko et al. 2018). Among them, RETREG1 (also known as FAM134B) is related to the impairment of pain and temperature sensation in humans (Kurth et al. 2009). RPL7 encodes a component of 60S ribosomal subunit and showed a fourfold up-regulation in the skin of freeze tolerant frogs (Wu et al. 2018). SLC8A1 encodes for solute carrier family 8 member A1 that is predominantly expressed in human heart and its mutations was found to be significantly associated with blood pressure rising during salt load (Liu et al. 2018). Another study on cold adaptation of Hereford and the Kazakh Whiteheaded cattle bred in Siberia for several decades identified a single candidate genomic region by both genome-wide association study (GWAS) and scan for selective sweeps, where MSANTD4 and GRIA4 were annotated. It was believed that both genes contribute to the cold-stress resistance phenotype due to their indirect involvement in the cold shock response (MSANTD4) and body thermoregulation (GRIA4) (Igoshin et al. 2019).
In northern Swedish cattle (Fjäll or Swedish mountain cattle) that also live in areas with a subarctic cold climate, signatures of positive selection were found near AQP3 and AQP7 (Ghoreishifar et al. 2020). AQP3 and AQP7 are mapped at the same chromosomal location as an aquaporin cluster and they encode the water channel protein aquaporin 3 and a member of the aquaporin family of water-selective membrane channels, respectively. AQP7 facilitates water, glycerol, and urea transport, and thus may play an important role in thermoregulation in the form of perspiration. Mongolian and Yanbian cattle in northern China with an annual average temperature of 2–6 °C harbor a substitution in PRDM16 (p.P779L), which maintains brown adipocyte formation by boosting thermogenesis-related gene expression, indicating its vital role in cold tolerance (Yan et al. 2022).In northern Chinese cattle, LCORL has also been identified as a candidate gene for larger body size and greater height that may reflect local environmental effects (Lango Allen et al. 2010).
Adaptation to tropical regions
Temperature increases lead to changes in forage quality and exacerbate livestock susceptibility to pests and diseases (Berman 2011). These factors cause physiological or behavioral changes in livestock, driving their adaptation to high-temperature environments. The body temperature is coordinated and controlled by the balance between metabolic heat production and loss (Bernabucci et al. 2014). Heat stress occurs when animals are exposed to high temperatures and cannot dissipate sufficient endogenous heat in time (Koch et al. 2019). Heat stress directly affects the food intake, growth, milk yield, and health conditions of domestic animals (e.g., heat shock), resulting in losses to production (Silva et al. 2021). Therefore, the study of thermal adaptation in cattle has become an important topic of research. Indicine cattle have several phenotypes that reflect adaptations to tropical environments, including their hump, large ears, and excess skin.
The hump, large ears and excess skin of indicine cattle
The hump is a muscular structure located above the withers on the dorsal region of the thoracic cage of indicine cattle; it is more prominent in males than females. The biological importance of the indicine hump remains unclear. Its abundant fat has inspired speculations regarding a role in energy storage in times of starvation, whereas the biomechanical relevance of the hump to the stabilization of the scapula suggests artificial selection for animal performance during draft (Utsunomiya et al. 2019). To date, no specific genomic region or gene has been implicated in the development of the hump.
Most indicine cattle carry large ears that are either spear tip-shaped or pendulous. Excess skin is usually present across the entire ventral midline, especially around the neck (throatlatch), chest (dewlap), and navel. A recent haplotype-based GWAS revealed that navel size was strongly associated with copy number variation at intron 3 of the high-mobility group AT-hook 2 gene (HMGA2) (Aguiar et al. 2018). The available evidence is not yet conclusive but suggests that structural variants (SVs) of HMGA2 may explain the excess skin and ear morphology of indicine cattle, similar to findings in pigs (Li et al. 2012) and dogs (Boyko et al. 2010).
Skin morphology of indicine cattle
Cutaneous evaporation is the main avenue by which cattle dissipate heat, with the involvement of sweat glands and other skin components. A comparison of the skin morphology revealed denser, larger, and baggier sweat glands in indicine cattle with smaller capillary surface areas of hair follicles than those in taurine cattle, whereas the differences in skin morphology in their crossbreds correlated with the proportion of taurine ancestry (Jian et al. 2014). Likely the combination of large ears and excess skin with high density and effective sweat glands provides a smart, adaptive tolerance of indicine cattle to heat stress.
Positively selected genes associated with thermotolerance
Heat tolerance is a well-known characteristic of indicine cattle (Hansen 2004) and a prerequisite for indicine survival in hot climates. Genomic selection studies on African and Asian indicine cattle have identified a large number of candidate genes associated with heat tolerance. A subset of these genes (such as HSPA4, HSPA9, DNAJC18, and SOD1) are under selection (Bahbahani et al. 2017; Kim et al. 2017, 2020).
The prolactin signaling pathway not only is involved in lactation but also affects the hair morphology and thermoregulatory phenotype of cattle. Mutations in the 11th exon of the prolactin receptor (PRLR) have been shown to have a major effect on the slick-hair phenotype of cattle (Flórez Murillo et al. 2021; Littlejohn et al. 2014; Porto-Neto et al. 2018). The first slick mutation was found in the Senepol cattle, a tropically adapted breed of mostly European cattle descent, resulting in truncation of the C-terminal region of the protein involved in STAT5 activation during prolactin signaling (Littlejohn et al. 2014). Cattle with an extremely short and slick-hair coat show strong thermotolerance to withstand hot weather (Dikmen et al. 2014; Olson et al. 2003). Interestingly, the analysis of African cattle genomes also revealed a significant selective signal in prolactin releasing hormone gene PRLH, of which a missense mutation (p. Arg76His) in its exon 2 was highly conserved in African indicine cattle (73%) but absent in commercial taurine breeds, indicating its selective advantage by regulating prolactin expression relevant to thermotolerance in African indicine cattle (Kim et al. 2017).
Heat stress increases sympathetic nerve activity in kidneys, muscle, and skin (Rowell 1990). A genomic region with the access of indicine ancestry (92.44%) was found on Bos taurus chromosome (BTA) 13:57.15–57.65 Mb (Bahbahani et al. 2017; Kim et al. 2017, 2020), where GNAS complex locus gene is annotated. This gene is related to water reabsorption through mediating the antidiuretic hormone arginine vasopressin in aquaporin-2 water channels and subsequently contributing to the water conservation pathway of kidney (Boone and Deen 2008). This finding suggested that this specific indicine haplotype contributes to the local adaptation of African humped cattle to arid climate (Kim et al. 2020).
Adaptation to endemic pathogens
Pathogenic burden is an important driver of adaptation to tropical environments in cattle. Bovine trypanosomiasis, a vector-borne parasitic infection caused by Trypanosoma spp., has long been a constraint on cattle farming in sub-Saharan Africa. Some African taurine cattle such as N'Dama, can withstand infection by Trypanosoma congolense. Genetic analysis has revealed selective sweeps at TICAM1 and ARHGAP15 loci in African taurine cattle, which were linked to previously identified quantitative trait loci (Noyes et al. 2011). Another selection signature in African taurine cattle is located in the upstream of CARD11 (Kim et al. 2020), which is essential for the signaling of T and B cells in the innate and adaptive immune systems (Hara et al. 2003; Pomerantz et al. 2002). Furthermore, CARD11 has been found to be differentially expressed between the trypanotolerant (N’Dama) and the trypanosusceptible (Boran) breeds (Noyes et al. 2011).
Candidate selective loci on BTA7 (MATR3, MZB1, and STING1) and BTA3 (ATG4B) with the excess of indicine ancestry were identified in both African humped as well as Asian and American-Australian indicine cattle, suggesting their possible contribution to genetic resistance to ticks and tick-borne diseases such as East Coast fever (Kim et al. 2020). STING1 regulates the production of intracellular DNA-mediated type I interferon and is thus essential for host defense against DNA pathogens (Ishikawa et al. 2009).
Adaptation to ultraviolet exposure
Coat color variation may contribute to the adaptation of cattle to tropical/subtropical or high-latitude environments. Many indicine breeds, such as Nellore, Tharparkar, Bhagnari, Dajal, Hariana, and Guzerat, have light colors that can reflect a large proportion of incident solar radiation (Hansen 2004). The mixture of short, thick, densely arranged white/gray and dark hairs that cover the black skin of indicine cattle provides reflectance at short light wavelengths. Other studies have shown positive selection for the melanocortin 1 receptor (MC1R) gene in indicine cattle (Brahman, Nellore, and Gir), implying that their light coat color has played an important role in adaptation to tropical environments (Mei et al. 2018; Xu et al. 2015). The uniform white to cream coat of Borgou cattle is likely the result of artificial selection on the candidate gene SILV (Flori et al. 2014). This gene encodes a type I integral membrane protein in the premelanosome matrix (PMEL17), which is essential for melanosome development and is responsible for lightening or diluting the base color defined by the MC1R in some cattle breeds (Kühn and Weikard 2007; Schmutz and Dreger 2013).
The Nellore breed has been strongly selected for white coat, but bulls of this breed generally exhibit darker hair, ranging from light gray to black, on the head, neck, hump, and knees. GWAS has shown that this darkness is associated with a deletion of 1,155 bp followed by a small SINE-1 insertion (more than 150 bp) between the 1B and 1C noncoding exons of ASIP (Trigo et al. 2021). ASIP plays a crucial role in decreasing eumelanin and increasing pheomelanin production by blocking MC1R (Barsh et al. 2000; Cieslak et al. 2011). Thus, this SV of ASIP may cause darker coat pigmentation on specific parts of the body by decreasing the expression of ASIP and consequently increasing the production of eumelanin.
The common denotation of yellow cattle for all indigenous Chinese cattle refers to its predominant light to dark brown color. Using a whole-genome scan for genetic differentiation and association analyses with both environmental and morphological covariables, several coat color and pigmentation genes (KITLG, LEF1, and MCM6) were identified in Chinese cattle and considered to be involved in UV protection (Gao et al. 2017). Black Angus and Wagyu are typical black-coat taurine cattle breeds, while Zhoushan is an endangered black-coat indicine breed in southern China. The identification of a shared genomic region between Zhoushan and Angus cattle shows that the dark coat color of Zhoushan cattle may be related to the p.F195L mutation in MC1R (Jiang et al. 2021).
Although many genetic variants associated with cattle coat color have been identified (Mei et al. 2018; Trigo et al. 2021), little is known about the genetic basis of light coat color in indicine cattle in Asia and Africa. Indeed, coat color in Asian and African indicine cattle exhibits high variability, and the genetic basis of adaptations involving coat color requires further study.
Adaptation to high altitude
High altitudes (> 2,500 m above sea level) in regions, such as the Qinghai-Tibetan Plateau, the Rocky Mountains of the USA, and the Simien Mountains Plateau of Ethiopia, can cause hypoxia due to an insufficient supply of oxygen to vital organs. However, cattle populations have thrived in these regions for thousands of years as a result of various physiological adaptations to hypoxic environments (Newman et al. 2015; Wang et al. 2021; Wu et al. 2020).
EPAS1 encodes a subunit of the HIF transcription factor and is a key gene for hypoxia adaptation in Tibetans (Beall et al. 2010; Simonson et al. 2010; Yi et al. 2010). Recently, this gene was found to have evolved under positive selection in Tibetan cattle (top 5% ranking), corroborating the previously reported convergence of genetic adaptation to high altitude in dogs and humans (Wu et al. 2020). Some highly differentiated nonsynonymous SNPs were found in EPAS1 of Tibetan cattle, which likely contribute to their local adaptation (Wu et al. 2020).
Similarly, a strong high association of double variants in the oxygen degradation domain of EPAS1 has been found in Angus cattle in the Rocky Mountains, where they suffer from high-altitude pulmonary hypertension (HAPH) (Newman et al. 2015). These variants likely represent gain-of-function mutations that are prevalent in Angus cattle found at low altitude, but may be pathogenic under hypoxic conditions at high altitudes (Newman et al. 2015).
Short stature in adult Tibetan cattle, which have an average height of less than 110 cm, is another distinctive phenotype thought to contribute to their adaptation to the Qinghai-Tibetan Plateau. HMGA2 has been identified as a candidate gene associated with the high-altitude adaptation of humans and domestic animals (Kader et al. 2015; Weedon et al. 2008, 2007). This gene was found to be positively selected for in Tibetan cattle (Wu et al. 2020). Notably, a nonsynonymous SNP (p.A64P in HMGA2) has a higher frequency in Tibetan cattle than in other cattle populations. ADH7, another gene that is associated with short stature in Tibetan cattle (Wu et al. 2020), has undergone positive selection in Tibetan cattle and is correlated with human weight and the body mass index (Weedon et al. 2008).
Potentially selected genes related to environmental adaptation in other local cattle populations
Adaptation to local climate conditions is also being studied in other cattle populations. Chinese cattle still serve as a major labor force in agricultural production and are well known for their endurance and adaptive ability (Randhawa et al. 2016).
Genomic selection signatures of cattle breed in northern (Mongolian cattle) and southern China (Minnan cattle) identified several adaptive genes related to local environmental challenges, such as DVL2, HSPA4, and CDHR4 (Mei et al. 2021). DVL2 plays an important role in limiting hair growth and links to the hair follicle cycle (Gutiérrez-Gil et al. 2017). The missense mutations in DVL2 show significant north–south population stratification and might impact both fur growth in different cattle breeds and adaptation to different climates (Mei et al. 2021). A region on BTA3 that includes the signaling lymphocytic activation molecule family (SLAMF) genes SLAMF1, CD84, and SLAMF6 might be associated with high disease resistance in native Chinese cattle breeds, such as Jiaxian Red, Bashan, and Bohai Black cattle (Ma et al. 2022; Sun et al. 2021; Xia et al. 2021). SLAMF receptors are involved in the regulation and interconnection of innate and adaptive immune responses.
Adaptive introgression
Yak introgression into Tibetan taurine cattle
Yak (Bos grunniens) are thought to have inhabited the Tibetan Plateau for millions of years and have high altitude adaptations, such as enlarged lungs and hearts. In contrast, domestic taurine cattle were introduced to the Tibetan Plateau by humans only a few thousand years ago (Chen et al. 2015). Taurine cattle that were not adapted to the Tibetan Plateau suffered from severe pulmonary hypertension in the early period (Will et al. 1975). Yak introgression into Tibetan cattle genomes partially facilitated cattle adaptation to high altitude (Chen et al. 2018a; Wu et al. 2018). Several adaptive introgressed genes have been identified that are related to the response-to-hypoxia pathway (for example, COPS5, IL1A, IL1B, MMP3, EGLN1, EGLN2, HIF3α, RYR2, and SDHD).
Introgression of banteng-like sequences into East Asian indicine cattle
East Asian indicine cattle are unique in caring for significant exotic ancestry, which is related to and can be modeled as gene flow from banteng (Bos javanicus) (Chen et al. 2018a, 2018b). However, while the banteng provides a good genetic match, it is not the precise source of East Asian indicine cattle (Sinding et al. 2021), indicating that more wild Bos diversity needs to be sequenced to fully explain their evolution. Nevertheless, using the Javan banteng as a reference is informative, and it has been inferred that between 2.38 and 3.84% of the southeastern Chinese indicine genome is of banteng ancestry (Chen et al. 2018a). Analysis of nonsynonymous substitutions in the introgression region led to the identification of the genes, such as T2R12, TAS2R9, and TAS2R6. These are homologues of bitter taste receptors in humans and giant pandas (Meyerhof et al. 2010; Zhao et al. 2013) may serve similar functions in East Asian indicine cattle. Although a clear correlation between bitterness and toxicity has not been established, it is generally believed that this taste ability prevents mammals from intoxication by avoiding ingestion of potentially harmful food constituents (Meyerhof et al. 2010). In addition, several introgressed genes conducive to the local adaptation of East Asian indicine cattle to the hot and humid tropical climate have been detected (Chen et al. 2018a). For example, several heat-shock protein (HSP) genes, including HSPA1A, HSPB8, HSPA8, HSPA4, HSPB2 and HSF2, are involved in key cellular defense mechanisms during exposure to hot environments. Genes related to hair cell differentiation and blood circulation, such as ATOH, GNA14, VPS13 and KIF2B, also play important roles in temperature adaptation (Chen et al. 2018a).
Introgression of banteng segmenst into Indonesian indicine cattle
Indonesia is home to Bali cattle, a local domesticated version of the Javan banteng, either or both of which are admixed into local Indonesian indicine breeds such as Galekan and Madura (Mohamad et al. 2009; Sudrajad et al. 2020), to the extent that this admixture likely has adaptive implications, hopefully future research will clarify this interesting question.
New approaches to exploring genetic adaptation
Unearthing cattle adaptation with ancient DNA (aDNA)
aDNA sequencing provides a historical record of genomic variation. It provides new possibilities for identifying introgressions of wild stock and for studying domestication at the genomic level. Comparisons of early domestic cattle genomes in the Fertile Crescent to the genomes of their aurochs progenitors revealed diverse origins with separate introgressions of wild stock, such as British and Moroccan aurochs introgression into Neolithic Balkans and Levantine cattle, indicating that genetic exchange among early domestic groups and wild progenitors widely contributed to the development of domestic cattle (Verdugo et al. 2019). A genome comparison of British aurochs with modern European cattle revealed a number of genes associated with neurobiology, growth, metabolism, and immunobiology that show evidence of having undergone positive selection within the time since cattle domestication ~ 10,000 years ago. The analysis further showed significant introgression from the British aurochs into modern British and Irish cattle (Park et al. 2015). The environmental differences between Europe and Southwest Asia where cattle originated are notable. It is likely that the adaptation of European Neolithic cattle to a cold and wet Europe involved local aurochs introgression. Mitochondrial data and archaeological evidence revealed that East Asian aurochs, which belonged to the C haplogroup, were distributed in northern China during the Holocene and overlapped with early domestic cattle for millennia, possibly also contributing to the formation of modern East Asian cattle, a possibility that future research will hopefully clarify (Brunson et al. 2016; Cai et al. 2018; Zhang et al. 2013). Together, these findings suggest that additional aurochs populations have contributed to local cattle, which calls for future research into aurochs genomics. Furthermore, Verdugo et al. (2019) identified rapid and widespread introgression of indicine cattle in southwest and central Asia ~ 4,200 years ago, which was likely associated with climate change that led to the preferential use of arid-adapted indicine bulls for breeding. aDNA therefore has incredible potential to reveal not only aurochs evolution and the origin of cattle but also the transitions and breeding transformation of cattle, and much history has yet to be explored.
Adaptive potential arising from structural variation (SV)
Compared to smaller variants, SVs of at least 50 base pairs (bp) in length often have more extreme consequences (Chiang et al. 2017) and thus may have made substantial contributions to cattle adaptive evolution. For example, an additional copy of FADS2P1 has been under positive selection in Brahman cattle (Low et al. 2020). It is a pleiotropic gene involved in the biosynthesis of unsaturated fatty acids, lipid homeostasis, inflammatory response, and promotion of myocyte growth and cell signaling. Its additional copy in indicine cattle may very well modulate water permeability and heat loss from skin by regulating the composition of fatty acids in the cell membranes (Low et al. 2020). However, SVs are extremely challenging to detect via short-read sequencing technologies. An accurate identification and characterization of SVs on the genome requires long-read sequencing technologies, novel computational approaches (Ho et al. 2020), and the availability of pangenome that represents the genome variations of a wide panel of cattle (Crysnanto et al. 2021; Leonard et al. 2022; Talenti et al. 2022; Zhou et al. 2022; Gong et al. 2022).
Integration of phenotypic, genetic, and functional data into adaptation studies
In the study of cattle environmental adaptability, phenotypic traits are easily distinguished. “Omics”-based analysis, such as transcriptomics and epigenomics, facilitates the study of functional variations that affect nonobvious or intermediate phenotypes. Furthermore, the integration of genome-wide selection scans with GWAS helps map putative regions of functional influence. This is particularly important in studies of indigenous populations, in which obtaining large sample sizes is challenging. Last, histological analyses or functional experiments can directly establish the links between candidate adaptive variants and phenotypes. For example, analyses combining GWAS and functional validation of mutations suggest that PRL and PRLR affect thermoregulatory and hair-morphology phenotypes in cattle (Flórez Murillo et al. 2021; Littlejohn et al. 2014; Porto-Neto et al. 2018).
Conclusions and perspectives
After domestication, cattle spread around the world with human migrations. Selection pressures in response to regional conditions have affected global cattle genome diversity. The collection of global genome-wide population genetic data has led to the discovery of several examples of local adaptations to diverse environments. Further progress will depend on research in several areas as follows:
High-coverage long-read whole-genome sequencing of local diverse populations can be used to construct pangenomes, which will include a large part of the global repertoire of SNPs and SVs.
Whole-genome sequencing of aurochs and ancient cattle will enable hypotheses of the genetic consequences of recent artificial and natural selection in domestic cattle to be tested.
Extending current GWAS-based methods to detect polygenic adaptations in combination with climate and environmental data is an important future direction.
Detailed phenotyping will be stimulated by artificial intelligence approaches (Liang et al. 2021) and/or on integrative omics datasets (Peng et al. 2020).
These lines of progress are expected to illuminate the mode and tempo of local adaptation as large numbers of cattle settled in all inhabited continents.
Acknowledgements
We thank the High-Performance Computing (HPC) center of Northwest A&F University (NWAFU) for providing computing resources.
Abbreviations
- BP
Before present
- GWAS
Genome-wide association study
- BTA
Bos taurus chromosome
- aDNA
Ancient DNA
- SV
Structural variant
- Bp
Base pairs
- Mb
Megabases
- DCMS
Decorrelated composite of multiple signals
- Pi
Nucleotide diversity
- FST
Fixation index
- iHS
Integrated haplotype score
- Hp
Pooled heterozygosity
- XP-EHH
Cross-population extended haplotype homozygosity
- XP-CLR
Cross-population composite likelihood ratio
- PBS
Population branch statistics
- XtX statistics
Population differentiation
- θπ ratio
Genetic diversity ratio
- hapFLK
Haplotype-based statistic
Authors’ contributions
N.C. and X.X. conceptualized the study and wrote the initial draft of the manuscript; X.X., N.C., and F.W. prepared the figures; Q.H. and Z.A. helped with English language editing; and K.Q., Y.W., M.S.S., J.A.L., J.H., and C.L. revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the earmarked fund of the CARS-37, Foreign Young Talents Program (QN2022172008L) and a grant from the National Natural Science Foundation of China (31872317) to C.L.; the fellowship of the China Postdoctoral Science Foundation (2021T140564 and 2020M683587), Shaanxi Youth Science and Technology New Star (2022KJXX-77), the Natural Science Basic Research Program of Shaanxi (2021JQ-137), the National Natural Science Foundation of China (32102523), the Fundamental Research Funds for the Central Universities and High-end Foreign Experts Recruitment Plan (G2022172032L) to N.C.; the Scientific Research Fund of the Department of Education of Yunnan (2022J0830) to K.Q.; the Carlsberg Foundation (CF20-0355) to M.S.S.
Availability of data and materials
Not applicable.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chuzhao Lei, Email: leichuzhao1118@nwafu.edu.cn.
Ningbo Chen, Email: ningbochen@nwafu.edu.cn.
References
- Achilli A, Olivieri A, Pellecchia M, Uboldi C, Colli L, Al-Zahery N, Accetturo M, Pala M, Hooshiar Kashani B, Perego UA, Battaglia V, Fornarino S, Kalamati J, Houshmand M, Negrini R, Semino O, Richards M, Macaulay V, Ferretti L, Bandelt HJ, Ajmone-Marsan P, Torroni A. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr Biol. 2008;18(4):R157–158. doi: 10.1016/j.cub.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Aguiar TS, Torrecilha RBP, Milanesi M, Utsunomiya ATH, Trigo BB, Tijjani A, Musa HH, Lopes FL, Ajmone-Marsan P, Carvalheiro R, Neves HHR, do Carmo AS, Hanotte O, Sonstegard TS, Garcia JF, Utsunomiya YT. Association of copy number variation at intron 3 of HMGA2 with navel length in Bosindicus. Front Genet. 2018;9:627. doi: 10.3389/fgene.2018.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajmone-Marsan P, Garcia JF, Lenstra J. On the origin of cattle: how aurochs became domestic and colonized the world. Evol Anthropol. 2010;19:148–157. doi: 10.1002/evan.20267. [DOI] [Google Scholar]
- Bahbahani H, Tijjani A, Mukasa C, Wragg D, Almathen F, Nash O, Akpa GN, Mbole-Kariuki M, Malla S, Woolhouse M, Sonstegard T, Van Tassell C, Blythe M, Huson H, Hanotte O. Signatures of selection for environmental adaptation and zebu × taurine hybrid fitness in East African Shorthorn zebu. Front Genet. 2017;8:68. doi: 10.3389/fgene.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh G, Gunn T, He L, Schlossman S, Duke-Cohan J. Biochemical and genetic studies of pigment-type switching. Pigment Cell Res. 2000;13:48–53. doi: 10.1034/j.1600-0749.13.s8.10.x. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, Montgomery HE, Pan H, Robbins PA, Shianna KV, Tam SC, Tsering N, Veeramah KR, Wang W, Wangdui P, Weale ME, Xu Y, Xu Z, Yang L, Zaman MJ, Zeng C, Zhang L, Zhang X, Zhaxi P, Zheng YT. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman A. Invited review: Are adaptations present to support dairy cattle productivity in warm climates? J Dairy Sci. 2011;94(5):2147–2158. doi: 10.3168/jds.2010-3962. [DOI] [PubMed] [Google Scholar]
- Bernabucci U, Biffani S, Buggiotti L, Vitali A, Lacetera N, Nardone A. The effects of heat stress in Italian Holstein dairy cattle. J Dairy Sci. 2014;97(1):471–486. doi: 10.3168/jds.2013-6611. [DOI] [PubMed] [Google Scholar]
- Boone M, Deen PM. Physiology and pathophysiology of the vasopressin-regulated renal water reabsorption. Pflugers Arch. 2008;456(6):1005–1024. doi: 10.1007/s00424-008-0498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM, Cargill M, Auton A, Reynolds A, Elkahloun AG, Castelhano M, Mosher DS, Sutter NB, Johnson GS, Novembre J, Hubisz MJ, Siepel A, Wayne RK, Bustamante CD, Ostrander EA. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8(8):e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunson K, Zhao X, He N, Dai X, Rodrigues A, Yang D. New insights into the origins of oracle bone divination: Ancient DNA from Late Neolithic Chinese bovines. J Archaeol Sci. 2016;74:35–44. doi: 10.1016/j.jas.2016.08.008. [DOI] [Google Scholar]
- Buggiotti L, Yurchenko AA, Yudin NS, Vander Jagt CJ, Vorobieva NV, Kusliy MA, Vasiliev SK, Rodionov AN, Boronetskaya OI, Zinovieva NA, Graphodatsky AS, Daetwyler HD, Larkin DM. Demographic history, adaptation, and NRAP convergent evolution at amino acid residue 100 in the world northernmost cattle from Siberia. Mol Biol Evol. 2021;38(8):3093–3110. doi: 10.1093/molbev/msab078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Zhang N, Zhu S, Chen Q, Wang L, Zhao X, Ma X, Royle TCA, Zhou H, Yang DY. Ancient DNA reveals evidence of abundant aurochs (Bos primigenius) in Neolithic Northeast China. J Archaeol Sci. 2018;98:72–80. doi: 10.1016/j.jas.2018.08.003. [DOI] [Google Scholar]
- Cao Y, Xia X, Hou J, Chen N, Zhao X, Chen S, Dang R, Huang Y, Chen H, Lei C. Y-chromosomal haplogroup distributions in Chinese cattle. Anim Genet. 2019;50(4):412–413. doi: 10.1111/age.12804. [DOI] [PubMed] [Google Scholar]
- Chen N, Lei C. The origins and utilization history of Chinese cattle as revealed by DNA analysis (in Chinese) Quaternary Sciences. 2021;41:92–100. doi: 10.11928/j.issn.1001-7410.2022.01.08. [DOI] [Google Scholar]
- Chen S, Lin BZ, Baig M, Mitra B, Lopes RJ, Santos AM, Magee DA, Azevedo M, Tarroso P, Sasazaki S, Ostrowski S, Mahgoub O, Chaudhuri TK, Zhang YP, Costa V, Royo LJ, Goyache F, Luikart G, Boivin N, Fuller DQ, Mannen H, Bradley DG, Beja-Pereira A. Zebu cattle are an exclusive legacy of the South Asia neolithic. Mol Biol Evol. 2010;27(1):1–6. doi: 10.1093/molbev/msp213. [DOI] [PubMed] [Google Scholar]
- Chen FH, Dong GH, Zhang DJ, Liu XY, Jia X, An CB, Ma MM, Xie YW, Barton L, Ren XY, Zhao ZJ, Wu XH, Jones MK. Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P. Science. 2015;347(6219):248–250. doi: 10.1126/science.1259172. [DOI] [PubMed] [Google Scholar]
- Chen N, Cai Y, Chen Q, Li R, Wang K, Huang Y, Hu S, Huang S, Zhang H, Zheng Z, Song W, Ma Z, Ma Y, Dang R, Zhang Z, Xu L, Jia Y, Liu S, Yue X, Deng W, Zhang X, Sun Z, Lan X, Han J, Chen H, Bradley DG, Jiang Y, Lei C. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat Commun. 2018;9(1):2337. doi: 10.1038/s41467-018-04737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Huang J, Zulfiqar A, Li R, Xi Y, Zhang M, Dang R, Lan X, Chen H, Ma Y, Lei C. Population structure and ancestry of Qinchuan cattle. Anim Genet. 2018;49(3):246–248. doi: 10.1111/age.12658. [DOI] [PubMed] [Google Scholar]
- Chen N, Fu W, Zhao J, Shen J, Chen Q, Zheng Z, Chen H, Sonstegard TS, Lei C, Jiang Y. BGVD: An integrated database for bovine sequencing variations and selective signatures. Genom Proteom Bioinf. 2020;18(2):186–193. doi: 10.1016/j.gpb.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Shen J, Hanif Q, Chen N, Huang Y, Dang R, Lan X, Chen H, Lei C. Whole genome analyses revealed genomic difference between European taurine and East Asian taurine. J Anim Breed Genet. 2021;138(1):56–68. doi: 10.1111/jbg.12501. [DOI] [PubMed] [Google Scholar]
- Chiang C, Scott AJ, Davis JR, Tsang EK, Li X, Kim Y, Hadzic T, Damani FN, Ganel L, Montgomery SB, Battle A, Conrad DF, Hall IM. The impact of structural variation on human gene expression. Nat Genet. 2017;49(5):692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M, Reissmann M, Hofreiter M, Ludwig A. Colours of domestication. Biol Rev. 2011;86(4):885–899. doi: 10.1111/j.1469-185X.2011.00177.x. [DOI] [PubMed] [Google Scholar]
- Crysnanto D, Leonard AS, Fang ZH, Pausch H. Novel functional sequences uncovered through a bovine multiassembly graph. Proc Natl Acad Sci U S A. 2021;118(20):e2101056118. doi: 10.1073/pnas.2101056118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubric-Curik V, Novosel D, Brajkovic V, Rota Stabelli O, Krebs S, Sölkner J, Šalamon D, Ristov S, Berger B, Trivizaki S, Bizelis I, Ferenčaković M, Rothammer S, Kunz E, Simčič M, Dovč P, Bunevski G, Bytyqi H, Marković B, Brka M, Kume K, Stojanović S, Nikolov V, Zinovieva N, Schönherz AA, Guldbrandtsen B, Čačić M, Radović S, Miracle P, Vernesi C, Curik I, Medugorac I. Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evol Appl. 2022;15(4):663–678. doi: 10.1111/eva.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brøndum RF, Liao X, Djari A, Rodriguez SC, Grohs C, Esquerré D, Bouchez O, Rossignol MN, Klopp C, Rocha D, Fritz S, Eggen A, Bowman PJ, Coote D, Chamberlain AJ, Anderson C, VanTassell CP, Hulsegge I, Goddard ME, Guldbrandtsen B, Lund MS, Veerkamp RF, Boichard DA, Fries R, Hayes BJ. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46(8):858–865. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- Dikmen S, Khan FA, Huson HJ, Sonstegard TS, Moss JI, Dahl GE, Hansen PJ. The SLICK hair locus derived from Senepol cattle confers thermotolerance to intensively managed lactating Holstein cows. J Dairy Sci. 2014;97(9):5508–5520. doi: 10.3168/jds.2014-8087. [DOI] [PubMed] [Google Scholar]
- Edwards CJ, Ginja C, Kantanen J, Pérez-Pardal L, Tresset A, Stock F, Gama LT, Penedo MC, Bradley DG, Lenstra JA, Nijman IJ. Dual origins of dairy cattle farming–evidence from a comprehensive survey of European Y-chromosomal variation. PLoS ONE. 2011;6(1):e15922. doi: 10.1371/journal.pone.0015922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felius M, Beerling M-L, Buchanan DS, Theunissen B, Koolmees PA, Lenstra JA. On the history of cattle genetic resources. Diversity. 2014;6(4):705–750. doi: 10.3390/d6040705. [DOI] [Google Scholar]
- Felius M, Koolmees PA, Theunissen B, Consortium ECGD, Lenstra JA On the breeds of cattle—historic and current classifications. Diversity. 2011;3(4):660–692. doi: 10.3390/d3040660. [DOI] [Google Scholar]
- Flórez Murillo JM, Landaeta-Hernández AJ, Kim ES, Bostrom JR, Larson SA, Pérez O'Brien AM, Montero-Urdaneta MA, Garcia JF, Sonstegard TS. Three novel nonsense mutations of prolactin receptor found in heat-tolerant Bos taurus breeds of the Caribbean Basin. Anim Genet. 2021;52(1):132–134. doi: 10.1111/age.13027. [DOI] [PubMed] [Google Scholar]
- Flori L, Thevenon S, Dayo GK, Senou M, Sylla S, Berthier D, Moazami-Goudarzi K, Gautier M. Adaptive admixture in the West African bovine hybrid zone: insight from the Borgou population. Mol Ecol. 2014;23(13):3241–3257. doi: 10.1111/mec.12816. [DOI] [PubMed] [Google Scholar]
- Gao Y, Gautier M, Ding X, Zhang H, Wang Y, Wang X, Faruque MO, Li J, Ye S, Gou X, Han J, Lenstra JA, Zhang Y. Species composition and environmental adaptation of indigenous Chinese cattle. Sci Rep. 2017;7(1):16196. doi: 10.1038/s41598-017-16438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Flori L, Riebler A, Jaffrézic F, Laloé D, Gut I, Moazami-Goudarzi K, Foulley JL. A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics. 2009;10:550. doi: 10.1186/1471-2164-10-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreishifar SM, Eriksson S, Johansson AM, Khansefid M, Moghaddaszadeh-Ahrabi S, Parna N, Davoudi P, Javanmard A. Signatures of selection reveal candidate genes involved in economic traits and cold acclimation in five Swedish cattle breeds. Genet Sel Evol. 2020;52(1):52. doi: 10.1186/s12711-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Yang P, Fang W, Li R, Jiang Y (2022) Building a cattle pan-genome using more de novo assemblies. J Genet Genomics 49(9):906-8. 10.1016/j.jgg.2022.01.003 [DOI] [PubMed]
- Gutiérrez-Gil B, Esteban-Blanco C, Wiener P, Chitneedi PK, Suarez-Vega A, Arranz JJ. High-resolution analysis of selection sweeps identified between fine-wool Merino and coarse-wool Churra sheep breeds. Genet Sel Evol. 2017;49(1):81. doi: 10.1186/s12711-017-0354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen PJ. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim Reprod Sci. 2004;82–83:349–360. doi: 10.1016/j.anireprosci.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D’Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18(6):763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020;21(3):171–189. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoshin AV, Yurchenko AA, Belonogova NM, Petrovsky DV, Aitnazarov RB, Soloshenko VA, Yudin NS, Larkin DM. Genome-wide association study and scan for signatures of selection point to candidate genes for body temperature maintenance under the cold stress in Siberian cattle populations. BMC Genet. 2019;20(Suppl 1):26. doi: 10.1186/s12863-019-0725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian W, Duangjinda M, Vajrabukka C, Katawatin S. Differences of skin morphology in Bos indicus, Bos taurus, and their crossbreds. Int J Biometeorol. 2014;58(6):1087–1094. doi: 10.1007/s00484-013-0700-9. [DOI] [PubMed] [Google Scholar]
- Jiang L, Kon T, Chen C, Ichikawa R, Zheng Q, Pei L, Takemura I, Nsobi LH, Tabata H, Pan H, Omori Y, Ogura A. Whole-genome sequencing of endangered Zhoushan cattle suggests its origin and the association of MC1R with black coat colour. Sci Rep. 2021;11(1):17359. doi: 10.1038/s41598-021-96896-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A, Li Y, Dong K, Irwin DM, Zhao Q, He X, Liu J, Pu Y, Gorkhali NA, Liu X, Jiang L, Li X, Guan W, Zhang Y, Wu DD, Ma Y. Population variation reveals independent selection toward small body size in Chinese Debao pony. Genome Biol Evol. 2015;8(1):42–50. doi: 10.1093/gbe/evv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantanen J, Edwards CJ, Bradley DG, Viinalass H, Thessler S, Ivanova Z, Kiselyova T, Cinkulov M, Popov R, Stojanović S, Ammosov I, Vilkki J. Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus) Heredity. 2009;103(5):404–415. doi: 10.1038/hdy.2009.68. [DOI] [PubMed] [Google Scholar]
- Kim J, Hanotte O, Mwai OA, Dessie T, Bashir S, Diallo B, Agaba M, Kim K, Kwak W, Sung S, Seo M, Jeong H, Kwon T, Taye M, Song KD, Lim D, Cho S, Lee HJ, Yoon D, Oh SJ, Kemp S, Lee HK, Kim H. The genome landscape of indigenous African cattle. Genome Biol. 2017;18(1):34. doi: 10.1186/s13059-017-1153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kwon T, Dessie T, Yoo D, Mwai OA, Jang J, Sung S, Lee S, Salim B, Jung J, Jeong H, Tarekegn GM, Tijjani A, Lim D, Cho S, Oh SJ, Lee HK, Kim J, Jeong C, Kemp S, Hanotte O, Kim H. The mosaic genome of indigenous African cattle as a unique genetic resource for African pastoralism. Nat Genet. 2020;52(10):1099–1110. doi: 10.1038/s41588-020-0694-2. [DOI] [PubMed] [Google Scholar]
- Koch F, Thom U, Albrecht E, Weikard R, Nolte W, Kuhla B, Kuehn C. Heat stress directly impairs gut integrity and recruits distinct immune cell populations into the bovine intestine. Proc Natl Acad Sci U S A. 2019;116(21):10333–10338. doi: 10.1073/pnas.1820130116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Weikard R. An investigation into the genetic background of coat colour dilution in a Charolais x German Holstein F2 resource population. Anim Genet. 2007;38(2):109–113. doi: 10.1111/j.1365-2052.2007.01569.x. [DOI] [PubMed] [Google Scholar]
- Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, Baets J, Senderek J, Topaloglu H, Farrell SA, Nürnberg G, Nürnberg P, De Jonghe P, Gal A, Kaether C, Timmerman V, Hübner CA. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41(11):1179–1181. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segrè AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Mägi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, König IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Müller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpeläinen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Paré G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietiläinen KH, Pouta A, Ridderstråle M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kähönen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimäki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tönjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Grönberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Völzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, Barton L, Climer Vigueira C, Denham T, Dobney K, Doust AN, Gepts P, Gilbert MT, Gremillion KJ, Lucas L, Lukens L, Marshall FB, Olsen KM, Pires JC, Richerson PJ, Rubio de Casas R, Sanjur OI, Thomas MG, Fuller DQ. Current perspectives and the future of domestication studies. Proc Natl Acad Sci U S A. 2014;111(17):6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra JA, Ajmone-Marsan P, Beja-Pereira A, Bollongino R, Bradley DG, Colli L, De Gaetano A, Edwards CJ, Felius M, Ferretti L, Ginja C, Hristov P, Kantanen J, Lirón JP, Magee DA, Negrini R, Radoslavov GA. Meta-analysis of mitochondrial DNA reveals several population bottlenecks during worldwide migrations of cattle. Diversity. 2014;6(1):178–187. doi: 10.3390/d6010178. [DOI] [Google Scholar]
- Leonard AS, Crysnanto D, Fang ZH, Heaton MP, Vander Ley BL, Herrera C, Bollwein H, Bickhart DM, Kuhn KL, Smith TPL, Rosen BD, Pausch H. Structural variant-based pangenome construction has low sensitivity to variability of haplotype-resolved bovine assemblies. Nat Commun. 2022;3(1):3012. doi: 10.1038/s41467-022-30680-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Xiao S, Wei N, Zhang Z, Huang R, Gu Y, Guo Y, Ren J, Huang L, Chen C. Fine mapping of a QTL for ear size on porcine chromosome 5 and identification of high mobility group AT-hook 2 (HMGA2) as a positional candidate gene. Genet Sel Evol. 2012;44(1):6. doi: 10.1186/1297-9686-44-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Miao J, Wang X, Chang T, An B, Duan X, Xu L, Gao X, Zhang L, Li J, Gao H. Application of ensemble learning to genomic selection in Chinese Simmental beef cattle. J Anim Breed Genet. 2021;138(3):291–299. doi: 10.1111/jbg.12514. [DOI] [PubMed] [Google Scholar]
- Littlejohn MD, Henty KM, Tiplady K, Johnson T, Harland C, Lopdell T, Sherlock RG, Li W, Lukefahr SD, Shanks BC, Garrick DJ, Snell RG, Spelman RJ, Davis SR. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat Commun. 2014;5(1):5861. doi: 10.1038/ncomms6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WY, Tearle R, Liu R, Koren S, Rhie A, Bickhart DM, Rosen BD, Kronenberg ZN, Kingan SB, Tseng E, Thibaud-Nissen F, Martin FJ, Billis K, Ghurye J, Hastie AR, Lee J, Pang AWC, Heaton MP, Phillippy AM, Hiendleder S, Smith TPL, Williams JL. Haplotype-resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nat Commun. 2020;11(1):2071. doi: 10.1038/s41467-020-15848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Cheng H, Liu Y, Sun L, Chen N, Jiang F, You W, Yang Z, Zhang B, Song E, Lei C. Assessing genomic diversity and selective pressures in Bohai Black cattle using whole-genome sequencing data. Animals (basel) 2022;12(5):665. doi: 10.3390/ani12050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Wang H, Liao Q, Wang L, Cheng G, Wang H, Zhao C, Zhao S, Song J, Guang X, Liu GE, Li A, Wu X, Wang C, Fang X, Zhao X, Smith SB, Yang W, Tian W, Gui L, Zhang Y, Hill RA, Jiang Z, Xin Y, Jia C, Sun X, Wang S, Yang H, Wang J, Zhu W, Zan L. Genetic architecture and selection of Chinese cattle revealed by whole genome resequencing. Mol Biol Evol. 2018;35(3):688–699. doi: 10.1093/molbev/msx322. [DOI] [PubMed] [Google Scholar]
- Mei C, Gui L, Hong J, Raza SHA, Aorigele C, Tian W, Garcia M, Xin Y, Yang W, Zhang S, Zan L. Insights into adaption and growth evolution: a comparative genomics study on two distinct cattle breeds from Northern and Southern China. Mol Ther Nucleic Acids. 2021;23:959–967. doi: 10.1016/j.omtn.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35(2):157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Mohamad K, Olsson M, van Tol HT, Mikko S, Vlamings BH, Andersson G, Rodríguez-Martínez H, Purwantara B, Paling RW, Colenbrander B, Lenstra JA. On the origin of Indonesian cattle. PLoS ONE. 2009;4(5):e5490. doi: 10.1371/journal.pone.0005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JH, Holt TN, Cogan JD, Womack B, Phillips JA, Li C, Kendall Z, Stenmark KR, Thomas MG, Brown RD, Riddle SR, West JD, Hamid R. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat Commun. 2015;6(1):6863. doi: 10.1038/ncomms7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes H, Brass A, Obara I, Anderson S, Archibald AL, Bradley DG, Fisher P, Freeman A, Gibson J, Gicheru M, Hall L, Hanotte O, Hulme H, McKeever D, Murray C, Oh SJ, Tate C, Smith K, Tapio M, Wambugu J, Williams DJ, Agaba M, Kemp SJ. Genetic and expression analysis of cattle identifies candidate genes in pathways responding to Trypanosoma congolense infection. Proc Natl Acad Sci U S A. 2011;108(22):9304–9309. doi: 10.1073/pnas.1013486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TA, Lucena C, Chase CC, Jr, Hammond AC. Evidence of a major gene influencing hair length and heat tolerance in Bos taurus cattle. J Anim Sci. 2003;81(1):80–90. doi: 10.2527/2003.81180x. [DOI] [PubMed] [Google Scholar]
- Park SD, Magee DA, McGettigan PA, Teasdale MD, Edwards CJ, Lohan AJ, Murphy A, Braud M, Donoghue MT, Liu Y, Chamberlain AT, Rue-Albrecht K, Schroeder S, Spillane C, Tai S, Bradley DG, Sonstegard TS, Loftus BJ, MacHugh DE. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol. 2015;16:234. doi: 10.1186/s13059-015-0790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Wang K, Chen Z, Cao Y, Gao Q, Li Y, Li X, Lu H, Du H, Lu M, Yang X, Liang C. MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 2020;48(D1):D1085–D1092. doi: 10.1093/nar/gkz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pardal L, Sánchez-Gracia A, Álvarez I, Traoré A, Ferraz JBS, Fernández I, Costa V, Chen S, Tapio M, Cantet RJC, Patel A, Meadow RH, Marshall FB, Beja-Pereira A, Goyache F. Legacies of domestication, trade and herder mobility shape extant male zebu cattle diversity in South Asia and Africa. Sci Rep. 2018;8(1):18027. doi: 10.1038/s41598-018-36444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, Martinez R, Bruford MW, Orozco-terWengel P. Domestication of cattle: Two or three events? Evol Appl. 2019;12(1):123–136. doi: 10.1111/eva.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21(19):5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto-Neto LR, Bickhart DM, Landaeta-Hernandez AJ, Utsunomiya YT, Pagan M, Jimenez E, Hansen PJ, Dikmen S, Schroeder SG, Kim ES, Sun J, Crespo E, Amati N, Cole JB, Null DJ, Garcia JF, Reverter A, Barendse W, Sonstegard TS. Convergent evolution of slick coat in cattle through truncation mutations in the prolactin receptor. Front Genet. 2018;9:57. doi: 10.3389/fgene.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa IA, Khatkar MS, Thomson PC, Raadsma HW. A meta-assembly of selection signatures in cattle. PLoS ONE. 2016;11(4):e0153013. doi: 10.1371/journal.pone.0153013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell LB. Hyperthermia: a hyperadrenergic state. Hypertension (dallas, Tex: 1979) 1990;15(5):505–507. doi: 10.1161/01.hyp.15.5.505. [DOI] [PubMed] [Google Scholar]
- Schmutz SM, Dreger DL. Interaction of MC1R and PMEL alleles on solid coat colors in Highland cattle. Anim Genet. 2013;44(1):9–13. doi: 10.1111/j.1365-2052.2012.02361.x. [DOI] [PubMed] [Google Scholar]
- Silva PS, Hooper HB, Manica E, Merighe GKF, Oliveira SA, Traldi AS, Negrão JA. Heat stress affects the expression of key genes in the placenta, placental characteristics, and efficiency of Saanen goats and the survival and growth of their kids. J Dairy Sci. 2021;104(4):4970–4979. doi: 10.3168/jds.2020-18301. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, Ge R. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329(5987):72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Sinding MS, Ciucani MM, Ramos-Madrigal J, Carmagnini A, Rasmussen JA, Feng S, Chen G, Vieira FG, Mattiangeli V, Ganjoo RK, Larson G, Sicheritz-Pontén T, Petersen B, Frantz L, Gilbert MTP, Bradley DG. Kouprey (Bos sauveli) genomes unveil polytomic origin of wild Asian Bos. iScience. 2021;24(11):103226. doi: 10.1016/j.isci.2021.103226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudrajad P, Subiharta S, Adinata Y, Lathifah A, Lee JH, Lenstra JA, Lee SH. An insight into the evolutionary history of Indonesian cattle assessed by whole genome data analysis. PLoS ONE. 2020;15(11):e0241038. doi: 10.1371/journal.pone.0241038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Qu K, Liu Y, Ma X, Chen N, Zhang J, Huang B, Lei C. Assessing genomic diversity and selective pressures in Bashan cattle by whole-genome sequencing data. Anim Biotechnol. 2021;11:1–12. doi: 10.1080/10495398.2021.1998094. [DOI] [PubMed] [Google Scholar]
- Talenti A, Powell J, Hemmink JD, Cook EAJ, Wragg D, Jayaraman S, Paxton E, Ezeasor C, Obishakin ET, Agusi ER, Tijjani A, Marshall K, Fisch A, Ferreira BR, Qasim A, Chaudhry U, Wiener P, Toye P, Morrison LJ, Connelley T, Prendergast JGD. A cattle graph genome incorporating global breed diversity. Nat Commun. 2022;13(1):910. doi: 10.1038/s41467-022-28605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigo BB, Utsunomiya ATH, Fortunato A, Milanesi M, Torrecilha RBP, Lamb H, Nguyen L, Ross EM, Hayes B, Padula RCM, Sussai TS, Zavarez LB, Cipriano RS, Caminhas MMT, Lopes FL, Pelle C, Leeb T, Bannasch D, Bickhart D, Smith TPL, Sonstegard TS, Garcia JF, Utsunomiya YT. Variants at the ASIP locus contribute to coat color darkening in Nellore cattle. Genet Sel Evol. 2021;53(1):40. doi: 10.1186/s12711-021-00633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truszkowska GT, Bilińska ZT, Muchowicz A, Pollak A, Biernacka A, Kozar-Kamińska K, Stawiński P, Gasperowicz P, Kosińska J, Zieliński T, Płoski R. Homozygous truncating mutation in NRAP gene identified by whole exome sequencing in a patient with dilated cardiomyopathy. Sci Rep. 2017;7(1):3362. doi: 10.1038/s41598-017-03189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya YT, Milanesi M, Fortes MRS, Porto-Neto LR, Utsunomiya ATH, Silva M, Garcia JF, Ajmone-Marsan P. Genomic clues of the evolutionary history of Bos indicus cattle. Anim Genet. 2019;50(6):557–568. doi: 10.1111/age.12836. [DOI] [PubMed] [Google Scholar]
- Verdugo MP, Mullin VE, Scheu A, Mattiangeli V, Daly KG, Maisano Delser P, Hare AJ, Burger J, Collins MJ, Kehati R, Hesse P, Fulton D, Sauer EW, Mohaseb FA, Davoudi H, Khazaeli R, Lhuillier J, Rapin C, Ebrahimi S, Khasanov M, Vahidi SMF, MacHugh DE, Ertuğrul O, Koukouli-Chrysanthaki C, Sampson A, Kazantzis G, Kontopoulos I, Bulatovic J, Stojanović I, Mikdad A, Benecke N, Linstädter J, Sablin M, Bendrey R, Gourichon L, Arbuckle BS, Mashkour M, Orton D, Horwitz LK, Teasdale MD, Bradley DG. Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. Science. 2019;365(6449):173–176. doi: 10.1126/science.aav1002. [DOI] [PubMed] [Google Scholar]
- Wang X, Ju Z, Jiang Q, Zhong J, Liu C, Wang J, Hoff JL, Schnabel RD, Zhao H, Gao Y, Liu W, Wang L, Gao Y, Yang C, Hou M, Huang N, Regitano LCA, Porto-Neto LR, Decker JE, Taylor JF, Huang J. Introgression, admixture, and selection facilitate genetic adaptation to high-altitude environments in cattle. Genomics. 2021;113(3):1491–1503. doi: 10.1016/j.ygeno.2021.03.023. [DOI] [PubMed] [Google Scholar]
- Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39(10):1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldenegodguad M, Popov R, Pokharel K, Ammosov I, Ming Y, Ivanova Z, Kantanen J. Whole-genome sequencing of three native cattle breeds originating from the northernmost cattle farming regions. Front Genet. 2018;9:728. doi: 10.3389/fgene.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will DH, Hicks JL, Card CS, Alexander AF. Inherited susceptibility of cattle to high-altitude pulmonary hypertension. J Appl Physiol. 1975;38(3):491–494. doi: 10.1152/jappl.1975.38.3.491. [DOI] [PubMed] [Google Scholar]
- Wollenberg Valero KC, Pathak R, Prajapati I, Bankston S, Thompson A, Usher J, Isokpehi RD (2014) A candidate multimodal functional genetic network for thermal adaptation. PeerJ 2: e578. doi: 10.7717/peerj.578 [DOI] [PMC free article] [PubMed]
- Wu DD, Ding XD, Wang S, Wójcik JM, Zhang Y, Tokarska M, Li Y, Wang MS, Faruque O, Nielsen R, Zhang Q, Zhang YP. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol. 2018;2(7):1139–1145. doi: 10.1038/s41559-018-0562-y. [DOI] [PubMed] [Google Scholar]
- Wu DD, Yang CP, Wang MS, Dong KZ, Yan DW, Hao ZQ, Fan SQ, Chu SZ, Shen QS, Jiang LP, Li Y, Zeng L, Liu HQ, Xie HB, Ma YF, Kong XY, Yang SL, Dong XX, Esmailizadeh A, Irwin DM, Xiao X, Li M, Dong Y, Wang W, Shi P, Li HP, Ma YH, Gou X, Chen YB, Zhang YP. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl Sci Rev. 2020;7(6):952–963. doi: 10.1093/nsr/nwz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Qu K, Zhang G, Jia Y, Ma Z, Zhao X, Huang Y, Chen H, Huang B, Lei C. Comprehensive analysis of the mitochondrial DNA diversity in Chinese cattle. Anim Genet. 2019;50(1):70–73. doi: 10.1111/age.12749. [DOI] [PubMed] [Google Scholar]
- Xia X, Yao Y, Li C, Zhang F, Qu K, Chen H, Huang B, Lei C. Genetic diversity of Chinese cattle revealed by Y-SNP and Y-STR markers. Anim Genet. 2019;50(1):64–69. doi: 10.1111/age.12742. [DOI] [PubMed] [Google Scholar]
- Xia X, Zhang S, Zhang H, Zhang Z, Chen N, Li Z, Sun H, Liu X, Lyu S, Wang X, Li Z, Yang P, Xu J, Ding X, Shi Q, Wang E, Ru B, Xu Z, Lei C, Chen H, Huang Y. Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data. BMC Genomics. 2021;22(1):43. doi: 10.1186/s12864-020-07340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Bickhart DM, Cole JB, Schroeder SG, Song J, Tassell CP, Sonstegard TS, Liu GE. Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol Biol Evol. 2015;32(3):711–725. doi: 10.1093/molbev/msu333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang L, Zhu B, Zhang W, Wang Z, Chen Y, Zhang L, Gao X, Gao H, Liu GE, Li J. Genome-wide scan reveals genetic divergence and diverse adaptive selection in Chinese local cattle. BMC Genomics. 2019;20(1):494. doi: 10.1186/s12864-019-5822-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CL, Lin J, Huang YY, Gao QS, Piao ZY, Yuan SL, Chen L, Ren X, Ye RC, Dong M, Zhang HL, Zhou HQ, Jiang XX, Jin WZ, Zhou XM, Yan CG. Population genomics reveals that natural variation in PRDM16 contributes to cold tolerance in domestic cattle. Zool Res. 2022;43(2):275–284. doi: 10.24272/j.issn.2095-8137.2021.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Asan NP, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Huasang LJ, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolund L, Kristiansen K, Li Y, Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, Wang J. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko AA, Daetwyler HD, Yudin N, Schnabel RD, Vander Jagt CJ, Soloshenko V, Lhasaranov B, Popov R, Taylor JF, Larkin DM. Scans for signatures of selection in Russian cattle breed genomes reveal new candidate genes for environmental adaptation and acclimation. Sci Rep. 2018;8(1):12984. doi: 10.1038/s41598-018-31304-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Paijmans JL, Chang F, Wu X, Chen G, Lei C, Yang X, Wei Z, Bradley DG, Orlando L, O'Connor T, Hofreiter M. Morphological and genetic evidence for early Holocene cattle management in northeastern China. Nat Commun. 2013;4:2755. doi: 10.1038/ncomms3755. [DOI] [PubMed] [Google Scholar]
- Zhang K, Lenstra JA, Zhang S, Liu W, Liu J. Evolution and domestication of the Bovini species. Anim Genet. 2020;51(5):637–657. doi: 10.1111/age.12974. [DOI] [PubMed] [Google Scholar]
- Zhao S, Zheng P, Dong S, Zhan X, Wu Q, Guo X, Hu Y, He W, Zhang S, Fan W, Zhu L, Li D, Zhang X, Chen Q, Zhang H, Zhang Z, Jin X, Zhang J, Yang H, Wang J, Wang J, Wei F. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 2013;45(1):67–71. doi: 10.1038/ng.2494. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang L, Han X, Han J, Hu Y, Li F, Xia H, Peng L, Boschiero C, Rosen BD, Bickhart DM, Zhang S, Guo A, Van Tassell CP, Smith TPL, Yang L, Liu GE. Assembly of a pangenome for global cattle reveals missing sequences and novel structural variations, providing new insights into their diversity and evolutionary history. Genome Res. 2022 doi: 10.1101/gr.276550.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.