Abstract
The surface proteins (SU) of murine type-C retroviruses have a central hypervariable domain devoid of cysteine and rich in proline. This 41-amino-acid region of Friend ecotropic murine leukemia virus SU was shown to be highly tolerant of insertions and deletions. Viruses in which either the N-terminal 30 amino acids or the C-terminal 22 amino acids of this region were replaced by the 7-amino-acid sequence ASAVAGA were fully infectious. Insertions of this 7-amino-acid sequence at the N terminus, center, and the C terminus of the hypervariable domain had little effect on envelope protein (Env) function, while this insertion at a position 10 amino acids following the N terminus partially destabilized the association between the SU and transmembrane subunits of Env. Large, complex domains (either a 252-amino-acid single-chain antibody binding domain [scFv] or a 96-amino-acid V1/V2 domain of HIV-1 SU containing eight N-linked glycosylation sites and two disulfides) did not interfere with Env function when inserted in the center or C-terminal portions of the hypervariable domain. The scFv domain inserted into the C-terminal region of the hypervariable domain was shown to mediate binding of antigen to viral particles, demonstrating that it folded into the active conformation and was displayed on the surface of the virion. Both positive and negative enrichment of virions expressing the V1/V2 sequence were achieved by using a monoclonal antibody specific for a conformational epitope presented by the inserted sequence. These results indicated that the hypervariable domain of Friend ecotropic SU does not contain any specific sequence or structure that is essential for Env function and demonstrated that insertions into this domain can be used to extend particle display methodologies to complex protein domains that require expression in eukaryotic cells for glycosylation and proper folding.
The external proteins of enveloped viruses mediate binding to and penetration of the host cell. Retroviral envelope proteins (Env) consist of a peripheral, receptor-binding surface protein (SU) subunit and a membrane-spanning transmembrane protein (TM) subunit that contains an N-terminal fusion domain. They are synthesized as a single polypeptide that is proteolytically processed into the mature Env complex (31). In the type-C murine leukemia virus (MuLV) and related viruses, the N- and C-terminal sequences of SU are independent globular domains (20, 35), with receptor-binding activity residing in the N-terminal domain (2–4, 10, 25, 29). The recently determined crystal structure of the receptor-binding N-terminal domain of an ecotropic MuLV SU suggests that a conserved core of β sheets in an immunoglobulin fold provides the structural underpinning for presenting the receptor-binding site assembled from sequences that vary among receptor classes (7). Many of these Envs contain a labile disulfide bond between SU and TM (17, 23, 28, 32–35, 52) that involves a pair of cysteines present in a highly conserved structural motif near the beginning of the C-terminal domain of SU and that may be important in Env function (39). Connecting the N- and C-terminal globular domains of SU is a region that is rich in proline. This proline-rich region can be divided into two domains by sequence comparisons: an N-terminal domain of 12 residues that is highly conserved among MuLV SUs and somewhat conserved among a broader group of viruses and a C-terminal domain that is hypervariable. Deletion of the conserved proline-rich domain results in failure of processed Env complex to be incorporated into virions, while the hypervariable domain tolerates significant deletions and small insertions, some of which weaken the association between SU and TM (53).
In this report, the function of the hypervariable domain linking the N-terminal receptor-binding domain and the highly conserved C-terminal domain of MuLV SUs was further investigated by constructing a series of small and large insertions and deletions in this region of Friend ecotropic MuLV (Fr-MuLV). Insertions into the N-terminal portion of the hypervariable domain destabilized the interaction between SU and TM, sometimes sufficiently to interfere with viral growth. In contrast, the C-terminal portion of the hypervariable domain was found to be extremely tolerant of modification, including the insertion of large sequences containing N-linked glycosylation sites and internal disulfide bonds. These modified Envs retained full function, and the inserted sequences were exposed at the surface of viral particles, where they were efficiently recognized by antibodies and other ligands directed against the inserted sequences. Furthermore, it was demonstrated that virions carrying such insertions could be physically selected out of mixed populations, thus enabling a novel retroviral particle display system suitable for eukaryotic sequences that cannot be expressed in bacterial systems. Similar insertions may also prove to have relevance for redirecting the cell specificity of the virus, allowing targeting of retroviral gene therapy delivery to cells of choice.
MATERIALS AND METHODS
Viruses and cell lines.
The MuLV env was from clone 57 Fr-MuLV (26). MuLV was expressed from a chimeric Fr-MuLV 2 long terminal repeat colinear genomic plasmid (pLRB303 for wild-type virus) containing most non-env sequences from the FB29 clone (15). Mouse NIH 3T3 fibroblasts were maintained as previously described (14). SEC-CHO, a CHO cell line that secretes a truncated, soluble form of the HIVHXB2 Env precursor, gp140, and its cleavage product, gp120, was obtained from Judith White and maintained as described previously (51). Mutant viruses were expressed by transfecting the genomic viral plasmids into 3T3 cells by using Lipofectamine (GibcoBRL). Insertion mutations, introducing NheI, Eco47III, NgoMI, and NarI restriction sites and encoding a 7-amino-acid sequence, ASAVAGA (5′-GCT AGC GCT GTT GCC GGC GCC-3′), were constructed at each of the sites indicated in Fig. 1 by PCR overlap mutagenesis (11). Human monoclonal antibody (MAb) 5145a recognizes a CD4 binding site epitope on human immunodeficiency virus type 1 (HIV-1) SU (gp120) (38). A 252-amino-acid 5145a scFv gene fragment with a (Gly4Ser)3 sequence linking the heavy- and light-chain variable domains (12) was constructed by PCR overlap mutagenesis from clones provided by Ellen Murphy and cloned into various insertion site plasmids on NheI and NgoMI ends, retaining the AS dipeptide N-terminal to the scFv domain and the AGA tripeptide C-terminal to it. The 96-amino-acid gp120 V1V2 domain of the CaseA2 HIV-1 sequence, which has been described previously (37), was inserted between residues 273 and 274 by using NheI and NarI restriction sites, retaining the AS dipeptide N-terminal to the V1V2 domain and the GA dipeptide C-terminal to it.
FIG. 1.
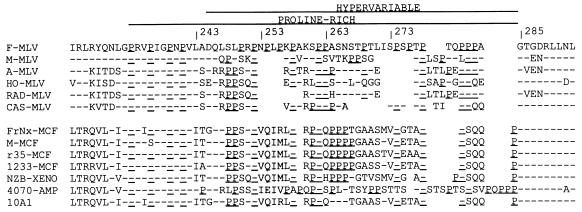
Sequence conservation near the proline-rich domain of MuLV SUs. Residues matching that of the Fr-MuLV sequence are indicated with a hyphen; Pro residues are underlined; gaps introduced for alignment have been left blank. The first group of sequences are from ecotropic envs; the second group are from envs of other receptor classes. F-MLV (16); M-MLV (45); A-MLV (18); HO-MLV (49); RAD-MLV (22); CAS-MLV (42); FrNx-MCF (1); M-MCF (5); r35-MCF (41); 1233-MCF (46); NZB-XENO (27); 4070-AMP and 10A1 (30).
Immunoassays.
Goat anti-Rauscher gp70 serum and goat anti-Rauscher p30 serum were obtained from Quality Biotech (Camden, N.J.). Rat MAb 10BA10 specific for Fr-MuLV p12gag (14) and mouse MAb SC258, provided by Abbott Laboratories and specific for a conformational epitope in the V1V2 domain of HIV-1 gp120 (24, 54), have been previously described. Viral infection was detected by immunofluorescence assay (IFA) by using 10BA10 as previously described (14). Following transfection with a plasmid expressing a noninfectious virus, no increase in Gag+ cells is seen by IFA beyond 18 h posttransfection, indicating that all successfully transfected cells express detectable Gag by this time point (14). Specific infectivity was examined by determining the percent of cells producing p12gag 18 h following a standard infection protocol by using serial dilutions of virus containing culture supernatants with similar amounts of p30gag. The most-concentrated sample was a 1:20 dilution of culture supernatant. Viral proteins were characterized by radioimmunoprecipitation (RIP) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography as previously described (36). Radioisotopes were obtained from New England Nuclear.
Enrichment procedures.
Pansorbin cells (Calbiochem), prepared for RIP, were washed five times with 10 volumes of PBS and then stored at 4°C as a 10% suspension. His6-tagged protein A was prepared as described previously (40) from the expression plasmid kindly provided by Tim Hunt of the Imperial Cancer Research Fund. Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) was washed three times in PBS and resuspended to a 50% slurry in PBS. One-half volume of His6 protein A at 1.5 mg/ml was added to the washed Ni2+-NTA agarose slurry, followed by the addition of 3 volumes of PBS and overnight incubation at 4°C. Culture supernatants containing either wild-type or V1V2-SU virus were mixed in proportions to give either an excess of wild-type virus for positive enrichment experiments or an excess of V1V2-SU virus for negative enrichment experiments. A 0.5-ml portion of the virus mixture was incubated with MAb SC258 at 37°C for 1 h. For positive enrichment, the virus mixture was used to suspend 0.05 ml of packed Ni2+-NTA agarose with prebound His6 protein A and rotated at room temperature for 1 h. The Ni2+-NTA agarose was washed twice with 0.5 ml of PBS by pelleting and then suspended in 0.2 ml of 10 mM EDTA in PBS for 5 min at room temperature. The Ni2+-NTA agarose was removed by centrifugation, and 0.2 ml of 40 mM MgCl2 was added immediately. For negative enrichment, the virus mixture was used to suspend 0.01 ml of packed Pansorbin and rotated at room temperature for 1 h. Pansorbin was then removed by centrifugation. Aliquots of unseparated virus mixtures (starting materials), Pansorbin supernatants (negatively enriched sample), and Ni2+-NTA agarose eluates (positively enriched samples) were used to infect 3T3 cells, and virus growth was monitored by IFA. When the cultures were fully infected, [35S]cysteine-labeled culture supernatants were prepared and analyzed by RIP with goat anti-gp70 serum followed by SDS-PAGE and autoradiography. The amount of each SU was quantitated on a Molecular Dynamics PhosphorImager.
RESULTS
The hypervariable domain of Fr-MuLV SU is tolerant of insertions and deletions.
Comparison of the proline-rich central domains of murine type-C retroviral envelope genes (env) indicates that the first four of these prolines constitute a motif conserved among these envs, while the following region (residues 244 to 284 in Fr-MuLV) is hypervariable even within receptor classes (Fig. 1). To examine the tolerance of the hypervariable domain to modification, a 7-amino-acid insert, ASAVAGA, a sequence expected to have little intrinsic structure, was placed at five sites across this region. Mutants in which the ASAVAGA sequence replaced residues 244 to 273 or 264 to 285 were also constructed.
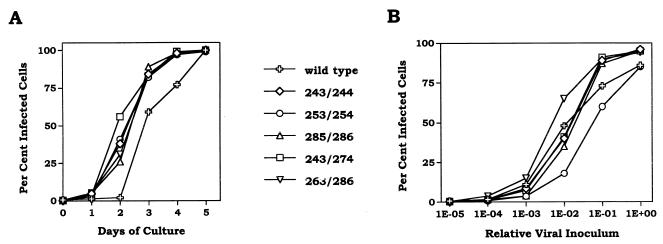
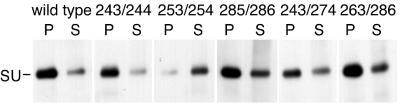
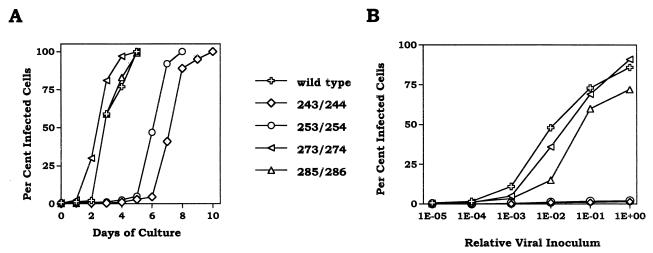
All of the ASAVAGA insertion and substitution mutants grew normally. Growth curves following transfection of plasmids expressing selected mutant viruses are presented in Fig. 2A. Differences of 1 day or less in initial growth were attributable to small differences in transfection efficiency. The specific infectivities of the virus present at the end of these growth curves were also similar to that of wild type (Fig. 2B). Despite these normal growth characteristics, examination of the envelope proteins associated with virus particles revealed that insertion of ASAVAGA following residue 253 significantly destabilized the interaction between SU and TM (Fig. 3). Cultures resulting from the above-mentioned transfections were labeled with [35S]cysteine, and particle-associated proteins were separated from soluble proteins in culture media by pelleting virus. In each case, essentially all of the core protein, p30gag, was found in the viral pellet (data not shown). The majority of wild-type SU was associated with the viral pellet. This was also the case for all of the ASAVAGA mutants except the 253/254 insertion, for which most of the SU was soluble protein found in the supernatant fraction (Fig. 3A). Interestingly, particle association of the 243/274 ASAVAGA substitution mutant SU was close to normal, despite deletion of the 253/254 region that was sensitive to insertion.
FIG. 2.
Growth characteristics of ASAVAGA insert mutants. (A) 3T3 cells were transfected with the expression plasmid for the indicated viruses, and slides were prepared for IFA daily until viral infection reached 100%. Day 0 represents data from 18 h posttransfection, at which time between 0.1 and 0.7% of cells were expressing Gag. (B) Serial dilutions of culture supernatants from the ends of the growth curves in panel A were infected into 3T3 cells, and the percentage of infected cells was determined 18 h later by IFA.
FIG. 3.
Particle association of ASAVAGA insert SUs. 3T3 cells producing the indicated viruses were labeled with [35S]cysteine, and culture supernatants were separated into soluble (S) and particulate (P) fractions by sedimentation. Samples were analyzed by RIP with hyperimmune anti-gp70 serum, followed by SDS-PAGE and autoradiography.
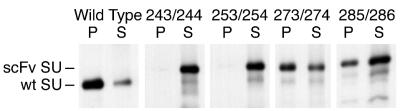
To further explore the degree of tolerance for insertions within the hypervariable domain, large insertions consisting of a single-chain antibody binding domain (scFv), derived from human MAb 5145a that recognizes a CD4 binding site epitope on the HIV-1 SU (gp120), were constructed. As shown in Fig. 4A, insertion of the scFv domain was well tolerated at the 273 and 285 insertion sites but not at the 243 and 253 insertion sites, where a significant growth delay resulted. The growth defect of the 243/244 and 253/254 scFv insertion mutants correlated with severe decreases in the specific infectivity of viral particles (Fig. 4B). The low specific infectivity of these virions indicated that the virus present at the end of these growth curves was mutant, despite the rapid spread of infection following the 4-day lag. The apparent discrepancy between this eventual rapid spread (Fig. 4A) and the extremely low specific infectivity of the virions (Fig. 4B) may reflect a contribution of cell-to-cell infection to viral spread in culture and/or the additional opportunity for shedding of SU afforded by the handling of viral supernatants in the specific infectivity experiment. These growth defects were consistent with the greatly reduced amounts of particle-associated SU found for the 243/244 and 253/254 scFv insertion mutants (Fig. 5). A small decrease in the amount of particle-associated SU was also seen for the scFv 285/286 insertion mutant, at the C-terminal boundary of the hypervariable domain (Fig. 5). There was a small amount of material in the supernatants of the scFv insert viruses that migrated similarly to wild-type SU. It appeared to be a C-terminal fragment of the mutant SU, since in each case its degree of particle association matched that of the intact scFv SU. Although the scFv insertion was not tested following residue 263, other large insertions (such as V1V2 and V4C4 domains of HIV-1 SU) at this site did not affect virus growth (data not shown). Taken together, these data show that the hypervariable domain of MuLV SU is highly tolerant of insertion and deletion, particularly in its central region.
FIG. 4.
Growth characteristics of 5145A scFv insert mutants. (A) 3T3 cells were transfected with the expression plasmid for the indicated viruses, and slides were prepared for IFA daily until viral infection reached 100%. Day 0 represents data from 18 h posttransfection, at which time between 0.1 and 0.7% of cells were expressing Gag. (B) Serial dilutions of culture supernatants from the ends of the growth curves in panel A were infected into 3T3 cells, and the percentage of infected cells was determined 18 h later by IFA.
FIG. 5.
Particle association of 5145A scFv insert SUs. 3T3 cells producing the indicated viruses were labeled with [35S]cysteine, and culture supernatants were separated into soluble (S) and particulate (P) fractions by sedimentation. Samples were analyzed by RIP with hyperimmune anti-gp70 serum, followed by SDS-PAGE and autoradiography.
Foreign sequences inserted within the hypervariable domain express active conformational structures that are exposed on the virus particle.
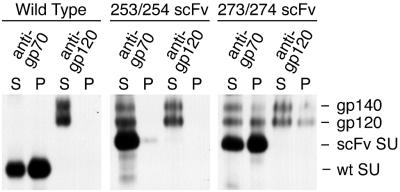
To determine whether the 5145a scFv domain inserted into the hypervariable domain of SU folded properly and was exposed on the surface of the virus particle, the ability of soluble and particle-associated SUs containing this insert to bind antigen was investigated. SEC-CHO, a CHO cell line that expresses a form of HIV-1 Env that is truncated at the boundary between the ectodomain of TM and its transmembrane domain, was used as the source of antigen. These cells secrete both the primary translation product, gp140, and gp120, the product of cleavage at the normal site between SU and TM (51). Culture supernatant of SEC-CHO labeled with [35S]cysteine was mixed with culture supernatant of MuLV-producing 3T3 cells also labeled with [35S]cysteine, particle-associated and soluble proteins were separated by centrifugation, and samples were immunoprecipitated with serum specific for MuLV SU (gp70) or for HIV-1 SU (gp120) (Fig. 6). In the wild-type control, MuLV SU was precipitated from both particulate and soluble fractions by using the anti-gp70 serum, while HIV-1 SU was precipitated only from the soluble fraction and only with the anti-gp120 antiserum. These results demonstrated that HIV-1 SU does not associate with wild-type MuLV SU or with any other component on the surface of MuLV particles. For the 273/274 scFv insertion mutant, the distribution of MuLV SU between particulate and soluble fractions, detected by immunoprecipitation with the anti-gp70 serum, was similar to that of wild type, as expected. However, unlike the results for wild-type virus, HIV-1 SU was detected in the particulate fraction containing the 273/274 scFv insert virions by immunoprecipitation with anti-gp120 serum. This association of HIV-1 SU with the mutant virus was dependent on the association of the scFv insert SU with virions, since it was not seen for the 253/254 scFv insertion mutant that contained only a trace of MuLV SU in the particulate fraction due to its defect in SU-TM interaction. Consistent with these data, a large portion of the HIV-1 SU was coprecipitated with the MuLV SU by the anti-gp70 serum from all fractions containing both scFv insert SU and HIV-1 SU. In contrast, coprecipitation of MuLV SU with HIV-1 SU with the anti-gp120 serum was not detected in any sample, presumably reflecting higher specific radioactivity and lower concentration for the HIV-1 SU than for the MuLV SU. These data clearly indicated that the 5145a scFv expressed within the hypervariable domain of MuLV SU efficiently bound antigen both on the surface of intact virions and free in solution.
FIG. 6.
gp120 binding by 5145a scFv insert SUs. 3T3 cells producing the indicated viruses and SEC-CHO cells secreting HIV-1 gp120 and gp140 were labeled with [35S]cysteine. Culture supernatants were mixed as indicated, and virus particles were separated from soluble proteins by sedimentation. Samples were analyzed by RIP with hyperimmune anti-gp70 serum or human anti-HIV-1 serum, followed by SDS-PAGE and autoradiography.
Foreign sequences inserted within the hypervariable domain provide the basis for a retroviral particle display system.
The efficient expression of inserted sequences on the surface of intact retroviral particles suggested the possibility of using such inserts for a retroviral display system. MuLV expressing the V1/V2 domain of HIV-1 gp120 as a 273/274 insert was used to demonstrate that particles expressing inserted sequences could be separated based on the binding activities of the inserts. The V1/V2 domain used consists of 96 amino acids and contains two disulfide bonds and eight signals for N-linked glycosylation (50). It presents a number of linear and conformational epitopes recognized by available MAbs (data not shown). The wild-type SU and V1/V2-bearing SU are easily resolved by SDS-PAGE due to a difference of about 30 kDa in apparent molecular size, allowing quantitation of the ratio of the two viruses present before and after separation.
Methods for selectively depleting (negative enrichment) or recovering (positive enrichment) V1/V2-expressing particles from mixtures with wild-type particles using anti-V1/V2 MAbs were established. In a negative enrichment, the desired viruses are those that are not bound by a specific antibody. This was achieved by removing virus particles bound to MAb SC258, specific for a conformational epitope expressed on the V1/V2 insert, on standard Pansorbin cells. A mixture of wild-type and V1/V2-chimeric virus was incubated with SC258 and then with Pansorbin, and unbound viruses were recovered following centrifugation. The initial virus mixture and the virus recovered after separation were amplified by infection into 3T3 cells, and [35S]cysteine-labeled supernatants were analyzed by immunoprecipitation with anti-gp70 serum (Fig. 7A). The ratio between V1/V2 SU and wild-type SU in the starting mixture was 5.3:1 after amplification, while after depletion and amplification it was 1:77, overall a 410-fold enrichment for the nonreactive virus or depletion of the reactive virus. The epitope seen by SC258 requires correct glycosylation and disulfide-bond formation of the V1/V2 domain (54). Thus, the successful depletion of V1/V2 SU virus with SC258 demonstrated that, like scFv domains, the V1/V2 domain is both correctly folded and exposed on the surface of virus particles when inserted into the hypervariable domain of MuLV SU.
FIG. 7.
Separation of retroviral particles with a MAb specific for an insert in SU. Mixtures of wild-type and V1/V2CaseA2 273/274 chimeric viruses were subjected to negative enrichment with MAb SC258 at 5 μg/ml on Pansorbin (A) or positive enrichment with SC258 at the indicated concentrations and His6-protein (A) on Ni2+-NTA resin (B). Virus mixtures before and after enrichment were expanded in 3T3 cells, and [35S]cysteine-labeled culture supernatants were analyzed by RIP with hyperimmune anti-gp70 serum, followed by SDS-PAGE and autoradiography. S, starting mixture; FT, virus not removed by Pansorbin; E20, virus eluted from Ni2+-NTA when SC258 was used at 20 μg/ml; E5, virus eluted from Ni2+-NTA when SC258 was used at 5 μg/ml; E1, virus eluted from Ni2+-NTA when SC258 was used at 1 μg/ml.
Positive enrichment requires recovery of infectious virus from the bound state. Standard conditions used to disrupt antibody-antigen complexes, such as extremes of pH or high concentrations of chaotropic agents, are lethal to MuLV (data not shown). To overcome this problem, a recombinant protein A containing a six-histidine affinity tag (40) was used. This provided a system in which the binding of antibody to a solid support, Ni2+-NTA resin, was reversible under mild conditions. Viruses complexed with MAb were adsorbed on Ni2+-NTA resin carrying His6-protein A, washed, and eluted with 10 mM EDTA. Recovered viruses were amplified by infection into 3T3 cells following immediate addition of MgCl2, and the ratios of V1/V2 SU to wild-type SU in labeled supernatants from mixtures before and after separation were compared (Fig. 7B). The ratio of chimeric to wild-type SU increased from 1:2.7 to 12:1, overall a 32-fold enrichment for reactive virus when SC258 was used at 20 μg/ml. Similar results were obtained with as little as 1 μg of SC258 per ml.
DISCUSSION
A central proline-rich and hypervariable domain is a conserved structural feature of all classes of MuLV Env (30). This study demonstrates that a large fraction of this hypervariable domain in the Fr-MuLV SU (at least the N-terminal three-fourths and the C-terminal one-half) can be deleted without significant effect on Env function and that inserts containing either 252 amino acids or 96 amino acids and eight N-linked glycosylation sites are well tolerated in the C-terminal portion of this domain. Related studies on the hypervariable domain of the amphotropic MuLV SU in an otherwise ecotropic env have recently been reported (53). In that study, progressive deletions from the C terminus of the hypervariable domain had little effect on viral growth until over 60% of the domain was removed, and tolerance for small insertions was demonstrated.
In the Fr-MuLV SU studied here, the hypervariable domain consists of 41 amino acids, residues 244 to 284. The N-terminal section of this domain appeared to be more sensitive to insertion than the C-terminal region. Seven-amino-acid insertions (ASAVAGA) were well tolerated at the beginning of the domain (between residues 243 and 244), but large insertions were not (Fig. 2 and 4). Even small insertions had a significant deleterious effect when they were placed 10 residues from this end (between residues 253 and 254) (Fig. 3). In contrast to the relative sensitivity of the N-terminal region of the hypervariable domain, even large inserts had no effect when they were placed following residues 263 or 273 and only a minor effect when placed following residue 285 at the C-terminal boundary of the domain (Fig. 5). In all cases, the biochemical defect associated with the insertions was destabilization of the interaction between SU and TM, but the Envs appeared to fold and be processed efficiently (Fig. 3 and 5). This was consistent with the elevated shedding of SU reported for other alterations in the hypervariable domain (53), in the conserved proline-rich domain (8, 53), and at a highly conserved glycan attachment site in the adjacent, N-terminal region of the C-terminal domain (at residue 302 in Fr-MuLV SU) (19). These observations suggest that the hypervariable domain is situated between sites in the end of the N-terminal domain and beginning of the C-terminal domain of SU that are involved in its interaction with TM.
Despite the sensitivity of the 253/254 site within the hypervariable domain to even the small insertion, substitution of residues 244 to 273 with the same seven-residue sequence had little or no impact on Env function. The 7-amino acid sequence could also substitute for residues 264 to 285 without deleterious effect. The ability to delete all regions of the hypervariable domain argues strongly that this domain does not contain any specific sequence or structure that is essential for Env function. This conclusion is consistent with the extensive sequence and length differences seen for this domain in natural isolates. Hypervariable domains containing as few as 30 residues have been reported (42), and the maximum deletion examined here retained 12 residues of the domain and had an additional 7 residues of foreign sequence. A structural requirement for a spacer between the globular domains of SU seems likely, given the loss of viral titer reported for deletions that retained fewer than 18 residues of the amphotropic hypervariable domain (53). These data are most consistent with a view of the linker as a flexible domain that allows the specific interactions among the N- and C-terminal domains of SU and TM needed to assemble and maintain the active structure of the Env complex. Only changes that interfere with these interactions external to the hypervariable domain would impair envelope function.
Not only are large insertions well tolerated within the hypervariable domain, but coherent structural domains that are inserted can fold into native conformations and can be effectively presented on the surface of the retroviral particle. An SU with an scFv insertion, which itself contains no internal disulfide bonds and carries no glycans, was able to bind antigen when on virus particles (Fig. 6); and an SU with an insert of the 96-amino-acid V1/V2 domain of HIV-1 gp120, which contains two disulfide bonds and eight N-linked glycans, allowed removal of virus particles from suspension by using a MAb directed against a conformational epitope in the V1/V2 domain (Fig. 7A).
These properties of insertions in the hypervariable domain of MuLV SU allowed development of a retroviral particle display system. Bacteriophage particle display systems are not suitable for expression of protein domains whose proper folding is dependent on the glycosylation or other activity found only in eukaryotic cells. An analogous system based on expression in mammalian cells would allow enrichment for variants of such domains. Two types of enrichments might be performed with such a particle display system. Isolation of a sequence with a desired binding activity requires a positive enrichment, in which particles that bind to a specific ligand are preferentially recovered. Isolation of variant sequences that have lost the ability to bind to a specific ligand requires a negative enrichment or depletion protocol in which particles that bind are preferentially removed. Methods for both types of enrichment were demonstrated for MuLV particles carrying the V1/V2 insert in SU, using the MAb directed against a conformational epitope on the insert (Fig. 7). Greater than 30-fold positive enrichment or 400-fold negative enrichment was achieved in a single step of selection and amplification, suggesting that as few as four cycles of enrichment would allow isolation of sequences present in a library at 10−6. Cycling the enrichment procedure should not present a problem, since the 273/274 site insertions are extremely stable, showing no accumulation of deleted genomes after five cycles of passage through 3T3 cells (data not shown). As constituted, the retroviral particle display system might allow directed modification of complex immunogens that present both desirable and undesirable epitopes, enriching against modified sequences that present the undesirable epitopes and for sequences that continue to express the desirable epitopes in alternation. This system could also be used to isolate small glycopeptides that interact specifically with particular ligands.
An ongoing problem in the use of retroviral vectors for human gene therapy is the lack of target cell specificity afforded by the amphotropic MuLV Env used in most systems (43). Much effort has therefore been put into engineering retroviral Envs to express binding activities that can be used to direct infection to cells of choice, the most successful of which used a 16-residue collagen-binding peptide inserted into an avian retroviral Env (48). Previous attempts with large inserts or substitutions used sites in the N terminus of SU. These constructs lost normal Env function, often required wild-type Env for incorporation into virions, and resulted in low transducing efficiencies (6, 9, 13, 21, 44, 47). The tolerance of the hypervariable domain of SU to large insertions that present new binding activities on the particle surface suggests that expression of ligands at this site in SU may lead to more efficient targeted vector delivery. This use of scFvs would provide a powerful method for targeting a wide range of cell types (47).
ACKNOWLEDGMENTS
This work was supported by NIH/CFAR subgrant P30 AI-27742 to S.C.K. and by DOD grant no. 94-0910003 and NIH/NIAID grant no. RO1-AI34217 to A.P.
REFERENCES
- 1.Adachi A, Spector S A, Kitamura N, Nakanishi S, Niwa O, Matsuyama M, Ishimoto A. Characterization of the env gene and long terminal repeat of molecularly cloned Friend mink cell focus-inducing virus DNA. J Virol. 1984;50:813–821. doi: 10.1128/jvi.50.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini J-L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J-L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J L, Kayman S C, Pinter A, Heard J M, Danos O. Role of N-linked glycosylation in the activity of the Friend murine leukemia virus SU protein receptor-binding domain. Virology. 1994;202:496–499. doi: 10.1006/viro.1994.1369. [DOI] [PubMed] [Google Scholar]
- 5.Bosselman R A, Straaten F V, Beveren C V, Verma I M, Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982;44:19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosset F-L, Morling F J, Takeuchi Y, Weiss R A, Collins M K L, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 8.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Kasahara N, Kan Y W. Ligand-directed retroviral targeting of human breast cancer cells. Proc Natl Acad Sci USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heard J M, Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Huston J S, Levinson D, Mudgett-Hunter M, Tai M-S, Novotny J, Margolies M N, Ridge R J, Bruccoleri R E, Haber E, Crea R, Oppermann H. Protein engineering of antibody sites: recovery of specific activity in an anti-digoxin single chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 14.Kayman S, Kopelman R, Kinney D, Projan S, Pinter A. Mutational analysis of N-linked glycosylation sites of the Friend murine leukemia virus envelope proteins. J Virol. 1991;65:5323–5332. doi: 10.1128/jvi.65.10.5323-5332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayman S C, Wu Z, Revesz K, Chen H-C, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 domains of HIV-1 gp120 by fusion glycoproteins containing fragments of gp120. J Virol. 1994;68:400–410. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch W, Zimmerman W, Oliff A, Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984;49:828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leamnson R N, Shander M H M, Halpern M S. A structural protein complex in Moloney leukemia virus. Nature. 1977;227:680–685. doi: 10.1016/0042-6822(77)90318-x. [DOI] [PubMed] [Google Scholar]
- 18.Lenz J, Crowther R, Straceski A, Haseltine W. Nucleotide sequence of the Akv env gene. J Virol. 1982;42:519–529. doi: 10.1128/jvi.42.2.519-529.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Pinter A, Kayman S C. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the post-cleavage envelope complex. J Virol. 1997;71:7012–7019. doi: 10.1128/jvi.71.9.7012-7019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linder M, Linder D, Hahnen J, Schott H-H, Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992;203:65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- 21.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S, Cosset F-L, Piechaczyk M. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merregaert J, Janowski M, Reddy E P. Nucleotide sequence of a Radiation Leukemia Virus genome. Virology. 1987;158:88–102. doi: 10.1016/0042-6822(87)90241-8. [DOI] [PubMed] [Google Scholar]
- 23.Montelaro R C, Sullivan S J, Bolognesi D P. An analysis of type-C retrovirus polypeptides and their associations in the virion. Virology. 1978;84:19–31. doi: 10.1016/0042-6822(78)90215-5. [DOI] [PubMed] [Google Scholar]
- 24.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S-W, Fung M S, Traincard F, Pincus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan R A, Nussbaum O, Muenchau D D, Shu L, Couture L, Anderson W F. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:4712–4721. doi: 10.1128/jvi.67.8.4712-4721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliff A I, Hager G L, Change E H, Scolnick E M, Chan H W, Lowy D R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980;33:475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Neill R R, Buckler C E, Theodore T S, Martin M A, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opstelten D-J E, Wallin M, Garoff H. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J Virol. 1998;72:6537–6545. doi: 10.1128/jvi.72.8.6537-6545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ott D, Rein A. Basis for receptor specificity of nonecotropic leukemia virus surface glycoprotein gp70(SU) J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinter A. Functions of murine leukemia virus envelope products in leukemogenesis. In: Hanafusa H, Pinter A, Pullman M, editors. Retroviruses and disease. San Diego, Calif: Academic Press; 1989. pp. 20–39. [Google Scholar]
- 32.Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15(E) complexes in AKR MuLV. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 33.Pinter A, Fleissner E. Characterization of oligomeric complexes of murine and feline leukemia virus envelope and core components formed upon crosslinking. J Virol. 1979;30:157–165. doi: 10.1128/jvi.30.1.157-165.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinter A, Honnen W J. Topography of murine leukemia virus envelope proteins: characterization of transmembrane components. J Virol. 1983;46:1056–1060. doi: 10.1128/jvi.46.3.1056-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinter A, Honnen W J. Characterization of structural and immunological properties of specific domains of Friend ecotropic and dualtropic murine leukemia virus gp70s. J Virol. 1984;49:452–458. doi: 10.1128/jvi.49.2.452-458.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinter A, Honnen W J. O-linked glycosylation of retroviral envelope gene products. J Virol. 1988;62:1016–1021. doi: 10.1128/jvi.62.3.1016-1021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinter A, Honnen W J, Kayman S C, Troshev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–1811. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 38.Pinter A, Honnen W J, Racho M E, Tilley S A. A potent, neutralizing human monoclonal antibody against a unique epitope overlapping the CD4-binding site of HIV-1 gp120 that is broadly conserved across North American and African virus isolates. AIDS Res Hum Retroviruses. 1993;9:985–996. doi: 10.1089/aid.1993.9.985. [DOI] [PubMed] [Google Scholar]
- 39.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon R Y C, Hunt T. Reversible immunoprecipitation using histidine- or glutathione S-transferase-tagged staphylococcal protein A. Anal Biochem. 1994;218:26–33. doi: 10.1006/abio.1994.1137. [DOI] [PubMed] [Google Scholar]
- 41.Quint W, Boelens W, Wezenbeck P V, Maandag E R, Berns A. Generation of AKR mink cell focus-forming virus: nucleotide sequence of the 3′ end of a somatically acquired AKR-MCF. Virology. 1984;136:425–434. doi: 10.1016/0042-6822(84)90178-8. [DOI] [PubMed] [Google Scholar]
- 42.Rassart E, Nelbach L, Jolicoeur P. Cas-Br-E murine leukemia virus: sequencing of the paralytogenic region of its genome and derivation of specific probes to study its origin and the structure of its recombinant genomes in leukemic tissues. J Virol. 1986;60:910–919. doi: 10.1128/jvi.60.3.910-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmons B, Gunzburg W H. Targetting of retroviral vectors for gene therapy. Hum Gene Ther. 1993;4:129–141. doi: 10.1089/hum.1993.4.2-129. [DOI] [PubMed] [Google Scholar]
- 44.Schnierle B S, Moritz D, Jeschke M, Groner B. Expression of chimeric envelope proteins in helper cells and integration into Moloney murine leukemia virus particles. Gene Ther. 1996;3:334–342. [PubMed] [Google Scholar]
- 45.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 46.Sijts E J, Leupers C J, Mengede E A, Loenen W A, van den Elsen P J, Melief C J. Cloning of the MCF1233 murine leukemia virus and identification of sequences involved in viral tropism, oncogenicity and T cell epitope formation. Virus Res. 1994;34:339–349. doi: 10.1016/0168-1702(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 47.Somia N V, Zoppe M, Verma I M. Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc Natl Acad Sci USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J-M, Danos O, Verdier G, Cosset F-L. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voytek P, Kozak C A. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology. 1989;173:58–67. doi: 10.1016/0042-6822(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 50.Wang N, Zhu T, Ho D D. Sequence diversity of V1 and V2 domains of gp120 from human immunodeficiency virus type 1: lack of correlation with viral phenotype. J Virol. 1995;69:2708–2715. doi: 10.1128/jvi.69.4.2708-2715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss C D, White J M. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1993;67:7060–7066. doi: 10.1128/jvi.67.12.7060-7066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witte O N, Tsukamoto-Adey A, Weissman I L. Cellular maturation of oncornavirus glycoproteins: topological arrangement of precursor and product forms in cellular membranes. Virology. 1977;76:539–553. doi: 10.1016/0042-6822(77)90236-7. [DOI] [PubMed] [Google Scholar]
- 53.Wu B W, Cannon P M, Gordon E M, Hall F L, Anderson W F. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol. 1998;72:5383–5391. doi: 10.1128/jvi.72.7.5383-5391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z, Kayman S C, Revesz K, Chen H C, Warrier S, Tilley S A, McKeating J, Shotton C, Pinter A. Characterization of neutralization epitopes in the V2 region of HIV-1 gp120: role of conserved glycosylation sites in the correct folding of the V1/V2 domain. J Virol. 1995;69:2271–2278. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]