Abstract
Nitrogen is one of the most important nutrient for plant growth and development; it is strongly associated with a variety of abiotic stress responses. As sessile organisms, plants have evolved to develop efficient strategies to manage N to support growth when exposed to a diverse range of stressors. This review summarizes the recent progress in the field of plant nitrate (NO3-) and ammonium (NH4+) uptake, which are the two major forms of N that are absorbed by plants. We explore the intricate relationship between NO3-/NH4+ and abiotic stress responses in plants, focusing on stresses from nutrient deficiencies, unfavorable pH, ions, and drought. Although many molecular details remain unclear, research has revealed a number of core signaling regulators that are associated with N-mediated abiotic stress responses. An in-depth understanding and exploration of the molecular processes that underpin the interactions between N and abiotic stresses is useful in the design of effective strategies to improve crop growth, development, and productivity.
Keywords: Nitrate, Ammonium, Uptake, Signaling, Abiotic stress
Introduction
Nitrogen is an essential macronutrient for plants, where its availability is a determinant of plant productivity (Chen et al. 2020). Nitrate (NO3-) and ammonium (NH4+) are the two major forms of N that are absorbed by plants; however, both forms are in short supply in agricultural and natural ecosystems (Crawford and Forde 2002). To achieve sufficient crop production levels and satisfy the global food demands, more than 110 Tg of N fertilizer is applied annually to crops; as such, the global demand for agricultural N fertilizer continues to escalate (Schroeder et al. 2013). However, the excessive input of N fertilizer and the inappropriate application of fertilization methods results in low N use efficiency (NUE), where 50–70% of the applied N fertilizer is lost to the surrounding environment, causing serious environmental problems, such as soil acidification and the eutrophication of water (Guo et al. 2010; McAllister et al. 2012; Kissel et al. 2020).
Plants live in dynamic and complex environments that often contain sources of stress (Zhu 2016). As plants are sessile organisms, they are unable to select their growth environment, and are limited to adapting to such environments. While N is an essential macronutrient for plant growth and development, it is also closely associated with plant adaptations to various abiotic stressors. For example, the competition or coordination between NO3-/NH4+ and other ions across the plasmalemma, affects plant resilience to stressors such as salt, potassium deficiency, and heavy metal toxicity; plants with low resilience require more fertilizer compared with plants with high resilience (Zhu 2016). As N is considered the most important nutrient for plant growth from a quantitative perspective, plants have evolved efficient strategies to manage N levels in response to various complex stressors (Nacry et al. 2013). As such, understanding the interactions between N and abiotic stress in plants is crucial to optimize the use of N fertilizers, while keeping the balance between application and the adverse effects of abiotic stresses. This understanding is important for improving modern agricultural systems and developing sustainable agricultural practices. This review briefly summarizes the process of NO3-/NH4+ uptake in plants and discusses the roles of these two forms of N in relation to different abiotic stressors, including other nutrient deficiencies, unfavorable pH, ionic stress, and drought.
Nitrogen uptake
Molecular basis of nitrate uptake
Plants have developed two NO3- uptake systems to better adapt to the fluctuating availability of NO3- in soils: a high-affinity transport system (HATS) acting at low external NO3- levels, while a low-affinity transport system (LATS) operating at high NO3- levels (Crawford and Glass 1998; Forde 2000; Lejay and Gojon 2018). In Arabidopsis, two families of transporters, the nitrate transporter 1 or peptide transporter (NRT1/PTR/NPF) and nitrate transporter 2 (NRT2), play a role in root NO3- uptake (Wang et al. 2018).
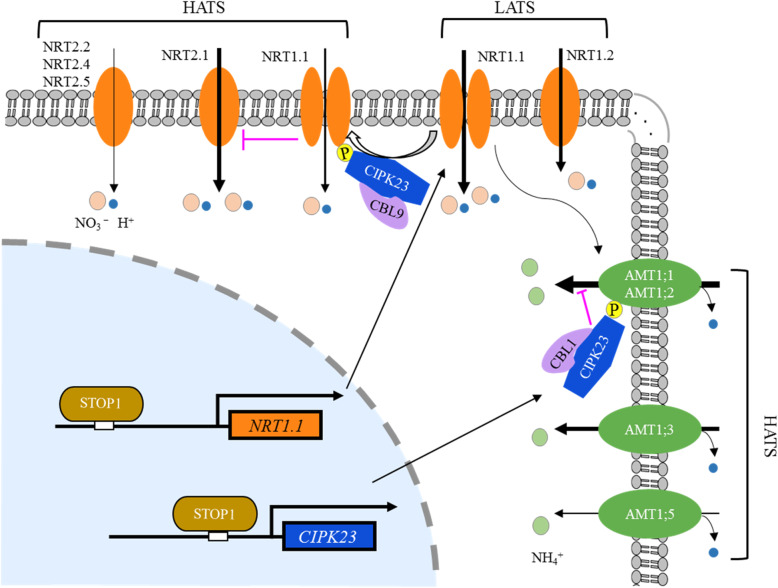
In Arabidopsis plants, NRT1.1 (also known as CHL1 or NPF6.3), was the first transporter that was identified in root NO3- uptake and is responsible for most low-affinity NO3- uptake in NO3--sufficient growth conditions (Tsay et al. 1993; Huang et al. 1996). Subsequent studies have reported that > 75% of the high-affinity NO3- uptake in plants was also contributed by NRT1.1 (Wang et al. 1998; Liu et al. 1999). In contrast, recent studies questioned this contribution, as reduced HATS influxes were not observed in the nrt1.1 mutant under low NO3- conditions (compared to wild-type plants) (Touraine and Glass 1997; Muños et al. 2004; Remans et al. 2006). These contradicting findings have obfuscated the role of NRT1.1 in root NO3- uptake. Recently, Ye et al. (2019) clarified that the critical factor in these contradictory conclusions was the varying extent of interference in NO3- uptake by NRT2.1 and NRT2.2. An nrt1.1/2.1/2.2 triple deletion mutant was generated to evaluate the role of NRT1.1 in high-affinity NO3- uptake. The difference in NO3- uptake between the nrt1.1/2.1/2.2 and nrt2.1/2.2 mutants showed that NRT1.1 contributed to ~ 12% of the high-affinity NO3- uptake in Arabidopsis (Ye et al. 2019). The switch from the low-affinity to high-affinity mode of NRT1.1 was regulated by the phosphorylation of NRT1.1 on the T101 residue (Liu and Tsay 2003). Ho et al. (2009) found that the calcineurin B-like interacting protein kinase, CIPK23, was responsible for phosphorylation in response to low NO3- cues, in which the process required the action of CBL9. NRT1.2 is another NRT1 transporter is expressed in epidermal cells and root hairs; it also absorbs NO3- from soils, though it is only directly involved in constitutive low-affinity NO3- uptake (Huang et al. 1999).
As opposed to NRT1 transporters, all NRT2 genes encode for high-affinity NO3- transporters, including NRT2.1, NRT2.2, NRT2.4, and NRT2.5, which are expressed in the roots of plants (Wang et al. 2018; Fig. 1). Among these genes, NRT2.1 is the major contributor to high-affinity NO3- uptake, and its disruption reduces HATS activity levels by up to 72% (Li et al. 2007). NRT2.2 exhibits similar expression patterns and properties to those of NRT2.1 (Li et al. 2007); however, its disruption in the nrt2.1 mutant only reduces HATS activity levels by 8% (Li et al. 2007), suggesting that there is only a marginal contribution by NRT2.2 to HATS. Two other NRT2 transporters, NRT2.4 and NRT2.5, were expressed only in response to extreme N starvation, making only minor contributions to NO3- uptake (Kiba et al. 2012; Lezhneva et al. 2014; Liu et al. 2020a).
Fig. 1.
Schematic of membrane transporters involved in the root uptake of nitrate and ammonium in Arabidopsis thaliana. The diagram represents an idealized root cell, disregarding developmental differentiation. Major signaling pathways regulating the expression and biochemical activity of these transporters are also included. Proteins are grouped as high-affinity transport systems (HATS) or lowaffinity transport systems (LATS), based on their affinity for the substrate. The contribution of transporters to N uptake is illustrated by the width of the solid lines. NRT1.1 are LATS and HATS, depending on phosphorylation by the CIPK23-CBL complex. The magenta lines indicate negative regulation
Molecular basis of ammonium uptake
The Arabidopsis genome has six ammonium transporters (AMT), all of which encode high-affinity NH4+ transporters and their expression is upregulated under N limitation (Yuan et al. 2007). Among these six genes, AMT1;4 is expressed in the shoots, while the other five genes (i.e., AMT1;1, AMT1;2, AMT1;3, AMT1;5, and AMT2;1), are expressed in the roots. To date, there has been no evidence of the contribution of AMT2;1 to high-affinity NH4+ influx (Sohlenkamp et al. 2002; Yuan et al. 2007). However, under NH4+ supply, AMT2;1 is mainly expressed in the pericycle and may contribute to the root-to-shoot translocation of NH4+ (Giehl et al. 2017); as such, NH4+ uptake is largely mediated by other AMT1s. AMT1;1, AMT1;3, and AMT1;5 are mainly expressed in the root tips and epidermal cells to uptake NH4+ from the soil (Loqué et al. 2006; Yuan et al. 2007), whereas AMT1;2 is localized in the endodermis and cortex to transport apoplastic NH4+ into the cell (Neuhäuser et al. 2007). Studies on NH4+ influx in Arabidopsis mutants have shown that AMT1;1, AMT1;2, and AMT1;3, collectively contribute to ~ 90% of the overall high-affinity NH4+ uptake capacity, while AMT1;5 mediates the remaining capacity (Loqué et al. 2006; Yuan et al. 2007). These results confirm that AMT1;1, AMT1;2, and AMT1;3 are major contributors to high-affinity NH4+ uptake, demonstrating that plants utilize different NH4+ transporters for effective NH4+ uptake under low N availability.
As high NH4+ concentrations are toxic, AMTs in Arabidopsis are efficiently deactivated by phosphorylation to prevent toxicity under high NH4+ availability (Lanquar et al. 2009). Neuhäuser et al. (2007) demonstrated that external NH4+ promotes the phosphorylation of a conserved threonine residue in the cytosolic C-terminal domain of AMT1 proteins. Subsequently, Straub et al. (2017) found that CIPK23 physically interacted with and phosphorylated AMT1;1 and AMT1;2. Moreover, they reported that the inhibiting effect of CIPK23 on AMT1 activity was CBL1-dependent and CBL9-independent (Straub et al. 2017). This is contrary to NRT1.1, which was found to be phosphorylated by CIPK23, in a CBL9-dependent manner (Ho et al. 2009); the CBL-mediated specificity may be attributable to this difference.
Roles of nitrogen in plant adaptation to nutrient deficiency
Phosphate deficiency
Phosphorus (P) is another essential macronutrient required for plant growth. Several studies have shown that the N and P uptake processes interact with each other, and require coordination to achieve optimal growth and nutritional balance in an environment with fluctuating nutrient availability (Gusewell 2004; Kant et al. 2011; Hu and Chu 2020). In most cases, N uptake in various plant species reduces under phosphate (Pi) deficiency when compared to Pi sufficiency (Lee 1982; Rufty et al. 1990; Wang et al. 2020); this is most likely to maintain the balance between N and P (Ueda et al. 2020).
In recent years, interaction mechanisms between N and P, particularly NO3- and Pi, have been studied extensively in Arabidopsis and rice. A previous study showed that nitrogen limitation adaptation (NLA) and micro-RNA827 were involved in maintaining NO3--dependent Pi homeostasis in Arabidopsis (Kant et al. 2011). NO3--inducible GARP-type transcriptional repressor 1 (NIGT1) proteins were initially identified as mediators of NO3- responses in rice (Sawaki et al. 2013); subsequently, NIGT1/HRS1 was found to integrate N and P signals in Arabidopsis (Medici et al. 2015). The expression of NIGT1 was induced by NO3- supply in an NRT1.1-dependent manner and was inhibited by Pi deficiency (Medici et al. 2015). NIGT1 was also found to repress NO3- uptake in response to Pi deficiency by directly modulating NRT2.1 and NRT2.4 expression (Kiba et al. 2018; Maeda et al. 2018). Furthermore, AtNIGT1 proteins modulate Pi starvation signaling and uptake by directly repressing the expression of SPX (Ueda et al. 2020); this inhibits the master regulator, phosphate starvation response 1 (PHR1), in response to Pi starvation (Rubio et al. 2001). A recent report has shown that NIGT1.2 can directly downregulate the transcription of the NO3- transporter gene NRT1.1 and upregulate the expression of the phosphate transporter1;1 (PHT1;1) and phosphate transporter1;4 (PHT1;4) Pi transporter genes by binding to their promoters; this will promote Pi uptake and inhibits NO3- influx during Pi deficiency (Wang et al. 2020). In rice plants, OsNRT1.1B, which is a functional homolog of AtNRT1.1, also modulates optimal NO3--phosphate acquisition (Hu et al. 2019). The repressor protein, OsSPX4, is able to interact with OsNLP3 and OsPHR2 to inhibit the NO3- and Pi starvation responses, respectively (Hu et al. 2019). Interestingly, OsNRT1.1B uses the plasma membrane-localized E3 ubiquitin ligase, NBIP1 and OsSPX4, to form a complex that promotes OsSPX4 ubiquitination and degradation in an NO3--dependent manner (Hu et al. 2019). Thus, OsNLP3 and OsPHR2 may be released and translocated to the nucleus, transducing N and P signals (Hu et al. 2019).
Unlike NO3-, the application of NH4+ fertilizers has been known to improve soil Pi uptake in agriculture (Thomson et al. 1993); however, the underlying basis linking NH4+ and Pi signals remains unclear. Recently, the transcription factor, sensitive to proton rhizotoxicity 1 (STOP1), has been found to coordinate NH4+ and Pi acquisition in Arabidopsis (Tian et al. 2021). NH4+ uptake mediated by AMTs induces rapid acidification in the rhizosphere in response to Pi deficiency. This triggers the accumulation of STOP1 in the nucleus and the subsequent excretion of organic acids by the cell, which helps to solubilize P from insoluble Pi sources (Tian et al. 2021). Interestingly, NH4+ absorption was downregulated by the protein kinase, CIPK23, whose expression was directly controlled by STOP1 when NH4+ reached toxic levels (Tian et al. 2021). Collectively, Tian et al. (2021) demonstrates that STOP1 plays a key role in coordinating NH4+ and P signals. The next challenge is determining how plants detect fluctuating environmental conditions to activate STOP1 accumulation and trigger the associated molecular and physiological responses.
Potassium deficiency
Potassium is another essential macronutrient for plant growth and development, alongside N and P. The absorption and translocation of K+ and NO3- are positively correlated in plants (Blevins et al. 1978; Triplett et al. 1980; Coskun et al. 2017; Li et al. 2017); the presence of K+ increases NO3- uptake and assimilation in wheat seedlings (Blevins et al. 1978), in turn, NO3- promotes K+ uptake and root-to-shoot translocation (Triplett et al. 1980). Recently, Fang et al. (2020) showed that NRT1.1 was upregulated at the transcriptional and post-transcriptional levels in response to low-K stress. They demonstrated that NO3- uptake by NRT1.1 in the root epidermis-cortex, favored K+ uptake, playing an important role in improving plant tolerance to low-K stress. The uptake of K+ across the plasmalemma of the root cortex cells was coupled with proton (H+) efflux mediated by H+-ATPase (Zhang et al. 2017). The optimum pH for plasmalemma H+-ATPase activity in plant roots was found to be ∼6.2–6.5 (Cowan et al. 1993; Zhu et al. 2009); lowering the pH of growth medium markedly reduced root K+ uptake (Fang et al. 2020). The NRT1.1-mediated NO3- uptake by the cell was accompanied by the co-transport of extracellular H+, which alkalizes the rhizosphere (Marschner 1995; Fang et al. 2016). Thus, the NRT1.1-mediated H+/NO3- symport of epidermis-cortex cells reduces K+ uptake-coupled H+ efflux, maintaining a suitable pH in the rhizosphere to optimize H+-ATPase activity for K+ uptake transporters (e.g., AKT1, HAK5, and KUP7), and enhance root K+ uptake (Fang et al. 2020). However, it remains unclear how NRT1.1 is regulated in response to low-K+ stress. The process described above is likely to be a general (as opposed to specific), mechanism to regulate NRT1.1 during K+ uptake; it also plays a role in the root uptake of similar ions coupled to the H+ efflux/influx.
In addition to K uptake, root-to-shoot K translocation is regulated by NRTs; NRT1.5 is a low-affinity NO3- transporter that has been identified as a major component involved in this process (Lin et al. 2008). The nrt1.5 mutants presented disturbed root-to-shoot K allocation (Drechsler et al. 2015; Li et al. 2017). Further investigations showed that while NRT1.5 is a NO3- transporter, it can also be an H+/K+ antiporter; NRT1.5-mediated K+ transportation into the xylem is independent of NO3- transport (Li et al. 2017; Du et al. 2019). In addition to its expression in the root epidermis-cortex, NRT1.1 is also expressed in the root central vasculature, where it plays a role in the coordination of K+/NO3- translocation (Fang et al. 2020). However, unlike NRT1.5, NRT1.1 is unable to directly transport K+ and its improved K+ translocation in the central vasculature is also dependent on pH regulation.
Iron deficiency
Iron is an essential micronutrient for plant growth and development. The bioavailable Fe in soils, particularly calcareous soils, often fails to meet plant needs, resulting in Fe deficiency and reduced crop yields (Guerinot and Yi 1994; Rodríguez-Celma et al. 2019). In agriculture, the application of NO3--N fertilizers often aggravates symptoms of chlorosis induced by Fe deficiency (Zhao and Ling 2007). This may be attribute to the inhibition of Fe3+-chelated reductase activity in roots by NO3- supply (Nikolic et al. 2007). As previously discussed, cellular NO3- uptake is coupled with extracellular H+ influx to alkalize the rhizosphere (Marschner 1995; Fang et al. 2016). The alkalized rhizosphere may directly restrict cellular Fe uptake and translocation, reducing Fe accumulation in young leaves. Additionally, the loss of the NRT1.1 function enhances plant tolerance to Fe deficiency (Liu et al. 2015), confirming the negative effect of NO3- on Fe nutrition in plants. However, the total Fe accumulation in nrt1.1 mutants plants was reduced along with lower expression levels of Fe-acquisition genes (e.g., IRT1, FRO2, and FIT) in response to Fe deficiency (Muños et al. 2004; Mao et al. 2014; Liu et al. 2015). These results suggest that NRT1.1-regulated Fe deficiency responses may not be associated with reduced Fe uptake, and may instead be relating to an impaired FIT-dependent Fe deficiency signaling pathway. Regardless, it is still difficult to determine the specific role of NRT1.1-mediated NO3- uptake in the Fe deficiency response because of the pleiotropic functions of NRT1.1. One possible explanation may be that NRT1.1 indirectly stimulates Fe depletion during NO3- assimilation in plants, as Fe is required as a metal cofactor in the NR assimilation pathway and NR activity increases in the nrt1.1 mutant under Fe-deficient conditions (Liu et al. 2015); there is still little clarity as to how nrt1.1 mutants increase NR activity.
In contrast, NH4+ supply has been reported to promote Fe uptake, as NH4+ uptake induces H+ release from the cell and acidifies the rhizosphere (Mengel and Geurtzen 1988; Kosegarten et al. 1999). Recently, Coleto et al. (2021) reported that the uptake of excess NH4+ by roots also affected Fe homeostasis in Arabidopsis through an unknown mechanism. This was based on the observed altered gene expression in response to Fe uptake and deficiency under high NH4+ supply relative to NO3- supply. If this impact exists, the effect of NH4+ on Fe homeostasis may be partially independent of pH regulation; however, further research is required to support this hypothesis.
Notably, some studies have found that Fe concentrations in chlorotic leaves are equal to or (in some cases), greater than those in green leaves (Kosegarten et al. 1999; López-Millán et al. 2000). This suggests that other than the restriction of Fe acquisition by roots, there may be other mechanisms that play in NO3--related Fe deficiency chlorosis. To date, various studies have shown that the chlorosis-inducing effect of NO3- may be associated with the inactivation of physiological Fe in leaf apoplasts, as NO3- results in high apoplastic pH (Hoffmann et al. 1994; Kosegarten and Englisch 1994; Mengel et al. 1994). Additionally, Fe deficiency chlorosis cannot be treated by replacing NO3- with NH4+; in contrast to NO3-, NH4+ acidifies leaf apoplasts without any external Fe supply (Aktas and Van Egmond 1979; Mengel and Geurtzen 1988; Kosegarten et al. 1999; López-Millán et al. 2000). Therefore, N-regulated apoplastic pH may play an important role in Fe deficiency responses. Specifically, there may be a central hub that regulates apoplastic pH by modulating the balance between NO3- and NH4+ uptake, in response to Fe deficiency. Further studies are required to test this hypothesis and identify potential candidates involved in these pathways.
Sulfur and molybdenum homeostasis
Sulfur is an essential constituent of enzymes that participate in N metabolism (Scherer 2008), and S addition increases NUE and biomass in plants (Kaur et al. 2011; Rais et al. 2013; Scherer 2001; Swamy et al. 2005; Carciochi et al. 2020; Salvagiotti et al. 2009; Salvagiotti and Miralles 2008). However, the application of N fertilizer aggravates S deficiency and the extent of this aggravation varies from different forms of N (Clarkson et al., 1989). Although S deficiency reduces NO3- uptake and assimilation, it had a reduced impact on NH4+ uptake (Clarkson et al., 1989). This indicates that NH4+ may be a better N source for plant growth under S deficiency compared to reduced N supply. Furthermore, De Bona et al. (2011) found that NO3- supply increased NO3- accumulation and asparagine in plants as a response to S deficiency when compared with NH4+-N supply as urea, thus repressing nitrate reductase (NR) activity.
Contrary to the positive interactions between N and S, an antagonistic interaction was observed between N and molybdenum (Rietra et al. 2017). Mo acts as catalytic center in NR, and Mo deficiency often leads to N deficiency (Rana et al. 2020). Unlike most elements, Mo bioavailability increases with soil pH (Wichard et al. 2009), and the uptake of N may theoretically regulate plant Mo deficiency responses based on the different effects that various forms of N have on the pH in the rhizosphere. This assumption is supported by the finding that N supply as NH4+ decreased the Mo content in cabbage (Domagała-Świątkiewicz and Sady 2012). Further evidence is required to fully test this hypothesis.
Roles of nitrogen in plant adaptations to H+ and alkali stresses
Acidic soils are widespread, spanning approximately half of the global arable land (Kochian et al. 2015). Acidic soils with high H+ concentrations are highly toxic, inhibiting plant growth and development (Schubert and Mengel 1990; Iuchi et al. 2007). The H+ in acidic soils is also linked to many other stress factors, such as aluminum (Al3+) toxicity and Pi deficiency (Sawaki et al. 2009; Kochian et al. 2015). Human activities exacerbate soil acidification, particularly the use of N fertilizers including urea and NH4+ (Guo et al. 2010; Kissel et al. 2020). The alkalization of the rhizosphere as a result of NO3- uptake is critical to counteract H+ stress. This conclusion is supported by Fang et al. (2016) who observed an increase in NO3- uptake with H+ stress through the specific upregulation of NRT1.1 activity; this in turn, alleviated H+ stress by increasing the pH in the rhizosphere. By contrast, although H+ stress also stimulates the expression of other NRTs, their disruptive function failed to reduce H+ stress tolerance (Fang et al. 2016). This may be potentially because NRT1.1 is responsible for the majority of NO3- transport (Wang et al. 2018; Fang et al. 2021). Notably, the growth of nlp7 mutants, which disrupts NO3- detection whilst exhibits normal NO3- uptake activity levels (Castaings et al. 2009; Marchive et al. 2013), was similar to that of Col-0 plants at low pH (Fang et al. 2016). Furthermore, the growth of chl1–9 mutants, which disrupts NO3- uptake activity but exhibits normal NO3- detection (Ho et al. 2009), was considerably lower than that of Col-0 plants and was similar to the NRT1.1-null mutants (Fang et al. 2016). These findings demonstrate that NO3- transport activity, as opposed to NO3- signaling, stimulates H+ resistance.
Recently, Ye et al. (2021) found that the low pH-related spatial expression pattern of NRT1.1 in Arabidopsis roots requires the action of the C2H2-type transcription factor, STOP1. The nrt1.1 and stop1 mutants, and the nrt1.1 stop1 double mutant, exhibited a similar phenotype that was hypersensitive to low pH. This indicates that STOP1 and NRT1.1 function in the same pathway in H+ tolerance. Molecular assays revealed that STOP1 directly activates NRT1.1 by binding to its promoter, enhancing the NO3- uptake of NRT1.1 (Ye et al. 2021). This improves the NUE of plants and creates a favorable pH in the rhizosphere for root growth by decreasing H+ concentrations. CIPK23 which regulates the NO3- uptake affinity of NRT1.1 via phosphorylation on the T101 residue (Ho et al. 2009), is also a key target gene of STOP1 (Sadhukhan et al. 2019; Tian et al. 2021). Additionally, NH4+ transport controlled by STOP1-CIPK23 may acidify the rhizosphere when only NH4+ is supplied (Tian et al. 2021). However, neither NH4+ nor NO3- uptake mediated by STOP1-CIPK23, resulted in significant changes in terms of H+ tolerance (Ye et al. 2021). Therefore, the STOP1-NRT1.1 module is likely to serve as the primary mechanism for plant adaptation to acidic environments. Further studies are needed to elucidate how roots avoid excess H+ accumulation in the cytoplasm, after stimulating H+-coupled NO3- uptake by NRT1.1.
Alkalinized soils are widespread across the earth, in which there is > 434 million ha of alkaline soils in the world (Wang et al. 2008) and > 70% of the land in northeast China is alkaline (Kawanabe and Zhu, 1991). Alkali stress may inhibit NO3- uptake and assimilation in plants (Yang et al. 2007; Yang et al. 2008; Wang et al. 2011; Wang et al. 2012). Based on physiological and tandem mass tag-based proteomic analyses, Zhao et al. (2019) found that increased N uptake and assimilation promoted plant tolerance to alkali stress, but the underlying mechanism remain unclear. To date, there has been little research on the mechanism underpinning plant adaptation to acidic or alkali stresses. This may be because both forms of stress are consistently accompanies with other unfavorable stresses, such as Al3+ toxicity in acidic soils and salt stress in alkaline soils; these are the issues that attract research attention. Indeed, acidic and alkali stresses, as opposed to the accompanying stresses, have been found to have a destructive effect on plants (Yang et al. 2008; Wang et al. 2011; Ye et al. 2021). Therefore, determining the interaction mechanism between N nutrition and unfavorable pH stresses may be hugely significant to improve plant growth under unfavorable pH stresses and helpful in understanding the accompanying stresses.
Roles of nitrogen in plant adaptations to ionic stress
Ammonium toxicity
Although NH4+ is one of the predominant N sources in many natural ecosystems, excess NH4+ is toxic to plants (von Wirén et al. 2000; Britto and Kronzucker 2002). Compared to plants growing in high-NO3- environments, plants growth in under high-NH4+ conditions exhibit several distinct toxicity symptoms, such as stunted root systems and leaf chlorosis (Britto and Kronzucker 2002; Li et al. 2014). Previous studies have shown that the excretion of H+ and general cation uptake suppression are the major contributors to the impaired growth from high NH4+ concentrations (von Wirén et al. 2000; Li et al. 2014). Interestingly, NH4+ toxicity symptoms may be reduced through the concurrent presence of small amounts of NO3- (Roosta and Schjoerring 2007; Hachiya et al. 2011). The role of NO3- in alleviating NH4+ toxicity is partially attributed to the increase in the pH in the rhizosphere and stimulation of cation uptake during NO3- uptake (Hachiya et al. 2011; Hachiya and Noguchi 2011). Surprisingly, the NRT1.1-null mutants in Arabidopsis showed a higher resistance to high NH4+ than wild-type plants, suggesting that NRT1.1 alleviates NH4+ toxicity, independent of NO3- uptake (Hachiya et al. 2011; Hachiya and Noguchi 2011). Jian et al. (2018) proposed that NH4+ toxicity is related to the NRT1.1-mediated signaling process as the NRT1.1P492L point mutant chl1–9 displayed symptoms that were similar to the wild-type plants under high-NH4+ conditions. Additional experimental data are required to clarify the exact signaling controlled by NRT1.1 in NH4+ tolerance. Another plausible explanation of the role of NO3- in counteracting NH4+ toxicity is that it inhibits chloride (Cl-) uptake via competition between NO3- and Cl-. The presence of NH4+ improves Cl- uptake to maintain balanced charge in the roots; this process is significantly inhibited by NO3- (Liu et al. 2020). In addition to being an NO3- transporter, NRT1.1 also exhibits Cl- permeability in Arabidopsis and the Xenopus oocyte system (Wen et al. 2017; Liu et al. 2020). Therefore, the enhanced NH4+ tolerance of the nrt1.1 mutant may be associated with their reduced capacity for Cl- uptake under high-NH4+ conditions when compared to wild-type plants.
Salt stress
High levels of salt stress negatively influence plant growth and crop productivity. In recent decades, it has been widely acknowledged that the inhibition of nutrient uptake via competition among sodium and other nutritional ions is a major contributor to high salt stress (Tang at el. 2011; Hessini et al. 2013). NO3- application has been demonstrated to increase root uptake and xylem loading of Na+, increasing salinity-driven root inhibition (Álvarez-Aragón et al. 2016; Álvarez-Aragón and Rodríguez-Navarro 2017). Based on kinetic data of NO3--dependent Na+ uptake at various Na+ concentrations, Álvarez-Aragón and Rodríguez-Navarro (2017) proposed that Na+ may be co-transported with NO3-. Although the co-transport of Na+ and NO3- has also been described in Zostera marina and Suaeda physophora (García-Sánchez et al. 2000; Yuan et al. 2010), the transporters that are involved have not yet been identified. Nitrate transporters such as NRT1.1 may be involved in this pathway as Na+ is partially deficient in NRT1.1-null mutants only in the presence of NO3-, when compared to wild-type plants (Álvarez-Aragón and Rodríguez-Navarro 2017).
Several studies have found that NH4+ exacerbates salt stress more than NO3-; this is observed only in a limited number of species including pea (Pisum sativum L.), poplar (Populus simonii), and wheat (Triticum aestivuni L.) (Lewis et al. 1989; Frechilla et al. 2001; Meng et al. 2016). In a recent study, Liu et al. (2020) found that when NH4+ was the sole N source, the loss of the NRT1.1 function improved the salt stress tolerance of plants. Further investigation revealed that excess Cl-, as opposed to Na+, may be responsible for the hypersensitivity to salt in wild-type Arabidopsis, with NH4+ as the sole N source (Liu et al. 2020). Consistent with this finding, AtNRT1.1 and its homolog ZmNPF6.4 have Cl- permeability in the Xenopus oocyte system; their activity were observed to be considerably inhibited by NO3- (Wen et al. 2017). Liu et al. (2020) also showed that the disruptive function of NRT1.1 in nrt1.1 mutants reduces the transmembrane Cl- influx rate in NH4+-treated Arabidopsis. Therefore, enhanced Cl- uptake by NRT1.1 in wild-type plants may be a mechanism to induce salt hypersensitivity in plants under high-NH4+ conditions. Although AtNRT1.1 specifically recognizes NO3- and chlorate (ClO3-) which have similar structures (Parker and Newstead 2014), these results raise the question of how AtNRT1.1 recognizes structurally different substrates of NO3- and Cl-. Further studies are required to fully elucidate how NRT1.1 balances NO3- and Cl- uptake in response to salt stress based on environmental NO3- and NH4+ concentrations.
Heavy metal stress
Soil heavy metal contamination has become a critical environmental issue because of its adverse ecological effects. Cadmium is one of the most toxic heavy metals in the environment. Studies have shown that NH4+ application enhances Cd uptake compared to the application of NO3-; this may be due to a decrease in soil pH (Florjin et al. 1992; Sarwar et al. 2010; Zaccheo et al. 2006). NH4+-increased Cd uptake may also be associated with NH4+ interactions with pectate and protein, as well as cell wall polymerization in the roots of Kandelia obovata (Chai et al. 2018). By contrast, several other studies have demonstrated that Cd uptake is enhanced by NO3- in many species, such as Arabidopsis, rice, potato tubers, and rape (Eriksson 1990; Maier et al. 2002; Hassan et al. 2008; Sarwar et al. 2010). In a hydroponic systems, Xie et al. (2009) found that NO3--treated Thlaspi caerulescens plants accumulated more Cd than NH4+-treated plants, despite the pH of the NH4+ solution being lower. Luo et al. (2012) reported that in pH-buffered hydroponic culture, NO3--treated plants accumulate more Cd than NH4+-treated plants, where the upregulation of Fe uptake was responsible for NO3--facilitated Cd accumulation. Within a soil system, Jalloh et al. (2009) showed that rice plants fed NO3- had higher Cd concentrations than plants fed NH4+. These findings indicate that, in addition to changing the pH in the rhizosphere, NO3- may regulate Cd uptake in plants, through NO3- transporters; this potential has been supported by subsequent evidence. Mao et al. (2014) revealed that in the presence of NO3-, the functional disruption of NRT1.1 reduces Cd uptake via a synergistic mechanism involving the simultaneous uptake of NO3-, thus enhancing Cd tolerance. In a recent study, Guan et al. (2021) found that NRT2.1 contributed substantially to facilitate Cd uptake under low-NO3- conditions by controlling NO3- uptake, further suggests that NO3- uptake exacerbates the adverse effects of Cd stress on plants.
In addition to NO3- uptake, Cd resistance in plants is also associated with NO3- allocation. For example, NRT1.8, which removes NO3- from xylem vessels, is strongly stimulated by Cd2+ stress; the disruption of NRT1.8 increases plant sensitivity to Cd2+ stress in an NO3--dependent manner (Li et al. 2010). By contrast, NRT1.5 which transports NO3- into the xylem, is strongly downregulated by Cd2+ stress; as such, it retains NO3- in the roots and contributes to Cd2+ tolerance in a similar mechanism to NRT1.8 (Chen et al. 2012). This demonstrates that plant tolerance to Cd2+ stress is regulated by NO3- reallocation to roots, mediated by NRT1.8 and NRT1.5 (Chen et al. 2012). This contrasting expression pattern of NRT1.8 and NRT1.5 in response to stress may be a result of crosstalk between ethylene (ET) and jasmonic acid (JA) signaling pathways (Zhang et al. 2014). The NRT1.1-regulated expression of NRT1.5 and NRT1.8 in roots may also contribute to Cd2+ detoxification (Gojon and Gaymard 2010; Jian et al. 2019). Jian et al. (2019) found that NRG2 operates downstream of NRT1.1 to regulate Cd2+/NO3- allocation and Cd stress tolerance. Critical factors resulting in the discrepancy described by Mao et al. (2014) and Jian et al. (2019) may be due to the variable NO3- and Fe concentrations in the growth medium, as both affect Cd uptake (He et al. 2017).
Zinc (Zn) is an essential nutrient for living organisms, though it may cause phytotoxicity when concentrations exceed requirements. The application of NO3- enhances Zn uptake in wheat roots (Erenoglu et al. 2011; Kutman et al. 2011). Additionally, Pan et al. (2020) demonstrated that a disruption in NRT1.1 reduced Zn accumulation in Arabidopsis; as such, the growth of the nrt1.1 mutant increased under Zn stress, indicating that the NRT1.1-mediated NO3- uptake pathway may play an important role in modulating Zn accumulation and tolerance to Zn stress. However, the role of other NRTs in NO3--induced Zn accumulation in plants remains unclear. By contrast, NO3- decreases Pb uptake in roots and NRT1.1 enhances Pb2+ resistance in Arabidopsis (Zhu et al. 2019). Under Pb2+ stress, NRT1.1 induces NO3- uptake, which decreases the bioavailability of Pb by preventing acidification in the rhizosphere, thus reducing Pb uptake by the roots.
Roles of nitrogen in plant adaptations to drought stress
Drought stress is a serious threat to plant life and productivity (Ding et al. 2015; Saud et al. 2017). NO3- and NH4+ concentrations have distinct effects on plant performance under drought stress; the application of NH4+ mitigates the impact of drought on plant growth, while NO3- has the opposite effect (Gao et al. 2010; Yang et al. 2012; Ding et al. 2015; Saud et al. 2017). The role of NH4+ in enhancing the drought tolerance of rice is associated with improved water uptake due to an increase in root numbers and surface area (Li et al. 2009). The decrease in the aerenchyma formation may also contribute to NH4+-enhanced drought tolerance (Yang et al. 2012). Ding et al. (2015) showed that the increased expression of root aquaporin also contributes to enhanced drought tolerance in rice plants under high-NH4+ conditions. Currently, there is a lack of evidence that NH4+ uptake is directly involved in plant drought responses.
The effect of NO3- on plant drought responses is associated with NO3- transport/assimilation. Under drought stress, many genes involved in NO3- transport/assimilation (including NRT2.5, GOGAT, GS, and AS), are repressed (Nagy et al. 2013; Singh and Ghosh 2013; Goel and Singh 2015; Duan et al. 2016). The disruptive function of genes responsible for NO3- uptake or assimilation pathways improves plant drought response. For example, Guo et al. (2003) found that NRT1.1 which is also highly expressed in guard cells, decreased plant resistance to drought stress. Notably, the reduced stomatal aperture of the Arabidopsis nrt1.1 mutant was not the result of effects on abscisic acid (ABA) responses, rather, of impaired NO3- uptake by guard cells and NO3--induced membrane depolarization (Guo et al. 2003). Additionally, mutations in genes encoding NR (NIA1 and NIA2) also exhibited a drought-resistant phenotype; this may be the result of the dual function of smaller mutants and their enhanced sensitivity to ABA (Lozano-Juste and León 2010; Chen et al. 2016). Studies have also reported that the role of NO3- in maintaining an open stomata to fix more carbon dioxide (CO2) for NO3- assimilation may contribute to higher transpiration rates in leaves under drought conditions (Guo et al. 2007; Shi et al. 2014; Ren et al. 2015). Recently, Han et al. (2021) reported that OsNR1.2 loss-of-function mutants were more tolerant to drought stress than wild-type rice under NO3--sufficient conditions, confirming that the suppression of N assimilation contributes to the survival of rice crops under drought stress. Further investigation revealed that the inhibition of the OsNR1.2 expression and the suppression of N assimilation in response to drought stress is associated with a C2H2 zinc-finger transcription factor, known as drought and salt tolerance (DST), which plays a role in H2O2 and cytokinin homeostasis (Huang et al. 2009; Li et al. 2013). Under drought stress, the expression of DST is downregulated; this action directly inhibits the activation of DST to its target genes, OsNR1.2 and OsPrx24, thereby facilitate stomata closure via preventing N assimilation and inducing H2O2 accumulation in the stomatal apparatus, respectively.
In addition to NO3- transport and assimilation, NO3- signaling also contributes to drought stress tolerance. The disruption of the NIN-like protein 7 (NLP7) in nlp7 mutants led to the impaired transduction of the NO3- signal, resulting in lower transpiration and extended survival under drought stress (Castaings et al. 2009). Many NLP7- and NRT1.1-dependent genes are differentially expressed in response to drought or ABA treatment, suggesting that disruptions in NO3- signaling may prompt changes in drought-responsive gene expressions (Araus et al. 2020). These results suggest that NO3- plays a role in drought response by regulating the activity of genes involved in NO3- uptake/assimilation and signaling, acting through or independent of the ABA pathway.
Concluding remarks and perspectives
The role of N in various abiotic stress responses has been attracting increasing attention, and there has been considerable progress in understanding these mechanisms. This review explored the effects of NO3-/NH4+, particularly NO3- uptake, on plant tolerance to different abiotic stressors. These stressors includes nutrient deficiency, unfavorable pH, ionic stress, and drought; the effects of these stressors were investigated in both physiological and molecular terms. Advancing current knowledge on plant regulation of various abiotic stress responses via N is critical to design strategies to improve crop growth, development, and productivity.
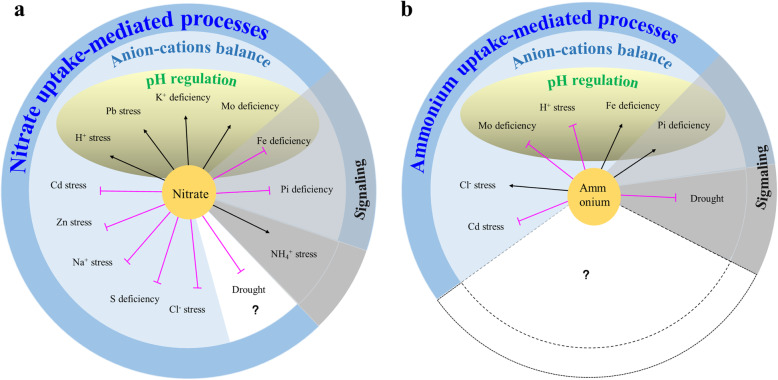
As N is always quantitatively required by plants, N uptake may affect the uptake of other ions via a common cation-anion balance mechanism (Narcy et al. 2013; Mao et al. 2014). This competition or coordination mechanism appears to substantially contribute to stress responses mediated by N. For example, NO3- uptake facilitates the synergetic transport of cations (such as H+, K+, Na+, Cd2+, and Zn2+) while it inhibits the uptake of anions (such as Cl- and SO4-), playing a role in ~ 90% of the stress responses (Fig. 2a). Among these cation-anion balance mechanisms, the presence of H+/NO3- plays a major role in plant tolerance to stresses as it effectively increases the pH in the rhizosphere, affecting the bioavailability of many elements (Marschner 1995; Fang et al. 2016; Zhu et al. 2019). The H+/NH4+ antiport also changes the pH in the rhizosphere, and may theoretically play a role in many abiotic stress responses (Fig. 2b). To date, only the Pi (Tian et al. 2021) and Fe deficiency responses (Mengel and Geurtzen 1988; Kosegarten et al. 1999) have been associated with the antiport of H+/NH4+, while the role of NH4+ in other stress responses largely remains unclear. This may be because NH4+ is toxic and NH4+ uptake is lower than NO3- uptake; further research is required to clarify the role of NH4+ in plant stress responses. The cation-anion balance mechanism may theoretically depend on the cooperation between anion and cation transporters/channels. However, to date, none of the protein-protein interactions involved in this process have been identified.
Fig. 2.
Schematic of the roles of nitrate (NO3.-)/ammonium (NH4.+) in plant responses to different stresses: (a) NO3- uptake plays a role in ~ 90% of the NO3--mediated abiotic stress responses, while only NH4 + toxicity is mediated by NO3 - signaling independent to NO3- uptake. Pi and Fe deficiency responses were mediated by NO3- uptake and NO3- signaling. The anion-cation balance mechanism contributes to abiotic stress responses mediated by NO3- uptake. NO3- uptake facilitates the synergetic transport of cations (e.g., H+, K+, Na+, Cd2+, and Zn2+), while inhibiting the uptake of anions (e.g., S, Cl-, and Pi), acting in 90% of the stress responses mediated by NO3-. The presence of H+/NO3- contributes to half of the stresses mediated by the cation-anion balance mechanism, as it effectively increases the pH in the rhizosphere; this affects the bioavailability of many elements. Although NO3- uptake exacerbates drought stress, the underlying mechanism remains unclear; (b) to date, only seven types of abiotic stresses are mediated by NH4 +, while the role of NH4 + in mediating responses to other types of stress is yet to be identified. Among the known NH4 + -mediated abiotic stress responses, ~ 80% is mediated by NH4 + uptake. Plant responses to Pi deficiency and drought stress may require the normal function of NH4 + signaling. With the exception of Cl- and Cd2+ stresses, the other four NH4 + uptake-mediated abiotic stresses (i.e., H+ stress, Pi deficiency, Mo deficiency, and Fe deficiency) were all associated with the antiport of H+/NH4+, which also changes the pH in the rhizosphere. Black arrows demonstrate the positive regulation of stress reduction responses, while the magenta lines indicate negative regulation
Plants are constantly exposed to abiotic stresses under various combinations and their response to one stress may be affected by the presence of other stresses. Thus, plant responses to multiple stresses are not just the simple summations of their responses to each individual stress (Bouain et al. 2019). For example, both Pi or Fe deficiency stress inhibits the growth of primary roots (Gruber et al. 2013; Gutierrez-Alanis et al. 2018), while this effect is eliminated when these stresses are combined as Pi-deficient root elongation is associated with the overaccumulation of Fe (Ward et al. 2008; Müller et al. 2015; Müller et al. 2015; Dong et al. 2017). Similarly, the inhibition of NO3- uptake in the nrt1.1 mutant leads to greater salt stress sensitivity under NO3- supply, while it has the opposite effect on salt stress when NH4+ is the main N source (Álvarez-Aragón et al. 2016; Álvarez-Aragón and Rodríguez-Navarro 2017; Liu et al. 2020b). These examples illustrate how plants respond to combined stress. As N is in short supply in most agricultural and natural systems, it is important to explore plant mechanisms that control growth by integrating and responding to N deficiency signals alongside other stress signals.
Finally, N allocation, distribution, and metabolism may also respond to abiotic stresses. For example, NRT1.5 and NRT1.8 participate in root-to-shoot NO3-/Na+ or NO3-/Cd transport (Chen et al. 2012). Incorporating these findings may enhance the current understanding of N-modulated abiotic stress responses.
Acknowledgements
We apologize to those whose work is not cited due to space limitations. We would like to thank Editage (www.editage.cn) for English language editing.
Abbreviations
- N
Nitrogen
- NO3
Nitrate
- NH4+
Ammonium
- NUE
Nitrogen use efficiency
- HATS
high-affinity transport systems
- LATS
low-affinity transport systems
- NRT1/PTR/NPF
Nitrate Transporter 1/Peptide Transporter
- NRT2
Nitrate Transporter 2
- NRT
Nitrate Transporter
- NR
Nitrate Reductase
- AMT
Ammonium Transporter
- CBL
Calcineurin B-Like
- CIPK23
Calcineurin B-like Interacting Protein Kinase 23
- DST
Drought and Salt Tolerance
- P
Phosphorus
- Pi
Phosphate
- NLA
Nitrogen Limitation Adaptation
- NIGT1
GARP-type Transcriptional Repressor 1
- PHR1
Phosphate Starvation Response 1
- PHT1;1
Phosphate Transporter 1;1
- PHT1;4
Phosphate Transporter1;4
- S
Sulfur
- Mo
molybdenum
- STOP1
Sensitive To Proton Rhizotoxicity 1
- K
Potassium
- Fe
Iron
- Cl-
Chloride
- ClO3-
Chlorate
- H+
Proton
- Al3+
Aluminum
- Na+
Sodium
- NLP7
NIN-like protein 7
- Cd
Cadmium
- ET
Ethylene
- JA
Jasmonic acid
- Zn
Zinc
- Pb2+
Lead
- ABA
abscisic acid
Authors’ contributions
JYY and WHT wrote the manuscript. WHT and CWJ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Zhejiang Province Natural Science Foundation (grant no. LZ21D010001).
Availability of data and materials
Not applicable.
Declarations
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wen Hao Tian, Email: tianwenhao@caas.cn.
Chong Wei Jin, Email: jincw@zju.edu.cn.
References
- Álvarez-Aragón R, Rodríguez-Navarro A. Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. 2017;91:208–219. doi: 10.1111/tpj.13556. [DOI] [PubMed] [Google Scholar]
- Álvarez-Aragón R, Haro R, Benito B, Rodríguez-Navarro A. Salt intolerance in Arabidopsis: shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta. 2016;243:97–114. doi: 10.1007/s00425-015-2400-7. [DOI] [PubMed] [Google Scholar]
- Araus V, Swift J, Alvarez JM, Henry A, Coruzzi GM. A balancing act: how plants integrate nitrogen and water signals. J Exp Bot. 2020;71:4442–4451. doi: 10.1093/jxb/eraa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas M, Egmond VF. Effect of nitrate nutrition on iron utilization by an Fe-efficient and an Fe-inefficient soybean cultivar. Plant Soil. 1979;51:257–274. doi: 10.1007/BF02232888. [DOI] [Google Scholar]
- Blevins DG, Barnett NM, Frost WB. Role of potassium and malate in nitrate uptake and translocation by wheat seedlings. Plant Physiol. 1978;62:784–788. doi: 10.1104/pp.62.5.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ. NH4+ toxicity in higher plants: a critical review. J Plant Physiol. 2002;159:567–584. doi: 10.1078/0176-1617-0774. [DOI] [Google Scholar]
- Bouain N, Krouk G, Lacombe B, Rouached Getting to the root of plant mineral nutrition: combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci. 2019;24(6):542–552. doi: 10.1016/j.tplants.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou J, Daniel-Vedele F, Fernandez E, Meyer C, Krapp A. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- Carciochi WD, Sadras VO, Pagani A, Ciampitti IA. Co-limitation and stoichiometry capture the interacting effects of nitrogen and sulfur on maize yield and nutrient use efficiency. Eur J Agron. 2020;113:125973. doi: 10.1016/j.eja.2019.125973. [DOI] [Google Scholar]
- Chai MW, Li RY, Shen XX, Tam NFY, Zan QJ, Li RL. Does ammonium nitrogen affect accumulation, subcellular distribution and chemical forms of cadmium in Kandelia obovata? Ecotox Environ Safe. 2018;162:430–437. doi: 10.1016/j.ecoenv.2018.07.031. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 2012;159:1582–1590. doi: 10.1104/pp.112.199257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KE, Chen HY, Tseng CS, Tsay YF. Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat Plants. 2020;6:1126–1135. doi: 10.1038/s41477-020-00758-0. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Wang Y, Wang JW, Babla M, Zhao C, Garcia-Mata C, Sani E, Differ C, Mak M, Hills A, Amtmann A, Blatt MR. Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol. 2016;209:1456–1469. doi: 10.1111/nph.13714. [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Saker LR, Purves JV. Depression of nitrate and ammonium transport in barley plants with diminished sulphate status. Evidence of co-regulation of nitrogen and sulphate intake. J Exp Bot. 1989;40:953–963. doi: 10.1093/jxb/40.9.953. [DOI] [Google Scholar]
- Coskun D, Britto DT, Kronzucker HJ. The nitrogen-potassium intersection: membranes, metabolism, and mechanism. Plant Cell Environ. 2017;40:2029–2041. doi: 10.1111/pce.12671. [DOI] [PubMed] [Google Scholar]
- Coleto I, Bejarano I, Marin-Pena AJ, Medina J, Rioja C, Burow M, Marino D. Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol. 2021;229:1021–1035. doi: 10.1111/nph.16918. [DOI] [PubMed] [Google Scholar]
- Cowan DSC, Clarkson DT, Hall JL. A comparison between the ATPase and proton pumping activities of plasma membranes isolated from the stele and cortex of Zea mays roots. J Exp Bot. 1993;44:983–989. doi: 10.1093/jxb/44.5.983. [DOI] [Google Scholar]
- Crawford NM, Forde BG. Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book. 2002;1:e11. doi: 10.1199/tab.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. doi: 10.1016/S1360-1385(98)01311-9. [DOI] [Google Scholar]
- De Bona FD, Fedoseyenko D, von Wirén N, Monteiro FA. Nitrogen utilization by sulfur-deficient barley plants depends on the nitrogen form. Environ Exp Bot. 2011;74:237–244. doi: 10.1016/j.envexpbot.2011.06.005. [DOI] [Google Scholar]
- Dong JS, A. Pineros M, Li XX, Yang HB, Liu Y, S. Murphy A, V. Kochian L, Liu D. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol Plants. 2017;10:244–259. doi: 10.1016/j.molp.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Ding L, Gao CM, Li YR, Li YR, Zhu YY, Xu GH, Shen QR, Kaldenhoff R, Kai L, Guo SW. The enhanced drought tolerance of rice plants under ammonium is related to aquaporin (AQP) Plant Sci. 2015;234:14–21. doi: 10.1016/j.plantsci.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Domagała-Świątkiewicz I, Sady W. Effect of nitrogen fertilization on cu, Mn, Zn, Fe, B and Mo availability in commercially grown white head cabbage. J Elem. 2012;15:455–465. doi: 10.5601/jelem.2010.15.3.455-465. [DOI] [Google Scholar]
- Drechsler N, Zheng Y, Bohner A, Nobmann B, von Wirén N, Kunze R, Rausch C. Nitrate-dependent control of shoot K homeostasis by NPF7.3/NRT1.5 and SKOR in Arabidopsis. Plant Physiol. 2015;169(4):1152–2015. doi: 10.1104/pp.15.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du XQ, Wang FL, Li H, Jing S, Yu M, Li JG, Wu WH, Kudla J, Wang Y. The transcription factor MYB59 regulates K+/NO3- translocation in the Arabidopsis response to low K+ stress. Plant Cell. 2019;31:699–714. doi: 10.1105/tpc.18.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JF, Tian H, Gao YJ. Expression of nitrogen transporter genes in roots of winter wheat (Triticum aestivum L.) in response to soil drought with contrasting nitrogen supplies. Crop Pasture Sci. 2016;67:128–136. doi: 10.1071/CP15152. [DOI] [Google Scholar]
- Erenoglu EB, Kutman UB, Ceylan Y, Yildiz B, Cakmak I. Improved nitrogen nutrition enhances root uptake, root-to-shoot translocation and remobilization of zinc (Zn) in wheat. New Phytol. 2011;189:438–448. doi: 10.1111/j.1469-8137.2010.03488.x. [DOI] [PubMed] [Google Scholar]
- Eriksson JE. Effect of nitrogen-containing fertilizers on solubility and plant uptake of cadmium. Water Air Soil Pollut. 1990;49:355–368. doi: 10.1007/BF00507075. [DOI] [Google Scholar]
- Fang XZ, Tian WH, Liu XX, Lin XY, Jin CW, Zheng SJ. Alleviation of proton toxicity by nitrateuptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol. 2016;211:149–158. doi: 10.1111/nph.13892. [DOI] [PubMed] [Google Scholar]
- Fang XZ, Liu XX, Zhu YX, Ye JY, Jin CW. The K+ and NO3- interaction mediated by NITRATE TRANSPORTER1.1 ensures better plant growth under K+-limiting conditions. Plant Physiol. 2020;184:1900–1916. doi: 10.1104/pp.20.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang XZ, Fang SQ, Ye ZQ, Liu D, Zhao KL, Jin CW (2021) NRT1.1 dual-affinity nitrate transport/signalling and its roles in plant abiotic stress resistance. Front. Plant Sci 1817. 10.3389/fpls.2021.715694 [DOI] [PMC free article] [PubMed]
- Florjin PJ, Nelemans JA, Van-Beusichem ML. The influence of the form of nitrogen nutrition on uptake and distribution of cadmium in lettuce varieties. J Plant Nutrition. 1992;15:2405–2416. doi: 10.1080/01904169209364483. [DOI] [Google Scholar]
- Forde BG. Nitrate transporters in plants: structure, function and regulation. Netherlands: Elsevier B. V; 2000. pp. 219–235. [DOI] [PubMed] [Google Scholar]
- Frechilla S, Lasa B, Ibarretxe L, Lamsfus C, Aparicio-Tejo P. Pea responses to saline stress is affected by the source of nitrogen nutrition (ammonium or nitrate) Plant Growth Regul. 2001;35:171–179. doi: 10.1023/A:1014487908495. [DOI] [Google Scholar]
- Gao YX, Li Y, Yang XX, Li HJ, Shen QR, Guo SW. Ammonium nutrition increases water absorption in rice seedlings (Oryza sativa L.) under water stress. Plant Soil. 2010;331:193–201. doi: 10.1007/s11104-009-0245-1. [DOI] [Google Scholar]
- García-Sánchez MJ, Jaime MP, Ramos A, Sanders D, Fernández JA. Sodium-dependent nitrate transport at the plasma membrane of leaf cells of the marine higher plant Zostera marina L. Plant Physiol. 2000;122:879–886. doi: 10.1104/pp.122.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl R, Laginha AM, Duan F, Rentsch D, Yuan L, von Wiren N. A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol Plant. 2017;10:1449–1460. doi: 10.1016/j.molp.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Goel P, Singh AK. Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS ONE. 2015;10:143645. doi: 10.1371/journal.pone.0143645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Gaymard F. Keeping nitrate in the roots: an unexpected requirement for cadmium tolerance in plants. J Mol Cell Biol. 2010;2:299–301. doi: 10.1093/jmcb/mjq019. [DOI] [PubMed] [Google Scholar]
- Guan MY, Chen MM, Cao ZZ (2021) NRT2.1, a major contributor to cadmium uptake controlled by high-affinity nitrate transporters. Ecotox environ safe 218: 112269. 10.1016/j.ecoenv.2021.112269 [DOI] [PubMed]
- Guo FQ, Young J, Crawford NM. The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell. 2003;15:107–117. doi: 10.1105/tpc.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding WTG, Vitousek PM, Zhang FS. Significant acidification in major Chinese croplands. Science. 2010;327:1008–1010. doi: 10.1126/science.1182570. [DOI] [PubMed] [Google Scholar]
- Guo SW, Kaldenhoff R, Uehlein N, Sattelmacher B, Brueck H. Relationship between water and nitrogen uptake in nitrate- and ammonium-supplied Phaseolus vulgaris L. plants. J Plant Nutr Soil SC. 2007;170:73–80. doi: 10.1002/jpln.200625073. [DOI] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, Wiren NV. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013;163:161–179. doi: 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusewell S. N:P ratios in terrestrial plants: variation and functional significance. New Phytol. 2004;164:243–266. doi: 10.1111/j.1469-8137.2004.01192.x. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cardenas-Torres L, Herrera-Estrella L. Adaptation to phosphate scarcity: tips from arabidopsis roots. Trends Plant Sci. 2018;23:721–730. doi: 10.1016/j.tplants.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Hachiya T, Noguchi K. Mutation of NRT1.1 enhances ammonium/low pH-tolerance in Arabidopsis thaliana. Plant signal. Behav. 2011;6:706–708. doi: 10.4161/psb.6.5.15068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Mizokami Y, Miyata K, Tholen D, Watanabe CK, Noguchi K. Evidence for a nitrate-independent function of the nitrate sensor NRT1.1 in Arabidopsis thaliana. J Plant Res. 2011;124:425–430. doi: 10.1007/s10265-010-0385-7. [DOI] [PubMed] [Google Scholar]
- Han ML, Lv QY, Zhang J, Wang T, Zhang CX, Tan RJ, Wang TL, Zhong LY, Gao YQ, Chao ZF, Li QQ, Chen GY, Shi Z, Lin HX, Chao DY (2021) Decreasing nitrogen assimilation under drought stress by suppressing DST-mediated activation of nitrate reductase 1.2 in rice. Mol Plant. 10.1016/j.molp.2021.09.005 [DOI] [PubMed]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009;23:1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MJ, Shafi M, Zhang G, Zhu Z, Qaisar M. The growth and some physiological responses of rice to cd toxicity as affected by nitrogen form. Plant Growth Regul. 2008;54:125–132. doi: 10.1007/s10725-007-9235-6. [DOI] [Google Scholar]
- He XL, Fan SK, Zhu J, Guan MY, Liu XX, Zhang YS, Jin CW. Iron supply prevents cd uptake in Arabidopsis by inhibiting IRT1 expression and favoring competition between Fe and cd uptake. Plant Soil. 2017;416:453–462. doi: 10.1007/s11104-017-3232-y. [DOI] [Google Scholar]
- Hessini K, Hamed KB, Gandour M, Mejri M, Abdelly C, Cruz C. Ammonium nutrition in the halophyte Spartina alterniflora under salt stress: evidence for a priming effect of ammonium? Plant Soil. 2013;370:163–173. doi: 10.1007/s11104-013-1616-1. [DOI] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Flänker R, Mengel K. Measurements of pH in the apoplast of sunflower leaves by means of fluorescence. Physiol Plantarum. 1994;84:146–153. doi: 10.1111/j.1399-3054.1992.tb08777.x. [DOI] [Google Scholar]
- Hu B, Chu CC. Nitrogen-phosphorus interplay: old story with molecular tale. New Phytol. 2020;225:1455–1460. doi: 10.1111/nph.16102. [DOI] [PubMed] [Google Scholar]
- Hu B, Jiang ZM, Wang W, Qiu YH, Zhang ZH, Liu YQ, Li AF, Gao XK, Liu LC, Qian YW, et al. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants. 2019;5:401–413. doi: 10.1038/s41477-019-0384-1. [DOI] [PubMed] [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF. CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell. 1996;8:2183–2191. doi: 10.1105/tpc.8.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell. 1999;11:1381–1392. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. PNAS. 2007;104:9900–9905. doi: 10.1073/pnas.0700117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalloh MA, Chen JH, Zhen FR, Zhang GP. Effect of different N fertilizer forms on antioxidant capacity and grain yield of rice growing under cd stress. J Hazard Mater. 2009;162:1081–1085. doi: 10.1016/j.jhazmat.2008.05.146. [DOI] [PubMed] [Google Scholar]
- Jian SF, Liao Q, Song HX, Liu Q, Lepo JE, Guan CY, Zhang JH, Ismail AM, Zhang ZH. NRT1.1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 2018;178:1473–1488. doi: 10.1104/pp.18.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian SF, Luo JS, Liao Q, Liu Q, Guan CY, Zhang ZH (2019) NRT1.1 regulates nitrate allocation and cadmium tolerance in Arabidopsis. Front. Plant Sci 10. 10.3389/fpls.2019.00384 [DOI] [PMC free article] [PubMed]
- Kant S, Peng MS, Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011;7:e1002021. doi: 10.1371/journal.pgen.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Chandna R, Pandey R, Abrol YP, Iqbal M, Ahmad A. Sulfur starvation and restoration affect nitrate uptake and assimilation in rapeseed. Protoplasma. 2011;248:299–311. doi: 10.1007/s00709-010-0171-3. [DOI] [PubMed] [Google Scholar]
- Kawanabe S, Zhu T. Degeneration and conservational trial of Aneurolepidium chinense grassland in northern China. J Japan Grassland Sci. 1991;37:91–99. doi: 10.14941/grass.37.91. [DOI] [Google Scholar]
- Kiba T, Feria-Bourrellier A, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, Krapp A. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 2012;24:245–258. doi: 10.1105/tpc.111.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Inaba J, Kudo T, Ueda N, Konishi M, Mitsuda N, Takiguchi Y, Kondou Y, Yoshizumi T, Ohme-Takagi M, et al. Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell. 2018;30:925–945. doi: 10.1105/tpc.17.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel DE, Bock BR, Ogles CZ (2020) Thoughts on acidification of soils by nitrogen and sulfur fertilizers. Agrosystems Geosciences Environ 3. 10.1002/agg2.20060
- Kosegarten HU, Hoffmann B, Mengel K. Apoplastic pH and Fe3+ reduction in intact sunflower leaves. Plant Physiol. 1999;121:1069–1079. doi: 10.1104/pp.121.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosegarten H, Englisch G. Effect of various nitrogen forms on the pH in leaf apoplast and on iron chlorosis of glycine max L. J Plant Nutr Soil Sci. 1994;157:401–405. doi: 10.1002/JPLN.19941570602. [DOI] [Google Scholar]
- Kochian LV, Pineros MA, Liu J, Magalhaes JV. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- Kutman UB, Yildiz B, Cakmak I. Effect of nitrogen on uptake, remobilization and partitioning of zinc and iron throughout the development of durum wheat. Plant Soil. 2011;342:149–164. doi: 10.1007/s11104-010-0679-5. [DOI] [Google Scholar]
- Lanquar V, Loque D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wiren N, Frommer WB. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell. 2009;21:3610–3622. doi: 10.1105/tpc.109.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB. Selectivity and kinetics of ion uptake by barley plants following nutrient deficiency. Ann Bot. 1982;50:429–449. doi: 10.1093/oxfordjournals.aob.a086383. [DOI] [Google Scholar]
- Lejay L, Gojon A. In: Root nitrate uptake. Advances in botanical research. Maurel C, editor. Amsterdam: Elsevier; 2018. pp. 139–169. [Google Scholar]
- Lewis OAM, Leidi EO, Lips SH. Effect of nitrogen source on growth response to salinity stress in maize and wheat. New Phytol. 1989;111:155–160. doi: 10.1111/j.1469-8137.1989.tb00676.x. [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Kiba T, Feria-Bourrellier A, Lafouge F, Boutet-Mercey S, Zoufan P, Sakakibara H, Daniel-Vedele F, Krapp A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014;80:230–241. doi: 10.1111/tpj.12626. [DOI] [PubMed] [Google Scholar]
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y. NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell. 2017;29:2016–2026. doi: 10.1105/tpc.16.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, Li L, Schroeder JI, Gassmann W, Gong JM. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell. 2010;22:1633–1646. doi: 10.1105/tpc.110.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WB, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass ADM. Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 2007;143:425–433. doi: 10.1104/pp.106.091223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li G, Kronzucker HJ, Baluška F, Shi W. Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci. 2014;19:107–114. doi: 10.1016/j.tplants.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Gao YX, Ding L, Shen QR, Guo SW. Ammonium enhances the tolerance of rice seedlings (Oryza sativa L.) to drought condition. Agric Water Manag. 2009;96:1746–1750. doi: 10.1016/j.agwat.2009.07.008. [DOI] [Google Scholar]
- Li SY, Zhao BR, Yuan DY, Duan MJ, Qian Q, Tang L, et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. PNAS. 2013;110:3167–3172. doi: 10.1073/pnas.1300359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XX, Zhu YX, Fang XZ, Ye JY, Du WX, Zhu QY, Lin XY, Jin CW. Ammonium aggravates salt stress in plants by entrapping them in a chloride over-accumulation state in an NRT1.1-dependent manner. Sci Total Environ. 2020;746:141244. doi: 10.1016/j.scitotenv.2020.141244. [DOI] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. Switching between the two action modes ofthe dual-affnity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Cui HQ, Li AN, Zhang M, Teng YB. The nitrate transporter NRT1.1 is involved in iron deficiency responses in Arabidopsis. J Plant Nutr Soil Sci. 2015;178:601–608. doi: 10.1002/jpln.201400480. [DOI] [Google Scholar]
- Liu RR, Jia T, Cui B, Song J. The expression patterns and putative function of nitrate transporter 2.5 in plants. Plant Signal Behav. 2020;15(12):1815980. doi: 10.1080/15592324.2020.1815980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 2010;152:891–903. doi: 10.1104/pp.109.148023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Abadía A, Abadía J. Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport1. Plant Physiol. 2000;124(2):873–884. doi: 10.1104/pp.124.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, Ishiyama K, Takahashi H, von Wirén N. Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 2006;48:522–534. doi: 10.1111/j.1365-313X.2006.02887.x. [DOI] [PubMed] [Google Scholar]
- Luo BF, Du ST, Lu KX, Liu WJ, Lin XY, Jin CW. Iron uptake system mediates nitrate-facilitated cadmium accumulation in tomato (Solanum lycopersicum) plants. J Exp Bot. 2012;63:3127–3136. doi: 10.1093/jxb/ers036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier NA, McLaughlin MJ, Heap M, Butt M, Smart MK. Effect of nitrogen source and calcitic lime on soil pH and potato yield, leaf chemical composition, and tuber cadmium concentrations. J Plant Nutr. 2002;25:523–544. doi: 10.1081/PLN-120003380. [DOI] [Google Scholar]
- Mao QQ, Guan MY, Lu KX, Du ST, Fan SK, Ye YQ, Lin XY, Jin CW. Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol. 2014;166:934–944. doi: 10.1104/pp.114.243766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. [Google Scholar]
- Maeda Y, Konishi M, Kiba T, Sakuraba Y, Sawaki N, Kurai T, Ueda Y, Sakakibara H, Yanagisawa S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat Commun. 2018;9:1376. doi: 10.1038/s41467-018-03832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CH, Beatty PH, Good AG. Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J. 2012;10:1011–1025. doi: 10.1111/j.1467-7652.2012.00700.x. [DOI] [PubMed] [Google Scholar]
- Meng S, Su L, Li YM, Wang YJ, Zhang CX, Zhao Z. Nitrate and ammonium contribute to the distinct nitrogen metabolism of Populus simonii during moderate salt stress. PLoS ONE. 2016;11:150354. doi: 10.1371/journal.pone.0150354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel K, Geurtzen G. Relationship between iron chlorosis and alkalinity in Zeu mays. Plant Physiol. 1988;72:460–465. doi: 10.1111/j.1399-3054.1988.tb09151.x. [DOI] [Google Scholar]
- Mengel K, Planker R, Hoffmann B. Relationship between leaf apoplast pH and iron chlorosis of sunflower (Helianthus annuus L.) J Plant Nutr. 1994;17:1053–1065. doi: 10.1080/01904169409364787. [DOI] [Google Scholar]
- Munos S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, L. Moore K, Hause G, Dinesh DC, Burstenbinder K, Abel S. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Development Cell. 2015;33:216–230. doi: 10.1016/j.devcel.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Nacry P, Bouguyon E, Gojon A. Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil. 2013;370:1–29. doi: 10.1007/s11104-013-1645-9. [DOI] [Google Scholar]
- Neuhauser B, Dynowski M, Mayer M, Ludewig U. Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 2007;143:1651–1659. doi: 10.1104/pp.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M, Cesco S, Römheld V, Varanini Z, Pinton R. Short-term interactions between nitrate and iron nutrition in cucumber. Funct Plant Biol. 2007;34:402. doi: 10.1071/FP07022. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Németh E, Guóth A, Bona L, Wodala B, Pécsváradi A. Metabolic indicators of drought stress tolerance in wheat: glutamine synthetase isoenzymes and rubisco. Plant Physiol Bioch. 2013;67:48–54. doi: 10.1016/j.plaphy.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Pan W, You Y, Weng Y, Shentu J, Lu Q, Xu Q, Liu H, Du ST. Zn stress facilitates nitrate transporter 1.1-mediated nitrate uptake aggravating Zn accumulation in Arabidopsis plants. Ecotox Environ Safe. 2020;190:110104. doi: 10.1016/j.ecoenv.2019.110104. [DOI] [PubMed] [Google Scholar]
- Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature. 2014;507:68–72. doi: 10.1038/nature13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana MS, Bhantana P, Sun XC et al (2020) Molybdenum as an essential element for crops: an overview. Biomed J Sci Tech Res 24, 18535. 10.26717/BJSTR.2020.24.004104
- Rais L, Masood A, Inam A, Khan N. Sulfur and nitrogen co-ordinately improve photosynthetic efficiency, growth and proline accumulation in two cultivars of mustard under salt stress. J Plant Biochem Physiol. 2013;1:101. doi: 10.4172/jpbp.1000101. [DOI] [Google Scholar]
- Rietra RPJJ, Heinen M, Dimkpa CO, Bindraban PS. Effects of nutrient antagonism and synergism on yield and fertilizer use efficiency. Commun Soil Sci Plant Analysis. 2017;48:1895–1920. doi: 10.1080/00103624.2017.1407429. [DOI] [Google Scholar]
- Ren BB, Wang M, Chen YP, Sun GM, Li Y, Shen QR, Guo SW. Water absorption is affected by the nitrogen supply to rice plants. Plant Soil. 2015;396:397–410. doi: 10.1007/s11104-015-2603-5. [DOI] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. PNAS. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosta HR, Schjoerring JK. Effects of ammonium toxicity on nitrogen metabolism and elemental profile of cucumber plants. J Plant Nutr. 2007;30:1933–1951. doi: 10.1080/01904160701629211. [DOI] [Google Scholar]
- Rodríguez-Celma J, Connorton JM, Kruse I, Green RT, Franceschetti M, Chen Y, Cui Y, Ling H, Yeh K, Balk J. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. PNAS. 2019;116:17584–17591. doi: 10.1073/pnas.1907971116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufty TW, Mackown CT, Israel DW. Phosphorus stress effects on assimilation of nitrate. Plant Physiol. 1990;94:328–333. doi: 10.1104/pp.94.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwar N, Malhi SS, Zia MH, Naeem A, Bibi S, Farid G. Role of mineral nutrition in minimizing cadmium accumulation by plants. J Sci Food Agric. 2010;90:925–937. doi: 10.1002/jsfa.3916. [DOI] [PubMed] [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, Koyama H. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009;150:281–294. doi: 10.1104/pp.108.134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaki N, Tsujimoto R, Shigyo M, Konishi M, Toki S, Fujiwara T, Yanagisawa S. A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 2013;54:506–517. doi: 10.1093/pcp/pct007. [DOI] [PubMed] [Google Scholar]
- Saud S, Fahad S, Yajun C, Ihsan MZ, Hammad HM, Nasim W, Amanullah J, Arif M, Alharby H. Effects of nitrogen supply on water stress and recovery mechanisms in Kentucky bluegrass plants. Front Plant Sci. 2017;8:983. doi: 10.3389/fpls.2017.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvagiotti F, Miralles DJ. Radiation interception, biomass production and grain yield as affected by the interaction of nitrogen and sulfur fertilization in wheat. Europ J Agron. 2008;28:282–290. doi: 10.1016/j.eja.2007.08.002. [DOI] [Google Scholar]
- Salvagiotti F, Castellarín JM, Miralles DJ, Pedrol HM. Sulfur fertilization improves nitrogen use efficiency in wheat by increasing nitrogen uptake. Field Crops Res. 2009;113:170–177. doi: 10.1016/j.fcr.2009.05.003. [DOI] [Google Scholar]
- Scherer HW. Sulphur in crop production — invited paper. Europ J Agron. 2001;14:81–111. doi: 10.1016/S1161-0301(00)00082-4. [DOI] [Google Scholar]
- Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK, Tsay YF, Sanders D. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert SSE, Mengel K. Effect of low pH of the root medium on proton release, growth, and nutrient uptake of field beans (Vicia faba) Plant Soil. 1990;124:239–244. doi: 10.1007/BF00009266. [DOI] [Google Scholar]
- Scherer HW. Impact of sulfur on N2 fixation of legumes. In: Khan NA, Singh S, Umar S, editors. Sulfur assimilation and abiotic stresses in plants. Berlin, Heideiberg: Springer; 2008. pp. 43–54. [Google Scholar]
- Shi JC, Yasuor H, Yermiyahu U, Zuo Q, Ben-Gal A. Dynamic responses of wheat to drought and nitrogen stresses during re-watering cycles. Agr Water Manage. 2014;146:163–172. doi: 10.1016/j.agwat.2014.08.006. [DOI] [Google Scholar]
- Singh KK, Ghosh S. Regulation of glutamine synthetase isoforms in two differentially drought-tolerant rice (Oryza sativa L.) cultivars under water deficit conditions. Plant Cell Rep. 2013;32:183–193. doi: 10.1007/s00299-012-1353-6. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 2002;130:1788–1796. doi: 10.1104/pp.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Ludewig U, Neuhäuser B. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell. 2017;29:409–422. doi: 10.1105/tpc.16.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy U, Wang M, Tripathy JN, Kim S, Hirasawa M, Knaff DB, Allen JP. Structure of spinach nitrite reductase: implications for multi-electron reactions by the iron-sulfur:siroheme cofactor. Biochemistry. 2005;44:16054–16063. doi: 10.1021/bi050981y. [DOI] [PubMed] [Google Scholar]
- Wichard T, Mishra B, Myneni SCB, Bellenger J, Kraepiel AML. Storage and bioavailability of molybdenum in soils increased by organic matter complexation. Nat Geosci. 2009;2:625–629. doi: 10.1038/NGEO589. [DOI] [Google Scholar]
- Tang ZH, Liu YJ, Guo XR, Zu YG. The combined effects of salinity and nitrogen forms on Catharanthus roseus: the role of internal ammonium and free amino acids during salt stress. J Plant Nutr Soil Sci. 2011;174:135. doi: 10.1002/jpln.200900354. [DOI] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA and Crawford NM (1993) The herbicide sensitivity gene cm.1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72(5): 705–713. 10.1016/0092-8674(93)90399-B. [DOI] [PubMed]
- Tian WH, Ye JY, Cui MQ, Chang JB, Liu Y, Li GX, Wu YR, Xu JM, Harberd NP, Mao CZ, Jin CW, Ding ZJ, Zheng SJ. A transcription factor STOP1-centered pathway coordinates ammonium and phosphate acquisition in Arabidopsis. Mol Plant. 2021;14(9):1554–1568. doi: 10.1016/j.molp.2021.06.024. [DOI] [PubMed] [Google Scholar]
- Thomson CJ, Marschner H, Romheld V. Effect of nitrogen-fertilizer form on pH of the bulk soil and rhizosphere, and on the growth, phosphorus, and micronutrient uptake of bean. J Plant Nutr. 1993;16:493–506. doi: 10.1080/01904169309364548. [DOI] [Google Scholar]
- Touraine B, Glass A. NO3- and ClO3- fluxes in the chl1-5 mutant of Arabidopsis thaliana. Does the CHL1-5 gene encode a low-affinity NO3- transporter? Plant Physiol. 1997;114(1):137–144. doi: 10.1104/pp.114.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett EW, Barnett NM, Blevins DG. Organic acids and ionic balance in xylem exudate of wheat during nitrate or sulfate absorption. Plant Physiol. 1980;65:610–613. doi: 10.1104/pp.65.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Kiba T, Yanagisawa S. Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 2020;102:448–466. doi: 10.1111/tpj.14637. [DOI] [PubMed] [Google Scholar]
- von Wirén N, Gazzarrini S, Gojon A, Frommer WB. The molecular physiology of ammonium uptake and retrieval. Curr Opin Plant Biol. 2000;3:254–261. doi: 10.1016/S1369-5266(00)80074-6. [DOI] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt D, G. Raghothama K. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang HF, Chen Y, Sun MM, Wang Y, Chen YF. The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in Arabidopsis and maize. Plant Cell. 2020;32:3519–3534. doi: 10.1105/tpc.20.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Cheng YH, Chen KE, Tsay YF. Nitrate transport, signaling, and use efficiency. Annual Rev Plant Biol. 2018;69:85–122. doi: 10.1146/annurev-arplant-042817-040056. [DOI] [PubMed] [Google Scholar]
- Wang H, Wu Z, Chen Y, Yang C, Shi D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.) Plant Soil Environ. 2011;57:286–294. doi: 10.17221/36/2011-PSE. [DOI] [Google Scholar]
- Wang YC, Ma H, Liu GF, Xu CX, Zhang DW, Ban QY. Analysis of gene expression profile of Limonium bicolor under NaHCO 3 stress using cDNA microarray. Plant Mol Biol Rep. 2008;26:241–254. doi: 10.1007/s11105-008-0037-4. [DOI] [Google Scholar]
- Wang H, Wu Z, Han J, Zheng W, Yang C. Comparison of ion balance and nitrogen metabolism in old and young leaves of alkali-stressed rice plants. PLoS ONE. 2012;7:e37817. doi: 10.1371/journal.pone.0037817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZY, Tyerman SD, Dechorgnat J, Ovchinnikova E, Dhugga KS, Kaiser BN. Maize NPF6 proteins are homologs of Arabidopsis CHL1 that are selective for both nitrate and chloride. Plant Cell. 2017;29:2581–2596. doi: 10.1105/tpc.16.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HL, Jiang RF, Zhang FS, McGrath SP, Zhao FJ. Effect of nitrogen form on the rhizosphere dynamics and uptake of cadmium and zinc by the hyperaccumulator Thlaspi caerulescens. Plant Soil. 2009;318:205–215. doi: 10.1007/s11104-008-9830-y. [DOI] [Google Scholar]
- Yang XX, Li Y, Ren BB, Ding L, Gao CM, Shen QR, Guo SW. Drought-induced root aerenchyma formation restricts water uptake in rice seedlings supplied with nitrate. Plant Cell Physiol. 2012;53:495–504. doi: 10.1093/pcp/pcs003. [DOI] [PubMed] [Google Scholar]
- Yang CW, Chong JN, Li CY, Kim CM, Shi DC, Wang DL. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil. 2007;294:263–276. doi: 10.1007/s11104-007-9251-3. [DOI] [Google Scholar]
- Yang CW, Shi DC, Wang D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.) Plant Growth Regul. 2008;56:179–190. doi: 10.1007/s10725-008-9299-y. [DOI] [Google Scholar]
- Ye JY, Tian WH, Jin CW. A reevaluation of the contribution of NRT 1.1 to nitrate uptake in Arabidopsis under low-nitrate supply. FEBS Lett. 2019;593(15):2051–2059. doi: 10.1002/1873-3468.13473. [DOI] [PubMed] [Google Scholar]
- Ye JY, Tian WH, Zhou M, Zhu QY, Du WX, Zhu YX, Liu XX, Lin XY, Zheng SJ, Jin CW. STOP1 activates NRT1.1-mediated nitrate uptake to create a favorable rhizospheric pH for plant adaptation to acidity. Plant Cell. 2021;33:3658–3674. doi: 10.1093/plcell/koab226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan LX, Loque D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wiren N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 2007;19:2636–2652. doi: 10.1105/tpc.107.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JF, Tian CY, Feng G. Effects of sodium on nitrate uptake and osmotic adjustment of Suaeda physophora. J Arid Land. 2010;2(3):190–196. doi: 10.3724/SP.J.1227.2010.00190. [DOI] [Google Scholar]
- Zaccheo P, Crippa L, Pasta VDM. Ammonium nutrition as a strategy for cadmium mobilisation in the rhizosphere of sunflower. Plant Soil. 2006;283:43–56. doi: 10.1007/s11104-005-4791-x. [DOI] [Google Scholar]
- Zhang GB, Yi HY, Gong JM. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell. 2014;26:3984–3998. doi: 10.1105/tpc.114.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Wei J, Li DX, Kong XY, Rengel Z, Chen LM, Yang Y, Cui XM, Chen Q. The role of the plasma membrane H+-ATPase in plant responses to aluminum toxicity. Front Plant Sci. 2017;8:1757. doi: 10.3389/fpls.2017.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Ling HQ. Effects of pH and nitrogen forms on expression profiles of genes involved in iron homeostasis in tomato. Plant Cell Environ. 2007;30:518–527. doi: 10.1111/j.1365-3040.2007.01638.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Liu JH, Jia RZ, Bao S, Chen XJ. Physiological and TMT-based proteomic analysis of oat early seedlings in response to alkali stress. J Proteome. 2019;193:10–26. doi: 10.1016/j.jprot.2018.12.018. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fang XZ, Dai YJ, Zhu YX, Chen HS, Lin XY, Jin CW. Nitrate transporter 1.1 alleviates lead toxicity in Arabidopsis by preventing rhizosphere acidification. J Exp Bot. 2019;70:6363–6374. doi: 10.1093/jxb/erz374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YY, Di TJ, Xu GH, Chen X, Zeng HQ, Yan F, Shen QR. Adaptation of plasma membrane H1-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ. 2009;32:1428–1440. doi: 10.1111/j.1365-3040.2009.02009.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.