Abstract
Plants are sessile organisms that evolve with a flexible signal transduction system in order to rapidly respond to environmental changes. Drought, a common abiotic stress, affects multiple plant developmental processes especially growth. In response to drought stress, an intricate hierarchical regulatory network is established in plant to survive from the extreme environment. The transcriptional regulation carried out by transcription factors (TFs) is the most important step for the establishment of the network. In this review, we summarized almost all the TFs that have been reported to participate in drought tolerance (DT) in plant. Totally 466 TFs from 86 plant species that mostly belong to 11 families are collected here. This demonstrates that TFs in these 11 families are the main transcriptional regulators of plant DT. The regulatory network is built by direct protein-protein interaction or mutual regulation of TFs. TFs receive upstream signals possibly via post-transcriptional regulation and output signals to downstream targets via direct binding to their promoters to regulate gene expression.
Supplementary Information
The online version contains supplementary material available at 10.1007/s44154-022-00048-z.
Keywords: Plant, Drought tolerance, Transcription factor, Regulatory network, Direct target
Introduction
Sessile plants constantly deal with adverse environmental changes in their entire lifetime through a dynamic responsive system. Perceiving various stresses by specific sensors immediately triggers intracellular signals, which are transmitted into the nucleus to induce transcriptional reprogramming for cellular and physiological reactions. Drought is one of the most detrimental abiotic stresses for plant growth. A few review papers focusing on general mechanism of drought resistance, drought-responding long-distance signaling, ABA-dependent and -independent phosphorylation networks in cellular signal transduction have been published (Fang and Xiong 2015; Gong et al. 2020b; Takahashi et al. 2018; Yoshida et al. 2014). Transcriptional regulators including transcription factors (TFs), Mediators and chromatin regulators that are involved in drought tolerance (DT) have also been summarized (Chang et al. 2020; Chong et al. 2020; Han and Wagner 2014; Kim et al. 2010; Takahashi et al. 2018). However, only a small proportion of reported TFs that are related to DT were mentioned in the reviews. Decades of research has revealed hundreds of DT-related TFs in different plant species, which will be discussed in this paper.
TFs, binding specific DNA elements, regulate gene expression by directly affecting binding affinity of RNA polymerase II (Pol II) to core promoters or recruiting chromatin regulators to change local chromatin accessibility (Spitz and Furlong 2012). There are at least 56 families of TFs in plants (Jin et al. 2017b), many of which are plant-specific, implying that the distinctive regulatory networks have been established in plants to transmit cellular signal for development and stress response. Some of plant TF families are large families containing more than one hundred members such as, MYB, bZIP, ERF, WRKY, bZIP, ZnF and NAC (Jin et al. 2017b). However, it is not clear whether all of plant TF families are involved in stress response. In other words, is there a possibility that only some specific TF families or subfamilies are responsible? To address this question, we collected nearly all the reported TFs (466 TFs from 86 plant species), whose roles in DT have been functionally studied (Table S1) by forward or reverse genetic methods. We found that these TFs mainly fell into 11 families including NAC, ERF, WRKY, bZIP, MYB, HD-ZIP, ZnF, bHLH, ASR, NF-Y and HSF. They act as either positive or negative regulators of DT. Overexpression of positive regulators enhances drought resistance, and mutation or silencing of these genes decreases drought resistance. By contrast, negative regulators influence DT in an opposite way. It is worth noting that a number of genes from some species were functionally analyzed by ectopic expression in the model organisms like Arabidopsis, rice, tobacco, etc. This may not truly reflect the intrinsic DT mechanism in these species. The post-transcriptional regulations of TFs, which could possibly serve as the approaches to receive upstream signals, and the direct downstream target genes of TFs are also reviewed in this paper.
DT-related TFs
Most TF families are divided into several classes or groups based on phylogenetic analysis. In order to see if specific classes of each family TFs would be responsible for drought response, we sorted out the DT-related TFs and found that more TFs in some classes have been reported to be involved in DT than the others, which is exemplified by ATAF subgroup of NAC family and group A of bZIP family (Table S1).
NAC family
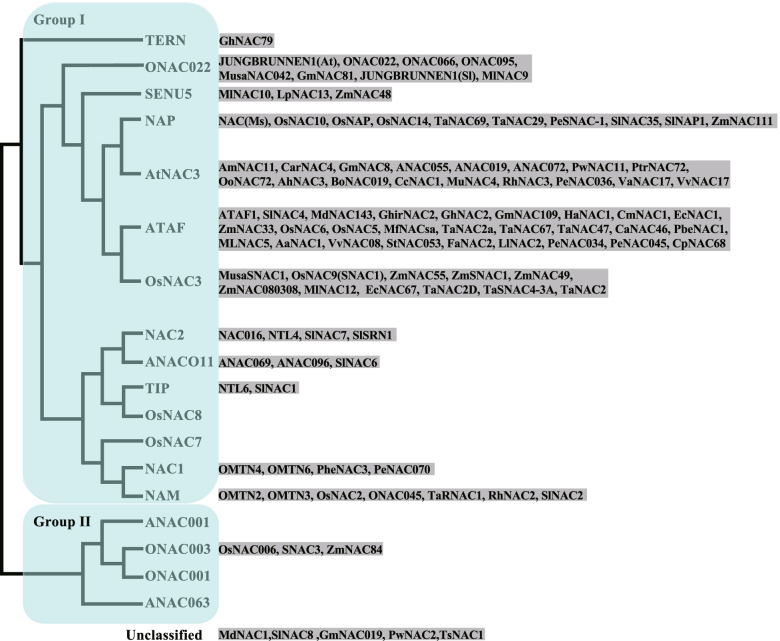
Typical NAC [No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), Cup-shaped cotyledon (CUC)] family TFs contain a conserved N-terminal NAC domain that is involved in DNA binding and dimerization, and a potential C-terminal transcriptional regulatory (TR) domain which has either activator or repressor function (Puranik et al. 2012). NAC proteins bind to various stress responsive and non-responsive NACRS (NAC recognition sequence) to regulate downstream gene expression. NAC TFs can be classified into two large groups which are further divided into 18 subgroups (Ooka et al. 2003). Members in subgroups NAP, AtNAC3, ATAF, and OsNAC3 are designated as Stress-associated NAC (SNAC). Indeed, we found that the reported NAC TFs related to DT are more enriched in these four subgroups than the others (Fig. 1). One hundred five NAC genes belonging to 14 subgroups (including unclassified) were collected in this paper and 64 of them fall into four SNAC subgroups (Fig. 1, Table S1). Specifically, 27 NAC genes from 22 plant species are included in the ATAF subgroup (Table S1), suggesting functional conservation of this subgroup genes in drought response across the plant kingdom. Seventeen of the one hundred four NAC genes negatively regulate DT while the others are positive regulators. In addition, NAC genes in group II containing four subgroups are unlikely to regulate DT as few of them have been characterized to exert the function.
Fig. 1.
NAC family TFs involved in drought tolerance. The phylogenetic tree was drawn according to (Ooka et al. 2003). 105 NAC genes belonging to 14 subgroups (including unclassified) have been studied to regulate drought tolerance
ERF family
ERF family TFs belong to AP2/ERF (APETALA2/ethylene-responsive element binding factors) superfamily which contain an AP2/ERF domain (Nakano et al. 2006). The ERF family is classified into two groups: group A (CBF/DREB proteins) and group B (ERF proteins) (Sakuma et al. 2002). Each group can be divided into six subgroups (A-1 to A-6 and B-1 to B-6). Group A proteins recognize A/GCCGAC (DRE/CRT; Dehydration-Responsive or C-Repeat element) whereas Group B proteins bind to the GCC-Box. 67 ERF genes assigned to all the subgroups except B-5 were discovered to play important roles in DT (Fig. 2, Table S1). It has long been considered that subgroup A-1 (CBF/DREB1) and subgroup A-2 (DREB2) ERF proteins are conserved regulators for improving abiotic stress tolerance (Agarwal et al. 2006). The sole member of subgroup A-3 ABI4 is an ABA-dependent transcriptional regulator which is associated with the specific CE1 element [CACC(G)], acting as either an activator or a repressor of gene expression (Wind et al. 2013). However, mutation of ABI4 reduces plant resistance to drought (Khan et al. 2020). In addition to these prominent DT genes, the other ERF TFs including group B proteins could equally contribute to DT (Fig. 2). Interestingly, overexpression of the homologs of SHN1 from seven plant species could confer DT in these plants, suggesting this gene to be a common potential DT regulator in plants (Table S1). Besides, only 1 of 67 ERF proteins, OsEBP89 in rice, has been proved to be a negative regulator of DT, as mutation of OsEBP89 leads to enhanced plant resistance to drought (Table S1) (Zhang et al. 2020b).
Fig. 2.
ERF family TFs involved in drought tolerance. The phylogenetic tree was drawn according to (Sakuma et al. 2002). 67 ERF genes assigned to all the subgroups except B-5 were discovered to play important roles in drought tolerance
WRKY family
WRKY family TFs possess one or two WRKY domains, about 60 amino acid residues with the WRKYGQK sequence followed by a C2H2 or C2HC zinc finger motif (Wu et al. 2005). The cognate binding site of WRKY domain is the W box (TTGACC/T) which could be recognized by many WRKY TFs. However, a few studies reported that some WRKY proteins also bound to non-W box cis-elements (Rushton et al. 2010). Sixty-five WRKY genes covering all the seven WRKY subfamilies have been revealed to participate in DT, and relatively more genes in subgroup I, IIc and III have been characterized (Fig. 3, Table S1). Thirteen WRKY genes in subgroup I, IIc, IId and II are negatively related to DT.
Fig. 3.
WRKY family TFs involved in drought tolerance. The phylogenetic tree was drawn according to (Rushton et al. 2010). 65 WRKY genes covering all the seven WRKY subfamilies have been revealed to participate in drought tolerance
bZIP family
bZIP (the basic leucine zipper) TFs are defined by a basic region for DNA-binding and a leucine zipper motif for dimerization (Jakoby et al. 2002). They preferentially bind to DNA sequences with an ACGT core such as the A-box (TACGTA), C-box (GACGTC) and G-box (CACGTG) (Jakoby et al. 2002). According to the basic region and additional conserved motifs, 13 groups of bZIP proteins were defined (Droge-Laser et al. 2018). The ABA signaling-engaged ABI5, ABF1, ABF2/AREB1, ABF3, ABF4/AREB2 and AREB3 belong to group A. Notably, 37 out of 58 bZIP proteins that have been identified to play important roles in DT fall in this group (Fig. 4, Table S1). Overexpression of AtABF3 in Arabidopsis, Medicago and cotton increases plant DT (Kerr et al. 2018; Wang et al. 2016b; Yoshida et al. 2010), demonstrating conserved function of this group genes in plant response to drought. However, the homologs of ABI5 seem to function distinctly in different plant species for DT. Both overexpression of wheat Wabi5 in tobacco and ectopic expression of Arabidopsis AtABI5 in cotton could enhance plant resistance to drought (Kobayashi et al. 2008; Mittal et al. 2014), whereas drought-tolerant phenotype was observed in barley hvabi5.d mutant carrying G1751A transition (Collin et al. 2020). The negative role of HvABI5 in barley DT was explained by the proposal issued by authors that it might be involved in the feedback regulation of ABA biosynthesis. Additional six bZIP genes were testified to be negative regulators of DT, as plants with overexpression of these genes is more sensitive to drought treatment (Table S1).
Fig. 4.
bZIP family TFs involved in drought tolerance. The phylogenetic tree was drawn according to (Droge-Laser et al. 2018). 58 bZIP proteins identified to play roles in drought tolerance fall in nine groups
MYB family
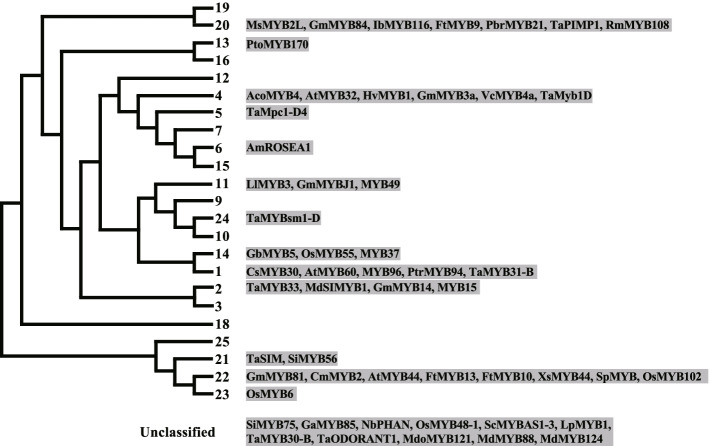
MYB (myeloblastosis) proteins are identified through a highly conserved DNA-binding domain: the MYB domain, which consists of up to four imperfect amino acid sequence repeats (R) of about 52 amino acids (Dubos et al. 2010). They can be divided into four classes based on the number of adjacent repeats: 4R-MYB, R1R2R3-type MYB (3R-MYB), R2R3-MYB and MYB-related. Plant MYB genes mostly encode R2R3-MYB, which bind to MYB-core [C/T]NGTT[G/T] and AC-rich elements (Millard et al. 2019). Plant R2R3-MYB can be divided into 25 subgroups according to MYB domain and C-terminal motifs (Millard et al. 2019). To date, functional studies on 54 R2R3-MYBs assigned to 13 subgroups as well as unidentified group have been performed for DT (Fig. 5, Table S1). These studies unraveled that seven R2R3-MYB genes were negatively engaged in plant tolerance to drought.
Fig. 5.
MYB family TFs involved in drought tolerance. The phylogenetic tree was drawn according to (Dubos et al. 2010). Functional studies on 54 R2R3-MYBs assigned to 13 subgroups as well as unidentified group have been performed for drought tolerance
HD-zip
HD-Zip (homeodomain-leucine zipper) protein is composed of a homeodomain (HD) and an immediately downstream leucine zipper motif (LZ), and classified into four subfamilies on the basis of HD-Zip domain conservation and additional conserved motifs (Ariel et al. 2007). It seems that HD-Zip TFs in each subfamily have distinct DNA-binding specificity (Ariel et al. 2007). HD-Zip I and HD-Zip II proteins forming dimers recognize CAAT(A/T)ATTG and CAAT(C/G)ATTG respectively. The binding sites of HD-Zip IV proteins are more divergent despite TAAA core is present in their target sequences. HD-Zip III protein binding site is less characterized than the other subfamilies. Eighteen HD-Zip genes encoding 11 HD-Zip I, four HD-Zip II and three HD-Zip IV proteins are implicated in plant DT (Table S1), echoing the importance of HD-Zip I ZFs in the regulation of plant drought stress response. Among these genes, rice Oshox22 and Arabidopsis ABIG1, HAT1 and HAT3 negatively regulate drought response (Zhang et al. 2012; Tan et al. 2018; Liu et al. 2016). Furthermore, HAT1 and HTA3 function redundantly since plant DT is not affected by single mutation of them while dramatically increased by double mutation (Tan et al. 2018).
ZnF family
ZnF (zinc finger) proteins are classified into several different types based on the number and order of the Cys and His residues that bind the Zinc ion in the secondary structure of the finger (Ciftci-Yilmaz and Mittler 2008). C2H2-type is one of the most abundant ZnF proteins in eukaryotes, which is mainly classified into three sets (A, B and C) (Ciftci-Yilmaz and Mittler 2008). Plant-specific Set C can be further divided into three subsets (C1, C2 and C3). We found 15 C2H2 zinc finger proteins involved in DT, all of which belong to C1 subset and most are concentrated in subclass C1–2i (Table S1). Two genes, Rice DST and soybean GmZFP3, are negative regulators of DT (Cui et al. 2015; Huang et al. 2009; Zhang et al. 2016). In addition to C2H2 zinc finger proteins, seven C3H type zinc finger proteins, three Di-19 family zinc finger proteins, two BBX family proteins and four others are also implicated in the regulation of DT (Table S1).
bHLH family
bHLH (basic/helix-loop-helix) family proteins contain the conserved bHLH domain, which consists of N-terminal basic region (15 to 20 residues rich in basic amino acids) for DNA-binding and HLH region (two amphipathic a-helices linked by a loop region) for protein-protein interaction (Toledo-Ortiz et al. 2003). The so-called core E-box hexanucleotide consensus sequence 5′-CANNTG-3′ is recognized by the bHLH proteins (Toledo-Ortiz et al. 2003). Based on phylogenetic analysis the plant bHLH proteins could be classified into 32 subfamilies (Carretero-Paulet et al. 2010). Twenty-four bHLH genes belonging to 13 subfamilies have been functionally characterized for DT (Table S1). Only three BEE genes in Arabidopsis play negative roles in DT, mutation of which simultaneously (triple mutant) enhanced drought resistance.
ASR family
The ASR (abscisic acid, stress, ripening induced) protein, albeit absent in Arabidopsis, was initially screened from tomato leaves under water-stress conditions (Gonzalez and Iusem 2014). It might have either chaperone-like function or transcription factor activity (Gonzalez and Iusem 2014). The latter has been proved by its ability to bind DNA directly. Several downstream target genes regulated by ASR proteins in some species have also been identified in vivo, further supporting its regulatory role in gene transcription. The ASR-binding DNA motif identified in rice and tomato is conserved with consensus core sequence GCCCA (Arenhart et al. 2014; Ricardi et al. 2014). Seventeen ASR proteins have been reported to be involved in DT regulation, all of which are positive regulators (Table S1).
NF-Y family
Nuclear factor Y (NF-Y) TF is formed by three subunits: NF-YA, NF-YB and NF-YC (Chaves-Sanjuan et al. 2021). The histone fold domain (HFD) of NF-YB and NF-YC mediates their heterodimerization, which produces a molecular scaffold for NF-YA interaction. The NF-Y DNA target is the CCAAT box, recognized by NF-YA. The heterodimer of NF-YB and NF-YC can also bind DNA but in a non-sequence-specific manner (Chaves-Sanjuan et al. 2021). All of the studied NF-Y TFs including seven NF-YAs, four NF-YBs and one NF-YC are positive regulators of DT (Table S1).
HSF family
HSF (Heat shock factor) proteins, which bind the conserved cis-acting (5′-nGAAn-3′) heat shock elements (HSE), transcriptionally regulate heat shock protein (Hsp) genes to play a central role in the heat stress response (Nagaraju et al. 2015). Numerous publications document that HSP also affects other abiotic stresses as well as biotic stress. HSF genes can be grouped into three classes: A, B and C. Nine class A HSF proteins participate in the regulation of DT (Table S1). Simultaneous mutation of Arabidopsis HSFA6a and HSFA6b leads to the enhancement of drought resistance while single mutation does not, suggesting cooperative and negative effect of these two genes on DT regulation (Wenjing et al. 2020).
Others
Genes in some families, whose members are always considered as important developmental regulators like WOX, KNOX, GT2, BES/BZR and GRAS, also function as DT regulators (Table S1). Some of them directly regulate the expression of genes involved in drought response or genes encoding enzymes for scavenging reactive oxygen species (ROS). And the others developmentally control plant architecture to influence DT. For example, repression of SDD1 by Arabidopsis GTL1 contributes to high abaxial stomatal density resulting in lower water use efficiency (Yoo et al. 2010). Overexpression of PagKNAT2/6b causes shorter internode length and smaller leaf size by inhibiting GA biosynthesis (Song et al. 2021) but improves drought resistance.
Regulatory network established by TFs
In response to drought, the regulatory network could be built by direct interaction of TFs to regulate common targets and mutual regulation of TFs to amplify or compromise drought signal. The direct interaction or mutual regulation summarized here has been experimentally evidenced by protein-protein interaction assays or protein-DNA binding assays in vitro or in vivo. The NAC family proteins are the most reported TFs to associate with other factors, involving both hetero-dimerization within family and interaction with other family TFs such as ERF family (LlDREB1, PbeDREB1, PbeDREB2A and DREB2A in Picea (P.) wilsonii), bZIP family (ABF2, ABF3 and ABF4), ZnF family (GmDi19–3), ZFHD family (TsHD1 and ZFHD4) (Table 1). Interestingly, by binding to each other GmNAC81 and GmNAC30 synergistically either activate or repress common target genes expression, which would depend on the conformational assembly of the two TFs at their binding site (Mendes et al. 2013). Majority of TFs interaction mediate their cooperative regulation of target genes except TINY and BES1 in Arabidopsis. Although interacting with each other, TINY and BES1 oppositely regulate a significant set of drought-induced and growth-related genes by inhibiting each other’s activities under different conditions (Xie et al. 2019). Under normal condition, BES1 promotes growth-related genes and represses drought responsive genes while TINY’s activity is inhibited. Under drought condition, TINY is induced to activate drought response and inhibit plant growth by counteracting BES1 functions. Three WRKY proteins (WRKY46, WRKY54 and WRKY70) also interact with BES1 but in a cooperative way to regulate BR-mediated plant growth and drought response (Chen et al. 2017).
Table 1.
The interaction proteins of drought-responsive transcription factors
| Function of interaction protein | TF name | Species | Interaction protein | References |
|---|---|---|---|---|
| TFs | ANAC096 | Arabidopsis | ABF2 and ABF4 | (Xu et al. 2013) |
| GmNAC81 | Soybean | GmNAC30 | (Mendes et al. 2013) | |
| PeSNAC-1 | Moso bamboo | PeSNAC-2/4 and PeNAP-1/4/5 | (Hou et al. 2020) | |
| GmNAC8 | Soybean | GmDi19–3 | (Yang et al. 2020a) | |
| PwNAC11 | Picea wilsonii | ABF3, DREB2A | (Yu et al. 2021) | |
| HaNAC1 | Haloxylon ammodendron | AtNAC32 | (Gong et al. 2020a) | |
| PbeNAC1 | Pyrus | PbeDREB1, PbeDREB2A | (Jin et al. 2017a) | |
| LlNAC2 | Lily | LlDREB1, ZFHD4 | (Yong et al. 2019b) | |
| TsNAC1 | Thellungiella halophile | TsHD1 | (Liu et al. 2019a) | |
| TINY | Arabidopsis | BES1 | (Xie et al. 2019) | |
| GmWRKY27 | Soybean | GmMYB174 | (Wang et al. 2015) | |
| WRKY46 | Arabidopsis | BES1 | (Chen et al. 2017) | |
| WRKY54 | Arabidopsis | BES1 | (Chen et al. 2017) | |
| WRKY70 | Arabidopsis | BES1 | (Chen et al. 2017) | |
| TaHDZipI-5 | Wheat | TaHDZipI-3 | (Yang et al. 2018) | |
| DST | Rice | DCA1 | (Cui et al. 2015) | |
| CmBBX19 | Chrysanthemum | CmABF3 | (Xu et al. 2020) | |
| PdNF-YB21 | Poplar | PdFUS3 | (Zhou et al. 2020) | |
| Kinases | NTL6 | Arabidopsis | SnRK2.8 | (Kim et al. 2012) |
| ZmNAC84 | Maize | ZmCCaMK | (Zhu et al. 2016) | |
| AtERF7 | Arabidopsis | PKS3, AtSin3 | (Song et al. 2005) | |
| OsEBP89 | Rice | SnRK1alph | (Zhang et al. 2020b) | |
| RAP2.6 | Arabidopsis | CDK8 and SnRK2.6 | (Zhu et al. 2020b) | |
| OsWRKY30 | Rice | OsMPK3, OsMPK4, OsMPK7, OsMPK14, OsMPK20–4, and OsMPK20–5, | (Shen et al. 2012) | |
| GhWRKY59 | Cotton | GhMAP3K15-GhMKK4-GhMPK6 | (Li et al. 2017a) | |
| OsWRKY55 | Rice | OsMPK7, OsMPK9, OsMPK20–1, and OsMPK20–4 | (Huang et al. 2021) | |
| ABF1 | Arabidopsis | AtCPK4, AtCPK11 | (Zhu et al. 2007) | |
| AREB1 | Arabidopsis | SRK2D/SnRK2.2 | (Yoshida et al. 2015) | |
| AREB2 | Arabidopsis | SRK2D/SnRK2.2, AtCPK4, AtCPK11 | (Yoshida et al. 2015; Zhu et al. 2007) | |
| ABF3 | Arabidopsis | SRK2D/SnRK2.2, AtCPK6 | (Yoshida et al. 2015; Zhang et al. 2020a) | |
| ABI5 | Arabidopsis | AtCPK6 | (Zhang et al. 2020a) | |
| OsbZIP23 | Rice | SAPK2 | (Zong et al. 2016) | |
| OsbZIP46CA1 | Rice | SAPK6 | (Chang et al. 2017) | |
| CbABF1 | Cryophyte | CbSnRK2.6 | (Yue et al. 2019) | |
| OsbZIP62 | Rice | SAPKs | (Yang et al. 2019) | |
| FtbZIP5 | Buckwheat | FtSnRK2.6 | (Li et al. 2020) | |
| MYB44 | Arabidopsis | MPK3 | (Persak and Pitzschke 2014) | |
| HAT1 | Arabidopsis | SnRK2.3 | (Tan et al. 2018) | |
| Di19 | Arabidopsis | CPK11 | (Liu et al. 2013) | |
| TINY | Arabidopsis | BIN2 | (Xie et al. 2019) | |
| WRKY46 | Arabidopsis | BIN2 | (Chen et al. 2017) | |
| WRKY54 | Arabidopsis | BIN2 | (Chen et al. 2017) | |
| WRKY70 | Arabidopsis | BIN2 | (Chen et al. 2017) | |
| SlVOZ1 | Tomato | SlOST1 | (Chong et al. 2022) | |
| UPS | ABI5 | Arabidopsis | DWA1/DWA2, KEG, ABD1 | (Chen et al. 2013; Lee et al. 2010; Seo et al. 2014) |
| ABF1 | Arabidopsis | KEG | (Chen et al. 2013) | |
| ABF3 | Arabidopsis | KEG | (Chen et al. 2013) | |
| DREB2A | Arabidopsis | DRIP1 | (Qin et al. 2008) | |
| MdNAC143 | Apple | MdBT2 | (Ji et al. 2020) | |
| AtERF53 | Arabidopsis | RGLG2 | (Cheng et al. 2012) | |
| OsWRKY11 | Rice | ubiquitin-proteasome | (Lee et al. 2018) | |
| CaATBZ1 | Capsicum Annuum | CaASRF1 | (Joo et al. 2019b) | |
| CaDILZ1 | Capsicum Annuum | CaDSR1 | (Lim et al. 2018) | |
| ROC4 | Rice | DHS | (Wang et al. 2018) | |
| Others | CcNAC1 | Jute | KCS | (Zhang et al. 2021) |
| PwNAC2 | Picea wilsonii | PwRFCP1 | (Zhang et al. 2018) | |
| ABI4 | Arabidopsis | PWR, HDA9 | (Khan et al. 2020; Baek et al. 2020) | |
| OsDRAP1 | Rice | OsCBSX3 | (Huang et al. 2018) | |
| MeWRKY20 | Cassava | MeHSP90s | (Wei et al. 2020) | |
| PtrAREB1–2 | Poplar | ADA2b-GCN5 | (Li et al. 2019a) | |
| GmMYB81 | Soybean | GmSGF14l | (Bian et al. 2020) | |
| ZFP182 | Rice | ZIURP1 | (Huang et al. 2012) | |
| IbC3H18 | Sweetpotato | IbPR5 | (Zhang et al. 2019) | |
| MfNACsa | Medicago | APT1 | (Duan et al. 2017) |
Although DREB proteins including DREB1 and DREB2 are considered as master regulators of both drought and cold response, their direct downstream genes proved experimentally have seldom been reported. By contrast, DREB proteins are likely to serve as common targets regulated by multiple family TFs like NAC (JUNGBRUNNEN1 and ONAC066), WRKY (TaWRKY19 and GhWRKY59), bZIP (AREB1, AREB2 and AtABF3), ZnF (Os12g38960, Os03g32230 and Os11g47630), MYB (AtMYB32), bHLH (ZjICE2, ZmbHLH124 and ZmPTF1), NF-Y (GmNFYA5) and WOX (OsWOX13) (Table 2). Among these TFs, only three ZnF family proteins and AtMYB32 are negative regulators of DREB genes (Figueiredo et al. 2012; Li et al. 2021b). The activation of DREB genes by AREB1, AREB2 and AtABF3 couples ABA-dependent and ABA-independent pathways in response to drought (Kim et al. 2011). In addition, ABA-dependent bZIP family regulators could be regulated by various TFs. For example, AREB1 is activated by AtWRKY63 while repressed by drought-responsive NAC016 to form a feed-forward loop (Ren et al. 2010; Sakuraba et al. 2015). AtMYB32 is responsible for the activation of ABI5 while AtWRKY40 and AtRAV1 are the negative regulators of ABI5 (Li et al. 2021b; Liu et al. 2012; Feng et al. 2014).
Table 2.
The direct downstream genes of drought-responsive transcription factors
| Function of downstream genes | Protein name | Species | Direct target genes | Transcriptional activity | References |
|---|---|---|---|---|---|
| TFs | JUNGBRUNNEN1 | Tomato | DREB1, DREB2 | Activation | (Thirumalaikumar et al. 2018) |
| ONAC066 | Rice | OsDREB2A | Activation | (Yuan et al. 2019) | |
| NAC016 | Arabidopsis | AREB1 | Repression | (Sakuraba et al. 2015) | |
| TaWRKY2 | Wheat | STZ | Activation | (Niu et al. 2012) | |
| TaWRKY19 | Wheat | DREB2A | Activation | (Niu et al. 2012) | |
| GmWRKY27 | Soybean | GmNAC29 | Repression | (Wang et al. 2015) | |
| MdWRKY31 | Apple | MdRAV1 | Repression | (Zhao et al. 2019) | |
| GhWRKY91 | Cotton | GhWRKY17 | Activation | (Gu et al. 2019) | |
| AtWRKY63 | Arabidopsis | ABF2 | Activation | (Ren et al. 2010) | |
| GhWRKY59 | Cotton | GhDREB2 | Activation | (Li et al. 2017a) | |
| OsWRKY55 | Rice | OsAP2–39 | Activation | (Huang et al. 2021) | |
| CaWRKY70 | Chickpea | CaHDZ12 | Repression | (Sen et al. 2017) | |
| AREB1 | Arabidopsis | DREB2A | Activation | (Kim et al. 2011) | |
| AREB2 | Arabidopsis | DREB2A | Activation | (Kim et al. 2011) | |
| AtABF3 | Arabidopsis | DREB2A | Activation | (Kim et al. 2011) | |
| PtrAREB1–2 | Poplar | PtrNAC006, PtrNAC007, PtrNAC120 | Activation | (Li et al. 2019a) | |
| OsABF1 | Rice | OsbZIP23, OsbZIP46, OsbZIP72 | Activation | (Zhang et al. 2017) | |
| AtMYB32 | Arabidopsis | ABI3, ABI4, ABI5, CBF4 |
Activation/ Repression |
(Li et al. 2021b) | |
| AtWRKY40 | Arabidopsis | ABI5 | Repression | (Liu et al. 2012) | |
| AtHB13 | Arabidopsis | JUNGBRUNNEN1 | Activation | (Ebrahimian-Motlagh et al. 2017) | |
| Os12g38960 | Rice | OsDREB1B | Repression | (Figueiredo et al. 2012) | |
| Os03g32230 | Rice | OsDREB1B | Repression | (Figueiredo et al. 2012) | |
| Os11g47630 | Rice | OsDREB1B | Repression | (Figueiredo et al. 2012) | |
| ZjICE2 | Zoysia Japonica | ZjDREB1 | Activation | (Zuo et al. 2020) | |
| ZmbHLH124 | Maize | ZmDREB2A | Activation | (Wei et al. 2021) | |
| ThMYC6 | Tamarix Hispida | ThbZIP1 | Activation | (Ji et al. 2013) | |
| ZmPTF1 | Maize | CBF4, ATAF2/NAC081, NAC30 | Activation | (Li et al. 2019b) | |
| ZmNF-YA3 | Maize | bHLH92 | Repression | (Su et al. 2018) | |
| GmNFYA5 | Soybean | GmDREB2, GmbZIP1 | Activation | (Ma et al. 2020) | |
| SiARDP | Foxtail Millet | SiASR4 | Activation | (Li et al. 2016) | |
| OsWOX13 | Rice | OsDREB1A, OsDREB1F | Activation | (Minh-Thu et al. 2018) | |
| ABA metabolism and signaling | ATAF1 | Arabidopsis | NCED3 | Activation | (Jensen et al. 2013) |
| GhirNAC2 | Cotton | GhNCED3a/3c | Activation | (Shang et al. 2020) | |
| WRKY57 | Arabidopsis | NCED3 | Activation | (Jiang et al. 2012) | |
| MeWRKY20 | Cassava | MeNCED5 | Activation | (Wei et al. 2020) | |
| MaWRKY80 | Banana | MaNCED | Activation | (Liu et al. 2020a) | |
| PbrWRKY53 | Pyrus | PbrNCED1 | Activation | (Liu et al. 2019b) | |
| OsbZIP23 | Rice | OsPP2C49, OsNCED4 | Activation | (Zong et al. 2016) | |
| MdMYB88 | Apple | NCED3 | Activation | (Xie et al. 2021) | |
| HAT1 | Arabidopsis | ABA3 and NCED3 | Repression | (Tan et al. 2018) | |
| ZmPTF1 | Maize | NCED | Activation | (Li et al. 2019b) | |
| PdNF-YB21 | Poplar | PdNCED3 | Activation | (Zhou et al. 2020) | |
| GmWRKY54 | Soybean | PYL8, SRK2A | Activation | (Wei et al. 2019) | |
| GhWRKY21 | Cotton | GhHAB | Activation | (Wang et al. 2020) | |
| ATHB7 | Arabidopsis | PP2C, PYL5, PYL8 | Activation | (Valdes et al. 2012) | |
| ATHB12 | Arabidopsis | PP2C, PYL5, PYL8 | Activation | (Valdes et al. 2012) | |
| OsABF1 | Rice | OsPP48, OsPP108 | Activation | (Zhang et al. 2017) | |
| bHLH122 | Arabidopsis | CYP707A3 | Repression | (Liu et al. 2014b) | |
| OsNAC2 | Rice | OsSAPK1 | Repression | (Shen et al. 2017) | |
| ROS-related | NTL4 | Arabidopsis | AtrbohC, AtrbohE | Activation | (Lee et al. 2012) |
| ZmNAC84 | Maize | SOD2 | Activation | (Han et al. 2021) | |
| NtERF172 | tobacco | NtCAT | Activation | (Zhao et al. 2020) | |
| TaBZR2 | Wheat | TaGST1 | Activation | (Cui et al. 2019) | |
| GmMYB84 | Soybean | GmRBOHB-1 and GmRBOHB-2 | Activation | (Wang et al. 2017) | |
| Aquaporins | TG | Arabidopsis | AtTIP1;1, AtTIP2;3, AtPIP2;2 | Activation | (Zhu et al. 2014) |
| ASR1 | Tomato | Solyc10g054820 | Activation | (Ricardi et al. 2014) | |
| LEA proteins | OsNAC2 | Rice | OsLEA3 | Repression | (Shen et al. 2017) |
| OsWRKY11 | Rice | RAB21 | Activation | (Lee et al. 2018) | |
| TabHLH49 | Wheat | WZY2 | Activation | (Liu et al. 2020b) | |
| Wax biosynthesis | OsWR1 | Rice | OsLACS2, OsFAE1’-L | Activation | (Wang et al. 2012) |
| PeSHN1 | Poplar | LACS2 | Activation | (Meng et al. 2019) | |
| MYB94 | Arabidopsis | wax biosynthetic genes | Activation | (Lee et al. 2016b) | |
| MYB96 | Arabidopsis | wax biosynthetic genes | Activation | (Lee et al. 2016b; Seo et al. 2011) | |
| Polyamine biosynthesis | PtrNAC72 | Poncirus trifoliata | PtADC | Repression | (Wu et al. 2016) |
| FcWRKY70 | Fortunella Crassifolia | FcADC | Activation | (Gong et al. 2015) | |
| PbrMYB21 | Pyrus | PbrADC | Activation | (Li et al. 2017b) | |
| OsHSFA3 | Rice | OsADC | Activation | (Zhu et al. 2020a) | |
| Others | TaNAC69 | Wheat | chitinase, ZIM, glyoxalase I | Activation | (Xue et al. 2011) |
| ANAC019 | Arabidopsis | ERD1 | Activation | (Tran et al. 2004) | |
| ANAC055 | Arabidopsis | ERD1 | Activation | (Tran et al. 2004) | |
| ANAC072 | Arabidopsis | ERD1 | Activation | (Tran et al. 2004) | |
| PwNAC11 | Picea wilsonii | ERD1 | Activation | (Yu et al. 2021) | |
| OsNAC6 | Rice | NICOTIANAMINE SYNTHASE | Activation | (Lee et al. 2017) | |
| ZmNAC49 | Maize | ZmMUTE | Repression | (Xiang et al. 2021) | |
| RhNAC2 | Rose | RhEXPA4 | Activation | (Dai et al. 2012) | |
| OsERF48 | Rice | OsCML16 | Activation | (Jung et al. 2017) | |
| OsERF71 | Rice | OsCINNAMOYL-COENZYME A REDUCTASE1 | Activation | (Lee et al. 2016a) | |
| RAP2.6 | Arabidopsis | RD29A, COR15A | Activation | (Zhu et al. 2020b) | |
| AtWRKY53 | Arabidopsis | QQS | Activation | (Sun and Yu 2015) | |
| SbWRKY30 | Sorghum | SbRD19 | Activation | (Yang et al. 2020b) | |
| TaAREB3 | Wheat | RD29A, RD29B, COR15A, COR47 | Activation | (Wang et al. 2016a) | |
| OsbZIP71 | Rice | OsNHX1, COR413-TM1 | Activation | (Liu et al. 2014a) | |
| GmMYB14 | Soybean | GmBEN1 | Activation | (Chen et al. 2021) | |
| LlMYB3 | Lily | LlCHS2 | Activation | (Yong et al. 2019a) | |
| OsTF1L | Rice | poxN/PRX38, Nodulin protein, DHHC4, CASPL5B1, AAA-type ATPase. | Activation | (Bang et al. 2019) | |
| Di19 | Arabidopsis | PR1, PR2, PR5 | Activation | (Liu et al. 2013) | |
| PagKNAT2/6b | Poplar | PagGA20ox1 | Repression | (Song et al. 2021) | |
| GTL1 | Arabidopsis | SDD1 | Repression | (Yoo et al. 2010) | |
| ANAC096 | Arabidopsis | RD29A | Activation | (Xu et al. 2013) | |
| TaWRKY2 | Wheat | RD29B | Activation | (Niu et al. 2012) | |
| TaWRKY19 | Wheat | Cor6.6 | Activation | (Niu et al. 2012) | |
| AREB1 | Arabidopsis | RD29B | Activation | (Uno et al. 2000) | |
| AREB2 | Arabidopsis | RD29B | Activation | (Uno et al. 2000) | |
| GmWRKY54 | Soybean | CIPK11, CPK3 | Activation | (Wei et al. 2019) | |
| SlVOZ1 | Tomato | SFT | Activation | (Chong et al. 2022) |
Post-transcriptional regulation of TFs
In addition to transcriptional regulation by each other, TFs could subject post-transcriptional regulation including phosphorylation carried out by different kinds of kinases, degradation of TFs by ubiquitin-proteasome system (UPS) (Table 1), translocation of TFs from the cytoplasm to the nucleus, and post-transcriptional regulation by miRNA.
Phosphorylation of TFs
In ABA-dependent pathway ABA signal is transmitted to TFs like AREB1, AREB2 and ABF3 through phosphorylation of them by SNF1-Related Protein Kinase (SnRK)2 Protein Kinases (Yoshida et al. 2015). The phosphorylation of TFs contributes to full activation of their transcriptional activities. Calcium dependent protein kinases, CPK4 and CPK11 or CPK6, were also reported to mediate ABA responsive phosphorylation of ABFs and/or ABI5 to enhance transcriptional activities (Zhang et al. 2020a; Zhu et al. 2007). The phosphorylation of bZIP TFs by SnRK2 kinases has been observed in rice (OsbZIP23,OsbZIP46CA1,OsbZIP62) (Chang et al. 2017; Yang et al. 2019; Zong et al. 2016), Cryophyte (CbABF1) (Yue et al. 2019) and buckwheat (FtbZIP5) (Li et al. 2020). Most of these bZIP proteins belong to the same group as ABFs, suggesting the conserved mechanism for signal transmission in plants. Besides, some TFs in other families could also be substrates of the SnRK2 kinases although the effect of phosphorylation of these TFs seems to be different. The NAC TF NTL6 in Arabidopsis is phosphorylated by SnRK2.8, which however is required for the entrance of NTL6 into the nucleus (Kim et al. 2012). Phosphorylation of the Arabidopsis HD-Zip TF HAT1 by SnRK2.3 both destabilize and repress promoter-binding activity of HAT1 for negatively regulating HAT1 in response to drought (Tan et al. 2018). In rice, the fact that OsSnRK1α interacts and phosphorylates ERF family TF OsEBP89 has also been observed, despite that the effect of phosphorylation remains to be determined (Zhang et al. 2020b). Recently, it was demonstrated that phosphorylation of SlVOZ1 by SlOST1 promoted both stability and nuclear translocation of SlVOZ1 in tomato (Chong et al. 2022). In addition to SnRK2 and CPK, other kinases such as MAK kinases, GSK3-like kinases (BIN2), could be responsible for phosphorylation of TFs to affect their stability and transcriptional activity (Table 1).
Degradation of TFs
UPS is an important pathway for post-transcriptional regulation of gene expression and involved in several hormone signal transductions like GA, auxin, Brassinosteroids, strigolactone by degrading transcriptional repressors that are also negative regulators of signaling. The specificity of UPS is determined by E3 ligases which interact with and attach ubiquitins to target proteins. We found that most TFs regulated by UPS are positive regulators of DT and accordingly the corresponding E3 ligases are negatively engaged in DT (Table 1, Table S1). One exception is that a DT repressor CaAIBZ1 that is a bZIP TF is ubiquitinated by the E3 ligase CaASRF1 (Joo et al. 2019a). CaASRF1 is induced by drought and targets CaAIBZ1 for ubiquitination, thus it positively modulates ABA signaling and drought response. However, two genes encoding E3 ligases, MdBT2 and CaDSR1, are also induced by drought although acting as negative regulators of DT (Ji et al. 2020; Lim et al. 2018). The induction of these genes might be dedicated for attenuating drought response by destabilizing TFs to generate a feedback loop. A feed-forward loop could also be established by translocation of E3 ligase, represented by RGLG2, from the plasma membrane to the nucleus induced by stress (Cheng et al. 2012).
Translocation
Translocation of TFs from the plasma membrane to the nucleus in response to drought also occurs. Under unstressed conditions, MfNACsa is targeted to the plasma membrane through S-palmitoylation at Cys26 in the endoplasmic reticulum/Golgi (Duan et al. 2017). Under drought stress, MfNACsa translocates to the nucleus through de-S-palmitoylation mediated by the thioesterase MtAPT1, whose encoding gene is rapidly induced by dehydration stress. NTL6 contain strong α-helical transmembrane motifs (TMs) in their C-terminal regions and are predicted to be membrane-associated (Kim et al. 2007). ABA and cold treatment induce NTL6 release from membrane, suggesting that both cleavage and phosphorylation of NTL6 are prerequisite for NTL6 to enter the nucleus to exert its regulatory function (Kim et al. 2007). Indeed, in Arabidopsis more than 190 TFs are predicted to be membrane-bound transcription factors (MTFs) (Seo et al. 2008). They are dormant by associated with the intracellular membranes and activated by proteolytic cleavage that might be stress responsive. This strategy could potentially transmitted stress signals from outside into the nucleus, but more evidences are required to explain how stress triggers proteolytic cleavage.
Post-transcriptional regulation by microRNAs
MicroRNAs (miRNAs) are encoded by endogenous genes, whose initial products are designated primary miRNAs (pri-miRNAs) (Yu et al. 2017). Pri-miRNAs are then cleaved to generate precursor-miRNAs (pre-miRNAs), which are finally processed to mature miRNAs. miRNAs repress gene expression by cleavage of RNAs or inhibition of translation, playing important roles in plant development and stress response. Genome-wide identification of differentially expressed miRNAs in response to drought has been performed in various plant species (Ren et al. 2021; Yu et al. 2020; Fan et al. 2020; Pegler et al. 2019; Liu et al. 2018; Ferdous et al. 2017; Hamza et al. 2016; Xie et al. 2015; Yin et al. 2014; Xie et al. 2014; Wang et al. 2014; Zhou et al. 2010), which would not be discussed here. Functional analysis of miRNAs targeting TFs involved in DT mainly focus on miR164 and miR169. The targets of miR164 are NAC family TFs belonging to NAC1 and NAM subgroup (Fang et al. 2014). Their targets seem to play similar roles in drought resistance. Overexpression of OMTN2, OMTN3, OMTN4, and OMTN6 in rice leads to enhanced drought sensitivity at the reproductive stage, suggesting their negative effects on drought resistance. The contradicting conclusion concerning the function of OsNAC2 were drawn by two groups. One group showed that OsNAC2-overexpressing transgenic plants exhibited drought sensitive phenotype while silencing of OsNAC2 enhanced tolerance to drought (Shen et al. 2017). The other group revealed that overexpression of a miR164b-resistant OsNAC2 mutant gene improved DT (Jiang et al. 2019). The negative effect on DT has also been observed for PeNAC070, a miR164 target in Populus euphratica (Lu et al. 2017). The miR169 or its targets, NF-YA genes, have been characterized in Arabidopsis, tomato, rapeseed and soybean (Ni et al. 2013; Li et al. 2008; Li et al. 2021a; Zhang et al. 2011). Arabidopsis NFYA5, soybean GmNFYA3 and rapeseed NF-YA8 are positive regulators of plant tolerance to drought stress. However, overexpression of tomato miR169 and soybean miR169c also enhanced DT (Yu et al. 2019; Zhang et al. 2011), demonstrating complicated roles of miR169 in the regulation of drought stress response.
Downstream targets of TFs
Regulatory hub established by TFs receive input signals triggered by drought from outside of the nucleus and transmit output signals by regulating downstream gene expression for reaction. Here we summarized the direct downstream genes of TFs, whose promoter could be bound by drought-induced TFs in vitro or in vivo, and the genes which were only examined to transcriptionally change responding to mutation or altered expression of TF genes are not included. The function of the direct downstream genes entails ABA biosynthesis and signaling, ROS-related, aquaporin, LEA proteins, wax biosynthesis and others (Table 2). In particular, osmotic adjustment (OA) is an important biochemical mechanism helping plants to acclimate to drought conditions. Several organic compounds like sugars, proline, betaines and inorganic ions contributes to OA (Turner 2018). However, the TFs that have been experimentally proved to direct regulate the genes related to accumulation of these solutes in response to drought cannot be found out. The direct regulators of P5CS1, encoding a rate-limiting enzyme in proline biosynthesis, have been reported for their roles in salt tolerance (Dai et al. 2018; Verma et al. 2020).
ABA metabolism and signaling
The biosynthesis of ABA is rapidly induced by drought stress to elicit series of responses such as stomatal closure. It is generally accepted that in higher plants ABA is synthesized through carotenoids pathway by multi-step enzymatic reactions, in which NCEDs function as the key rate-limiting enzymes. ABA can be degraded by hydroxylation which is mediated by CYP707A subfamily monooxygenases (Ma et al. 2018). Until now various families TFs in different plant species have been reported to directly activate expression of NCED genes to increase ABA level, which are represented by NAC (ATAF1, GhirNAC2), WRKY (WRKY57, MeWRKY20, MaWRKY80, PbrWRKY53), MYB (MdMYB88), bZIP (OsbZIP23), bHLH (ZmPTF1) and NF-Y (PdNF-YB21) (Table 2). The bZIP TF, HAT1, was identified to be the repressor of NCED3 in Arabidopsis (Tan et al. 2018). The fact that mutation or over-expression of HAT1 affects plant sensitivity to ABA indicates that it also negatively regulates ABA signaling although the target gene is not clear. Induction of elevated ABA level by stresses could also be achieved via repressing expression of CYP707A. bHLH122, strongly induced by drought, NaCl and osmotic stresses but not ABA treatment, directly binds to the promoter of CYP707A3 to repress its expression (Liu et al. 2014b).
ABA signaling is transmitted by ABA receptors RCARs/PYR1/PYLs, PP2Cs, SnRK2s and specific TFs (Zhu 2016). In the absence of ABA, PP2Cs interact with SnRK2s to inhibit their activity by dephosphorylation of multiple Ser/Thr residues in the activation loop. ABA perception by the RCAR/PYR1/PYL proteins releases the PP2C-mediated inhibition of SnRKs’ activity The released SnRK2s are activated through autophosphorylation and subsequently phosphorylate downstream substrates including TFs as described above. GmWRKY54, AtHB7 and AtHB12 are directly involved in the activation of PYL genes. The SnRK2 gene is activated by GmWRKY54 in soybean and repressed by OsNAC2 in rice. Although PP2C genes are negative regulators of ABA signaling, they can be transcriptionally stimulated by multiple TFs induced by drought or ABA such as OsbZIP23, OsABF1, GhWRKY21, AtHB7 and AtHB12 (Table 2). The stimulation of PP2C genes was proposed to form a feedback loop for mitigating the intensity of ABA signal.
ROS-related
Under normal conditions, the basal level of Reactive oxygen species (ROS), which induces redox signals that regulates numerous cellular processes, is balanced by generation in various subcellular organelles such as chloroplasts, mitochondria, peroxisomes and apoplast and scavenging by antioxidative systems involving enzymes like SOD, CAT, APX and non-enzymatic antioxidants such as ascorbate (ASC), glutathione (GSH) and carotenoids (Farooq et al. 2019; Waszczak et al. 2018). Under stress conditions, increased ROS concentrations incurs cellular damage by oxidizing macromolecular such as proteins, DNA and lipids (Farooq et al. 2019). The plasma membrane-localized NADPH oxidases termed respiratory burst oxidase homologs (RBOHs) are the major ROS producers in apoplast (Waszczak et al. 2018). In Arabidopsis, NTL4, a NAC TFs, is responsible for coupling ROS production to drought-induced leaf senescence via facilitating expression of AtrbohC and AtrbohE directly (Lee et al. 2012). However, in soybean a hypothesis was issued that up-regulation of GmRBOHB-1 and GmRBOHB-2 by GmMYB84 improves DT by inducing SOD/POD/CAT activity due to the increased H2O2 contents (Wang et al. 2017). The elevated production of ROS scavenging enzymes SOD and CAT can be induced by drought stress through transcriptional activation of TFs. For example, ZmNAC84 directly promotes ZmSOD2 expression to enhance maize DT (Han et al. 2021). NtERF172 positively regulates NtCAT expression to confer tobacco drought resistance (Zhao et al. 2020). In addition, T. aestivum glutathione s-transferase-1 (TaGST1), which functions positively in scavenging drought-induced ROS, is activated by TaBZR2, a positive regulator of BR signaling (Cui et al. 2019).
Aquaporins
Aquaporins (AQPs) are transmembrane channels for water and some solutes transportation. Based on their intracellular locations and sequence similarities, AQPs can be divided into seven subfamilies, among which the plasma membrane intrinsic proteins (PIPs) and tonoplast intrinsic proteins (TIPs) are considered to be the major AQPs mediated water uptake in roots (Afzal et al. 2016). However, the role of PIPs and TIPs in DT is intricate. Although most of PIP genes are down-regulated in response to drought, silencing of PIP genes in different plant species decreased drought resistance possibly resulted from decreased root hydraulic conductivity (Lpr) or transpiration rates (Afzal et al. 2016). This is supported by the evidence that overexpression of AQP genes enhances drought resistance. Three Aquaporin Genes: AtTIP1;1, AtTIP2;3, and AtPIP2;2 are directly activated by a ERF gene, TG (Zhu et al. 2014). The vitrified leaf phenotype was observed in both 35S:TG and 35S:AtTIP1;1 plants possibly as a result of excess water accumulated in the intercellular spaces. Whether the vitrified leaf phenotype is related to enhanced DT is yet to be clarified. The aquaporin gene, Solyc10g054820.1, was screened as a direct target of tomato ASR1 by ChIP-seq experiment. Expression of this gene in ASR1-silenced lines is reduced (Ricardi et al. 2014).
LEA proteins
Late embryogenesis abundant (LEA) proteins were first identified in cotton (Gossypium hirsutum) seeds and were also found to be accumulated under stress conditions. Their precise roles in DT remain unknown, but they could serve as chaperones to protect macromolecules or as hydrophilic proteins to retain water (Bies-Etheve et al. 2008). LEA proteins were classified into nine groups based on the phylogenetic analysis in Arabidopsis (Hundertmark and Hincha 2008). Two dehydrin group genes, RAB21 and WZY2, are directly activated by OsWRKY11 and TabHLH49 in rice and wheat respectively (Table 2). OsNAC5 and OsNAC2 antagonistically regulate expression of OsLEA3, which belongs to LEA_4 group (Table 2). Overexpression of OsLEA3–1 and OsLEA3–2, also LEA_4 members, improved drought resistance, suggesting the functional importance of these proteins in plant DT (Duan and Cai 2012; Xiao et al. 2007).
Wax biosynthesis
The cuticle on the surface of plant, an extracellular hydrophobic layer, mainly contains cutin and cuticular wax (Lewandowska et al. 2020). Under drought conditions, it becomes thicker to reduce water loss accompanied with greater wax deposition. Cuticular wax is composed of very-long-chain fatty acids (VLCFAs), their esters, alcohols, aldehydes, alkanes and ketones. Wax biosynthesis consists of de novo biosynthesis of C16 and C18 fatty acids, fatty acid elongation (FAE) and wax production including alcohol-forming pathway and alkane-forming pathway (Yeats and Rose 2013; Lewandowska et al. 2020). In Arabidopsis, MYB96 and MYB94 redundantly regulate wax biosynthesis by targeting multiple wax biosynthetic genes such as KCS1, KCS2, CER2, CER6, CER10, KCR1 for FAE, CER3 and WSD1 in alcohol-forming pathway and CER4 in alkane-forming pathway (Lee et al. 2016b). LACS2, catalyzing formation of long-chain-acyl-CoA, is conservatively activated by SHN1 homologs in rice (OsWR1) and Populus x euramericana (PeSHN1) (Table 2).
Polyamine biosynthesis
Polyamines, including putrescine (Put), spermidine (Spd), and spermine (Spm), play important roles in plant development and stress tolerance (Shi and Chan 2014). The function of polyamines in abiotic stress tolerance may involve ion homeostasis, keeping water status, ROS homeostasis, Osmotic balance and other responses. The genes encoding enzymes involved in polyamine biosynthetic pathway are induced by various stresses. The arginine decarboxylase (ADC) catalyzes the first step of polyamine biosynthesis. Expression of ADC genes in Fortunella Crassifolia,Pyrus Betulaefolia and rice is promoted by FcWRKY70,PbrMYB21,OsHSFA3 respectively (Table 2). PtrNAC72 acts as a transcriptional repressor of PtADC to negatively regulate polyamine level in response to drought (Table 2).
Others
The other direct targets of drought-induced TFs are listed in Table 2, mainly including genes encoding lignin biosynthetic enzymes, proteases, GA biosynthetic enzyme, BR catabolic enzyme, expansin, stomatal development regulator, RD29A, RD29B and so on. Most of these regulations are found only in one species although how they are related to DT are elucidated. More evidences are required to prove whether they are common targets regulated by drought in different plant species. Interestingly, recent studies revealed that direct activation of SINGLE FLOWER TRUSS (SFT) by SlVOZ1 is essential for drought-accelerated flowering, known as a drought escape (DE) response (Chong et al. 2022).
Concluding remarks
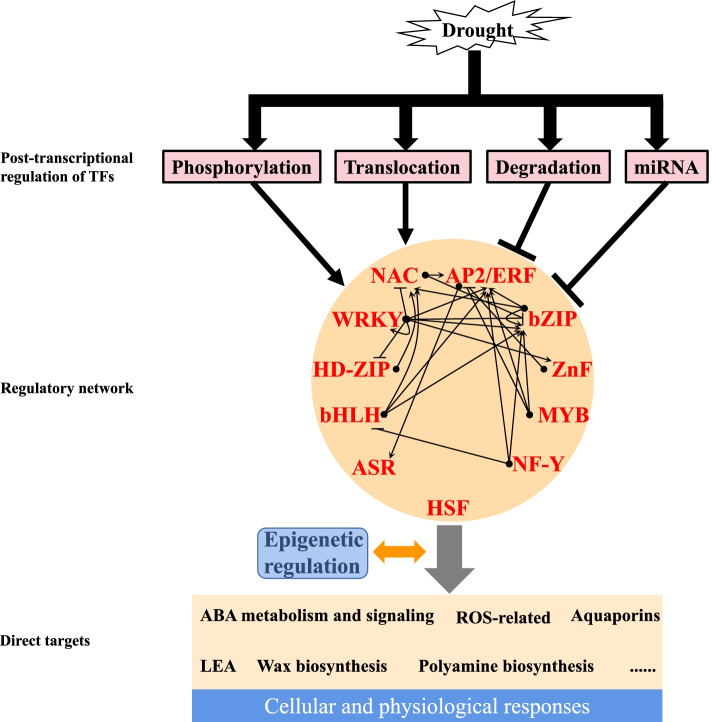
In this review, we summarized the reported TFs that have been functionally analyzed for their roles in DT in multiple higher plants. These TFs are mainly distributed in 11 families, which form the regulatory network to receive signals possibly via post-transcriptional regulation and trigger cellular and physiological reactions by direct regulating expression of downstream genes (Fig. 6). There is still a long way to go for completing the interactive map of TFs. For the role of TFs in the transmission of drought signal, the key question is how they receive the signal from outside the nucleus. The phosphorylation of TFs could serve as one way but definitely not the only way. Besides, whether or not the drought-responsive pioneer TFs, which have the ability to recognize DNA sequence motifs exposed on the surface of a nucleosome (Zaret 2020), exist in plant is yet to be determined. As a matter of fact, the drought-responsive TF has been reported to interact with histone acetyltransferase (HAT) to regulate chromatin state of target genes for activation of these genes (Li et al. 2019a). The priming chromatin state is crucial for reactivation of genes during repeated stress, thus serving as epigenetic stress memory (Baurle and Trindade 2020). It is of great importance to understand how TFs and epigenetic regulators cooperated to mediate drought stress memory. Finally, identification and characterization of the TF genes that specifically induced by drought and could confer DT but do not affect plant development would provide theoretical basis for genetic improvement of crop drought resistance.
Fig. 6.
The diagram of drought signaling transmitted by regulatory network. Drought-induced post-transcriptional regulation of transcription factors (TFs) may function to transmit the signal to TFs by activating TFs such as phosphorylation and translocation of TFs, or to mitigate intensity of the signal by compromising TFs such as ubiquitin-proteasome system (UPS) mediated protein degradation and microRNA (miRNA). The regulatory network is built by mutual regulation of TFs (activation or repression) and interaction of TFs that is not shown in the figure to trigger cellular and physiological responses by directly regulating related genes. The regulation of TFs and downstream genes here refers to direct binding of TFs to promoters of target genes which has been experimentally proved to regulate their expression. How TFs and epigenetic regulation that mediates stress memory cooperate for fine tuning of drought-responsive genes will be of great interest in the future
Supplementary Information
Additional file 1: Table S1. The list of transcription factors (TFs) functionally involved in drought tolerance.
Acknowledgments
Not applicable.
Abbreviations
- TF
Transcription factor
- DT
Drought tolerance
- NAC
No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), Cup-shaped cotyledon (CUC)
- ERF
Ethylene-responsive element binding factor
- bZIP
Basic leucine zipper
- HD-Zip
Homeodomain-leucine zipper
- ZnF
Zinc finger
- bHLH
Basic/helix-loop-helix
- ASR
Abscisic acid, stress, ripening induced
- NF-Y
Nuclear factor Y
- HSF
Heat shock factor
Authors’ contributions
YH: Conceptualization, Investigation, Writing – original draft, Writing – review & editing; XC: Investigation, Visualization; XS: Funding acquisition, Writing – review & editing. The author(s) read and approved the final manuscript.
Funding
This work was supported by Research Initiation Fund for High-level Talents of China Three Gorges University.
Availability of data and materials
Not applicable.
Declarations
Competing interests
All authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongfeng Hu, Email: huyongfeng@ctgu.edu.cn.
Xiaoliang Chen, Email: 980790457@qq.com.
Xiangling Shen, Email: shenxl1982@hotmail.com.
References
- Afzal Z, Howton TC, Sun Y, Mukhtar MS (2016) The roles of Aquaporins in plant stress responses. J Dev Biol 4(1). 10.3390/jdb4010009 [DOI] [PMC free article] [PubMed]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Arenhart RA, Bai Y, de Oliveira LF, Neto LB, Schunemann M, Maraschin Fdos S, Mariath J, Silverio A, Sachetto-Martins G, Margis R, Wang ZY, Margis-Pinheiro M. New insights into aluminum tolerance in rice: the ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol Plant. 2014;7(4):709–721. doi: 10.1093/mp/sst160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-zip family. Trends Plant Sci. 2007;12(9):419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Baek D, Shin G, Kim MC, Shen M, Lee SY, Yun DJ. Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front Plant Sci. 2020;11:143. doi: 10.3389/fpls.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang SW, Lee DK, Jung H, Chung PJ, Kim YS, Choi YD, Suh JW, Kim JK. Overexpression of OsTF1L, a rice HD-zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol J. 2019;17(1):118–131. doi: 10.1111/pbi.12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurle I, Trindade I. Chromatin regulation of somatic abiotic stress memory. J Exp Bot. 2020;71(17):5269–5279. doi: 10.1093/jxb/eraa098. [DOI] [PubMed] [Google Scholar]
- Bian S, Jin D, Sun G, Shan B, Zhou H, Wang J, Zhai L, Li X. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene. 2020;753:144803. doi: 10.1016/j.gene.2020.144803. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67(1–2):107–124. doi: 10.1007/s11103-008-9304-x. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Nguyen BH, Xie Y, Xiao B, Tang N, Zhu W, Mou T, Xiong L. Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in Rice. Front Plant Sci. 2017;8:1102. doi: 10.3389/fpls.2017.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YN, Zhu C, Jiang J, Zhang H, Zhu JK, Duan CG. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol. 2020;62(5):563–580. doi: 10.1111/jipb.12901. [DOI] [PubMed] [Google Scholar]
- Chaves-Sanjuan A, Gnesutta N, Gobbini A, Martignago D, Bernardini A, Fornara F, Mantovani R, Nardini M. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J. 2021;105(1):49–61. doi: 10.1111/tpj.15038. [DOI] [PubMed] [Google Scholar]
- Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in Brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29(6):1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yang H, Fang Y, Guo W, Chen H, Zhang X, Dai W, Chen S, Hao Q, Yuan S, Zhang C, Huang Y, Shan Z, Yang Z, Qiu D, Liu X, Tran LP, Zhou X, Cao D. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol J. 2021;19(4):702–716. doi: 10.1111/pbi.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Liu H, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013;75(6):965–976. doi: 10.1111/tpj.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012;158(1):363–375. doi: 10.1104/pp.111.189738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L, Guo P, Zhu Y (2020) Mediator complex: a pivotal regulator of ABA signaling pathway and abiotic stress response in plants. Int J Mol Sci 21(20). 10.3390/ijms21207755 [DOI] [PMC free article] [PubMed]
- Chong L, Xu R, Huang P, Guo P, Zhu M, Du H, Sun X, Ku L, Zhu JK, Zhu Y. The tomato OST1–VOZ1 module regulates drought-mediated flowering. Plant Cell. 2022;34(5):2001–2018. doi: 10.1093/plcell/koac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65(7–8):1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin A, Daszkowska-Golec A, Kurowska M, Szarejko I. Barley ABI5 (abscisic acid INSENSITIVE 5) is involved in abscisic acid-dependent drought response. Front Plant Sci. 2020;11:1138. doi: 10.3389/fpls.2020.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LG, Shan JX, Shi M, Gao JP, Lin HX. DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in Rice. PLoS Genet. 2015;11(10):e1005617. doi: 10.1371/journal.pgen.1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XY, Gao Y, Guo J, Yu TF, Zheng WJ, Liu YW, Chen J, Xu ZS, Ma YZ. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019;180(1):605–620. doi: 10.1104/pp.19.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Zhang C, Jiang X, Kang M, Yin X, Lu P, Zhang X, Zheng Y, Gao J. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 2012;160(4):2064–2082. doi: 10.1104/pp.112.207720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Wang M, Gong X, Liu JH. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018;219(3):972–989. doi: 10.1111/nph.15240. [DOI] [PubMed] [Google Scholar]
- Droge-Laser W, Snoek BL, Snel B, Weiste C. The Arabidopsis bZIP transcription factor family-an update. Curr Opin Plant Biol. 2018;45(Pt A):36–49. doi: 10.1016/j.pbi.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One. 2012;7(9):e45117. doi: 10.1371/journal.pone.0045117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell. 2017;29(7):1748–1772. doi: 10.1105/tpc.17.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Ebrahimian-Motlagh S, Ribone PA, Thirumalaikumar VP, Allu AD, Chan RL, Mueller-Roeber B, Balazadeh S. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-zip I transcription factor AtHB13. Front Plant Sci. 2017;8:2118. doi: 10.3389/fpls.2017.02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Liu Y, Du H, Kuang T, Zhang Y. Identification of drought-responsive miRNAs in Hippophae tibetana using high-throughput sequencing. 3. Biotech. 2020;10(2):53. doi: 10.1007/s13205-019-2045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. 2014;65(8):2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci. 2015;72(4):673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq MA, Niazi AK, Akhtar J, Saifullah FM, Souri Z, Karimi N, Rengel Z. Acquiring control: the evolution of ROS-induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol Biochem. 2019;141:353–369. doi: 10.1016/j.plaphy.2019.04.039. [DOI] [PubMed] [Google Scholar]
- Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014;80(4):654–668. doi: 10.1111/tpj.12670. [DOI] [PubMed] [Google Scholar]
- Ferdous J, Sanchez-Ferrero JC, Langridge P, Milne L, Chowdhury J, Brien C, Tricker PJ. Differential expression of microRNAs and potential targets under drought stress in barley. Plant Cell Environ. 2017;40(1):11–24. doi: 10.1111/pce.12764. [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Barros PM, Cordeiro AM, Serra TS, Lourenco T, Chander S, Oliveira MM, Saibo NJ. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot. 2012;63(10):3643–3656. doi: 10.1093/jxb/ers035. [DOI] [PubMed] [Google Scholar]
- Gong L, Zhang H, Liu X, Gan X, Nie F, Yang W, Zhang L, Chen Y, Song Y, Zhang H. Ectopic expression of HaNAC1, an ATAF transcription factor from Haloxylon ammodendron, improves growth and drought tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2020;151:535–544. doi: 10.1016/j.plaphy.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Gong X, Zhang J, Hu J, Wang W, Wu H, Zhang Q, Liu JH. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015;38(11):2248–2262. doi: 10.1111/pce.12539. [DOI] [PubMed] [Google Scholar]
- Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, Li J, Ding Y, Shi Y, Wang Y, Yang Y, Guo Y, Zhu JK. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63(5):635–674. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- Gonzalez RM, Iusem ND. Twenty years of research on Asr (ABA-stress-ripening) genes and proteins. Planta. 2014;239(5):941–949. doi: 10.1007/s00425-014-2039-9. [DOI] [PubMed] [Google Scholar]
- Gu L, Ma Q, Zhang C, Wang C, Wei H, Wang H, Yu S. The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front Plant Sci. 2019;10:1352. doi: 10.3389/fpls.2019.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza NB, Sharma N, Tripathi A, Sanan-Mishra N. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr Patterns. 2016;20(2):88–98. doi: 10.1016/j.gep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Han SK, Wagner D. Role of chromatin in water stress responses in plants. J Exp Bot. 2014;65(10):2785–2799. doi: 10.1093/jxb/ert403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Yan J, Xiang Y, Zhang A. Phosphorylation of ZmNAC84 at Ser-113 enhances the drought tolerance by directly modulating ZmSOD2 expression in maize. Biochem Biophys Res Commun. 2021;567:86–91. doi: 10.1016/j.bbrc.2021.06.026. [DOI] [PubMed] [Google Scholar]
- Hou D, Zhao Z, Hu Q, Li L, Vasupalli N, Zhuo J, Zeng W, Wu A, Lin X. PeSNAC-1 a NAC transcription factor from moso bamboo (Phyllostachys edulis) confers tolerance to salinity and drought stress in transgenic rice. Tree Physiol. 2020;40(12):1792–1806. doi: 10.1093/treephys/tpaa099. [DOI] [PubMed] [Google Scholar]
- Huang J, Sun S, Xu D, Lan H, Sun H, Wang Z, Bao Y, Wang J, Tang H, Zhang H. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.) Plant Mol Biol. 2012;80(3):337–350. doi: 10.1007/s11103-012-9955-5. [DOI] [PubMed] [Google Scholar]
- Huang K, Wu T, Ma Z, Li Z, Chen H, Zhang M, Bian M, Bai H, Jiang W, Du X (2021) Rice transcription factor OsWRKY55 is involved in the drought response and regulation of plant growth. Int J Mol Sci 22(9). 10.3390/ijms22094337 [DOI] [PMC free article] [PubMed]
- Huang L, Wang Y, Wang W, Zhao X, Qin Q, Sun F, Hu F, Zhao Y, Li Z, Fu B, Li Z. Characterization of transcription factor gene OsDRAP1 conferring drought tolerance in Rice. Front Plant Sci. 2018;9:94. doi: 10.3389/fpls.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009;23(15):1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, b ZIPRG bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Lindemose S, de Masi F, Reimer JJ, Nielsen M, Perera V, Workman CT, Turck F, Grant MR, Mundy J, Petersen M, Skriver K. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 2013;3:321–327. doi: 10.1016/j.fob.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Liu G, Liu Y, Zheng L, Nie X, Wang Y. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 2013;13:151. doi: 10.1186/1471-2229-13-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji XL, Li HL, Qiao ZW, Zhang JC, Sun WJ, Wang CK, Yang K, You CX, Hao YJ. The BTB-TAZ protein MdBT2 negatively regulates the drought stress response by interacting with the transcription factor MdNAC143 in apple. Plant Sci. 2020;301:110689. doi: 10.1016/j.plantsci.2020.110689. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhou L, Chen W, Ye N, Xia J, Zhuang C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice (N Y) 2019;12(1):76. doi: 10.1186/s12284-019-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yu D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant. 2012;5(6):1375–1388. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- Jin C, Li KQ, Xu XY, Zhang HP, Chen HX, Chen YH, Hao J, Wang Y, Huang XS, Zhang SL. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front Plant Sci. 2017;8:1049. doi: 10.3389/fpls.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(D1):D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H, Lim CW, Lee SC. A pepper RING-type E3 ligase, CaASRF1, plays a positive role in drought tolerance via modulation of CaAIBZ1 stability. Plant J. 2019;98(1):5–18. doi: 10.1111/tpj.14191. [DOI] [PubMed] [Google Scholar]
- Joo H, Lim CW, Lee SC. Roles of pepper bZIP transcription factor CaATBZ1 and its interacting partner RING-type E3 ligase CaASRF1 in modulation of ABA signalling and drought tolerance. Plant J. 2019;100(2):399–410. doi: 10.1111/tpj.14451. [DOI] [PubMed] [Google Scholar]
- Jung H, Chung PJ, Park SH, Redillas M, Kim YS, Suh JW, Kim JK. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol J. 2017;15(10):1295–1308. doi: 10.1111/pbi.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TCC, Abdel-Mageed H, Aleman L, Lee J, Payton P, Cryer D, Allen RD. Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum) Plant Cell Environ. 2018;41(5):898–907. doi: 10.1111/pce.12906. [DOI] [PubMed] [Google Scholar]
- Khan IU, Ali A, Khan HA, Baek D, Park J, Lim CJ, Zareen S, Jan M, Lee SY, Pardo JM, Kim WY, Yun DJ. PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in Arabidopsis. Front Plant Sci. 2020;11:623. doi: 10.3389/fpls.2020.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010;33(4):604–611. doi: 10.1111/j.1365-3040.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, Yamaguchi-Shinozaki K. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011;52(12):2136–2146. doi: 10.1093/pcp/pcr143. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Park MJ, Seo PJ, Song JS, Kim HJ, Park CM. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem J. 2012;448(3):353–363. doi: 10.1042/BJ20120244. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35(1):203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi F, Maeta E, Terashima A, Takumi S. Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant. 2008;134(1):74–86. doi: 10.1111/j.1399-3054.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- Lee DK, Chung PJ, Jeong JS, Jang G, Bang SW, Jung H, Kim YS, Ha SH, Choi YD, Kim JK. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol J. 2017;15(6):754–764. doi: 10.1111/pbi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Jung H, Jang G, Jeong JS, Kim YS, Ha SH, Do Choi Y, Kim JK. Overexpression of the OsERF71 transcription factor alters Rice root structure and drought resistance. Plant Physiol. 2016;172(1):575–588. doi: 10.1104/pp.16.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Cha J, Choi C, Choi N, Ji HS, Park SR, Lee S, Hwang DJ. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice (N Y) 2018;11(1):5. doi: 10.1186/s12284-018-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell. 2010;22(6):1716–1732. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012;70(5):831–844. doi: 10.1111/j.1365-313X.2012.04932.x. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim HU, Suh MC. MYB94 and MYB96 additively activate Cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 2016;57(11):2300–2311. doi: 10.1093/pcp/pcw147. [DOI] [PubMed] [Google Scholar]
- Lewandowska M, Keyl A, Feussner I. Wax biosynthesis in response to danger: its regulation upon abiotic and biotic stress. New Phytol. 2020;227(3):698–713. doi: 10.1111/nph.16571. [DOI] [PubMed] [Google Scholar]
- Li F, Li M, Wang P, Cox KL, Jr, Duan L, Dever JK, Shan L, Li Z, He P. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017;215(4):1462–1475. doi: 10.1111/nph.14680. [DOI] [PubMed] [Google Scholar]
- Li J, Dong Y, Li C, Pan Y, Yu J. SiASR4, the target gene of SiARDP from Setaria italica, improves abiotic stress adaption in plants. Front Plant Sci. 2016;7:2053. doi: 10.3389/fpls.2016.02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Duan Y, Sun N, Wang L, Feng S, Fang Y, Wang Y. The miR169n-NF-YA8 regulation module involved in drought resistance in Brassica napus L. Plant Sci. 2021;313:111062. doi: 10.1016/j.plantsci.2021.111062. [DOI] [PubMed] [Google Scholar]
- Li K, Xing C, Yao Z, Huang X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol J. 2017;15(9):1186–1203. doi: 10.1111/pbi.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhao H, Wang X, Kang J, Lv B, Dong Q, Li C, Chen H, Wu Q. Tartary buckwheat transcription factor FtbZIP5, regulated by FtSnRK2.6, can improve salt/drought resistance in transgenic Arabidopsis. Int J Mol Sci. 2020;21(3):1123. doi: 10.3390/ijms21031123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin YJ, Wang P, Zhang B, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu X, Wang Z, Dai X, Yu J, Zhou C, Liu B, Wang JP, Chiang VL, Li W. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell. 2019;31(3):663–686. doi: 10.1105/tpc.18.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhong M, Qu L, Yang J, Liu X, Zhao Q, Liu X, Zhao X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021;310:110983. doi: 10.1016/j.plantsci.2021.110983. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu C, Zhang Y, Wang B, Ran Q, Zhang J. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J Exp Bot. 2019;70(19):5471–5486. doi: 10.1093/jxb/erz307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CW, Baek W, Lee SC. Roles of pepper bZIP protein CaDILZ1 and its interacting partner RING-type E3 ligase CaDSR1 in modulation of drought tolerance. Plant J. 2018;96(2):452–467. doi: 10.1111/tpj.14046. [DOI] [PubMed] [Google Scholar]
- Liu C, Ma H, Zhou J, Li Z, Peng Z, Guo F, Zhang J. TsHD1 and TsNAC1 cooperatively play roles in plant growth and abiotic stress resistance of Thellungiella halophile. Plant J. 2019;99(1):81–97. doi: 10.1111/tpj.14310. [DOI] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol. 2014;84(1–2):19–36. doi: 10.1007/s11103-013-0115-3. [DOI] [PubMed] [Google Scholar]
- Liu G, Li B, Li X, Wei Y, He C, Shi H. MaWRKY80 positively regulates plant drought stress resistance through modulation of abscisic acid and redox metabolism. Plant Physiol Biochem. 2020;156:155–166. doi: 10.1016/j.plaphy.2020.09.015. [DOI] [PubMed] [Google Scholar]
- Liu H, Yang Y, Liu D, Wang X, Zhang L. Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat. BMC Plant Biol. 2020;20(1):259. doi: 10.1186/s12870-020-02474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yu H, Zhao G, Huang Q, Lu Y, Ouyang B. Identification of drought-responsive microRNAs in tomato using high-throughput sequencing. Funct Integr Genomics. 2018;18(1):67–78. doi: 10.1007/s10142-017-0575-7. [DOI] [PubMed] [Google Scholar]
- Liu T, Longhurst AD, Talavera-Rauh F, Hokin SA, Barton MK (2016) The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence. Elife 5. 10.7554/eLife.13768 [DOI] [PMC free article] [PubMed]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014;201(4):1192–1204. doi: 10.1111/nph.12607. [DOI] [PubMed] [Google Scholar]
- Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH, Chen YF. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant. 2013;6(5):1487–1502. doi: 10.1093/mp/sst031. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang T, Lin Z, Gu B, Xing C, Zhao L, Dong H, Gao J, Xie Z, Zhang S, Huang X. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol J. 2019;17(9):1770–1787. doi: 10.1111/pbi.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot. 2012;63(18):6371–6392. doi: 10.1093/jxb/ers293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Dun H, Lian C, Zhang X, Yin W, Xia X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol Biochem. 2017;115:418–438. doi: 10.1016/j.plaphy.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Yu TF, Li XH, Cao XY, Ma J, Chen J, Zhou YB, Chen M, Ma YZ, Zhang JH, Xu ZS. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020;20(1):123. doi: 10.1186/s12870-020-02337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Cao J, He J, Chen Q, Li X, Yang Y (2018) Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int J Mol Sci 19(11). 10.3390/ijms19113643 [DOI] [PMC free article] [PubMed]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci U S A. 2013;110(48):19627–19632. doi: 10.1073/pnas.1311729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Cao Y, Li H, Bian Z, Wang D, Lian C, Yin W, Xia X. PeSHN1 regulates water-use efficiency and drought tolerance by modulating wax biosynthesis in poplar. Tree Physiol. 2019;39(8):1371–1386. doi: 10.1093/treephys/tpz033. [DOI] [PubMed] [Google Scholar]
- Millard PS, Kragelund BB, Burow M. R2R3 MYB transcription factors - functions outside the DNA-binding domain. Trends Plant Sci. 2019;24(10):934–946. doi: 10.1016/j.tplants.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Minh-Thu PT, Kim JS, Chae S, Jun KM, Lee GS, Kim DE, Cheong JJ, Song SI, Nahm BH, Kim YK. A WUSCHEL Homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in Rice. Mol Cells. 2018;41(8):781–798. doi: 10.14348/molcells.2018.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A, Gampala SS, Ritchie GL, Payton P, Burke JJ, Rock CD. Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J. 2014;12(5):578–589. doi: 10.1111/pbi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju M, Reddy PS, Kumar SA, Srivastava RK, Kishor PB, Rao DM. Genome-wide scanning and characterization of Sorghum bicolor L. Heat Shock Transcription Factors Curr Genomics. 2015;16(4):279–291. doi: 10.2174/1389202916666150313230812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140(2):411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82(1–2):113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35(6):1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Pegler JL, Oultram JMJ, Grof CPL, Eamens AL (2019) Profiling the abiotic stress responsive microRNA landscape of Arabidopsis thaliana. Plants (Basel) 8(3). 10.3390/plants8030058 [DOI] [PMC free article] [PubMed]
- Persak H, Pitzschke A. Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress. Int J Mol Sci. 2014;15(2):2517–2537. doi: 10.3390/ijms15022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20(6):1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Zhang H, Shi X, Ai X, Dong J, Zhao X, Zhong C, Jiang C, Wang J, Yu H. Genome-wide identification of key candidate microRNAs and target genes associated with Peanut drought tolerance. DNA Cell Biol. 2021;40(2):373–383. doi: 10.1089/dna.2020.6245. [DOI] [PubMed] [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63(3):417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardi MM, Gonzalez RM, Zhong S, Dominguez PG, Duffy T, Turjanski PG, Salgado Salter JD, Alleva K, Carrari F, Giovannoni JJ, Estevez JM, Iusem ND. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. 2014;14:29. doi: 10.1186/1471-2229-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell. 2015;27(6):1771–1787. doi: 10.1105/tpc.15.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Chakraborty J, Ghosh P, Basu D, Das S. Chickpea WRKY70 regulates the expression of a homeodomain-leucine zipper (HD-zip) I transcription factor CaHDZ12, which confers abiotic stress tolerance in transgenic tobacco and chickpea. Plant Cell Physiol. 2017;58(11):1934–1952. doi: 10.1093/pcp/pcx126. [DOI] [PubMed] [Google Scholar]
- Seo KI, Lee JH, Nezames CD, Zhong S, Song E, Byun MO, Deng XW. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell. 2014;26(2):695–711. doi: 10.1105/tpc.113.119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim SG, Park CM. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008;13(10):550–556. doi: 10.1016/j.tplants.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23(3):1138–1152. doi: 10.1105/tpc.111.083485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X, Yu Y, Zhu L, Liu H, Chai Q, Guo W. A cotton NAC transcription factor GhirNAC2 plays positive roles in drought tolerance via regulating ABA biosynthesis. Plant Sci. 2020;296:110498. doi: 10.1016/j.plantsci.2020.110498. [DOI] [PubMed] [Google Scholar]
- Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, Wang X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol. 2012;80(3):241–253. doi: 10.1007/s11103-012-9941-y. [DOI] [PubMed] [Google Scholar]
- Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep. 2017;7:40641. doi: 10.1038/srep40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Chan Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J Integr Plant Biol. 2014;56(2):114–121. doi: 10.1111/jipb.12128. [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17(8):2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhao Y, Wang J, Lu MZ (2021) The transcription factor KNAT2/6b mediates changes in plant architecture in response to drought via downregulating GA20ox1 in Populus alba x P. glandulosa. J Exp Bot. 10.1093/jxb/erab201 [DOI] [PubMed]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Su H, Cao Y, Ku L, Yao W, Cao Y, Ren Z, Dou D, Wang H, Ren Z, Liu H, Tian L, Zheng Y, Chen C, Chen Y. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J Exp Bot. 2018;69(21):5177–5189. doi: 10.1093/jxb/ery299. [DOI] [PubMed] [Google Scholar]
- Sun Y, Yu D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015;34(8):1295–1306. doi: 10.1007/s00299-015-1787-8. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Kuromori T, Sato H, Shinozaki K. Regulatory gene networks in drought stress responses and resistance in plants. Adv Exp Med Biol. 2018;1081:189–214. doi: 10.1007/978-981-13-1244-1_11. [DOI] [PubMed] [Google Scholar]
- Tan W, Zhang D, Zhou H, Zheng T, Yin Y, Lin H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018;14(4):e1007336. doi: 10.1371/journal.pgen.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalaikumar VP, Devkar V, Mehterov N, Ali S, Ozgur R, Turkan I, Mueller-Roeber B, Balazadeh S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol J. 2018;16(2):354–366. doi: 10.1111/pbi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC. Turgor maintenance by osmotic adjustment: 40 years of progress. J Exp Bot. 2018;69(13):3223–3233. doi: 10.1093/jxb/ery181. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97(21):11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AE, Overnas E, Johansson H, Rada-Iglesias A, Engstrom P. The homeodomain-leucine zipper (HD-zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol Biol. 2012;80(4–5):405–418. doi: 10.1007/s11103-012-9956-4. [DOI] [PubMed] [Google Scholar]
- Verma D, Jalmi SK, Bhagat PK, Verma N, Sinha AK. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020;287(12):2560–2576. doi: 10.1111/febs.15157. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, Man WQ, Zhang JS, Chen SY. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83(2):224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- Wang J, Li Q, Mao X, Li A, Jing R. Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int J Biol Sci. 2016;12(2):257–269. doi: 10.7150/ijbs.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Yan Y, Zhang S, Li H, Gao Z, Wang C, Guo X (2020) GhWRKY21 regulates ABA-mediated drought tolerance by fine-tuning the expression of GhHAB in cotton. Plant Cell Rep. 10.1007/s00299-020-02590-4 [DOI] [PubMed]
- Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017;58(10):1764–1776. doi: 10.1093/pcp/pcx111. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wan L, Zhang L, Zhang Z, Zhang H, Quan R, Zhou S, Huang R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol Biol. 2012;78(3):275–288. doi: 10.1007/s11103-011-9861-2. [DOI] [PubMed] [Google Scholar]
- Wang YG, An M, Zhou SF, She YH, Li WC, Fu FL. Expression profile of maize microRNAs corresponding to their target genes under drought stress. Biochem Genet. 2014;52(11–12):474–493. doi: 10.1007/s10528-014-9661-x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Su G, Li M, Ke Q, Kim SY, Li H, Huang J, Xu B, Deng XP, Kwak SS. Overexpressing Arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol Biochem. 2016;109:199–208. doi: 10.1016/j.plaphy.2016.09.020. [DOI] [PubMed] [Google Scholar]
- Wang Z, Tian X, Zhao Q, Liu Z, Li X, Ren Y, Tang J, Fang J, Xu Q, Bu Q. The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates Cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in Rice. Plant Cell. 2018;30(1):228–244. doi: 10.1105/tpc.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjarvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018;69:209–236. doi: 10.1146/annurev-arplant-042817-040322. [DOI] [PubMed] [Google Scholar]
- Wei S, Xia R, Chen C, Shang X, Ge F, Wei H, Chen H, Wu Y, Xie Q (2021) ZmbHLH124 identified in maize recombinant inbred lines contributes to drought tolerance in crops. Plant Biotechnol J. 10.1111/pbi.13637 [DOI] [PMC free article] [PubMed]
- Wei W, Liang DW, Bian XH, Shen M, Xiao JH, Zhang WK, Ma B, Lin Q, Lv J, Chen X, Chen SY, Zhang JS. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and ca(2+) signaling pathways in transgenic soybean. Plant J. 2019;100(2):384–398. doi: 10.1111/tpj.14449. [DOI] [PubMed] [Google Scholar]
- Wei Y, Liu W, Hu W, Yan Y, Shi H. The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol. 2020;226(2):476–491. doi: 10.1111/nph.16346. [DOI] [PubMed] [Google Scholar]
- Wenjing W, Chen Q, Singh PK, Huang Y, Pei D. CRISPR/Cas9 edited HSFA6a and HSFA6b of Arabidopsis thaliana offers ABA and osmotic stress insensitivity by modulation of ROS homeostasis. Plant Signal Behav. 2020;15(12):1816321. doi: 10.1080/15592324.2020.1816321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC. ABI4: versatile activator and repressor. Trends Plant Sci. 2013;18(3):125–132. doi: 10.1016/j.tplants.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Wu H, Fu B, Sun P, Xiao C, Liu JH. A NAC transcription factor represses putrescine biosynthesis and affects drought tolerance. Plant Physiol. 2016;172(3):1532–1547. doi: 10.1104/pp.16.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12(1):9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Sun X, Bian X, Wei T, Han T, Yan J, Zhang A. The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J Exp Bot. 2021;72(4):1399–1410. doi: 10.1093/jxb/eraa507. [DOI] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115(1):35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- Xie F, Stewart CN, Jr, Taki FA, He Q, Liu H, Zhang B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol J. 2014;12(3):354–366. doi: 10.1111/pbi.12142. [DOI] [PubMed] [Google Scholar]
- Xie F, Wang Q, Sun R, Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot. 2015;66(3):789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Bao C, Chen P, Cao F, Liu X, Geng D, Li Z, Li X, Hou N, Zhi F, Niu C, Zhou S, Zhan X, Ma F, Guan Q. Abscisic acid homeostasis is mediated by feedback regulation of MdMYB88 and MdMYB124. J Exp Bot. 2021;72(2):592–607. doi: 10.1093/jxb/eraa449. [DOI] [PubMed] [Google Scholar]
- Xie Z, Nolan T, Jiang H, Tang B, Zhang M, Li Z, Yin Y. The AP2/ERF transcription factor TINY modulates Brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell. 2019;31(8):1788–1806. doi: 10.1105/tpc.18.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhao X, Aiwaili P, Mu X, Zhao M, Zhao J, Cheng L, Ma C, Gao J, Hong B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020;103(5):1783–1795. doi: 10.1111/tpj.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, do Hyeon Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell. 2013;25(11):4708–4724. doi: 10.1105/tpc.113.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant. 2011;4(4):697–712. doi: 10.1093/mp/ssr013. [DOI] [PubMed] [Google Scholar]
- Yang C, Huang Y, Lv W, Zhang Y, Bhat JA, Kong J, Xing H, Zhao J, Zhao T. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020;293:110442. doi: 10.1016/j.plantsci.2020.110442. [DOI] [PubMed] [Google Scholar]
- Yang S, Xu K, Chen S, Li T, Xia H, Chen L, Liu H, Luo L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019;19(1):260. doi: 10.1186/s12870-019-1872-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Luang S, Harris J, Riboni M, Li Y, Bazanova N, Hrmova M, Haefele S, Kovalchuk N, Lopato S. Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol J. 2018;16(6):1227–1240. doi: 10.1111/pbi.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chi X, Guo F, Jin X, Luo H, Hawar A, Chen Y, Feng K, Wang B, Qi J, Yang Y, Sun B. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J Plant Physiol. 2020;246-247:153142. doi: 10.1016/j.jplph.2020.153142. [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. The formation and function of plant cuticles. Plant Physiol. 2013;163(1):5–20. doi: 10.1104/pp.113.222737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Gao J, Liu M, Qin C, Zhang W, Yang A, Xia M, Zhang Z, Shen Y, Lin H, Luo C, Pan G. Genome-wide analysis of water-stress-responsive microRNA expression profile in tobacco roots. Funct Integr Genomics. 2014;14(2):319–332. doi: 10.1007/s10142-014-0365-4. [DOI] [PubMed] [Google Scholar]
- Yong Y, Zhang Y, Lyu Y (2019a) A MYB-related transcription factor from Lilium lancifolium L. (LlMYB3) is involved in anthocyanin biosynthesis pathway and enhances multiple abiotic stress tolerance in Arabidopsis thaliana. Int J Mol Sci 20(13):3195. 10.3390/ijms20133195 [DOI] [PMC free article] [PubMed]
- Yong Y, Zhang Y, Lyu Y. A stress-responsive NAC transcription factor from Tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int J Mol Sci. 2019;20(13):3225. doi: 10.3390/ijms20133225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell. 2010;22(12):4128–4141. doi: 10.1105/tpc.110.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38(1):35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61(4):672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Yu L, Zhou L, Liu W, Huang P, Jiang R, Tang Z, Cheng P, Zeng J. Identification of drought resistant miRNA in Macleaya cordata by high-throughput sequencing. Arch Biochem Biophys. 2020;684:108300. doi: 10.1016/j.abb.2020.108300. [DOI] [PubMed] [Google Scholar]
- Yu M, Liu J, Du B, Zhang M, Wang A, Zhang L (2021) NAC transcription factor PwNAC11 activates ERD1 by interaction with ABF3 and DREB2A to enhance drought tolerance in transgenic Arabidopsis. Int J Mol Sci 22(13). 10.3390/ijms22136952 [DOI] [PMC free article] [PubMed]
- Yu Y, Jia T, Chen X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017;216(4):1002–1017. doi: 10.1111/nph.14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ni Z, Wang Y, Wan H, Hu Z, Jiang Q, Sun X, Zhang H. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019;285:68–78. doi: 10.1016/j.plantsci.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Yuan X, Wang H, Cai J, Bi Y, Li D, Song F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019;19(1):278. doi: 10.1186/s12870-019-1883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Zhang G, Zhao Z, Yue J, Pu X, Sui M, Zhan Y, Shi Y, Wang Z, Meng G, Zhao Z, An L. A cryophyte transcription factor, CbABF1, confers freezing, and drought tolerance in tobacco. Front Plant Sci. 2019;10:699. doi: 10.3389/fpls.2019.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Pioneer transcription factors initiating gene network changes. Annu Rev Genet. 2020;54:367–385. doi: 10.1146/annurev-genet-030220-015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li C, Liu J, Lv Y, Yu C, Li H, Zhao T, Liu B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J Exp Bot. 2017;68(16):4695–4707. doi: 10.1093/jxb/erx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tong J, Xu Z, Wei P, Xu L, Wan Q, Huang Y, He X, Yang J, Shao H, Ma H. Soybean C2H2-type zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis. Front Plant Sci. 2016;7:325. doi: 10.3389/fpls.2016.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Huang S, Zhang C, Li D, Wu Y, Deng J, Shan S, Qi J. Overexpression of CcNAC1 gene promotes early flowering and enhances drought tolerance of jute (Corchorus capsularis L.) Protoplasma. 2021;258(2):337–345. doi: 10.1007/s00709-020-01569-y. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cui X, Guo Y, Luo C, Zhang L. Picea wilsonii transcription factor NAC2 enhanced plant tolerance to abiotic stress and participated in RFCP1-regulated flowering time. Plant Mol Biol. 2018;98(6):471–493. doi: 10.1007/s11103-018-0792-z. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao X, Zhi Y, Li X, Zhang Q, Niu J, Wang J, Zhai H, Zhao N, Li J, Liu Q, He S. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019;223(4):1918–1936. doi: 10.1111/nph.15925. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu D, Yang B, Liu WZ, Mu B, Song H, Chen B, Li Y, Ren D, Deng H, Jiang YQ. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J Exp Bot. 2020;71(1):188–203. doi: 10.1093/jxb/erz432. [DOI] [PubMed] [Google Scholar]
- Zhang S, Haider I, Kohlen W, Jiang L, Bouwmeester H, Meijer AH, Schluepmann H, Liu CM, Ouwerkerk PB. Function of the HD-zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol. 2012;80(6):571–585. doi: 10.1007/s11103-012-9967-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett. 2011;33(2):403–409. doi: 10.1007/s10529-010-0436-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li J, Chen S, Ma X, Wei H, Chen C, Gao N, Zou Y, Kong D, Li T, Liu Z, Yu S, Luo L. An APETALA2/ethylene responsive factor, OsEBP89 knockout enhances adaptation to direct-seeding on wet land and tolerance to drought stress in rice. Mol Gen Genomics. 2020;295(4):941–956. doi: 10.1007/s00438-020-01669-7. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Hu RS, Liu D, Liu X, Wang J, Xiang XH, Li YY. The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol J. 2020;18(12):2444–2455. doi: 10.1111/pbi.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Qi CH, Jiang H, You CX, Guan QM, Ma FW, Li YY, Hao YJ. The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic Res. 2019;6:66. doi: 10.1038/s41438-019-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61(15):4157–4168. doi: 10.1093/jxb/erq237. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang Y, Wang X, Han X, An Y, Lin S, Shen C, Wen J, Liu C, Yin W, Xia X. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020;227(2):407–426. doi: 10.1111/nph.16524. [DOI] [PubMed] [Google Scholar]
- Zhu D, Wu Z, Cao G, Li J, Wei J, Tsuge T, Gu H, Aoyama T, Qu LJ. TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol Plant. 2014;7(4):601–615. doi: 10.1093/mp/sst152. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu MD, Zhang M, Gao DJ, Zhou K, Tang SJ, Zhou B, Lv YM. Rice OsHSFA3 gene improves drought tolerance by modulating polyamine biosynthesis depending on abscisic acid and ROS levels. Int J Mol Sci. 2020;21(5):1857. doi: 10.3390/ijms21051857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, Xu YH, Zhang XY, Zhang DP. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19(10):3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang P, Guo P, Chong L, Yu G, Sun X, Hu T, Li Y, Hsu CC, Tang K, Zhou Y, Zhao C, Gao W, Tao WA, Mengiste T, Zhu JK. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020;228(5):1573–1590. doi: 10.1111/nph.16787. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yan J, Liu W, Liu L, Sheng Y, Sun Y, Li Y, Scheller HV, Jiang M, Hou X, Ni L, Zhang A. Phosphorylation of a NAC transcription factor by a calcium/calmodulin-dependent protein kinase regulates abscisic acid-induced antioxidant defense in maize. Plant Physiol. 2016;171(3):1651–1664. doi: 10.1104/pp.16.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016;171(4):2810–2825. doi: 10.1104/pp.16.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo ZF, Kang HG, Hong QC, Park MY, Sun HJ, Kim J, Song PS, Lee HY. A novel basic helix-loop-helix transcription factor, ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ROS scavenging. Plant Mol Biol. 2020;102(4–5):447–462. doi: 10.1007/s11103-019-00957-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The list of transcription factors (TFs) functionally involved in drought tolerance.
Data Availability Statement
Not applicable.