Abstract
We studied the involvement of the human T-cell leukemia virus type 1 (HTLV-1) Gag matrix protein in the cell-to-cell transmission of the virus using missense mutations of the basic amino acids. These basic amino acids are clustered at the N terminus of the protein in other retroviruses and are responsible for targeting the Gag proteins to the plasma membrane. In the HTLV–bovine leukemia virus genus of retroviruses, the basic amino acids are distributed throughout the matrix protein sequence. The HTLV-1 matrix protein contains 11 such residues. A wild-type phenotype was obtained only for mutant viruses with mutations at one of two positions in the matrix protein. The phenotypes of the other nine mutant viruses showed that the basic amino acids are involved at various steps of the replication cycle, including some after membrane targeting. Most of these nine mutations allowed normal synthesis, transport, and cleavage of the Gag precursor, but particle release was greatly affected for seven of them. In addition, four mutated proteins with correct particle release and envelope glycoprotein incorporation did not however permit cell-to-cell transmission of HTLV-1. Thus, particle release, although required, is not sufficient for the cell-to-cell transmission of HTLV-1, and the basic residues of the matrix protein are involved in steps that occur after viral particle release.
The Gag proteins of retroviruses form the viral core and are sufficient to govern assembly and release of viral particles from infected cells in the absence of envelope glycoproteins (7; for reviews, see references 20 and 46). These functions are performed by Gag precursor polyproteins, which, upon budding of the particles, are cleaved by the viral protease to produce the matrix (MA), the capsid (CA), and the nucleocapsid (NC), the mature proteins that compose the virion.
In the late stages of replication of type C retroviruses, which include lentiviruses and viruses of the human T-cell leukemia virus–bovine leukemia virus (HTLV-BLV) genus, the MA domain anchors the Gag polyproteins to the plasma membrane. The proteins are anchored to the membrane by N-terminal myristylation (2, 17, 32, 33) and ionic interactions involving, at least in lentiviruses, an N-terminal cluster of basic residues in the MA domain and the acidic plasma membrane surface (40, 51–53). However, some type C retroviruses, such as the avian Rous sarcoma virus (RSV), have MA proteins with no myristylation signal (see Fig. 1A). In such cases, the Gag proteins are bound to the plasma membrane via the N-terminal half of the MA, which contains basic residues (25, 41). The final stages in the production of retroviruses include association of Gag polyproteins with each other, leading to the activation of the virally encoded protease, processing of the precursor polyproteins, and the final assembly and budding of virus particles (20, 46).
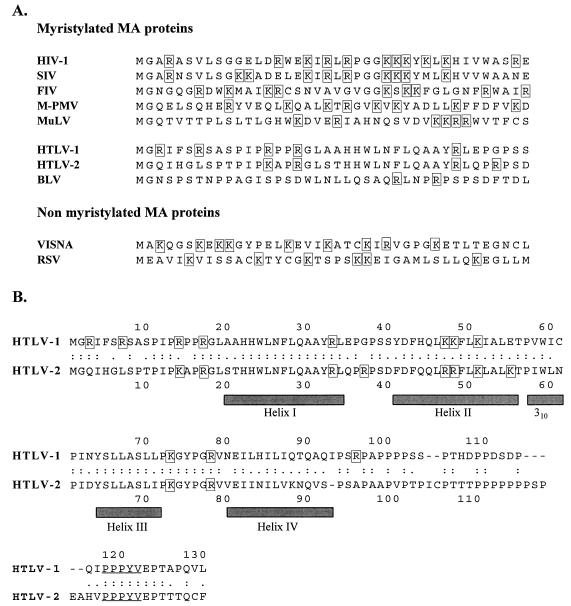
FIG. 1.
Sequence alignment for retroviral Gag polyproteins. The basic amino acids are boxed throughout. (A) The 43 N-terminal amino acids of the MA of several retroviruses. Abbreviations: SIV, simian immunodeficiency virus; FIV, feline immunodeficiency virus; M-PMV, Mason-Pfizer monkey virus; MuLV, murine leukemia virus; VISNA, Visna-Maedi virus. (B) Sequence alignment for the HTLV-1 and HTLV-2 matrix proteins. The HTLV-1 sequence is taken from Seiki et al. (38), and the HTLV-2 sequence is taken from Shimotohno et al. (39). The shaded bars indicate the helices predicted from the NMR structure analysis of the HTLV-2 matrix protein (4). The PPPYV motif is underlined.
In addition to its roles in the late steps of the replication cycle in the producing cell, the MA of the human immunodeficiency virus type 1 (HIV-1) is also involved in the early stages of the viral life cycle (23, 31, 43) and may participate in the nuclear targeting of the infecting virus (3, 15, 16; however, see references 12 and 13). It has been suggested that this early function of the MA is specific to the lentiviruses which, unlike the other retroviruses (35), replicate in nondividing cells (3, 42). However, the MA of RSV is also required, after budding, for viral infection, and the nature of its function at this stage is unknown (28).
Nothing is known about the molecular determinants of Gag protein functions in the HTLV-BLV genus of type C retroviruses. The Gag proteins are believed to have similar functions in all retroviruses, but the sequences of the HTLV-BLV Gag polyproteins have no obvious conserved motifs. In particular, the MA of HTLV-BLV retroviruses does not contain an N-terminal cluster of basic amino acids, similar to that of lentiviruses (see Fig. 1A). However, the MA proteins of both BLV and HTLV-2 adopt a conformation very similar to that of primate lentiviruses, as shown by structural determination by nuclear magnetic resonance (NMR) spectroscopy (4, 24). The structure determined suggested that some of the basic amino acids of the HTLV-2 MA, although scattered throughout the primary structure of the MA, form a conformational cluster exposed at the surface of the protein, similar to the N-terminal basic domain of the lentiviral MA. The HTLV-2 and HTLV-1 MA proteins are both myristylated (27), and their amino acid sequences are 58% identical. They are therefore likely to have similar structures. Eight of the 11 basic amino acids present in the MA of HTLV-2 are also present in the MA of HTLV-1, which also has 11 basic amino acids (see Fig. 1B).
In this study, we mutated each of the basic residues of the HTLV-1 MA to determine their roles in viral transmission and to elucidate the functions of the MA in infection by retroviruses of the HTLV-BLV genus. We studied whether these mutations affected late or early steps in the replication cycle of HTLV-1 by examining the intracellular synthesis, processing, distribution, and membrane binding of the Gag proteins as well as the virion production and the infectivity of viruses with mutated MA proteins. We found that none of the basic residues of the HTLV-1 MA were essential for intracellular processing or membrane binding of the Gag precursor protein, but most were involved at subsequent stages, including virus production and the early events required for infection by HTLV-1. Thus, the HTLV-1 MA has a key function required for infection in addition to its involvement in late stages of the replication cycle of the virus.
MATERIALS AND METHODS
Cell lines.
COS-1 and HeLa cells were obtained from the American Type Culture Collection. B5 cells were a gift from D. Waters (Frederick Cancer Research and Development Facility, Frederick, Md.). 293-TSA cells (19) were obtained from J. Neyton (Ecole Normale Supérieure, Paris, France). All cells were grown in Dulbecco’s modified Eagle medium containing 5% fetal calf serum, at 37°C in a 5% CO2 atmosphere.
Plasmids.
The XMT plasmid (8), which contains a complete infectious HTLV-1 provirus, was a gift from D. Derse (National Cancer Institute, Frederick, Md.). The pCS-HTLV-neo plasmid is an HTLV-1 proviral clone with the env gene replaced by the neomycin resistance gene under the control of the simian virus 40 promoter; it has been described elsewhere, as has the CMV-ENV HTLV-1 envelope expressor (6).
Site-directed mutagenesis.
Oligonucleotide-directed mutagenesis of the sequence encoding the HTLV-1 MA protein was performed with the pGEM-5MA plasmid, by the Kunkel method. The pGEM-5MA construct contains a 934-bp NdeI-NcoI fragment of the gag gene from the XMT proviral clone (positions 319 to 1253 in Seiki’s sequence [38]) inserted into the pGEM-5Zf(+) vector (Promega). The mutated gag fragments were then excised and inserted into the XMT and pCS-HTLV-neo plasmids, sequenced, and used to transfect cells. The mutants were named according to the pattern X amino acid position-Y, where X and Y are the wild-type and replacement amino acids, respectively, and amino acid position 1 corresponds to the initiator methionine of the HTLV-1 MA protein. The triple mutant Lys47,48,51-Ile plasmid, has each of the three lysine residues, at positions 47, 48, and 51, replaced by an isoleucine residue.
Western blotting of HTLV-1 Gag proteins.
Cell lysates and supernatants were collected 48 h after transient transfection of COS-1 cells with the mutated MA proviral constructs (5). Virus pellets were obtained from the transfected cell supernatant by centrifugation at 3,000 × g for 15 min, filtration of the supernatant through filters with 0.45-μm pores, and centrifugation in an SW41 Beckman rotor at 25,000 rpm for 2 h. Cell lysates and virus pellets were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting to detect the Gag proteins, according to standard procedures. Immunoblotting was performed with a pool of sera from HTLV-1-infected individuals as the primary antibody (a gift from Y. Coste, CRTS, Montpellier, France), and 125I-labeled protein A (Amersham) as the second-step reagent.
For each mutated protein, the cleavage index of the Gag precursor, which is the percentage cleavage of the mutated Gag precursor relative to that of the wild-type protein, was calculated as follows, after determining the radioactivity of each band from the transfected cell lysate, with a phosphorimager (Molecular Dynamics):
 |
The virus particle production index is the percentage of particle release for the MA mutant virus relative to that of the wild-type virus. It was calculated after determining the radioactivity of the CA band from the virus pellets, as follows: radioactivity of the CA band of the mutated protein/radioactivity of the CA band of the wild type protein × 100.
Radioimmunoprecipitation of pulse-labeled HTLV-1 Gag proteins.
Forty-eight hours after transient transfection of COS-1 cells with the mutated MA proviral constructs (5), the cells were starved for 1 h in methionine- and cysteine-free medium and pulse-labeled for 15 min at 37°C with Promix labeling medium (1.5 mCi/ml) containing [35S]cysteine and [35S]methionine (Amersham). Cells were then subjected to a chase period of 1 to 12 h in complete Dulbecco’s modified Eagle medium. At the end of the chase period, the cells were lysed and the Gag proteins were immunoprecipitated with a pool of sera from HTLV-1-infected individuals. Immunoprecipitates were electrophoresed in SDS–13% polyacrylamide gels and visualized by autoradiography.
Immunofluorescence detection of HTLV-1 Gag proteins.
Twenty-four hours after transient transfection of HeLa cells with the mutated MA proviral constructs by the calcium phosphate precipitation method, the cells were treated with trypsin and transferred to microwell slides, in which they were incubated for 24 h at 37°C. The cells were then washed in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min at room temperature, followed by quenching with 0.1 M glycine in PBS for 15 min at room temperature. Permeabilization and saturation were achieved by incubating the cells for 2 h in PBS containing 0.05% saponin and 0.2% bovine serum albumin, and all subsequent steps were performed in this buffer at room temperature. A 1:800 dilution of the primary antibody directed against the HTLV-1 MA (11) (a gift from C. Desgranges, INSERM U271, Lyon, France) was added to each well, and the mixture was incubated for 1 h. The cells were then washed five times and incubated for 1 h with a 1:300 dilution of goat anti-mouse–cyanin 3 conjugate (Jackson Immunoresearch Laboratories). The slides were washed five times and mounted in Mowiol 4-88 (Calbiochem). Fluorescent images of equatorial slices were examined with a confocal microscope (model MRC-1000; Bio-Rad).
Envelope glycoprotein incorporation into virions.
The incorporation of the envelope glycoproteins into virions was evaluated for each mutated Gag protein, as described previously (6). Cells transiently transfected with the proviral constructs were metabolically labeled, virions were collected from the cell supernatants, purified on discontinuous sucrose gradients, lysed, and immunoprecipitated with a pool of sera from HTLV-1-infected individuals.
Membrane binding assay.
The binding of HTLV-1 Gag precursor proteins to the cell membrane was studied in a cell fractionation assay. Forty-eight hours after transfection with provirus constructs, HeLa cells (3 × 106) were detached with trypsin, washed three times in PBS, and collected by centrifugation. Cells were ruptured by three freeze-thaw cycles, using dry ice plus ethanol for freezing and a 37°C water bath for thawing, suspended in 500 μl of TBS buffer (0.15 M NaCl, 10 mM Tris [pH 7.4]), containing protease inhibitors (2 mg/ml) [4-(2-aminoethyl)-benzenesulfonyl fluoride (Interchim, Montluçon, France); Complete (1 tablet/50 ml) (Boehringer Mannheim)]. The solution was centrifuged at 2,500 × g for 10 min and at 9,000 × g for 30 min, at 4°C, to remove the unbroken cells and nuclei. The salt concentration was brought to 1 M NaCl, and the soluble fraction was separated from the membrane fraction by centrifugation at 100,000 × g for 1 h at 4°C. The pellet was suspended in 500 μl of TBS buffer containing 1% Triton X-100; 50 μl of 10% Triton X-100 was added to the supernatant cytosolic fraction. HTLV-1-specific proteins were then characterized by immunoblotting, using 30 μl of each fraction.
Infectivity assay.
The abilities of the mutated Gag proteins to mediate the cell-to-cell transmission of HTLV-1 were evaluated by a quantitative assay described elsewhere (6). COS-1 cells were cotransfected with the pCS-HTLV-neo provirus constructs with mutated MA (0.75 μg) and the HTLV-1 envelope expressor CMV-ENV (0.75 μg). At 1 day after transfection, the cells were treated with 10 μg of mitomycin per ml (Amétycine; Laboratoires Choay, Paris, France) for 3 h at 37°C to stop growth. The cells were then washed five times with PBS, treated with trypsin, and seeded with B5 cells. After 2 days of coculture, half of the cells were transferred to selection medium containing 125 μg of G418 sulfate (Geneticin; Gibco) per ml. G418 sulfate-resistant colonies were counted after 2 to 3 weeks. The infectivity index was calculated as described previously (6, 36). This index is the percentage of viral transmission with the mutated MA provirus relative to that with the wild-type virus.
RESULTS
Function of the HTLV-1 MA basic amino acids in cell-to-cell transmission.
We investigated whether the basic amino acids of the HTLV-1 MA are important primarily for anchoring Gag polyproteins to the plasma membrane or are involved at various steps of the HTLV-1 replication cycle, by replacing each of the 11 basic residues of the MA separately with a nonconservative amino acid (Fig. 1B and Table 1). The MA protein of HTLV-1 has an N-terminal consensus sequence for myristylation [methionine-glycine-X-X-X-(Serine/Threonine)] (Fig. 1) and is known to be myristylated (27). Therefore, we also studied the phenotype of an HTLV-1 virus with a conservative substitution of the corresponding glycine residue (Gly2-Ala), which was expected to have defective membrane anchoring.
TABLE 1.
Phenotypes of HTLV-1 MA mutantsa
| MA protein | Precursor cleavage index (%) | Particle production index (%) | Infectivity index (%) |
|---|---|---|---|
| Wild type | 100 | 100 | 100 |
| Gly2-Ala | 7 | 1 | 0 |
| Arg3-Leu | 38 | 20 | 0 |
| Arg7-Leu | 50 | 17 | 61 |
| Arg14-Leu | 58 | 16 | 3 |
| Arg17-Leu | 40 | 8 | 6 |
| Arg33-Leu | 63 | 44 | 0 |
| Lys47-Ile | 53 | 40 | 0 |
| Lys48-Ile | 61 | 7 | 2 |
| Lys51-Ile | 72 | 39 | 0 |
| Lys74-Ile | 35 | 28 | 0 |
| Arg79-Leu | 17 | 0 | 0 |
| Arg97-Leu | 73 | 70 | 50 |
| Lys47,48,51-Ile | 0 | 0 | 0 |
Data are the means of at least two independent transfections. The precursor cleavage and virus particle production indices were calculated as described in Materials and Methods, and the infectivity index was calculated as described elsewhere (6).
HTLV-1 is transmitted almost exclusively via cell-to-cell contact in vivo (9, 26) and in vitro (30, 48). The effect of the mutated MA proteins on the cell-to-cell transmission of HTLV-1 was investigated by an assay that we have previously described (6) for quantitatively evaluating viral transmission in one round of infection, using transcomplementation of a proviral construct with an env deletion and an HTLV-1 envelope expressor.
As expected from results with other mutated retroviral MA proteins, there was no viral transmission with the HTLV-1 MA with no myristylation site (Gly2-Ala) (Table 1).
Only 2 of the 11 single substitutions of basic MA residues (Arg7-Leu and Arg97-Leu) allowed the cell-to-cell transmission of HTLV-1 (Table 1). Mutations of the remaining nine basic residues completely abolished viral infectivity. Therefore, all but two of the basic amino acids of the MA protein were individually required for the correct transmission of HTLV-1.
As substitution of most of the basic amino acids of the HTLV-1 MA protein abolished viral transmission, we investigated which step of viral transmission was affected by each of the mutations. We therefore investigated, for each MA mutant, the production of viral particles, including quantitative evaluation of particle release and glycoprotein incorporation into the particles. The intracellular processing, distribution, and membrane binding of the mutated Gag proteins in the transfected cells were also studied to identify possible defects of the Gag proteins in the cells that produced them.
Function of HTLV-1 MA basic amino acids in virus particle production.
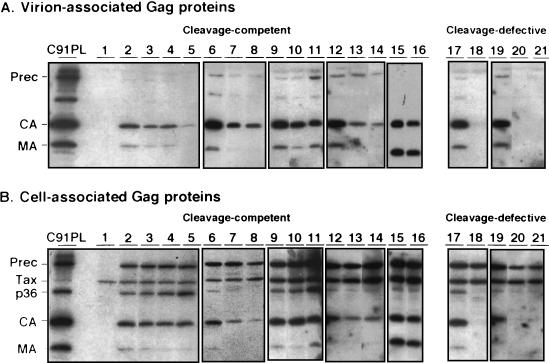
We investigated whether the low level of infectivity observed with most MA proteins with mutated basic residues was due to a defect in virus particle production, by quantifying the amount of mature Gag proteins in viral pellets from the supernatants of cells transfected with the mutated MA proviral constructs and comparing it to the amount obtained with the wild-type provirus. The profiles of the virion-associated Gag proteins are shown in Fig. 2A, and the calculated particle production indices are given in Table 1.
FIG. 2.
Western-blotting of the HTLV-1 Gag proteins from virus pellets (A) and transfected-cell lysates (B). C91PL, lysate of HTLV-1-infected cells; lane 1, negative control; lanes 2, 6, 9, 12, 15, 17, 19, wild-type positive control; lane 3, Arg33-Leu; lane 4, Lys47-Ile; lane 5, Lys48-Ile; lane 7, Arg7-Leu; lane 8, Arg14-Leu; lane 10, Lys51-Ile; lane 11, Lys74-Ile; lane 13, Arg3-Leu; lane 14, Arg17-Leu; lane 16, Arg97-Leu; lane 18, Gly2-Ala; lane 20, Arg79-Leu; lane 21, Lys47,48,51-Ile. Prec, Gag precursor; Tax, transactivator protein.
The Gly2-Ala myristylation mutant had the expected phenotype: it produced no virus particles (Fig. 2A, lane 18).
Three of the nine mutated MA proviruses that were not infectious had a severe defect in virus particle production, because no viral protein was detected in the supernatant of transfected cells. These mutants were Arg17-Leu, Lys48-Ile, and Arg79-Leu. The lack of cell-to-cell transmission of the corresponding proviruses is therefore presumably due to their inability to produce virus particles.
The remaining six mutated MA viruses that were not infectious had low (Arg3-Leu and Arg14-Leu) or substantial (Arg33-Leu, Lys47-Ile, Lys51-Ile, and Lys74-Ile) levels of virus particle production, but their levels were lower than that of the wild-type virus in all cases (Table 1 and Fig. 2A). The amounts of particle production obtained with these constructs were similar to, or greater than, that of the Arg7-Leu mutant, which nevertheless allowed viral transmission. Therefore, the lack of infection by these six mutated MA viruses does not result only from reduced levels of virus particle production.
Envelope glycoprotein incorporation into virions of MA mutants with virus particle production but no viral transmission.
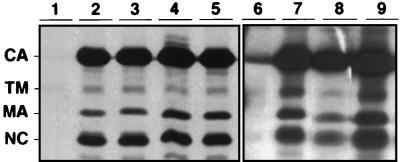
For the six mutated MA viruses with low infectivity despite virus particle production, we investigated whether a defect in envelope glycoprotein incorporation into virions was responsible for the lack of viral transmission. Incorporation was assessed by immunoprecipitation of the mature TM glycoprotein from virions purified from supernatants of cells transfected with the proviral constructs (Fig. 3 and data not shown), in the absence of the glycoprotein precursor. As we have reported previously (6), the level of wild-type glycoprotein incorporation was low, the exposure times of the gels were very long, probably reflecting instability of the glycoproteins at the virion surface. However, glycoproteins were incorporated with the six mutated MA constructs at a level similar to that of the wild-type provirus. This excludes a major incorporation defect as the reason for the low infectivity of these viruses. Instead, the lack of infectivity of these mutants suggests that the HTLV-1 MA protein may be involved in viral entry at a step that occurs after the virus particle production and glycoprotein incorporation.
FIG. 3.
Envelope glycoprotein incorporation into virions with mutated MA. The presence of the TM glycoprotein indicates glycoprotein incorporation into the virions. Lanes 1 and 6, negative control; lanes 2 and 7, wild-type positive control; lane 3, Arg3-Leu; lane 4, Arg33-Leu; lane 5, Lys51-Ile; lane 8, Arg7-Leu; lane 9, Lys47-Ile.
Function of HTLV-1 MA basic amino acids in intracellular processing.
Our results suggest that six of the basic residues which are essential for viral transmission are not essential for late steps of the replication cycle, because virus particles were produced when each of them was mutated. If this is so, the intracellular processing of the corresponding mutated Gag precursors should be normal. Conversely, the three mutations preventing particle production might also prevent precursor cleavage. We further investigated the mutated constructs, by studying the intracellular synthesis and processing of the Gag proteins (Fig. 2B and Table 1).
The Gag precursor with no myristylation site (Gly2-Ala) was not cleaved (Fig. 2B, lane 18). This has also been observed with various other myristylation-negative retroviral Gag protein mutants (2, 33, 37, 43).
In contrast, most of the Gag precursors with single mutations of basic amino acids of the MA were correctly processed. These included, as expected, the proteins with mutations allowing substantial virus particle production but also, more surprisingly, most of those proteins with mutations resulting in a defect in virus particle production.
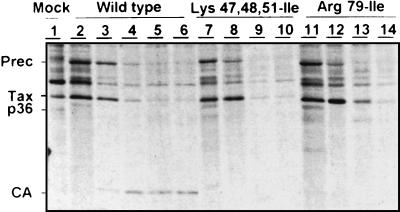
One mutant (Arg79-Leu), however, had greatly impaired processing of the Gag precursor protein into its mature derivatives, and the levels of intracellular accumulation of Gag proteins for this mutant were consistently lower than those for the wild type (Fig. 2B, lane 20). As the amount of Gag precursor protein was less than that in the wild type, we investigated whether the corresponding mutation was associated with instability of the Gag precursor protein, by pulse-chase experiments and immunoprecipitation of the Gag proteins (Fig. 4). Immediately after pulse-labeling, similar amounts of Gag precursor proteins were immunoprecipitated from cells transfected with the wild-type provirus and the Arg79-Leu mutant, suggesting that similar amounts of these two proteins were synthesized. However, the half-life of the Arg79-Leu Gag precursor was shorter than that of the wild-type Gag precursor. Thus, the arginine residue at position 79 of the MA is important for the stability of the Gag precursor protein.
FIG. 4.
Immunoprecipitation of HTLV-1 Gag proteins from transfected cell lysates after pulse-chase experiments. After a 30-min pulse with radioactive medium, a chase with nonradioactive medium was performed for various amounts of time. Lanes 2, 7, and 11, no chase; lanes 3, 8, 12, 1-h chase; lanes 4, 9, and 13, 4-h chase; lanes 5, 10, and 14, 8-h chase; lane 6, 12-h chase. Wild type, wild-type Gag proteins; Prec, Gag precursor; Tax, transactivator protein.
Function of HTLV-1 MA basic amino acids in determining intracellular distribution and binding of Gag proteins to the cell membrane.
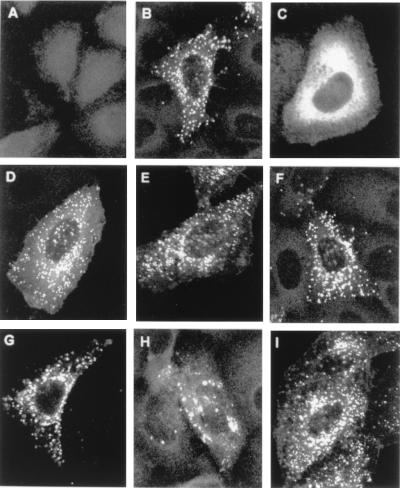
Cleavage of the Gag precursor is believed to occur at the plasma membrane during and soon after budding (21). Therefore, our results suggested that the intracellular targeting of most of the Gag proteins with mutated basic residues was normal, including that of the mutants with defects in virus particle production. This was tested by in situ immunofluorescence analysis of the distribution of the Gag proteins in transfected cells (Fig. 5 and data not shown). There was typical punctate membrane staining in cells transfected with the wild-type provirus (Fig. 5B), similar to that reported for other retroviral Gag proteins (18, 43, 51). The Gly2-Ala mutation was the only mutation to affect the intracellular distribution of the Gag proteins, giving diffuse cytoplasmic staining (Fig. 5C). The intracellular distributions of the Gag proteins with mutations of basic amino acids of the MA were similar to that of the wild-type protein (Fig. 5D to I). This was particularly so for the mutated proteins with profound release defects, Arg7-Leu, Arg17-Leu, and Lys48-Ile (Fig. 5D, F, and H).
FIG. 5.
Immunofluorescence staining of HTLV-1 Gag proteins in transfected cells. (A) Negative control; (B) wild-type provirus; (C) Gly2-Ala; (D) Arg7-Leu; (E) Arg14-Leu; (F) Arg17-Leu; (G) Arg33-Leu; (H) Lys48-Ile; (I) Arg97-Leu.
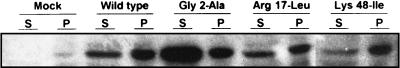
We further examined the membrane binding capacity of the mutants with the most marked defect in virus release using a direct approach based on cell fractionation. We found that the amounts of Gag precursor in the pelletable membrane-containing fraction (Fig. 6, P fractions) in the wild type and the Arg17-Leu or Lys48-Ile mutants were similar. In contrast, the Gly2-Ala mutation greatly impaired the association of the Gag precursor with membranes, as shown by the increased amount of mutated Gag precursor in the soluble fraction (Fig. 6, Gly2-Ala).
FIG. 6.
Binding of HTLV-1 Gag precursor proteins to membranes of transfected cells. Transfected HeLa cells were lysed and fractionated by differential centrifugation to give supernatant (S) and membrane pellet (P) fractions. Proteins from the fractions were subjected to SDS-PAGE and immunoblotted with HTLV-1 infected patient serum.
These results suggest that the basic residues of the HTLV-1 MA protein are important for infectivity at various stages of the viral replication cycle, but do not play a major role, at least individually, in targeting the Gag precursor to the plasma membrane.
A combination of three basic residue mutations results in defective intracellular processing of the Gag precursor.
Studies on the NMR structure of the HTLV-2 matrix protein (4) suggest that the basic amino acids of HTLV-2, and probably of HTLV-1, form a plaque exposed at the surface of the Gag precursor that is required for stable association with the plasma membrane. None of the single mutations of the basic amino acids of the HTLV-1 MA affected the intracellular distribution of the Gag proteins, so we mutated a combination of three basic residues predicted to be exposed at the surface of the MA, and studied the phenotype of the resulting provirus.
Like the Gly2-Ala and the Arg79-Leu mutants, the Gag precursor with the triple mutation (Lys47,48,51-Ile) was not cleaved at all (Fig. 2B and Table 1). Like the Arg79-Leu mutant, the triple-mutated precursor was unstable, with small amounts of precursor protein accumulated in the transfected cells (Fig. 2B, lane 21) and a half-life shorter than that of the wild type (Fig. 4). No particle production was observed, and this mutant was not infectious, as expected (Table 1). We could not determine the intracellular distribution of the triple-mutated Gag protein, because we were unable to detect an immunofluorescence signal above background, probably due to the instability of the Gag precursor. The lack of cleavage of the Lys47,48,51-Ile Gag precursor, however, suggests that there was disruption of a process required for cleavage, such as protein folding, anchoring to the plasma membrane, or both.
DISCUSSION
In this study, we investigated whether the basic amino acids of the MA protein were important for infectivity of HTLV-1. Only 2 of the 11 basic amino acids of the HTLV-1 MA could be mutated with no loss of infectivity. These two amino acids are at positions 7 and 97 of the MA protein, and neither of the equivalent positions in HTLV-2 is occupied by a basic residue (Fig. 1B), strongly suggesting that these residues are not critical for the function of the MA protein. The high frequency of nonfunctional proteins was unexpected, however, because similar mutagenesis of the MA basic residues of HIV results in no loss of infectivity (14), and in RSV, a large region of the MA is dispensable for infectivity (25). Studies in progress in our laboratory indicate that mutation of other (nonbasic) residues of the MA also cause marked infectivity defects. We have previously reported the low tolerance to mutagenesis of the env gene of HTLV-1 (29). HTLV-1 strains are highly conserved throughout the world, and this conservation may be partly due to selection against viruses with mutations because of their lack of infectivity. The low tolerance of HTLV-1 to mutagenesis may be typical of viruses that have evolved for a long time in their hosts.
The late steps in the replication cycle of type C retroviruses include the assembly and release of virions at the plasma membrane. These steps require the synthesis of the Gag polyproteins and their transport to the membrane budding site, where precursor cleavage by the viral protease starts. We investigated whether impairment of any of these late steps in the cell producing the virions could account for the low infectivity of most of our MA mutants with single substitutions of basic residues. The only mutated protein with no detectable precursor cleavage despite correct accumulation of the Gag precursor was Gly2-Ala, in which the myristylation site was eliminated. Thus, as for other myristylated retroviral Gag proteins (2, 33, 37, 43), processing of the HTLV-1 Gag precursor requires myristylation of the MA domain. The Gly2-Ala Gag precursor also failed to associate with membranes, again as observed with other retroviruses (32, 43). Thus, its cleavage-defective phenotype is probably due to defective activation of the protease initiated at the plasma membrane during virus release (21). Gly2-Ala was the only mutated protein with impaired membrane association. Immunofluorescence studies showed that the intracellular distributions of all the Gag proteins with single mutations of the basic residues were similar to that of the wild-type protein. Moreover, direct evaluation of membrane association by cell fractionation showed that two mutations causing profound defects of particle release and infectivity, Arg17-Leu and Lys48-Ile, did not alter the binding of the precursor to membranes. In HIV-1, both the N-terminal basic amino acids and myristylation are involved in Gag protein binding to the plasma membrane (40, 51, 52). Our results demonstrate that myristylation of the HTLV-1 Gag precursor is critical for membrane targeting of the Gag proteins, but none of the basic residues alone are important for this step.
It has been suggested, based on the NMR structure of the MA of HTLV-2 (4), that the basic residues of helix II, the lysines at positions 47, 48, and 51 of the HTLV-1 MA, form a structural motif exposed at the surface of the molecule that interacts with the lipid bilayer. We therefore constructed the Lys47,48,51-Ile triple mutant to determine whether a combination of mutations could impair interaction with membranes. We were unable to study the intracellular targeting of the protein with the triple mutation however, because it had a very short half-life. This was also the case for the single mutation of the arginine residue at position 79, an amino acid that is very likely to be exposed at the surface of the MA (4). MA mutated proteins with small deletions in other retroviruses have been reported to be similarly unstable (1, 34), showing that the MA plays a key role in the correct folding and stability of Gag polyproteins.
Our study shows that virus particle release is a limiting step in the late events of HTLV-1 replication, because most of the mutated MA proviruses released fewer particles into the cell supernatant than did the wild type, despite the correct intracellular processing of the Gag proteins. In RSV, a large deletion of the C-terminal part of the MA does not prevent budding (25), whereas in this study, only one mutation, that of the arginine residue at position 97, allowed near-normal particle release. The HTLV-1 MA basic residues thus play a key role either in budding or in the pinching off of particles from the plasma membrane. We attempted to determine whether the MA mutations causing profound defects of particle release also resulted in intracellular accumulation of particles by studying our mutants by electron microscopy (data not shown). There were very few cell-associated virus particles, with both the wild-type and the mutated proteins, and there was no accumulation of budding particles at the plasma membrane of cells expressing mutated proteins defective for virus release. Although these negative results should be regarded with caution, they suggest that an effect on particle release is not due to a late-release defect. In RSV, mutations of the PPPYV proline-rich motif of a small p2b protein, located between the MA and CA in the Gag polyprotein, impair particle release (45, 47). The motif is conserved among a variety of retroviral Gag polyproteins, including those of HTLV-1 (Fig. 1B). Mutation of the basic residues of HTLV-1 MA may impair virus release indirectly, by affecting the exposure of the PPPYV motif, or the basic residues may be involved more directly in efficient release, perhaps together with the PPPYV motif. Recent evidence shows that the release of Mason-Pfizer monkey virus particles involves ATP hydrolysis and suggests that Gag polyproteins interact with cellular components in this process (44). If such interactions occur in other retroviruses, including HTLV-1, then the MA basic amino acids may be involved in their mediation.
Correct particle release is probably required for HTLV-1 transmission, because we did not obtain any transmission-competent viruses with no particle production. HTLV-1 transmission in vitro (30, 48) and in vivo (9, 26) requires contact with infected cells, cell-free particles are very weakly (or not) infectious, possibly because they are very fragile. Cell-to-cell transmission, which also occurs for other retroviruses, is more efficient than cell-free transmission. It could occur via a budding particle, with simultaneous pinching off from the plasma membrane of the producing cell on one side and microfusion with the plasma membrane of the target cell on the other or via virions just released into the intercellular space. Our results suggest that the molecular determinants of particle release are also important for cell-to-cell transmission. However, the viral transmission of the Arg7-Leu mutant was more efficient than expected based on its virus particle production. This mutant MA virus may be intrinsically more infectious than the wild-type virus, compensating for its low levels of virus release.
Four MA proteins with mutated basic residues did not allow the cell-to-cell transmission of HTLV-1, despite correct particle release. We verified that the incorporation of envelope glycoproteins into virus particles was not impaired. In HIV-1, mutations in the MA disturb glycoprotein incorporation (10, 50), probably because the MA has to accommodate the long cytoplasmic tail of the glycoproteins. In HTLV-1, however, the cytoplasmic domain of the glycoproteins is short, and incorporation probably does not require the two proteins to be particularly compatible (unpublished results). Thus, as an incorporation defect cannot account for the lack of infectivity of these four mutants, the MA of HTLV-1 is probably involved, via its basic amino acids, in early steps of the replication cycle.
The MA is involved in post-budding events in lentiviruses (22, 31, 49). The mature MA protein of HIV-1 adopts a conformation different from that of the MA domain in Gag polyproteins (53). This may make possible a myristyl switching mechanism, regulating membrane affinity and allowing the N-terminal basic sequence to serve as a nuclear targeting signal. The involvement of lentiviral MA in nuclear targeting is a matter of debate (12, 13), but it is clear that the MA of lentiviruses is involved in early events of the replication cycle. This property of the MA may not be specific to lentiviruses, which replicate in nondividing cells, and thus require processes allowing the penetration of the virus core into the nucleus. Results with RSV (28) and our results with HTLV-1 suggest that the involvement of the matrix protein in early steps of the replication cycle is a general property of retroviruses. The MA of retroviruses thus has a dual role. First, as part of the Gag polyproteins, the MA targets the forming virus to the plasma membrane and facilitates its budding. Second, as the mature MA protein, it enables the virus to enter and replicate in the target cell. Retrovirus transmission requires that the MA be efficient at both functions.
ACKNOWLEDGMENTS
We thank Claude Desgranges (INSERM U271, Lyon, France) for kindly donating the anti-MA monoclonal antibody, Y. Coste (CRTS, Montpellier, France) for providing sera from HTLV-1-infected individuals, and Isabelle Bouchaert for excellent assistance with confocal microscopy. The English text was edited by Owen Parkes.
This work was supported by grants from the Association Nationale pour la Recherche sur le SIDA (Paris, France) and from the Association pour la Recherche sur le Cancer (Villejuif, France) as well as by equipment grants from the Ligue Départementale des Yvelines (Versailles, France) and the Fondation pour la Recherche Médicale (Paris, France).
REFERENCES
- 1.Bonefeld Jørgensen E C, Pedersen F S, Jørgensen P. Matrix protein of Akv murine leukemia virus: genetic mapping of regions essential for particle formation. J Virol. 1992;66:4479–4487. doi: 10.1128/jvi.66.7.4479-4487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen A M, Massiah M A, Turner B G, Sundquist W I, Summers M F. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J Mol Biol. 1996;264:1117–1131. doi: 10.1006/jmbi.1996.0700. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B R. Use of eukaryotic expression technology in the functional expression of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 6.Delamarre L, Rosenberg A R, Pique C, Pham D, Dokhélar M-C. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J Virol. 1997;71:259–266. doi: 10.1128/jvi.71.1.259-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The Gag precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derse D, Mikovits J, Waters D, Brining S, Ruscetti F. Examining the molecular genetics of HTLV-I with an infectious molecular clone of the virus and permissive cell culture systems. J Acquired Immune Defic Syndr. 1996;12:1–5. doi: 10.1097/00042560-199605010-00001. [DOI] [PubMed] [Google Scholar]
- 9.Donegan E, Lee H, Operskalski E A, Shaw G M, Kleinman S H, Busch M P, Stevens C E, Schiff E R, Nowicki M J, Hollingsworth C G, Mosley J W The Transfusion Safety Study Group. Transfusion transmission of retroviruses: human T-lymphotropic virus types I and II compared with human immunodeficiency virus type 1. Transfusion. 1994;34:478–483. doi: 10.1046/j.1537-2995.1994.34694295061.x. [DOI] [PubMed] [Google Scholar]
- 10.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebersold A, Noraz N, Grange J, Gasmi M, Grange M P, Souche S, Mamoun R, Desgranges C. Production and characterization of a monoclonal antibody directed against HTLV-1 p19: use in a specific capture enzyme immunoassay. Hybridoma. 1998;12:185–195. doi: 10.1089/hyb.1993.12.185. [DOI] [PubMed] [Google Scholar]
- 12.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 16.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 17.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen M, Jelinek L, Jones R S, Stegeman-Olsen J, Barklis E. Assembly and composition of intracellular particles formed by Moloney murine leukemia virus. J Virol. 1993;67:5163–5174. doi: 10.1128/jvi.67.9.5163-5174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison T, Graham F, Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977;77:319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- 20.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 21.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y-M, Tang X-B, Cimakasky L M, Hildreth J E K, Yu X-F. Mutations in the matrix protein of human immunodeficiency virus type 1 inhibit surface expression and virion incorporation of viral envelope glycoproteins in CD4+ T lymphocytes. J Virol. 1997;71:1443–1452. doi: 10.1128/jvi.71.2.1443-1452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews S, Mikhailov M, Burny A, Roy P. The solution structure of the bovine leukaemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO J. 1996;15:3267–3274. [PMC free article] [PubMed] [Google Scholar]
- 25.Nelle T D, Wills J W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okochi K, Sato H, Hinuma Y. A retrospective study on transmission of adult T-cell leukemia virus by blood transfusion: seroconversion in recipients. Vox Sang. 1984;46:245–253. doi: 10.1111/j.1423-0410.1984.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 27.Ootsuyama Y, Shimotohno K, Miwa M, Oroszlan S, Sugimura T. Myristylation of gag protein in human T-cell leukemia virus type-I and type-II. Jpn J Cancer Res. 1985;76:1132–1135. [PubMed] [Google Scholar]
- 28.Parent L J, Wilson C B, Resh M D, Wills J W. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J Virol. 1996;70:1016–1026. doi: 10.1128/jvi.70.2.1016-1026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pique C, Tursz T, Dokhélar M-C. Mutations introduced along the HTLV-I envelope gene result in a non-functional protein: a basis for envelope conservation? EMBO J. 1990;9:4243–4248. doi: 10.1002/j.1460-2075.1990.tb07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popovic M, Sarin P S, Robert-Gurroff M, Kalyanaraman V S, Mann D, Minowada J, Gallo R C. Isolation and transmission of human retrovirus (human T-cell leukemia virus) Science. 1983;219:856–859. doi: 10.1126/science.6600519. [DOI] [PubMed] [Google Scholar]
- 31.Reicin A S, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rein A, McClure R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee S S, Hunter E. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J Virol. 1990;64:4383–4389. doi: 10.1128/jvi.64.9.4383-4389.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg A R, Delamarre L, Pique C, Pham D, Dokhélar M-C. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J Virol. 1997;71:7180–7186. doi: 10.1128/jvi.71.10.7180-7186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz A M, Rein A. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J Virol. 1989;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimotohno K, Takahashi Y, Shimizu N, Gojobori T, Golde D W, Chen I S Y, Miwa M, Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc Natl Acad Sci USA. 1985;82:3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spearman P, Wang J J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto N, Okada M, Koyanagi Y, Kannagi M, Hinuma Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science. 1982;217:737–739. doi: 10.1126/science.6980467. [DOI] [PubMed] [Google Scholar]
- 49.Yu X, Yu Q-C, Lee T-H, Essex M. The C-terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]