Abstract
Significance:
Stress granules (SGs) are biomolecular condensates that form upon global translation suppression during stress. SGs are enriched in translation factors and messenger RNAs (mRNAs), which they may sequester away from the protein synthesis machinery. While this is hypothesized to remodel the functional transcriptome during stress, it remains unclear whether SGs are a cause, or simply a consequence, of translation repression. Understanding the function of SGs is particularly important because they are implicated in numerous diseases including viral infections, cancer, and neurodegeneration.
Recent Advances:
We synthesize recent SG research spanning biological scales, from observing single proteins and mRNAs within one cell to measurements of the entire transcriptome or proteome of SGs in a cell population. We use the emerging understanding from these studies to suggest that SGs likely have less impact on global translation, but instead may strongly influence the translation of individual mRNAs localized to them.
Critical Issues:
Development of a unified model that links stress-induced RNA-protein condensation to regulation of downstream gene expression holds promise for understanding the mechanisms of cellular resilience.
Future Directions:
Therefore, upcoming research should clarify what influence SGs exert on translation at all scales as well as the molecular mechanisms that enable this. The resulting knowledge will be required to drive discovery in how SGs allow organisms to adapt to challenges and support health or go awry and lead to disease. Antioxid. Redox Signal. 39, 390–409.
Keywords: stress granules, biomolecular condensates, translation, RNP granules, stress response
Introduction
Protein biosynthesis is one of the most energy-intensive processes in the cell (Buttgereit and Brand, 1995; Li et al., 2014). It is therefore unsurprising that inhibition of translation is a crucial response of cells to a wide variety of stressors. A less foreseeable response to stress and translation suppression is the formation of microscopically visible foci of nontranslating RNA and proteins known as stress granules (SGs). SGs are part of a growing group of biomolecular condensates—large membraneless structures with distinct compositions from their surroundings that display a range of material properties similar to liquids, gels, or solids (Alberti et al., 2019). Biomolecular condensates include other ribonucleoprotein (RNP) granules such as transport granules in neuronal cells, which mediate subcellular localization of RNAs (Ainger et al., 1993; Kiebler and Bassell, 2006; Wilhelm and Vale, 1993) and germ granules, which store RNAs during development in germ line cells (Hegner, 1911; Schisa, 2014).
Besides these cell-type-specific examples, many biomolecular condensates are found in the nucleus, including superenhancers, speckles and paraspeckles, Cajal bodies, and the nucleolus (Sabari et al., 2020). Constitutive RNP granules are also found in the cytoplasm in the form of processing bodies (P-bodies), which are enriched in nontranslating messenger RNAs (mRNAs) and components of the RNA degradation and silencing machinery (Hubstenberger et al., 2017; Sheth and Parker, 2003). In contrast to most of these condensates, SGs are induced during specific challenges, including oxidative stress, heat shock, endoplasmic reticulum (ER) stress, and other insults that result in global translation repression.
The functional importance of most biomolecular condensates is enigmatic. Varied hypotheses propose that localization of biomolecules to condensates increases the effective concentration of components to increase their reaction rate or specificity, sequesters them away from outside factors to inhibit other reactions, and/or buffers cellular changes in the concentration of components (Alberti et al., 2019; Banani et al., 2017). However, the links between the physical properties of condensates and their biological outcomes are challenging to determine experimentally.
In the case of SGs, their formation displays characteristics of liquid–liquid phase separation (LLPS), resulting from high levels of exclusive but reversible intermolecular interactions between their components (Protter and Parker, 2016). Condensation of mRNAs into SGs has been proposed to suppress their translation, protect them from degradation, and allow rapid resumption of translation after stress removal (Ivanov et al., 2019). However, linking these functional outcomes to mRNA localization in SGs is challenging and is an active area of research.
Understanding the function of SGs is relevant for research areas across fundamental biology and human health. As a large and inducible condensate, SGs have been used to develop theoretical frameworks describing phase separation of biomolecules (Sanders et al., 2020) and to study the role of condensation in organismal fitness and adaptation to stress (Riback et al., 2017). Most importantly, SGs have emerged as key players in pathogenesis of multiple diseases, including cancer (Song and Grabocka, 2020), viral infections (Eiermann et al., 2020; McCormick and Khaperskyy, 2017), neurodegeneration (Wolozin and Ivanov, 2019), and aging (Cao et al., 2020).
Each of these separate topics has been reviewed in detail recently in the publications cited above. To progress in these areas, it is essential to address the gap in knowledge of how SGs are important for regulating translation during stress. In this study, we summarize current understanding of SGs as consequences and causes of translation repression, and in doing so highlight where more investigation is needed to understand their cellular function.
Induction and Assembly of Stress Granules
The first step in SG assembly is the inhibition of translation initiation upon acute stress. Temporally, SGs form almost immediately following the translational repression of bulk or representative mRNAs (Kedersha et al., 2000; Khong and Parker, 2018; Moon et al., 2019; Wheeler et al., 2016), which typically occurs within tens of minutes depending on the type of stress (Wheeler et al., 2016). In the vast majority of SG-forming circumstances, translation shutdown occurs through phosphorylation of eIF2α. Phosphorylated eukaryotic translation initiation factor 2α (P-eIF2α) inhibits the activity of its guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) (Pavitt et al., 1997; Scorsone et al., 1987). Limiting eIF2B activity reduces the assembly of the ternary complex containing eIF2α in the eIF2 complex, GTP, and the initiator tRNAiMet (Kedersha et al., 1999), and leads to the accumulation of mRNAs bound to an incomplete 48S preinitiation complex (PIC).
Components of the PIC, but not large ribosomal subunits or eIF2α itself, are highly enriched in SGs (Kedersha et al., 2002). Notably, P-eIF2α-mediated SG formation can be disrupted by increasing the activity of the eIF2B complex to drive ternary complex formation (Sidrauski et al., 2015; Sidrauski et al., 2013; Tsai et al., 2018). Phosphorylation of eIF2α is therefore a primary driver of translation initiation suppression leading to SG formation.
Phosphorylation of eIF2α to inhibit translation is performed by any of the four distinct kinases in the first step of the integrated stress response (ISR) (Fig. 1) (reviewed in English et al., 2022 and Costa-Mattioli and Walter, 2020). Numerous studies have evaluated the requirements of eIF2α kinases for SG formation, which are described in Table 1. For instance, treatment with arsenite is one of the most commonly used models for SG assembly in cell culture, and arsenite-induced SGs result, in part, from phosphorylation of eIF2α by heme-regulated inhibitor kinase (HRI) (McEwen et al., 2005). Another eIF2α kinase, protein kinase R (PKR), senses intracellular double-stranded RNAs, resulting in SG formation during some viral infections, although many viruses also have mechanisms to inhibit SG assembly (White and Lloyd, 2012).
FIG. 1.
Diverse stresses and multiple signaling pathways cause inhibition of translation initiation and formation of stress granules. A variety of stressors can activate any of four kinases for the key initiation factor eIF2α. This phosphorylation event enhances binding of the eIF2 complex to eIF2B, preventing GDP/GTP exchange and eIF2 release, and therefore limiting formation of the ternary complex containing GTP, the initiator Met-tRNAiMet, and eIF2. The ternary complex is essential for start codon recognition and is typically a component of the 48S PIC containing the mRNA, 40S ribosome subunit, and various other initiation factors. Among these is the 5′ cap binding complex, eIF4F. Inactivation of the kinase mTOR during conditions such as starvation leads to hypophosphorylated 4E-BP, which in turn binds and sequesters one of the cap binding complex components, eIF4E. This prevents assembly of the cap binding complex, inhibiting PIC formation and translation initiation. In both of these cases, translationally repressed mRNAs are recruited to stress granules, along with select PIC components. 4E-BP, eukaryotic translation initiation factor 4E-binding protein; eIF2B, eukaryotic translation initiation factor 2B; eIF4E, eukaryotic translation initiation factor 4E; mRNA, messenger RNA; mTOR, mammalian target of rapamycin; PIC, preinitiation complex.
Table 1.
Numerous Stresses Drive Formation of Stress Granules in Mammalian Cells Through eIF2α and mTOR Signaling Pathways
| Stress | Integrated stress response |
mTOR | References | |||
|---|---|---|---|---|---|---|
| HRI | PKR | PERK | GCN2 | |||
| Arsenite | Y | Y | Aulas et al. (2017); Fay et al. (2021); Fournier et al., (2013); McEwen et al. (2005); Sfakianos et al. (2018); Szaflarski et al. (2016) | |||
| Bortezomib | Y | Y | Fournier et al. (2013); Fournier et al. (2010) | |||
| Erythroid differentiation | Y | Ghisolfi et al. (2012) | ||||
| Hepatitis C virus and interferon-alpha | Y | Ruggieri et al. (2012) | ||||
| Measles virus, hantavirus, Respiratory syncytial virus, and yellow fever virus | Y | Beauclair et al. (2020); Christ et al. (2020); Lindquist et al. (2011); Okonski and Samuel (2013) | ||||
| G3BP1 overexpression | Y | Reineke and Lloyd (2015) | ||||
| ADAR1 depletion | Y | Corbet et al. (2022) | ||||
| 2′,5′-Oligoadenylate | Y | Manivannan et al. (2020) | ||||
| Small RNase L-cleaved RNAs | Y | Manivannan et al. (2020) | ||||
| Lapatinib | Y | Adjibade et al. (2020) | ||||
| Thapsigargin | Y | Aulas et al. (2017) | ||||
| Sorafenib | Y | Adjibade et al. (2015) | ||||
| Carbon monoxide | Y | Chen et al. (2019) | ||||
| Bisphenol A | Y | Fay et al. (2021) | ||||
| Vinorelbine | Y | Y | Schwed-Gross et al. (2022); Szaflarski et al. (2016) | |||
| MG-132 | Y | Y | Alvarez-Castelao et al. (2020); Aulas et al. (2017); Jiang and Wek (2005); Mazroui et al. (2007); Yerlikaya et al. (2008) | |||
| Ultraviolet light | Y | Ying and Khaperskyy (2020) | ||||
| Cortisone | Y | Schwed-Gross et al. (2022) | ||||
| Chronic nutrient deprivation | Y | Y | Y | Kuo et al. (2020); Reineke et al. (2018) | ||
| Dihydrocapsaicin | Y | De et al. (2022) | ||||
| Selenite | Y | Fujimura et al. (2012) | ||||
| Cold | Y | Hofmann et al. (2012) | ||||
| ATP depletion | Y | Wang et al. (2022) | ||||
| Hydrogen peroxide | Y | Emara et al. (2012) | ||||
Stresses for which specific signaling pathways have been defined as major contributors to SG formation through genetics and/or chemical genetics assays are indicated as “Y.” This includes all four eIF2α kinases in the integrated stress response pathway, as well as mTOR signaling. Signaling pathways upstream (e.g., AMPK) and downstream (e.g., 4E-BP) of mTOR are included.
4E-BP, eukaryotic translation initiation factor 4E-binding protein; AMPK, AMP-activated protein kinase; GCN2, general control nonderepressible 2; HRI, heme-regulated inhibitor kinase; mTOR, mammalian target of rapamycin; PERK, PKR-related endoplasmic reticulum associated kinase; PKR, protein kinase R; SG, stress granule.
The PKR-related endoplasmic reticulum-associated kinase (PERK) is activated by accumulation of unfolded proteins, and contributes to SG formation under circumstances such as ER stress (Aulas et al., 2017). The fourth eIF2α kinase, general control nonderepressible 2 (GCN2), is activated by a variety of stressors including amino acid starvation, and helps trigger SG assembly during processes as diverse as proteasome inhibition and ultraviolet radiation damage (Mazroui et al., 2007; Ying and Khaperskyy, 2020). Therefore, the ISR converges diverse stress circumstances into similar responses to drive inhibition of translation initiation and SG formation.
Various regulatory, physical, and experimental circumstances that do not target eIF2α can also lead to SG formation. Assembly of SGs in the classic model stress condition of heat shock, for instance, does not require P-eIF2α in flies and yeast, although mammalian cells do require P-eIF2α (Farny et al., 2009; Grousl et al., 2009). One alternative signaling event that may induce SGs is inhibition of the kinase mammalian target of rapamycin (mTOR), which itself is activated by diverse stimuli from nutrient availability to growth hormones. Loss of mTOR activity leads to sequestration of the mRNA cap-binding protein eukaryotic translation initiation factor 4E (eIF4E) by unphosphorylated eukaryotic translation initiation factor 4E-binding protein (4E-BP), resulting in decreased translation initiation that could contribute to SG assembly (Fig. 1).
Interestingly, in the P-eIF2α-independent induction of SGs by hydrogen peroxide, there is a unique requirement for signaling through the eIF4E cap protein, which may be performed by mTOR (Emara et al., 2012; Frydryskova et al., 2016). Disruption of the eIF4E-mTOR pathway also inhibits SG formation in arsenite, heat, and other stresses (Fournier et al., 2013). Inactivation of mTOR appears sufficient to cause SG formation in some cases, including severe ATP depletion in human cells (Wang et al., 2022) and nutrient deprivation in nematodes (Kuo et al., 2020).
Finally, assembly of condensates that share features with SGs, including a requirement for polysome runoff and the presence of several key SG RNA-binding proteins (RBPs), can be induced by osmotic stress (Bounedjah et al., 2012; Zeng et al., 2020). In these cases, it is likely that condensation is promoted by increased intermolecular interactions resulting from decreased cellular volume and crowding forces in the cytoplasm (Jalihal et al., 2020). However, for formation of canonical SGs during stress, repression of translation is the primary initiator.
Not all modes of translation inhibition lead to assembly of SGs, which tells us much about how they form (Fig. 2). Stalling elongating ribosomes on transcripts using small molecules such as cycloheximide or emetine, for instance, does not lead to SG formation (Kedersha et al., 1999). Instead, these inhibitors prevent SG assembly during stress (Kedersha et al., 1999; Mollet et al., 2008), while another inhibitor of translation elongation that allows ribosome removal, puromycin, does not. In contrast, inhibition of translation using small molecules such as hippuristanol and pateamine A that target eukaryotic translation initiation factor 4A (eIF4A) to prevent initiation or assembly of the eIF4F 5′ cap binding complex induces formation of SG-like structures (Dang et al., 2006; Kedersha and Anderson, 2007; Mazroui et al., 2006).
FIG. 2.
Different modes of translation inhibition promote or prevent interaction networks required for stress granule assembly. Small-molecule inhibition of translation initiation at multiple points by pateamine A or hippuristanol leads to stress granule formation. However, inhibition of elongation by any of three small molecules does not lead to stress granule formation. Among these, puromycin allows ribosome removal from the mRNA, and does not prevent stress granule formation upon stress. The ribosome-vacant mRNA then participates in a network of interactions with “hub” RBPs (hub defined as binding valency n ≥ 3) as well as other RNAs. This exclusive and interconnected network leads to phase separation and stress granule assembly. When ribosomes remain on mRNA, such as during normal translation or when trapped with inhibitors cycloheximide or emetine, interactions with RBPs and other RNAs are prevented or decreased. This, as well as inhibition of RBPs by “cap” proteins, conformational changes, or PTMs, prevents stress granule assembly. PTM, posttranslational modification; RBP, RNA-binding protein.
These distinct results of disrupting initiation versus elongation have been recapitulated using genetic depletion of various factors from these steps including the cap-binding protein eIF4E and large ribosomal subunits, respectively (Mokas et al., 2009). Mechanisms of translation elongation inhibition that trap mRNAs within polysomes are therefore proposed to limit the condensation of mRNAs into SGs (Kedersha et al., 2002). Indeed, there is evidence that select transcripts require removal of stalled or slowed ribosomes via a stress-activated ribosome quality control (saRQC) pathway to partition into SGs during arsenite or heat stress (Moon et al., 2020). These observations together suggest a model where the suppression of translation initiation and ribosome runoff of transcripts drives the formation of SGs.
A theoretical framework used to describe SG assembly is an interaction network containing “node” or “hub” biomolecules participating in three or more interactions with component proteins/RNAs, “bridges” that interact with two other molecules, and inhibitory “caps” that interact with only one molecular partner (Sanders et al., 2020) (Fig. 2). The formation of SGs, in particular, is dependent upon high-valency hub RBPs, which facilitate the phase transition to condensates in a manner dependent upon their own concentration and that of their RNA binding partners, primarily translationally repressed mRNAs (Guillén-Boixet et al., 2020; Van Treeck and Parker, 2018; Yang et al., 2020). After removal of polysomes from the coding sequence, the naked mRNA becomes a key multivalent hub available for interaction with these RBPs to drive SG condensation (Fig. 2) (Guillén-Boixet et al., 2020).
Furthermore, as RNA is capable of forming phase-separated structures at high concentrations in vitro, RNA-RNA interactions likely also play a key role in SG assembly (Van Treeck et al., 2018). These concepts explain the essential requirement for forms of translation suppression that lead to mRNAs freed from polysomes to drive SG formation, and the bias toward long RNAs being enriched in SGs (discussed in Composition and Physicial Properties of SGs), as both maximize the ability of an mRNA to form intermolecular interactions and nucleate SGs.
While RNA is an essential hub for SG assembly, many multivalent RBPs play crucial roles in SG assembly (Table 2). The functions and properties of these proteins are an area of intense study, as mutations in many SG RBPs lead to neurodegenerative disease. Central among these hub proteins are G3BP1 and G3BP2, which are indispensable for canonical SG formation upon most stresses (Kedersha et al., 2016). The essential role of G3BP1/2 in nucleating SGs results from their high valency, in particular from their RNA-binding capability and self-dimerization, both of which are required for SG formation (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020).
Table 2.
Characteristics of Key RNA Binding Proteins That Localize to Stress Granules
| RBP name | IDR/LCD | Prion-like | Self- interaction | Interactions with other SG RBPs | Disruption decreases SGs? | SGs form with overexpression? | Associated genetic diseases | Identified roles in translation | References |
|---|---|---|---|---|---|---|---|---|---|
| G3BP1/2 | + | − | Dimer | UBAP2/2L, Caprin | ++ | Yes | Neurodevelopmental disease | Promotes translation via ribosome recycling in unstressed cells | Huang et al. (2020), Jia et al. (2022); Meyer et al. (2020); Reineke et al. (2012); Yang et al. (2020) |
| UBAP2/2L | + | − | Dimer | G3BP1/2, FMRs/FXRs | ++ | Yes | Neurodevelopmental disease | Promotes translation of bound mRNAs in unstressed cells | Cirillo et al. (2020); Huang et al. (2020); Jia et al. (2022); Luo et al. (2020) |
| TIA-1/TIAR | + | + | Oligomer | n/a | + | Yes | ALS, FTD, Welander distal myopathy | Inhibits translation in unstressed cells, regulates mRNAs with 5′ terminal oligopyrimidine tracts | Damgaard and Lykke-Andersen (2011); Gilks et al. (2004); Hackman et al. (2013); Hirsch-Reinhagen et al. (2017); Waris et al. (2014) |
| hnRNPA1 | + | + | Oligomer | n/a | −/+ | No | ALS, FTD, multisystem proteinopathy, multiple hereditary neuropathies, and myopathies | Regulates cap-independent translation in a transcript-specific manner | Anees et al. (2021); Beijer et al. (2021); Bonnal et al. (2005); Cammas et al. (2007); Feng et al. (2022); Gui et al. (2019); Guil et al. (2006); Roy et al. (2014) |
| FMR/FXRs | + | + | Dimer | UBAP2/2L | − | No | Fragile X syndrome, fragile X tremor/ataxia syndrome, premature ovarian failure | Inhibits translation initiation and elongation in context-dependent manners | Dolen et al. (2007); Guo et al. (2014); Hagerman et al. (2017); Laggerbauer et al. (2001); Li et al. (2001); Sanders et al. (2020); Sopova et al. (2019); Sullivan et al. (2011) |
| Caprin | + | − | Dimer | G3BP1/2 | − | No | Neurodevelopmental disease | Inhibits translation Promotes translation of synaptic proteins through RNA transport |
Pavinato et al. (2022); Sanders et al. (2020); Shiina et al. (2010); Solomon et al. (2007) |
| TDP-43 | + | + | Di/Oligomer | ATXN2, hnRNP A/Bs | −/+ | n/a | ALS, ALS-FTD, frontotemporal lobar degeneration | Promotes translation of specific mRNAs, and at axons through RNA transport Inhibits translation by sequestration in RNP granules |
Altman et al. (2021); Briese et al. (2020); Buratti et al. (2005); Nagano et al. (2020); Nonaka and Hasegawa (2018); Sreedharan et al. (2008) |
| FUS | + | + | Oligomer | n/a | n/a | n/a | ALS, ALS-FTD, hereditary essential tremor | Promotes local translation within RNP granules Cytoplasmic FUS mutants are associated with translation suppression |
Kamelgarn et al. (2018); Lopez-Erauskin et al. (2018); Shorter (2017); Yasuda et al. (2013) |
| ATXN2/L | + | − | Dimer | TDP-43, G3BP1 | −/+ | Yes | Spinocerebellar ataxia type 2, ALS, late-onset Parkinson's disease | Promotes translation of specific mRNAs, ablation decreases global translation rate | Baumer et al. (2014); Elden et al. (2010); Fittschen et al. (2015); Kaehler et al. (2012); Li et al. (2022); Yamashita et al. (2014) |
The presence of IDR or LCD domains, prion-like behavior, multimerization behavior, and demonstrated interactions with other SG RNA binding proteins is listed for key RBPs found in SGs. Also shown is the impact of genetic disruption or overexpression on SG formation, associated genetic diseases, and identified roles in translation for each. Proteins with IDR/LCD or prion-like domains are indicated with “+,” while those proteins that do not have these regions are indicated with “−.” SG formation is: ++: prevented, +: greatly decreased; −/+: decreased in some studies; or −: unaffected by genetic disruption of the indicated proteins.
ALS, amyotrophic lateral sclerosis; FTD, frontotemporal dementia; IDR, intrinsically disordered region; LCD, low-complexity domain; mRNA, messenger RNA; n/a, not available; RBP, RNA-binding protein; RNP, ribonucleoprotein; TDP-43, TAR DNA binding protein; TIA-1, TIA1 cytotoxic granule associated RNA binding protein; TIAR, TIA1 cytotoxic granule associated RNA binding protein like 1; n/a, data not available.
Formation of SGs and similar structures can be induced purely by overexpression of G3BP1 (Kedersha and Anderson, 2007; Reineke et al., 2012), driving G3BP1-G3BP1 interactions using the light-induced oligomerization cryptochrome 2 (Cry2) domains (Zhang et al., 2019), or by addition of recombinant G3BP1 to cell lysates (Freibaum et al., 2021). Interaction of G3BP1 with the 40S ribosome is important for formation of SGs, and likely determines some of the specificity for initiation-stalled mRNAs in SGs (Kedersha et al., 2016). Similar to several other SG-associated RBPs such as hnRNPA1 and FMR1, G3BP1/2 contain multiple intrinsically disordered regions (IDRs), which are frequently reported as drivers of phase separation through promiscuous protein–protein interactions (Posey et al., 2018). Interestingly, while having defined regulatory roles and interaction partners, these IDRs alone are not required for SG formation (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020).
A more recently recognized central node protein is ubiquitin associated protein 2 like (UBAP2L), which has RNA- and G3BP1-binding activities essential for SG formation, and self-dimerizes similarly to G3BP1 (Huang et al., 2020; Sanders et al., 2020). Similar to G3BP1, overexpression of UBAP2L is sufficient to induce SG assembly in nonstressed conditions (Huang et al., 2020). Induction of SG assembly by increasing protein expression is also true for another group of key SG-associated RBPs, TIA1 cytotoxic granule associated RNA binding protein (TIA-1) and TIA1 cytotoxic granule associated RNA binding protein like 1 (TIAR) (Gilks et al., 2004; Kedersha and Anderson, 2007; Kedersha et al., 1999). Similarly to the IDRs found in other SG nucleator proteins, the TIA proteins contain low complexity prion-like domains whose aggregation propensity is an important interaction contributing to SG assembly (Gilks et al., 2004). Tethering studies have shown that rather than binding of any one specific SG RBP, the presence of many interactions is likely the major factor that determines the ability of a given RNA to be sequestered within an SG (Khong et al., 2017; Matheny et al., 2021; Moon et al., 2019). These data show that a variety of RBPs collaborate to form the interaction network that condenses RNAs into SGs.
To prevent aberrant granule formation, the protein–protein, protein-RNA, and RNA-RNA interactions that lead to SG formation must be inhibited in the absence of stress (Van Treeck and Parker, 2018). The primary mechanism for this appears to be that actively translating mRNAs are coated in polysomes and therefore the concentration of free mRNA available to form interactions is low. For G3BP1 in particular, increased RNA concentration leads to a conformational switch from low to high valency states, allowing it to initiate SG assembly (Yang et al., 2020). The development of conformations and interactions leading to RNA condensation into SGs may also be actively modulated by certain enzymes. The highly abundant eIF4A RNA helicase inhibits SG assembly in a manner not entirely dependent on its ATPase activity, suggesting that its RNA binding alone can disrupt important RNA-RNA interactions leading to condensation (Tauber et al., 2020).
Activity of protein-specific chaperones has also been found to inhibit SG assembly, potentially by remodeling protein–protein or protein-RNA interactions (Jain et al., 2016). Changes from an inhibited to active state of G3BP1 are mediated by binding of its inhibitor protein and “cap” USP10 (ubiquitin-specific peptidase 10) as well as some viral proteins to prevent SG formation (Kedersha et al., 2016; Panas et al., 2015). These studies show that, while translation suppression and the resulting interactions with nontranslating mRNAs are the key drivers of SG assembly, targeted regulation of SG formation does occur.
Posttranslational modifications (PTMs) have been extensively identified as molecular regulators of interactions leading to SG formation (Hofweber and Dormann, 2019). For instance, the autoinhibitory conformation of G3BP1's IDR domains that is controlled both by RNA- and USP10-binding appears to be modulated by multiple phosphorylation events (Guillén-Boixet et al., 2020; Yang et al., 2020), although the exact effects of these modifications are unclear (Panas et al., 2019). The arginine-glycine-glycine (RGG) motifs and RNA recognition motif (RRM) domains of G3BP1 and UBAP2L contain methylated arginines, which could interfere with RNA binding, and decreasing or preventing these modifications increases SG formation (Huang et al., 2020; Tsai et al., 2016).
A wide variety of other protein PTMs have been shown to influence SG assembly, including glycosylation (Ohn et al., 2008), ubiquitination (Mazroui et al., 2007), sumoylation (Jongjitwimol et al., 2016), acetylation (Jedrusik-Bode et al., 2013), and poly(ADP)-ribosylation (Leung et al., 2011). However, the signaling events leading to these modifications, what components are being modified, and the impacts of these PTMs on SG formation are not well-understood. Furthermore, the longer timescales required for these modifications to be generated upon stress may suggest that they are involved less in inducing SG formation as they are in granule stability, disassembly, or are secondary impacts from disrupted cell signaling pathways (Hofweber and Dormann, 2019).
As RNA and protein interactions accumulate following suppression of translation initiation, one model posits that the assembly of SGs occurs in two distinct steps. In this model, a stable “core” nucleating structure of ∼100–200 nm (Jain et al., 2016; Niewidok et al., 2018) forms first, followed by recruitment of a more dynamic “shell” to one or multiple cores. The formation of mature SGs may then occur through fusion of multiple of these structures, a process possibly facilitated by cytoskeletal movement of RNP complexes, as inhibition of microtubule dynamics limits formation of large SGs (Ivanov et al., 2003; Nadezhdina et al., 2010). The existence of the central cores is largely supported by data from live and fixed human cell fluorescence microscopy assays relying on imaging of SG proteins tagged with fluorescent proteins that show areas of increased RBP density and decreased dynamics within the granule (Jain et al., 2016; Niewidok et al., 2018) (Fig. 3).
FIG. 3.
Stress granules are dynamic condensates with discrete substructures. Stress granules display properties of liquid–liquid phase separation within the cell, including spherical shape and the ability to merge and split. Many stress granule components are characterized by rapid movement of molecules within, as well as frequent exchange with surroundings, which is proposed to indicate the presence of a highly dynamic “shell” layer. In contrast, numerous putative “core” regions within one stress granule are smaller, more densely packed, have less internal movement, and slower exchange with the surroundings. Substantial variability in, and biphasic distribution of, molecule lifetimes within the stress granule may also indicate these two distinct core and shell states.
Other live cell microscopy experiments show that a subpopulation of mRNAs interact stably with the SG for at least 10 minutes (the time limit of this imaging method), suggesting regions of variable dynamics with the granule (Moon et al., 2019). The existence of cores is also indicated by purification of G3BP1- and poly(A) RNA-containing structures from stressed cells, which contain many SG components but are smaller and much more stable than SGs within the cell (Jain et al., 2016).
Beyond evidence for this substructure itself, several lines of evidence suggest that cores form first, followed by recruitment of the shell, rather than cores condensing within the mature SG: (1) cores can be purified at or before the time of appearance of microscopically visible SGs, (2) the core structure in SGs appears similar in size and dynamicity through the entire lifetime of the granule, and (3) proximity mapping indicates that SG nucleator proteins maintain similar interaction networks both pre- and poststress, suggesting they exist as “seeds” even before stress (Markmiller et al., 2018; Marmor-Kollet et al., 2020; Wheeler et al., 2016). Distinct cores containing either G3BP1 or UBAP2L can be observed within an SG, and genetic disruption of these RBPs suggests that UBAP2L acts upstream of G3BP1, perhaps indicating a role in early core formation (Cirillo et al., 2020).
Correlations between proximity interactomes of multiple SG proteins pre- and post-stress have enabled definition of a putative set of proteins in the SG “seed,” including G3BP1, TIA1, and UBAP2L (Marmor-Kollet et al., 2020). However, one caveat to these proximity mapping experiments is that G3BP1, and likely other RBPs, can rapidly enter and exit the putative SG shell (as measured by fluorescence recovery after photobleaching [FRAP] assays) (Kedersha et al., 2005) Therefore, these G3BP1 interactomes likely include proteins that interact with the large fraction of G3BP1 present in the cytoplasm. While these lines of evidence suggest a substructure and ordered formation of SGs, further data are needed to identify the mechanisms of core assembly and to indicate what consequences they have for SG condensation and function.
Composition and Physical Properties of Stress Granules
The assembled SGs are present as microscopically visible foci of RNA and protein, typically with diameters of ∼1 μm (Moon et al., 2019) (Fig. 3). Canonical SGs are droplet-like, displaying many characteristics of LLPS, such as a spherical shape, rapid exchange of components with surroundings (Buchan and Parker, 2009; Kedersha et al., 2005), and ability to fuse and split (Kroschwald et al., 2015). For instance, single-molecule imaging techniques have shown that individual proteins diffuse within SGs and mRNAs dynamically interact with granules for seconds-minutes; behaviors characteristic of phase-separated liquids and suggestive of the “shell” component (Moon et al., 2019; Niewidok et al., 2018). These and other methods also observe biochemically purifiable, less dynamic “core” structures within SGs, as described above (Jain et al., 2016; Khong et al., 2017; Namkoong et al., 2018).
Both RNAs and proteins in SGs have been observed to rapidly exchange with the surrounding solvent and nearby P-bodies in live cells, although there is substantial variability between and within molecules in terms of lifetime in the SG or P-body (Buchan and Parker, 2009; Kedersha et al., 2005; Moon et al., 2019). These observations show that SGs are composed of both dynamic and stable components, rather than strictly sequestering all their protein and RNA components for long timescales during stress.
The majority of RNA content observed in SGs is mRNAs (Fig. 4), with estimates ranging from ∼80% of the RNA content in biochemically purified cores (Khong et al., 2017) to >99% of RNA species in the SG using in vivo proximity labeling by editing followed by sequencing (van Leeuwen et al., 2021). It should be noted, however, that these proportions are reported after removal of ribosomal RNA (rRNA) before RNA sequencing, and thus, rRNA may comprise a large fraction of SG RNAs. The mRNAs in SGs comprise a relatively small fraction (∼10%–13%) of the total mRNA content of the cell, and, while nearly all coding transcripts appear to be represented in SGs, the relative partitioning of a given mRNA to the granule varies greatly (Khong et al., 2017).
FIG. 4.
The RNA and protein composition of stress granules. Transcriptomic studies of purified stress granule cores suggest that they contain ∼80% mRNAs and 20% noncoding RNAs. While the vast majority of cellular mRNA species are present in the stress granule, relative enrichment varies greatly. A primary determinant of mRNA localization to stress granules is the low translation efficiency and ribosome occupancy, leaving sites available for interactions with RNA binding proteins. In contrast, mRNAs that continue to translate during stress may be excluded. A further important factor determining RNA localization to stress granules is the RNA length, with greater length providing more interaction sites and therefore increased valency “n.” Proteins present in stress granules are greatly enriched in RNA binding functions and include translation factors. Also present are various helicase and chaperone proteins. Proteins within stress granules are enriched in (and tend to have longer) IDRs, LCDs, and prion-like domains, all of which are implicated in phase separation. IDR, intrinsically disordered region; LCD, low-complexity domain.
These results are based primarily upon SG cores purified by centrifugation and G3BP1 affinity purification, and, while validated using single molecule fluorescent in-situ hybridization (smFISH) of multiple transcript components and overall poly(A) RNA signal, the in vivo mRNA composition of more dynamic SGs may differ substantially (Khong et al., 2017). Furthermore, other results suggest contamination with polysome-bound mRNAs when using similar centrifugation-based methods and imply that affinity purification is vital for achieving an SG-specific transcriptome or proteome (Matheny et al., 2019). Within SG mRNAs, an attractive idea is that specific functional classes of genes would be enriched to facilitate their coordinated regulation. While various transcriptomic studies have reported that mRNAs encoded by genes involved in metabolism (Matheny et al., 2019), oncogenesis (Namkoong et al., 2018), and ATP binding (van Leeuwen et al., 2021) are enriched in SGs, no consistent functional overrepresentations have been identified between analyses.
Instead, the mRNAs in SGs resulting from multiple stressors are heavily biased toward increased length and decreased translational efficiency (Khong et al., 2017; Namkoong et al., 2018; van Leeuwen et al., 2021; Van Treeck et al., 2018). In addition, mRNAs with longer open reading frames are sequestered within SGs for longer durations (Moon et al., 2019). This corresponds well with the proposed valency model of SG assembly, as both longer mRNAs and those with fewer ribosomes will present more sites for interactions with other mRNAs or nucleating RBPs. Conversely, mRNAs with very high translational efficiency localize to SGs only at very low levels (Matheny et al., 2019). Single-molecule imaging further shows that actively translating mRNA reporters have short interaction times with SGs and do not enter into a more long-lived “locked” state with them (Moon et al., 2019).
Interestingly, noncoding RNAs (ncRNA) are a relatively minor component of SGs, at 0.4% of distinct RNA species as measured by proximity transcriptomics (van Leeuwen et al., 2021) and ∼20% of bulk RNA content in purified SG cores (Khong et al., 2017). This also represented only ∼0.6% of nonribosomal ncRNAs (Khong et al., 2017), and suggests that translation repression is a determinant of targeting to SG rather than simply not being bound by ribosomes. The high level of consistency between many reports makes it likely that these two factors, translation efficiency and length, are the primary determinants of RNA enrichment within SGs (Fig. 4).
The targeting of specific RNAs to SGs could in principle also be due to specific interactions with RBPs that phase separate during stress. Studies of the SG transcriptome have not, however, uncovered any enrichment for particular RBP sequence motifs in SG RNAs (Khong et al., 2017; Matheny et al., 2021). The transcriptome of SG cores purified from yeast is very similar to RNA condensates formed in vitro using purified cellular RNAs (Van Treeck et al., 2018), suggesting that specific RBP-RNA interactions are not strictly necessary for transcript selection in SGs. Even loss of key hub proteins G3BP1/2 does not perturb the localization of their target transcripts during stresses that induce SGs in a G3BP1/2-independent manner (Matheny et al., 2021).
One possible exception is AU-rich elements (AREs), which are overrepresented in some SG RNAs (Namkoong et al., 2018), and could mediate interactions with SG RBPs such as TIA-1, TIAR, and human antigen R (HuR) (Waris et al., 2014). These AREs, and another RBP binding motif, pumilio response elements, are present in high numbers on several SG-enriched transcripts such as the NORAD long noncoding RNA (lncRNA). However, disruption of these sequence elements or depletion of the pumilio RBP does not entirely abrogate localization of target RNAs to SGs (Matheny et al., 2021; Namkoong et al., 2018).
Modification of RNAs with N6-methyladenosine (m6A) has also been proposed to target transcripts to, and even to explain the RNA length bias in, SGs (Fu and Zhuang, 2020; Ries et al., 2022). A recent study, however, found that knockout of the key methyltransferase responsible for m6A modification did not affect partitioning of m6A mRNAs to SGs, and that tethering of up to 25 molecules of the m6A binding protein YTH domain-containing family proteins (YTHDF) to an mRNA had only a modest impact on its SG localization (Khong et al., 2022). These observations combine to suggest that the sum of many protein-RNA or RNA-RNA interactions, more of which are present in long transcripts, rather than targeting by a specific RBP binding site or modification, is responsible for the enrichment of most transcripts within the SG (Matheny et al., 2021).
Sequence features of mRNAs that determine their translation status under stress do appear to influence their localization to SG. For instance, many transcripts encoding protein biosynthesis factors contain an mTOR-sensitive 5′ terminal oligopyrimidine (TOP) motif. When mTOR is inactivated during stress, TIA proteins bind to TOP sequences to assist in translational arrest of these mRNAs, possibly by targeting them to SGs (Damgaard and Lykke-Andersen, 2011). Conversely, some mRNAs are selectively translated during the canonical SG-forming ISR pathway, and are therefore likely to be largely excluded from SGs (Fig. 4). These include the stress response transcription factor activating transcription factor 4 (ATF4), which upon decreased translation initiation activity is relieved of translation inhibition via upstream open reading frame (uORF) elements (Vattem and Wek, 2004).
While endogenous ATF4 transcripts can be observed colocalizing with SGs (Adjibade et al., 2015; Mateju et al., 2020), fluorescence in situ hybridization analysis revealed this as only ∼20% of all ATF4 mRNAs (Adjibade et al., 2015). Heat shock protein mRNAs are largely absent from sedimented heat shock granules in plant cells and are instead present in polysome fractions (Nover et al., 1989). In addition, an analysis of nascent proteins captured using bio-orthogonal noncanonical amino acid tagging (BONCAT) revealed that the mRNAs encoding proteins that are generated during stress, including several heat shock protein mRNAs, are generally translated and depleted from SGs (Baron et al., 2019). These observations continue to reinforce the importance of translation repression as a determinant of mRNA partitioning to SGs.
The RNA content of canonical SGs is quite consistent across stress circumstances, cell types, and species that have so far been investigated. Similar categories of RNAs and many of the same individual transcripts are enriched or depleted in SGs between arsenite, heat shock, ER stress, and sorbitol-induced granules as measured by smFISH and transcriptomics (Khong et al., 2017; Matheny et al., 2021). Comparable mRNA enrichment profiles are found in SGs even when differing stress circumstances (heat shock and ER stress) change the cytoplasmic transcriptome, and high similarity is observed between SG transcriptomes from mouse and human cell lines (Namkoong et al., 2018). In Drosophila, an ∼60% overlap in SG transcriptomes was observed between the S2 cell line and differentiated neurons (van Leeuwen et al., 2021) showing that, while neuron-specific SG transcripts exist, much of the core SG RNA content is conserved.
Finally, using purified cytoplasmic RNA from yeast to assemble phase-separated granules in vitro also recapitulates many of the features of both the yeast and human SG transcriptome (Van Treeck et al., 2018). These observations suggest that the SG transcriptome across cell types and organisms shares certain features that underlie SG assembly and properties, including the increased length and decreased translation efficiency biases (Khong et al., 2017). Therefore, it is highly likely that different SG RNA contents between stress stimuli and cell types result primarily from differences in the bulk transcriptome of the cell before SG condensation.
Some of the protein content of SGs can be predicted from their mode of assembly and RNA content. For instance, the translationally stalled mRNAs necessarily contain initiation factors, small subunit ribosomal proteins, and cytoplasmic poly(A) binding protein (PABP) (Fig. 4) (Kedersha et al., 2002). As described earlier, characteristic RBPs such as G3BP1/2, Caprin (Kedersha et al., 2016), TIAR/TIA-1 (Kedersha et al., 1999), FMR1 (Mazroui et al., 2002), UBAP2L (Cirillo et al., 2020), and many others are present in intact SGs as determined by immunofluorescence staining, fluorescent fusion protein reporters, and proteomic assays. A wealth of proteomic data have revealed hundreds to thousands of other SG protein constituents with varying levels of confidence (collected in a database here: (Millar et al., 2023)). Unlike for its mRNAs, enrichment of one specific function is very apparent for the protein fraction of SGs: RNA interaction.
Predictably, proteins with RNA binding domains and a variety of annotated functions in RNA metabolism and regulation are highly enriched among the SG proteomes across all analyses (Jain et al., 2016; Marmor-Kollet et al., 2020; Youn et al., 2019). This includes at least ∼40 RBPs, which have been confirmed to be SG components in multiple cell types and stress circumstances as part of a high-throughput immunofluorescence microscopy screen (Markmiller et al., 2018). Thus, many of the protein components of SGs are involved in RNA metabolism.
Yet, approximately half of the proteins in various published SG proteomes are not annotated as RBPs (Jain et al., 2016). This remainder includes various translation system components, DNA and RNA helicases, chaperones, DEAD box proteins, and proteostasis factors, the potential functions of which are discussed further in the disassembly section below. Whether RBPs or not, SG proteins tend to have a higher fraction of IDRs that are longer than the proteome average (Markmiller et al., 2018; Youn et al., 2019). This agrees well with the observation that IDRs and low complexity domains are key participants in biomolecular phase separation (Posey et al., 2018). Similarly, prion-like domains, which can facilitate protein aggregation, are also enriched within SGs (Youn et al., 2019).
In contrast to the SG transcriptome, the SG proteome has been observed to vary between cell types, disease states, and stressors (Aulas et al., 2017; Markmiller et al., 2018; Marmor-Kollet et al., 2020). A literature meta-analysis comparing these and other data sets to identify a central set of high-confidence SG proteins has returned primarily the same characteristic RBPs including G3BP1/2, UBAP2L, and PABP (Youn et al., 2019). Many putative SG proteins and non-RBPs that vary between conditions and measurement types, especially those identified by proximity labeling proteomics, await further verification. Therefore, it remains to be established whether common determinants of protein recruitment to SGs beyond RNA binding exist, and what the functional consequences of their localization within SGs are.
Regulation of Translation by Stress Granules
SGs may cause global or transcript-specific translation suppression (Fig. 5). One mechanism by which SGs could trigger global translation suppression is by inducing eIF2α phosphorylation in a form of positive feedback. Formation of SGs by overexpression of G3BP1 alone induces P-eIF2α in human cells (Reineke et al., 2012). Intriguingly, P-eIF2α was not induced upon G3BP1 overexpression in mouse embryonic fibroblasts lacking PKR, suggesting that SGs can trigger PKR activation and thus eIF2α phosphorylation to globally repress translation initiation activity (Reineke et al., 2012). Cells lacking PKR with such G3BP1-driven SGs maintained global translation activity as assessed by the accumulation of puromycinylated proteins (Reineke et al., 2012). SGs may therefore play a role in mediating global translation suppression through activation of PKR.
FIG. 5.
Stress granules can regulate translation at global and transcript-specific levels. Formation of stress granules has been shown to activate PKR through an unknown mechanism, thereby causing global translation inhibition. Sequestration of a portion of cellular mRNAs and translation factors may also contribute to global translation repression. However, the proportion of these components relative to total cellular supply is likely minimal, because only ∼10% of total mRNA is highly enriched in stress granules and are expected to be present in a 1:1 stoichiometry of mRNA to most translation initiation factors. Induced stress granule formation using overexpression or forced oligomerization of key stress granule hubs (e.g., G3BP1) suggests that RNA-protein interactions may compete to some extent with the translation machinery. Finally, while translating mRNAs interact only transiently with stress granules, translationally repressed mRNAs can enter long-lived associations with stress granules that likely sequester them away from the translation machinery. PKR, protein kinase R.
A second possibility is that SGs can cause translational repression in an eIF2α-independent mechanism. In support of this idea, the formation of optogenetically induced SG-like condensates via light-dependent dimerization of G3BP1 called OptoGranules requires the release of mRNAs from polysomes (Zhang et al., 2019). This is evidenced by the observation that cycloheximide treatment, a translation elongation inhibitor that traps mRNAs within polysomes, blocked the formation of these light-induced G3BP1-driven granules (Zhang et al., 2019). In contrast to SGs driven by G3BP1 overexpression, eIF2α phosphorylation does not occur upon OptoGranule formation, nor is P-eIF2α required for OptoGranules to form, as pretreatment of cells with the eIF2B activator integrated stress response inhibitor (ISRIB) does not impair OptoGranule formation (Zhang et al., 2019). This study did not demonstrate that OptoGranules reduce global translation. However, the results do suggest that polysome collapse on the mRNAs contained in the OptoGranule is a consequence of light-induced, G3BP1-driven assembly, and imply that SG formation alone can drive release of mRNAs from polysomes to suppress translation.
A potential mechanism for SG-mediated translation suppression is that G3BP1 oligomerization and/or G3BP1-driven OptoGranules could outcompete the translation machinery for mRNAs by sequestration. However, because only 10%–13% of the transcriptome is estimated to be stably sequestered within SGs (Khong et al., 2017), while the majority of mRNAs would be predicted to interact transiently with them (Moon et al., 2019), OptoGranules would likely need to be fundamentally different from stress-induced granules for this competition to occur (and they do not appear to be). A second possibility is that G3BP1 plays a role in translation that is perturbed upon OptoGranule assembly.
In support of this possibility, a recent study demonstrated a role for G3BP1 and its binding partner USP10 in deubiquitination of 40S subunit proteins during ribosome-associated quality control to release them from mRNAs and rescue them to support translation (Meyer et al., 2020). G3BP1/2 knockout reduces polysomes, increases monosomes and 60S ribosomal subunits, and slows HEK 293 cell proliferation rates (Meyer et al., 2020). In addition, G3BP1 and G3BP2 cosediment with polysomes in these cells (Meyer et al., 2020). Therefore, it may be possible that G3BP1 sequestration into OptoGranules is itself the driver of translation suppression, rather than other aspects of condensate formation.
Another possible mechanism by which SGs may suppress translation activity during stress is by the sequestration of mRNAs and/or translation factors (Fig. 5). For this to be the case, either the majority of the mRNAs and translation factors would need to be sequestered within SGs, or key mRNAs or proteins necessary for global translation activity would need to be sequestered within them. The former is unlikely as only ∼10%–13% of RNAs are localized to SGs as measured by fluorescence in situ hybridization of polyadenylated transcripts and sequencing RNAs that copurify with stable SG “cores” (Khong et al., 2017).
Furthermore, while components of the PIC including 40S ribosomal subunits, eIF4F complex members, and other translation initiation factors as well as PABP colocalize with SGs (Kedersha et al., 2016; Kedersha et al., 2002; Kedersha et al., 1999; Moon et al., 2020), many of these would be expected to occur in a 1:1 stoichiometry with the SG mRNAs. Therefore, it appears unlikely that these translation components are sequestered in sufficient amounts to influence global translation outside the granule. It is, however, possible that more rRNA is sequestered to SGs than is currently indicated in the literature, as these are depleted from transcriptomic data sets of SG RNA composition. Notably, 60S ribosomal subunit components are generally not found in SGs, signifying that translation is unlikely to occur in SGs themselves (Kedersha et al., 2002; Moon et al., 2020).
Colocalization of translation factors with SGs in fixed cells also does not demonstrate whether these proteins are sequestered and therefore unavailable for use in active translation. For example, numerous FRAP experiments have demonstrated that there is some level of dynamic protein-SG interactions for virtually every fluorescent fusion protein reporter examined (reviewed in Buchan and Parker, 2009). As described above, cutting-edge live and fixed cell microscopy experiments revealed that each protein component examined likely exists in two phases—a static or dynamic phase—within SGs (Jain et al., 2016; Niewidok et al., 2018). Finally, while knockout of key SG nucleator proteins G3BP1/2 or UBAP2L impairs SG assembly, it does not prevent translation suppression upon stress (Cirillo et al., 2020; Kedersha et al., 2016). Therefore, the contribution of SGs to global translation suppression by sequestration of proteins or mRNAs appears to be minimal.
Single mRNP imaging studies revealed that mRNAs can spend long durations within SGs, and thus a portion of transcripts likely are sequestered away from the translation machinery (Fig. 5). Single mRNAs tagged with MS2 stem loops and visualized with HaloTag-MS2 coat protein interact in unstable and stable “docked” and “locked” states with SGs in human U-2 OS cells expressing GFP-G3BP1 (Moon et al., 2019). Evidence from prior studies suggests such “docking” and “locking” interactions are observed at the population level between numerous types of transcripts with SGs. For example, short (∼2 s) and long (∼1 min) recovery times were observed in FRAP experiments analyzing the mobility of GFP-tagged MS2 coat protein in cells expressing MS2 stem loops within β-Gal reporter mRNAs (Mollet et al., 2008).
In addition, FRAP experiments in arsenite-stressed cells microinjected with fluorescently labeled poly(U) probes to detect endogenous polyadenylated RNAs revealed a biphasic fluorescence recovery time of ∼40 and ∼275 s, again suggestive of distinct RNA pools “docking” and “locking” with SGs (Zhang et al., 2011). Therefore, accessibility of transcripts to the translation machinery outside the SG varies.
Interestingly, translating mRNA reporters transiently interact with SGs for at most several minutes, indicating that translation complexes are not likely to be stably sequestered within SGs and can only briefly “dock” with them (Mateju et al., 2020; Moon et al., 2020; Moon et al., 2019). Prior studies demonstrated that ribosome runoff is critical for SG formation through the use of the translation elongation inhibitor emetine, which traps ribosomes on transcripts, blocks SG assembly, and causes clearance of preformed SGs (Kedersha et al., 2000). Furthermore, ribosome stalling and trapping by cycloheximide slows mRNA accumulation within SGs, while puromycin increases mRNA localization to SGs during stress by causing ribosome release during elongation (Khong and Parker, 2018).
In addition, the length of the open reading frames of mRNAs correlates with the time before they become enriched in SGs after stress, supporting the notion that the ribosome runoff rate is a key determinant of mRNA recruitment to SGs (Khong and Parker, 2018). Another observation that supports the idea that ribosomes must run off and/or be removed from transcripts for their localization to SGs is that inhibition or genetic depletion of ribosome-associated quality control factors reduces the accumulation of specific mRNAs within SGs during arsenite or heat stress (Moon et al., 2020). Since translation repression and ribosome runoff appear to be a prerequisite for stable association of mRNAs within SGs, it follows that SGs likely keep many transcripts that have exited translation from re-entering the translating pool during stress.
Stress Granules Disassembly and Clearance
While the molecular mechanisms underlying assembly of SGs have become relatively well-understood, those determining granule disassembly or clearance have lagged behind. Multiple distinct pathways of RNA and protein remodeling and degradation have been identified as important for clearance of SGs (Hofmann et al., 2021), but no consensus mechanism has emerged. This process is particularly important to understand as impaired clearance of SGs is implicated in the formation of pathological aggregates in disease (Zhang et al., 2021). In laboratory model systems, SGs are readily cleared upon removal of stress, although the timing of disassembly varies during different stress contexts, just as it does for assembly. For instance, SGs resulting from 1-h arsenite stress in human U-2 OS cells start to break apart into smaller assemblies (potentially cores) 1 h after arsenite removal, with loss of microscopically visible foci by 1.5 h (Wheeler et al., 2016).
There appears to be some dependence on the duration of stress preceding disassembly, as another investigation using 30 min of arsenite treatment, also in U-2 OS cells, observed disassembly beginning 40 min after removal of stress (Marmor-Kollet et al., 2020), and longer periods of heat stress are associated with longer SG clearance times (Gwon et al., 2021). However, the contributions of cell type, type of stress, stress intensity, and stress duration to variation in SG disassembly rates have not been systematically determined.
One proposed mechanism for disassembly of SGs is active remodeling of biomolecules within the granule by various types of cellular machinery (Fig. 6). First among these are helicases, which could disrupt RNA-RNA and RNA-protein interactions to promote a dynamic equilibrium between SGs and their surroundings, shifts in which would drive disassembly (Jain et al., 2016). The crucial translation initiation factor eIF4A, for instance, has been identified as an inhibitor of SG assembly in vivo and in vitro, at least in part, through its helicase activity (Tauber et al., 2020). Inhibition of eIF4A's function in translation is itself sufficient to induce formation of noncanonical SGs (Mazroui et al., 2006). However, the ability of eIF4A to limit RNA recruitment to SGs may be independent of its role in translation, leading to the idea that it instead binds to mRNAs to interrupt their interactions with RBPs or other RNAs. Knockdown of the SG-localized minichromosome maintenance and RuvB-like DNA helicases also speeds SG disassembly in yeast, suggesting that diverse helicases may contribute to SG stability (Jain et al., 2016).
FIG. 6.
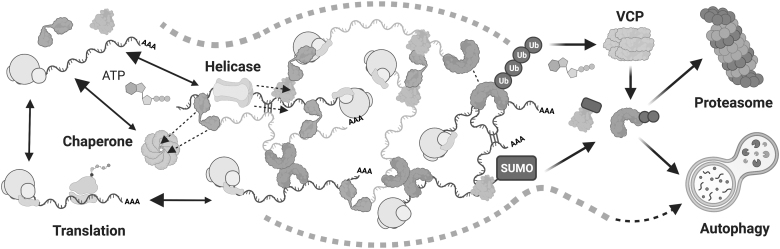
Disassembly and clearance of stress granules may occur by many distinct mechanisms. The action of ATPases such as helicases, chaperones, and VCP may dynamically disrupt interactions between, or extract components from, the stress granule. For mRNAs leaving the granule by these or other mechanisms, entering the translating pool would prohibit re-entry to the stress granule and promote disassembly. Ub modifications may be a mode of targeting stress granule proteins for removal, particularly during heat stress by VCP. The related SUMO modification may play a similar role in other conditions. Once removed, stress granule proteins may be degraded by the proteasome or autophagy pathways. It is also possible that autophagy clears intact stress granules or their cores. SUMO, small ubiquitin-related modifier; Ub, ubiquitin; VCP, valosin-containing protein.
A distinct type of remodeling protein, chaperones, has been implicated in maintaining SG dynamics and therefore influencing disassembly. Defects in specific yeast heat shock protein 40 (Hsp40)-class chaperones leads to impaired clearance of SGs (Walters et al., 2015), while mammalian heat shock protein 70 (Hsp70) family member heat shock protein A 1 A (HSPA1A) is required for disassembly of SGs during chronic proteasome inhibition (Ganassi et al., 2016). Linking these observations, depletion of cellular ATP using the glycolysis inhibitor 2-deoxyglucose and the electron transport chain inhibitor carbonyl cyanide m-chlorophenyl hydrazone (CCCP) reduces SG mobility, propensity to fuse in the cell, and results in longer recovery of GFP-G3BP1 within SGs from FRAP experiments (Jain et al., 2016). While this broad intervention likely has many off-target effects, it could indicate that the activity of ATP-requiring molecular machines increases dynamic movement of SG components, thus inhibiting SG assembly or promoting SG disassembly (Jain et al., 2016; Tauber et al., 2020).
Complementing the remodeling forces from helicases, chaperones, or other machinery is translation itself, which likely assists in SG disassembly. For instance, as mRNAs rapidly enter and exit SGs (Moon et al., 2019), the transcripts that form the basis of SGs may be outcompeted by translating polysomes and thus lead to SG disassembly (Fig. 6). This is supported by the observations that specific ribosome-trapping translation elongation inhibitors (e.g., cycloheximide or emetine) promote SG disassembly (Kedersha et al., 2000; Mollet et al., 2008). The translation-promoting small molecule ISRIB, which prevents inhibition of eIF2B by P-eIF2α, leads to rapid clearance of pre-existing SGs that are formed upon ISR activation (Sidrauski et al., 2015).
This observation suggests that translation alone can be sufficient to cause SG clearance. The exact order of events is unclear, however, as reporter mRNAs that localize to SGs appear to resume translation only after SGs are entirely disassembled (Moon et al., 2019). In long-term (10 hours) cold shock (10°C) of human cells, SGs are cleared within 10 minutes after return to normal growth temperature (37°C), while bulk translation activity as measured by polysomal ribosome accumulation increases 30 min after SG clearance (Hofmann et al., 2012). Overall, just as translation repression is required for SG assembly, the resumption of translation may also play a role in SG disassembly.
Another method proposed to drive SG clearance is the action of the AAA+ ATPase and ubiquitin segregase valosin-containing protein (VCP; also known as p97 or cell division control protein 48 [Cdc48]) (Fig. 6). Either chemical inhibition or genetic knockdown of VCP impairs clearance of mammalian SGs caused by heat shock or arsenite (Buchan et al., 2013; Gwon et al., 2021; Rodriguez-Ortiz et al., 2016), and yeast expressing a temperature-sensitive Cdc48 variant accumulates SGs (Buchan et al., 2013). Overexpression of disease-associated VCP mutants also impairs disassembly of SGs following heat or arsenite stress (Gwon et al., 2021; Rodriguez-Ortiz et al., 2016). Activation of VCP via phosphorylation by autophagic kinases promotes more rapid SG clearance following heat stress (Wang et al., 2019). Interestingly, VCP knockdown leads to smaller and more dispersed SGs forming during arsenite stress (Seguin et al., 2014). It is possible that freeing transcripts from stalled ribosomes during its role in a saRQC pathway underlies this role for VCP in SG formation (Moon et al., 2020). VCP is therefore important in several areas of the SG life cycle, likely through multiple distinct mechanisms.
An attractive proposed explanation for VCP's role in SG clearance is that ubiquitinated SG components are removed from the granule via its segregase activity. One specific piece of support for this mechanism is the finding that G3BP1 is ubiquitinated during heat shock, and this modification is required for its interaction with VCP (Gwon et al., 2021). Furthermore, ubiquitination of G3BP1 and VCP activity are both required for SG clearance in these circumstances. Notably, ubiquitination of G3BP1 was not observed during arsenite or sorbitol stress, nor is binding of G3BP1 to a specific VCP adaptor protein, Fas associated factor family member 2 (FAF2), which occurs during heat stress recovery (Gwon et al., 2021). These results suggest that the mechanism of VCP in SG disassembly varies between stresses.
Some of these processes may occur independent of ubiquitination, as one report found that ubiquitination and the related process of neddylation are not required for SG disassembly after arsenite stress (Markmiller et al., 2019), despite the requirement of VCP for SG clearance in this stress (Rodriguez-Ortiz et al., 2016). Inhibiting the related PTM of SUMOylation has also been found to impair SG disassembly after arsenite stress, and the small ubiquitin-related modifier (SUMO)-conjugating machinery appears to be recruited to these SGs during clearance (Marmor-Kollet et al., 2020) (Fig. 6). Overall, the roles of VCP, ubiquitination, and related modifications in both SG assembly and disassembly are likely complex, context dependent, and require further study.
While the exact role of these PTMs in SG disassembly is itself an unresolved area, the importance of the downstream pathways of autophagy and the proteasome in granule clearance is more so. In the case of SG assembly, contradictory reports exist as to whether inhibition of autophagy, lysosome fusion, or the proteasome impairs SG formation (Moon et al., 2020; Seguin et al., 2014) or triggers the ISR to induce SG formation (Mazroui et al., 2007; Suraweera et al., 2012). Early reports of VCP's role in SG disassembly found evidence that intact SGs were targeted for degradation by autophagy, as VCP-dependent accumulation of SG components was observed in vacuolar compartments in baker's yeast, and knockout of autophagy genes impaired SG clearance after heat shock in both yeast and mouse fibroblasts (Buchan et al., 2013).
It should be noted that, while frequently associated with proteasome function, the fate of VCP substrates after ubiquitin-dependent extraction varies from degradative to nondegradative pathways (Meyer and Weihl, 2014). So, while reports of VCP's role in SG disassembly suggest the involvement of the proteasome or autophagy in clearing granules, this may not be the case in every stress scenario.
The direct evidence for the role of these pathways in SG dissolution comes from reports that small-molecule inhibitors of lysosome function, autophagy, and/or the proteasome cause defects in SG disassembly (Rodriguez-Ortiz et al., 2016; Turakhiya et al., 2018). However, another study using the same small-molecule inhibitors found only a minor contribution of autophagy in SG disassembly, although this was after SG formation induced by long-term proteasome inhibition (Ganassi et al., 2016). It is also difficult to understand how targeting of SG components to the proteasome or autophagic processes would be required when the tightly linked process of ubiquitination is not (Markmiller et al., 2019).
The contribution of autophagy likely varies between cell type, stressor, stress duration, and severity of stress, as inhibitors of late-stage autophagy have been shown to impair SG disassembly only following prolonged, rather than brief, heat stress (Gwon et al., 2021), and variable stress stimuli are used throughout the above reports. Even if SGs themselves are not targeted for degradation through autophagy, this pathway may be required for signaling events such as phosphorylation of VCP, which could lead to SG disassembly by other proposed mechanisms (Wang et al., 2019). In summary, SG clearance appears to be facilitated by several PTMs, proteostasis pathways, translation, and protein and RNA remodeling processes that may act together or individually predominate in specific stress conditions.
Conclusions
SGs form both as a result of translation suppression during stress, and may themselves serve as key mediators of translation regulation. Technological advances in omics approaches, molecular genetics, and microscopy have yielded unprecedented insights into the mechanisms that lead to SG formation and their composition. We now know that assembly of SGs is driven by additive, multivalent interactions of diverse RBPs with nontranslating mRNAs (Khong et al., 2022; Sanders et al., 2020; Van Treeck et al., 2018). These forces seem to define much of the composition of SGs, as SGs are enriched in long, poorly translated mRNAs (Khong et al., 2017; van Leeuwen et al., 2021) and a similar subset of RBPs with many intrinsically disordered domains, even between distinct cell types and stressors (Markmiller et al., 2018; Millar et al., 2023).
In this study, we have synthesized multiple lines of evidence to show that the sequestration of mRNAs within SGs likely affects translation repression of individual transcripts (Kedersha et al., 2002; Moon et al., 2019). However, SG formation is not required for global translation suppression (Cirillo et al., 2020; Kedersha et al., 2016). It therefore remains unclear whether SGs sequester or outcompete translation components to make widespread impacts on protein synthesis as has been previously hypothesized (Ivanov et al., 2019; Protter and Parker, 2016).
Many outstanding questions remain regarding SG function. This is particularly true in their roles in regulating translation. One major unresolved question is, if SGs are not required for bulk translation shutoff during stress, then do they serve other roles such as buffering gene expression and/or dictating the specificity of translation during stress or recovery from it? Understanding how SGs interact with translation can then inform investigations into the roles of SGs in physiological and pathological contexts. For instance, what consequences do the potential posttranscriptional regulatory functions of SGs have within tissues, organs, and organisms? What are the ways in which SGs form or persist to lead to disease? Do these aberrant SGs then contribute to disease through dysregulation of translation, or other mechanisms? Overall, uncovering the mechanistic details of SG assembly, composition, properties, and disassembly holds great promise for deepened understanding of how cells adapt to challenges and will lend insight into key areas in human health.
Acknowledgments
The authors thank members of the Moon laboratory for critical reading of the article. Figures were created with BioRender.com
Abbreviations Used
- 4E-BP
eukaryotic translation initiation factor 4E-binding protein
- ALS
amyotrophic lateral sclerosis
- AMPK
AMP-activated protein kinase
- ARE
AU-rich element
- ATF4
activating transcription factor 4
- Cdc48
cell division control protein 48
- eIF2α
eukaryotic translation initiation factor 2α
- eIF2B
eukaryotic translation initiation factor 2B
- eIF4A
eukaryotic translation initiation factor 4A
- eIF4E
eukaryotic translation initiation factor 4E
- ER
endoplasmic reticulum
- FRAP
fluorescence recovery after photobleaching
- FTD
frontotemporal dementia
- GCN2
general control nonderepressible 2
- HRI
heme-regulated inhibitor kinase
- IDR
intrinsically disordered region
- ISR
integrated stress response
- ISRIB
integrated stress response inhibitor
- LCD
low complexity domain
- LLPS
liquid–liquid phase separation
- m6A
N6-methyladenosine
- mRNA
messenger RNA
- mTOR
mammalian target of rapamycin
- n/a
not available
- ncRNA
noncoding RNA
- PABP
poly(A) binding protein
- P-body
processing body
- P-eIF2α
phosphorylated eukaryotic translation initiation factor 2α
- PERK
PKR-related endoplasmic reticulum associated kinase
- PIC
preinitiation complex
- PKR
protein kinase R
- PTM
posttranslational modification
- RBP
RNA-binding protein
- RNP
ribonucleoprotein
- rRNA
ribosomal RNA
- saRQC
stress-activated ribosome quality control
- SG
stress granule
- SUMO
small ubiquitin-related modifier
- TDP-43
TAR DNA binding protein
- TIA-1
TIA1 cytotoxic granule associated RNA binding protein
- TIAR
TIA1 cytotoxic granule associated RNA binding protein like 1
- TOP
5′ terminal oligopyrimidine
- Ub
ubiquitin
- UBAP2L
ubiquitin associated protein 2 like
- USP10
ubiquitin specific peptidase 10
- VCP
valosin-containing protein
Authors' Contributions
M.B. drafted the majority of the article, prepared the figures, edited and revised the article, and approved the final version of the article. S.L.M. drafted parts of the article, edited and revised the article, and approved the final version.
Author Disclosure Statement
The authors declare no competing financial interests.
Funding Information
This work was supported by the LiveLikeLou Foundation Career Development Award, the Dystonia Medical Research Foundation, an NIH R35 award (R35GM146711) to S.L.M., and an NIH IRACDA fellowship (K12GM11172) to M.B.
References
- Adjibade P, Simoneau B, Ledoux N, et al. Treatment of cancer cells with lapatinib negatively regulates general translation and induces stress granules formation. PLoS One 2020;15(5):e0231894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjibade P, St-Sauveur VG, Quevillon Huberdeau M, et al. Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget 2015;6(41):43927–43943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Morgan F, et al. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol 1993;123(2):431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019;176(3):419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman T, Ionescu A, Ibraheem A, et al. Axonal TDP-43 condensates drive neuromuscular junction disruption through inhibition of local synthesis of nuclear encoded mitochondrial proteins. Nat Commun 2021;12(1):6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Castelao B, Tom Dieck S, Fusco CM, et al. The switch-like expression of heme-regulated kinase 1 mediates neuronal proteostasis following proteasome inhibition. Elife 2020;9:e52714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anees A, Salapa HE, Thibault PA, et al. Knock-down of heterogeneous nuclear ribonucleoprotein A1 results in neurite damage, altered stress granule biology, and cellular toxicity in differentiated neuronal cells. eNeuro 2021;8(6); doi: 10.1523/ENEURO.0350-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 2017;130(5):927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, et al. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017;18(5):285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Matheny T, Lin Y-C, et al. Quantitative proteomics identifies proteins that resist translational repression and become dysregulated in ALS-FUS. Hum Mol Genet 2019;28(13):2143–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer D, East SZ, Tseu B, et al. FTLD-ALS of TDP-43 type and SCA2 in a family with a full ataxin-2 polyglutamine expansion. Acta Neuropathol 2014;128(4):597–604. [DOI] [PubMed] [Google Scholar]
- Beauclair G, Streicher F, Chazal M, et al. Retinoic acid inducible gene I and protein kinase R, but not stress granules, mediate the proinflammatory response to yellow fever virus. J Virol 2020;94(22):e00403-20; doi: 10.1128/JVI.00403-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijer D, Kim HJ, Guo L, et al. Characterization of HNRNPA1 mutations defines diversity in pathogenic mechanisms and clinical presentation. JCI Insight 2021;6(14); doi: 10.1172/jci.insight.148363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Pileur F, Orsini C, et al. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem 2005;280(6):4144–4153. [DOI] [PubMed] [Google Scholar]
- Bounedjah O, Hamon L, Savarin P, et al. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: New role for compatible osmolytes. J Biol Chem 2012;287(4):2446–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M, Saal-Bauernschubert L, Lüningschrör P, et al. Loss of Tdp-43 disrupts the axonal transcriptome of motoneurons accompanied by impaired axonal translation and mitochondria function. Acta Neuropathol Commun 2020;8(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, et al. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013;153(7):1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 2009;36(6):932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 2005;280(45):37572–37584. [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 1995;312 (Pt 1):163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas A, Pileur F, Bonnal S, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell 2007;18(12):5048–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jin X, Liu B. The involvement of stress granules in aging and aging-associated diseases. Aging Cell 2020;19(4):e13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Joe Y, Park J, et al. Carbon monoxide induces the assembly of stress granule through the integrated stress response. Biochem Biophys Res Commun 2019;512(2):289–294. [DOI] [PubMed] [Google Scholar]
- Christ W, Tynell J, Klingström J. Puumala and Andes orthohantaviruses cause transient protein kinase R-dependent formation of stress granules. J Virol 2020;94(3):e01168–19; doi: 10.1128/JVI.01168-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L, Cieren A, Barbieri S, et al. UBAP2L forms distinct cores that act in nucleating stress granules upstream of G3BP1. Curr Biol 2020;30(4):698.e6–707.e6. [DOI] [PubMed] [Google Scholar]
- Corbet GA, Burke JM, Bublitz GR, et al. dsRNA-induced condensation of antiviral proteins modulates PKR activity. Proc Natl Acad Sci U S A 2022;119(33):e2204235119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. The integrated stress response: From mechanism to disease. Science 2020;368(6489):eaat5314; doi: 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 2011;25(19):2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low W-K, et al. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 2006;281(43):32870–32878. [DOI] [PubMed] [Google Scholar]
- De K, Jayabalan AK, Mariappan R, et al. Dihydrocapsaicin induces translational repression and stress granule through HRI-eIF2α phosphorylation axis. Biochem Biophys Res Commun 2022;588:125–132. [DOI] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BSS, et al. Correction of fragile X syndrome in mice. Neuron 2007;56(6):955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiermann N, Haneke K, Sun Z, et al. Dance with the devil: Stress granules and signaling in antiviral responses. Viruses 2020;12(9):984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim H-J, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010;466(7310):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Fujimura K, Sciaranghella D, et al. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem Biophys Res Commun 2012;423(4):763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AM, Green KM, Moon SL. A (dis)integrated stress response: Genetic diseases of eIF2α regulators. Wiley Interdiscip Rev RNA 2022;13(3):e1689. [DOI] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA 2009;15(10):1814–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Columbo D, Cotter C, et al. Bisphenol A promotes stress granule assembly and modulates the integrated stress response. Biol Open 2021;10(1):bio057539; doi: 10.1242/bio.057539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou J, Lin Y, et al. hnRNP A1 in RNA metabolism regulation and as a potential therapeutic target. Front Pharmacol 2022;13:986409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittschen M, Lastres-Becker I, Halbach MV, et al. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 2015;16(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M-J, Coudert L, Mellaoui S, et al. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol Cell Biol 2013;33(11):2285–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M-J, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int 2010;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Messing J, Yang P, et al. High-fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. J Cell Biol 2021;220(3):e202009079; doi: 10.1083/jcb.202009079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydryskova K, Masek T, Borcin K, et al. Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules. BMC Mol Biol 2016;17(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhuang X. m6A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol 2020;16(9):955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, Anderson P. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res 2012;40(16):8099–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, Mateju D, Bigi I, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell 2016;63(5):796–810. [DOI] [PubMed] [Google Scholar]
- Ghisolfi L, Dutt S, McConkey ME, et al. Stress granules contribute to α-globin homeostasis in differentiating erythroid cells. Biochem Biophys Res Commun 2012;420(4):768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 2004;15(12):5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frýdlová I, et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci 2009;122(Pt 12):2078–2088. [DOI] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 2020;181(2):346.e17–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Long JC, Cáceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol 2006;26(15):5744–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Luo F, Li Y, et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 2019;10(1):2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Qin Y, Jiao X, et al. FMR1 premutation is an uncommon explanation for premature ovarian failure in han chinese. PLoS One 2014;9(7):e103316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwon Y, Maxwell BA, Kolaitis R-M, et al. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 2021;372(6549):eabf6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackman P, Sarparanta J, Lehtinen S, et al. Welander distal myopathy is caused by a mutation in the RNA-binding protein TIA1. Ann Neurol 2013;73(4):500–509. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Hazlett HC, et al. Fragile X syndrome. Nat Rev Dis Primers 2017;3:17065. [DOI] [PubMed] [Google Scholar]
- Hegner RW. The germ cell determinants in the eggs of chrysomelid beetles. Science 1911;33(837):71–72. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Pottier C, Nicholson AM, et al. Clinical and neuropathological features of ALS/FTD with TIA1 mutations. Acta Neuropathol Commun 2017;5(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, et al. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 2012;23(19):3786–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Kedersha N, Anderson P, et al. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res 2021;1868(1):118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Dormann D. Friend or foe-post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem 2019;294(18):7137–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Chen Y, Dai H, et al. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ 2020;27(1):227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 2017;68(1):144.e5–157.e5. [DOI] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 2019;11(5):a032813; doi: 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PA, Chudinova EM, Nadezhdina ES. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp Cell Res 2003;290(2):227–233. [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 2016;164(3):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalihal AP, Pitchiaya S, Xiao L, et al. Multivalent proteins rapidly and reversibly phase-separate upon osmotic cell volume change. Mol Cell 2020;79(6):978.e5–990.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik-Bode M, Studencka M, Smolka C, et al. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci 2013;126(Pt 22):5166–5177. [DOI] [PubMed] [Google Scholar]
- Jiang H-Y, Wek RC. Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem 2005;280(14):14189–14202. [DOI] [PubMed] [Google Scholar]
- Jia X, Zhang S, Tan S, et al. De novo variants in genes regulating stress granule assembly associate with neurodevelopmental disorders. Sci Adv 2022;8(33):eabo7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongjitwimol J, Baldock RA, Morley SJ, et al. Sumoylation of eIF4A2 affects stress granule formation. J Cell Sci 2016;129(12):2407–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehler C, Isensee J, Nonhoff U, et al. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS One 2012;7(11):e50134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamelgarn M, Chen J, Kuang L, et al. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc Natl Acad Sci U S A 2018;115(51):E11904–E11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol 2007;431:61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, et al. Evidence that ternary complex (eIF2-GTP-tRNAi Met)–deficient preinitiation complexes are core constituents of mammalian stress granules. MBoC 2002;13(1):195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 2000;151(6):1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, et al. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 2016;212(7):845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005;169(6):871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, et al. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 1999;147(7):1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Huynh TN, et al. Limited effects of m6A modification on mRNA partitioning into stress granules. Nat Commun 2022;13(1):3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 2017;68(4):808.e5–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Parker R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J Cell Biol 2018;217(12):4124–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron 2006;51(6):685–690. [DOI] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 2015;4:e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-T, You G-T, Jian Y-J, et al. AMPK-mediated formation of stress granules is required for dietary restriction-induced longevity in Caenorhabditis elegans. Aging Cell 2020;19(6):e13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, et al. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet 2001;10(4):329–338. [DOI] [PubMed] [Google Scholar]
- Leung AKL, Vyas S, Rood JE, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 2011;42(4):489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, et al. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014;157(3):624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zeng L, Zhao H, et al. ATXN2-mediated translation of TNFR1 promotes esophageal squamous cell carcinoma via m6A-dependent manner. Mol Ther 2022;30(3):1089–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res 2001;29(11):2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist ME, Mainou BA, Dermody TS, et al. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 2011;413(1):103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Erauskin J, Tadokoro T, Baughn MW, et al. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron 2018;100(4):816–830.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo E-C, Nathanson JL, Tan FE, et al. Large-Scale tethered function assays identify factors that regulate mRNA stability and translation. Nat Struct Mol Biol 2020;27(10):989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivannan P, Siddiqui MA, Malathi K. RNase L amplifies interferon signaling by inducing protein kinase r-mediated antiviral stress granules. J Virol 2020;94(13):e00205-20; doi: 10.1128/JVI.00205-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Fulzele A, Higgins R, et al. Active protein neddylation or ubiquitylation is dispensable for stress granule dynamics. Cell Rep 2019;27(5):1356.e3–1363.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 2018;172(3):590.e13–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor-Kollet H, Siany A, Kedersha N, et al. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol Cell 2020;80(5):876.e6–891.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Eichenberger B, Voigt F, et al. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 2020;183(7):1801.e13–1812.e13; doi: 10.1016/j.cell.2020.11.010 [DOI] [PubMed] [Google Scholar]
- Matheny T, Rao BS, Parker R. Transcriptome-wide comparison of stress granules and P-bodies reveals that translation plays a major role in RNA partitioning. Mol Cell Biol 2019;39(24):e00313-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny T, Van Treeck B, Huynh TN, et al. RNA partitioning into stress granules is based on the summation of multiple interactions. RNA 2021;27(2):174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, et al. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell 2007;18(7):2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot M-E, Tremblay S, et al. Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet 2002;11(24):3007–3017. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau M-E, et al. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. MBoC 2006;17(10):4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 2017;17(10):647–660. [DOI] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, et al. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 2005;280(17):16925–16933. [DOI] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Morozov P, et al. The G3BP1-family-USP10 deubiquitinase complex rescues ubiquitinated 40S subunits of ribosomes stalled in translation from lysosomal degradation. Mol Cell 2020;77(6):1193.e5–1205.e5. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: Connecting cellular function to disease pathogenesis. J Cell Sci 2014;127(Pt 18):3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SR, Huang JQ, Schreiber KJ, et al. A new phase of networking: The molecular composition and regulatory dynamics of mammalian stress granules. Chem Rev 2023; doi: 10.1021/acs.chemrev.2c00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, et al. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell 2009;20(11):2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, et al. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 2008;19(10):4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol 2019;21(2):162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Stasevich TJ, et al. Coupling of translation quality control and mRNA targeting to stress granules. J Cell Biol 2020;219(8):e202004120; doi: 10.1083/jcb.202004120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdina ES, Lomakin AJ, Shpilman AA, et al. Microtubules govern stress granule mobility and dynamics. Biochim Biophys Acta 2010;1803(3):361–371. [DOI] [PubMed] [Google Scholar]
- Nagano S, Jinno J, Abdelhamid RF, et al. TDP-43 transports ribosomal protein mRNA to regulate axonal local translation in neuronal axons. Acta Neuropathol 2020;140(5):695–713. [DOI] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, et al. Systematic characterization of stress-induced RNA granulation. Mol Cell 2018;70(1):175.e8–187.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewidok B, Igaev M, Pereira da Graca A, et al. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J Cell Biol 2018;217(4):1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Hasegawa M. TDP-43 prions. Cold Spring Harb Perspect Med 2018;8(3); doi: 10.1101/cshperspect.a024463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Cytoplasmic Heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 1989;9(3):1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, et al. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 2008;10(10):1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonski KM, Samuel CE. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol 2013;87(2):756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Kedersha N, Schulte T, et al. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol 2019;218(7):2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Schulte T, Thaa B, et al. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 2015;11(2):e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavinato L, Delle Vedove A, Carli D, et al. CAPRIN1 haploinsufficiency causes a neurodevelopmental disorder with language impairment, ADHD and ASD. Brain 2022; doi: 10.1093/brain/awac278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Yang W, Hinnebusch AG. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol 1997;17(3):1298–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey AE, Holehouse AS, Pappu RV. Chapter One - phase Separation of Intrinsically Disordered Proteins. In: Intrinsically Disordered Proteins. Methods in Enzymology. (Rhoades E. ed). Academic Press: USA; 2018; pp. 1–30. [DOI] [PubMed] [Google Scholar]
- Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol 2016;26(9):668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Cheema SA, Dubrulle J, et al. Chronic starvation induces noncanonical pro-death stress granules. J Cell Sci 2018;131(19):jcs220244; doi: 10.1242/jcs.220244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Dougherty JD, Pierre P, et al. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol Biol Cell 2012;23(18):3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 2015;89(5):2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 2017;168(6):1028.e19–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Pickering BF, Poh HX, et al. m6A governs length-dependent enrichment of mRNAs in stress granules. bioRxiv 2022;2022.02.18.480977; doi: 10.1101/2022.02.18.480977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ortiz CJ, Flores JC, Valenzuela JA, et al. The myoblast C2C12 transfected with mutant valosin-containing protein exhibits delayed stress granule resolution on oxidative stress. Am J Pathol 2016;186(6):1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Durie D, Li H, et al. hnRNPA1 couples nuclear export and translation of specific mRNAs downstream of FGF-2/S6K2 signalling. Nucleic Acids Res 2014;42(20):12483–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A, Dazert E, Metz P, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 2012;12(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem Sci 2020;45(11):961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 2020;181(2):306.e28–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa JA. Effects of stress and aging on ribonucleoprotein assembly and function in the germ line. Wiley Interdiscip Rev RNA 2014;5(2):231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwed-Gross A, Hamiel H, Faber GP, et al. Glucocorticoids enhance chemotherapy-driven stress granule assembly and impair granule dynamics, leading to cell death. J Cell Sci 2022;135(14):jcs259629; doi: 10.1242/jcs.259629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorsone KA, Panniers R, Rowlands AG, et al. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J Biol Chem 1987;262(30):14538–14543. [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, et al. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 2014;21(12):1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos AP, Mellor LE, Pang YF, et al. The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ 2018;25(10):1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003;300(5620):805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Tokunaga M. RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J Biol Chem 2010;285(31):24260–24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. Liquidizing FUS via prion-like domain phosphorylation. EMBO J 2017;36(20):2925–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2013;2:e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, et al. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife 2015;4:e05033; doi: 10.7554/eLife.05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S, Xu Y, Wang B, et al. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 2007;27(6):2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M-S, Grabocka E.. Stress Granules in Cancer. Springer Berlin Heidelberg: Berlin, Heidelberg, 2020; pp. 1–28. [Google Scholar]
- Sopova JV, Koshel EI, Belashova TA, et al. RNA-Binding protein FXR1 is presented in rat brain in amyloid form. Sci Rep 2019;9(1):18983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008;319(5870):1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SD, Welt C, Sherman S. FMR1 and the continuum of primary ovarian insufficiency. Semin Reprod Med 2011;29(4):299–307. [DOI] [PubMed] [Google Scholar]
- Suraweera A, Münch C, Hanssum A, et al. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 2012;48(2):242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaflarski W, Fay MM, Kedersha N, et al. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 2016;7(21):30307–30322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, et al. Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 2020;180(3):411.e16–426.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JC, Miller-Vedam LE, Anand AA, et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science 2018;359(6383):eaaq0939; doi: 10.1126/science.aaq0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W-C, Gayatri S, Reineke LC, et al. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem 2016;291(43):22671–22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhiya A, Meyer SR, Marincola G, et al. ZFAND1 recruits p97 and the 26S proteasome to promote the clearance of arsenite-induced stress granules. Mol Cell 2018;70(5):906.e7–919.e7. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, VanInsberghe M, Battich N, et al. Identification of the stress granule transcriptome via RNA-editing in single cells and in vivo. bioRxiv 2021;2021.06.21.449212; doi: 10.1101/2021.06.21.449212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 2018;174(4):791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A 2018;115(11):2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 2004;101(31):11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, et al. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 2015;21(9):1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Maxwell BA, Joo JH, et al. ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol Cell 2019;74(4):742.e8–757.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian X, Kim HB, et al. Intracellular energy controls dynamics of stress-induced ribonucleoprotein granules. Nat Commun 2022;13(1):5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris S, Wilce MCJ, Wilce JA. RNA recognition and stress granule formation by TIA proteins. Int J Mol Sci 2014;15(12):23377–23388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, et al. Distinct stages in stress granule assembly and disassembly. Elife 2016;5:e18413; doi: 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol 2012;20(4):175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD. RNA on the move: The mRNA localization pathway. J Cell Biol 1993;123(2):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci 2019;20(11):649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita C, Tomiyama H, Funayama M, et al. Evaluation of polyglutamine repeats in autosomal dominant parkinson's disease. Neurobiol Aging 2014;35(7):1779.e17–21. [DOI] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis R-M, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 2020;181(2):325.e28–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Zhang H, Loiselle D, et al. The RNA-binding protein fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol 2013;203(5):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerlikaya A, Kimball SR, Stanley BA. Phosphorylation of eIF2alpha in response to 26S proteasome inhibition is mediated by the haem-regulated inhibitor (HRI) kinase. Biochem J 2008;412(3):579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Khaperskyy DA. UV damage induces G3BP1-dependent stress granule formation that is not driven by mTOR inhibition-mediated translation arrest. J Cell Sci 2020;133(20):jcs248310; doi: 10.1242/jcs.248310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-Y, Dyakov BJA, Zhang J, et al. Properties of stress granule and P-body proteomes. Mol Cell 2019;76(2):286–294. [DOI] [PubMed] [Google Scholar]
- Zeng W-J, Lu C, Shi Y, et al. Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. Biochim Biophys Acta Mol Cell Res 2020;1867(10):118795. [DOI] [PubMed] [Google Scholar]
- Zhang J, Okabe K, Tani T, et al. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J Cell Sci 2011;124(Pt 23):4087–4095. [DOI] [PubMed] [Google Scholar]
- Zhang P, Fan B, Yang P, et al. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 2019;8:e39578; doi: 10.7554/eLife.39578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gu J, Sun Q. Aberrant stress granule dynamics and aggrephagy in ALS pathogenesis. Cells 2021;10(9):2247; doi: 10.3390/cells10092247 [DOI] [PMC free article] [PubMed] [Google Scholar]