Abstract
The long control region (LCR) of human papillomavirus type 16 (HPV-16) has a size of 850 bp (about 12% of the viral genome) and regulates transcription and replication of the viral DNA. The 5′ segment of the LCR contains transcription termination signals and a nuclear matrix attachment region, the central segment contains an epithelial cell-specific enhancer, and the 3′ segment contains the replication origin and the E6 promoter. Here we report observations on the chromatin organization of this part of the HPV-16 genome. Treatment of the nuclei of CaSki cells, a cell line with 500 intrachromosomal copies of HPV-16, with methidiumpropyl-EDTA-Fe(II) reveals nucleosomes in specific positions on the LCR and the E6 and E7 genes. One of these nucleosomes, which we termed Ne, overlaps with the center of the viral enhancer, while a second nucleosome, Np16, overlaps with the replication origin and the E6 promoter. The two nucleosomes become positioned on exactly the same segments after in vitro assembly of chromatin on the cloned HPV-16 LCR. Primer extension mapping of DNase I-cleaved chromatin revealed Np16 to be positioned centrally over E6 promoter elements, extending into the replication origin. Ne covers the center of the enhancer but leaves an AP-1 site, one of the strongest cis-responsive elements of the enhancer, unprotected. Np16, or a combination of Np16 and Ne, represses the activity of the E6 promoter during in vitro transcription of HPV-16 chromatin. Repression is relieved by addition of Sp1 and AP-1 transcription factors. Sp1 alters the structure of Np16 in vitro, while no changes can be observed during the binding of AP-1. HPV-18, which has a similar arrangement of cis-responsive elements despite its evolutionary divergence from HPV-16, shows specific assembly in vitro of a nucleosome, Np18, over the E1 binding site and E6 promoter elements but positioned about 90 bp 5′ of the position of Np16 on the homologous HPV-16 sequences. The chromatin organization of the HPV-16 and HPV-18 genomes suggests important regulatory roles of nucleosomes during the viral life cycle.
Human papillomavirus type 16 (HPV-16) is the clinically most prevalent virus among the approximately 30 HPV types that cause genital cancer or genital warts (32, 85), and due to this significant medical importance, the viral life cycle and gene expression have been extensively researched. Despite efforts to study the contributions of known cis-responsive elements (for a review, see reference 49), some of the most interesting regulatory properties of HPV-16 are not yet understood. These poorly elucidated phenomena include transcriptional changes during the differentiation of stratified epithelia, the regulation of transcription during latent infection, genomic copy number control, and partition. Similar phenomena are regulated in other viruses and some cellular genes by the organization of DNA into chromatin and by interactions of the DNA with complex nuclear structures, such as the nuclear matrix (see reference 70 for references). We have begun to study the intranuclear organization of HPV-16 genomes (70) in the hope of explaining some of these aspects of the HPV-16 life cycle.
Our research focuses on the long control region (LCR) of papillomaviruses, which contains most of the cis-responsive elements regulating papillomavirus biology. The LCR of HPV-16 has a length of 850 bp and is organized similarly to the LCRs of all other genital HPVs (49). The 3′ portion of the LCR contains the replication origin (8) and the E6 promoter, which has four cis-responsive elements, namely, one binding site for the transcription factor Sp1, two binding sites for the viral factor E2, and a TATA box (16, 19, 23, 24, 71). Two additional E2 binding sites are located approximately 140 and 550 bp 5′ of the E6 promoter and flank the central section of the HPV-16 LCR, which contains the epithelial cell-specific enhancer (17, 18, 26). The enhancer, the E6 promoter, and the replication origin are collectively flanked by two nuclear matrix attachment regions (MARs), one located in the 5′ region of the LCR and the other one located in the E6 gene itself (70). Here, we report on the nucleosomal organization of the LCR between and overlapping with these two MARs.
The chromosomes of eukaryotes and of double-stranded DNA viruses of eukaryotes generally do not exist in vivo as naked DNA but rather exist in form of chromatin, i.e., associated with nucleosomes. Nucleosomes consist of nucleosomal cores, which are 146-bp lengths of DNA wound around octameric histone complexes. The internucleosomal stretches of DNA are called linker DNA and have lengths of 40 to 55 bp. The genomes of double-stranded DNA viruses, such as simian virus 40 (SV40) and adenoviruses, are organized in the form of nucleosomes both in the viral capsids and in the nuclei of infected cells (20, 37, 45, 77). The nucleosomal organization of cellular and viral DNAs has traditionally been seen as a requirement to store the bulky genomes. More recently, it has become clear that nucleosomes can be specifically positioned on regulatory elements, thereby constituting an important mechanism of gene regulation (for a review and references, see reference 81).
Most often this nucleosomal organization of eukaryotic genes interferes negatively with gene expression (for a review, see reference 28), as efficient initiation of transcription requires binding sites for transcription factors and RNA polymerase II to be free of nucleosomes (36), although elongation through nucleosomes is possible (67). When such sites are inaccessible, the structure or position of preexisting nucleosomes may be altered by specific enzymatic chromatin-remodeling activities, such as the SWI-SNF complex (reference 79 and references therein), nucleosomal rearrangement factor (75), chromatin accessibility complex (76), ACF (33), or the histone acetyltransferase activities associated with some transcription factors (for a review, see reference 82).
Such regulatory mechanisms have been found to play a role in the life cycle of viruses. This is particularly well documented for the DNA genome of the retrovirus mouse mammary tumor virus (MMTV). The long terminal repeat (LTR) of MMTV is organized in the form of six nucleosomes. Mutual influences between one of these nucleosomes and certain transcription factors determine promoter access and transcriptional activation (2, 72). In another retrovirus, human immunodeficiency virus type 1 (HIV-1), a nucleosome blocking the transcriptional start site has to be remodeled for activation of the virus promoter (63). In the case of SV40, it has been reported that most of the regulatory region is normally free of nucleosomes (4, 37). A nucleosome can form, however, adjacent to this region and overlapping with a late promoter element (55). In addition, nucleosomes can be assembled in vitro overlapping with the Sp1 binding sites of the early promoter, with flanking nucleosomes possibly being positioned due to neighbor effects (34). Antagonistic interactions between nucleosomes and Sp1 may reduce promoter access of the transcriptional apparatus (36, 41). Access of the replication machinery to the SV40 origin of replication is also blocked by nucleosomes, and this can be overcome by nucleosome remodeling in the presence of T antigen (1, 57).
In the case of papillomaviruses, chromatin organization has been established for bovine papillomavirus type 1 (BPV-1) in situ (59, 78), but it is still unclear whether nucleosomes persist on BPV-1 DNA packaged in viral particles (38). Chromatin packaging interferes with BPV-1 replication (42), although the exact nucleosomal organization has not been studied. Here we report on the chromatin organization of the genomes of HPV-16 and HPV-18 and on the functional consequences of precisely positioned nucleosomes whose positions overlap with transcription and replication elements of the viral LCRs.
MATERIALS AND METHODS
Bacterial strains, plasmid constructs, biochemicals, and cell lines.
Escherichia coli JM109 (recA) was used for all DNA cloning experiments as well as for propagation and expression of the histidine tag vectors pQE31-E2 (71) and pDS-6xHIS:YY1 (64). The transcription factors E2 and YY1 were prepared in our lab as described below, while AP-1, Sp1, and TATA binding protein (TBP) were commercially obtained from Promega, Madison, Wis. A segment of the HPV-16 genome that stretches from the position 7150 to 100 (47) and includes the complete LCR was isolated by PCR and cloned into the SrfI site of pCR-ScriptT SK(+) (Stratagene, San Diego, Calif.). To measure the effect on luciferase gene expression of this HPV-16 segment, the pCR-ScriptT SK(+) construct was cleaved with SacI and KpnI and the excised HPV-16 DNA was reinserted into the SacI- and KpnI-cleaved vector pGL3-basic (Promega) to yield the construct pHPV-16-Luc. For the analysis of HPV-16 chromatin structures in vivo, we used the cervical cancer-derived cell line CaSki, which contains about 500 endogenous HPV-16 genomes (3), under standard conditions in Dulbecco modified Eagle medium with 10% fetal calf serum.
Expression of recombinant proteins in E. coli.
For the expression of histidine-tagged HPV-16 E2 and cellular YY1 proteins, 400 ml of Luria-Bertani medium was inoculated with 10 ml of an overnight culture of JM109 containing pQE31-E2 or pDS-6xHIS:YY1, respectively, and incubated at room temperature until the bacterial suspension reached an optical density (600 nm) of 0.6. The culture was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), grown for another 4 h, pelleted, and resuspended in 25 ml of lysis buffer (20 mM HEPES [pH 7.2], 100 mM KCl, 5 mM MgCl2, 0.5% Triton X-100, 0.1% Nonidet P-40, 100 U of DNase I per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1% aprotinin, 5 mg of leupeptin per ml, and 1 mg of lysozyme per ml). After cell lysis by sonication, 500 mM NaCl and 20 mM imidazole (Sigma, St. Louis, Mo.) were added. The lysate was centrifuged and passed over an Ni2+-nitrilotriacetic acid column (Qiagen, Düsseldorf, Germany). The column was equilibrated at 4°C with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 18 mM Na2HPO4, 1.47 mM KH2PO4) which contained 20 mM imidazole and 1 mM dithiothreitol (DTT). To remove unspecific binding proteins, the column was washed with PBS containing increasing amounts of imidazole (30 to 80 mM). The final elution of the fusion protein was performed with PBS containing 150, 180, and 200 mM imidazole, respectively, in a volume of 5 ml or less. To remove residual imidazole, the fractions containing the fusion protein (examined by sodium dodecyl sulfate [SDS]-polyacrylamide gel electrophoresis) were dialyzed against transcription buffer (60 mM KCl, 6.25 mM MgCl2, 10% glycerol, 20 mM HEPES [pH 7.8], 0.2 mM PMSF, and 3 mM DTT).
Preparation of transcription extracts.
For in vitro transcription experiments, nuclear extracts were prepared from HeLa cells as described previously (21). Briefly, nuclei were isolated in a buffer containing 20 mM HEPES (pH 7.9), 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 0.5 mM DTT. Soluble proteins were precipitated with ammonium sulfate, redissolved in and dialyzed against transcription buffer, and stored in aliquots in liquid nitrogen.
In vitro transcription reactions.
Transcription from pHPV-16-Luc in the form of either naked circular plasmid DNA or in vitro-assembled chromatin was assayed by primer extension as described previously (11). Briefly, 250 ng of DNA was incubated with HeLa cell nuclear extract in the presence of 1 mM ribonucleoside triphosphates in transcription buffer in a volume of 100 μl. The reaction was stopped after 1 h at 30°C by proteinase K treatment followed by incubation with RNase-free DNase I (Boehringer) to destroy the template DNA. The reaction product was monitored by primer extension with an oligonucleotide complementary to the luciferase gene (5′-AGTGATGTCCACCTCGATATGTGCATCTGTAAAAGC-3′).
Preparation of Drosophila S190 extract for assembly of chromatin.
The preparation of the S190 extract was carried out as described previously (7, 35). Drosophila strain Canton-S wild-type embryos were harvested during five successive 2-h intervals to ensure that a high fraction of embryos would be in the preblastodermal stage. To arrest further development, the embryos were transferred into 0.7% NaCl–0.05% Triton X-100 and kept on ice. After extensive washing with water, the embryos were dechorionated by treatment with a 1:1 mixture of bleach and water for 90 s at room temperature. After being rinsed with water, the embryos were allowed to settle in 0.7% NaCl–0.05% Triton X-100 on ice for 5 min. This step was repeated twice. Finally, the embryos were resuspended in 0.7% NaCl, followed by two further washing steps in the same solution. Two additional washing steps were performed in extract buffer (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM β-glycerophosphate, 1 mM DTT, and 0.2 mM PMSF). The embryos in extract buffer were transferred into a 15-ml Dounce homogenizer and allowed to settle, and the supernatant was aspirated. The embryo pellet was homogenized with 10 strokes of pestle B and 10 strokes of pestle A. The homogenate was centrifuged in a 15-ml Falcon tube at 5,000 rpm in a Sorvall RT600RT B rotor. After the centrifugation, the cytoplasmic fraction was collected and the MgCl2 concentration was adjusted from 1.5 to 6.5 mM. The supernatant was clarified by ultracentrifugation in Beckmann 11- by 34-mm tubes in a TLS-55-rotor at 42,000 rpm for 90 min. The yellow cytoplasmic phase was harvested by puncturing the tube between the solid pellet and the upper lipid layer and frozen in aliquots in liquid nitrogen.
Chromatin assembly of HPV-16 DNA in vitro.
For the assembly of pHPV-16-Luc into chromatin, we mixed 150 μl of transcription buffer containing 10 mM β-glycerophosphate and 1 mM EDTA with 500 ng of circular plasmid DNA, 15 μl of Drosophila S190 extract (corresponding to a protein content of 20 μg/ml), and an ATP-regenerating system consisting of 40 mM creatine phosphate, 3 mM ATP, 1.5 ng of creatine kinase, and 3 mM MgCl2. After assembly of chromatin for 5 h at 30°C, each sample was split for structural and functional analyses. In contrast to findings reported by others (63), the S190 extract was sufficient for nucleosomal organization of the HPV-16 templates (as proven by treatment with micrococcal nuclease), and it was not necessary to supplement the reaction mixture with exogenous core or linker histones.
Preparation of nuclei.
For the analysis of the chromatin organization of the HPV-16 genome in vivo, cultured CaSki cells were washed twice with ice-cold PBS, harvested with a rubber policeman, centrifuged in a Sorvall RT600RT B rotor at 3,000 rpm for 5 min, and resuspended in a buffer containing 15 mM Tris-HCl (pH 7.4), 60 mM KCl, 15 mM NaCl, 3 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, and 0.25 M sucrose. After addition of Nonidet P-40 (final concentration, 0.2%), the cells were homogenized in a Dounce homogenizer with 10 strokes of a type S pestle. The nuclei were purified from cellular debris by centrifugation as described above. The nuclear pellet was resuspended in the same buffer for the analysis of the chromatin structure.
MPE treatment of CaSki nuclei.
Methidiumpropyl-EDTA-Fe(II) (MPE) cleavage was by a published protocol (13). Nuclei from CaSki cells were resuspended in 800 μl of reaction buffer (15 mM Tris-HCl [pH 7.4], 60 mM KCl, 15 mM NaCl, 1 mM EDTA, 0.25 M sucrose) supplemented with 2 mM DTT and 2 mM H2O2. The reaction was initiated with 50 μl of an MPE mixture that was prepared by mixing equal volumes of 1 mM MPE and 1 mM Fe(NH4)2(SO4)2 · 6H2O with subsequent 1:10 dilution in reaction buffer. Aliquots of the reaction were taken when the reaction was stopped after 5, 10, 15, 20, 40, and 60 min with 100 μl of 0.5 mM bathophenantrolinedisulfonate (Sigma), followed by incubation until the color of the samples changed. After overnight treatment with proteinase K, the samples were extracted twice with phenol, precipitated and washed with ethanol, and treated with 100 μg of RNase A per ml. Next, the DNA was reextracted with phenol-chloroform-isoamylalcohol (25:24:1) and precipitated with ethanol. As a control, purified genomic DNA was treated in a similar way.
Micrococcal nuclease treatment of in vitro-assembled chromatin.
pHPV-16-Luc DNA (250 ng) was assembled into chromatin with Drosophila S190 extract, supplemented with 3 mM CaCl2, and digested with 1 to 5 U of micrococcal nuclease (Sigma) at room temperature. The reactions were stopped after 1.5 min by addition of 200 μl of STOP buffer (75 mM EDTA, 900 mM ammonium acetate), and the products were digested with RNase A (100 μg/ml) for 20 min at 37°C and treated with 500 μg of proteinase K for 3 h.
Micrococcal nuclease digestion of CaSki nuclei.
CaSki nuclei were isolated and resuspended in 400 μl of reaction buffer containing 15 mM Tris-HCl (pH 7.4), 60 mM KCl, 15 mM NaCl, 1 mM EDTA, 0.25 M sucrose, and 2 mM DTT, supplemented with 3 mM CaCl2. The reaction was started with 1 to 5 U of micrococcal nuclease (Boehringer), allowed to proceed for 5 min, and stopped with 400 μl of STOP buffer. The samples were treated with 100 μg of RNase A per ml for 20 min and digested with 500 μg of proteinase K overnight. The DNA was purified by standard procedures and finally subjected to indirect end-labelling analysis as described below with the restriction enzyme NcoI. As a control, purified genomic CaSki DNA was treated in a similar fashion, except that micrococcal nuclease was used in a 1:100 dilution.
DNase I treatment of in vitro-assembled HPV-16 chromatin.
The rotational and translational phasing of nucleosomes on S190-assembled pHPV-16-Luc plasmids was determined by DNase I treatment and subsequent primer extension analysis. Samples from the chromatin assembly reaction mixture were split into two equal aliquots and assayed for transcriptional activity and nucleosomal positioning. Up to 70 ng of DNase I (Boehringer) was added to the samples, and the reactions were stopped after 1 min of incubation at room temperature with 100 μl of a buffer containing 450 mM sodium acetate, 0.1% SDS, and 10 mM EDTA. The DNA was recovered by standard procedures.
Low-resolution analysis of chromatin structure by indirect end-labelling.
For the in vivo analyses of HPV-16 chromatin structures, 10 μg of MPE-modified DNA from CaSki cells was digested with NcoI or EcoRI, purified, and loaded on 1.5% agarose gels. The gels were run at 50 V for 20 h, blotted on Hybond-N membranes (Amersham, Buckinghamshire, United Kingdom), and probed with 32P-labelled fragments of HPV-16 DNA (High Prime Labelling Kit; Boehringer). For the analysis of chromatin structures assembled in vitro on pHPV-16-Luc plasmids, the DNA was digested with micrococcal nuclease, purified, and digested to completion with ScaI. The DNA fragments were blotted on Hybond-N membranes and probed with a 32P-end-labelled oligonucleotide complementary to the luciferase-gene (5′-AGTGATGTCCACCTCGATATGTGCATCTGTAAAAGC-3′). After hybridization by standard protocols, membranes were washed in 0.1 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 10% SDS for 20 minutes at 65°C and autoradiographed with intensifying screens.
Primer extension footprinting of in vitro-assembled chromatin.
High-resolution DNase I footprinting of nucleosomes assembled in vitro on pHPV-16-Luc was performed by mapping cleavage sites with primer extension analysis (DNase I primer extension footprinting [52]). DNase I-treated DNA was purified by standard procedures and used as a template for the extension reaction with Vent (exo-) polymerase (New England Biolabs). The primer (5′-GATCGCAGATCTCGAGCCCGGGCTAGCACG-3′) was derived from the luciferase vector. A typical reaction mixture contained 100 ng of template DNA and 105 cpm of labelled oligonucleotide in a reaction volume of 50 μl. After annealing of the primer at 63°C for 30 min, the enzyme mix containing deoxyribonucleotides triphosphates was added for a 30-min reaction at 72°C. The reaction was stopped by phenol-chloroform-isoamylalcohol extraction, and the DNA was purified by ethanol precipitation. Pellets were dissolved in formamide loading buffer, and the samples were denatured and loaded onto a denaturing 6% polyacrylamide gel.
Restriction site accessibility assay.
To investigate the accessibility of a restriction site for the enzyme PinAI at genomic position 55 within the HPV-16 promoter, chromatin was assembled with S190 extract in the presence or absence of Sp1 as described above. The chromatin was purified on S400 Sephacryl mini-gel filtration columns (600 μl), 10 U of the restriction enzyme PinAI was added, and the samples were incubated for 1 h at 37°C. The DNA was purified and subsequently digested to completion with 10 U of the restriction enzyme ScaI. As a control, pure pHPV-16-Luc DNA was digested to completion either with ScaI alone or by subsequent treatment with ScaI and PinAI. After purification, the DNA fragments were loaded onto a 1% agarose gel and electrophoresed in 0.5× Tris-borate-EDTA buffer. The DNA was blotted onto a Hybond N membrane and probed with a 32P-labelled ScaI-KpnI fragment derived from pGL3. The membrane was hybridized, washed, and autoradiographed by standard procedures.
RESULTS
The HPV-16 LCR in CaSki cells is organized in the form of specifically positioned nucleosomes.
Since details of the chromatin organization of papillomaviruses have never been studied, we decided to investigate whether in vivo the LCR of HPV-16 is occupied by nucleosomes. For these experiments, we chose the cell line CaSki, which harbors about 500 chromosomally integrated HPV-16 genomes, since an elevated copy number is a prerequisite to obtain sufficiently strong signals in an indirect end-labelling analysis to investigate chromatin structures.
Low-resolution analysis of chromatin structure can be achieved by a combination of treatment of nuclei with micrococcal nuclease or MPE and subsequent indirect end-labelling analysis. Micrococcal nuclease and MPE cleave chromatin preferentially in the linker DNA between core nucleosomes. For many experiments, MPE cleavage is the technique of choice, as it cleaves naked DNA randomly, while micrococcal nuclease shows sequence preferences (30, 58).
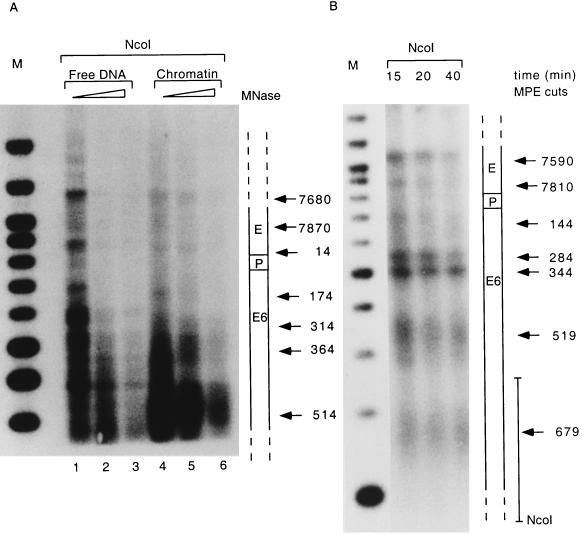
Figure 1 shows the results of experiments in which CaSki nuclei were treated either with micrococcal nuclease or with MPE, followed by purification of the DNA and cleavage with the restriction enzyme NcoI, which cuts at the end of the E7 gene. The blotted DNA was probed with labelled DNA fragments adjacent to this restriction site as shown at the right of Fig. 1B. This analysis revealed that in the case of the micrococcal nuclease, cuts were centered roughly on genomic positions 7680, 7870, 14, 174, 314, 364, and 514. The cuts obtained with chromatin differed from those obtained with free DNA and indicated nucleosomal organization, although sequence-preferential cleavage with free DNA confounds an unequivocal interpretation (Fig. 1A). Cleavage with MPE identified cuts centered roughly on genomic positions 7590, 7810, 144, 284, 344, 519, and 679, with the first three cuts falling within the LCR or about 50 bp downstream (144 bp) and the last four cuts falling within the E6 and E7 genes. The cuts at positions 519 and 679 are less precise than the other five, and the DNA fragments in this size range form a broader smear, possibly due to less-conserved nucleosomal positioning in the E6 and E7 genes (Fig. 1B). The cuts obtained with these two techniques indicated many similar sites, suggesting identical nucleosomal linkers despite the limitation of this crude mapping technique. We also mapped MPE cleavage from the 5′ side with a probe adjacent to an EcoRI site at position 7454 for a more precise resolution of nucleosomal cuts in the regions of the enhancer and promoter, and we identified preferential cuts at positions 7627, 7827, and 101 (data not shown). It should be stressed that these experimental data are based on low-resolution technology, and each of these cuts is identified with a precision of only 10 to 50 nucleotides, depending on the distance from the probe.
FIG. 1.
The LCR and the E6 gene of the HPV-16 genome in CaSki cells are nucleosomally organized. Nuclei were treated with increasing amounts of micrococcal nuclease (MNase) for 5 min (A) or of MPE for 10 to 40 min (B), and total genomic DNA was purified, restricted with NcoI separated by agarose gel electrophoresis, blotted onto nylon membranes, and processed with a radioactive probe close to the NcoI site, as indicated on the right of panel B. Arrows mark fragments with increased accessibility to micrococcal nuclease or MPE. Their sizes were estimated by comparison with a 100-bp size marker (lanes M). A scheme on the right of each panel shows the position of each cleavage site within the LCR of HPV-16.
Despite these limitations, our results show high accessibility of the chromatin to micrococcal nuclease and to MPE in two regions of the LCR, namely, at about bp 7590 to 7680 and 7810 to 7870. The former region harbors the 5′ part of the epithelial cell-specific enhancer, and the latter harbors the 3′ portion, including binding sites for the transcription factors AP-1 and TEF-1. A third cleavage site, between positions 101 and 144, lies immediately 3′ of the LCR and within the E6 gene. These three cleavage points indicate the precise positioning of two nucleosomes, one overlapping with the center of the enhancer, and one upstream of E6, overlapping with the HPV-16 E6 promoter and replication origin.
Specific positioning of in vitro-assembled nucleosomes on the HPV-16 LCR.
Precise positions of nucleosomes can be determined by properties of nucleotide sequences which influence physical properties, such as the curvature of this DNA. To investigate this possibility in the case of the HPV-16 genome, we asked whether nucleosomes assembled on the HPV-16 LCR in vitro would occupy positions similar to those observed on the HPV-16 LCR in CaSki cells. From among the techniques that are available for the in vitro assembly of chromatin (for a review, see reference 22), we chose a method based on cytoplasmic extracts from Drosophila embryos termed S190 (7). In the presence of an ATP-regenerating system, these extracts establish physiologically spaced nucleosomes irrespective of the source of the DNA, a consequence of the fact that histone proteins are highly conserved between Drosophila and vertebrates. The resulting chromatin structure is best analyzed by use of micrococcal nuclease, which cleaves the linker DNA between adjacent nucleosomes, followed by restriction cleavage with an enzyme cutting remotely from the DNA segment of interest and indirect end labelling with a probe that hybridizes close to these restriction cuts.
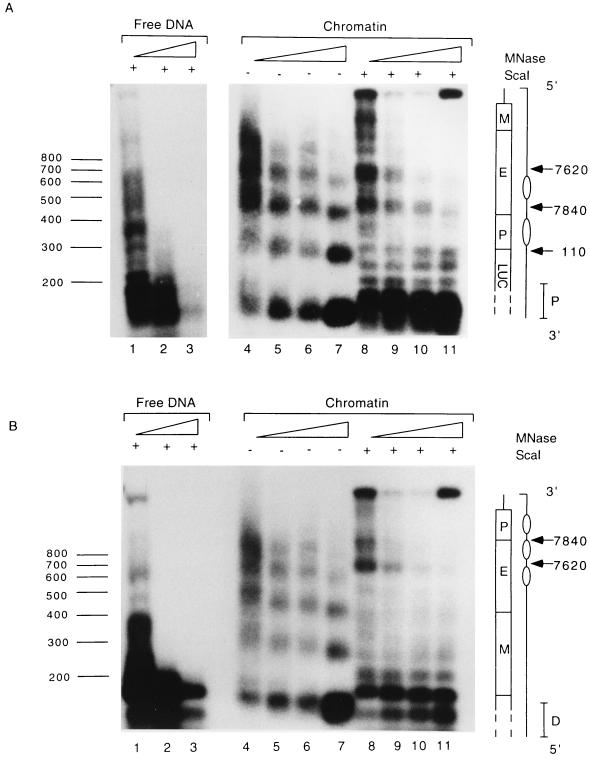
The outcome of this experiment (Fig. 2) indicated an approximately 180-bp spacing of nuclesomes assembled on the HPV-16 LCR in vitro, as apparent by regularly arranged fragments after micrococcal nuclease treatment. To map the precise positions of these nucleosomes, the purified DNA fragments were cut with the restriction enzyme ScaI, which has two cleavage sites in the vector sequences flanking the HPV-16 LCR. One ScaI site is at position 254 of the vector, downstream of the HPV-16 LCR, and the other one is at position 4717, upstream of the insert. As the micrococcal nuclease cleaves naked DNA in a nonrandom manner (30), we included in this experiment a direct comparison between nucleosome-free and chromatin-associated DNA.
FIG. 2.
Nucleosomes assembled in vitro on the LCR and the E6 gene of HPV-16 DNA are found in positions similar to those detected in CaSki cells. pHPV-16-Luc DNA was assembled into chromatin with Drosophila S190 extract and treated with 1, 2, 4, and 5 U of micrococcal nuclease (MNase) (lanes 4 to 7 and 8 to 11 of each panel). The nuclease-treated DNA was purified and fractionated on agarose gels without further treatment by restriction endonucleases (lanes 4 to 7 of each panel) or after digestion with ScaI (lanes 8 to 11). The DNA was blotted, and the blots were processed either with a proximal probe (P) (A) or with a distal probe (D) (B). As a control, lanes 1 to 3 of each panel show the digestion patterns of micrococcal nuclease-treated and ScaI-restricted free DNAs. The numbers on the left indicate the sizes of the fragments in base pairs. A schematic presentation on the right of each panel identifies within the HPV-16 LCR the MAR (M), the enhancer (E), the E6 promoter (P), the genomic positions of micrococcal nuclease-sensitive sites (arrows), and the predicted positions of nucleosomes (ovals).
When the micrococcal nuclease treatment of the HPV-16 chromatin was combined with ScaI cleavage and indirect end labelling with probes hybridizing close to the upstream or the downstream ScaI site, we observed regularly spaced and precisely positioned nucleosomes, with the strongest cuts of the micrococcal nuclease mapping around positions 7840 and 7620 of the HPV-16 LCR. Neither of these cleavage points appears after micrococcal nuclease treatment of naked DNA. These data indicate that in vitro-assembled nucleosomes occupy on the HPV-16 LCR positions very similar or identical to those observed in CaSki cells in vivo.
A nucleosome is specifically positioned over elements of HPV-16 p97.
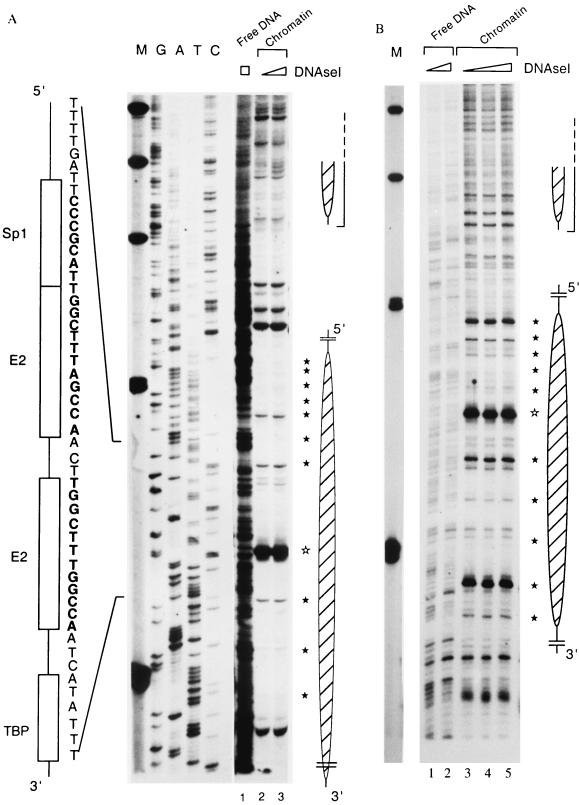
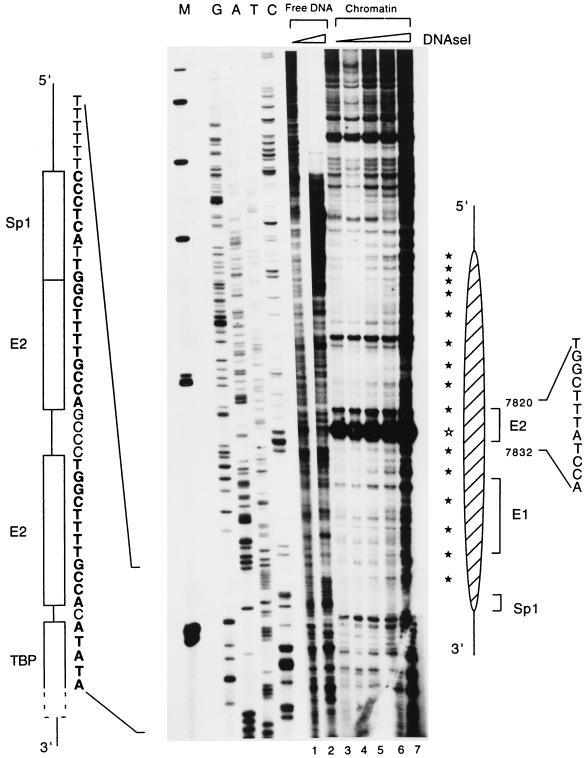
To improve the limited resolution of the experiments shown in Fig. 1 and 2, we footprinted the histone-DNA interactions in the region of the HPV-16 E6 promoter p97. Chromatin was assembled from the HPV-16 LCR and the Drosophila S190 extract, the samples were digested with DNase I, and the cleavage sites were identified by primer extension. Figure 3 shows the outcomes of two similar footprint experiments. Figure 3A includes a sequence ladder to map the locations of specific signals with the 3′ side of the HPV-16 LCR. The most obvious feature (lanes 2 and 3) is the strong protection of a segment that contains the binding site of the promoter factor Sp1, the two E2 binding sites, and the TATA box. The nucleosomal organization of this DNA is indicated by a 10-bp periodicity of DNase I-accessible sites due to the rotational phasing within the nucleosome. This periodicity is particularly clear in Fig. 3B, which, however, permits less clear mapping of the boundaries of the nucleosome. Such a 10-bp periodicity is not visible when free DNA is treated in a similar way (Fig. 3A, lane 1, and B, lanes 1 and 2). A particularly strong DNase I-hypersensitive site overlaps with the promoter-proximal binding site for the E2 protein. This increased accessibility may indicate the dyad axis of the nucleosome (73) centered over this E2 site. With a 5′ border approximately at position 7838, the nucleosome also covers the replication origin with the E1 binding site.
FIG. 3.
A specifically positioned nucleosome covers the promoter of HPV-16. (A) The HPV-16 LCR cloned in pHPV-16-Luc was assembled into chromatin with Drosophila S190 extracts and treated with increasing amounts of DNase I, and the resulting fragments were purified and assayed by primer extension. Lanes 2 and 3, footprints originating from two nucleosomes that overlap with the promoter and the enhancer (large and small oval shapes on the right). Weak 10-bp-spaced bands (filled stars) indicate DNase I accessibility due to the rotational phasing of the nucleosomes, and a strong hypersensitive site (open star) suggests the center of the dyad symmetry of the nucleosome. As controls, lane 1 shows DNase I treatment of free HPV-16 LCR DNA and the left side of the panel indicates a sequencing ladder. Lane M, size marker with bands at 500, 400, 300, 200, and 100 bp. Symbols and nucleotides on the left identify the four cis-responsive elements of the E6 promoter, namely, binding sites for Sp1, the viral factor E2, and TBP. (B) Footprint obtained in a similar experiment and permitting similar interpretations. It highlights the 10-bp periodicity but does not permit clear mapping of the extent of nucleosomal protection.
Transcriptional repression by a nucleosome positioned over the HPV-16 promoter is alleviated by the transcription factors AP-1 and Sp1.
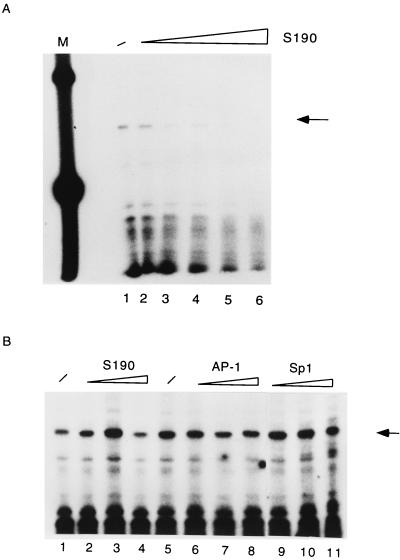
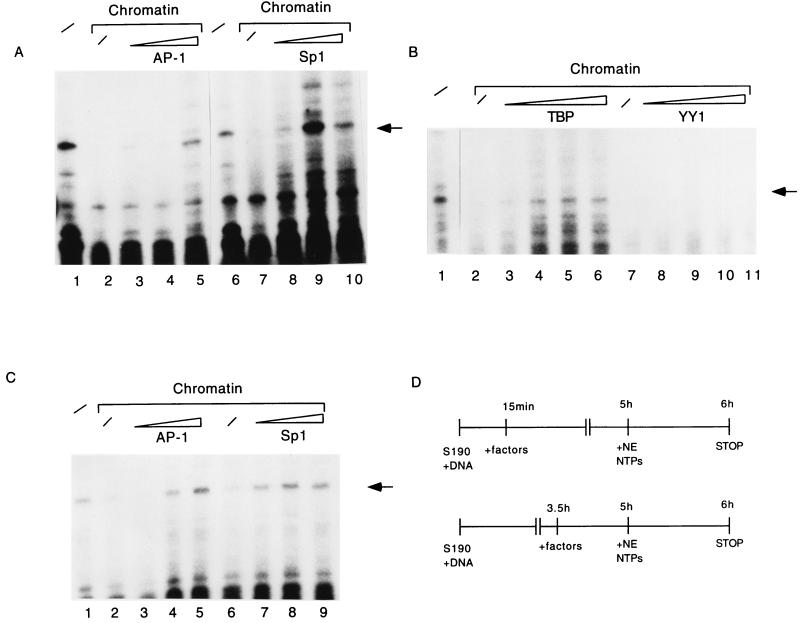
There are numerous examples of strong repressive influences of chromatin on gene expression (28, 36, 43), while a few genes are also known to be activated by nucleosomes (44, 54, 56, 60, 69). Since accessibility of cis-responsive elements is a necessary prerequisite for the formation of the preinitiation complex, we asked whether the occupation of parts of the LCR of HPV-16 by nucleosomes would have consequences for the activity of the E6 promoter. Figure 4A illustrates that in vitro transcription starting from the E6 promoter of the free pHPV-16-Luc DNA is gradually repressed by increasing amounts of Drosophila S190 extract and chromatin assembly (compare lane 1 with lanes 2 to 6). To ensure that repression of transcriptional activity was due to chromatin, we added the S190 extract concomitantly with the transcription extract. The outcome of this experiment is shown in Fig. 4B (lanes 2 to 4). We conclude that nonhistone components in the S190 extract do not interfere with accurate transcription from the HPV-16 promoter. As a further control, we ensured that the factors Sp1 and AP-1 were not limiting in the basic transcriptional system. Lanes 6 to 11 of Fig. 4B confirm that an excess of AP-1 and Sp1 does not superstimulate transcription of nonchromatin templates.
FIG. 4.
Nucleosomal assembly leads to transcriptional repression of the HPV-16 promoter. (A) Analyses of in vitro transcription from free pHPV-16-Luc DNA (lane 1) or the same plasmid assembled into nucleosomes by increasing amounts of Drosophila S190 extract (lanes 2 to 6). Lane M, 100-bp ladder serving as size standard. The specific transcript initiated at the HPV-16 E6 promoter is indicated by an arrow on the right. (B) The Drosophila S190 extract does not contain nonhistone components that might inhibit in vitro transcription, as shown by the addition of 20, 50, and 70 μl of S190 extract (lanes 2 to 4, respectively) at the beginning of an in vitro transcription reaction with nucleosome-free DNA (lane 1). As a further control, we ensured that the factors Sp1 and AP-1 are not limiting in the basic transcriptional system. Lanes 6 to 11 confirm that an excess of AP-1 and Sp1 does not superstimulate transcription of nonchromatin templates.
Next, we asked whether transcription factors that are known to activate HPV-16 expression would be able to relieve this repression when they are added shortly after initiation of the assembly process. For this we chose the transcription factor AP-1, which binds fp9e (27), a site 3′ of the enhancer, at a position about 70 bp upstream of the nucleosome that occupies the HPV-16 promoter in vitro and between the two nucleosomes mapped on the LCR in vivo. Similarly, we measured the effects of the factors Sp1, E2, YY1, and TBP, whose binding sites overlap with the promoter proximal nucleosome. There was an activation of transcription from the chromatin templates by AP-1 and Sp1 (Fig. 5A, lanes 3 to 5 and 8 to 10), whereas YY1 (Fig. 5B, lanes 8 to 11) and E2 (data not shown) did not relieve the nucleosomal repression. In addition, there was weak activation by TBP, which was visible only after exposure of the autoradiograph for 2 days (Fig. 5B, lanes 3 to 6).
FIG. 5.
Relief of nucleosomal transcriptional repression by trans-acting factors. (A) AP-1 (lanes 3 to 5) and Sp1 (lanes 8 to 10), added 15 min after initiation of chromatin assembly on pHPV-16-Luc, activate in vitro transcription from the E6 promoter. Specific transcripts were detected by primer extension (arrow). The same transcript is generated from free DNA in the absence of chromatin and an excess of any additional factor (lanes 1 and 6) but is repressed by chromatin alone (lanes 2 and 7). (B) Under the same conditions as used for panel A, TBP (lanes 3 to 6) marginally induces a nucleosomally organized E6 LCR, while YY1 (lanes 8 to 11) fails to do so. Lane 1, transcription of nucleosome-free DNA; lanes 2 and 7, repression of transcription by nucleosomes. To detect the weak signals of the transcriptional induction by TBP, this blot was exposed five times longer than the blot shown in panel A. YY1 has a strong binding site within the promoter sequence covered by the nucleosome, which does not lead to transcriptional repression in transfection experiments, while additional YY1 sites remote from the promoter and possibly protected by the upstream nucleosome may negatively interfere with transcription independent from the chromatin state of the HPV-16 LCR (50). (C) AP-1 (lanes 3 to 5) and Sp1 (lanes 7 to 9) can activate in vitro transcription from the E6 promoter even when added at a late stage of chromatin assembly. The conditions of this experiment resembled those for panel A, but the transcription factors were added 3.5 h after initiation of chromatin assembly. Lane 1, transcription from naked DNA, lanes 2 and 6, transcription from chromatin in the absence of additional factors. (D) Schematic diagram of the experiments in panels A and C. NTP, nucleoside triphosphate.
The experiments shown in Fig. 5A were performed under conditions in which formation of chromatin and transcription factor binding could each occur, mimicking conditions in which competition for transcription factor binding sites and nucleosomal positioning may take place. To investigate whether these factors may also counteract nucleosomal repression after a more stable establishment of chromatin, we took a similar experimental approach but added the transcription factors 3.5 h after initiating the assembly of chromatin, such that the establishment of chromatin is nearly complete (Fig. 5D). Figure 5C shows that AP-1 and Sp1 maintained the ability to activate repressed transcription under these circumstances (lanes 3 to 5 and 7 to 9), while the basal transcription factor TBP had lost its ability to do so (data not shown).
Rearrangement of the promoter-bound nucleosome by Sp1.
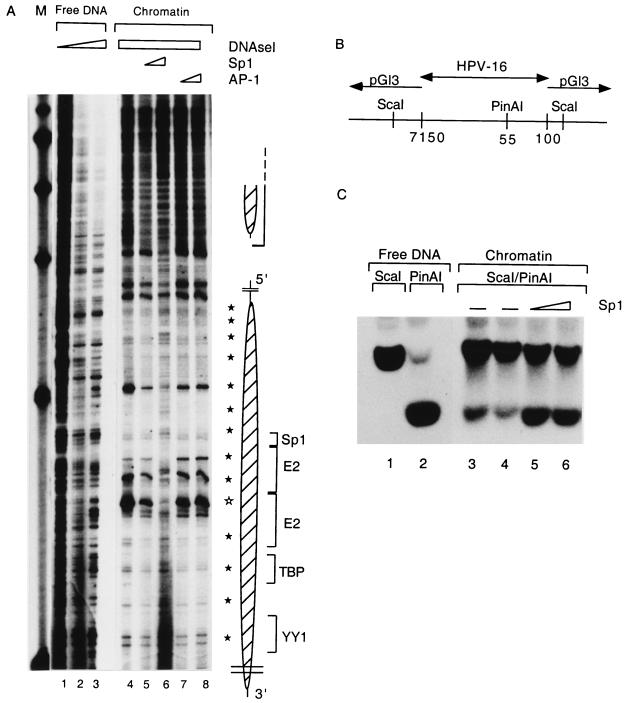
We next asked by which mechanisms AP-1 and Sp1 may relieve nucleosomal repression of p97. It is unlikely that both transcription factors act by the same mechanism, as the principal AP-1 binding site of HPV-16 (fp9e [27]) does not overlap with either of the nucleosomes attached to the enhancer or to p97 sequences, while Sp1 binds this promoter at nucleotide sequences fully protected by the nucleosome. Figure 6A shows that an increase in the concentration of Sp1 even at a late stage of the chromatin assembly process led to a change of the nucleosomal footprint as well as the hypersensitivity around the E2 binding site. In addition, the nucleosomal 10-bp pattern became less pronounced (lanes 5 and 6). These changes became even more pronounced when the transcription extract was added to the prebound Sp1 (data not shown). In contrast to Sp1, AP-1 did not affect the structure of the nucleosomal footprint (Fig. 6A, lanes 7 and 8).
FIG. 6.
Sp1 can rearrange the structure of the E6 promoter-bound nucleosome. (A) pHPV-16-Luc DNA was assembled into chromatin, subjected to DNase I treatment, and processed as described for Fig. 3 to visualize the nucleosomal footprints overlapping with the HPV-16 E6 promoter (lanes 4 to 8) and enhancer (large and small oval shapes, respectively, on the right). Weak 10-bp-spaced bands (filled stars) indicate DNase I accessibility due to the rotational phasing of the nucleosomal core, and a strong hypersensitive site (open star) is most likely the center of the dyad symmetry of the nucleosome. As controls, lanes 1 to 3 show DNase I treatment of free HPV-16 LCR DNA. A nucleosome (Np16) protects four cis-responsive elements of the E6 promoter, namely, binding sites for Sp1, the viral factor E2, TBP, and a YY1 site (brackets on the right of the oval shape). Sp1 (lanes 5 and 6) or AP-1 (lanes 7 and 8) was added 3.5 h after start of the chromatin assembly and in the case of Sp1 resulted in the reemergence of the cleavage pattern of the free DNA and the disappearance of nucleosome-specific DNase I-hypersensitive sites. Lane M, markers. (B) Schematic representation of an HPV-16 LCR clone from position 7150 to 100 in the vector pGI3. The enzyme ScaI cleaves in vector sequences on both sides of the cloned segment; PinAI cleaves at position 55 in the promoter sequences of HPV-16. (C) Southern blot of a ScaI digest (lane 1) and a ScaI-PinAI double digest (lane 2). The ScaI-PinAI fragment is released inefficiently in the presence of chromatin (lanes 3 and 4) but efficiently with increasing Sp1 concentrations (lanes 5 and 6). This blot was processed with a radioactive probe specific for the ScaI-PinAI fragment.
As an additional control for the rearrangement of this nucleosome by Sp1, we monitored the accessibility of a restriction site in the nucleosomally protected region in the absence and presence of Sp1. The restriction enzyme PinAI cleaves the hexanucleotide ACCGGT, which occurs in the HPV-16 promoter at the genomic position 55 and forms the 3′ part of the promoter-proximal E2 binding site. Two ScaI sites in the vector pGL3 (Fig. 6B) permit excision of the cloned HPV-16 LCR segment, which can be further cleaved with PinAI (Fig. 6C, lanes 1 and 2). This cleavage occurs very inefficiently when the HPV-16 DNA is organized as chromatin (Fig. 6C, lanes 3 and 4, lower band) but efficiently in the presence of Sp1 (lanes 5 and 6). We conclude that the PinAI site becomes more accessible by binding of Sp1 to a site 30 bp 5′ of the PinAI site with concomitant rearrangement or displacement of the nucleosome positioned over both sequence elements.
A specifically positioned nucleosome binds the HPV-18 replication origin after assembly of chromatin in vitro.
The nucleotide sequences of the LCRs of HPV-16 and HPV-18 show little similarity but conserve most cis-responsive elements. On the 3′ side within the LCR, there are three binding sites for the E2 protein, one of the replication factor E1, and one each for Sp1 and TBP (31, 49). It is of interest to ask whether the chromatin organization in both viruses may also be conserved. We addressed this question by assembling chromatin in vitro on the HPV-18 LCR and examining by primer extension footprinting the position of a potential nucleosome forming on this segment. Figure 7 (lanes 3 to 7) shows that a nucleosome occupies a highly specific position in the 3′ segment of the HPV-18 LCR. This nucleosome is positioned about 90 bp 5′ from the equivalent position of the HPV-16 promoter, with the center of symmetry apparently overlapping with the third E2 binding site upstream of the HPV-18 E6 promoter. The downstream end of the nucleosome occupies the E1 binding site and extends to the Sp1 binding site, leaving the two promoter-proximal E2 binding sites and the TATA box unprotected.
FIG. 7.
A specifically positioned nucleosome covers the replication origin of HPV-18 and extends into E6 promoter sequences. The cloned HPV-18 LCR was assembled into chromatin with Drosophila S190 extracts and treated with increasing amounts of DNase I, and the resulting fragments were assayed by primer extension. Lanes 3 to 7 show, with increasing DNase I treatment, the footprint of a nucleosome overlapping the replication origin (distal E2 and E1 binding site), indicated by a large hatched oval on the right. Weak 10-bp-spaced bands (filled stars) indicate DNase I accessibility due to the rotational phasing of the nucleosomal organization, and a strong hypersensitivity site (open star) suggests the center of the dyad symmetry of the nucleosome. As controls, lanes 1 and 2 show DNase I treatment of free HPV-18 LCR DNA, and the left side shows a sequencing ladder of this sequence. Lane M, size marker with bands at 500, 400, 300, 200, and 100 bp. Symbols and nucleotides on the right identify the third E2 binding site from the E6 promoter, the E1 binding site, and one of the four cis-responsive elements of the E6 promoter, namely, the binding site for Sp1.
DISCUSSION
Nucleosomal organization of the L1-LCR-E6-E7 segment of HPV-16.
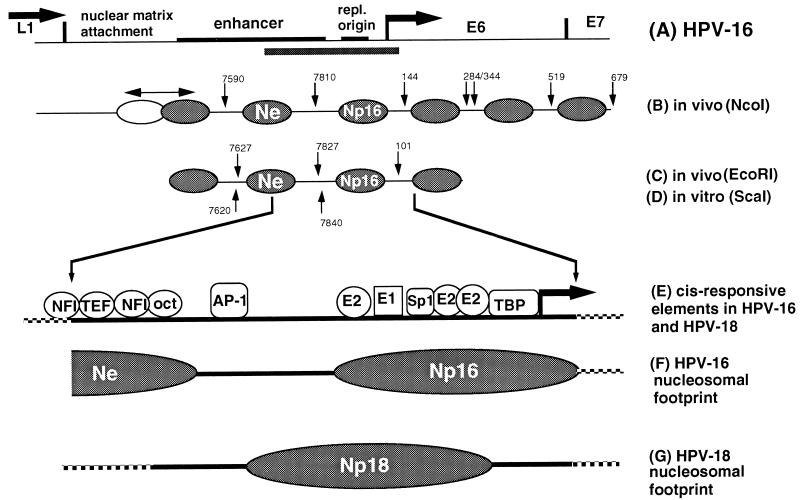
Figure 8 summarizes the positions of nucleosomes on a 1.5-kb segment of HPV-16 (about 20% of the viral genome) from the end of the L1 gene to the center of the E7 gene. In CaSki cells (Fig. 8B and C), three nucleosomes are located overlapping with the LCR. MPE cleavage in vivo suggests the presence of three additional nucleosomes within the E6 gene and the 5′ part of E7. Multiple cleavage sites in the nucleosomal linker DNA suggest loose positioning of some of these nucleosomes. Nucleosomal positioning is not precise in the 5′ segment of the LCR, since a nucleosome overlapping with the MAR and the 5′ side of the enhancer protects an unusually long DNA segment, as to be expected from nucleosomes forming on nuclear MARs (10). One nucleosome (Ne) overlaps with the center of the enhancer, and another (Np16) overlaps with the replication origin and the E6 promoter. The position of at least one of these nucleosomes, Np16, seems to be determined by intrinsic properties of the HPV-16 DNA sequences, as micrococcal nuclease mapping of in vitro-assembled nucleosomes indicates a position of Np16 similar or identical to that determined by MPE mapping of CaSki chromatin (Fig. 8D).
FIG. 8.
Nucleosomal organization of the LCR of HPV-16. (A) Genomic segment of HPV-16 with a size of approximately 1.5 kb. The LCR has a size of 850 bp and includes an MAR, the epithelial cell-specific enhancer, the replication origin (repl. origin), and the E6 promoter (arrow) and is flanked by the L1 and E6 genes. The bar below the 3′ side of the enhancer, the replication origin, and the promoter highlights the genomic fragment that has been studied in both HPV-16 and HPV-18 by footprint analysis to map specifically positioned nucleosomes. (B) MPE-hypersensitive sites in HPV-16 chromatin in CaSki cells permit the mapping of the positions of six nucleosomes relative to an NcoI site, which is positioned downstream of this segment. A double-headed arrow above the leftmost nucleosome, which overlaps with the MAR, indicates a region of protection whose size exceeds that for the normal protection by a nucleosome, possibly due to alternative positions of a nucleosome. The nucleosome (Ne) between the MPE cuts at position 7590 and 7810 overlaps with the epithelial cell-specific enhancer, and the third nucleosome (Np), between positions 7810 and 144, overlaps with the replication origin and the E6 promoter. Three additional nucleosomes are positioned within the E6 gene and at the 5′ end of the E7 gene, respectively. (C and D) The positions of four nucleosomes on the HPV-16 LCR in CaSki cells mapped with an upstream probe (hybridizing close to the EcoRI site at position 7456) are in agreement with the data obtianed with the NcoI probe, and at least two of these nucleosomes are positioned identically after assembly of chromatin in vitro (data not shown). (E and F) One of these nucleosomes was footprinted, and its center apparently overlaps with two E2 binding sites at the E6 promoter. The protection of this nucleosome includes the TBP binding site, the Sp1 binding site, and the binding site for the replication factor E1. (G) In HPV-18, which has a similar arrangement of elements of the replication origin and the E6 promoter, a nucleosome (Np18) it positioned about 90 bp further upstream from the E6 promoter, extending to and partially protecting the promoter elements. The HPV-16 DNA, and possibly also the DNA of HPV-18, may position in vitro the Ne nucleosome, which overlaps with the core of the epithelial cell-specific enhancer, but leave the 3′ side of the enhancer with a strong AP-1 site unprotected.
Most nucleosomes that exist in eukaryotic and viral chromatin are probably not specifically positioned (65) and most often assume random positions by sliding freely along any particular DNA segment (46, 53). It is well known, however, that the sequences of particular nucleotide segments can affect physical properties, like their curvature, and these properties may determine the precise position of nucleosomes (25, 29). Surprisingly, in the case of the majority of the few carefully studied genes, nucleosomes were found to occupy specific positions relative to regulatory DNA sequences (for reviews, see references 6 and 80). Paradigms for this scenario have been the promoter on the LTR of MMTV (2, 14, 58, 72, 73) and the promoters of the Xenopus 5S rRNA genes (reference 61 and references therein). Additional examples include the promoters for the Xenopus vitellogenin B1 gene (60), the human U6 gene (69), rat tyrosine aminotransferase (12), Drosophila hsp26 and hsp27 (44), Xenopus TFIIIA (54), and HIV-1 (52, 62) and the late promoter of SV40 (55). For some of these genes it has been proposed that only one or a few nucleosomes are specifically positioned, due to underlying structural properties of the DNA, while neighboring nucleosomes may be arranged in a regular array relative to a specifically positioned nucleosome, thereby also binding a constant segment.
Replication origin and E6 promoter sequences of HPV-16 and HPV-18 are occupied by specifically positioned nucleosomes.
The nucleosome Np16, and Np18 in the equivalent genomic segment of HPV-18, could be mapped with reasonable precision by primer extension footprinting experiments in vitro (Fig. 3 and 7). It is technically challenging to determine the precise borders of a nucleosomal footprint in the manner of a footprint generated by a DNA-bound transcription factor, but a 10-bp spacing of DNA fragments reveals the extent of a nucleosomal organization, and several bands originating from strong DNase I hypersensitivity within Np16 and Np18 most likely identify the center of dyad symmetry of each nucleosome. Interestingly, both DNase I-hypersensitive sites overlap with E2 binding motifs: in the case of Np16 with the promoter-proximal E2 binding site (Fig. 8E and F) and in the case of Np18 with the third E2 binding site, 90 bp upstream and within the homologous elements of HPV-18. While the exact positions of these nucleosomes differ for the two viruses in vitro, similar cis-responsive elements, namely, the E1 binding site necessary for replication and the Sp1 binding site that activates the E6 promoter, fall in the 146-bp segment protected by the nucleosomal core. These overlaps suggest a nucleosomal effect on replication as well as transcription in both viruses, but knowledge of details of these functional consequences beyond the repression of HPV-16 transcription studied here has to await future investigations.
Nucleosomal repression of the HPV-16 E6 promoter and derepression by AP-1 and Sp1.
Our in vitro transcription experiments document that usage of the E6 promoter is impeded as a consequence of the nucleosomal organization of the HPV-16 LCR, most likely due to impaired accessibility of the promoter sequences by Sp1 and the basic transcription machinery. This explanation is strengthened by the observation that an excess of Sp1 can functionally overcome this repression, apparently by altering or displacing the promoter nucleosome Np16. At this time we can offer only this very general explanation, as research on the interaction between nucleosomes and transcription factors on other promoters has shown that physical displacement or competitive binding applies to only some of these promoters (68), while other factor-nucleosome interactions are governed by more complex mechanisms (74). For example, in the case of a nucleosome blocking the MMTV LTR promoter, a complex interaction between the glucocorticoid receptor and other transcription factors with the nucleosomal remodeling activity of the SWI-SNF complex is required to permit access to cognate binding sites (66). In another case, a nucleosome on the promoter of the Xenopus oocyte 5S rRNA gene can slide freely, but it is forced into a specific position by interacting with histone H1, which recognizes a specific nucleotide sequence of this promoter. As a consequence, H1 overexpression represses this promoter (61).
There is a viral precedent for repression due to interference between nucleosomes and Sp1. Sp1 has six binding sites at the SV40 early promoter; however, nucleosomally organized SV40 DNA is bound by Sp1 with 10- to 20-fold-lower affinity than naked DNA. Access to these sites by Sp1 involves formation of a ternary Sp1-nucleosome-DNA complex. It was observed that activation by Sp1 involves stepwise nucleosomal disassembly, particularly when aided by the addition of nucleosplasmin, and removal of H2A-H2B dimers by the SWI-SNF complex enables transcription factors to bind (15, 41).
Some aspects of the abolition of nucleosomal repression by AP-1 have become clear with the detection of histone acetyltransferase activity of CREB binding protein, a transcriptional cofactor of AP-1. Acetylation destabilizes the interaction between histones and DNA, alters the shape of the octamer (5), and can open the way for increased accessibility by transcription factors (48, 83, 84).
There is also evidence for a change of chromatin under the influence of the papillomavirus transcription factor E2 (40). As this factor represses the HPV E6 promoter due to displacement of Sp1 and TBP (19, 71) irrespective of nucleosomal biology, we did not investigate whether its presence alters the chromatin structures of the HPV-16 and HPV-18 LCRs.
Potential consequences of the HPV chromatin organization for replication, transcription, and the viral life cycle.
The presently available technology does not permit studies of in which parts of the HPV life cycle transcriptional control by nucleosomes may take place, as this would require information about the HPV chromatin state in very small cell populations, such as basal, suprabasal, or differentiated layers of squamous epithelia, which even epithelial raft cultures (51) may not provide. We suggest, however, that chromatin actually does repress HPV-16 transcription in CaSki cells, as the signals that we observed after MPE cleavage suggest that the majority of the 500 HPV-16 genomes exhibit specifically positioned Ne and Np16 nucleosomes on the HPV-16 enhancer and promoter. This may explain the well-discussed enigma that SiHa and CaSki cells, which contain 1 and 500 HPV-16 genomes, respectively, show similar viral transcription levels (3, 9). It is tempting to speculate about whether the single integrated HPV-16 genome in SiHa cells may show the same or a different nucleosomal positioning. We have attempted to answer this question but have found that the analysis of a single genomic copy exceeded the sensitivity of the available technology.
HPV-16 shows analogies with HIV-1 as to the transcriptional regulation by chromatin. The DNA genome of HIV-1 has one nucleosome specifically positioned overlapping with the transcription start and another one in a remote upstream position. Remodeling of HIV-1 chromatin occurs in response to mitogenic signals and is mechanistically influenced by multiple transcription factors, but it apparently results in only a small fraction of all viral templates being active in any round of transcription. It is likely that chromatin represses transcription during latency in vivo, although details of these mechanisms were gleaned from in vitro studies (39, 62, 63).
Our research did not address the possibility of HPV-16 and HPV-18 replication being modulated by chromatin. This is quite likely, because Np16 and Np18 overlap with the respective E1 binding sites. Without knowledge of nucleosomal positioning, it has been reported that nucleosomes can repress BPV-1 replication and that this repression can be relieved by E2 and acidic transcription factors (42). A careful mechanistic study of replication control by chromatin has shown that the exact distance between a nucleosome and the yeast replication origin ARS1 determines accessibility to the replication machinery (65), and in vitro studies of SV40 replication pointed to the ability of the chromatin accessibility complex to stimulate nucleosomally repressed replication (1).
ACKNOWLEDGMENT
We are grateful to Robin M. Watts for discussions and critical reading of the manuscript.
REFERENCES
- 1.Alexiadis V, Varga-Weisz P D, Bonte E, Becker P B, Gruss C. In vitro chromatin remodeling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer T K, Cordingley M G, Wolford R G, Hager G L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batson S C, Rimsky S, Sundseth R, Hansen U. Association of nucleosome-free regions and basal transcription factors with in vivo-assembled chromatin templates active in vitro. Nucleic Acids Res. 1993;21:3459–3468. doi: 10.1093/nar/21.15.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer W R, Hayes J J, White J H, Wolffe A P. Nucleosome structural changes due to acetylation. J Mol Biol. 1994;236:685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 6.Becker P B. The establishment of active promoters in chromatin. Bioessays. 1994;16:541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- 7.Becker P B, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard, H. U., D. Apt, and B. Gloss. Unpublished observations.
- 10.Bode J, Stengert-Iber M, Kay V, Schlake T, Dietz-Pfeilstetter A. Scaffold/matrix-attached regions: topological switches with multiple regulatory functions. Crit Rev Eukaryot Gene Expr. 1996;6:115–138. doi: 10.1615/critreveukargeneexpr.v6.i2-3.20. [DOI] [PubMed] [Google Scholar]
- 11.Bungert J, Kober L, Düring F, Seifart K H. Transcription factor eUSF is an essential component of isolated transcription complexes on the duck histone H5 gene and it mediates the interaction of TFIID with a TATA-deficient promoter. J Mol Biol. 1992;22:885–898. doi: 10.1016/0022-2836(92)90250-n. [DOI] [PubMed] [Google Scholar]
- 12.Carr K D, Richard-Foy H. Glucocorticoids locally disrupt an array of positioned nucleosomes on the rat tyrosine aminotransferase promoter in hepatoma cells. Proc Natl Acad Sci USA. 1990;87:9300–9304. doi: 10.1073/pnas.87.23.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartwright I L, Hertzberg R P, Dervan P B, Elgin S C. Cleavage of chromatin with methidiumpropyl-EDTA-iron(II) Proc Natl Acad Sci USA. 1983;80:3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci USA. 1997;94:2885–2890. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Li B, Workman J L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong T, Apt D, Gloss B, Isa M, Bernard H U. The enhancer of human papillomavirus-16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NFI, and AP1 participate in the epithelial specific transcription. J Virol. 1991;65:5933–5943. doi: 10.1128/jvi.65.11.5933-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cripe T P, Haugen T H, Turk J P, Tabatabai F, Schmid P G, Dürst M, Gissmann L, Roman A, Turek L P. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demeret C, Desaintes C, Yaniv M, Thierry F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J Virol. 1997;71:9343–9349. doi: 10.1128/jvi.71.12.9343-9349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dery C V, Toth M, Brown M, Horvath J, Allaire S, Weber J M. The structure of adenovirus chromatin in infected cells. J Gen Virol. 1985;66:2671–2684. doi: 10.1099/0022-1317-66-12-2671. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J D, Lebowitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimitrov S, Wolffe A P. Chromatin and nuclear assembly: experimental approaches towards the reconstruction of transcriptionally active and silent states. Biochim Biophys Acta. 1995;1260:1–13. doi: 10.1016/0167-4781(94)00182-3. [DOI] [PubMed] [Google Scholar]
- 23.Dollard S C, Broker T R, Chow L T. Regulation of the human papillomavirus type 11 E6 promoter by viral and host transcription factors in primary human keratinocytes. J Virol. 1993;67:1721–1726. doi: 10.1128/jvi.67.3.1721-1726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.FitzGerald P C, Simpson R T. Effects of sequence alterations in a DNA segment containing the 5S RNA gene from Lythechinus variegatus on positioning of a nucleosome core particle in vitro. J Biol Chem. 1985;260:15318–15324. [PubMed] [Google Scholar]
- 26.Gloss B, Bernard H U, Seedorf K, Klock G. The upstream regulatory region of the human papillomavirus-16 contains an E2 protein independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 1987;6:3735–3743. doi: 10.1002/j.1460-2075.1987.tb02708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gloss B, Chong T, Bernard H U. Numerous nuclear proteins bind the long control region of human papillomavirus type 16: a subset of 6 of 23 DNase I-protected segments coincides with the location of the cell-type-specific enhancer. J Virol. 1989;63:1142–1152. doi: 10.1128/jvi.63.3.1142-1152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 29.Hayes J J, Tullius T D, Wolffe A P. The structure of DNA in a nucleosome. Proc Natl Acad Sci USA. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoerz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoppe-Seyler F, Butz K. Activation of human papillomavirus type 18 E6-E7 oncogene expression by transcription factor Sp1. Nucleic Acids Res. 1992;20:6701–6706. doi: 10.1093/nar/20.24.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. 64. Human papillomaviruses. Lyon, France: International Agency for Research on Cancer; 1995. [PMC free article] [PubMed] [Google Scholar]
- 33.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 34.Jeong S W, Stein A. DNA sequence affects nucleosome ordering on replicating plasmids in transfected COS-1 cells and in vitro. J Biol Chem. 1994;269:2197–2205. [PubMed] [Google Scholar]
- 35.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 36.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 37.Kondoleon S K, Kurkinen N A, Hallick L M. The SV40 nucleosome-free region is detected throughout the virus life cycle. Virology. 1989;173:129–135. doi: 10.1016/0042-6822(89)90228-6. [DOI] [PubMed] [Google Scholar]
- 38.Larsen P M, Storgaard L, Fey S J. Proteins present in bovine papillomavirus particles. J Virol. 1987;61:3596–3601. doi: 10.1128/jvi.61.11.3596-3601.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laughlin M A, Zeichner S, Kolson D, Alwine J C, Seshamma T, Pomerantz R J, Gonzalez-Scarano F. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology. 1993;196:496–505. doi: 10.1006/viro.1993.1505. [DOI] [PubMed] [Google Scholar]
- 40.Lefebvre O, Steger G, Yaniv M. Synergistic transcriptional activation by the papillomavirus E2 protein occurs after DNA binding and correlates with a change in chromatin structure. J Mol Biol. 1997;266:465–478. doi: 10.1006/jmbi.1996.0807. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Adams C C, Workman J L. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994;269:7756–7763. [PubMed] [Google Scholar]
- 42.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorch Y, La Pointe J W, Kornberg R D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 44.Lu Q, Wallrath L L, Elgin S C R. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 1995;14:4738–4746. doi: 10.1002/j.1460-2075.1995.tb00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcus-Sekura C J, Carter B J. Chromatin-like structure of adeno-associated virus DNA in infected cells. J Virol. 1983;48:79–87. doi: 10.1128/jvi.48.1.79-87.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meersseman G, Pennings S, Bradbury E M. Mobile nucleosomes, a general behavior. EMBO J. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers G, Bernard H U, Delius H, Favre M, Icenogle J, van Ranst M, Wheeler C. Human papillomaviruses. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 48.Ng K W, Ridgway P, Cohen D R, Tremethick D J. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 1997;16:2072–2085. doi: 10.1093/emboj/16.8.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Connor M, Chan S Y, Bernard H U. Transcription factor binding sites in the long control regions of genital HPVs. In: Meyers G, Bernard H U, Delius H, Baker C, Icenogle J, Halpern A, Wheeler C, editors. Human papillomaviruses, 1995 compendium, part III-A. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. 21–40. [Google Scholar]
- 50.O’Connor M J, Tan S H, Tan C H, Bernard H U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker J N, Zhao W, Askins K J, Broker T R, Chow L T. Mutational analysis of differentiation-dependent human papillomavirus type-18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. 1997;8:751–762. [PubMed] [Google Scholar]
- 52.Pazin M J, Sheridan L P, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 53.Pazin M J, Bhargava P, Geiduschek E P, Kadonaga J T. Nucleosome mobility and the maintenance of nucleosome positioning. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- 54.Pfaff S L, Taylor W L. Xenopus TFIIIA gene transcription is dependent on cis-element positioning and chromatin structure. Mol Cell Biol. 1998;18:3811–3818. doi: 10.1128/mcb.18.7.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Powers J H, Bina M. In vitro assembly of a positioned nucleosome near the hypersensitive region in simian virus 40 chromatin. J Mol Biol. 1991;221:795–803. doi: 10.1016/0022-2836(91)80176-u. [DOI] [PubMed] [Google Scholar]
- 56.Quivy J P, Becker P. The architecture of the heat-inducible Drosophila hsp27 promoter in nuclei. J Mol Biol. 1996;256:249–263. doi: 10.1006/jmbi.1996.0083. [DOI] [PubMed] [Google Scholar]
- 57.Ramsperger U, Stahl H. Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J. 1995;14:3215–3225. doi: 10.1002/j.1460-2075.1995.tb07324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richard-Foy H, Hager G L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rösl F, Waldeck W, Zentgraf H, Sauer G. Properties of intracellular bovine papillomavirus chromatin. J Virol. 1986;58:500–507. doi: 10.1128/jvi.58.2.500-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schild C, Claret F X, Wahli W, Wolffe A P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sera T, Wolffe A P. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus 5S rRNA genes. Mol Cell Biol. 1998;18:3668–3680. doi: 10.1128/mcb.18.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 63.Sheridan P L, Mayall T P, Verdin E, Jones K A. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 65.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 66.Spangenberg C, Eisfeld K, Stünkel W, Luger K, Flaus A, Richmond T J, Truss M, Beato M. The mouse mammary tumour virus promoter positioned on a tetramer of histones H3 and H4 binds nuclear factor 1 and OTF1. J Mol Biol. 1998;278:725–739. doi: 10.1006/jmbi.1998.1718. [DOI] [PubMed] [Google Scholar]
- 67.Studitsky V M, Kassavetis G A, Geiduschek E P, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 68.Stünkel W, Kober I, Kauer M, Taimor G, Seifart K H. Human TFIIIA alone is sufficient to prevent nucleosomal repression of a homologous 5S gene. Nucleic Acids Res. 1995;23:109–116. doi: 10.1093/nar/23.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stünkel W, Kober I, Seifart K H. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol. 1997;17:4397–4405. doi: 10.1128/mcb.17.8.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan S H, Bartsch D, Schwarz E, Bernard H U. Nuclear matrix attachment regions of human papillomavirus type 16 point toward conservation of these genomic elements in all genital papillomaviruses. J Virol. 1998;72:3610–3622. doi: 10.1128/jvi.72.5.3610-3622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan S H, Leong L E C, Walker P, Bernard H U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truss M, Bartsch J, Schelbert A, Hache R J G, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Truss M, Bartsch J, Mows C, Chavez S, Beato M. Chromatin structure of the MMTV promoter and its changes during hormonal induction. Cell Mol Neurobiol. 1996;16:85–101. doi: 10.1007/BF02088169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 75.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 76.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 77.Vayda M E, Rogers A E, Flint S J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983;11:441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waldeck W, Rösl F, Zentgraf H. Origin of replication in episomal bovine papilloma virus type 1 DNA isolated from transformed cells. EMBO J. 1984;3:2173–2178. doi: 10.1002/j.1460-2075.1984.tb02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 80.Wolffe A P. The role of transcription factors, chromatin structure, and DNA replication in 5s RNA gene regulation. J Cell Sci. 1994;107:2055–2063. doi: 10.1242/jcs.107.8.2055. [DOI] [PubMed] [Google Scholar]
- 81.Wolffe A P. Chromatin, structure and function. San Diego, Calif: Academic Press; 1995. [Google Scholar]
- 82.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 83.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y A. p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 85.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]