Abstract

Genetically encoded optical sensors and advancements in microscopy instrumentation and techniques have revolutionized the scientific toolbox available for probing complex biological processes such as release of specific neurotransmitters. Most genetically encoded optical sensors currently used are based on fluorescence and have been highly successful tools for single-cell imaging in superficial brain regions. However, there remains a need to develop new tools for reporting neuronal activity in vivo within deeper structures without the need for hardware such as lenses or fibers to be implanted within the brain. Our approach to this problem is to replace the fluorescent elements of the existing biosensors with bioluminescent elements. This eliminates the need of external light sources to illuminate the sensor, thus allowing deeper brain regions to be imaged noninvasively. Here, we report the development of the first genetically encoded neurotransmitter indicators based on bioluminescent light emission. These probes were optimized by high-throughput screening of linker libraries. The selected probes exhibit robust changes in light output in response to the extracellular presence of the excitatory neurotransmitter glutamate. We expect this new approach to neurotransmitter indicator design to enable the engineering of specific bioluminescent probes for multiple additional neurotransmitters in the future, ultimately allowing neuroscientists to monitor activity associated with a specific neurotransmitter as it relates to behavior in a variety of neuronal and psychiatric disorders, among many other applications.
Keywords: bioluminescence, bioluminescent, optogenetics, neurotransmitter, neuroimaging, optical sensors
Introduction
Optical biosensors have proven incredibly useful to researchers in a wide variety of fields for the study of neuronal and cellular activity.1,2 These probes generate changes in light emission intensity and/or wavelength in response to physiological events such as calcium influx, membrane voltage changes, or the presence of a ligand such as a neurotransmitter.3−7 Currently, nearly all available biosensors rely on fluorescence, leaving much room for improvement and further development of new approaches to better report changes in these cellular dynamics. One major improvement to such indicators would be to eliminate the need for an excitation light source, which is necessary to excite fluorescent reporters and is the limiting factor for imaging depth.
Bioluminescence is produced by an enzyme (luciferase) that catalyzes the oxidation of its specific substrate called luciferin, resulting in the emission of light. Different forms of biological light production exist in multiple domains in nature, including beetles, worms, bacteria, and marine organisms.8,9 Bioluminescence has been used for a variety of imaging applications, such as the quantification of gene expression over time and for imaging calcium dynamics or voltage to record neuronal function or to track other events within cells.10−14 Although bioluminescence has been used in research for decades, scientists are still rapidly improving luciferases and synthetic luciferins.15,16
Before genetically encoded indicators were available, the standard method to analyze specific neurotransmitters in vivo was to collect cerebrospinal fluid from within the brain via microdialysis or cyclic voltammetry, both of which offer poor time resolution (multiple minutes) and have decreasing performance in longitudinal studies. In the past several years, fluorescent genetically encoded neurotransmitter indicators such as iGluSnFR, dLight, and GRAB-DA6,7,17 have enabled the detection of neurotransmitters on a faster time scale. However, for recording activity in deep brain regions, for example, with fiber photometry, the implantation of an optical fiber into the brain is required to measure the output of these fluorescent probes. To circumvent the need of an illumination source, we are developing a series of bioluminescent Genetically Encoded Neurotransmitter Indicators (bGENIs) to provide neuroscientists with a set of tools that can be used in lieu of physical collection and fluorescence detection approaches. The goal is to overcome the depth limits associated with fluorescent imaging due to the excitation light not being able to penetrate the tissue to excite the fluorophore within deep brain regions without tissue heating and phototoxicity. There have been several recent achievements for deep brain imaging with bioluminescent sensors. Red-shifted bioluminescent calcium sensors have been used to image activity through the skull, 5 mm depth in stationary mice, and cortical activity imaging through the skull in freely behaving mice with off-the-shelf hardware without requiring an implant.18,19 These new red-shifted luciferase–luciferin pairs that enable noninvasive deep brain imaging can be utilized for future biosensor engineering.
We have made significant progress in developing a BioLuminescent Indicator of the Neurotransmitter Glutamate (BLING; Figure 1). Since glutamatergic neurotransmission is directly implicated in behavior, movement, mental health, pain perception, and addiction, BLING can potentially be used in a variety of drug discovery applications and used to record glutamatergic activity in experimental animal models. In the future, the design and engineering approach we describe here can be expanded to encompass other neurotransmitters and small-molecule analytes as has been done with fluorescent neurotransmitter sensors in recent years by substituting various analyte binding domains.5,20,21 Moreover, we expect BLING can be adapted to act on light-sensitive proteins (optogenetic actuators) for BioLuminescent-OptoGenetics (BL-OG).22
Figure 1.

Schematic of the BioLuminescent Indicator of the Neurotransmitter Glutamate (BLING) with a split luciferase (blue) and glutamate sensing domain (tan) displayed on the cell surface.
Results
In the first step, we created and tested 3 versions of BLING constructs based on the truncated periplasmic glutamate binding protein (Glt1) from the FRET-based glutamate sensor SuperGluSnFr23 flanked by short flexible linkers with a split luciferase “half” on each terminal. Initially, we chose various marine luciferases that have been engineered into ultrabright variants, do not require a cofactor to produce light, and have previously validated split sites. We created BLING 0.1 consisting of the Gaussia luciferase (GLuc) variant M43L, M110L, referred to as “Slow Burn GLuc,” which is brighter than native GLuc and has glow kinetics instead of flash kinetics, i.e., sustained light emission instead of rapid decay in light emission.24,25 We used the split site 105–106 previously used to successfully engineer calcium indicators,26 including the 17 AA native secretion signal from GLuc for surface display with a PDGFRβ membrane anchor. BLING 0.2 was created similarly using NanoLuc split at 66–67, which has been used to successfully engineer calcium indicators,27 with an Igk leader sequence for cell surface display. BLING 0.3 was created with NanoLuc large and small bits split at 159–160, which has also previously been used to generate calcium indicators.28 All initial BLING variants produced bioluminescence with their respective optimal luciferin, native CTZ for Gluc or h-CTZ and Furimazine for Nluc, retained the kinetic characteristics of their luciferase, with BLING 0.2, NanoLuc split at 66–67, having the largest response to glutamate and being the brightest sensor by multiple orders of magnitude (Figure 2A,C).
Figure 2.
(A) Protein maps of the three initial BLING designs that were tested. From top to bottom: slow burn Gaussia luciferase split at 105–106 including the native Gaussia secretion peptide on the N terminal instead of the IGK leader secretion sequence; Nanoluc split at 66–67, Nanoluc split at 159–160 all with Glt1 glutamate binding protein and PDGFRβ transmembrane domain to anchor the sensor to the extracellular side of the membrane. (B) Protein map of BLING 1.0 with variable linkers. (C) Results from the initial BLING constructs in response to 1 mM glutamate; n = 4. (D) Responses from the BLING linker library of ∼400 variants of BLING 0.2 with variable linkers tested, X axis is the rank order of variants.
Following the testing of our initial 3 variants, we aimed to improve the top-performing BLING design through linker optimization. BLING 0.2 was selected for further engineering, and the linkers were replaced with 3 amino acid variable linkers that code for A, S, or P at each position (Figure 2B and Supporting File 1). This combination of linker variants was used because they have been shown to produce diverse functionalities due to varying levels of rigidity/flexibility while significantly limiting the number of clones that need to be screened.28 From this library, the top-performing variant was selected for further characterization (Figure 2D). As a result of linker optimization, we were able to generate a BLING variant (BLING 1.0, Addgene plasmid: 171647) that has a robust response to glutamate addition when expressed in mammalian cells (Figure 3A). BLING 1.0, derived from BLING 0.2, consistently outperforms the parental construct by 2-fold in terms of response to glutamate while maintaining its brightness. We next sought to determine the dose-dependent response of BLING 1.0 when expressed in mammalian cells using a photon counting plate reader (Tecan Spark). The responses to background levels of glutamate were minimal, and we observed a 10.8% change in response to 10 μM with a 3-fold greater response to 100 μM with a 31.8% change in luminescence (Figure 3B).
Figure 3.
(A) Example responses of BLING 1.0 to 1 mM glutamate addition recorded with a plate reader from a single well of a 96-well plate of HEK cells expressing BLING 1.0. (B) Dose-dependent response of BLING when using a plate reader with concentrations ranging from 0.1 μM to 10 mM. n = 3.
Since BLING 1.0 successfully reports changes in extracellular glutamate levels when recorded on the population level, we next sought to determine if changes in extracellular glutamate can be observed at the single-cell level. For this, we used live-cell bioluminescence microscopy using HEK cells expressing BLING and perfusing varying concentrations of glutamate into the cell imaging chamber. We found BLING 1.0 to report changes in extracellular glutamate with responses up to 310% at the single-cell level to 1 mM glutamate, average response of 109.6% (Figure 4A,C and Supporting Movie). We also determined that this sensor reports glutamate in a dose-dependent manner, responding to physiological levels of glutamate and outperforming BLING 0.2 (Figure 4B). Furimazine was used for live-cell imaging because it provides a more stable, consistent signal than h-CTZ, which was used for the high-throughput screening assays.
Figure 4.
(A) Example trace of a single-cell ROI showing the perfusion of Furimazine starting at 60 s and the infusion of 1 mM glutamate (final concentration) at 120 s. (B) Dose responses of BLING 0.2 and the improved variant BLING 1.0, comparing responses of single cells to different concentrations of glutamate. N = 4 for BLING 0.2, N = 8 for BLING 1.0. (C) Image of background bioluminescence of BLING 1.0 and with 1 mM glutamate. * = p < 0.05, Two-way analysis of variance (ANOVA), Bonferroni post hoc test.
We next sought to determine if BLING 1.0 can successfully report changes in glutamatergic activity in vivo with an acute seizure model induced by local bicuculine injection. BLING 1.0 was expressed with AAV in the sensory cortex 2 mm below the surface of the skull (Figure 5A). For imaging, h-CTZ was delivered intraperitoneally, and rats were imaged with an IVIS Spectrum. In this preliminary proof-of-concept experiment, we were able to detect increases in luminescence following seizure induction in three out of six rats (Figure 5B,C,D) with an increase in luminescence over 100% and five out of six with a measurable increase in luminescence. Seizure activity was visible (muscle twitching) in three of the rats with detectable increases in light emission and not noted in the three with lower changes in light emission.
Figure 5.
(A) Schematic of the experimental design for expression of BLING 1.0 (blue circle) 2 mm deep within the sensory cortex, with bicuculline injected locally to induce an acute seizure. (B) Example images of bioluminescence before and after seizure induction. (C) Example trace of bioluminescent intensity in response to acute seizure induction. (D) Intensity changes following Bicuculline injection.
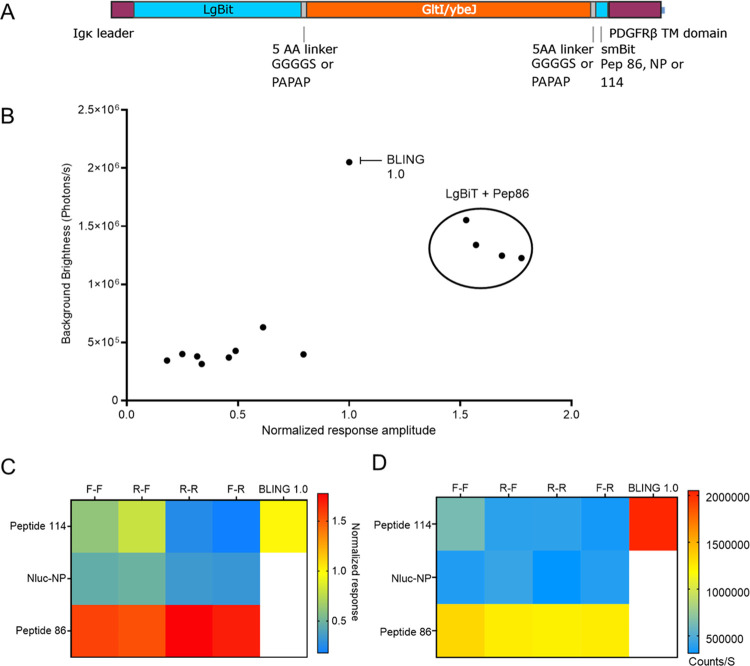
In an effort to continue improving the BLING family of sensors, we sought to create variants with lower levels of background luminescence and greater responses to glutamate. For this, we created and tested BLING variants containing either flexible or rigid linkers in combination with three different variants of the small C terminal fragment, SmBiT with the large N terminal fragment that was evolved from NanoLuc for improved protein stability, termed 11s29 to create 12 new variants. This approach was based on our results from the randomized linker screening used to generate BLING 1.0 where we discovered that the top five variants generated all contained similar linker compositions with the first linker being composed of amino acids with flexible properties and the second linker containing primarily prolines with rigid properties. We also tested three C terminal peptide variants of NanoLuc in combination with the flexible or rigid linkers which have varied affinities to the N terminal of the luciferase ranging from ∼1 nM to ∼200 μM. We found that the variant described in Dixon et al., which has the highest affinity (peptide 86), resulted in four BLING variants that all responded to glutamate with 1.5–2 times the delta as BLING 1.0 with 1 mM glutamate while having lower background light emission prior to glutamate addition with minimal effect from linker composition. We found that BLINGs created with the other two peptides, the native NanoLuc peptide and peptide 114, the medium- and lowest-affinity variants, all had lower responses to glutamate presentation but were more affected by the linker composition (Figure 6). BLINGs containing the peptide 114 with either flexible–flexible or rigid–flexible linkers compositions had 0.61 and 0.79 times the response of BLING 1.0, while variants containing native peptide with flexible–flexible or rigid–flexible linkers had 0.46 and 0.49 times the response and much lower background luminescence. Variants containing the native NanoLuc peptide or peptide 114 with rigid–rigid or rigid–flexible linker compositions all performed poorly. As can be seen in Figure 6, we created a new sub-family of BLINGs that is based on LgBiT and SmBiT variants with lower luminescence background yet display a greater increase in luminescence upon glutamate binding. Some of these BLINGs may prove more useful for imaging as they are still very bright and have greater responses while some variants may not have any use for imaging given that they are much dimmer but could be useful as optogenetic actuators since they may be able to avoid activation of light-sensitive elements in their off state. Taken together, we have described here a new family of synthetic biosensors that allow glutamate visualization both in vitro and in vivo and have created a diverse subset of sensors that can be used as new starting points for further evolution and since these sensors produce their own light, they may have applications beyond neurotransmitter sensing.
Figure 6.
(A) Protein map of the 12 new BLING variants that were tested that contain the 11s variant of NanoLuc called LgBiT for the N terminal half of the luciferase with either a flexible or rigid linker flanking the Glt1 glutamate binding protein and three C terminal variants with varying affinities to LgBiT Peptide 86, native peptide or pep 114 and PDFGRb transmembrane domain to anchor the sensor to the extracellular side of the membrane. (B) Scatter plot of the diversity of new BLING constructs response to glutamate normalized to BLING 1.0 vs baseline brightness for each combination of the three different C terminal peptides and combinations of flexible or rigid linkers. (C) Heatmap of the NanoBiT-based BLINGS response normalized to BLING 1.0. (D) Heatmap of background brightness of NanoBiT-based BLINGs.
One such application is the utilization of BLING as a switch for activating gene circuits. By producing light upon substrate administration, the BLING can activate opsins for downstream signaling and consequently avoid crosstalk with cellular signaling pathways. Thus, we tested the response of one of the new variants, consisting of LgBiT (S11), rigid linker, Glt1, rigid linker, pep 86, termed BLING 1.1, which had the largest response to glutamate (Figure 6C), changes in glutamate in three cell lines by measuring luciferase activity levels before and after addition of glutamate in three mammalian cell lines: HeLa, Human cervical cancer; MDA-MB-231, human breast cancer, and 9L rat glioblastoma. For all experimental groups, the addition of glutamate to a final concentration of 1 mM resulted in a rapid increase in brightness, with values ranging between 37 and 100% depending on the cell line (Figure 7A,B). These results emphasize that due to the fast kinetics of BLING, it can serve as a putative switch of light-sensitive gene circuits in a large array of cells. We also tested this new BLING variant in primary hippocampal cultures for response to a train of four electrical field simulations and found that it successfully reports synaptically released glutamate in cultured neurons (Figure 7C,D).
Figure 7.
Activation of BLING 1.1 upon addition of glutamate in three cell lines: HeLa, MDA, and 9L. (A) Kinetic behavior of BLING 1.1 before and after glutamate stimulation. Increases in extracellular glutamate induce a conformational change that restores the enzymatic activity of the split luciferase, resulting in a rapid increase in brightness. (B) Percentage increase in brightness following glutamate addition. (C) Hippocampal neuronal culture expressing BLING 1.1 (5× mag) before and after four trains of electrical stimulation pseudo-colored as a heatmap to show change in luminescence. (D) Change in light emission of the full field of view in counts per second following four trains of electrical field stimulation (vertical black lines).
Discussion
Here, we present the first genetically encoded bioluminescent neurotransmitter indicator, which reports changes in extracellular glutamate via changes in luminescence intensity. This was achieved by building on previous work done to engineer various fluorescent glutamate indicators based on the same periplasmic glutamate binding protein (Glt1), SuperGluSnFR, and iGluSnFR.7,23 In this study, we replaced the two fluorescent proteins from the FRET-based SuperGluSnFR with N and C terminal fragments of various marine luciferases. These indicators work well to report changes in extracellular glutamate when expressed in cultured cells. This new tool opens the possibility for high-throughput drug screening in cells as our bioluminescent indicator is highly sensitive when used in a plate reader compared to fluorescent indicators, which generally do not perform as well when recorded at the level of whole cell cultures, i.e., in bulk measurements.
Most importantly, we anticipate that BLING will perform well for a variety of molecular imaging applications in deep brain structures and have already successfully used BLING 1.0 to report changes in brain activity while imaging through significantly more material than what can be accomplished with fluorescent techniques. In our case, we report seizure through 2 mm of the brain and the rat skull with a similar success rate as described by Tian et al. with a red-shifted calcium sensor. We also believe we can significantly improve on these in vivo results when using more sophisticated imaging hardware such as bioluminescent mini scopes that can be used for recording activity in freely behaving animals30 and adopting red-shifted luciferases to increase imaging depths. This class of sensors will immediately benefit ongoing research efforts to study the mechanisms that give rise to a wide array of neuronal and psychiatric disorders and provide researchers with significantly improved approaches to study neuronal activity at the level of the cell, network, and behaving animals longitudinally. Furthermore, since these reporters produce their own light, they can potentially be used as activators for light-sensitive proteins to carry out a variety of downstream functions within a cell. For example, sensors such as BLING can replace the intact luciferases in current BioLuminescent-OptoGenetic (BL-OG) constructs22,25,31 so the light-sensitive ion channels open in response to glutamate resulting in activity-dependent excitation or inhibition. Additionally, now that we have created BLING variants with diverse properties such as varying levels of background luminescence and responses to their neurotransmitter (Figure 6) we can create a variety of neuromodulators based on these sensors tailored to specific applications. These can then be used to improve our prior and ongoing work in neurodegenerative disorders such as spinal cord injury and Parkinson’s disease by allowing the noninvasive current stimulation of neurons to be dependent on endogenous activity.22,25,32−34 These new BLINGs also present ample opportunity as multiple starting points for further evolution to create new BLINGs with greater responses to glutamate and lower background. Specifically improving BLINGs to have higher affinity to glutamate, in the 100–500 micromolar range to respond to fluctuations in glutamate on the lower end of the physiological spectrum and by improving membrane trafficking and trafficking the sensor specifically to the synaptic cleft as was recently done with fluorescent glutamate sensors35
In conclusion, we were able to successfully engineer a bioluminescent indicator for the neurotransmitter glutamate by adapting sensing domains and split luciferases that have previously been used with success for fluorescent glutamate sensors and bioluminescent calcium sensors. The most optimized BL glutamate sensors we report are already capable of reporting changes in extracellular glutamate and will serve as excellent starting points for engineering derivatives with even higher brightness, dynamic range, and for other neurotransmitters. We expect that this indicator will be useful for imaging brain activity within deep brain regions and that we can use this approach to engineer a variety of other neurotransmitter sensors. We also expect this line of neurotransmitter sensors to be adaptable for a variety of highly selective optogenetic actuators that are dependent on a specific neurotransmitter.
Methods
Sensor Design
The initial three BLING variants were constructed using Glt1, IgK leader, and PDFGRβ sequences described in ref (23) (GenBank EU42295) synthesized as a Gblock or oligos used for PCR and assembled into a pcDNA 3.1 vector using Gibson Assembly (NEB HiFi) with their respective luciferase fragments. BLING 0.1 consisting of the sbGluc using the split site 105–106,26 including the 17 AA native secretion signal. BLING 0.2 consisting of NanoLuc split at 66–67,27 with an Igk leader sequence for cell surface display. BLING 0.3 was created with NanoLuc large and small bits, split at 159–160.28,29 (Figure 2A and Supporting Files 2–4). The additional BLING variants were cloned from synthetic fragments encoding the 11s variant of NanoLuc and the three affinity variants of the C terminal peptide: peptide 86, peptide 114, and native peptide described in ref (29). HEK cells plated on poly-d-lysine-coated white 96-well plates grown to 50–70% confluency were transfected with 0.5 μL of lipofectamine 2000 per mL Opti MEM with 100 ng DNA per well using 20 μL of the transfection mix per well. Initial testing was done using 5 μM hCTZ (Nanolight Technologies #301) in FluoroBrite media (Thermo), media changed 15 min prior to reading to allow the reaction to stabilize, and plates read using a BioTek Cytation 5 or Tecan Spark, injecting 10 μL of 20 mM glutamate stock into 190 μL media for a final concentration of 1 mM.
Library Construction and Screening
Assembly products were digested with DpnI to eliminate any plasmid template material, eliminating background colonies and electroporated into Top10 cells (Thermo). Colonies were then grown in deep 96-well plates in 1.5 mL of LB media overnight and mini-prepped in 96-well format (Biobasic #B814152-0005). Each plasmid was transfected into HEK cells grown in poly-d-lysine-coated white 384-well plates in quadruplicate to generate an average for each variant tested. A transfection master mix was prepared with 21 mL of Opti MEM with 100 μL of lipofectamine 2000, distributed into four 96-well PCR plates, 50 μL per well and 5 μL DNA from the 96-well mini-preps was added to the transfection master mix with mini-prep DNA yields ranging from 100 to 200 ng/μL. Testing was done two days later using 5 μM hCTZ in FluoroBrite media, changed 15 min prior to reading and plates read using a BioTek Cytation 5, injecting 5 μL of 20 mM glutamate stock into 95 μL of media for a final concentration of 1 mM.
HeLa and MDA-MB-231 cell lines were cultured using Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) with penicillin–streptomycin antibiotics at 37 °C under 5% CO2. 9 L cells were grown using DMEM containing 10% FBS and geneticin (G418 Sulfate) antibiotic at the same temperature and CO2 conditions.
Characterization
The top BLING variant from the linker library was termed BLING 1.0 (Addgene plasmid: 171647) and further characterized and compared to the parent construct, BLING 0.2. HEK cells plated on white 96-well plates, grown to 50–70% confluency were transfected with 0.5 μL of lipofectamine 2000 per mL of Opti MEM with 100 ng DNA diluted in 0.5 mL of lipofectamine mix and 20 μL of the transfection mix used per well. Measurements were taken with 5 μM hCTZ in FluoroBrite media, changed 15 min prior to reading on a Tecan spark for bioluminescence. The concentration-dependent experiment was done in Hank’s balanced salt solution (HBSS) with magnesium and calcium with 10 mM HEPES buffer and 1 μM hCTZ as we found these conditions to provide less variability in measurements.
For microscopy experiments, HEK cells were seeded in 12-well plates at 8 × 105 per well, transfected the following day using 4 μL of lipofectamine 2000 in 100 μL of Opti MEM and 2 μg DNA in 100 mL of Opti MEM, incubated overnight, trypsinized (TrypLE, Thermo), and plated on poly-d-lysine-coated 18 mm coverslips (NeuVitro), and imaged one to three days later. Imaging was done using a Zeiss A1 Axioscope, 5 × 0.17 NA objective, Andor iXon 888 EMCCD camera, EM gain of 600, 4 × 4 binning with an open optical path and microscope within a dark box. Imaging was done in a perfusion chamber with artificial cerebral spinal fluid (ACSF) as described in ref (31) using 1 μM furimazine (Promega) and heated to 37 °C. ACSF was continually perfused followed by ACSF with furimazine and the respective concentration of glutamate followed by a washout with ACSF. The same ROI was used for all concentrations, and statistical analysis was done using a repeated-measures two-way ANOVA with Bonferroni post hoc, n = 8 per group. Images were analyzed using ImageJ for background subtraction and despeckling to reduce noise, and ROIs were selected manually for quantification. Experiments in primary neuron cultures were carried out following the same procedure. Hippocampal neurons were purchased from Brain Bits and cultured according to their protocol. Imaging was carried out three weeks later. Electrical stimulation was a 1 s long train at 100 Hz, 2.45 V with a square waveform.
All animal work was approved by Michigan State University’s Institutional Animal Care and Use Committee. BLING 1.0 was cloned into an AAV expression vector with a synapsin promoter for expression in neurons and AAV was made and purified in-house as previously described.33 Sprague Dawley rats were injected with 2 μL of 0.5 × 1012 copies/mL at a rate of 0.5 μL per minute with a 33G world precision syringe at 0 mm AP 3.5 mm R, and 2 mm ventral of the cortical surface and the needle was left in place for 5 min after infusion and slowly retracted. Imaging was done at least three weeks later with an IVIS spectrum (PerkinElmer). Animals were injected with water-soluble in vivo h-CTZ (Nanolight Technologies) 2 mg/kg intraperitoneally 1–2 h prior to imaging and checked for the onset of bioluminescence. Once bioluminescence was detectable, the incision was reopened and a custom-made cannula was inserted through the small burr hole from the prior surgery for bicuculline administration. Then, a baseline recording of 10 min was acquired and 10 μL of 10 mM bicuculline was slowly injected over 5 min. Images were captured using large binning, fstop of 1, with 1 min exposures. Images were analyzed with Living Image software with an ROI over the entire skull.
Acknowledgments
Figures contain material created with BioRender. A.A.G. acknowledges financial support from the NSF 2027113, NIH/NINDS: R01-NS098231; R01-NS104306 NIH/NIBIB: R01 EB031008; R01EB030565; R01EB031936 and P41-EB024495. E.D.P. was supported by the PerkinElmer Postdoctoral Fellowship.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00687.
Author Present Address
# College of Medicine, Central Michigan University. MI, USA
Author Contributions
E.D.P. and A.A.G. designed the experiments and wrote the paper. E.D.P., A.P.L., E.A.F.C., A.J.F., E.L.C., G.G.L., C.J.G., Z.A.T., and R.O. carried out experiments and analysis. A.A.G., U.H., and N.C.S. supervised the project.
The authors declare the following competing financial interest(s): E.D.P. has a patent pending for bioluminescent neurotransmitter sensors. The authors have no other competing financial interest.
Supplementary Material
References
- Lu R.; Liang Y.; Meng G.; et al. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat. Methods 2020, 17, 291–294. 10.1038/s41592-020-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Zou P.; Cohen A. E. Voltage imaging with genetically encoded indicators. Curr. Opin. Chem. Biol. 2017, 39, 1–10. 10.1016/j.cbpa.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland S.; Yang H. H.; Pan M. M.; et al. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 2017, 6, e25690 10.7554/eLife.25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F.; Marshall J. D.; Yang Y.; et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014, 17, 884–889. 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.; Hires S. A.; Mao T.; et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 2009, 6, 875–881. 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T.; Cho J. R.; Merten K.; et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 2018, 360, eaat4422 10.1126/science.aat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin J. S.; Borghuis B. G.; Tian L.; et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 2013, 10, 162–170. 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.; Hastings J. W. BIOLUMINESCENCE. Annu. Rev. Cell Dev. Biol. 1998, 14, 197–230. 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- Martini S.; Haddock S. H. D. Quantification of bioluminescence from the surface to the deep sea demonstrates its predominance as an ecological trait. Sci. Rep. 2017, 7, 45750 10.1038/srep45750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora M.; Giorgi C.; Bononi A.; et al. Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat. Protoc. 2013, 8, 2105–2118. 10.1038/nprot.2013.127. [DOI] [PubMed] [Google Scholar]
- Granatiero V.; Patron M.; Tosatto A.; Merli G.; Rizzuto R. Using targeted variants of aequorin to measure Ca2+ levels in intracellular organelles. Cold Spring Harbor Protoc. 2014, 2014, pdb.prot072843 10.1101/pdb.prot072843. [DOI] [PubMed] [Google Scholar]
- Thomou T.; Mori M. A.; Dreyfuss J. M.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S.; Tsutsui H.; Suzuki K.; et al. Genetically encoded bioluminescent voltage indicator for multi-purpose use in wide range of bioimaging. Sci. Rep. 2017, 7, 42398 10.1038/srep42398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P.; Griffin N. M.; Thakur D.; et al. An Autonomous Molecular Bioluminescent Reporter (AMBER) for Voltage Imaging in Freely Moving Animals. Adv. Biol. 2021, 5, 2100842 10.1002/adbi.202100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano S.; Sugiyama M.; Hama H.; et al. Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939. 10.1126/science.aaq1067. [DOI] [PubMed] [Google Scholar]
- Abe M.; Nishihara R.; Ikeda Y.; et al. Near-Infrared Bioluminescence Imaging with a through-Bond Energy Transfer Cassette. ChemBioChem 2019, 20, 1919–1923. 10.1002/cbic.201900149. [DOI] [PubMed] [Google Scholar]
- Sun F.; Zeng J.; Jing M.; et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 2018, 174, 481–496. 10.1016/j.cell.2018.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Zhang Y.; Li X.; et al. A luciferase prosubstrate and a red bioluminescent calcium indicator for imaging neuronal activity in mice. Nat. Commun. 2022, 13, 3967 10.1038/s41467-022-31673-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.; Walker J. R.; Hall M. P.; et al. An optimized bioluminescent substrate for non-invasive imaging in the brain. Nat. Chem. Biol. 2023, 19, 731–739. 10.1038/s41589-023-01265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin J. S.; Shimoda Y.; Magloire V.; et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 2019, 16, 763–770. 10.1038/s41592-019-0471-2. [DOI] [PubMed] [Google Scholar]
- Borden P. M.et al. A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies bioRxiv, 2020 10.1101/2020.02.07.939504. [DOI]
- Ikefuama E. C.; Kendziorski G. E.; Anderson K.; et al. Improved Locomotor Recovery in a Rat Model of Spinal Cord Injury by BioLuminescent-OptoGenetic (BL-OG) Stimulation with an Enhanced Luminopsin. Int. J. Mol. Sci. 2022, 23, 12994 10.3390/ijms232112994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hires S. A.; Zhu Y.; Tsien R. Y. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 4411–4416. 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J. P.; Patel K. G.; Manthiram K.; Swartz J. R. Multiply mutated Gaussia luciferases provide prolonged and intense bioluminescence. Biochem. Biophys. Res. Commun. 2009, 389, 563–568. 10.1016/j.bbrc.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Park S. Y.; Song S.; Palmateer B.; et al. Novel luciferase-opsin combinations for improved luminopsins. J. Neurosci. Res. 2020, 98, 410–421. 10.1002/jnr.24152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. B.; Sato M.; Tao H. Circularly Permutated Bioluminescent Probes for Illuminating Ligand-Activated Protein Dynamics. Bioconjugate Chem. 2008, 19, 2480–2486. 10.1021/bc800378a. [DOI] [PubMed] [Google Scholar]
- Suzuki K.; Kimura T.; Shinoda H.; et al. Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat. Commun. 2016, 7, 13718 10.1038/ncomms13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana I.; Hossain M. N.; Suzuki K.; Matsuda T.; Nagai T. Genetically Encoded Fluorescence/Bioluminescence Bimodal Indicators for Ca2+ Imaging. ACS Sens. 2019, 4, 1825–1834. 10.1021/acssensors.9b00531. [DOI] [PubMed] [Google Scholar]
- Dixon A. S.; Schwinn M. K.; Hall M. P.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Celinskis D.; Friedman N.; Koksharov M.. et al. Miniaturized Devices for Bioluminescence Imaging in Freely Behaving Animals. In 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society; IEEE, 2020; pp 4385–4389. [DOI] [PubMed]
- Berglund K.; Clissold K.; Li H. E.; et al. Luminopsins integrate opto- and chemogenetics by using physical and biological light sources for opsin activation. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E358 10.1073/pnas.1510899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenchak J. R.; Palmateer B.; Dorka N.; et al. Bioluminescence-driven optogenetic activation of transplanted neural precursor cells improves motor deficits in a Parkinson’s disease mouse model. J. Neurosci. Res. 2020, 98, 458–468. 10.1002/jnr.24237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E. D.; Sharkey E. D.; Pal A.; et al. Restoring Function After Severe Spinal Cord Injury Through BioLuminescent-OptoGenetics. Front. Neurol. 2022, 12, 792643 10.3389/fneur.2021.792643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. A.; Whitehead B. J.; Petersen E. D.; et al. Behavioral context improves optogenetic stimulation of transplanted dopaminergic cells in unilateral 6-OHDA rats. Behav. Brain Res. 2023, 441, 114279 10.1016/j.bbr.2022.114279. [DOI] [PubMed] [Google Scholar]
- Aggarwal A. Glutamate indicators with improved activation kinetics and localization for imaging synaptic transmission. Nat. Methods 2023, 20, 925–934. 10.1101/2022.02.13.480251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.