Abstract
Computer-aided Diagnosis (CADx) based on explainable artificial intelligence (XAI) can gain the trust of radiologists and effectively improve diagnosis accuracy and consultation efficiency. This paper proposes BI-RADS-Net-V2, a novel machine learning approach for fully automatic breast cancer diagnosis in ultrasound images. The BI-RADS-Net-V2 can accurately distinguish malignant tumors from benign ones and provides both semantic and quantitative explanations. The explanations are provided in terms of clinically proven morphological features used by clinicians for diagnosis and reporting mass findings, i.e., Breast Imaging Reporting and Data System (BI-RADS). The experiments on 1,192 Breast Ultrasound (BUS) images indicate that the proposed method improves the diagnosis accuracy by taking full advantage of the medical knowledge in BI-RADS while providing both semantic and quantitative explanations for the decision.
Keywords: Breast cancer, computer-aided diagnosis (CADx), explainable artificial intelligence, multi-task learning
I. INTRODUCTION
The Breast cancer is the most common cancer in women and causes the second-highest number of deaths among all cancers [1]. Early discovery and treatment can prevent breast cancer from becoming severe and significantly increase the survival rate [2]. Breast ultrasound is a highly effective imaging method for diagnosing breast cancer. It is non-invasive, painless, and does not involve exposure to radiation [3].
Artificial Intelligence (AI) technology is rapidly advancing, and many people anticipate that Computer-Aided Diagnosis (CADx) systems will have a significant impact on diagnosing breast cancer using ultrasound, particularly in areas with a shortage of medical resources [4]. In recent years, CADx systems have demonstrated competitive or even superior performance compared to human physicians [5] while providing increased reproducibility [6]. However, the widespread acceptance of CADx systems for sonography has been limited by the lack of transparency and explainability in these systems [7].
For breast cancer, the consequences of diagnostic errors can be severe [8], with delayed or missed diagnoses potentially delaying treatment and endangering patients’ lives. Conversely, a misdiagnosis can result in heavy emotional and financial burdens on patients. Therefore, both physicians and patients require an understanding of the internal mechanism and decision-making process of the CADx system before accepting a diagnosis.
As the importance of transparency and explainability in CADx systems has been increasingly recognized, researchers have developed methods to make these systems more explainable [9], [10], [11]. These methods can be broadly categorized into two groups. The first group introduces explainable or trackable components in the model, which are used to make decisions [11], [12], [13]. However, these methods often involve trade-offs between performance and explainability. Additionally, some methods [9], [14], [15] generate visual explanations based on attention mechanisms, but these explanations are not always well-accepted by physicians [16], [17]. The second group of methods use post-hoc analysis to interpret existing models [18] and have no impact on the performance, but these explanations may be difficult for patients and clinicians to understand, and further research is needed to connect them to medical knowledge.
This paper presents a novel network architecture called BI-RADS-Net-V2 for identifying breast cancer in ultrasound images. The system consists of three key components: a core classifier that predicts the type of mass (benign or malignant), a multi-branched network that functions as a semantic explainer by predicting Breast Imaging-Reporting and Data System (BI-RADS) descriptors (as detailed in Section II-A), and a quantitative explainer that approximates the classifier’s decision by combining the BI-RADS descriptors and providing clear explanations. The proposed model offers several advantages. By providing semantic explanations based on BI-RADS, the output can be easily understood and accepted by physicians and radiologists, as the BI-RADS descriptors are based on morphological features they use daily. The multi-task learning framework allows for medical knowledge in BI-RADS to enhance the classifier’s generalization ability, leading to better performance than single-task models. Additionally, the quantitative explainer provides insights into the inner workings of the classifier for each sample, allowing for a clearer understanding of the importance of different BI-RADS descriptors in the diagnostic process.
The main contributions of this paper are as follows.
A complete CAD system that concurrently outputs the tumor class, BI-RADS likelihood of malignancy, the BI-RADS descriptors, and the contributions of each descriptor.
A network architecture with a regression branch to handle the inherent noise in the ground-truth labels for the BI-RADS categories caused by inter-observer variability.
Increased tumor classification accuracy via learning feature representations related to clinical descriptors; and
The capacity to assess uncertainties in the model outputs for individual BUS images based on (dis)agreement in the predictions by the different model branches.
The remaining content of this paper is organized as follows. Section II reviews the current CAD for BUS images and XAI research. Section III describes our BIRADS-Net model. Section IV presents the experimental results on a combined dataset and analyzes the results; and finally, section V summarizes the paper and discusses future work.
II. RELATED WORK
A. BI-RADS
BI-RADS is a risk assessment system that standardizes the assessment, reporting, and training for breast imaging diagnosis. Published and trademarked by the American College of Radiology, BI-RADS has played an essential role in breast cancer diagnosis and reporting worldwide. The system applies to ultrasound, mammography, and MRI. BI-RADS summarizes a mass finding for breast ultrasound by one of the seven assessment categories (see Table 1). Except for categories 0 (incomplete) and 6 (biopsy-proven malignancy), the other categories correspond to the different odds of malignancy. Category 1 (no mass detected), 2 (benign), and 3 (risk malignancy 0-2%) indicate a meager chance of cancer, and category 5 indicates an extremely high risk of cancer. Category 4 is divided into three subcategories. Category 4a indicates mass finding with low risk from 2% to 10%, and category 4b indicates intermediate risk, 10% to 50%. Category 4c indicates moderate risk (50% to 95%) of malignancy. Clinically, follow-up is usually recommended for categories 3 and 4a, while categories 4c and 5 usually require biopsy examination.
TABLE 1.
The categories of BI-RADS assessments.
| Category | Assessment | Likelihood of Malignancy | Management |
|---|---|---|---|
| 0 | Incomplete | NA | Additional imaging required |
| 1 | Negative | No cancer detected | Annual screening |
| 2 | Benign | 0% | Annual screening |
| 3 | Probably benign | 0-2% | Follow-up in 6 months |
| 4A | Suspicious | 2-10% | Tissue diagnosis |
| 4B | Suspicious | 10-50% | Tissue diagnosis |
| 4C | Suspicious | 50-95% | Tissue diagnosis |
| 5 | Malignant | 95% | Tissue diagnosis |
| 6 | Biopsy-proven malignancy | Cancer present | Surgical excision |
BI-RADS standardizes diagnosis through pre-defined representative descriptors. The pre-defined breast sonographic image lexicon includes six morphologic features of solid mass findings: shape, orientation, margin, boundary, internal echo pattern, and posterior acoustic features. According to the BI-RADS lexicon, the shape of a mass could be oval, round, or irregular; the orientation could be parallel or not parallel to the skin; the margin features of a mass include circumscribed, microlobulated, indistinct, angular, and spiculated; the echo pattern inside the mass could be anechoic, hyperechoic, isoechoic, hypoechoic, and complex; the boundary features include abrupt interface and echogenic halo; the posterior acoustic features include shadowing, combined, enhancement, and no posterior acoustic features. The BI-RADS lexicon covers the most critical breast ultrasound image features for diagnosis. Some of these features could effectively identify benign mass from malignant mass and are accepted by doctors worldwide.
There are apparent advantages in building a diagnosis system based on a proven and effective knowledge system. Doctors and radiologists use BI-RADS daily for diagnosis. Automated diagnostic systems based on BI-RADS are more similar to the diagnostic thinking of physicians, and end-users can easily understand the BI-RADS-based explanations given by the system.
The descriptors in the BI-RADS lexicon are highly discriminative, and the medical knowledge in them can effectively improve the accuracy and generalization ability of the system. Based on the above reasons, we introduce BI-RADS into the CAD system and explain the system decisions based on BI-RADS.
B. EXPLAINABLE ARTIFICIAL INTELLIGENCE
As modern machine learning techniques achieve extraordinary success in a growing number of fields, the short-comings of machine learning algorithms, especially deep network models, in terms of lack of transparency and interpretability, are increasingly drawing the attention of researchers [19], [20]. XAI is becoming a popular research area in recent years. Došilović et al. [21] categorize the approaches to transparency and explainability into integrated and post-hoc methods. The former uses transparent, human-understandable information to construct models that can effectively explain the decision-making process. However, the usage of these models is associated with a trade-off between transparency and performance [22], [23]. The post-hoc methods extract information from existing models without impacting performance. Due to sample space and methodological limitations, there is a risk that these methods may not fully reflect the characteristics of the model and may produce misleading interpretations.
The requirements for interpretability vary by application type. Samek et al. [24] divided the interpretation methods into explaining learned representations [10], [25], [26], [27], [28], [29], explaining individual predictions [30], [31], [32], [33], explaining model behavior [34], and explaining with representative examples [18], [35]. The widespread use of neural network models has contributed significantly to developing the first class of methods. However, due to the complexity of modern machine learning methods, the effective explanation is still an open problem.
C. EXPLAINABLE ARTIFICIAL INTELLIGENCE IN HEALTHCARE
High-stakes applications such as medical image diagnosis require more explainability than general applications. The majority of the current work on XAI in medical image diagnosis employed model saliency to outline important regions in images that contributed the most to the model prediction [36], [37]. Accordingly, the attention mechanism in the neural network model is also used to label organs and tissues to be focused on from medical images [38]. Explainable models based on saliency or attention have a certain degree of explanatory power. However, there are still some limitations to the clinical meaning of these explanations and their acceptance [16], [17], [39].
Based on the Thyroid Imaging Reporting and Data System (TI-RADS), Zhang et al. [12] leveraged clinical features for XAI of thyroid nodules diagnosis. Another trend is concurrently processing medical images and creating textual reports similar to clinicians’ reports when interpreting medical images [40], [41]. Interpretable computer-aided diagnostic systems have great potential for application, while at the same time, interpretability issues in medical images present new challenges for the research community. These challenges are due to the tremendous diagnostic risks and include long-standing difficulties in the field of medical image processing, such as small sample size, low contrast, variety of image acquisition devices, and non-uniform image formats, among others.
The explainable ML algorithm for breast cancer CADx has been explored by researchers. Shen et al. [42] developed an interpretable ML classifier capable of producing pixel-level saliency maps to indicate the location of suspicious lesions in mammograms. Similarly, Wu et al. [43] proposed a convolutional network architecture called ‘DeepMiner,’ which used expert annotation to correspond the feature map of the last convolutional layer to the BI-RADS lexicon, thus giving a BI-RADS lexicon-based explanation while providing prediction about mass type. Kim et al. [9], [15] proposed a NN model that used the shape and margin features of the mass to produce a saliency map that justified the prediction given by the model. Due to the use of visualization-based interpretation methods, the above methods were weak in interpretation and not easily understood by end-users. At the same time, these methods utilized only a small portion of the medical knowledge in BI-RADS, and there was still great potential for BI-RADS-based interpretable diagnostic systems.
Although automatic breast ultrasound diagnosis systems have gained significant progress in the accuracy of recognition and segmentation, relatively little research has been done on interpretability. Shan et al. [10] designed a series of computational features based on BI-RADS and used a bottom-up approach for feature selection. After comparing several classifiers, the authors conclude that margin-based and orientation-based features have the most vital discriminative power. Zhang et al. [11] designed an interpretable BUS CAD system in which a pre-processing process was introduced to enhance the shape and margin features in the input BUS images. The authors then used a neural network based on auto encoder-decoder (AED) to predict tumor types and reconstruct the input images. The approach in [11] only considered shape and margin descriptors, and the system did not explicitly output the probabilities of these two descriptors as an interpretation of the prediction results. In addition, neural network models that can generate textual diagnostic reports of breast ultrasound images have been reported in the literature [44] and saliency-based methods for identifying interpretable salient regions in breast histopathology images [45]. Although some research exists, interpretable automatic BUS CAD is still an open field for further research and exploration.
III. PROPOSED EXPLAINABLE CADx SYSTEM FOR BREAST CANCER DIAGNOSIS
This section presents the proposed CADx system with integrated explainability, including the network structure, loss function, available dataset, and implementation details. We also present the evaluation metrics for the classifier, Explainer I, and Explainer II, respectively.
A. NETWORK ARCHITECTURE
The architecture of the proposed BI-RADS-Net-V2 is given in Figure 1. The architecture consists of a shared backbone network and three functional components. The three functional modules are a classifier that determines the category of the mass, a multi-task semantic explainer that predicts BI-RADS descriptors and likelihood of malignancy (BI-RADS assessment), and a quantitative explainer that predicts the contribution of each selected BI-RADS descriptor. To simplify the notation, in the rest of the paper, the semantic explainer and quantitative explainer are referred to as Explainer I and Explainer II, respectively.
FIGURE 1.
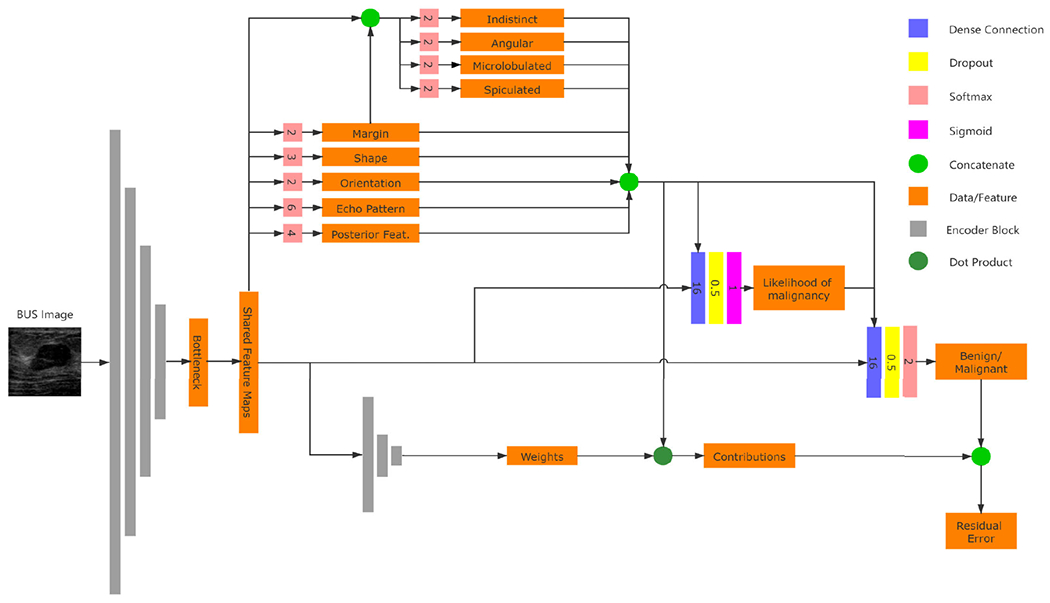
Network architecture of the proposed BI-RADS-Net-V2 for BUS CADx. The lines with arrows is the flow of data, and the numbers on softmax blocks are the dimension of the vectors.
The backbone network employs pre-trained convolutional layers and pooling layers to extract relevant features from the input BUS images, and then the feature maps are shared by the functional modules. In this subsection, we describe the specific structure of each of the three functional modules. The classifier is a convolutional neural network with binary outputs. For an input BUS image, the classifier predicts whether the mass finding contained in the image is benign or malignant. The classifier’s input consists of the features obtained by the backbone network and the judgments given by Explainer I, which are the BI-RADS assessment and descriptors. This design is because the BI-RADS assessment and descriptors contain high-level medical knowledge that helps the classifier make more accurate judgments.
1). SEMANTIC EXPLAINER
Explainer I consists of a regression branch that predicts the BI-RADS likelihood of malignancy, and a group of classifications branches that output the BI-RADS descriptors (see Table 2). In detail, the shape has 2 classes (parallel and not parallel), orientation has 3 classes, echo pattern has 6 classes, and posterior features has 4 classes. The margin can have multiple annotations. For instance, a tumor with a not circumscribed margin could be both indistinct and spiculated. Therefore, we employed a different approach to predict the margin descriptors. A margin branch predicts whether the margin is circumscribed or not, and afterward, four sub-branches are introduced to output binary values of margin sub-classes, including indistinct, angular, microlobulated, and spiculated.
TABLE 2.
BI-RADS Descriptors used in Explainer I.
| BI-RADS Descriptors | Descrptors Calss |
|---|---|
| Shape | Oval, Round, Irregular |
| Orientation | Parallel, Not Paralleled |
| Margin | Circumscribed, Not Circumscribed (Indistinct, Angular, Microlobulated, Spiculated) |
| Echo Pattern | Anechoic, Hypoechoic, Isoechoic, Hyperechoic, Complex cystic and solid, Heterogeneous |
| Posterior Features | No posterior features, Enhancement, Shadowing, Combined pattern |
The predictions of the multi-task branches are integrated with the shared feature map, and then the features are fed to the regression to predict the BI-RADS assessment. We use the likelihood of malignancy, a continuous value from 0% to 100%, to replace the discrete BI-RADS assessment. The likelihood of malignancy reflects the probability that the input BUS image contains a malignant tumor. The continuous likelihood values could be considered as the result of smoothing over the discrete labels. It is more robust to inter-observer variability than the discrete assessments and can reduce the impact of label noise. The tumor classification branch predicts the tumor type by integrating the BI-RADS descriptors, the likelihood of malignancy, and the shared feature map.
The objective of Explainer I is to explain the classification results semantically. Explainability is achieved by reporting the BI-RADS descriptors and likelihood of malignancy. We hold that this information would be beneficial and valuable to clinicians for interpreting BUS images. First, this information provides a link between the information processing by the CAD model and medical diagnosis by clinicians. Namely, clinical interpretation involves observing the shape, orientation, margin, echo pattern, and posterior features of masses, in combination with associated features (duct, skin changes), exceptional cases (implants), and considering additional information, such as the patient medical history, age, lifestyle, or known risk factors. Therefore, CAD systems that predict the BI-RADS descriptors can be valuable, as they can be related to the mental process undertaken by clinicians during BUS interpretation. Second, the provided information can be helpful for the reporting phase. Third, all CAD models inevitably make predicting errors (i.e., the accuracy on unseen images is always less than 100%). Evaluating the uncertainties in the ML predictions on individual BUS images is especially challenging: whenever there is a discrepancy between a clinician’s interpretation and the CAD tumor class prediction on an individual BUS image, the clinician might be suspicious about the CAD prediction. Providing explanations via the BI-RADS descriptors and the BI-RADS likelihood of malignancy can assist clinicians in understanding the level of uncertainties in the model’s output on individual BUS images. Subsequently, the provision of explainability using the BI-RADS lexicon can increase the trustworthiness of clinicians in the CAD systems.
The explanations given by Explainer I differ from the post-hoc explainability approaches for deep learning models, where explanations of the decision-making process for a model are provided after the training phase is completed. Instead, we use a single end-to-end deep learning model that furnishes explainability concurrently with the training/testing phases. We justify this approach because we relied on a clinically validated set of visual features—the BI-RADS descriptors—to explain BUS image analysis.
It is worth mentioning in a separate note that training independent NNs for the risk of malignancy and the BI-RADS descriptors may achieve similar performance. However, the output of these independent NNs is not considered an interpretation of the classifier because it uses different features. Explainer I shares the feature map with the classifier, providing Explainer I with the ability to explain the classifier. Independent neural networks, on the other hand, do not have the ability to explain.
2). QUANTITATIVE EXPLAINER
Explainer II constructs a quantitative explanation based on the classifier and Explainer I. The core idea of Explainer II is to approximate the classifier that is considered a ‘black-box’ with an explainable linear model. There have been methods with similar ideas applied to other image data [46], [47]. The output of Explainer I is categorized into benign favoring and malignant favoring groups. The benign favoring group includes 5 descriptors, and the malignant favoring group includes 11 descriptors. With the shared feature map as input, Explainer II predicts two weight vectors for the two groups, respectively. Then the dot products between the feature group and the predicted weight are calculated. In this way, Explainer II has two outputs, corresponding to benign and malignant. We expect the two outputs of Explainer II to be equal to the classifier output before the final SoftMax layer. The residual (see section III-B) is defined as the average differences between the explain II output and the classifier output on benign and malignant to reflect the similarity between Explainer II and the classifier. When the residual is small enough, Explainer II could be considered to have the same behavior pattern as the classifier, and the contribution of each descriptor could be evaluated by the corresponding weight.
Explainer II can be considered as a post-hoc method. It can use the same feature maps as the classifier and Explainer I, or not. However, experimental results (see section 4.4) proved that using shared feature map enhanced the explanation of Explainer II. Quantitative interpretation is critical in interpretable systems. It has been proved that different descriptors are not equally important in the diagnostic process. For example, the margin is a more significant feature in distinguishing malignant tumors from benign ones. Therefore, it is necessary to analyze the weights of the different descriptors in the classifier and check whether the weights given by the classifier match the clinical experience. In addition, quantitative analysis is an essential tool for our understanding of the inner workings of classifiers. In particular, when the classifier makes mistakes, the analysis of the quantitative explanations allows us to find the reasons for the errors and thus to clarify how to improve them.
B. LOSS FUNCTIONS
The training of BI-RADS-Net-V2 consists of two parts. The first part is to train the classifier and the Explainer I by using multi-task learning. In the multi-task model, Task 1 to 5 are the BI-RADS descriptors, Task 6 to 9 are the sub-classes for the margin BI-RADS descriptor, Task 10 is the BI-RADS likelihood of malignancy, and Task 11 is the tumor classification branch. For each task k, the network loss function is denoted by , where is the predicted value and is the ground-truth label (for classification) or value (for regression). Since the outputs of the likelihood of malignancy branch (Task 10) and the tumor classification branch (Task 11) both reflect the level of risk that the present tumor in the image is malignant, we added loss term to enforce the information shared between the two branches. The total loss is calculated as the weighted sum of all tasks, equation 1.
| (1) |
In the , the symbol denotes the weight coefficient of task i, K = 11 is the number of tasks, and is the weight coefficient for the term. Cross-entropy loss is used for the classification branches and mean-square error loss is used for the regression branch. The output of the classifier, which was denoted as in the above multi-task learning algorithm, was used as the ground truth when training Explainer II. The residual loss was calculated as equation 2.
| (2) |
where D is a vector that reflects the presence of the selected BI-RADS descriptors calculated based on Explainer I output, and and are the weight vectors for benignity and malignancy decisions, respectively. An efficient approximation of the classifier can be obtained by minimizing the residual loss.
C. DATASET AND IMPLEMENTATION DETAILS
1). DATASET
The proposed model was validated using 1,192 BUS images, which were obtained by combining two different datasets, BUSIS [48] and BUSI [49], into one dataset. The BUSIS dataset consists of 562 images, of which 306 images contain benign masses and 256 contain malignant tumors. For the BUSI dataset, we used a subset of 630 images that contain mass findings, of which 421 have benign masses, and 209 have malignant tumors. One BUS image that contains a malignant was excluded due to the incompleted BI-RADS label. Overall, the positive and negative samples in our experimental data are close to balance, as it consists of 727 benign (negative) and 465 malignant (positive) images. All images were annotated with ground-truth labels for the tumor class, BI-RADS assessment category, and BI-RADS descriptors. There are differences in acquisition equipment, imaging conditions, operators, and target populations between the two datasets described above. It is expected that these differences will lead to degradation of the system in terms of metrics such as classification accuracy. However, diverse data can enhance the robustness of the system and thus improve the performance of unobserved data. The details regarding the BUSIS and BUSI datasets are provided in the publications [48] and [49], respectively.
2). PRE-PROCESSING
During the experiment, the size of the input image was 256 by 256 pixels. Unlike generic object recognition tasks, directly adjusting the size and scale of the image can break the morphological features of the tumor, and the shape and orientation labels of some images would be incorrect (e.g., the shape of some tumors can change from oval to round when wide rectangular images are resized to square images). In order to prevent distortion of the morphological features of shape and orientation, the original BUS images were first cropped to the largest squared segment that encompasses the tumor, and afterward, the cropped segment was resized to 256×256 pixels.
Next, for the single-channel grayscale BUS images, we added two additional channels. One channel was obtained by performing histogram equalization to the gray channel, and another channel was obtained by applying smoothing to the gray channel. The experimental results show that this simple pre-preprocessing step was beneficial to improving the model performance [50]. We speculate that the reason for this result is that histogram equalization and smoothing reduced the variations across the images in BUSIS and BUSI datasets and resulted in a more uniformly distributed set of images.
3). CROSS-VALIDATION
We used a five-fold cross-validation method in our experiments. The total sample was randomly divided into five subsets of the same size. For each round of experiments, we used four subsets as the training set and the remaining one as the test set, i.e., 80% of the samples were used as training samples, and 20% were testing samples. In each round of experiments, 15% of the training samples were used as the validation data set. We observed the model’s performance on the validation dataset to adjust the model’s learning rate and determine when to stop training to avoid overfitting the model. The system performance was evaluated based on the average of the five experiments.
4). PARAMETER INITIALIZATION
The choice of backbone has a significant impact on system performance. We compared the performance differences resulting from different backbone choices. Since our data volume is relatively small, using migration learning can speed up the convergence and improve the system’s performance. Therefore, all backbone networks were initialized with pre-trained weights on the ImageNet database. On the other hand, all parameters, including the parameters of the backbone network, were updated during training to acquire unique features of BUS images from the training data.
5). DATA AUGMENTATION
To improve the accuracy of the model, we performed data augmentation on our BUS images. It is worth mentioning that not all transformations are available in order to maintain the morphological features of the tissues in the image and the positional relationships between organs. We applied various types of data augmentation techniques, including zoom (20%), width shift (10%), rotation (5 degrees), shear (20%), and horizontal flip. Up-down flip wasn’t involved because it changed the relative position of the tissues.
6). HYPERPARAMETERS
Hyperparameters in the training process were selected empirically. We set the batch size as 6. The models were trained by using the adaptive moment estimator optimized (Adam), with an initial learning rate of 10−5, which was reduced to 10−6 if the loss of the validation set did not reduce for 15 epochs. The training was stopped when the loss of the validation set did not reduce for 30 epochs to avoid over-fit. For the loss weight coefficients to , we adopted the following values: (0.2, 0.2, 0.2, 0.2, 0.2, 0.1, 0.1, 0.1, 0.1, 0.2, 0.5). That is, the largest weight was assigned to the tumor class branch. The weight for the loss term was set to 0.2 as well. Considering that the goal of Explainer II was to approximate the classifier, the hyperparameters set during training were the same as those of the classifier training.
7). EVALUATION METRICS
The performance of the classifier is evaluated using accuracy, sensitivity, specificity, and F1-score [51]. Explainer I includes both classification and regression branches. The classification branches are evaluated using accuracy, sensitivity, and specificity. The regression branch is evaluated using R-Square (equation 3), MSE (equation 4), and RMSE (equation 5), which are calculated as follows.
| (3) |
| (4) |
| (5) |
Explainer II is evaluated using the residual error, accuracy, and relative contribution. The residual error could be calculated according to equation 2. The smaller residual error reflects that Explainer II is a better approximation of the classifier and vice versa. The Accuracy of Explainer II w.r.t biopsy ground instead of classifier output reflects whether Explainer II learned adequate medical knowledge rather than simply fitting the classifier. Moreover, we defined a new matric named relative contribution to evaluate Explainer II. Raza et al. [52] summarized the reports and categorized the BI-RADS lexicons into three categories: favoring malignant, favoring benign, and undetermined features (see Table 3). Only the determinative descriptors are used in Explainer II. The relative contribution is calculated as follows (equation 6).
| (6) |
TABLE 3.
BI-RADS Descriptors favoring different masses.
| Descriptor | Favoring Benign | Favoring Malignant | Indenterminate |
|---|---|---|---|
| Shape of mass | Oval | Irregular, round | |
| Orientation of mass | Parallel to skin | Not parallel to skin | |
| Maring of mass | Circumscribed | Microlobulated, Indistinct, Angular, Spiculated | |
| Echo pattern | Anechoic, Hyperechoic | Complex Pattern | Isoechoic, Hypoechoic |
| Posterior features | Showing, Combined | Enhancement, No posterior features |
The relative contribution reflects whether the malignant favoring features and benign favoring features contribute to the malignant and benign decision, respectively. The benign favoring features should have more immense contributions than the malignant features for benign masses and vice versa.
IV. EXPERIMENTAL RESULTS
This section presents a series of experimental results to verify the impact of different elements on the performance of the system. These include pre-processing, the BI-RADS feature set used, the choice of different feature generators, how information is shared between different functional modules, and so forth. Besides, we analyzed the explanations given by the system for some typical cases, and the results corroborate the medical knowledge in BI-RADS, and there are some new findings.
A. DIAGNOSTIC PERFORMANCE
We divided the evaluation of BI-RADS-Net-V2 into two parts, diagnostic performance evaluation and explanation evaluation. The first part addresses the evaluation of the diagnostic performance of the system, which is the core of the system. The evaluation included the accuracy of tumor type classification, likelihood prediction of malignancy, and BI-RADS descriptor prediction.
As mentioned above, there are many factors in the experiment that have an impact on the performance. We designed an ablation study to evaluate the impact of the different components in the design of BI-RADS-Net-V2. The results are shown in Table 4 and Table 5.
TABLE 4.
The performance of tumor classification and BI-RADS assessment (in the form of likelihood of malignancy), regarding the impact of different components in the network design.
| Method | Tumor Class | Likelihood of Malignancy | |||||
|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | F1-Score | R 2 | MSE | RMSE | |
| BI-RADS-Net-V2 | 0.889 | 0.838 | 0.923 | 0.854 | 0.671 | 0.153 | 0.391 |
| Without Augmentation * | 0.868 | 0.789 | 0.919 | 0.832 | 0.648 | 0.159 | 0.399 |
| Without Pretraining * | 0.828 | 0.746 | 0.881 | 0.770 | 0.592 | 0.171 | 0.414 |
| Single Channel Images * | 0.817 | 0.726 | 0.875 | 0.754 | 0.582 | 0.173 | 0.416 |
| Without Image Cropping * | 0.799 | 0.711 | 0.855 | 0.733 | 0.528 | 0.184 | 0.429 |
| ResNet Backbone | 0.883 | 0.841 | 0.909 | 0.844 | 0.664 | 0.155 | 0.394 |
| EfficientNet-B6 Backbone | 0.856 | 0.826 | 0.904 | 0.809 | 0.667 | 0.154 | 0.392 |
The ablation steps are progressively applied, i.e., the model without augmentation is afterward evaluated without pretrained weights, etc.
TABLE 5.
The performance of BI-RADS descriptors prediction, regarding the impact of different components in the network design.
| Method | BI-RADS Descriptors | ||||
|---|---|---|---|---|---|
| Shape | Orientation | Margin | Echo Pat. | Post. Feat. | |
| BI-RADS-Net-V2 | 0.816 | 0.872 | 0.873 | 0.825 | 0.830 |
| Without Augmentation * | 0.832 | 0.848 | 0.855 | 0.804 | 0.828 |
| Without Pretraining * | 0.773 | 0.804 | 0.794 | 0.726 | 0.731 |
| Single Channel Images * | 0.764 | 0.809 | 0.792 | 0.720 | 0.739 |
| Without Image Cropping * | 0.755 | 0.788 | 0.774 | 0.716 | 0.729 |
| ResNet Backbone | 0.816 | 0.850 | 0.868 | 0.813 | 0.831 |
| EfficientNet-B6 Backbone | 0.819 | 0.858 | 0.847 | 0.795 | 0.826 |
The ablation steps are progressively applied, i.e., the model without augmentation is afterward evaluated without pretrained weights, etc.
The ablation study assesses the contributions by data augmentation, pre-trained network parameters on the ImageNet dataset, additional image channels with histogram equalization and smoothing, and cropping the original images to square-size segments. The results in Table 4 and Table 5 show that data augmentation, pre-trained weights, additional image channels, and image cropping all contribute to the system. Without pre-processing and trained from scratch, the model achieved accuracy slightly lower than 80.0%, 71.5% sensitivity, 85.5% specificity, and a 73.3% F1 score on the single channel ultrasound image. Removing the dependence of features on location by cropping the images to square increased the performance slightly to 81.7% accuracy, 72.6% sensitivity, 87.5% specificity, and 75.4% F1 score. Further, adding image channels created by smoothing and histogram equalization raised the performance to 82.8% accuracy, 74.6% sensitivity, 88.1% specificity, and 75.4% F1 score. Instead of training from scratch, the introduction of pre-trained weights helped the performance and achieved 86.8% accuracy, 78.9% sensitivity, 91.9% specificity, and 83.2% F1 score. Finally, the effect of data augmentation (detailed in subsection III-C) was pronounced. The final results indicate that the network achieved 88.9% accuracy, 83.8% sensitivity, 92.3% specificity, and 85.4% F1 score for mass type classification and over 80% accuracy for all five BI-RADS descriptors. Different backbone networks were also tested, the results in Table 4 and Table 5 present that the system with the VGG16 backbone outperformed the ResNet and EfficientNet-B6 backbones in most aspects. Due to the low resolution, brightness, and contrast of ultrasound images, a simpler structured network is more likely to produce better results.
With the hyperparameters determined, we compared the proposed method with a group of most current methods concerning the diagnostic performance. The compared methods include SHA-MAL [53], Ensemble Network [54], CNNSVM [55], and Dual Sampling Network [56]. The results are shown in Table 6.
TABLE 6.
Comparison with existing breast cancer diagnosis algorithms.
From the results in Table 6, it can be found that the BI-RADS-Net-V2 exhibits the highest accuracy and sensitivity and the next highest but very close specificity on the experimental data. This result demonstrates that medical knowledge in BI-RADS label information improves the classifier’s performance under the proposed multi-task learning algorithm.
We used a Wilcoxon signed rank test to validate the significance of our data, analyzing the distribution of metric values. Accuracy, sensitivity, specificity, and F1 score were calculated for each image, and we conducted a Wilcoxon signed rank test to compare each method against the proposed BI-RADS-Net-V2. The results of the hypothesis testing are presented in table 7. The cells with asterisks indicate rejection of the null hypothesis with a P-value < 0.05 Accordingly, for almost all metrics there is a statistically significant difference in the median values by the test models in comparison to BI-RADS-Net-V2.
TABLE 7.
Pair-wise Wilcoxon signed rank test (w.r.t. BI-RADS-Net-V2) of the per image performance of tumor classification metrics.
| Testing Method | Accuracy | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|
| SHA-MTL [53] | p<0.0001* | p<0.0001* | p=0.0022* | p=0.0006* |
| Ensemble Network [54] | p=0.0002* | p=0.0027* | p=0.0233* | p<0.0001* |
| CNNSVM [55] | p<0.0001* | p<0.0001* | p=0.0003* | p<0.0001* |
| Dual Sampling Network [56] | p<0.0001* | p<0.0001* | p<0.0001* | p<0.0001* |
Statistically significant difference, P-value < 0.05
B. SEMANTIC EXPLANATION
The semantic explanation can be evaluated from two perspectives. The first is correctness, i.e., the Explainer I must accurately identify the BI-RADS descriptors. Correctness is a fundamental prerequisite for semantic explanations to be effective. From the results in Table 5, we found that the accuracy of the network for shape, direction, margin, echo pattern, and posterior features is around 85% on average, based on the use of image cropping, enhancement, and the introduced pre-training weights. Comparing with the doctor’s conclusion, we believe the Explainer I output with the current accuracy can constitute a valid explanation.
The second way to evaluate the semantic explanation is perform case study. Figure 2 (b) and (d) give examples of the semantic explanations for benign and malignant tumors, respectively. For the benign mass in Figure 2 (b), the Explainer I predict the shape is oval, which is a benign favoring feature; the margin is circumscribed, which is also benign favoring. Besides the above two, the parallel orientation, hypoechoic pattern, and enhanced posterior features are all favoring benign. As a conclusion, Explainer I predict the likelihood of malignancy is 0.21%, and this matches the classifier decision and the clinical diagnosis.
FIGURE 2.
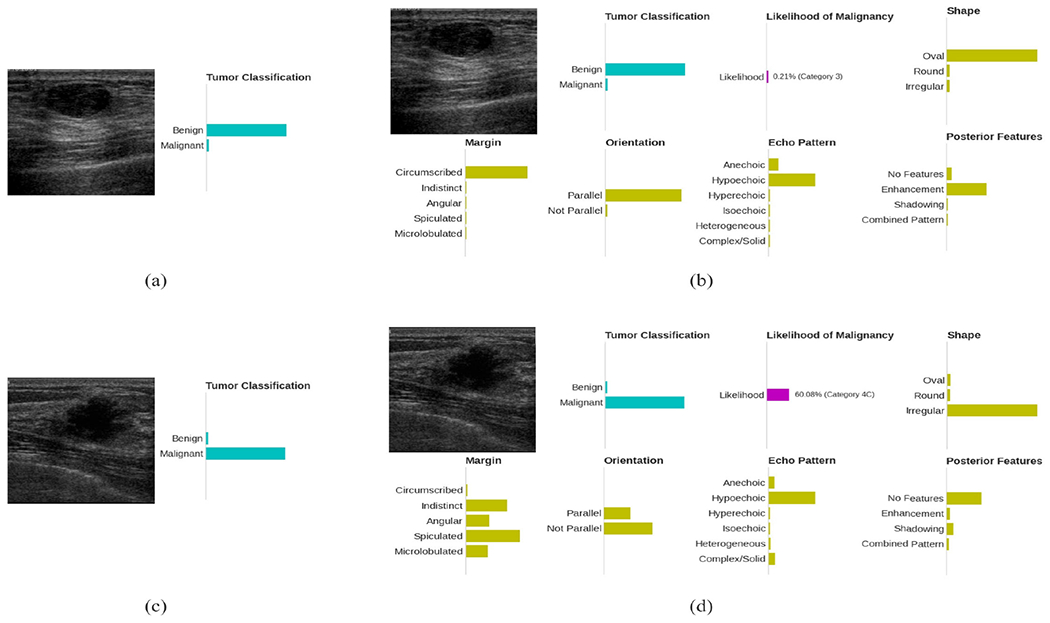
(a) Conventional BUS CAD system output for a benign mass finding. The bars in the sub-figures indicate the predicted class probabilities by the CAD systems.; (b) Output of the proposed explainable BUS CAD system for the same benign mass finding; (c) Conventional BUS CAD system output for a malignant mass finding; (d) Output of the proposed explainable BUS CAD system for the same malignant mass finding.
Another example, Explainer I is very certain that the tumor in Figure 2 (b) has an irregular shape and a group of malignant favoring margin descriptors. Although it isn’t very certain about the orientation (the ratio between height and width is very close due to the irregular shape), it still gives the likelihood of malignancy over 60%, which justifies the classifier decision. In summary, by validating the correctness of the BI-RADS descriptor prediction and the correlation between Explainer I output and the classifier decision, we can conclude that Explainer I could effectively justify the classifier decision and explains why the tumor was diagnosed as benign or malignant.
C. QUANTITATIVE EXPLANATION
Explainer II provides a quantitative explanation based on Explainer I. Explainer II is expected to have two essential characteristics, correctness and explainability. The explainability means the building blocks and calculations must be understandable for the end-users. The explainability is satisfied because Explainer II is a linear model based on BI-RADS descriptors. Correctness has multiple meanings. First, Explainer II should be a validated equivalent of the classifier, which is, the difference between Explainer II and the classifier should be minimized. And second, the explanation is required to match the medical knowledge. The correctness is evaluated using residual error, accuracy, and relative contribution.
When evaluating Explainer II, we binarized the Explainer I output and used 1 to indicate the presence of the BI-RADS descriptors and 0 otherwise. Overall, 15 determinative descriptors were used as the input of Explainer II, and the undetermined descriptors were ignored. Besides, an extra margin feature was added in Explainer II because the margin has the most positive correlation with the malignancy. The descriptor indicates whether the mass margin is circumscribed, and the not circumscribed cases include at least one from microlobulated, indistinct, angular, and spiculated. Overall, Explainer II used 11 malignant favoring and 5 benign favoring descriptors.
1). MODEL SELECTION
Three models were investigated. The first model was a single-layer MLP that took the Explainer I prediction as input, and the expected output was the classifier prediction. There was no activation function in the model. Meanwhile, the weights were restricted to be positive, and the bias was set as zero. It is easy to understand that the normalized weights are how vital the corresponding descriptor is for all training samples. The second model was a convolutional neural network with the encoder of the network initialized using the Imagenet weights. The model took the BUS images as input, and the output was a weight vector for the input image. The third model was similar to the second model, except that the backbone network was initialized using the classifier weights, and the parameters of the backbone network are not trainable. Compared to the second model, Explainer II shared the same feature with the classifier and Explainer I in the third model. The configuration added tougher restrictions but made the model more explainable. Five-folded cross-validation was used for evaluation. The averaged residual errors and deviations are presented in Figure 3.
FIGURE 3.
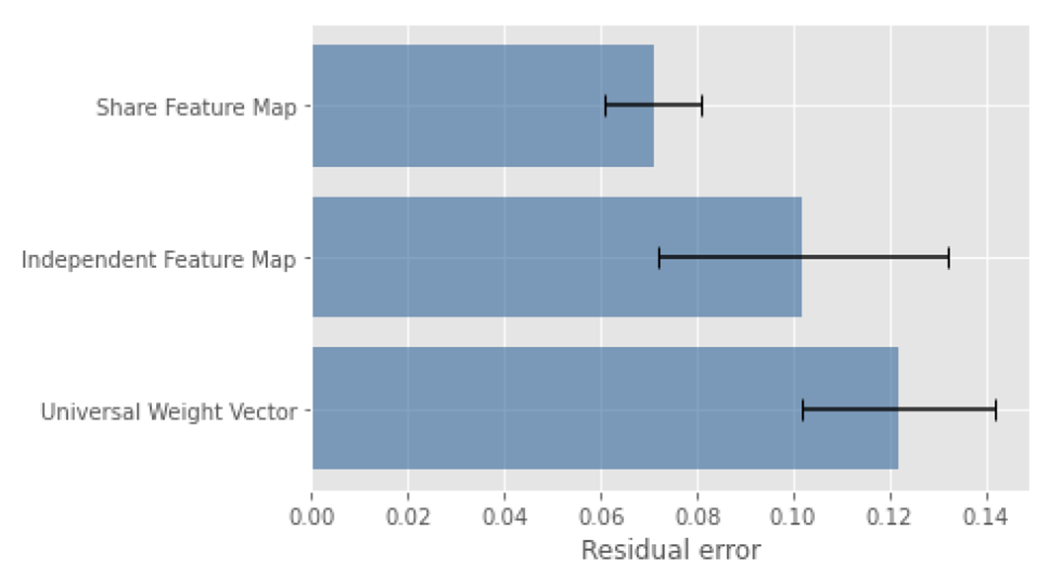
Residual Error using different models.
The third model, which shared the same feature map with the classifier, achieved significantly minimal residual error compared to the other two models. The results proved that the feature map played a key role when composing the classifier output. Intuitively, the second model had fewer restrictions and was expected to approximate the classifier better. However, the assumption is only feasible when a large number of samples are given.
Moreover, the residual error helps us understand the behaviors of the classifier. The MLP model learned one weight vector for all BUS images. The assumption behind the MLP model was that the classifier always puts fixed weights on descriptors and then integrates the contributions. The other two models assumed that the classifier used different weight vectors for different samples based on the input BUS images’ characteristics.
Besides the residual error, the weights learned using the MLP model should have minor variations if the assumption was valid. However, the experiments presented opposite results. The variations of the weights are relatively large (see Figure 4). The experimental results generally favored the second assumption that the classifier used different weights for different image samples.
FIGURE 4.
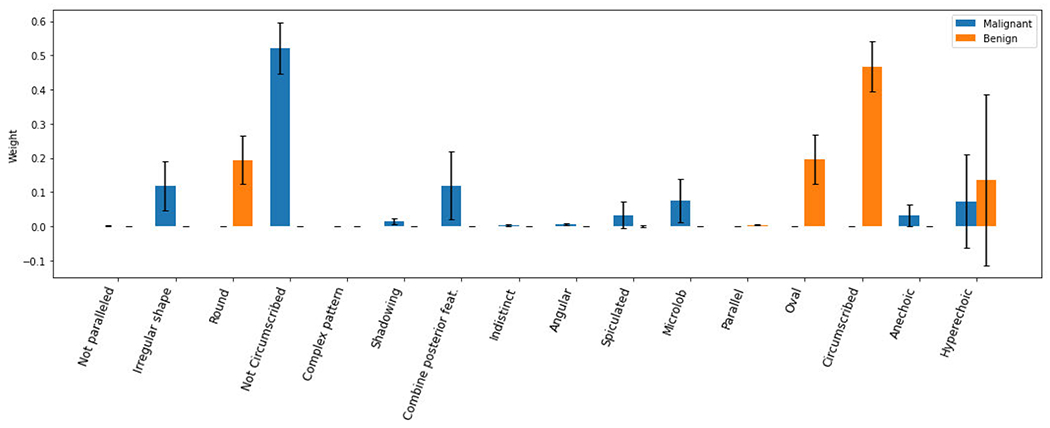
Average weights using MLP with errors.
2). BACKBONE
The backbone was another essential factor. The results above proved that using the shared feature map was the optimized option. Thus, we only compared the models using shared feature maps. The evaluated networks included VGG16, ResNet, and EfficientNet. The residual error, accuracy, and relative contribution of benign and malignant masses were calculated for each backbone. The results are presented in Table 8.
TABLE 8.
The correctness evaluation of explainer.
| Model | Residual Error | Accuracy (Classification) | Relative contribution | ||
|---|---|---|---|---|---|
| Explainer II | Performer | Benign | Malignant | ||
| VGG16 | 0.07 | 0.88 | 0.89 | 0.84 | 0.81 |
| ResNet52 | 0.08 | 0.87 | 0.88 | 0.83 | 0.78 |
| EfficentNet-B0 | 0.08 | 0.87 | 0.83 | 0.85 | 0.76 |
The VGG16 backbone that was initialized using classifier weight outperformed the ResNet and EfficentNet regarding the residual error, and all models achieved competitive accuracy. Besides, the model with a VGG16 backbone had a higher relative contribution regarding malignant masses. This result explained the result in Table 4 that the structure with VGG16 backbone had higher specificity and sensitivity. Explainer II with VGG16 backbone achieved minimal residual error, competitive classification accuracy, and a higher relative contribution on malignant masses.
3). CASE STUDY
Similar to the semantic explainer, we present a set of representative cases as the case study of Explainer II. Figure 5 (a) and (b) shows a true negative mass finding. Based on Explainer II output, we could find that the circumscribed margin and the oval shape contributed the most to the decision. Besides, the model put consideration on the parallel orientation by a smaller portion. It could be seen that the components matched the image feature and the biopsy result. Another example, Figure 5 (c) and (d) present a true positive sample. The model put heavyweights on the irregular shape and not circumscribed margin for the tumor, and the above two contribute the most to the final decision. These two figures prove that Explainer II could effectively explain the classifier output, and the explanation matched the clinical experiences.
FIGURE 5.
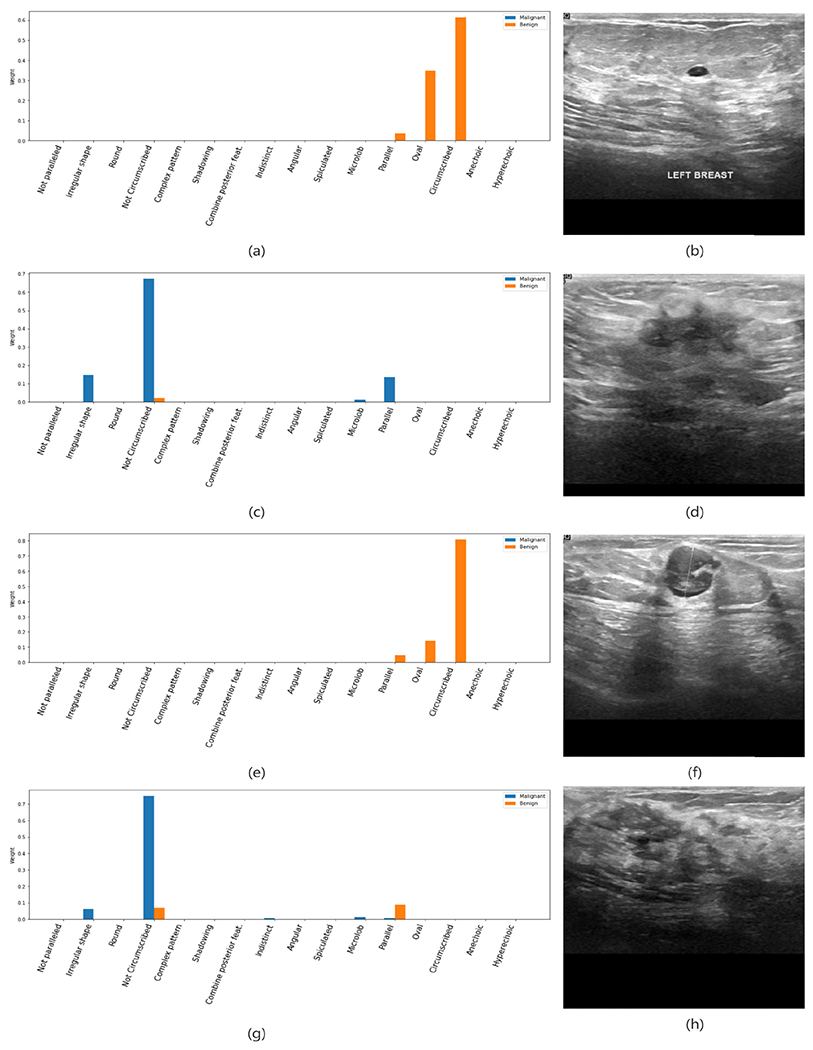
Representative output on the BUSI dataset. (a) and (b) are the output and BUS image of a true negative sample; (c) and (d) are the output and BUS image of a true positive example; (e) and (f) are the output and BUS image of a false positive example; (g) and (h) are the output and BUS image of a false negative example.
Meanwhile, we noticed that there were some errors in the explanations. For the malignant mass finding in Figure 5 (c) and (d), a small weight was put on the parallel orientation, which is a benign favoring feature. We believe these minor mismatches are from the BI-RADS features’ ambiguity and the training labels’ noise.
More than justifying the classifier output, another desired expectation of the explainers is to reveal the reasons when the classifier made a mistake. Figure 5 (e) and (f) show a false negative example. It could be found that the classifier made the wrong decision because that the malignant descriptors, including complex posterior feature and echo pattern, were undetected, and the benign favoring descriptors dominated the decision. This reminded us to introduce more similar samples into the training set to enhance Explainer I. Comparingly, Figure 5 (g) and (h) gives a more complex error, where the tissues around the mass formed a margin-like area that was considered as not circumscribed, and vague margin caused uncertainty of the shape descriptor, which should have a regular shape based on the clinicians. Thus, the mass was misclassified into the malignant class, where the biopsy result was benign. The explanation reveals that the image-based BI-RADS descriptors don’t cover all mass features even confirmed by its clinical applications. In general, the presented examples prove that Explainer II explanations could help end-users understand the foundation of the classifier’s decision and reveal the possible reasons for making mistakes.
Inspired by the above results, we calculate the average contribution for all involved BI-RADS descriptors. The results are presented in Figure 6 and Figure 7. The average contribution shows that the most benign favoring descriptors present strong distinguishing power, including the parallel orientation, oval shape, circumscribed margin. The anechoic and hyperechoic echo patterns didn’t affect much during the process. The malignant favoring descriptors, not circumscribed margin, shadowing posterior feature, and indistinct and microlobulated margin present reliable distinguishing power. The rest descriptors contributed similarly to both categories.
FIGURE 6.
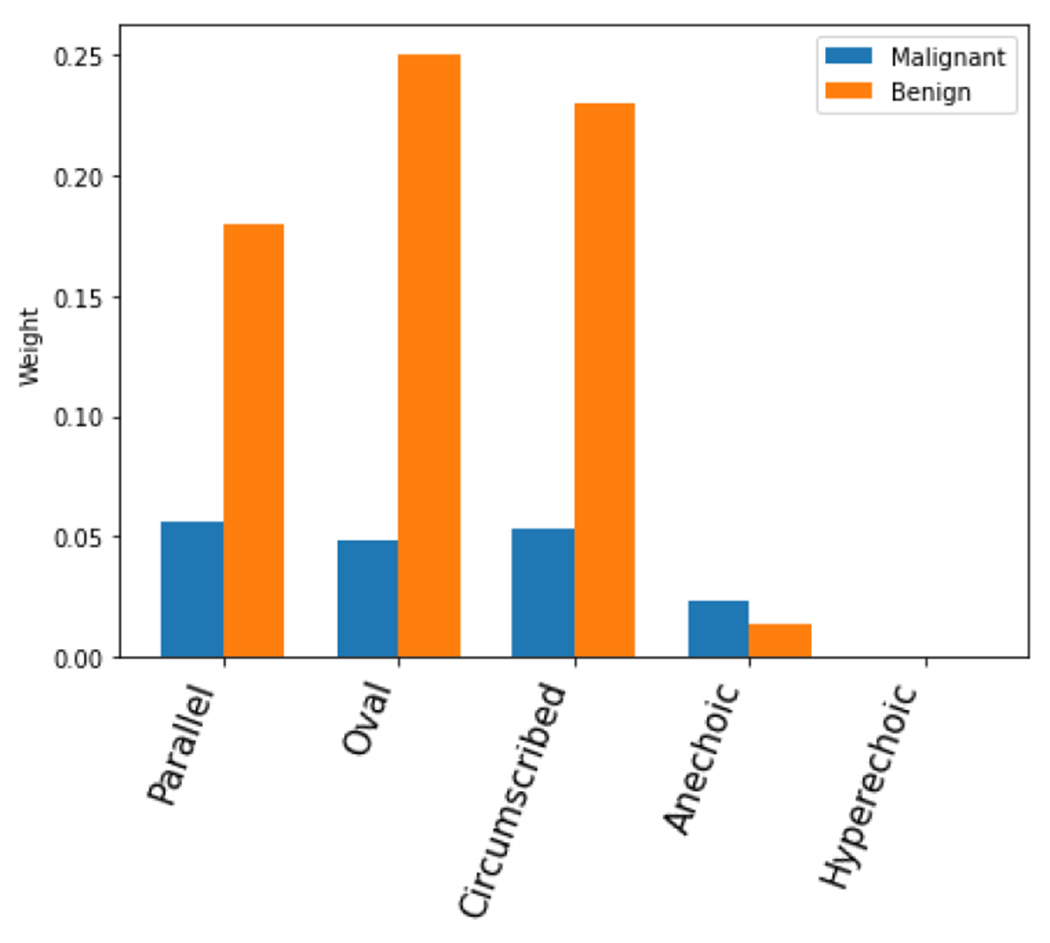
Average contributions of benign favoring descriptors.
FIGURE 7.
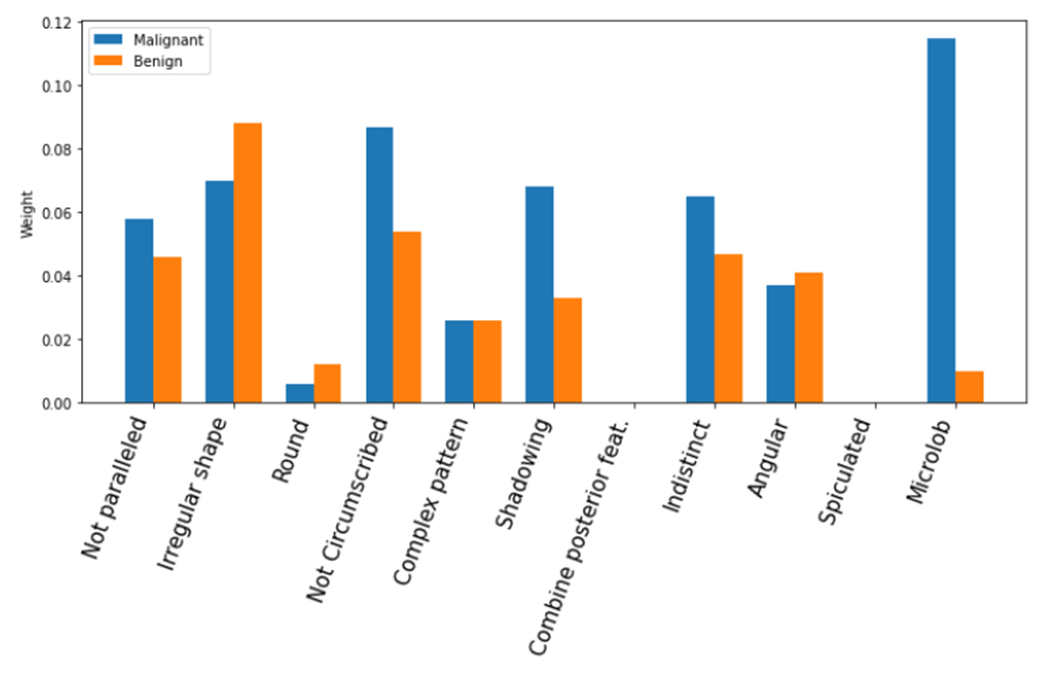
Average contribution of malignant favoring descriptors.
To summarize, we posit that explainability is task-dependent and audience-dependent, and therefore, requires ML models designed for specific tasks and targeted to end-users. For instance, the practical relevance of our proposed explainable model for BUS would diminish for other tasks because they employ different image features for representation learning. Likewise, our approach may not provide adequate explainability to a data scientist without medical knowledge or patients. In this aspect, our model is designed for providing explanations to and assisting BUS clinicians.
V. CONCLUSION AND FUTURE WORK
This paper designs an interpretable, deep network-based breast ultrasound diagnostic system, BI-RADS-Net-V2. This system provides reliable and efficient interpretation for end-users by introducing medical expertise in BI-RADS. The system has a high accuracy and is more likely to gain the trust of end-users, which facilitates the diffusion of the automated diagnostic system. It promotes the diffusion of early universal breast cancer screening. The experimental results demonstrate that the introduction of BI-RADS can enhance the generalization ability of the diagnostic model and improve the accuracy of the model under a multi-task learning framework. In addition, systematic BI-RADS descriptor prediction can effectively prove the correctness of the discriminative model. The quantitative explanation based on knowledge distillation, on the other hand, can analyze the causes of errors in the discriminative model and indicate the direction to improve the model performance.
The further work includes further expanding the BUS image dataset and collecting BI-RADS description sublevel labels. The analysis of existing samples is also used to increase the interpretability of the model by cross-corroborating with clinical BI-RADS diagnoses.
ACKNOWLEDGMENT
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award P20GM104420.
Biographies

BOYU ZHANG received the Ph.D. degree in computer science and technology from the Harbin Institute of Technology, Harbin, China, in 2015. He is currently a Computational Data Scientist with the Institute for Interdisciplinary Data Sciences, University of Idaho. His research interests include machine learning, computer vision, and explainable artificial intelligence, with a focus on the applications of machine learning techniques in wildlife epidemiology.

ALEKSANDAR VAKANSKI received the Ph.D. degree in mechanical and industrial engineering from Toronto Metropolitan University, Canada, in 2013. He is currently an Assistant Professor in industrial technology with the University of Idaho, Idaho Falls, ID, USA. His research work has been published in multiple journal articles and conference proceedings. His research interests include machine learning and mechatronics, with a focus on designing robust, secure, and trustworthy machine learning systems.

MIN XIAN (Member, IEEE) received the M.S. degree in pattern recognition and intelligence system from the Harbin Institute of Technology, Harbin, China, in 2011, and the Ph.D. degree in computer science from Utah State University, Logan, Utah, in 2017. He is an affiliate Professor and Doctorial Supervisor of the Bioinformatics and Computational Biology (BCB) program at the University of Idaho, an affiliate of the Center for Advanced Energy Studies (CAES), and a participating faculty of the Institute for Modeling Collaboration and Innovation (IMCI). He is leading projects on AI-enhanced cancer detection (NIH) and material characterization and development (DOE). He is an Associate Professor in the Department of Computer Science at the University of Idaho. He is currently the Director of the Machine Intelligence and Data Analytics (MIDA) laboratory, a research-oriented collaborative and synergistic core to impel interdisciplinary research. His research interests include artificial intelligence, machine learning, deep neural networks, adversarial learning, biomedical data analytics, material informatics, and digital image understanding. He is a guest editor at Healthcare and a session chair for the conference of the Association for the Advancement of Artificial Intelligence (AAAI).
Footnotes
The authors confirm that all human/animal subject research procedures and protocols are exempt from review board approval.
The associate editor coordinating the review of this manuscript and approving it for publication was Yizhang Jiang (0000-0002-4558-9803).
REFERENCES
- [1].US Cancer Statistics Data Visualizations Tool, Based on November 2017 Submission Data (1999–2015): US Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute, Centers for Disease Control and Prevention and National Cancer Institute, Bethesda, MD, USA, 2018, vol. 6. [Google Scholar]
- [2].Ruhl J, Callaghan C, Hurlbut A, Ries L, Adamo P, Dickie L, and Schussler N, Summary Stage 2018: Codes and Coding Instructions. Bethesda, MD, USA: National Cancer Institute, 2018. [Google Scholar]
- [3].Sehgal CM, Weinstein SP, Arger PH, and Conant EF, “A review of breast ultrasound,” J. Mammary Gland Biol. Neoplasia, vol. 11, no. 2, pp. 113–123, Apr. 2006. [DOI] [PubMed] [Google Scholar]
- [4].Houssami N, Kirkpatrick-Jones G, Noguchi N, and Lee CI, “Artificial intelligence (AI) for the early detection of breast cancer: A scoping review to assess AI’s potential in breast screening practice,” Expert Rev. Med. Devices, vol. 16, no. 5, pp. 351–362, May 2019. [DOI] [PubMed] [Google Scholar]
- [5].O’Connell AM, Bartolotta TV, Orlando A, Jung S, Baek J, and Parker KJ, “Diagnostic performance of an artificial intelligence system in breast ultrasound,” J. Ultrasound Med, vol. 41, no. 1, pp. 97–105, Jan. 2022. [DOI] [PubMed] [Google Scholar]
- [6].Wu G-G, Zhou L-Q, Xu J-W, Wang J-Y, Wei Q, Deng Y-B, Cui X-W, and Dietrich CF, “Artificial intelligence in breast ultrasound,” World J. Radiol, vol. 11, no. 2, p. 19, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chan H-P, Samala RK, and Hadjiiski LM, “CAD and AI for breast cancer—Recent development and challenges,” Brit. J. Radiol, vol. 93, no. 1108, Apr. 2020, Art. no. 20190580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elfgen C, Varga Z, Reeve K, Moskovszky L, Bjelic-Radisic V, Tausch C, and Güth U, “The impact of distinct triple-negative breast cancer subtypes on misdiagnosis and diagnostic delay,” Breast Cancer Res. Treatment, vol. 177, no. 1, pp. 67–75, Aug. 2019. [DOI] [PubMed] [Google Scholar]
- [9].Kim ST, Lee H, Kim HG, and Ro YM, “ICADX: Interpretable computer aided diagnosis of breast masses,” Proc. SPIE, vol. 10575, pp. 450–459, Feb. 2018. [Google Scholar]
- [10].Shan J, Alam SK, Garra B, Zhang Y, and Ahmed T, “Computer-aided diagnosis for breast ultrasound using computerized BI-RADS features and machine learning methods,” Ultrasound Med. Biol, vol. 42, no. 4, pp. 980–988, Apr. 2016. [DOI] [PubMed] [Google Scholar]
- [11].Zhang E, Seiler S, Chen M, Lu W, and Gu X, “BIRADS features-oriented semi-supervised deep learning for breast ultrasound computer-aided diagnosis,” Phys. Med. Biol, vol. 65, no. 12, Jun. 2020, Art. no. 125005. [DOI] [PubMed] [Google Scholar]
- [12].Zhang S, Du H, Jin Z, Zhu Y, Zhang Y, Xie F, Zhang M, Tian X, Zhang J, and Luo Y, “A novel interpretable computer-aided diagnosis system of thyroid nodules on ultrasound based on clinical experience,” IEEE Access, vol. 8, pp. 53223–53231, 2020. [Google Scholar]
- [13].Huang Y, Han L, Dou H, Luo H, Yuan Z, Liu Q, Zhang J, and Yin G, “Two-stage CNNs for computerized BI-RADS categorization in breast ultrasound images,” Biomed. Eng. OnLine, vol. 18, no. 1, pp. 1–18, Dec. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gu R, Wang G, Song T, Huang R, Aertsen M, Deprest J, Ourselin S, Vercauteren T, and Zhang S, “CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation,” IEEE Trans. Med. Imag, vol. 40, no. 2, pp. 699–711, Feb. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim ST, Lee J-H, Lee H, and Ro YM, “Visually interpretable deep network for diagnosis of breast masses on mammograms,” Phys. Med. Biol, vol. 63, no. 23, Dec. 2018, Art. no. 235025. [DOI] [PubMed] [Google Scholar]
- [16].Grimsley C, Mayfield E, and Bursten J, “Why attention is not explanation: Surgical intervention and causal reasoning about neural models,” in Proc. 12th Conf. Lang. Resour. Eval., 2020, pp. 1780–1790. [Google Scholar]
- [17].Jain S and Wallace BC, “Attention is not explanation,” 2019, arXiv:1902.10186. [Google Scholar]
- [18].Khanna R, Kim B, Ghosh J, and Koyejo S, “Interpreting black box predictions using Fisher kernels,” in Proc. 22nd Int. Conf. Artif. Intell. Statist., 2019, pp. 3382–3390. [Google Scholar]
- [19].Gunning D, Stefik M, Choi J, Miller T, Stumpf S, and Yang G-Z, “XAI-explainable artificial intelligence,” Sci. Robot, vol. 4, no. 37, 2019, Art. no. eaay7120. [DOI] [PubMed] [Google Scholar]
- [20].Adadi A and Berrada M, “Peeking inside the black-box: A survey on explainable artificial intelligence (XAI),” IEEE Access, vol. 6, pp. 52138–52160, 2018. [Google Scholar]
- [21].Došilovic FK, Brcic M, and Hlupic N, “Explainable artificial intelligence: A survey,” in Proc. 41st Int. Conv. Inf. Commun. Technol., Electron. Microelectron. (MIPRO), May 2018, pp. 210–215. [Google Scholar]
- [22].Jin Y and Sendhoff B, “Pareto-based multiobjective machine learning: An overview and case studies,” IEEE Trans. Syst. Man, Cybern. C, Appl. Rev, vol. 38, no. 3, pp. 397–415, May 2008. [Google Scholar]
- [23].Freitas AA, “A critical review of multi-objective optimization in data mining: A position paper,” ACM SIGKDD Explor. Newslett, vol. 6, no. 2, pp. 77–86, Dec. 2004. [Google Scholar]
- [24].Samek W, Montavon G, Vedaldi A, Hansen LK, and Müller K-R, Explainable AI: Interpreting, Explaining Visualizing Deep Learning, vol. 11700. Cham, Switzerland: Springer, 2019. [Google Scholar]
- [25].Bau D, Zhou B, Khosla A, Oliva A, and Torralba A, “Network dissection: Quantifying interpretability of deep visual representations,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 3319–3327. [Google Scholar]
- [26].Kim B, Wattenberg M, Gilmer J, Cai C, Wexler J, and Viegas F, “Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (TCAV),” in Proc. Int. Conf. Mach. Learn., 2018, pp. 2668–2677. [Google Scholar]
- [27].Nguyen A, Dosovitskiy A, Yosinski J, Brox T, and Clune J, “Synthesizing the preferred inputs for neurons in neural networks via deep generator networks,” in Proc. Adv. Neural Inf. Process. Syst., vol. 29, 2016, pp. 3387–3395. [Google Scholar]
- [28].Nguyen A, Yosinski J, and Clune J, “Understanding neural networks via feature visualization: A survey,” in Explainable AI: Interpreting, Explaining Visualizing Deep Learning. Cham, Switzerland: Springer, 2019, pp. 55–76. [Google Scholar]
- [29].Yosinski J, Clune J, Nguyen A, Fuchs T, and Lipson H, “Understanding neural networks through deep visualization,” 2015, arXiv:1506.06579. [Google Scholar]
- [30].Bach S, Binder A, Montavon G, Klauschen F, Müller K-R, and Samek W, “On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation,” PLoS ONE, vol. 10, no. 7, Jul. 2015, Art. no. e0130140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Arras L, Montavon G, Müller K-R, and Samek W, “Explaining recurrent neural network predictions in sentiment analysis,” 2017, arXiv:1706.07206. [Google Scholar]
- [32].Samek W, Montavon G, Vedaldi A, Hansen LK, and Müller K-R, Explainable AI: Interpreting, Explaining Visualizing Deep Learning, vol. 11700. Cham, Switzerland: Springer, 2019. [Google Scholar]
- [33].Montavon G, Samek W, and Müller K-R, “Methods for interpreting and understanding deep neural networks,” Digit. Signal Process, vol. 73, pp. 1–15, Feb. 2018. [Google Scholar]
- [34].Lapuschkin S, Wäldchen S, Binder A, Montavon G, Samek W, and Müller K-R, “Unmasking clever Hans predictors and assessing what machines really learn,” Nature Commun., vol. 10, no. 1, pp. 1–8, Mar. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Koh PW and Liang P, “Understanding black-box predictions via influence functions,” in Proc. Int. Conf. Mach. Learn., 2017, pp. 1885–1894. [Google Scholar]
- [36].Zhao G, Zhou B, Wang K, Jiang R, and Xu M, “Respond-CAM: Analyzing deep models for 3D imaging data by visualizations,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2018, pp. 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tang Z, Chuang KV, DeCarli C, Jin L-W, Beckett L, Keiser MJ, and Dugger BN, “Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline,” Nature Commun., vol. 10, no. 1, pp. 1–14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Qin Y, Kamnitsas K, Ancha S, Nanavati J, Cottrell G, Criminisi A, and Nori A, “Autofocus layer for semantic segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2018, pp. 603–611. [Google Scholar]
- [39].Wiegreffe S and Pinter Y, “Attention is not not explanation,” 2019, arXiv:1908.04626. [Google Scholar]
- [40].Zhang Z, Xie Y, Xing F, McGough M, and Yang L, “MDNet: A semantically and visually interpretable medical image diagnosis network,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 3549–3557. [Google Scholar]
- [41].Jing B, Xie P, and Xing E, “On the automatic generation of medical imaging reports,” 2017, arXiv:1711.08195. [Google Scholar]
- [42].Shen Y, Wu N, Phang J, Park J, Liu K, Tyagi S, Heacock L, Kim SG, Moy L, Cho K, and Geras KJ, “An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization,” Med. Image Anal, vol. 68, Feb. 2021, Art. no. 101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu J, Zhou B, Peck D, Hsieh S, Dialani V, Mackey L, and Patterson G, “DeepMiner: Discovering interpretable representations for mammogram classification and explanation,” 2018, arXiv:1805.12323. [Google Scholar]
- [44].Lee H, Kim ST, and Ro YM, “Generation of multimodal justification using visual word constraint model for explainable computer-aided diagnosis,” in Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support. Cham, Switzerland: Springer, 2019, pp. 21–29. [Google Scholar]
- [45].Couture HD, Marron JS, Perou CM, Troester MA, and Niethammer M, “Multiple instance learning for heterogeneous images: Training a CNN for histopathology,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2018, pp. 254–262. [Google Scholar]
- [46].Ribeiro MT, Singh S, and Guestrin C, “Why should I trust you?’ explaining the predictions of any classifier,” in Proc. 22nd ACM SIGKDD Int. Conf. Knowl. Discovery Data Mining, 2016, pp. 1135–1144. [Google Scholar]
- [47].Chen R, Chen H, Huang G, Ren J, and Zhang Q, “Explaining neural networks semantically and quantitatively,” in Proc. IEEE/CVF Int. Conf. Comput. Vis. (ICCV), Oct. 2019, pp. 9186–9195. [Google Scholar]
- [48].Xian M, Zhang Y, Cheng H-D, Xu F, Huang K, Zhang B, Ding J, Ning C, and Wang Y, A Benchmark for Breast Ultrasound Image Segmentation (BUSIS). London, U.K.: Infinite Study, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Al-Dhabyani W, Gomaa M, Khaled H, and Fahmy A, “Dataset of breast ultrasound images,” Data Brief, vol. 28, Feb. 2020, Art. no. 104863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ilesanmi AE, Chaumrattanakul U, and Makhanov SS, “A method for segmentation of tumors in breast ultrasound images using the variant enhanced deep learning,” Biocybern. Biomed. Eng, vol. 41, no. 2, pp. 802–818, Apr. 2021. [Google Scholar]
- [51].Yerushalmy J, Statistical Problems in Assessing Methods of Medical Diagnosis, With Special Reference to X-Ray Techniques. Washington, DC, USA: Public Health Reports, 1947, pp. 1432–1449. [PubMed] [Google Scholar]
- [52].Raza S, Goldkamp AL, Chikarmane SA, and Birdwell RL, “U.S. of breast masses categorized as BI-RADS 3, 4, and 5: Pictorial review of factors influencing clinical management,” Radio Graphics, vol. 30, no. 5, pp. 1199–1213, Sep. 2010. [DOI] [PubMed] [Google Scholar]
- [53].Zhang G, Zhao K, Hong Y, Qiu X, Zhang K, and Wei B, “SHA-MTL: Soft and hard attention multi-task learning for automated breast cancer ultrasound image segmentation and classification,” Int. J. Comput. Assist. Radiol. Surg, vol. 16, no. 10, pp. 1719–1725, Oct. 2021. [DOI] [PubMed] [Google Scholar]
- [54].Tanaka H, Chiu S-W, Watanabe T, Kaoku S, and Yamaguchi T, “Computer-aided diagnosis system for breast ultrasound images using deep learning,” Ultrasound Med. Biol, vol. 45, no. 23 , 2019, Art. no. 235013. [DOI] [PubMed] [Google Scholar]
- [55].Shia W-C and Chen D-R, “Classification of malignant tumors in breast ultrasound using a pretrained deep residual network model and support vector machine,” Computerized Med. Imag. Graph, vol. 87, Jan. 2021, Art. no. 101829. [DOI] [PubMed] [Google Scholar]
- [56].Xie J, Song X, Zhang W, Dong Q, Wang Y, Li F, and Wan C, “A novel approach with dual-sampling convolutional neural network for ultrasound image classification of breast tumors,” Phys. Med. Biol, vol. 65, no. 24, Dec. 2020, Art. no. 245001. [DOI] [PubMed] [Google Scholar]


