Abstract
Neurodevelopmental disorders, including autism spectrum disorder (ASD) and attention-deficit/hyper-activity disorder (ADHD), represent a group of conditions that manifest early in child development and produce impairments across multiple domains of functioning. Although a number of pharmacological and psychosocial treatments exist to improve the symptoms associated with these syndromes, treatment advances have lagged. The Precision Medicine Initiative was launched with the goal of revolutionizing medicine by progressing beyond the historical one-size-fits-all approach. In this review, we evaluate current research efforts to personalize treatments for ASD and ADHD. Most pharmacogenetic testing has focused on the cytochrome P450 enzyme family with a particular focus on CYP2D6 and CYP2C19, which are genes that produce an enzyme that acts as a key metabolizer of many prescribed medications. This article provides an update on the state of the field of pharmacogenetics and “therapy-genetics” in the context of ASD and ADHD, and it also encourages clinicians to follow US Food and Drug Administration recommendations regarding pharmacogenetic testing.
Neurodevelopmental disorders, including attention-deficit/hyperactivity disorder (ADHD) and autism spectrum disorder (ASD), have an early onset in life and produce pervasive impairments in personal, social, and academic functioning. ADHD is typified by inattention, hyperactivity, and impulsivity, whereas the hallmark features of ASD include difficulties in social communication and restricted or repetitive behaviors and interests. Current estimates indicate a prevalence of 9% for ADHD and 1.74% for ASD for children age 3 to 17 years, with boys being at substantially greater risk for both disorders. (1) Unfortunately, advances in the development of new treatments and the improvement of existing treatments have been slow.
The Precision Medicine (PM) initiative was introduced as an innovative approach to reform how human diseases are treated. Historically, pharmacological and psychosocial treatments have been based on a nomothetic, or “group” mean perspective. Under this one-size-fits-all approach, treatments may be successful for some patients but not for others. PM was introduced as an approach to be used by all medical and psychosocial interventions with the goal of considering individual differences in a person’s genes and environments. As such, efforts to develop methods for individualizing evidence-based treatments have increased in recent years.
With the completion of the Human Genome Project, the scientific community learned that the genetic architecture of humans was largely identical, with a small (0.9%) interindividual genetic variability accounting for the substantial diversity exhibited by humans. (2) The entrance of a vast array of new technologies has made PM a greater reality and provided clinical researchers an opportunity to connect a person’s personal genetic and environmental circumstances with their clinical profiles. (3) The introduction of two critical scientific innovations allowed for substantial progress within the field of PM: (1) single nucleotide polymorphism (SNP) genotyping and (2) microarray technologies. (4) The identification of polymorphisms is key given that allelic variation has been associated with human liability to disease states and pharmacological responsiveness. In this review, we evaluate current research efforts to personalize treatment of ASD and ADHD. We conclude with recommendations central to advancing personalized intervention science and provide reference resources for clinicians.
Genetic Tests for Diagnosis and Prognosis of ADHD and ASD
Most neurodevelopmental conditions do not have specific diagnostic genetic testing options available. Even still, genetic testing may provide substantial benefits and is a critical part of PM. People with neurodevelopmental disorders are more likely to have copy number variants, and identifying these genomic alterations may inform clinical management, elucidate the need for additional specialists, aid medication and dosing decisions, and identify incidental medically actionable findings. (5,6) The chromosomal microarray (CMA) is a first-tier clinical genetic test for children presenting with congenital abnormalities, intellectual disabilities, global and/or pervasive developmental delays, and ASD. CMAs are frontline tests because they perform genome-wide assessments of copy number variation and may detect regions of homozygosity and mosaicism. Even though many children with ASD have unique genomic profiles, clinicians can compare genomic profiles across patients to help predict symptom trajectory (Figure 1). The diagnostic success rate (here defined as identifying either pathogenic or probable pathogenic genomic variants related to testing indication among a mixed patient population with ASD, congenital abnormalities, developmental delays, and/or intellectual disability) for CMAs is approximately 15% to 20% for simple ASD cases and approximately 30% for complex ASD cases that include additional physical indications (eg, microcephaly). The only genetic testing option with a greater diagnostic success rate for ASD is whole-genome and whole-exome sequencing (28%-68%). (5) As the cost of sequencing continues to fall, whole-genome and whole-exome sequencing may replace CMAs as frontline tests because they provide greater resolution of genetic variation relevant to health and drug metabolism.
Figure 1.
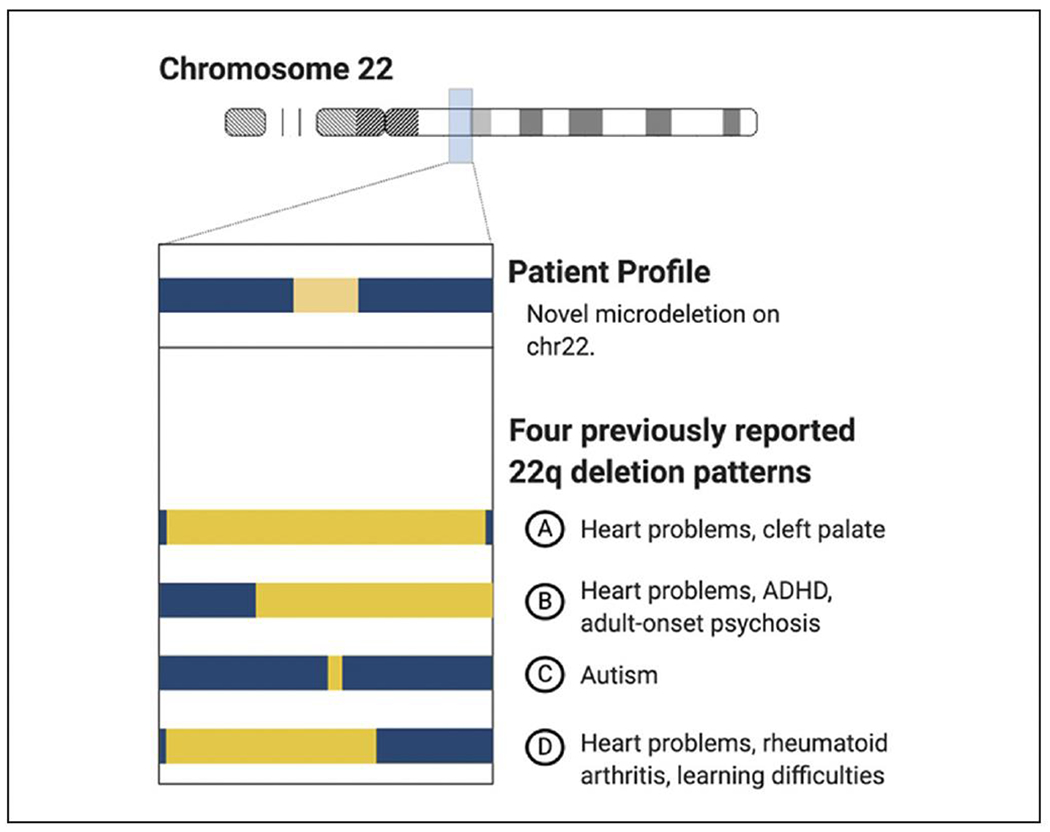
Utility of comparing chromosomal microarray results across patients. Even though patients often have unique genomic profiles of copy number changes, chromosomal microarray (CMA) results may still be informative.This figure shows four unique genomic profiles for a single chromosome.The top profile (A) is from a young child.The bottom three profiles (B, C, D) are results from a clinical database that pairs CMA results with medical records.The clinical team can use this data to assess the need to perform additional testing, to pursue symptom monitoring, or to consult with specialists such as a nephrologist. ADHD, attention-deficit/hyperactivity disorder.
Pharmacogenetic Testing
The purpose of pharmacogenetic testing is to predict which medications and dosages will work optimally for people based on the estimated efficiency of biological pathways salient to drug metabolism. Both genetic and environmental factors influence drug metabolism efficiency. Most of the over-the-counter and clinical pharmacogenetic testing options to estimate drug metabolism efficiency focus on genetic variations in cytochrome P450 enzyme family members. The most popular enzyme to study is CYP2D6, which acts as a major metabolizer of a substantial number of antidepressant medications and stimulants. (7) Multiple studies have reported that adjustments in medication selection and dose based on a person’s predicted CYP2D6 metabolism efficiency category have improved drug tolerability. (7) Unfortunately, categorizing a person’s CYP2D6 drug metabolism activity can be challenging (Figure 2). Each person inherits two copies of CYP2D6, and each copy may contain common, rare, and copy number variants. As a result, geneticists must categorize the CYP2D6 activity level of each inherited copy and then predict their average combined activity. Predicting CYP2D6 activity level can be further complicated by diet and other medications. Some molecules found in common foods (eg, grapefruit and broccoli) and medications (eg, fluoxetine, fluvoxamine, and paroxetine) can interfere with CYP2D6 drug metabolism. (7)
Figure 2.
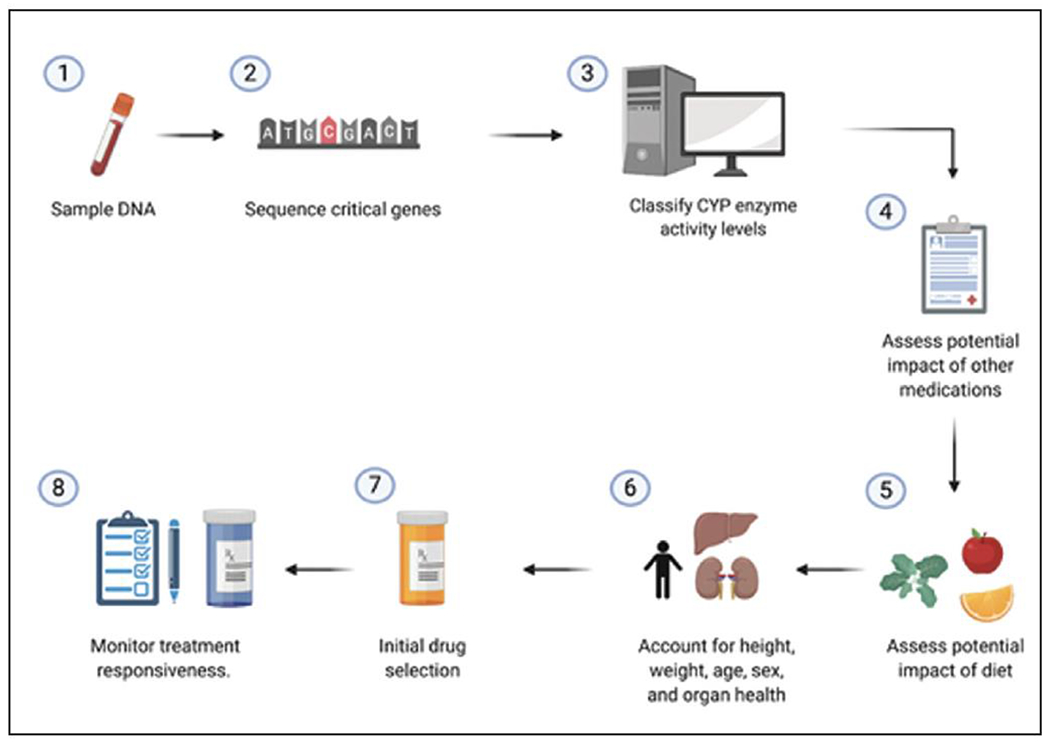
Workflow for precision medicine.(1) A DNA sample is obtained, most commonly from a blood draw or cheek swab. (2) A subset of cytochrome P450 genes are sequenced. Additional genetic variants at key genomic regulation sequences also may be sequenced. (3) The drug metabolism efficiency of each sequenced gene is estimated for both the maternally and paternally inherited alleles. Next, the combined drug metabolism efficacy is estimated. (4) Patient medical records must be checked to determine if and how any medications or vitamins they are taking inhibit cytochrome P450 activity. (5) Self-report information about patient diet must be collected to assess the impact of certain foods in the diet that may inhibit cytochrome P450 activity. (6) Key physical attributes must be assessed, including weight, age, organ health, and sex, when considering medication and dosage selection. (7) Initial medication and dose are prescribed. (8) Additional monitoring is still necessary to assess treatment responsiveness and drug tolerability.Changes in dosage and/or medication may be necessary because drug metabolism efficacy is not a good predictor of treatment responsiveness.
Overall, more testing is needed to understand the complex interplay between genetic background, drug metabolism, and treatment responsiveness. The US Food and Drug Administration (FDA) has recommended against the routine use of genomic testing to select medications to treat psychiatric disorders (8) and has issued pointed warning letters to genomic testing laboratories for advertising that these genetic panels provide “actionable: information, as this may mislead health care workers, patients, and their families and contribute to “inappropriate treatment decisions based on these test results, including inappropriate dosing adjustments, prescribing an ineffective therapy, and not prescribing a therapy that could benefit the patient.” (9) It is possible that as pharmacogenetic research progresses, FDA approval of pharmacogenetic testing for medications to treat the symptoms of psychiatric disorders may increase comparably to FDA-approved genetic testing to select chemotherapy treatments for cancer. In particular, more testing must be done in pediatric populations. Most of the safety and efficacy data for pharmacogenetic testing have come from studies of adults despite multiple lines of evidence that support the notion that children metabolize drugs so differently than adults that simply treating children as “small adults” is inappropriate.
Pharmacological Treatments ADHD
The research summarized in the ADHD and ASD sections of this article varies because the pharmacogenetics studies of ADHD are more advanced compared to those for ASD. For this reason, the ADHD section of this article focuses on studies with replicated findings, whereas the ASD section will present emerging research findings that should be considered preliminary. Stimulants and other attention-enhancing drugs
Methylphenidate.
Methylphenidate (MPH) is a stimulant frequently used to treat the symptoms of ADHD. MPH has undergone substantial pharmacogenetic research, and several genetic variants have been implicated in positive and negative responses to MPH. A recent meta-analysis found five genes associated with positive response to MPH treatment, including improvement of ADHD symptoms (10) (Table 1). Specifically, the genes with significant meta-analytic findings were DRD4 (48-bp VNTR), COMT (rs4680), ADRA4 (rs1800544), SL6A3 (40-bp VNTR), and SLC6A2 (rs28386840 and rs5569). Emerging evidence also suggests that specific genes (such as DRD4, SLC6A4, LPHN3, CES1, SNAP25) may be implicated in negative side effects observed in patients treated with MPH, such as sadness, appetite reduction, skin picking, nail biting, tics, and irritability. (11) However, the evidence for genetic variants associated with increased side effects is not as well established as evidence for positive treatment response.
Table 1:
Summary of Pharmacogenetic Findings
| Drug (symptoms/pediatric disorders treated) | Gene | Findings |
|---|---|---|
| Antipsychotics | ||
| Risperidone (irritability associated with ASD) | CYP2D6 | In patients with ASD (19) * Conflicting results for metabolism * Reduced weight gain in UMs vs EMs * Conflicting results for effect on prolactin levels * No association with insulin resistance |
| ABCB1 | In patients with ASD, treatment response associated with T allele of rs1128503 polymorphism. (19) No significant effect on insulin resistance (rs2032582 and rs1045642) (19) | |
| BDNF | In patients with ASD, the GA and AA genotypes are significantly associated with increased insulin-resistance(rs6265) (33) and prolactin increase (20) | |
| CNR1 | In patients with ASD, increased weight gain associated with T allele of rs806378 and the G allele of rs1049353 (19) | |
| DRD2 | In patients with ASD, conflicting results for effect on prolactin levels. (17) No significant effect on insulin resistance (rs4436578, rs1800497) (19) | |
| DRD3 | In patients with ASD, treatment response associated with the C allele of DRD3 Ser9Gly (rs6280) polymorphism (19) | |
| GHRL | No significant effect on insulin resistance (rs27647) (19) | |
| HTR2A | In patients with ASD, treatment response associated with A allele (rs6311) (19) | |
| HTR2C | In patients with ASD, risperidone-induced weight gain associated with CC genotype (rs3813929) (19) | |
| LEP | In patients with ASD, trend for insulin-resistance associated with AG and GG genotype (rs7799039), (31) weight gain significantly associated with the G allele (19) | |
| UGT1A1 | In patients with ASD, the AG genotype (rs10929302, in complete linkage disequilibrium with rs887829 and rs111741722) was significantly associated with hyperprolactinemia but did not survive correction for multiple testing (21) | |
| Aripiprazole (irritability associated with ASD) | CYP2D6 | Not studied in ASD; however, FDA-approved dosage guidelines recommend reduced dosage for CYP2D6 PRMs and those taking concomitant CYP3A4 inhibitors (19) |
| Stimulants and other attention-enhancing drugs | ||
| Methylphenidate (attention, impulsivity, hyperactivity, oppositional behavior, conduct problems, aggression/ADHD) | SLC6A2 | Meta-analysis of 3 studies shows rs28386840 T allele (compared to A/A) associated with improved response to MPH (10) Meta-analysis of 7 studies shows rs5569 G/G allele (compared to A) associated with improved response to MPH (10) |
| SLC6A3 | Meta-analysis of 16 studies shows 40-base pair VNTR 10-repeat variant (compared to others) associated with reduced efficacy of MPH (10) In patients with ASD, VNTR 9 allele carriers were associated with treatment response but the effect was not significant after multiple correction (19) | |
| SLC6A4 STin2 |
In patients with ASD, VNTR 10 allele carriers were associated with treatment response compared to VNTR 12 allele carriers but effect was not significant after correction for multiple testing (19) | |
| ADRA2A | In patients with ASD, rs1800544 CC allele carriers wereassociated with treatment response but not after correction for multiple testing (19) | |
| ADRA4 | Meta-analysis of 4 studies shows rs1800544 G allele (compared to C) associated with improved response to MPH (10) In patients with ASD, rs1800544 was not statistically associated with treatment response after correction for multiple testing (19) | |
| COMT | Meta-analysis of 7 studies shows rs4680 val/val allele (compared to met) is associated with improved treatment response to MPH (10) In patients with ASD, rs4680 met was associated with treatment response but not after correction for multiple testing (19) | |
| DRD1 | In patients with ASD, the rs5326 effects of CT and TT genotypes on treatment response were not significant after correction for multiple testing, (19) and the rs4867798 effects of CC and CT genotypes on treatment response were not significant after correction for multiple testing (19) | |
| DRD2 | In patients with ASD, those homozygous for the common allele had significantly increased intolerability (19) | |
| DRD3 | In patients with ASD, carriers of the CT and TT genotypes (rs6280) may have reduced tolerability compared to the CC genotype; effect of TT genotype on treatmentresponse was not significant after correction for multiple testing (19) | |
| DRD4 | Meta-analysis of 4 studies shows 48-base pair VNTR 4-repeat variant (compared to others) associated with improved response (10) In patients with ASD, rs11246226 was not associated with treatment response after correction for multiple testing (19) | |
| Amphetamine (attention, impulsivity, hyperactivity, oppositional behavior, conduct problems, aggression/ADHD) | COMT | Improved executive function/working memory/selective attention with val/val variant of rs4680 (13) |
| Atomoxetine (attention, impulsivity, hyperactivity, oppositional behavior/ADHD) | CYP2D6 | PRMs are more likely to have the treatment be effective, butare also more likely to have side effects. Intermediate effects for IMs and EMs. Opposite effects for UMs (14) In patients with ASD, no significant effect of metabolizer status on treatment response (24) |
| Antidepressants and mood stabilizers | ||
| Imipramine (hyperactivity, impulsivity/ADHD) | CYP2D6 | PRMs require lower dosage. Higher doses would be required for efficacy in UMs but this is contraindicated due to high plasma concentration potentially leading to cardiotoxicity (try another drug) (16) |
| Lithium (aggression) | SLC6A4 (5HTTLPR) | S/Sassociated with better response (symptom decrease) in bipolar patients (not tested with aggressive symptoms) (18) |
| BDNF | val/met associated with better response in patients who are bipolar (not tested with aggressive symptoms) (18) | |
| Escitalopram (depression, 12+ years) | SLC6A4 (5HTTLPR) | In patients with ASD, rs25531 S/S genotype was nota significant predictor of treatment response. (19) Small but significant effect of rs2020936-rs2020937 haplotype: reduced treatment effect of rs2020936 S/S only in absence of rs2020937 TT/TT diplotype (23) |
| HTR2A | In patients with ASD, rs7997012 AA genotype not associated with treatment response (19) | |
| CYP2C19 | In patients with ASD, no difference in treatment response. UMs had reduced tolerability to a fixed titration schedule (19) | |
| Fluvoxamine (obsessions and compulsions) | SLC6A4 (5HTTLPR) | In patients with ASD, treatment response significantly reduced in patients with S/S genotype (19) |
Amphetamine.
The FDA-approved amphetamine medications used in the treatment of ADHD include dextroamphetamine/amphetamine and lisdexamfetamine. Amphetamines are partially metabolized by the enzyme CYP2D6, although no genetic variants in the CYP2D6 gene have yet been associated with treatment response. Research is also lacking on the relevant effects of concomitant use of CYP2D6 inhibitors, although similar research on other classes of drugs metabolized by CYP2D6 indicate a strong possibility that CYP2D6 inhibitors, such as paroxetine or fluoxetine, may inhibit amphetamine metabolism, thereby increasing a patient’s blood levels of amphetamine. (12) In addition to the potential impact of CYP2D6 on drug metabolism, there is some evidence that COMT (rs4680) is associated with improved executive functioning, memory, and selective attention in patients with ADHD treated with amphetamine. (13)
Atomoxetine.
Atomoxetine is a selective norepinephrine reuptake inhibitor used to treat symptoms of ADHD. Atomoxetine was the first nonstimulant drug approved to treat ADHD. Atomoxetine is primarily metabolized by CYP2D6, and variations in the gene for this enzyme are associated with variations in therapeutic response. Brown et al. (14) reviewed the evidence for CYP2D6 variations impacting atomoxetine response. They found that people classified as “poor metabolizers” (PRMs) by genetic testing were more likely to experience improved symptoms, but they also were more likely to have cardiovascular side effects such as rapid heart rate and hypertension. Conversely, those classified as “ultra-rapid metabolizers” (UMs) were less likely to experience both symptom improvement and side effects. These responses are likely due to the metabolic profile of patients either increasing the exposure to the active compound (as in PRMs) or decreasing exposure (in UMs). The FDA-approved drug label for atomoxetine includes dosage adjustment recommendations for people who are PRMs for CYP2D6 or for children taking concomitant CYP2D6 inhibitors.
Alpha-2 agonists.
Two alpha-2 agonist medications frequently used as adjunctive treatments in children with ADHD are clonidine and guanfacine. No studies have investigated the pharmacogenetic outcomes of clonidine or guanfacine in children with ADHD. However, guanfacine is metabolized primarily by CYP3A4, so it is possible that variation in this gene may impact drug metabolism. The FDA-approved drug label for guanfacine includes dosage adjustment recommendations for patients taking concomitant CYP3A4 inhibitors and inducers.
Imipramine.
Imipramine is a tricyclic antidepressant used primarily to treat depression in adults. However, it also may be used as an adjunctive treatment for enuresis. In the past, like several other tricyclic antidepressants, it was occasionally used as a second-line treatment for the ADHD. (15) Imipramine is primarily metabolized by CYP2D6. Dean (16) reviewed the evidence for CYP2D6 variation impacting treatment response to imipramine and found that those who are classified as PRMs require lower doses to reduce negative side effects such as sedation, blurred vision, dry mouth, and orthostatic hypotension. Although those classified as UMs technically need higher doses for adequate treatment response, this is not suggested due to increased risk for cardiotoxicity. Therefore, it is generally recommended that patients who are UMs are prescribed an alternative treatment.
Lithium.
Lithium is approved by the FDA to treat pediatric bipolar disorder and may be prescribed off-label for the treatment of conduct disorder that is oftentimes associated with ADHD. (17) No studies have investigated the pharmacogenetic properties of lithium use in children with aggression. However, there is some evidence (although not adequately replicated) that variants in genes such as SLC6A4 and BDNF may influence treatment response in those with bipolar disorder. (18)
Autism Spectrum Disorder
To date, there are no FDA-approved treatments for the core features of ASD. Psychiatric treatment focuses, instead, on the amelioration of associated features including irritability, repetitive behaviors, anxiety, and attention. In the following text we review promising pharmacogenetic studies of FDA-approved medications for irritability associated with ASD, anxiety, and attention/hyperactivity that have been examined in samples of people with ASD.
Risperidone.
Risperidone is an atypical antipsychotic that is FDA-approved for the management of irritability in children and adolescents with ASD (age 5-16 years). Risperidone is the most extensively studied pharmacologic treatment for ASD, and it has received the most pharmacogenetic research of any treatment for ASD. Much research has focused on drug metabolism and the CYP2D6 genetic variability, and although there have been studies in people with and without ASD showing differences in drug metabolism based on genotype (PRMs and UMs), the effects are small and the FDA does not currently recommend adjustment of dosage based on CYP2D6 or any other genotype. Other genes have been studied in people with ASD taking risperidone to determine differences in treatment response, including ABCB1, DRD3, HTR2A, and HTR6, yet inconsistent findings and small effect sizes call into question the clinical value of these genes. Additional studies have examined the genetic effect on side effects associated with risperidone in children with ASD, including weight gain (CYP2D6, HTR2C, FTO, MC4R, LEP, CNR1, FAAH), prolactin elevation (CYP2D6, DRD2, BDNF, UGT1A1), and insulin-resistance (ABCB1, LEP, GHRL, DRD2, CYP2D6, BDNF). (19?21)
Aripiprazole.
Aripiprazole is another atypical antipsychotic that is FDA-approved for the management of irritability in youth with ASD, and the metabolism of this medication has been shown to be significantly impacted by CYP2D6 genotype, although these studies have not been conducted in patients with ASD. FDA-approved dosage recommendations based on the CYP2D6 genotype are included in the package insert and they recommend starting with one-half the usual dose for patients who are PRMs and one-quarter of the usual dose for those taking concomitant CYP3A4 inhibitors.
Metformin.
Insulin resistance and weight gain are common side effects of antipsychotic medications. Metformin has been shown to be effective in mitigating medication-induced weight gain and insulin resistance in children with ASD taking antipsychotic medications, and the genes ATM (C allele) and OCT1 (carriers of one or more variant alleles) were found to be predictors of response to metformin. (22)
Fluvoxamine.
Fluvoxamine, an FDA-approved treatment for obsessions and compulsions associated with obsessive-compulsive disorder in children age 8 years and older, has been examined in one genetic study of people with ASD. In this study, treatment response was significantly reduced in carriers of the S/S genotype. (19)
Escitalopram.
Escitalopram is FDA-approved to treat depression in children age 12 years and older as well as anxiety and depression in adults. The pharmacogenetic effects of escitalopram have been investigated in a few studies of people with ASD and have resulted in discrepant findings. Although general population studies have found pharmacogenetic effects of serotonin transporter (SLC6A4) and serotonin-2A receptor (HTR2A) genes, others have failed to find significant effects in a sample of patients with ASD. (19) Owley et al. (23) found a small effect for SLC6A4 in a forced-titration study of children with ASD in which patients with low 5-HT expression (ie, carriers of the S/S genotype who did not have the TT diplotype in intron 1 of the gene) had the smallest treatment response. A study of CYP2C19 found no difference in treatment response to escitalopram; however, people who are URMs had reduced tolerability to a fixed titration schedule. (19)
Methylphenidate.
One study has examined the pharmacogenomic effects of methylphenidate in patients with ASD. Several genes were nominally associated with treatment response, including DRD1, DRD3, DRD4, ADRA2A, SLC6A3, SLC6A4 STin2, and COMT, although none were significant after correction for multiple testing. DRD2 and DRD3 were significantly associated with tolerability, and these effects survived correction for multiple testing. (19)
Atomoxetine.
The FDA-approved drug label for atomoxetine includes dosage recommendations based on extensive research on the effects of CYP2D6 metabolism of the drug, treatment response, and tolerability or adverse reactions. It is worth noting that atomoxetine can be used in combination with stimulants for patients with both ADHD and ASD. For patients with only ASD, joint use of atomoxetine and stimulants constitutes an off-label treatment of attentional symptoms. The only study to examine the pharmacogenomics of atomoxetine in patients with ASD failed to find a significant effect of CYP2D6 metabolizer status on compliance and ADHD symptoms; however, effects on dosage or adverse events were not reported. (24)
Nonpharmacological Treatments
Although a number of medications exist to treat neurodevelopmental conditions, a number of parents/legal guardians are hesitant to use one or more medications to treat their child’s ADHD or ASD symptoms. For this reason, evidence-based psychosocial treatments may be preferred by some. Parent- and child-based behavioral therapies and behavior modification are evidence-based psychosocial treatments for ADHD and ASD. By their nature, behavioral therapies are highly personalized to the individual child to target specific deficits and/or difficulties. Similar to pharmacogenetics, “therapy-genetics” is an emerging field of research focusing on genetic markers as predictors of psychological therapy response. (25) For ADHD and ASD, there are no well-designed therapy-genetic studies. The phenotypic heterogeneity of these two disorders presents both challenges and opportunities for future gene-informed precision medicine given that treatment targets and markers of treatment response may be obscured by the presence of multiple subgroups. (26) To date, the state of the science is insufficient to make specific treatment recommendations based on specific genetic profiles; however, emerging evidence suggests there may be value in genetic testing to identify whether specific genetic syndromes such as copy number variants or chromosomal aneuploidies are present. For example, carriers of copy number variants of large effect may benefit less from psychosocial/behavior treatments (eg, social skills group treatment) compared to cases with milder phenotypic presentations or polygenic causes. (27)
A number of candidate gene association studies of ADHD and ASD have examined genetic variation in a preselected gene(s) of interest and their association with treatment response (eg, van den Hoofdakker et al. (28). In 2017, the National Advisory Mental Health Council Workgroup on Genomics (29) maintained that candidate gene studies should not be conducted given that candidate gene(s) are selected based on biological theories. As such, the Workgroup recommended that only genes stemming from well-powered genome-wide association studies be considered in association with phenotypes. Moreover, candidate gene-environment studies were deemed to be inadequately powered and plagued by poor operational definitions of environmental variables. (30) Thus, although the inclusion of genotype may enhance the clinician’s ability to personalize psychosocial treatments, only genes derived from genome-wide association studies should be considered.
Conclusions
The goal of the current review was to evaluate the current state of science with regard to the PM effort to improve treatments for ASD and ADHD. What this review highlights is that although there is significant interest amongst clinicians to use genetics to personalize pharmacological and psychosocial treatments, the field is currently in its infancy. At present, the only FDA-approved kits are those designed to test the drug metabolizing enzymes CYP2D6 and CYP2C19. A complete list of cleared or approved tests can be found on the FDA’s website (Table 2). Beyond these drug metabolizing enzymes, the FDA warns clinicians that, for most medications, the relationship between genetic variants and a medication’s effects have not been established. Clinicians are urged to read the FDA-approved drug label or the label of the FDA-cleared or approved genetic test for information regarding whether genetic information should be used to determine therapeutic treatment.
Table 2:
Internet-Based Resources for Clinicians About Precision Medicine
| Public Health Genomics and Precision Health Knowledge Base |
| https://phgkb.cdc.gov |
| US Food and Drug Administration |
| https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests |
| https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific |
| Clinical Pharmacogenetics Implementation Consortium |
| https://cpicpgx.org |
| National Center for Biotechnology Information Pharmacogenetics Resources for Clinical Care |
| https://youtu.be/4Mhzv8LQTa0 |
The PM initiative will face enormous challenges given the sheer complexity of genetic variation, including additive, gene-environment interaction, gene-gene interactions, (31) and genetic pleiotrophy. (32) Moreover, the developing child’s diverse genetic architecture alone is not sufficient to prescribe an individualized treatment plan. Indeed, the contribution of environmental factors to disease etiology and health must be considered. Synthesizing genetic variation with other genomic processes also may lead to the development of enhanced therapeutics tailored to individual patients.
In summary, although PM is a promising approach to determine individualized pharmacological and psychosocial treatments for child and adolescent mental illness, a deep understanding of genetics, biology, environment, and clinical heterogeneity is of paramount importance. The promise to deliver “the right treatment at the right time, every time, to the right person” is a substantial challenge to the field, but this effort has the potential to transform child and adolescent psychiatry.
REFERENCES
- 1.Zablotsky B, Black LI, Maenner MJ, et al. Prevalence and trends of developmental disabilities among children in the United States: 2009-2017. Pediatrics. 2019;144(4):e20190811. 10.1542/peds.2019-0811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novelli G Personalized genomic medicine. Intern Emerg Med. 2010;5(suppl 1):S81–S90. 10.1007/sll739-010-Q455-9 [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg GS, McCarthy JJ. Personalized medicine: revolutionizing drug discovery and patient care. Trends Biotechnol. 2001;19(12):491–496. 10.1016/SQ167-7799(01)01814-5 [DOI] [PubMed] [Google Scholar]
- 4.Jain KK Personalized medicine. Curr Opin Mol Ther. 2002;4(6):548–558. [PubMed] [Google Scholar]
- 5.Malinowski J, Miller DT, Demmer L, et al. ; ACMG Professional Practice and Guidelines Committee. Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability. Genet Med. 2020;22(6):986–1004. 10.1038/s41436-020-0771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waggoner D, Wain KE, Dubuc AM, et al. ; ACMG Professional Practice and Guidelines Committee. Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2018;20(10):1105–1113. 10.1038/s41436-018-004Q-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namerow LB, Walker SA, Loftus M, Bishop JR, Ruaño G, Malik S. Pharmacogenomics: an update for child and adolescent psychiatry. Curr Psychiatry Rep. 2020;22(5):26. 10.1007/sll920-020-Q1145-4 [DOI] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration. The FDA warns against the use of many genetic tests with unapproved claims to predict patient response to specific medications: FDA safety communication. Accessed March 9, 2021. https://www.fda.gov/medical-devices/safety-communications/fda-warns-againstuse-many-genetic-tests-unapproved-claimspredict-patient-response-specific
- 9.US Food and Drug Administration. FDA warning letter Inova Genomics Laboratory. Accessed March 9, 2021. https://www.fda.gov/inspections-complianceenforcement-and-criminal-investigations/warning-letters/inova-genomics-laboratory-577422-04042019.
- 10.Myer NM, Boland JR, Faraone SV. Pharmacogenetics predictors of methylphenidate efficacy in childhood ADHD. Mol Psychiatry. 2018;23(9):1929–1936. 10.1038/mp.2017.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joensen B, Meyer M, Aagaard L. Specific genes associated with adverse events of methylphenidate use in the pediatric population: a systematic literature review. J Res Pharm Pract. 2017;6(2):65–72. 10.4103/jrpp.JRPP_16_161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoretsanitis G, de Leon J, Eap CB, Kane JM, Paulzen M. Clinically significant drug-drug interactions with agents for attention-deficit/hyperactivity disorder. CNS Drugs. 2019;33(12):1201–1222. 10.1007/s40263-019-00683-7 [DOI] [PubMed] [Google Scholar]
- 13.Schacht JP COMT val158met moderation of dopaminergic drug effects on cognitive function: a critical review. Pharmacogenomics J. 2016;16(5):430–438. 10.1038/tpj.2016.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JT, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium guideline for cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin Pharmacol Ther. 2019;106(1):94–102. 10.1002/cpt.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banaschewski T, Roessner V, Dittmann RW, Santosh PJ, Rothenberger A. Nonstimulant medications in the treatment of ADHD. Eur Child Adolesc Psychiatry. 2004;13 (suppl 1):I102–I116. 10.1007/s00787-004-1010-x [DOI] [PubMed] [Google Scholar]
- 16.Dean L Imipramine therapy and CYP2D6 and CYP2C19 genotype. In: Pratt VM, McLeod HL, Rubinstein WS, et al. , eds. Medical Genetics Summaries. National Center for Biotechnology Information; 2012:29–43. [Google Scholar]
- 17.Malone RP, Delaney MA, Luebbert JF, Cater J, Campbell M. A double-blind placebo controlled study of lithium in hospitalized aggressive children and adolescents with conduct disorder. Arch Gen Psychiatry. 2000; 57(7):649–654. 10.1001/archpsyc.57.7.649. [DOI] [PubMed] [Google Scholar]
- 18.Pagani R, Gasparini A, Ielmini M, et al. Twenty years of lithium pharmacogenetics: a systematic review. Psychiatry Res. 2019;278:42–50. 10.1016/i.psvchres.2019.05.036 [DOI] [PubMed] [Google Scholar]
- 19.Brown JT, Eum S, Cook EH, Bishop JR. Pharmacogenomics of autism spectrum disorder. Pharmacogenomics. 2017;18(4):403–414. 10.2217/pgs-2016-0167 [DOI] [PubMed] [Google Scholar]
- 20.Correia CT, Almeida JP, Santos PE, et al. Pharmacogenetics of risperidone therapy in autism: association analysis of eight candidate genes with drug efficacy and adverse drug reactions. Pharmacogenomics J. 2010;10(5):418–430. 10.1038/tpi.2009.63 [DOI] [PubMed] [Google Scholar]
- 21.Hongkaew Y, Medhasi S, Pasomsub E, et al. UGT1A1 polymorphisms associated with prolactin response in risperidone-treated children and adolescents with autism spectrum disorder. Pharmacogenomics J. 2018;18(6):740–748. 10.1038/s41397-018-0Q31-7 [DOI] [PubMed] [Google Scholar]
- 22.Garfunkel D, Anagnostou EA, Aman MG, et al. Pharmacogenetics of metformin for medication-induced weight gain in autism spectrum disorder. J Child Adolesc Psychopharmacol. 2019;29(6):448–455. 10.1089/cap.2018.0171 [DOI] [PubMed] [Google Scholar]
- 23.Owley T, Brune CW, Salt J, et al. A pharmacogenetic study of escitalopram in autism spectrum disorders. Autism Res. 2010;3(1):1–7. 10.1002/aur.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrie ES, Pinsonneault JK, Sadee W, et al. Testing genetic modifiers of behavior and response to atomoxetine in autism spectrum disorder with ADHD. J Dev Phys Disabil. 2018;30(3):355–371. 10.1007/slQ882-018-9590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lester KJ, Eley TC. Therapygenetics: using genetic markers to predict response to psychological treatment for mood and anxiety disorders. Biol Mood Anxiety Disord. 2013;3(1):4. 10.1186/2045-5380-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benger M, Kinali M, Mazarakis ND. Autism spectrum disorder: prospects for treatment using gene therapy. Mol Autism. 2018;9(1):39. 10.1186/sl3229-018-0222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tammimies K, Li D, Rabkina I, et al. Association between copy number variation and response to social skills training in autism spectrum disorder. Sci Rep. 2019;9(1):9810. 10.1038/s41598-019-46396-l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Hoofdakker BJ, Nauta MH, Dijck-Brouwer DAJ, et al. Dopamine transporter gene moderates response to behavioral parent training in children with ADHD: a pilot study. Dev Psychol. 2012;48(2):567–574. 10.1037/a0026564 [DOI] [PubMed] [Google Scholar]
- 29.National Institute of Mental Health. Report of the National Advisory Mental Health Council Workgroup on Genomics. Accessed March 9, 2021. https://www.nimh.nih.gov/about/advisory-boards-and-groups/namhc/reports/report-of-the-national-advisorymental-health-council-workgroup-ongenomics.shtml#references
- 30.Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168(10):1041–1049. 10.1176/appi.ajp.2011.11020191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visscher PM Human complex trait genetics in the 21st century. Genetics. 2016;202(2):377–379. 10.1534/genetics.115.180513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visscher PM, Yang J. A plethora of pleiotropy across complex traits. Nat Genet. 2016;48(7):707–708. 10.1038/ng.3604 [DOI] [PubMed] [Google Scholar]
- 33.Sukasem C, Hongkaew Y, Ngamsamut N, et al. Impact of pharmacogenetic markers of CYP2D6 and DRD2 on prolactin response in risperidone-treated Thai children and adolescents with autism spectrum disorders. J Clin Psychopharmacol. 2016;36(2):141–146. 10.1097/JCP.000000000000Q474 [DOI] [PubMed] [Google Scholar]


