Abstract
Background
Carbapenemase-producing (CP) Escherichia coli (CP-Ec) are a global public health threat. We aimed to describe the clinical and molecular epidemiology and outcomes of patients from several countries with CP-Ec isolates obtained from a prospective cohort.
Methods
Patients with CP-Ec were enrolled from 26 hospitals in 6 countries. Clinical data were collected, and isolates underwent whole-genome sequencing. Clinical and molecular features and outcomes associated with isolates with or without metallo-β-lactamases (MBLs) were compared. The primary outcome was desirability of outcome ranking (DOOR) at 30 days after the index culture.
Results
Of the 114 CP-Ec isolates in Consortium on resistance against carbapenems in Klebsiella and other Enterobacterales-2 (CRACKLE-2), 49 harbored an MBL, most commonly blaNDM-5 (38/49, 78%). Strong regional variations were noted with MBL-Ec predominantly found among patients in China (23/49). Clinically, MBL-Ec were more often from urine sources (49% vs 29%), less often met criteria for infection (39% vs 58%, P = .04), and had lower acuity of illness when compared with non–MBL-Ec. Among patients with infection, the probability of a better DOOR outcome for a randomly selected patient with MBL-Ec as compared with non–MBL-Ec was 62% (95% CI: 48.2–74.3%). Among infected patients, non–MBL-Ec had increased 30-day (26% vs 0%; P = .02) and 90-day (39% vs 0%; P = .001) mortality compared with MBL-Ec.
Conclusions
Emergence of CP-Ec was observed with important geographic variations. Bacterial characteristics, clinical presentations, and outcomes differed between MBL-Ec and non–MBL-Ec. Mortality was higher among non-MBL isolates, which were more frequently isolated from blood, but these findings may be confounded by regional variations.
Keywords: multidrug resistance, carbapenem resistance, E. coli
Carbapenemase-producing Escherichia coli (CP-Ec) are concerning given the global prevalence of E. coli in many infection sites and limited available treatment options. We observed regional variation in CP-Ec, often among high-risk genotypes. Mortality was lower in those infected with metallo-β-lactamase producers.
Antimicrobial resistance is a major concern, presenting both a challenge to the treatment of patients today and an ever-growing threat to global public health. Carbapenem-resistant Enterobacterales (CRE), such as Escherichia coli, are particularly concerning because of their high level of antimicrobial resistance, limited current treatment options, and potential for widespread transmission via mobile genetic elements [1]. Carbapenem resistance can occur in the setting of carbapenemase production or via a combination of a β-lactamase with porin mutations [2, 3]. Carbapenem resistance due to carbapenemases is particularly worrisome as genes encoding these β-lactamases are often present on mobile genetic elements that facilitate further horizontal transmission. From their initial identification, carbapenemase-producing Enterobacterales have rapidly spread globally during the last 25 years [3].
Escherichia coli are the most common cause of Enterobacterales infections in various clinical settings and are often community acquired [4, 5]. Importantly, resistant E. coli is the organism responsible for most deaths due to antimicrobial resistance globally [6]. Thus, acquisition of carbapenem resistance among E. coli is of significant concern. While carbapenem resistance was initially described among hospital-acquired infections, a rising incidence of carbapenem-resistant E. coli infections not associated with hospitalization is increasingly reported [4].
Among the various carbapenemase-producing (CP) E. coli (CP-Ec), the presence of a metallo-β-lactamase (MBL; class B carbapenemase) is particularly worrisome because treatment options against these isolates are limited. Clinically important MBLs include the New-Delhi (NDM), Verona integron-encoded (VIM), and imipenemase (IMP) MBLs. Currently approved novel β-lactamase inhibitors such as avibactam, vaborbactam, and relebactam lack sufficient inhibitory activity against MBL to protect their companion β-lactams [7, 8]. Cefiderocol is one of the few agents with activity against MBL-producing Enterobacterales and early efficacy data to support its use [9]. A better understanding of the epidemiology and outcomes of MBL-Ec is needed to guide proper therapy. We conducted this study to describe the clinical and molecular epidemiology and outcomes of CP-Ec and to examine how these factors differed between MBL and non-MBL CP-Ec.
METHODS
Study Design
CRACKLE-2 was a multicenter, international, prospective cohort study [10]. The inclusion criteria and study design have been previously described [10]. Briefly, patients of any age admitted to the hospital with CRE clinical isolates consistent with the Centers for Disease Control and Prevention (CDC) definition from any site were consecutively enrolled. In the present analysis, patients enrolled from June 2017 to July 2018 were eligible for inclusion when the first qualifying CRE culture was positive for a carbapenem-resistant E. coli isolate, and at least 1 carbapenemase gene was present on whole-genome sequencing (WGS). The study was approved by each site's institutional review board, including a waiver for the requirement to obtain informed consent.
Study Procedures and Definitions
Clinical data including patient demographics, clinical characteristics, laboratory findings, antimicrobial treatments received, and outcomes were recorded from the electronic health record (EHR) by on-site investigators. We used clinical data to calculate 2 index scores to capture acute severity of illness and comorbidity: the Pitt bacteremia score (PBS) and the Charlson comorbidity index, respectively [11]. The PBS is a severity of acute illness index that ranges from 0 to 14 and has been previously validated in predicting mortality among gram-negative bacteremic and nonbacteremic infections [12, 13]. Higher scores indicate more severe illness; a score of 4 or higher indicates critical illness. The Charlson comorbidity index ranges from 0 to 37, with higher scores indicating more comorbid conditions and/or more severe comorbidities [11].
Infection was defined based on culture source as previously described [10] and detailed in the Supplementary Methods. We categorized isolates as either MBL-carbapenemase-producing E. coli (MBL-Ec) or as non–MBL-carbapenemase-producing E. coli (non–MBL-Ec), based on the presence of any MBL gene including blaNDM, blaVIM, or blaIMP detected by sequencing.
Whole-Genome Sequencing
Whole-genome sequencing was performed on all isolates at UTHealth, Houston, Texas, USA (HiSeq 4000, NextSeq 2000, and MiSeq; Illumina; San Diego, CA, USA); the Molecular Resource Facility, Rutgers, New Brunswick, New Jersey, USA (NextSeq 500; Illumina); the University of El Bosque, Bogota, Colombia (MiSeq, HiSeq 4000, and NextSeq 2000; Illumina); and BGI Genomics, BGI-Shenzhen, Shenzhen, China (HiSeq X; Illumina), as previously described [10]. Draft genomes were assembled by use of SPAdes, version 3.13.0. Escherichia coli multilocus sequence types (STs) were determined using mlst v2.22 (github.com/tseemann/mlst), whereas phylogroups, serotypes, and fimH types were examined using ClermonTyping v21.3 [14], ECTyper v1.0.0 [15], and FimTyper v1.0 [16], respectively.
Resistance genes were determined using AMRFinderPlus, version 3.10.20 [17], and ARIBA, version 2.14.6 [18]. Core genome phylogeny was generated by Snippy, version 4.6.0, and the single nucleotide polymorphisms (SNPs) in the prophages, repeated regions, and insertion elements were filtered as previously described [10]. A maximum likelihood phylogenetic tree was constructed in RAxML, version 8.2.4 [19]. The genomes sequenced in this study were deposited in GenBank (accession number PRJNA658369).
Outcomes
Mortality outcomes were captured through 90 days after discharge. We analyzed mortality at 30 and 90 days from index culture. Other relevant outcome data included the presence of clinical response, time from culture to discharge, and discharge disposition. We assessed mortality outcomes across the entire cohort and separately among those who met infection criteria. The remainder of the outcomes analyses were performed only on the subset of patients who met the study definition for infection.
The primary outcome measure was a desirability of outcome ranking (DOOR) [20], as previously described [10]. Briefly, at 30 days following the index culture, the following outcomes were assessed: (1) deleterious effects, including absence of clinical response, prolonged hospitalization (≥30 days after first positive culture or readmission within 30 days); (2) adverse events, including new renal failure and/or Clostridioides difficile infection; and (3) survival at 30 days after the index culture. The absence of clinical response was defined as no improvement or resolution of symptoms at either 30 days or at the time of discharge, when discharge occurred before 30 days from the index culture (details are shown in the Supplementary Methods). These data were used to categorize DOOR rankings at 30 days as (1) alive with no events, (2) alive with 1 event, (3) alive with 2 or more events, or (4) dead. Post hoc, to examine other factors potentially related to mortality outcomes, we assessed the association between age, STs, and bacteremia and the outcome of 30-day mortality.
Antimicrobial Therapy
The receipt of antimicrobial therapy with potential activity against CP-Ec was evaluated, during either empiric or definitive therapy. Additionally, the time to receipt of a presumed active antibiotic according to local susceptibility data was analyzed by MBL group. Details of these analyses are shown in the Supplementary Methods.
Statistical Analysis
We conducted descriptive statistics of patient demographics, clinical characteristics, molecular and bacterial characteristics, and outcomes. We examined patient demographics, clinical factors, molecular and bacterial characteristics, and outcomes by MBL status, using Pearson chi-square tests, and differences in distributions of continuous variables by MBL status using Kruskal-Wallis tests.
We calculated a DOOR probability by evaluating 30-day composite outcomes by MBL status using pairwise DOOR analyses. Each DOOR analysis estimates the probability that a randomly selected patient in 1 group would have a better overall outcome than a patient in the comparison group. A probability where the confidence interval (CI) crosses 50% implies no difference in the distribution of composite outcomes and is not considered statistically significant. Separately, 30- and 90-day mortality by MBL status was examined using Pearson chi-square analysis. We conducted all analyses in SAS, version 9.4 (SAS Institute Inc., Cary, NC, USA). Data visualizations of molecular bacterial characteristics were created using Tableau Desktop. CRACKLE-2 was registered with ClinicalTrials.gov (NCT03646227).
RESULTS
Clinical Epidemiology
From 1 June 2017 to 31 July 2018, 196 unique patients were enrolled with an eligible first culture positive for CRE E. coli that was confirmed using WGS. Of these, 114 (58%) unique patients from 26 hospitals in 6 countries had a carbapenemase gene detected and were included here (Table 1).
Table 1. .
Clinical Characteristics of Patients With Carbapenemase-Producing Escherichia coli (CP-Ec) Isolates
| Total Patients (n = 114) | MBL-Ec (n = 49) | Non–MBL-Ec (n = 65) | P a | |
|---|---|---|---|---|
| Age, median (Q1, Q3), y | 60 (42, 74) | 60 (37, 74) | 61 (45, 76) | .97 |
| Sex, male, n (%) | 57 (50%) | 21 (43%) | 36 (55%) | .19 |
| Countryb | <.0001c | |||
| Australia | 1 (1%) | 1 (2%) | 0 (0%) | |
| China | 24 (21%) | 23 (47%) | 1 (2%) | |
| Colombia | 26 (23%) | 5 (10%) | 21 (32%) | |
| Lebanon | 34 (30%) | 16 (33%) | 18 (28%) | |
| Singapore | 4 (4%) | 0 (0%) | 4 (6%) | |
| United States | 25 (22%) | 4 (8%) | 21 (32%) | |
| Charlson comorbidity index,d median (Q1, Q3) | 2 (0, 3) | 2 (0, 3) | 2 (1, 4) | .12 |
| Pitt bacteremia score,e median (Q1, Q3) | 2 (0, 3) | 0 (0, 2) | 2 (0, 3) | .005 |
| Admitted from, n (%) | .10f | |||
| Home | 80 (70%) | 32 (65%) | 48 (74%) | |
| Hospital transfer | 28 (25%) | 16 (33%) | 12 (18%) | |
| Long-term care | 4 (4%) | 0 (0%) | 4 (6%) | |
| Transfer from outside country? | 2 (2%) | 1 (2%) | 1 (2%) | |
| Location on day of positive culture, n (%) | .10 | |||
| Intensive care unit | 32 (28%) | 13 (27%) | 19 (29%) | |
| Medical/surgical ward | 62 (40%) | 29 (59%) | 33 (51%) | |
| Emergency room | 15 (13%) | 3 (6%) | 12 (18%) | |
| Other | 5 (4%) | 4 (8%) | 1 (2%) | |
| Days from admittance to positive culture,f median (Q1, Q3) | 6 (1, 17) | 6 (1, 15) | 6 (0, 21) | .67 |
| Culture source, n (%) | ||||
| Blood | 24 (21%) | 3 (6%) | 21 (32%) | .02f |
| Urine | 39 (34%) | 20 (41%) | 19 (29%) | |
| Respiratory | 10 (9%) | 6 (12%) | 4 (6%) | |
| Wound | 8 (7%) | 4 (8%) | 4 (6%) | |
| Non-wound abdominal | 12 (11%) | 6 (12%) | 6 (9%) | |
| Other | 21 (18%) | 10 (20%) | 11 (17%) | |
| Isolates meeting infection criteria, n (%) | 57 (50%) | 19 (39%) | 38 (58%) | .04 |
Abbreviations: MBL, metallo-β-lactamase gene; non–MBL, no metallo-β-lactamase gene present; Q, quartile.
Statistical tests performed include chi-square test for categorical variables, except in the case of low expected cell counts when Fisher's exact was used, and Kruskal-Wallis test for differences in distributions for continuous variables. P values compare isolates harboring MBL with those not harboring MBL.
Percentages may total >100% due to rounding.
Fisher's exact test conducted when any expected cell counts were <5.
A chronic comorbidity score ranging from 0 to 37, with higher scores indicating the presence of more comorbidities.
An acute severity of illness score, with higher scores indicating more severe illness.
Time to first positive culture indicates the number of days from admission to the collection date of the index culture, with 0 indicating that the index culture was obtained on the day of admission.
The median age of enrolled patients was 60 years (quartile [Q] 1: 42 years; Q3: 74 years), and half were male. Most patients (70%; 80/114) were admitted from home, and cultures with CP-Ec were isolated a median of 6 (Q1: 1; Q3: 7) days after admission. Isolates were from various sources, most commonly urine (34%; 39/114) or blood (21%, 24/114).
Forty-three percent of isolates (49/114) harbored a gene encoding for an MBL, whereas 57% (65/114) represented non–MBL-Ec. Patient demographics including age and sex were similar among MBL-Ec and non–MBL-Ec. Geographic variations were evident (Table 1) (P < .0001), with MBL-Ec found most commonly in isolates from China (23/24); non–MBL-Ec isolates predominating in the United States (21/25), Colombia (21/26), and Singapore (4/4); and both MBL-Ec (16/34; 47%) and non–MBL-Ec isolates (18/34; 53%) detected in Lebanon. Half of the isolates (57/114) accompanied a clinical episode that met the definition for infection. Compared with non–MBL-Ec isolates, patients with MBL-Ec were less likely to meet criteria for infection (39% vs 58%; P = .04). Patients harboring MBL-Ec isolates were less acutely ill at the time of first positive culture than those with non–MBL-Ec (median PBS: 0 [Q1: 0; Q3: 2] vs 2 [Q1: 0; Q3: 3]; P = .005) but had similar summary comorbidity scores (median Charlson comorbidity index: 2 [Q1: 0; Q3: 3] vs 2 [Q1: 1; Q3: 4]). Among patients meeting infection criteria, Pitt scores remained lower among those with MBL-Ec than those with non–MBL-Ec (median of 0 [Q1: 0; Q3: 2] vs median of 2 [Q1: 0; Q3: 3]; P = .0007).
The source of culture was associated with MBL status (Table 1) (P = .02). Isolates from blood were more likely to be non–MBL-Ec than MBL-Ec (32% vs 6%); conversely, isolates from urine were more often MBL-Ec (41% MBL-Ec vs 29% non–MBL-Ec).
Molecular Epidemiology
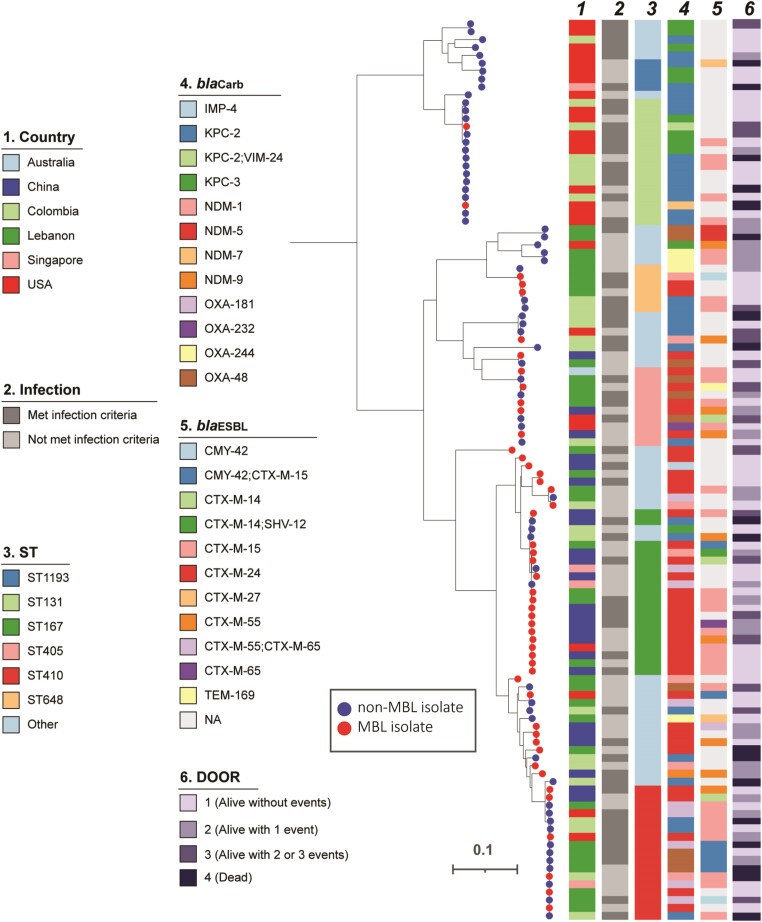
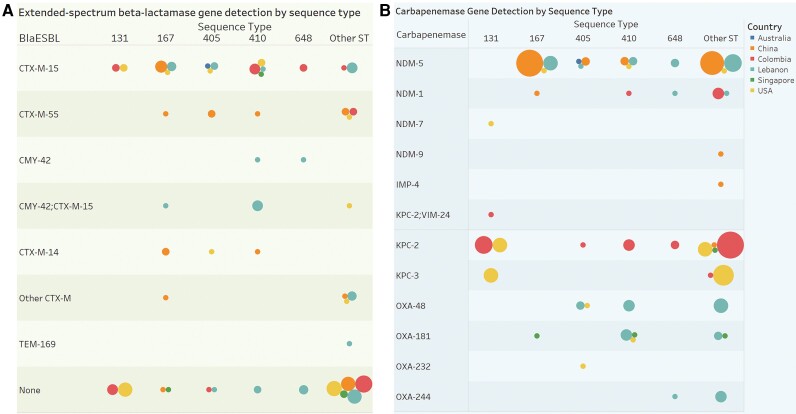
The bacterial population structure of isolates in this study is shown in Figure 1. Several regional variations were observed in ST and molecular characteristics including carbapenemases (Figure 2A) and extended-spectrum β-lactamase (ESBL) genes (Figure 2B) by country. Among MBLs harboring CP-Ec isolates, blaNDM was the most common carbapenemase (Table 2) (47/49; 96%), predominantly blaNDM-5 (38/49; 78%). Among non–MBL-Ec, blaKPC-2 was the most common carbapenemase gene (30/65; 46%). Sequence types also varied by MBL status (P < .0001). The high-risk ST167 was more common among MBL than non-MBL isolates (31% vs 2%), while ST131 was observed more frequently among non-MBL isolates (20% vs 4%). Serotypes also varied by MBL status; H4:025 was the most common and was associated with non-MBL isolates (18% vs 4%), whereas H9:O101 was more frequently observed among MBL isolates (22% vs 3%). Acquired ESBL genes were common among CP-Ec (59/114; 52%), with blaCTX-M-15 the most common among both MBL (16/49; 33%) and non-MBL (22/65; 34%) isolates.
Figure 1.
Phylogenetic population structures of CP-Ec isolates. blaCarb = carbapenemase gene detected. Abbreviations: CP-Ec, carbapenemase-producing Escherichia coli; DOOR, desirability of outcome ranking; IMP, imipenemase; KPC, Klebsiella pneumoniae complex; MBL, metallo-β-lactamase; NA: not applicable; NDM, New Delhi metallo-β-lactamase; OXA, OXA-type carbapenemase; ST, sequence type; VIM, Verona integron-encoded.
Figure 2.
Regional differences in molecular characteristics of carbapenemase-producing E. coli. A, Circles determined by country (color, see legend) and distinct count of subjects (size of circle) broken down by ST and carbapenemase. B, Circles determined by country (color) and distinct count of subjects (size) broken down by sequence type versus blaESBL. The CTX-M-14 group includes 1 subject with CTX-M-14 and SHV-12 detected. The other CTX-M group includes 1 individual with CTX-M-65, 1 with CTX-M-55 and CTX-M-65, 1 with CTX-M-24, and 2 with CTX-M-27. Abbreviations: ESBL, extended-spectrum β-lactamase; IMP, imipenemase; KPC, Klebsiella pneumoniae complex; MBL, metallo-β-lactamase; NDM, New Delhi metallo-β-lactamase; OXA, OXA-type carbapenemase; ST, sequence type; VIM, Verona integron-encoded.
Table 2. .
Bacterial Characteristics of Carbapenemase-Producing Escherichia coli Isolates
| Total (N = 114) | MBL (n = 49) | Non-MBL (n = 65) | P (α = .05)a | |
|---|---|---|---|---|
| Carbapenemaseb | ||||
| blaIMP-4 | 1 (1%) | 1 (2%) | 0 (0%) | |
| blaKPC-2 | 31 (27%) | 1 (2%)c | 30 (46%) | |
| blaKPC-3 | 11 (10%) | 0 (0%) | 11 (17%) | |
| blaNDM-5 | 38 (33%) | 38 (78%) | 0 (0%) | |
| blaNDM-1 | 7 (6%) | 7 (14%) | 0 (0%) | |
| blaNDM-7 | 1 (1%) | 1 (2%) | 0 (0%) | |
| blaNDM-9 | 1 (1%) | 1 (2%) | 0 (0%) | |
| blaOXA-48 | 10 (9%) | 0 (0%) | 10 (15%) | |
| blaOXA-181 | 9 (8%) | 0 (0%) | 9 (14%) | |
| blaOXA-232 | 1 (1%) | 0 (0%) | 2 (2%) | |
| blaVIM-24 | 1 (1%) | 1 (2%)c | 0 (0%) | |
| Sequence type | <.0001d | |||
| 410 | 17 (15%) | 6 (12%) | 11 (17%) | |
| 167 | 16 (14%) | 15 (31%) | 1 (2%) | |
| 131 | 15 (13%) | 2 (4%) | 13 (20%) | |
| 405 | 9 (8%) | 4 (8%) | 5 (8%) | |
| 648 | 6 (5%) | 3 (6%) | 3 (5%) | |
| 1193 | 4 (4%) | 0 (0%) | 4 (6%) | |
| 10 | 3 (3%) | 0 (0%) | 3 (5%) | |
| 101 | 3 (3%) | 1 (2%) | 2 (3%) | |
| 448 | 3 (3%) | 3 (6%) | 0 (0%) | |
| 46 | 3 (3%) | 2 (4%) | 1 (2%) | |
| 69 | 3 (3%) | 1 (2%) | 2 (3%) | |
| 1722 | 2 (2%) | 0 (0%) | 2 (3%) | |
| 354 | 2 (2%) | 0 (0%) | 2 (3%) | |
| 361 | 2 (2%) | 2 (4%) | 0 (0%) | |
| 617 | 2 (2%) | 1 (2%) | 1 (2%) | |
| 73 | 2 (2%) | 0 (0%) | 2 (3%) | |
| Other | 22 (19%) | 9 (18%) | 13 (20%) | |
| Serotype | .0019e | |||
| H4:025 | 14 (12%) | 2 (4%) | 12 (18%) | |
| H9:O101 | 13 (11%) | 11 (22%) | 2 (3%) | |
| H6:O102 | 10 (9%) | 5 (10%) | 5 (8%) | |
| H21:O8 | 8 (7%) | 1 (2%) | 7 (11%) | |
| H10:O101 | 4 (4%) | 1 (2%) | 3 (5%) | |
| H5:O75 | 4 (4%) | 0 (0%) | 4 (6%) | |
| H6:O45 | 4 (4%) | 1 (2%) | 3 (5%) | |
| H9:O8 | 4 (4%) | 2 (4%) | 2 (3%) | |
| Other | 53 (46%) | 26 (23%) | 27 (38%) | |
| ESBL not detected | 55 (48%) | 20 (41%) | 35 (54%) | |
| ESBL detectedf | 59 (52%) | 29 (59%) | 30 (46%) | .17 |
| blaCTX-M-15 | 38 (33%) | 16 (33%) | 22 (34%) | |
| blaCTX-M-55 | 10 (9%) | 8 (16%) | 2 (3%) | |
| blaCMY-42 | 8 (7%) | 3 (6%) | 5 (8%) | |
| blaCTX-M-14 | 4 (4%) | 3 (6%) | 1 (2%) | |
| blaCTX-M-24 | 2 (2%) | 0 (0%) | 2 (3%) | |
| blaCTX-M-27 | 2 (2%) | 0 (0%) | 2 (3%) | |
| blaCTX-M-65 | 1 (1%) | 1 (2%) | 0 (0%) |
Data are presented as n (%). Values displayed in table if containing at least 1% of the cohort for each category.
Abbreviations: ESBL, extended-spectrum β-lactamases; IMP, imipenemase; KPC, Klebsiella pneumoniae complex; MBL, metallo-β-lactamase; NDM, New Delhi metallo-β-lactamase; OXA, OXA-type carbapenemase; VIM, Verona integron-encoded.
Statistical tests performed include chi-square test unless otherwise noted. P values compare isolates harboring MBL with those not harboring MBL.
Among the entire cohort, there were additionally 1 of each of the following carbapenemases: blaNDM-7, blaNDM-9, blaOXA-232, blaIMP-4, blaVIM-24.
One isolate had both KPC-2 and VIM-24 classified as MBL; because of this, column totals to 50 carbapenemases detected among n = 49 patients.
For this analysis, all sequence types that were present in <10% of the total cohort were grouped into an “other” category for Fisher's exact analysis.
For this analysis, all serotypes that were present in <10% of the total cohort were grouped into a “other” category for Fisher's exact test.
Among the entire cohort, there were additionally the following acquired ESBLs detected: 2 blaCTX-M-24 and 2 blaCTX-M-27 in non-MBL isolates and blaCTX-M-65 detected in 1 MBL isolate.
Outcomes
Mortality at 30 days among all patients was 16% (18/114) and was higher in patients with non–MBL-Ec than those with MBL-Ec isolates (23% vs 6%; P = .02). All-cause 90-day mortality was 22% (25/114) and remained associated with non–MBL-Ec isolates (32% vs 8%; P = .003).
Patient outcomes among those who met the definition for infection (n = 57) are detailed in Table 3. Most had documented clinical response (39/57; 68%), and this was not associated with MBL status. Thirty-day mortality among infected patients was 18% (10/57) and was higher in patients with non–MBL-Ec than in those with MBL-Ec (10/38 vs 0/19; P = .02). At 90 days, 15 patients had died (26%), all of whom were infected with non–MBL-Ec (P = .001). In DOOR analysis (Table 3), the probability of a better outcome for a randomly selected patient with MBL-Ec compared with a patient with non–MBL-Ec was 62% (95% CI: 48.2–74.3%). Mortality varied by ST, with no deaths in high-risk clones ST131 or ST167 and 3 deaths in ST410. Age was associated with mortality, with a median (Q1, Q3) age of 66 (63, 78) years among those who died by 30 days, compared with a median age of 54 (37, 68) years among those alive at 30-day follow-up (P = .03). Among patients with bacteremia (n = 24), death by 30 days was not associated with MBL status (0/3 deaths in MBL-Ec, 5/21 deaths in non–MBL-Ec; P = .34).
Table 3.
Clinical Outcomes of Patients With Carbapenemase-Producing Escherichia coli (CP-Ec) Infection
| Total Patients (N = 57) | MBL-Ec (n = 19) | Non–MBL-Ec (n = 38) | P (α = 0.05)a | |
|---|---|---|---|---|
| Clinical response | 39 (68%) | 14 (74%) | 25 (66%) | .76 |
| Disposition after dischargeb | .06 | |||
| Home | 38 (67%) | 16 (84%) | 22 (58%) | |
| Other care facility | 5 (9%) | 2 (11%) | 3 (8%) | |
| Hospice or death | 14 (25%) | 1 (5%) | 13 (34%) | |
| Time from culture to discharge, median days (Q1, Q3) | 18 (7, 25) | 21 (6, 54) | 17.5 (8, 24) | .35c |
| 30-Day mortality | 10 (18%) | 0/19 (0%) | 10/38 (26%) | .02 |
| 90-Day mortality | 15 (26%) | 0/19 (0%) | 15/38 (39%) | .001 |
| DOOR outcomes (30 d) | 62.0% (48.2%-74.3%)d | |||
| Alive without events | 25 (44%) | 10 (53%) | 15 (39%) | |
| Alive with 1 event | 11 (19%) | 4 (21%) | 7 (18%) | |
| Alive with 2 or 3 events | 11 (19%) | 5 (26%) | 6 (16%) | |
| Dead | 10 (18%) | 0 (0%) | 10 (26%) |
Data are presented as n (%).
Abbreviations: DOOR, desirability of outcome ranking; MBL, metallo-β-lactamase; Q, quartile.
Statistical tests performed include chi-square test for categorical variables, except in the case of low expected cell counts when Fisher's exact was used, and Kruskal-Wallis test for differences in distributions for continuous variables. P values compare isolates harboring MBL with those not harboring MBL.
Disposition to “other care facility” includes 3 transfers to another hospital, 1 transfer to long-term care, 1 transfer to long-term acute care.
Kruskal-Wallis test.
DOOR probability (95% confidence interval), or the probability that a randomly selected patient in 1 group would have a better overall outcome than a randomly selected patient in the comparison group.
Among patients who met criteria for infection (n=57), receipt of antibiotics with potential activity against CP-Ec, receipt of and time to receipt of presumed effective antibiotic based on local susceptibility testing are summarized in Supplementary results.
DISCUSSION
In this large, multinational cohort of CP-Ec, isolates harboring MBLs were clustered in specific geographic regions and among specific bacterial genotypes. In this study, MBL-Ec isolates were more frequently isolated from urine, less frequently met criteria for infection, and patients infected with MBL-Ec demonstrated lower 30- and 90-day mortality than those infected with non–MBL-Ec. Using DOOR analyses, similar patient outcomes between MBL- and non–MBL-Ec were observed.
The presence of MBL in CP-Ec isolates was associated with several characteristics of the patients from whom they were identified. First, carbapenemase enzymes were strongly associated with the geographic origin of the source patient. In our analysis, carbapenem resistance in E. coli was mediated by MBLs in China. In contrast, in the United States, Colombia, and Singapore, carbapenem resistance by carbapenemases was primarily mediated by non–MBL-Ec. These data were largely consistent with previous studies [21–23]. Patients from Lebanon had both non-MBL and MBL carbapenemases. We cannot comment substantially on geographic variations in IMP or VIM as we had limited isolates with these MBL types; however, recent literature suggests that these carbapenemases are most prevalent in countries such as Greece, Taiwan, and Japan [24–26], where we did not enroll patients. Both globalization and community spread of CP-Ec in different geographic regions result in rapidly changing epidemiology; this necessitates a structured, prospective, and continuous approach to monitor trends in the geography of CP-Ec.
Second, carbapenemases were associated with specific background bacterial characteristics and ST of E. coli. We observed that MBL-Ec isolates were associated with ST167 and ST410, primarily harboring a blaNDM gene. The clustering of blaNDM-5, specifically on ST167 and ST410, has been recently described in both local and global studies [27, 28]. Conversely, non–MBL-Ec, particularly blaKPC, most frequently was harbored by ST131. These findings are consistent with earlier analyses; since the emergence of E. coli with blaKPC with ST131 was first described in the United States [29], several studies have described the global emergence of CP-Ec with blaKPC, facilitated by the efficient spread of ST131 [27, 30–32]. The underlying mechanisms leading to clustering of specific carbapenemases in specific STs are not fully elucidated and require further investigation. Additionally, the detection of ESBL genes in addition to carbapenemase genes was common among both MBL-Ec and non–MBL-Ec. The efficient carriage of both ESBL and carbapenemase by high-risk E. coli strains is of particular concern because of the high potential for rapid spread and dearth of therapeutic options. These data raise questions about the unique biological underpinnings of E. coli that allow for high-risk E. coli clones to efficiently acquire multiple multidrug-resistance genes.
Finally, the clinical scenarios associated with MBL-Ec differed from those with non–MBL-Ec. Patients with MBL-Ec less frequently met infection criteria than those with non–MBL-Ec. Correspondingly, patients with MBL-Ec had a lower acuity of illness and the culture source was more often from urine, whereas patients with non–MBL-Ec had a higher acuity of illness and more bloodstream infections. We observed increased mortality in patients with non–MBL-Ec that could not be completely explained by the higher rates of infection alone, since increased mortality among patients with non–MBL-Ec persisted, even when limiting analysis to those who met infection criteria. The reasons for these mortality differences are unclear. Bacteremia may be an important predictor of mortality due to the severity of illness with bacteremia compared with urinary tract infection, and this may be incompletely captured by the PBS. This study is limited to univariate analysis, but future studies should examine whether bacteremia remains an independent predictor of mortality from CP-Ec infections. Additionally, most patients in our study with MBL-Ec were from China, and these mortality differences are consistent with lower mortality seen in patients from China in an international cohort study of carbapenem-resistant Pseudomonas aeruginosa [33] and with outcomes from CRACKLE-2 Klebsiella pneumoniae isolates [10]. This suggests that differences in patient characteristics and healthcare systems and supportive care in various regions may confound the mortality differences observed here. Importantly, there may also exist a difference in MBL-Ec and non–MBL-Ec reservoirs, resulting in the emergence of community spread of MBL-Ec in certain parts of the world, suggested by recent work showing rapid increases in fecal colonization with MBL-Ec in China [34].
Current data comparing outcomes of CP-Ec by carbapenemase type are lacking. We observed collinearity between geographic regions, culture sources, acuity of illness, and MBL status; because of the small sample size and infrequent mortality in the MBL group, we were unable to determine if MBL carriage in E. coli was an independent predictor of outcomes. Future, larger studies are needed that can account for potential confounders to evaluate the impact of carbapenemase type on patient outcomes and confirm our findings.
Limitations
This study has several important limitations. First, while we recruited patients from various global regions, patients with CP-Ec originated from a limited number of centers in 6 countries. We recognize that the lack of inclusion of other regions with known high rates of multidrug antimicrobial resistance limits the generalizability to those global regions. Second, this was an observational study, and given the waived requirement to obtain informed consent, only data collected per standard of care were available. However, the waiver of informed consent allowed for consecutive inclusion of patients and minimized the risk of selection bias. Third, the DOOR analyses may have been underpowered to detect a true difference in the outcomes of MBL-Ec versus non–MBL-Ec isolates in this study since we only examined those who met criteria for infection, resulting in a small sample size.
Conclusions
In summary, this study describes the emergence of CP-Ec in many regions across the globe, including among high-risk strains of E. coli such as ST131, 167, and 410. Geographic, bacterial, and clinical features were associated with MBL-harboring CP-Ec. Patients with non-MBL isolates had higher mortality, but this may be confounded by differences in illness acuity and region. With limited novel therapeutic options in the pipeline, and the potential for further global dissemination of carbapenem resistance among high-risk E. coli strains, ongoing studies of CP-Ec epidemiology and outcomes are of paramount importance.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Angelique E Boutzoukas, Division of Infectious Diseases, Duke University, Durham, North Carolina, USA; Duke Clinical Research Institute, Duke University, Durham, North Carolina, USA.
Lauren Komarow, The Biostatistics Center, George Washington University, Rockville, Maryland, USA.
Liang Chen, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, New Jersey, USA.
Blake Hanson, Center for Infectious Diseases and Microbial Genomics, UTHealth, McGovern School of Medicine at Houston, Houston, Texas, USA.
Souha S Kanj, Division of Infectious Diseases, and Center for Infectious Diseases Research, American University of Beirut Medical Center, Beirut, Lebanon.
Zhengyin Liu, Infectious Disease Section, Department of Internal Medicine, Peking Union Medical College Hospital, Beijing, China.
Soraya Salcedo Mendoza, Servicio de Infectología, Organizacion Clinica General del Norte, Barranquilla, Colombia.
Karen Ordoñez, Department of Infectious Diseases, E.S.E. Hospital Universitario, San Jorge de Pereira, Pereira, Colombia.
Minggui Wang, Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, China.
David L Paterson, ADVANCE-ID, Saw Swee Hock School of Public Health, National University of Singapore, Singapore.
Scott Evans, The Biostatistics Center, George Washington University, Rockville, Maryland, USA.
Lizhao Ge, The Biostatistics Center, George Washington University, Rockville, Maryland, USA.
Abhigya Giri, The Biostatistics Center, George Washington University, Rockville, Maryland, USA.
Carol Hill, Duke Clinical Research Institute, Duke University, Durham, North Carolina, USA.
Keri Baum, Duke Clinical Research Institute, Duke University, Durham, North Carolina, USA.
Robert A Bonomo, Louis Stokes Cleveland Department of Veterans Affairs Medical Center, Cleveland, Ohio, USA; VA–Case Center for Antibiotic Resistance and Epidemiology (Case-VA CARES), Cleveland, Ohio, USA.
Barry Kreiswirth, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, New Jersey, USA.
Robin Patel, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, and Division of Public Health, Infectious Diseases, and Occupational Medicine, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA.
Cesar A Arias, Division of Infectious Diseases and Center for Infectious Diseases, Houston Methodist Hospital and Houston Methodist Research Institute, Houston, Texas, USA.
Henry F Chambers, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
Vance G Fowler, Jr, Division of Infectious Diseases, Duke University, Durham, North Carolina, USA; Duke Clinical Research Institute, Duke University, Durham, North Carolina, USA.
David van Duin, Division of Infectious Diseases, University of North Carolina, Chapel Hill, North Carolina, USA.
Multi-Drug Resistant Organism Network Investigators:
S Kanj Souha, Francois (Jeff) Jabbour Jean, Zhang Fujie, J Lok Judith, A Salata Robert, Stryjewski Martin, Di Castelnuovo Valentina, Millan Oñate Gutierrez Jose, Cober Eric, Richter Susan, J Anderson Deverick, Evans Beth, Hill Carol, R Cross Heather, Baum Keri, Arias Rebekka, G Fowler Vance, Jr, Ordoñez Karen, T Jacob Jesse, Li Linghua, N Kreiswirth Barry, Manca Claudia, Chen Liang, Desai Samit, Herc Erica, Cordova Ezequiel, Rioseco Maria, Vichez Samuel, L Sanchez Marisa, Valderrama Sandra, Figueroa Jairo, A Arias Cesar, Q Dinh An, Panesso Diane, Rydell Kirsten, T Tran Truc, Hu Fupin, Su Jiachun, Jiang Jianping, Wang Minggui, Xu Xiaogang, Yang Yang, M Munita Jose, Spencer Maria, Alenazi Thamer, A Bonomo Robert, H Marshall Steven, D Rudin Susan, Huskins Charles, Greenwood-Quaintance Kerry, Patel Robin, Schmidt-Malan Suzannah, Revolinski Sara, Wortmann Glenn, C Kalayjian Robert, Weston Greg, Ostrowsky Belinda, Patel Gopi, Eiras Daniel, Kim Angela, Garcia-Diaz Julia, Salcedo Soraya, J Farrell John, Liu Zhengyin, Henderson Andrew, L Paterson David, Xie Qing, S Kaye Keith, Gao Hainv, Yu Yunsong, Waters Mary, C Fries Bettina, Eilertson Brandon, Marimuthu Kalisvar, Lee Chew Kean, Smitasin Nares, Ananth Tambyah Paul, C Gallagher Jason, Peleg Anton, Leroi Marcel, Li Lanjuan, Komarow Lauren, Ge Lizhao, Evans Scott, McCarty Todd, F Chambers Henry, B Garner Omai, M Abbo Lilian, van Duin David, Lautenbach Ebbing, H Han Jennifer, Doi Yohei, Wong Darren, Hanson Blake, Reyes Jinnethe, Virginia Villegas Botero Maria, Diaz Lorena, Perez Federico, Banerjee Ritu, Dhar Sorabh, J Satlin Michael, and Zong Zhiyong
Notes
Author Contributions. D. v. D. led the protocol from which the study data are derived. A. E. B. and D. v. D. were responsible for overall analysis development, supervision of the project, and review of the final manuscript. V. G. F. and H. F. C. acquired funding for the study. A. E. B., L. K., L. C., C. H., and K. B. accessed the data in the study. D. v. D., M. W., C. A. A., and D. L. P. served as regional leads. A. E. B. developed the methods, performed the analysis with L. K., and generated the tables and figures. L. C. created the genomic visualizations. C. A. A., B. H., M. W., and B. K. oversaw sequencing activities and L. C. and C. H. conducted the bioinformatic analysis on the sequence results. R. P. and R. A. B. oversaw centralized susceptibility testing. All authors were involved with the scientific review and editing of the manuscript.
Acknowledgments . The investigators thank all the patients and their families and all contributing research fellows and clinical microbiological laboratory personnel. The investigators also thank Dr. Michael Satlin and Dr. Keith Kaye for their detailed review of an earlier version of this article.
Disclaimer. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award number UM1AI104681 and the US Eunice Kennedy Shriver National institute of child health and human development T32 training grant (1T32HD094671).
Multi-Drug Resistant Organism Network Investigators (listed by center alphabetically) . American University of Beirut Medical Center, Beirut, Lebanon: Souha S. Kanj, Jean Francois (Jeff) Jabbour; Beijing Ditan Hospital, Capital Medical University, Beijing, China: Fujie Zhang; Boston University, Boston, MA, USA: Judith J. Lok; Case Western, Cleveland, OH, USA: Robert A. Salata; Centro de Educacion Medica e Investigaciones Clinicas, Buenos Aires, Argentina: Martin Stryjewski, Valentina Di Castelnuovo; Centro Medico Imbanaco, Cali, Colombia: Jose Millan Oñate Gutierrez; Cleveland Clinic, Cleveland, OH, USA: Eric Cober, Susan Richter; Duke University, Durham, NC, USA: Deverick J. Anderson, Beth Evans, Carol Hill, Heather R. Cross, Keri Baum, Rebekka Arias, Vance G. Fowler, Jr; E.S.E Hospital Universitario, San Jorge de Pereira, Pereira, Colombia: Karen Ordoñez; Emory University, Atlanta, GA, USA: Jesse T. Jacob; Guangzhou Eighth People's Hospital, Guangzhou, China: Linghua Li; Hackensack Meridian Health, Nutley, NJ, USA: Barry N. Kreiswirth, Claudia Manca, Liang Chen, Samit Desai; Henry Ford Hospital, Detroit, MI, USA: Erica Herc; Hospital “Cosme Argerich” de Buenos Aires, Buenos Aires, Argentina: Ezequiel Cordova; Hospital de Puerto Montt, Puerto Montt, Chile: Maria Rioseco; Hospital Escuela Oscar Danilo Rosales Arguello, Leon, Nicaragua: Samuel Vichez; Hospital Italiano de Buenos Aires, Buenos Aires, Argentina: Marisa L. Sanchez; Hospital San Ignacio, Bogota, Colombia: Sandra Valderrama; Hospital Universitario Erasmo Meoz ESE, Cucuta, Colombia: Jairo Figueroa; Houston Methodist, Houston, TX, USA: Cesar A. Arias, An Q. Dinh, Diane Panesso, Kirsten Rydell, Truc T. Tran; Huashan Hospital, Fudan University, Shanghai, China: Fupin Hu, Jiachun Su, Jianping Jiang, Minggui Wang, Xiaogang Xu, Yang Yang; Instituto de Ciencias e Innovacion en Medicina, Clinica Alemana, Universidad del Desarrollo, Santiago, Chile: Jose M. Munita, Maria Spencer; King Abdulaziz Medical City, Riyadh, Saudi Arabia: Thamer Alenazi; Louis Stokes Cleveland Department of Veterans Affairs Medical Center, Cleveland, OH, USA: Robert A. Bonomo, Steven H. Marshall, Susan D. Rudin; Mayo Clinic, Rochester, MN, USA: Charles Huskins, Kerry Greenwood-Quaintance, Robin Patel, Suzannah Schmidt-Malan; Medical College of Wisconsin, Milwaukee, WI, USA: Sara Revolinski; MedStar Washington Hospital Center, Washington, DC, USA: Glenn Wortmann; MetroHealth Medical Center, Cleveland, OH, USA: Robert C. Kalayjian; Montefiore Medical Center, Albert Einstein College of Medicine, New York, NY, USA: Greg Weston; Montefiore Medical Center, Moses Campus, Bronx, NY, USA: Belinda Ostrowsky; Mount Sinai, New York, NY, USA: Gopi Patel; New York University Langone Medical Center, New York, NY, USA: Daniel Eiras; North Shore University Hospital, Manhasset, NY, USA: Angela Kim; Ochsner Clinic Foundation, New Orleans, LA, USA: Julia Garcia-Diaz; Organizacion Clinica General del Norte, Barranquilla, Colombia: Soraya Salcedo; OSF Saint Francis Medical Center, Peoria, IL, USA: John J. Farrell; Peking Union Medical College Hospital, Beijing, China: Zhengyin Liu; Princess Alexandra Hospital, Brisbane, Queensland, Australia: Andrew Henderson; Royal Brisbane and Women's Hospital, Brisbane, Queensland, Australia: David L. Paterson; Ruijin Hospital, Shanghai Jiaotong University, Shanghai, China: Qing Xie; Rutgers University, New Brunswick, NJ, USA: Keith S. Kaye; Shulan Hangzhou Hospital, Shulan Health, Hangzhou, China: Hainv Gao; Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China: Yunsong Yu; St Vincent's Hospital, Melbourne, Victoria, Australia: Mary Waters; Stony Brook University, Stony Brook, NY, USA: Bettina C. Fries; SUNY Downstate Medical Center, New York, NY, USA: Brandon Eilertson; Tan Tock Seng Hospital, Singapore: Kalisvar Marimuthu, Kean Lee Chew, Nares Smitasin, Paul Ananth Tambyah; Temple University Hospital, Philadelphia, PA, USA: Jason C. Gallagher; The Alfred Hospital, Melbourne, Victoria, Australia: Anton Peleg; The Austin Hospital, Heidelberg West, Victoria, Australia: Marcel Leroi; The First Affiliated Hospital of Medical School of Zhejiang University, Hangzhou, China: Lanjuan Li; The George Washington University, Washington, DC, USA: Lauren Komarow, Lizhao Ge, Scott Evans; The University of Alabama, Birmingham, AL, USA: Todd McCarty; The University of California San Francisco, San Francisco, CA, USA: Henry F. Chambers; The University of California Los Angeles, Los Angeles, CA, USA: Omai B. Garner; The University of Miami Miller School of Medicine and Jackson Health System, Miami, FL, USA: Lilian M. Abbo; The University of North Carolina, Chapel Hill, NC, USA: David van Duin; The University of Pennsylvania Health System, Philadelphia, PA, USA: Ebbing Lautenbach, Jennifer H. Han; The University of Pittsburgh School of Medicine, Pittsburgh, PA, USA: Yohei Doi; The University of Southern California, Los Angeles, CA, USA: Darren Wong; The University of Texas Health Science Center at Houston, Houston, TX, USA: Blake Hanson; Universidad El Bosque, Bogota, Colombia: Jinnethe Reyes, Maria Virginia Villegas Botero, Lorena Diaz; University Hospitals Cleveland Medical Center, Cleveland, OH, USA: Federico Perez; Vanderbilt University Medical Center, Nashville, TN, USA: Ritu Banerjee; Wayne State University, Detroit, MI, USA: Sorabh Dhar; Weill Cornell Medicine, New York–Presbyterian Hospital, New York, NY, USA: Michael J. Satlin; West China Hospital of Sichuan University, Chengdu, China: Zhiyong Zong.
Data sharing. Individual deidentified participant data (and supporting documentation, data dictionaries, and protocol) that underlie the results in this article can be made available to investigators following submission of a plan for data use, approval by the ARLG or designated entity, and execution of required institutional agreements. Provision might be contingent upon the availability of funding for data preparation and deidentification. More information can be found at https://arlg.org/request-data/. Sequences will be publicly available through the National Center for Biotechnology Information (accession number PRJNA658369; https://www.ncbi.nlm.nih.gov/bioproject/658369).
References
- 1. Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med 2015; 277:501–12. [DOI] [PubMed] [Google Scholar]
- 2. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017; 215(Suppl 1):S28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Cleve Clin J Med 2013; 80:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents 2017; 50:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paterson DL. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Contr 2006; 34(5, Suppl):S20–S8. [DOI] [PubMed] [Google Scholar]
- 6. Antimicrobial Resistance Collaborators . Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399:629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zasowski EJ, Rybak JM, Rybak MJ. The beta-lactams strike back: ceftazidime-avibactam. Pharmacotherapy 2015; 35:755–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnston BD, Thuras P, Porter SB, et al. Activity of imipenem-relebactam against carbapenem-resistant Escherichia coli isolates from the United States in relation to clonal background, resistance genes, coresistance, and region. Antimicrob Agents Chemother 2020; 64:e02408-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Timsit JF, Paul M, Shields RK, et al. Cefiderocol for the treatment of infections due to metallo-B-lactamase-producing pathogens in the CREDIBLE-CR and APEKS-NP phase 3 randomized studies. Clin Infect Dis 2022; 75:1081–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020; 20:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 12. Al-Hasan MN, Lahr BD, Eckel-Passow JE, Baddour LM. Predictive scoring model of mortality in gram-negative bloodstream infection. Clin Microbiol Infect 2013; 19:948–54. [DOI] [PubMed] [Google Scholar]
- 13. Henderson H, Luterbach CL, Cober E, et al. The Pitt bacteremia score predicts mortality in nonbacteremic infections. Clin Infect Dis 2020; 70:1826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. Clermontyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 2018; 4:e000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bessonov K, Laing C, Robertson J, et al. ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb Genom 2021; 7:000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roer L, Tchesnokova V, Allesoe R, et al. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 2017; 55:2538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feldgarden M, Brover V, Gonzalez-Escalona N, et al. AMRFinderplus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 2021; 11:12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunt M, Mather AE, Sanchez-Buso L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3:e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans SR, Rubin D, Follmann D, et al. Desirability of outcome ranking (DOOR) and response adjusted for duration of antibiotic risk (RADAR). Clin Infect Dis 2015; 61:800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuzon G, Naas T, Correa A, Quinn JP, Villegas M-V, Nordmann P. Dissemination of the KPC-2 carbapenemase in non-Klebsiella pneumoniae enterobacterial isolates from Colombia. Int J Antimicrob Agents 2013; 42:59–62. [DOI] [PubMed] [Google Scholar]
- 22. van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017; 8:460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tian X, Zheng X, Sun Y, et al. Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002-2017. Infect Drug Resist 2020; 13:501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yano H, Ogawa M, Endo S, et al. High frequency of IMP-6 among clinical isolates of metallo-beta-lactamase-producing Escherichia coli in Japan. Antimicrob Agents Chemother 2012; 56:4554–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsumura Y, Peirano G, Motyl MR, et al. Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob Agents Chemother 2017; 61:e02729-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumura Y, Peirano G, Devinney R, et al. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J Antimicrob Chemother 2017; 72:2249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peirano G, Chen L, Nobrega D, et al. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015-2017. Emerg Infect Dis 2022; 28:924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bibbolino G, Di Lella FM, Oliva A, et al. Molecular epidemiology of NDM-5-producing Escherichia coli high-risk clones identified in two Italian hospitals in 2017-2019. Diagn Microbiol Infect Dis 2021; 100:115399. [DOI] [PubMed] [Google Scholar]
- 29. Kim YA, Qureshi ZA, Adams-Haduch JM, Park YS, Shutt KA, Doi Y. Features of infections due to Klebsiella pneumoniae carbapenemase–producing Escherichia coli: emergence of sequence type 131. Clin Infect Dis 2012; 55:224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peirano G, Bradford PA, Kazmierczak KM, et al. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 2014; 20:1928–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ellaby N, Doumith M, Hopkins KL, Woodford N, Ellington MJ. Emergence of diversity in carbapenemase-producing Escherichia coli ST131, England, January 2014 to June 2016. Euro Surveill 2019; 24:1800627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piazza A, Principe L, Comandatore F, et al. Whole-genome sequencing investigation of a large nosocomial outbreak caused by ST131 H30Rx KPC-producing Escherichia coli in Italy. Antibiotics (Basel) 2021; 10:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reyes J, Komarow L, Chen L, et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 2023; 4:e159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen B, Berglund B, Wang S, et al. Rapid increase in occurrence of carbapenem-resistant Enterobacteriaceae in healthy rural residents in Shandong province, China, from 2015 to 2017. J Glob Antimicrob Resist 2022; 28:38–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.