Abstract
We describe the use of herpesvirus promoters to regulate the expression of a Sindbis virus replicon (SINrep/LacZ). We isolated cell lines that contain the cDNA of SINrep/LacZ under the control of a promoter from a herpesvirus early gene which requires regulatory proteins encoded by immediate-early genes for expression. Wild-type Sindbis virus and replicons derived from this virus cause death of most vertebrate cells, but the cells discussed here grew normally and expressed the replicon and β-galactosidase only after infection with a herpesvirus. Vero cell lines in which the expression of SINrep/LacZ was regulated by the herpes simplex virus type 1 (HSV-1) infected-cell protein 8 promoter were generated. One Vero cell line (V3-45N) contained, in addition to the SINrep/LacZ cDNA, a Sindbis virus-defective helper cDNA which provides the structural proteins for packaging the replicon. Infection of V3-45N cells with HSV-1 resulted in the production of packaged SINrep/LacZ replicons. HSV-1 induction of the Sindbis virus replicon and packaging and spread of the replicon led to enhanced expression of the reporter gene, suggesting that this type of cell could be used to develop sensitive assays to detect herpesviruses. We also isolated a mink lung cell line that was transformed with SINrep/LacZ cDNA under the control of the promoter from the human cytomegalovirus (HCMV) early gene UL45. HCMV carries out an abortive infection in mink lung cells, but it was able to induce the SINrep/LacZ replicon. These results, and those obtained with an HSV-1 mutant, demonstrate that this type of signal amplification system could be valuable for detecting herpesviruses for which a permissive cell culture system is not available.
Replicons derived from alphaviruses such as Sindbis virus are well established as useful vectors for gene expression (2, 3, 21, 36). We have been investigating the potential use of the Sindbis virus replicon as a tool for detection of other viruses, particularly those that grow to low titer and are not easily detected by plaque assay (27). We have also been interested in establishing cell lines that can be induced to produce functional Sindbis virus replicons. These two goals, detection of viruses and induction of a replicon, led to the studies presented here, in which two members of the herpesvirus family were used to induce a Sindbis virus replicon. Herpes simplex virus type 1 (HSV-1) is a member of this family which is easily detected, but because it has been well-characterized, it provided a useful model to test our concepts. In contrast, human cytomegalovirus (HCMV), because it replicates slowly and in a limited number of cell lines, is an example of a herpesvirus for which a better detection assay could be valuable (39).
There are several features of the Sindbis virus genome and its replication strategy that are important for understanding its development as a gene expression vector and its use in the systems to be described here (37, 41). The positive-strand RNA genome consists of a sequence of approximately 12 kb that is divided into two functional modules. The 5′ two-thirds codes for the nonstructural proteins, and the 3′ one-third codes for the structural proteins. In the infected cell only the nonstructural proteins, which are required for replication and transcription of the RNA, are translated from the genomic RNA. The structural proteins are translated from a subgenomic RNA that is colinear with the 3′ one-third of the genome. The structural proteins are not required for replication and transcription of the genomic RNA. cDNAs of Sindbis virus replicons have been genetically engineered by replacing genes for the structural proteins with a heterologous gene (3, 11, 35, 43). Under conditions in which the structural proteins are provided in trans by means of a defective RNA containing these genes, the replicon genome is packaged into virus-like particles (4). In the original studies, the engineered Sindbis virus cDNA was placed under the control of the SP6 bacteriophage promoter, the DNA was transcribed in vitro, and the RNA was transfected into susceptible cultured cells (43). More recently, there have been several studies in which Sindbis virus replicon cDNAs or defective cDNAs have been transcribed from cellular RNA polymerase II promoters after transfection of the DNA into cultured cells (9, 15, 16, 26, 29). Although it has been possible to obtain stable cell lines containing defective Sindbis virus cDNAs under the control of constitutive eukaryotic promoters (26), cells containing intact Sindbis virus replicons under the control of such promoters would not survive because expression of these replicons is cytopathic in vertebrate cells (12).
The herpesviruses are a family of complex DNA viruses. The best-studied member of this family is HSV-1, which has a 150-kb genome that contains over 80 genes (32). Based on their temporal expression and regulation during the viral replication cycle, HSV-1 genes are classified into immediate-early (alpha), early (beta), and late (gamma) genes (17), and this is dictated in part by their promoters. We chose to use the promoter of the HSV-1 UL29 gene, which encodes infected-cell protein 8 (ICP8), the HSV-1 major single-strand DNA-binding protein (22, 31). The 5′ end of the ICP8 transcript has been mapped (42). This allowed us to design a DNA construct in which the cDNA of the Sindbis virus replicon was linked to the HSV-1 ICP8 DNA promoter in such a way that transcription from this chimeric gene yielded an RNA transcript with a 5′ end compatible with a functional replicon. Essential features of the gene encoding ICP8 are that it is an early (beta) gene and that its expression is dependent on the regulatory proteins encoded by HSV-1 immediate-early genes such as ICP0 and ICP4 (24, 30, 42). For the HCMV studies, we also used a promoter from an early gene, UL45, which encodes a homolog of the HSV-1 large subunit of ribonucleotide reductase (6). Any gene under the control of a herpesvirus early promoter is not likely to be expressed in the absence of infection by the cognate herpesvirus. We describe our results obtained by using this viral regulatory system to control expression of a Sindbis virus replicon.
MATERIALS AND METHODS
Plasmids. (i) pICP8SINrep/LacZ.
The term ICP8 is used for all references to the UL29 gene or its promoter because it has been the more widely used designation. The ICP8 promoter sequence was amplified from purified HSV-1 genomic DNA by PCR by using synthetic oligonucleotides (DNAgency, Inc.) flanking this region. The primers were 30- and 35-mer oligonucleotides (upstream primer, 5′-GTT TGT CTG GCG GAT CCG GAC GGC GAG CTG-3′; downstream primer, 5′-CCA TGG CTC GAG GTA TGC GGT TGG TAT ATG TAC AC-3′). The 5′ and 3′ termini contained BspEI and XhoI sites, respectively, to facilitate cloning. The amplification was done in 20 cycles by using KlenTaq-LA enzyme (Wayne Barnes, Washington University, St. Louis, Mo.) in the presence of 2.2 M betaine (Sigma, St. Louis, Mo.) under the following conditions: 30 s at 94°C, 30 s at 60°C, and 1 min at 68°C. The PCR product was digested with BspEI and XhoI, and the product was then cloned into NgoMI- and XhoI-digested pSINrep/LacZ by placing a XhoI site at the 5′ terminus of the cDNA of pSINrep/LacZ (4). Plasmid and promoter identities were confirmed by restriction mapping and nucleotide sequencing. Plasmid DNAs were purified on CsCl gradients for use in transfections.
(ii) pUL45SINrep/LacZ and pUL45/LacZ.
The HindIII M fragment of the HCMV (Towne strain) genome was cloned into pBR322 to generate pCMHM. The UL45 promoter was isolated from pCMHM as a 657-bp SmaI fragment, which was cloned into pUC18. Finally, the UL45 promoter was cloned as an XmaIII to XhoI fragment in front of SINrep/LacZ cDNA or the lacZ gene to generate pUL45SINrep/LacZ and pUL45/LacZ, respectively.
(iii) p987DHBBneo.
This plasmid was made from pDHBB, a defective helper cDNA which contains the Sindbis virus structural protein genes downstream of the subgenomic RNA promoter (4). p987DHBBneo was constructed by positioning the cDNA of DHBB immediately 3′ of the Rous sarcoma virus promoter and inserting an IRES element-neo gene cassette downstream of the stop codon for the structural proteins in the 3′ nontranslated region of the DHBB genome (further details are available on request).
Cells and viruses.
Baby hamster kidney (BHK-21) cells, Vero cells (African green monkey kidney cells), and mink lung cells were obtained from the American Type Culture Collection (ATCC, Manassas, Va.) BHK cells were grown in alpha-minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum, nonessential amino acids, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Vero and mink lung cells were propagated in Dulbecco’s MEM (DMEM) containing 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
HSV-1 (KOS strain) was obtained from Mark Challberg (National Institutes of Health, Bethesda, Md.). Stocks of HSV-1 were grown and titered on Vero cells. HSV-1 mutant viruses d21 and dlx3.1 were obtained from Priscilla Schaffer (University of Pennsylvania), and d120 was obtained from Neal DeLuca (University of Pittsburgh). HCMV strains (Towne and AD169 strains) were obtained from Gregory Storch (Washington University). HCMV stocks were grown on MRC5 cells (Diagnostic Hybrids, Inc., Athens, Ohio) and titrated by an infectious foci assay by using indirect immunofluorescence and monoclonal antibody to an immediate-early antigen (Chemicon, Temecula, Calif.).
Transfection and selection of cell lines: (i) ICP8SINrep/LacZ-transformed V2-33, V2-34, and V3-45 cells.
Subconfluent (50 to 60% confluence) Vero cells in 35-mm-diameter dishes were transfected with pICP8SINrep/LacZ and a plasmid containing the hygromycin B gene under control of the simian virus 40 promoter (pMonHygro; a gift of Paul Hippenmeyer, Monsanto, St. Louis, Mo.) at a molar ratio of 5:1 or 10:1 by using 6 μl of Lipofectamine in Opti-MEM (Life Technologies, Gaithersburg, Md.) according to the manufacturer’s recommendations at 37°C for 5 h, at which time the media were changed to complete media. At 72 h posttransfection cells were split 1:10 in selective media containing 500 μg of hygromycin B/ml (Boehringer Mannheim, Indianapolis, Ind.), which were then replaced every 3 days. After 3 weeks, individual colonies were isolated and the hygromycin B concentration was reduced to 100 μg/ml.
(ii) ICP8SINrep/LacZ and helper cDNA doubly transformed V3-45N cells.
The doubly transformed replicon/helper cell line was isolated by transfecting V3-45 cells with p987DHBBneo by using 1 μg of DNA plus 6 μl of Lipofectamine per 35-mm-diameter dish for 5 h at 37°C in Opti-MEM. Seventy-two hours posttransfection, the cells were seeded in selective media containing 100 μg of hygromycin/ml and 5 mg of G418 (Life Technologies) per ml. Once the nontransformed control cells were all killed, the G418 concentration was decreased to 1 mg/ml, and 2 weeks later it was decreased to 400 μg/ml. Individual colonies were isolated and expanded in media containing hygromycin B (100 μg/ml) and G418 (400 μg/ml).
(iii) MLUL45SINrep/LacZ and MLUL45/LacZ cells.
These cell lines were isolated by cotransfecting mink lung cells with pUL45SINrep/LacZ (2 μg) and pMonHygro (0.1 μg) or pUL45/LacZ (1 μg) and pMonHygro (0.1 μg). Forty hours later the cells were treated with hygromycin B (500 μg/ml) for 7 days. After 3 weeks individual colonies were isolated, expanded in media containing hygromycin B (100 μg/ml), and evaluated for HCMV-inducible β-galactosidase.
RNA isolation.
MLUL45SINrep/LacZ cells (2 × 107 cells) were pretreated overnight with Turbotreat reagent (a proprietary medium additive that enhances HCMV gene expression in mink lung cells) (Diagnostic Hybrids, Inc.) (38) and were either mock infected or infected with HCMV (multiplicity of infection [MOI] of 2) in the presence of a 10-fold dilution of Turbotreat reagent. Forty hours postinfection cells were harvested on ice and resuspended in TRIZOL reagent (Life Technologies). RNA was isolated according to the manufacturer’s procedure. Polyadenylated RNA was then isolated by chromatography on oligodeoxyribosylthymine-cellulose or by immobilization on streptavidin magnetic particles as recommended by the manufacturer (Boehringer Mannheim). Three hundred sixty micrograms of total RNA and 6 to 12 μg of poly(A) RNA were obtained from 2 × 107 cells.
β-Galactosidase assays. (i) Colorimetric assay.
The β-galactosidase activity of cell extracts was measured by using the substrate chlorophenol red-β-d-galactopyranoside (CPRG; Boehringer Mannheim) at a final concentration of 5 mM in a 0.2 M potassium phosphate buffer, pH 7.8, with 1 mM MgCl2. Extracts were made in this buffer containing 1% Triton X-100 and 1 mM dithiothreitol. A sample of the extract (10 to 50 μl) was mixed with 50 μl of substrate in a microtiter plate well. After incubation for 1 to 2 h at room temperature, the optical density at a wavelength of 562 nM (OD562) was measured with a THERMOmax microplate reader by using SOFTmax software (Molecular Devices, Sunnyvale, Calif.). The OD562 of a control containing no extract was subtracted from all sample values. The assay was shown to be linear up to an OD562 of 3.0.
(ii) Histochemical staining.
Cell monolayers were washed one time with 1 ml of phosphate-buffered saline (PBS, pH 7.2) and then fixed in 2% formaldehyde and 0.4% glutaraldehyde in PBS for 5 min. After they were washed twice with PBS, cells were incubated at room temperature in staining solution (1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal; Sigma] per ml, 4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, and 2 mM MgCl2 in PBS).
Titration of SINrep/LacZ.
SINrep/LacZ particles were titrated by using the X-Gal histochemical stain and counting the number of blue cells. Serial dilutions of a stock of SINrep/LacZ were inoculated onto BHK cell monolayers in the wells (each 4 cm2) of 12-well dishes. Sixteen hours later the cells were fixed and histochemically stained for β-galactosidase. The number of blue cells was determined, and this was then correlated with OD562 readings from the colorimetric assay performed under a standard set of conditions with extracts from cells infected in parallel. The standard curve generated allowed us to determine the infectious particle (packaged replicon) titer in samples of media without the tedium of having to count cells under the microscope.
RESULTS
Induction of SINrep/LacZ by infection with HSV-1.
We have used two criteria in these studies to demonstrate that SINrep/LacZ could be induced by infection of cells with a herpesvirus. The first was HSV-1-dependent β-galactosidase activity. Induction alone does not establish that the replicon genome is intact and that translation of β-galactosidase is dependent on prior replication and transcription of the replicon. The second, and essential, criterion was that infectious, extracellular particles were produced when induction occurred in the presence of a Sindbis virus defective helper that provides the structural proteins for packaging.
SINrep/LacZ cDNA was placed under the control of the ICP8 promoter (see Materials and Methods and Fig. 1). The cloning strategy was designed to place the previously mapped transcription start site of the ICP8 promoter close to the 5′ end of the replicon (Fig. 1B). Before attempting to isolate inducible cell lines, we first asked if this construct was silent in uninfected cells and inducible by HSV-1. We tested both BHK cells and Vero cells in transient-transfection assays. Twenty-four hours after transfection with pICP8SINrep/LacZ the cells were infected with HSV-1, and β-galactosidase activity was measured after 18 h by histochemistry. A few BHK cells stained positively for β-galactosidase in the absence of infection with HSV-1, although the number of stained cells increased after infection. In contrast, Vero cells showed β-galactosidase activity only after infection with HSV-1 and were chosen for all further studies with HSV-1 (data not shown).
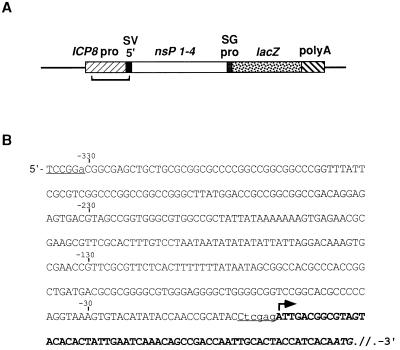
FIG. 1.
ICP8 promoter/Sindbis replicon cDNA chimeric construct. (A) Schematic diagram of ICP8SINrep/LacZ construct. The figure is not drawn to scale. pro, promoter; SV, Sindbis virus; nsP, nonstructural proteins; SG, subgenomic. The sequence shown in panel B is located in the bracketed region. (B) Sequence of the junction of the ICP8 promoter and the 5′ terminus of the Sindbis virus replicon genome. The arrow indicates the predicted start of transcription based on mapping studies of the ICP8 transcript (42). Uppercase nonboldface letters indicate HSV-1 sequences. Boldface letters indicate Sindbis virus sequences. Lowercase letters indicate bases introduced during cloning to create restriction sites BspEI and XhoI (underlined).
These results did not establish that HSV-1 was able to induce SINrep/LacZ in the Vero cells. It was possible that lacZ expression occurred independently of replicon activity because HSV-1 had been shown to transactivate a foreign gene even when the latter was placed within a defective Sindbis cDNA several kb downstream of a polymerase II promoter (26). To determine if a functional replicon was being transcribed in Vero cells, we included in the transfection mixture a defective helper plasmid (p987DHBBneo; described in Materials and Methods) capable of packaging the replicon. The protocol was further modified by the addition of acyclovir to block HSV-1 DNA synthesis and HSV-1 production. The absence of HSV-1 in the supernatant fluids of the transfected Vero cells made it possible to detect packaged Sindbis virus replicons in these samples without cytopathic effects attributable to HSV-1 progeny. Samples of media taken from variously treated Vero cells were inoculated onto naive BHK cells. β-Galactosidase-positive BHK cells were detected only with media obtained from Vero cells that had been cotransfected with pICP8SINrep/LacZ and p987DHBBneo and infected with HSV-1. No infectious particles capable of inducing β-galactosidase were detected in mock-infected Vero cells, HSV-1-infected Vero cells transfected without the defective helper plasmid, or HSV-1-infected Vero cells cotransfected with the defective helper plasmid and a control plasmid, pICP8LacZ, which contains the lacZ gene directly downstream of the ICP8 promoter. (These data are not shown, but see Fig. 3 for similar results obtained with stable cell lines.)
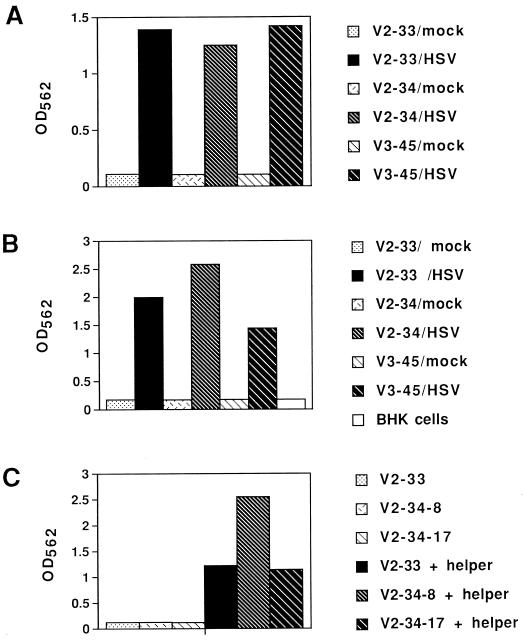
FIG. 3.
Complementation of the HSV-1-induced SINrep/LacZ replicon by a defective helper expressing the Sindbis virus structural proteins. (A) Induction of β-galactosidase activity in V2-33, V2-34, and V3-45 Vero cell lines was dependent on infection with HSV-1. Three independently transformed cell clones, V2-33, V2-34, and V3-45, were plated into the wells of a 24-well dish. Subconfluent monolayers (50% confluence) of the cells were transfected with the defective helper cDNA plasmid, p987DHBBneo. After 72 h, the cells were either mock infected or infected with HSV-1 at an MOI of 5 in the presence of acyclovir. At 30 h postinfection, cell extracts (10 μl) were assayed for β-galactosidase activity as described in Materials and Methods. (B) Production of SINrep/LacZ particles was dependent on HSV-1 infection of ICP8SINrep/LacZ-transformed Vero cell lines (V2-33, V2-34, and V3-45). A sample (200 μl) of the medium harvested from the Vero cell lines (assayed as described for panel A) was inoculated onto BHK cells in the wells of a 24-well dish. At 18 h postinfection of the BHK cells, lysis buffer (250 μl) was added to the cells and the extracts (50 μl) were assayed for β-galactosidase activity. (C) Production of SINrep/LacZ particles was dependent on transfection of the Vero cell lines with the defective helper plasmid. Subclones of V2-33 and V2-34 cells were infected with HSV-1 (MOI = 10). At 5 h postinfection the cells were either transfected with the helper plasmid p987DHBBneo or mock transfected. The media were collected 18 h later and inoculated onto BHK cells, and 18 h later β-galactosidase activity was assayed as described for panel B.
Isolation of a stable cell line of Vero cells with an HSV-1-inducible SINrep/LacZ replicon.
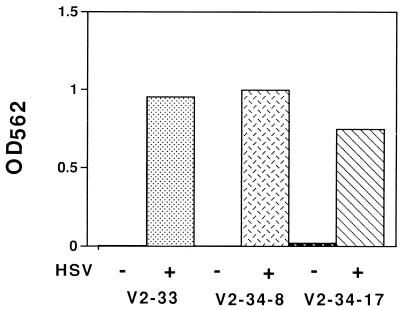
pICP8SINrep/LacZ and a plasmid carrying the gene for hygromycin B resistance (pMonHygro) were cotransfected into Vero cells, which were then selected for hygromycin B resistance (see Materials and Methods). A number of cell lines that exhibited positive staining for β-galactosidase in most of the cells only after infection with HSV-1 were selected. Cell extracts from three clones (V2-33, V2-34, and V3-45) had activity dependent upon infection of the cells with HSV-1 (data for V2-33 and V2-34 are shown in Fig. 2; data for V3-45 are not shown).
FIG. 2.
HSV-1-induced expression of β-galactosidase in Vero cells transformed with a Sindbis virus replicon cDNA under control of the ICP8 promoter. Several Vero cell clones transformed with pICP8SINrep/LacZ were either mock infected or infected with wild-type HSV-1 (MOI = 10) in the presence of acyclovir in the wells of a 6-well dish. At 24 h postinfection, 2 ml of lysis buffer was added and 10 μl of the extracts was assayed for β-galactosidase activity as described in Materials and Methods. V2-33 and V2-34 were independently isolated transformed clones; V2-34-8 and V2-34-17 were subclones from the original cloned V2-34 population. The results shown are the means of results from duplicate samples.
The second method for identifying replicons was to determine if functional SINrep/LacZ replicons could be packaged in these cells after they had been infected with HSV-1. We transfected V2-33, V2-34, and V3-45 cells with the helper plasmid, p987DHBBneo, and then infected them with HSV-1 or mock infected them. Thirty hours later, cell extracts were assayed for β-galactosidase (Fig. 3A). Samples from the media of these Vero cell cultures were inoculated onto BHK cells, and 18 h later β-galactosidase activity was measured in the BHK cell extracts (Fig. 3B). These data show that following transfection with a helper plasmid, HSV-1 induced the production of infectious particles that behaved like SINrep/LacZ particles. We also used a slightly different protocol, in which the Vero cells were first infected with HSV-1 and then were transfected with helper plasmid or mock transfected. Only samples of the media from HSV-infected cultures which had also been transfected with helper plasmid produced infectious particles, as shown by the β-galactosidase activity in extracts from BHK cells (Fig. 3C). Further support for the idea that these cells produced Sindbis virus replicon particles was provided by the observation that neutralizing antisera to Sindbis virus were able to substantially inhibit this activity (data not shown, but see Fig. 5).
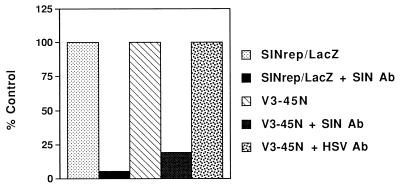
FIG. 5.
Effect of Sindbis virus-neutralizing antiserum on the β-galactosidase-inducing activity in the media of HSV-1-infected V3-45N cells. A sample (100 μl) from HSV-1-infected V3-45N cells was incubated with neutralizing rabbit antisera to Sindbis virus (SIN Ab; final dilution, 1:250) or with neutralizing human antisera to HSV (HSV Ab; final dilution, 1:400) in 500 μl for 1 h at room temperature. A sample of authentic SINrep/LacZ (titer of 2 × 108/ml) was diluted 1:1,000 and was also incubated with the SIN antiserum. After the incubation, a 20-μl sample was used to infect BHK cells in 12-well dishes. At 18 h postinfection, lysis buffer (200 μl) was added and 50 μl of the extract was assayed for β-galactosidase activity as described in Materials and Methods. The values obtained with samples not treated with antisera were designated as 100%.
Isolation of a Vero cell line that produces a packaged Sindbis virus replicon following HSV-1 infection.
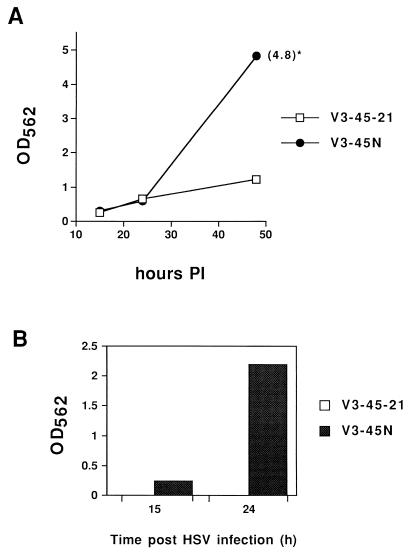
Our ability to detect packaged replicons in helper-transfected and HSV-1-infected Vero cells led us to attempt to isolate stable cell lines that carried both the SINrep/LacZ genome and the helper plasmid. To this end, we transfected V3-45 cells with p987DHBBneo and selected for neomycin-resistant colonies as described in Materials and Methods. A cell line, designated V3-45N, that induced HSV-1-dependent β-galactosidase activity and released infectious particles into the media was isolated. Extracts from a clone (V3-45-21) of the original V3-45 cells and those from the V3-45N cells had the same level of β-galactosidase activity at 24 h after infection with HSV-1 in the presence of acyclovir (Fig. 4A). As expected, V3-45-21 cells showed only a small increase in enzyme activity from 24 to 48 h. Although the initial MOI of HSV-1 infection was low (0.5), HSV-1 could not spread to uninfected cells because of the presence of acyclovir, and in the absence of a packaging helper, there would be no spread of the Sindbis virus replicon. V3-45N cells, however, had a substantial increase in β-galactosidase activity between 24 and 48 h, suggesting that the lacZ-containing replicon was spreading in these cultures. We found infectious particles (packaged replicons) in the media from HSV-1-infected V3-45N cells but not from HSV-1-infected V3-45-21 cultures (Fig. 4B). The particles released into the media from HSV-1-infected V3-45N cells were identified as Sindbis virus replicons by their ability to be neutralized by antisera directed against Sindbis virus but not by anti-HSV antiserum (Fig. 5).
FIG. 4.
Comparison of the Vero cell line that contains both pICP8SINrep/LacZ and the packaging helper plasmid (V3-45N) with the Vero cell line containing only the replicon plasmid (V3-45-21). V3-45N cells were isolated as described in Materials and Methods. V3-45-21 cells are a subclone of the original cloned V3-45 cells. β-Galactosidase assays were performed as described in Materials and Methods. (A) β-Galactosidase activity of V3-45N and V3-45-21 cell extracts 15, 24, and 48 h after infection with HSV-1 (MOI = 0.5). Extracts were made from the cells in the wells of a 24-well dish by using 250 μl of lysis buffer, and 10 μl of extract was used to assay β-galactosidase activity. The data point indicated with an asterisk (∗) was obtained by diluting the sample to obtain an OD562 reading within the linear range and multiplying the result by the dilution factor. (B) β-Galactosidase activity of BHK cells inoculated with medium removed from V3-45N or V3-45-21 cells 15 and 24 h after infection with HSV-1. A sample of the medium (50 μl of 2 ml) was inoculated onto BHK cells in the wells of a 24-well dish. At 18 h postinfection, the BHK cells were treated with 200 μl of lysis buffer and 10 μl of extract was assayed for β-galactosidase activity.
The spread of SINrep/LacZ in V3-45N cells indicated that these cells might provide a very sensitive means for detecting HSV-1. In preliminary experiments, less than 10 PFU of HSV-1 led to an eightfold induction of β-galactosidase activity (data not shown).
Induction of SINrep/LacZ by HSV-1 mutants.
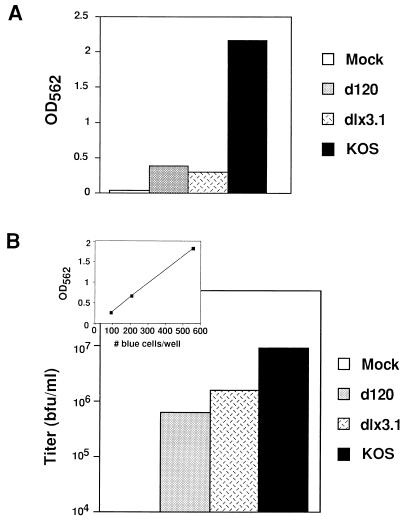
The HSV-1-encoded proteins ICP0 and ICP4, the products of immediate-early (alpha) genes, are the major transactivators of the ICP8 promoter, and they have been shown to have a synergistic effect on expression of ICP8 and other HSV-1 early genes (5, 10, 13, 42). To provide further evidence that SINrep/LacZ was under the control of the ICP8 promoter, we infected V3-45N cells with either of two mutant viruses, one containing a deletion in the gene for ICP0 (dlx3.1) (20) and the other containing a deletion in the gene for ICP4 (d120) (7). Both mutants induced much less β-galactosidase activity in V3-45N cells than wild-type virus (KOS) (Fig. 6A). The infectious titers of the SINrep/LacZ particles released from V3-45N cells in these experiments were calculated from a standard curve (shown in the inset of Fig. 6B and described in Materials and Methods). Although the mutant viruses were able to induce infectious replicon particles, the titers were significantly lower than those induced by wild-type virus (Fig. 6B). These results provide evidence that the regulation of SINrep/LacZ in these cells was similar to that of the native ICP8 gene, which requires both ICP0 and ICP4 for maximal expression.
FIG. 6.
Induction of the Sindbis virus replicon in V3-45N cells by HSV-1 mutants. V3-45N cells were infected with wild-type HSV-1 (KOS), dlx3.1 or d120 (MOI = 5). Wild-type HSV-1 (KOS) and dlx3.1-infected cells were treated with acyclovir (50 μM). At 24 h after infection, the medium (2 ml) were collected and cell extracts were prepared. A sample of the medium (200 μl) was inoculated onto BHK cells in the wells of a 24-well dish. After 18 h, cell extracts were prepared with 200 μl of lysis buffer and β-Galactosidase activity was assayed as described in Materials and Methods. (A) β-Galactosidase activity of V3-45N cell extracts. (B) Titers of SINrep/LacZ in media from V3-45N cells after infection with wild-type HSV-1 (KOS), infection with mutant viruses (d120 and dlx3.1), or mock infection. The titer of SINrep/LacZ, expressed as blue-forming units (bfu), was determined from a standard curve (see inset) generated as described in Materials and Methods.
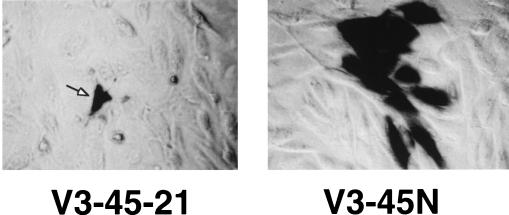
A mutant of HSV-1 that contains intact ICP0 and ICP4 genes should activate the ICP8 promoter even if it is unable to replicate. This type of mutant is equivalent to wild-type HSV-1 in the presence of acyclovir and would provide another means of activating SINrep/LacZ in the absence of HSV-1 replication. One such mutant of HSV-1, d21, which has a large deletion in the ICP8 gene (28), was able to induce β-galactosidase in V3-45N cells to essentially the same levels as those obtained with wild-type HSV-1 (data not shown). We also examined the distribution of β-galactosidase-positive cells in both V3-45N cells and the parental cells (V3-45-21) that were not capable of packaging the replicon. Three days after infection with d21, under conditions in which only a small number of cells in the monolayer were infected, foci of blue cells were seen in V3-45N cells, indicating that SINrep/LacZ particles had spread from a d21-infected cell to neighboring cells (Fig. 7). In V3-45-21 cells only individual blue cells were observed. Most of these cells looked as though they were dead or dying, as was expected since both d21 and SINrep/LacZ are capable of causing cell death. Many of the blue cells in the V3-45N cell population retained the appearance of viable cells. They most likely represented cells infected by SINrep/LacZ particles released from previously d21-induced cells and had been infected for a shorter period of time.
FIG. 7.
Photomicrographs of histochemically stained d21-infected V3-45-21 (nonpackaging) and V3-45N (packaging) cells. V3-45-21 and V3-45N cells were plated at 80% confluence in the wells of a six-well dish. The cells were infected with HSV mutant virus d21 (MOI = 0.001), and 3 days after infection the monolayers were fixed and stained for β-galactosidase.
A cell line with an HCMV-inducible Sindbis virus replicon.
We wished to extend the model of induction of SINrep/LacZ by HSV-1 to another member of the herpesvirus family, HCMV. The latter virus replicates well only in primary human cells which cannot be used to establish stable cell lines (23). HCMV infects mink lung cells, and although it does not complete its replication cycle in these cells, infected cells do express some of the immediate-early and early genes (14). Based on our results demonstrating that HSV-1 was able to induce SINrep/LacZ without undergoing a complete replication cycle (see above), we used mink lung cells to isolate cell lines analogous to those described for HSV-1.
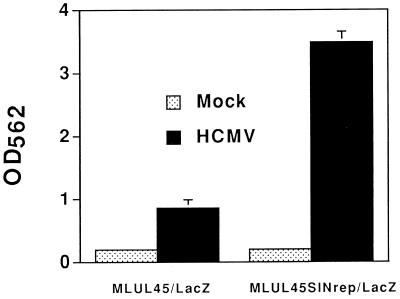
We constructed a plasmid (pUL45SINrep/LacZ) in which the cDNA of SINrep/LacZ was placed immediately downstream of and under the regulatory control of the HCMV promoter for the UL45 gene, an early gene that encodes a homolog of the HSV-1 ribonucleotide reductase large subunit (1, 6). As the 5′ terminus of the transcript from this gene has not been mapped, for this construct we estimated the start of transcription by using the apparent TATA sequence element. Preliminary tests with pUL45SINrep/LacZ in transient-transfection assays in mink lung cells showed that HCMV-infected cells, but not uninfected cells, produced β-galactosidase (data not shown). We cotransfected mink lung cells with pUL45SINrep/LacZ and pMonHygro and selected hygromycin-resistant colonies (see Materials and Methods). Isolated clonal cell lines were screened for HCMV-inducible β-galactosidase activity. One cell line (MLUL45SINrep/LacZ) showed many positive cells when it was stained for β-galactosidase activity only after infection with HCMV. The β-galactosidase activity induced in these cells was significantly higher than that observed in a control cell line (MLUL45/LacZ) transformed with a DNA construct in which the lacZ gene was directly under the regulatory control of the UL45 promoter (Fig. 8).
FIG. 8.
HCMV induction of β-galactosidase activity in mink lung cells transformed with pUL45SINrep/LacZ. Isolation of MLUL45SINrep/LacZ cells and MLUL45/LacZ cells is described in Materials and Methods. Each cell line was plated in the wells of a 24-well dish. The cells were infected with 50 μl of HCMV (AD169 strain; titer of 106 infectious focus units/ml) in the presence of 0.1× TurboTreat (Diagnostic Hybrids, Inc.). At 48 h after infection, the cells were treated with 200 μl of lysis buffer and 50 μl of extract was assayed for β-galactosidase activity as described in Materials and Methods. Results shown are the means of results from triplicate samples.
We were unable to obtain packaged replicons from the MLUL45SINrep/LacZ cells after transfection of the helper plasmid 987DHBBneo as we had done with HSV-1-infected V3-45 cells. We were not surprised by this result; one reason is that Sindbis virus and our standard packaged SINrep/LacZ replicate poorly in these cells. When mink lung cells were infected with Sindbis virus at a high MOI, the yields of virus were more than 100-fold lower than those obtained in BHK cells (data not shown). Other studies have shown that replicons that produce low levels of genomic RNA in BHK cells are packaged much less efficiently than the wild-type replicon (8). In addition, preliminary observations suggest that interferon induction may be contributing to the poor growth of Sindbis virus, and this could also affect our attempts to package replicons directly (34).
We used a different method to establish the presence of replicons in the HCMV-induced MLUL45SINrep/LacZ cells. We isolated polyadenylated RNA from HCMV-infected and mock-infected MLUL45SINrep/LacZ cells and then coelectroporated the RNA into BHK cells with in vitro-transcribed defective helper (DHEB) RNA (4). This is essentially our protocol for packaging of SINrep/LacZ transcripts, but in this case the (putative) replicon RNA came from MLUL45SINrep/LacZ cells. Samples of media harvested 24 h posttransfection with RNA isolated from MLUL45SINrep/LacZ cells or with in vitro-synthesized SINrep/LacZ RNA as a control were then assayed for the presence of packaged SINrep/LacZ particles by inoculating naive BHK cells. RNA obtained from the MLUL45SINrep/LacZ cells infected with HCMV contained SINrep/LacZ RNA that was packaged into infectious particles (Fig. 9). No packaged particles were detected when the RNA came from uninfected MLUL45SINrep/LacZ cells.
FIG. 9.
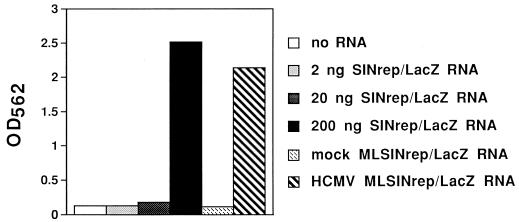
Production of SINrep/LacZ particles by using mRNA isolated from HCMV-infected MLUL45SINrep/LacZ cells. Poly(A)-selected RNAs were obtained from MLUL45SINrep/LacZ cells infected with HCMV or mock infected (Materials and Methods). The RNA samples were transfected into BHK cells by electroporation with defective helper RNA transcribed in vitro from pDHEB (4). Several different amounts (2, 20, and 200 ng) of SINrep/LacZ RNA, transcribed in vitro, were also electroporated into BHK cells with DHEB RNA as a positive control. After 24 h, media from the BHK cells were collected and samples were taken to infect naive BHK cells. Twenty-four hours later, the cells were treated with 250 μl of lysis buffer and 50 μl of extract was assayed for β-galactosidase activity as described in Materials and Methods.
DISCUSSION
The first goal of these studies was to determine if it was possible to obtain cell lines in which the Sindbis virus replicon was inducible. Most inducible systems can tolerate a certain amount of basal transcription, either because its level is several orders of magnitude lower than induced levels or because the level of basal transcription does not result in detectable levels of the gene product. If, however, the transcribed product is a replicon derived from a cytopathic RNA virus, any intact RNA molecules that found their way to the cytoplasm would initiate an autocatalytic cycle of RNA replication and transcription which would result in the inhibition of host protein synthesis and cell death. The generation of stable cell lines with an inducible RNA replicon requires, therefore, a high degree of regulatory stringency. We attribute our success to the use of tightly regulated herpesvirus early promoters. Early gene expression in herpesvirus-infected cells is dependent on certain immediate-early genes, the products of which are transcriptional transactivators of early genes (32). We took advantage of this herpesvirus gene regulatory system to make cell lines in which the cDNA of the Sindbis virus RNA replicon was transcriptionally inactive because of the absence of any immediate-early gene products. Infection with the herpesvirus then brought the necessary transactivators into the cell, and a DNA-dependent transcription process initiated an RNA-to-RNA amplification process. This system may be useful for any purpose which requires tightly regulated, high-level foreign gene expression.
Our second goal was to explore the feasibility of using the induction of the Sindbis virus replicon as a means of detecting and assaying DNA viruses, specifically herpesviruses. A number of cell lines that express a reporter gene in response to infection by a particular virus have been described, and these cell lines have been shown to be useful for detecting and quantifying viruses (25–27, 40). Certain viruses, however, replicate poorly in cultured cells, and these viruses would likely induce a weak signal from reporter cell lines. We had speculated that induction of a Sindbis virus replicon by the transcriptional transactivator of a poorly replicating virus could enhance the level of reporter gene expression by causing a shift from a state of low-level gene expression of the activating virus to a state of high-level gene expression of the Sindbis virus replicon. This in effect would provide an intracellular amplification of the initial virus-induced signal. Our results show that for both HSV-1 and HCMV, transactivation of the Sindbis virus replicon significantly enhanced the β-galactosidase signal. The ability to package the replicon in the HSV-1 system provided an additional amplification step by production of infectious replicon particles which allowed intercellular spread of the signal.
This type of signal amplification should be applicable to other replicons and to other herpesviruses. Sindbis and other alphavirus replicons have a wide host range (21, 43), but in some cell lines, for example, the mink lung cells described here, replication is inefficient. Replicons derived from other families of RNA viruses (18) could extend the use of this type of system to a much wider variety of cultured cell lines. The replication of SINrep/LacZ was triggered by a mutant of HSV-1, d21, that is unable to undergo replication and expresses only a subset of HSV-1 genes (28). It was also triggered by HCMV in a cell line that is able to carry out only some of the early steps in the viral replication cycle. These results, taken together, suggest that an inducible RNA replicon could be developed for detection of other viruses, including ones such as human herpesvirus type 8, for which a fully permissive cell culture system does not yet exist.
ACKNOWLEDGMENTS
We thank Hong Liu for expert technical assistance and Paul Hippenmeyer for plasmid pMonHygro. Thanks to Priscilla Schaffer for providing us with HSV mutant viruses d21 and dlx3.1 and to Neal DeLuca for providing d120. Special thanks to David Knipe for sharing information about the ICP8 transcript. We are grateful to Milton Schlesinger, David Leib, and Ilya Frolov for critical reading of the manuscript.
This work was supported by grant AI11377 from the National Institutes of Health.
REFERENCES
- 1.Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Chee M S, Hutchison C D, Kouzarides T, Martignetti J A, Preddie E, et al. The DNA sequence of the human cytomegalovirus genome. DNA Sequence. 1991;2:1–12. doi: 10.3109/10425179109008433. [DOI] [PubMed] [Google Scholar]
- 2.Berglund P, Sjöberg M, Garoff H, Atkins G J, Sheahan B J, Liljestöm P. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Bio/Technology. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 3.Bredenbeek P, Rice C M. Animal RNA virus expression systems. Semin Virol. 1992;3:297–310. [Google Scholar]
- 4.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C D, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryga, S., I. Frolov, and S. Schlesinger. Unpublished results.
- 9.Dubensky T W J, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, Jolly D J, Chang S M W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D. Trans-activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov I, Hoffman T A, Pragai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Alphavirus-based expression vectors: strategies and applications. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gleaves C A, Smith T F, Shuster E A, Pearson G R. Comparison of standard tube and shell vial culture techniques for the detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 1985;21:217–221. doi: 10.1128/jcm.21.2.217-221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hariharan M J, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J E, Karavodin L, Dubensky T W, Jr, Chang S M, Banks T A. DNA immunization against herpes simplex virus: enhanced efficacy using a Sindbis virus-based vector. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herweijer H, Latendresse J S, Williams P, Zhang G, Danko I, Schlesinger S, Wolff J A. A plasmid-based self-amplifying Sindbis virus vector. Hum Gene Ther. 1995;6:1161–1167. doi: 10.1089/hum.1995.6.9-1161. [DOI] [PubMed] [Google Scholar]
- 17.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson K L, Ball L A. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J Virol. 1997;71:3323–3327. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kääriäinen L, Ranki M. Inhibition of cell functions by RNA-virus infections. Annu Rev Microbiol. 1984;38:91–109. doi: 10.1146/annurev.mi.38.100184.000515. [DOI] [PubMed] [Google Scholar]
- 20.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 23.Mocarski E S., Jr . Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven; 1996. pp. 2447–2492. [Google Scholar]
- 24.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivo P D. Transgenic cell lines for detection of animal viruses. Clin Microbiol Rev. 1996;9:321–334. doi: 10.1128/cmr.9.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivo P D, Frolov I, Schlesinger S. A cell line that expresses a reporter gene in response to infection by Sindbis virus: a prototype for detection of positive strand RNA viruses. Virology. 1994;198:381–384. doi: 10.1006/viro.1994.1046. [DOI] [PubMed] [Google Scholar]
- 27.Olivo P D, Peeples M E, Collins P L, Schlesinger S. Detection and quantitation of human respiratory syncytial virus (RSV) using minigenome cDNA and a Sindbis virus replicon: a prototype assay for detecting negative-strand RNA viruses. Virology. 1998;251:198–205. doi: 10.1006/viro.1998.9419. [DOI] [PubMed] [Google Scholar]
- 28.Orberg P K, Schaffer P A. Expression of herpes simplex virus type 1 major DNA-binding protein, ICP8, in transformed cell lines: complementation of deletion mutants and inhibition of wild-type virus. J Virol. 1987;61:1136–1146. doi: 10.1128/jvi.61.4.1136-1146.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo J M, Dubensky T W., Jr DNA vaccines with a kick. Nat Biotechnol. 1998;16:517–518. doi: 10.1038/nbt0698-517. [DOI] [PubMed] [Google Scholar]
- 30.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn J P, McGeoch D J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985;13:8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 33.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger, S. Unpublished results.
- 35.Schlesinger S. Alphaviruses—vectors for the expression of heterologous genes. Trends Biotechnol. 1993;11:18–22. doi: 10.1016/0167-7799(93)90070-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlesinger S. RNA viruses as vectors for the expression of heterologous proteins. Mol Biotechnol. 1995;3:155–165. doi: 10.1007/BF02789111. [DOI] [PubMed] [Google Scholar]
- 37.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven; 1996. pp. 825–841. [Google Scholar]
- 38.Scholl, D. R. Personal communication.
- 39.Spector S A. Diagnosis of cytomegalovirus infection. Semin Hematol. 1990;27:11–16. [PubMed] [Google Scholar]
- 40.Stabell E C, Olivo P D. Isolation of a cell line for rapid and sensitive histochemical assay for the detection of herpes simplex virus. J Virol Methods. 1992;38:195–204. doi: 10.1016/0166-0934(92)90110-y. [DOI] [PubMed] [Google Scholar]
- 41.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su L, Knipe D M. Mapping the transcriptional initiation site of the herpes simplex virus type 1 ICP8 gene in infected and transfected cells. J Virol. 1987;61:615–620. doi: 10.1128/jvi.61.2.615-620.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]