Abstract
During herpes simplex virus (HSV) assembly, immature procapsids must expel their internal scaffold proteins, transform their outer shell to form mature polyhedrons, and become packaged with the viral double-stranded (ds) DNA genome. A large number of virally encoded proteins are required for successful completion of these events, but their molecular roles are poorly understood. By analogy with the dsDNA bacteriophage we reasoned that HSV DNA packaging might be an ATP-requiring process and tested this hypothesis by adding an ATP depletion cocktail to cells accumulating unpackaged procapsids due to the presence of a temperature-sensitive lesion in the HSV maturational protease UL26. Following return to permissive temperature, HSV capsids were found to be unable to package DNA, suggesting that this process is indeed ATP dependent. Surprisingly, however, the display of epitopes indicative of capsid maturation was also inhibited. We conclude that either formation of these epitopes directly requires ATP or capsid maturation is normally arrested by a proofreading mechanism until DNA packaging has been successfully completed.
Herpes simplex virus (HSV)-infected cells accumulate three different types of capsid in their nuclei. C capsids contain the viral DNA genome and are thought to be the precursors to enveloped, infectious viral particles. B capsids lack the viral genome but instead contain an electron translucent core of scaffolding proteins, including the major scaffold polypeptide ICP35 (11, 20, 25, 30). A capsids lack both DNA and scaffold proteins and are thought to be the dead-end products of a failed DNA packaging reaction. It is likely that A, B, and C capsids arise from the maturation of a short-lived unstable capsid precursor, termed the procapsid or large-cored B capsid (27, 32, 42, 45). The procapsid contains immature forms of the capsid scaffold proteins and has been considered analogous to the prohead of double-stranded (ds) DNA bacteriophage (27, 45).
Rapidly after its formation the spherical procapsid is thought to undergo a dramatic structural transformation, in which the surface shell angularizes into the polyhedral form characteristic of B and C capsids. The polyhedral capsid is considerably more stable than the procapsid and has a thicker surface shell with a much less open structure (27, 45). In addition, angularization is accompanied by the exposure of several new epitopes in the major capsid shell protein VP5 (9, 14, 21, 47). In vivo, the maturation of the procapsid usually correlates with proteolytic cleavage and the reorganization of the capsid scaffold proteins, including the most abundant scaffold polypeptide, ICP35 (11, 20, 25, 26, 30). The responsible protease, Pra, encoded by the UL26 gene, is itself a scaffold constituent and cleaves itself at two internal positions, known as the maturation and release sites, to generate the VP24 and VP21 polypeptides (6, 18–20, 26, 33, 34). UL26 proteolytic activity and self-cleavage at the release site are required for the conversion of the procapsid to the polyhedral form in vivo (14, 22, 32). The role of this cleavage in structural transformation, however, remains unclear (22, 27, 35).
Elegant studies using baculovirus expression systems both in vivo (40, 43, 46) and in vitro (27, 28, 45) have demonstrated that the major structural polypeptides of the HSV capsid are capable of spontaneous self-assembly, generating angular polyhedral B capsids in the absence of any other herpesvirus proteins. Strikingly, procapsids assembled in vitro are capable of maturing into polyhedral capsids lacking scaffolds, suggesting that scaffold ejection has also been reconstituted under these conditions (27). It is unclear whether, during the course of a normal HSV infection, such a spontaneous assembly and maturation are allowed to occur or whether additional regulatory pathways exist, for example, to couple polyhedral capsid formation to the machinery of DNA packaging and capsid envelopment.
The packaging of DNA into the maturing viral capsid requires the activity of at least seven HSV gene products: UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (1–3, 15, 16, 24, 29, 31, 37, 41, 51). Little is known of the molecular function of each of these proteins; however, it has been noted that the product of the HSV UL15 gene has some homology with the large subunit of the terminase complex responsible for cleavage and packaging of the phage T4 genome (4, 10, 13). UL15 is known to be present as multiple polypeptide species (2, 51) and interacts with capsids in a manner dependent on other components of the packaging apparatus (36, 53).
In dsDNA-containing bacteriophage, terminase-mediated DNA packaging is generally thought to be ATP dependent, and all in vitro phage DNA packaging systems require ATP (4, 7, 44, and references therein). Interestingly, the terminase-related region of UL15 includes a putative ATP binding motif essential for packaging (52); however, in the absence of an in vitro assay it has been difficult to test whether HSV DNA packaging is truly an ATP-dependent process. In the present study we address this question by making use of a synchronized HSV assembly assay recently established in our laboratory (8, 9). This experimental system is based upon the properties of the HSV mutant strain tsProt.A, which carries a reversible temperature-sensitive lesion in its UL26 protease gene. At the nonpermissive temperature of 39°C tsProt.A-infected cells accumulate procapsids (14, 32), and following a downshift to the permissive temperature of 31°C these procapsids mature, package DNA, and give rise to infectious particles in a single, synchronized wave (9). This assay system enables us to separate early events in HSV replication, such as entry, DNA synthesis, and gene expression, from the later events of capsid maturation, DNA packaging, and egress.
Here we report that when accumulated tsProt.A procapsids were released from their temperature block in the presence of diminished levels of cellular ATP, capsid scaffold cleavage proceeded normally but DNA packaging was completely inhibited. Unexpectedly, two VP5 epitopes characteristic of capsid maturation also failed to form, suggesting the inability of the procapsid shell to properly mature. In contrast, an HSV mutant strain lacking the essential packaging gene UL15 gave rise to capsids with normal antibody reactivity. Our findings suggest that DNA packaging is indeed ATP dependent but that, unexpectedly, so are some of the events in HSV capsid maturation.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and the UL15-complementing cell line M-3 were grown as previously described (8). Stocks of HSV strains tsProt.A, SC16, and the UL15 null mutant hr81-2 were prepared, and titers were determined as in earlier studies (8, 9).
Measurements of scaffold cleavage in total cell extracts and pelleted capsids.
Cell extracts were prepared and Western blotted as in earlier studies (9). Cleavage of ICP35c,d was monitored with the monoclonal antibody MCA406 as previously described (9). Cleavage of Pra at the carboxy-terminal maturation site and production of the VP24 amino-terminal cleavage product were monitored by Western blotting with an anti-VP24 rabbit polyclonal antiserum. To test the extent of scaffold cleavage within capsids, infected cells were incubated under appropriate conditions of temperature and ATP depletion and then washed in phosphate-buffered saline (PBS). All subsequent procedures were carried out at 4°C. Cells were collected by scraping in distilled water, and then Triton X-100 was added to a final concentration of 2%. Following overnight incubation on ice the mixture was sonicated and subjected to a 1,500-g clearing spin for 10 min. The supernatant was collected, and then the pellet was resuspended in TNE (500 mM NaCl, 1 mM EDTA, 10 mM Tris · Cl; pH 8.0) and centrifuged as before. After the supernatant had been collected the pellet was subjected to one further round of TNE extraction, and the three supernatants were combined. The pooled supernatants were layered on top of a 35% (wt/vol) sucrose–10 mM Tris · Cl (pH 8.0) cushion and then spun at 25,000 × g in a Beckman TLS55 swinging bucket rotor for 75 min. The pellet was resuspended in PBS and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Measurements of DNA packaging.
Southern blotting with the SQ junction fragment probe was performed as in earlier studies (9). Total and encapsidated infected cell DNA for Southern blot analysis was prepared as previously described (8). The trichloroacetic acid (TCA) precipitation assay used to measure DNA packaging was modified from an earlier published method (8) as follows. Vero cells were infected with HSV at multiplicity of infection (MOI) of 10 and then incubated at 39°C in Dulbecco modified Eagle medium (DMEM) containing 1% dialyzed newborn calf serum (NCS). After 2 h, [3H]thymidine (New England Nuclear) was added to a final concentration of 25 μCi/ml, and incubations were continued as required. At the appropriate times cells were rinsed twice in Tris-buffered saline (130 mM NaCl–20 mM KCl–25 mM Tris · Cl; pH 7.4) and then were frozen, thawed, collected by scraping, and sonicated. The resulting extracts were incubated in the presence of 2 mM MgCl2 and 70 U of DNase per ml for 2 h at 37°C, and then EDTA and SDS were added to final concentrations of 10 mM and 0.3%, respectively. Aliquots were spotted onto 24-mm-diameter glass fiber filters (GF/C; Whatman) and incubated in ice-cold TP buffer (5% TCA, 20 mM sodium pyrophosphate) for 5 min. After two further incubations in fresh TP buffer at 65°C for 5 min, filters were rinsed in 70% ethanol at room temperature for 3 min and then dried, and levels of TCA-precipitable radioactivity were determined by liquid scintillation counting.
Determining levels of protein synthesis.
Vero cells were infected with HSV tsProt.A at an MOI of 10 and then incubated at 39°C in DMEM containing 10% NCS. After 5.5 h cells were washed twice in prewarmed PBS and then starved of methionine and cysteine by incubation at 39°C in DMEM lacking these amino acids and containing 1% dialyzed NCS. After 1 h of starvation the cell culture medium was supplemented with 10 μCi of 35S-radiolabeled methionine and cysteine per ml (New England Nuclear) and also with either 20 μg of cycloheximide per ml or an ATP depletion cocktail, consisting of 25 mM sodium azide and 25 mM 2-deoxyglucose, or was mock treated. A control sample was recovered immediately after the addition of radiolabel to determine the level of unincorporated background radioactivity. The other samples were incubated for a further 0.5 h at 39°C and then shifted to 31°C, harvested after 40 or 150 min, washed twice in Tris-buffered saline, and collected by scraping. After freezing, thawing, and sonication cell extracts were adjusted to concentrations of 0.4% SDS and 10 mM EDTA, and aliquots were TCA precipitated on GF/C filters as described above.
Immunocytochemistry and electron microscopy.
Infected Vero or M-3 cells were processed for indirect immunocytochemistry or electron microscopy, exactly as described previously (9).
RESULTS
ATP depletion blocks production of new infectious progeny following release of the temperature block.
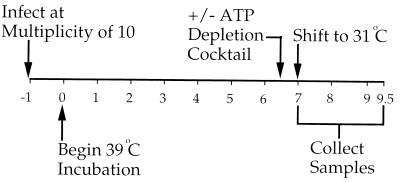
To determine the requirement for normal cellular levels of ATP in HSV assembly we used the experimental approach shown in Fig. 1. Vero cells were infected for 1 h at 37°C at an MOI of 10 with HSV strain tsProt.A, the residual input virus was inactivated by acid washing, and cells were incubated at the nonpermissive temperature of 39°C to accumulate a population of procapsids. After 6.5 h an ATP depletion cocktail consisting of 25 mM 2-deoxyglucose and 25 mM sodium azide was added or omitted, and after a further 30 min the infected cells were shifted to 31°C to allow HSV assembly to proceed. In earlier studies (9) it has been demonstrated that infectious virus assembly was essentially complete by 140 min of incubation under these conditions, so 150 min of incubation was selected as a convenient end point for these experiments. The concentrations of 2-deoxyglucose and sodium azide used in these studies are comparable to those routinely used for ATP depletion experiments (38, 48, 50), and its addition under these conditions did not result in increased levels of trypan blue sensitivity compared to that of untreated cells (data not shown) or lead to gross morphological changes (see Fig. 5 and 6).
FIG. 1.
General experimental design. Numbers correspond to the time (in hours) after completion of initial infection. An ATP depletion cocktail was added (+) or omitted (−) after 6.5 h of incubation at the nonpermissive temperature of 39°C, and cells were incubated for a further 0.5 h before being shifted to 31°C. At particular times samples of infected cells were collected and assayed as described in the text.
FIG. 5.
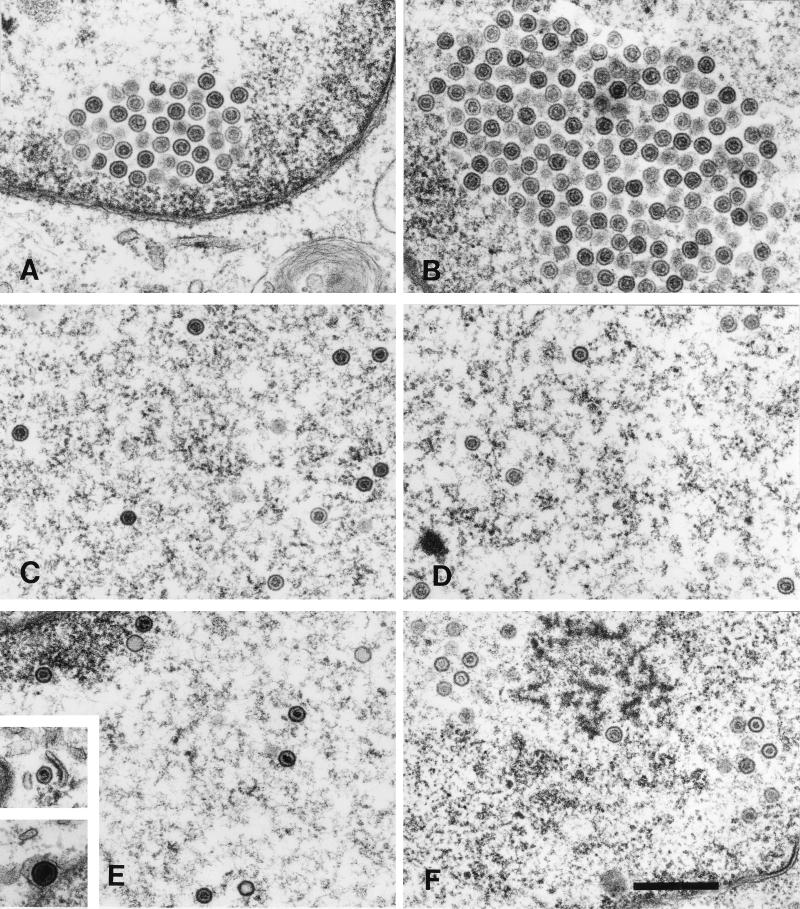
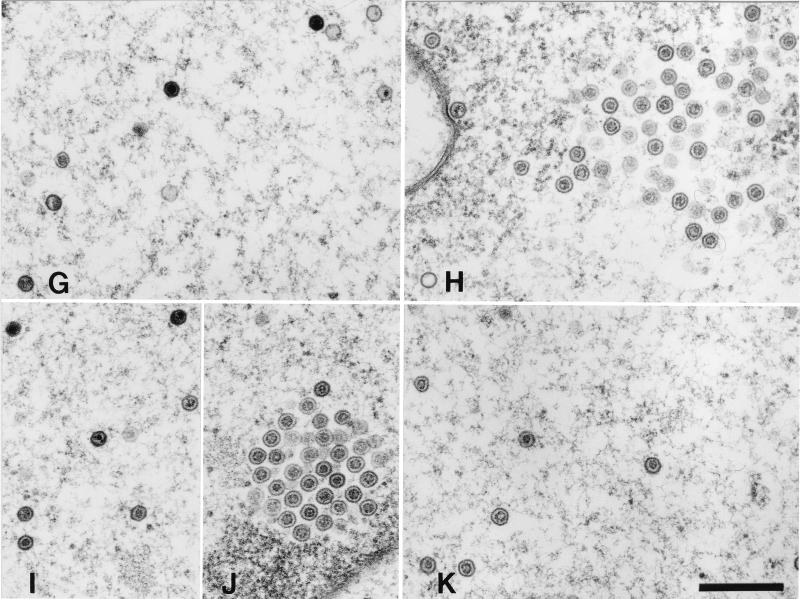
Ultrastructural analysis of capsid maturation in ATP-depleted and control cells. Vero cells were infected with tsProt.A and incubated (see Fig. 1). They were mock treated (left-hand panels) or depleted of ATP (right-hand panels) and then downshifted to 31°C. Samples were fixed in 2.5% glutaraldehyde immediately (A and B), after 40 min (C and D), or after 150 min (E to K) and processed for electron microscopy. (A and B) Clustered procapsids accumulating close to nuclear rim. (C and D) Dispersed nuclear capsids resembling B capsids. (E, G, I, and J) Mixture of A, B, and C capsids dispersed within the nucleus and also present within clusters. (Insets in E) Two images from the same field of cells, revealing a cytoplasmic C capsid (upper inset) and an enveloped C capsid lying between the nuclear membranes (lower inset). (I and J) Different regions of the same nucleus. (F, H, and K) ATP-depleted cells accumulate mainly B capsids. Bars, 500 nm.
FIG. 6.
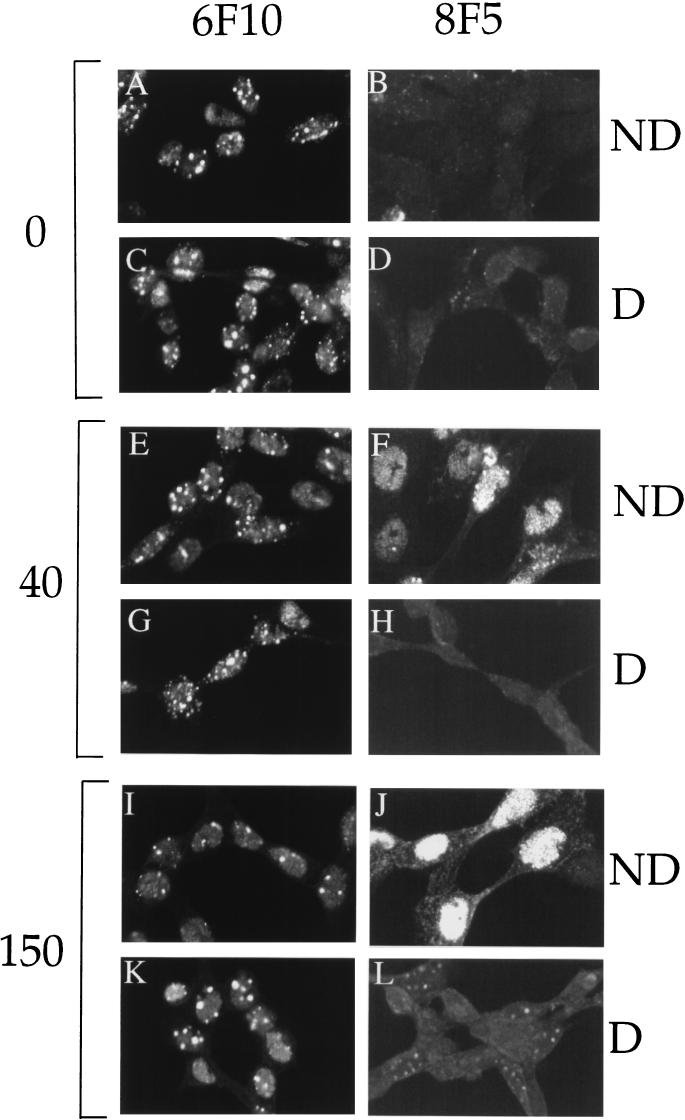
Immunoreactivity and nuclear distribution of maturing HSV capsids in ATP-depleted (D) or nondepleted (ND) cells. Vero cells were infected and incubated (see Fig. 1) and fixed following 0 min (A to D), 40 min (E to H), or 150 min (I to L) of incubation at 31°C. After fixation, samples were permeabilized, incubated with anti-VP5 monoclonal antibody 6F10 or 8F5 and a fluorescein isothiocyanate secondary antibody, and viewed by laser scanning confocal microscopy.
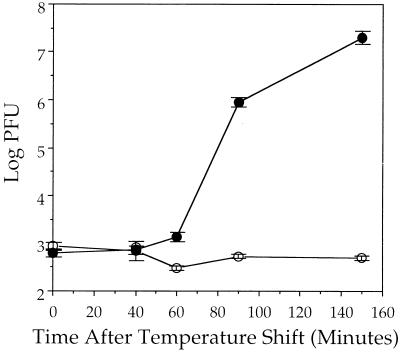
Figure 2 shows that when this protocol was followed and samples were assayed for PFU production at successive times after shifting to 31°C, there was a rapid burst of infectious virus yield in control cells, resulting in a 4-log increase in viral titer over the time course of the experiment. In contrast, cells depleted of ATP were unable to support the production of infectious particles. We conclude that one or more HSV assembly steps downstream of procapsid formation require normal levels of cellular ATP for completion.
FIG. 2.
Effect of ATP depletion on production of infectious virus. Vero cells were infected with tsProt.A and processed as shown in Fig. 1. Cells were recovered at particular times after shifting to the permissive temperature (indicated on the horizontal axis), and extracts were prepared. Levels of PFU present in each extract are represented on the logarithmic vertical axis. Plotted points represent the mean and standard deviation from the mean for triplicate plaque assays. Viral yields at zero time after the temperature shift derive from residual input virus (9). ●, control-treated cells; ○, ATP-depleted cells.
We previously showed that, following the shift to 31°C, a wave of infectious tsProt.A particles were assembled, even in the presence of a cycloheximide concentration sufficient to completely abolish PFU production in a wild-type HSV infection (9). This led us to conclude that the wave of infectious tsProt.A virions resulted from the assembly of presynthesized polypeptides. However, we could not discount the possibility that some low level of ongoing protein synthesis was necessary to permit infectious tsProt.A production, for example, to supply some essential polypeptide which is normally present in limiting amounts. If this were true, ATP depletion could be inhibiting PFU production, simply by acting as a more potent inhibitor of protein synthesis than cycloheximide. To test this possibility we performed an experiment similar to that depicted in Fig. 1 but added [35S]methionine and [35S]cysteine at the time of addition of cycloheximide or the ATP depletion cocktail. Following a further 30-min incubation at 39°C cells were downshifted to 31°C and incubated for 40 or 150 min, and crude cell extracts were prepared. The amount of protein synthesis which had occurred since the drug addition was then determined by measuring the amount of TCA-precipitable radioactivity present. As can be seen in Table 1, the ATP depletion cocktail inhibited protein synthesis by 82% when measured over the entire 31°C time course. By comparison, 20 μg of cycloheximide per ml, the concentration used in our standard HSV assembly assays (9), inhibited protein synthesis by about 94%. Since this concentration of cycloheximide nevertheless permits a 3-log increase in PFU production under these conditions (9), we conclude that the abolition of PFU production by ATP depletion is not due to its effect on protein synthesis. That the cells are able to maintain 18% of the normal level of protein synthesis is further evidence that the ATP depletion cocktail is not having a gross toxic effect during the course of this experiment.
TABLE 1.
TCA-precipitable 35S radioactivity and inhibition of protein synthesis
| Minutes after downshift | Mean (SD) cpm of 35S label
|
% Inhibition of protein synthesisa
|
|||
|---|---|---|---|---|---|
| Untreated | Cyclo-heximide added | ATP depleted | Cyclo-heximide added | ATP depleted | |
| −30b | 250 (6) | ||||
| 40 | 11,989 (432) | 768 (22) | 2,747 (279) | 93.6 | 77.1 |
| 150 | 19,975 (863) | 1,119 (102) | 3,595 (276) | 94.4 | 82 |
Calculated by comparing mean incorporated counts per minute with untreated control.
Sample collected 30 min before downshift, at time of addition of radiolabel.
ATP depletion does not block capsid scaffold cleavage.
We tested whether ATP depletion affected the earliest known event following the release of the temperature block: cleavage of the capsid scaffold polypeptides. The results shown in Fig. 3A demonstrate that when ICP35 cleavage was examined by Western blotting, the kinetics and extent of processing of full-length ICP35c,d to mature ICP35e,f were similar in both the presence and absence of the ATP depletion cocktail. Similarly, as shown in Fig. 3B the extent of cleavage of the full-length UL26 gene product Pra at the terminal maturation site, generating a slightly smaller polypeptide, and at the internal release site, generating the VP24 polypeptide, was also unaffected by ATP depletion.
FIG. 3.
ATP depletion does not affect the rate or extent of capsid scaffold cleavage. Vero cells were infected with tsProt.A and incubated as shown in Fig. 1. At 6.5 h postinfection cells were mock treated (ND, nondepleted control cells) or treated with an ATP depletion mixture (D, cells depleted of ATP), incubated for a further 0.5 h at 39°C, harvested, and processed as follows. (A) Cell extracts were prepared immediately or after successive times of incubation at 31°C (as indicated above the figure in minutes), subjected to SDS-PAGE, and Western blotted for ICP35. (B) Cell extracts were prepared immediately or after 150 min of incubation, subjected to SDS-PAGE, Western blotted, and probed with antisera specific for the amino-terminal portion of Pra. The positions of Pra and VP24 are indicated at the left of the figure. (C) Cell extracts were chilled, solubilized in Triton X-100, and subjected to several high-salt extractions in order to release viral capsids. Capsids were then pelleted through a sucrose cushion, and the pellet was subjected to SDS-PAGE, Western blotted, and probed for ICP35. The positions of Pra and ICP35 are indicated by the upper and lower pair of arrowheads, respectively. The uppermost of each pair of arrowheads indicates the position of the unprocessed form of the scaffold polypeptide. The autoradiograph is deliberately overexposed to show the low levels of uncleaved ICP35c,d and Pra in the capsid pellet. The reactive bands at the top of the gel appear to be capsid specific and may represent aggregated or multimeric Pra.
Although similar amounts of scaffold cleavage occurred in both control and ATP-depleted cells, it is unclear from these data how much processing occurred within capsids, since ICP35c,d cleavage occurs at similar rates whether or not capsids are able to assemble (12, 22). It is therefore possible that only a small proportion of total ICP35 is present within procapsids under our conditions. In this case, even if ATP depletion had prevented scaffold cleavage within capsids this inhibition would have been difficult to detect against a high background level of soluble ICP35c,d processing. To resolve this question we prepared a crude capsid fraction from infected cells by Triton X-100 solubilization, followed by two rounds of high-salt extraction from the nuclear pellet (see Materials and Methods). The capsids were then centrifuged through a sucrose cushion to separate them from soluble polypeptides, and the resulting pellet was blotted for the presence of ICP35. As can be seen in Fig. 3C, samples which had not been shifted to 31°C failed to yield any pelletable scaffold antigens. This is consistent with the cold sensitivity of the procapsid (27), which would be expected to disintegrate under these isolation conditions, and suggests that any pelleted immunoreactive material is truly capsid associated. In contrast, extracts prepared from cells which had been shifted to 31°C for 150 min did contain pelletable, cold-stable capsids, whether ATP had been depleted or not. Both ICP35 and Pra appeared to have been efficiently processed at the carboxy-terminal maturation site. This is in contrast to the composition of scaffold antigen in total cell extracts, where cleaved scaffolds are present at similar (Fig. 3A) or reduced (Fig. 3B) levels compared to their uncleaved precursors. We conclude that ATP depletion affects neither scaffold cleavage within capsids nor conversion of the procapsid to a cold-resistant structure.
ATP depletion blocks DNA packaging.
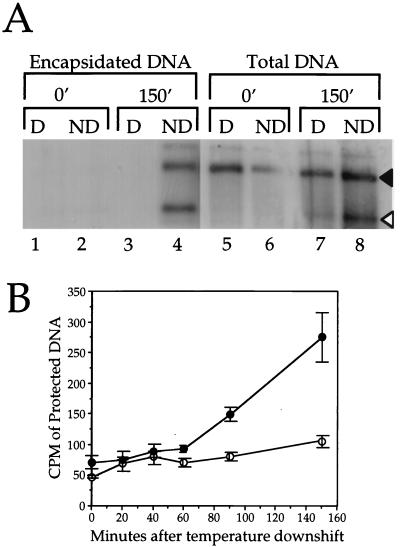
We found that ATP depletion resulted in a striking inhibition of DNA packaging (Fig. 4A). When ATP was depleted, little or no new DNA cleavage occurred by the 150-min time point, and no viral DNA became protected from DNase I digestion, suggesting that the DNA packaging machinery had failed. Although in this particular experiment the final levels of cleaved DNA in ATP-depleted cells appear slightly higher than the background level (Fig. 4A, lanes 5 and 7), this was not reproducible between experiments (data not shown). Figure 4B shows that similar results were obtained when we independently quantitated packaging by measuring the rate of production of DNase I-resistant [3H]thymidine-labeled TCA-precipitable DNA, as previously described (8). We conclude that, as for dsDNA-containing bacteriophage, the packaging of the HSV genome requires ATP. This requirement is unlikely to be at the level of DNA synthesis, since we have previously shown that under these conditions, DNA packaging does not require measurable levels of ongoing DNA replication (8).
FIG. 4.
DNA packaging is inhibited following ATP depletion. (A) Following the procedure summarized in Fig. 1, cell extracts were prepared after 0 or 150 min of incubation at 31°C, as indicated at the top of the figure. The extracts were used to prepare total infected cell DNA or first were treated with DNase I to permit isolation of only that DNA present in viral capsids. In each case the resulting purified DNA was digested with BamHI, electrophoresed on a 1.0% agarose gel, blotted onto a nylon membrane, and hybridized to a 32P-labeled probe corresponding to the SQ cleavage junction. The positions of the SQ and Q fragments are indicated by black and white arrowheads, respectively. D, cells depleted of ATP; ND, nondepleted control cells. (B) Results of an experiment similar to that in panel A, except that DNA synthesized during infection was labeled with [3H]thymidine, and the rate of production of DNase I-resistant, TCA-precipitable counts per minute was determined. Plotted points indicate the means and standard deviations from the mean for triplicate precipitates. ●, control-treated cells; ○, ATP-depleted cells.
Infected cells depleted of ATP accumulate nuclear B capsids.
In order to investigate assembly arrest at the ultrastructural level we performed electron microscopy at the time of release of the temperature block, after 40 min at 31°C and after 150 min at 31°C in either mock-treated or ATP-depleted cells (Fig. 5). As observed previously (9) under normal conditions large-cored B capsids (procapsids) were the sole species visible at the end of the 39°C incubation, irrespective of whether an ATP depletion cocktail was present or not (Fig. 5A and B). Similarly, by 40 min of 31°C incubation (a time sufficient to process most of the cleavable ICP35c,d [Fig. 3A and reference 9]), both ATP-depleted and control cells contained numerous nuclear B capsids both in clusters (data not shown and reference 9) and dispersed (Fig. 5C and D). Under both sets of conditions we observed structures resembling large-cored B capsids (procapsids) and small-cored B capsids (for example, in the top right-hand corners of Fig. 5C and D); however, it is extremely difficult to unambiguously discriminate between procapsids and small-cored B capsids by thin-section electron microscopy, and we did not attempt to quantitate these capsid forms at either 40 or 150 min of incubation. In both ATP-depleted (Fig. 5F, H, and K) and nondepleted (panels E, G, I, and J) cells there were numerous dispersed capsids and also some remaining capsid clusters which appeared to mainly be composed of immature procapsids (panels H and J). Nondepleted cells also contained numerous A and C capsids in the nucleoplasm (Fig. 5E, G, and I), in the perinuclear space, and more rarely in the cytoplasm (panel E, inset). In contrast, A and C capsids were extremely rare in ATP-depleted cells, although occasional examples could be found (in Fig. 5H, a C and an A capsid are visible at the left edge of the panel). The absence of A capsids is a common phenotype in viral mutants unable to encode an essential packaging function and is commonly interpreted to mean that the DNA packaging process could not be initiated. Although in the absence of the essential packaging factors UL28 and UL15 B capsids are able to be enveloped and traffic into the cytoplasm (2, 41), we observed no extranuclear B capsids in ATP-depleted cells.
Generation of mature capsid-specific epitopes is inhibited following ATP depletion.
To further examine the effects of ATP depletion on HSV assembly we investigated the distribution of viral capsids in normal and ATP-depleted cells by indirect immunocytochemistry, by using the anti-VP5 monoclonal antibodies 6F10 and 8F5. Although both of these antibodies are specific for VP5 when it resides within hexons, the 6F10 epitope is present both in procapsids and in mature capsids (27, 39), whereas the 8F5 epitope is only generated following the conversion of the procapsid to the polyhedral form (9, 14, 21, 23, 47). As shown in Fig. 6, 6F10-reactive VP5 was readily detectable in the nuclei of infected cells at the time of and up to at least 150 min following the shift to 31°C. There was no apparent difference in the pattern of 6F10 reactivity during the time course of incubation or when comparing ATP-depleted and nondepleted cells (Fig. 6A, C, E, G, I, and K). In contrast, reactivity to the 8F5 antibody was dramatically different between the ATP-depleted and nondepleted cells. Although 8F5 reactivity was readily detectable by 40 min after release of the temperature block in normal cells (as previously reported in reference 9) there was little immunoreactivity in ATP-depleted cells (Fig. 6F and H). Even incubation for 150 min failed to yield levels of 8F5 immunoreactivity above the background level (Fig. 6B and L). We obtained identical results with the anti-VP5 monoclonal antibody 5C (data not shown), which is also specific for mature hexons but recognizes an epitope at a site quite distinct from that of 8F5 (47). These results suggest that depletion of intracellular ATP results in an inhibition of capsid maturation.
Generation of the 8F5 epitope can occur in the absence of DNA packaging.
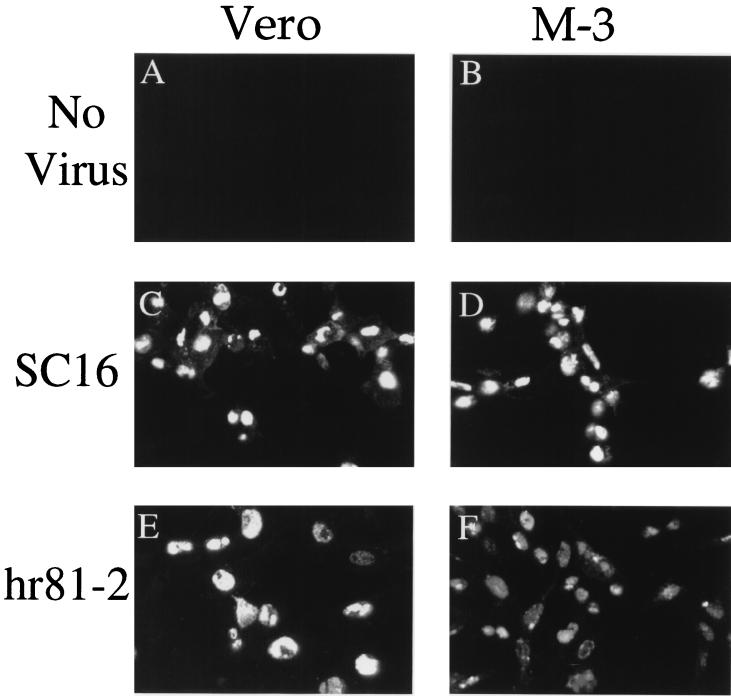
We were surprised to find that 8F5 failed to react with the capsids accumulated in ATP-depleted cells, since purified, unpackaged B capsids are known to react with 8F5 (47) and the capsids accumulated under our depletion conditions are cold resistant (Fig. 3C), suggesting that some degree of capsid shell maturation has occurred. Nevertheless, a simple interpretation of our data is that under our conditions, DNA packaging is a prerequisite for exposure of the 8F5 epitope. To directly test this possibility we examined the ability of packaging-defective viruses to react with 8F5. Vero cells or the UL15-expressing Vero cell line, M-3, was infected with the wild-type HSV strain SC16 or with the UL15 null mutant hr81-2. As shown in Fig. 7, infections carried out in the presence of wild-type levels of UL15 (panel C) or no UL15 (panel E) resulted in a similar level of 8F5 reactivity in infected cell nuclei. Similarly, 8F5 reactivity appeared unaffected when wild-type or UL15 null virions were provided with UL15 by the complementing cell line M-3 (Fig. 7D and F). In Fig. 7A and B are shown fields of uninfected Vero or M-3 cells, demonstrating the specificity of the 8F5 antibody. We conclude that the failure to package DNA is not sufficient to block the expression of the 8F5 epitope, consistent with earlier in vitro findings (47).
FIG. 7.
Immunoreactivity and nuclear distribution of maturing HSV capsids in the presence and absence of UL15. Vero cells or UL15-expressing M-3 cells were mock infected (A and B) or infected with HSV SC16 (C and D) or hr81-2 (E and F), incubated for 9.5 h at 37°C, fixed, and permeabilized. Samples were incubated with monoclonal antibody 8F5 and a fluorescein isothiocyanate secondary antibody and viewed by laser scanning confocal microscopy.
DISCUSSION
Studies of the assembly of dsDNA bacteriophage such as T4 and λ have revealed roles for ATP at several different steps in the packaging process. By analogy, we reasoned that ATP would be required for HSV DNA packaging, and in this study we have used the reversible temperature-sensitive mutant tsProt.A to test this prediction. As anticipated, the depletion of intracellular ATP resulted in a complete abolition of HSV DNA encapsidation and cleavage and prevented the assembly of any measurable levels of infectious virus. We conclude that ATP is required for some step in the process of HSV DNA packaging.
Since HSV capsid assembly can occur spontaneously in cell extracts lacking ATP, we anticipated that it would also occur normally under our conditions, giving rise to B capsids with a matured, polyhedral 8F5-reactive and 5C-reactive outer shell. When we examined the maturation of the viral capsids, we found that total cellular and capsid scaffold maturation did indeed appear to be normal in ATP-depleted cells and that the Pra and ICP35 polypeptides were proteolytically cleaved to the same extent as those in control cells. Furthermore, capsids became cold resistant, an event believed to accompany the transformation of the fragile procapsid shell to the mature, stable angular form. Unexpectedly, however, the accumulated capsids failed to react with the antibodies 8F5 and 5C, which recognize epitopes presented by mature angular capsids. Reactivity with the anti-VP5 antibody 6F10, however, which recognizes both procapsids and mature capsids, appeared to be normal. Although the ultrastructural examination of infected cell nuclei revealed the accumulation of apparently normal large- and small-cored B capsids in ATP-depleted cells (Fig. 5), it is extremely difficult to unambiguously discriminate between procapsids and small-cored B capsids by thin-section electron microscopy (27).
We have considered two possible models to explain these unexpected findings. The first is based on the observation made by Baines and colleagues, who demonstrated that in the absence of UL15 large numbers of B capsids were assembled and subsequently could traffic from the cell nucleus into the cytoplasm (2). Similar results were obtained by Tengelsen and coworkers for viruses lacking the packaging factor encoded by UL28 (41). These observations led to the suggestion that the packaging machinery could also serve as a proofreading apparatus which normally prevents unpackaged capsids from proceeding to later stages of assembly (2). Under our conditions of ATP depletion, DNA packaging fails to occur, but the proofreading apparatus could remain fully functional and might prevent the maturation of the capsid shell. In this model, UL15 deletion would disrupt the proofreading apparatus, allow complete capsid maturation, and lead to generation of the 8F5 epitope, as seen in Fig. 7. Note that this model requires UL15 only to be an essential component of the proofreading apparatus and not necessarily directly responsible for monitoring the completion of packaging. The model does require, however, that whatever stage in capsid maturation is blocked by the proofreading mechanism, it must lie after the formation of cold-resistant capsids and before the conformational changes which accompany 8F5 and 5C epitope formation. Further, it requires that it is the stable, presumably angularized capsid which is competent to package DNA and not the cold-sensitive, porous procapsid.
An alternative model, which we favor, is that 8F5 and 5C epitope formation requires ATP-dependent conformational changes subsequent to the ATP-independent generation of cold-resistant shells and independent of DNA packaging. One speculative possibility is that the ATP dependence lies in the recruitment of the capsid subunit VP26, which binds to the tips of VP5 when present in hexons but not in pentons (5, 54). In this scenario the 8F5 and 5C epitopes might be dependent on the formation of a VP26-VP5 complex. However, this model presumes that VP26 is normally absent from the procapsid and is recruited only following the release of the temperature block; unfortunately, the time of VP26 assembly into capsids is not known. A further limitation of this model is that the binding of VP26 to capsids does not require ATP in vitro (49). An alternative and interesting possibility is that the formation of the 5C and 8F5 epitopes may require the action of ATP-requiring chaperones, as has been suggested for the correct assembly of other animal virus capsids (17, 48).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI38265 to D.W.W. Core support was provided by NIH Cancer Center grant P30-CA13330.
HSV strain hr81-2 and the cell line M-3 were generous gifts from Sandra Weller, and the anti-VP24 antibody was kindly provided by Zhi Hong. We thank Jay Brown, Bill Newcomb, and Carol Harley for helpful discussions and Lily Huang for excellent technical assistance. We also gratefully acknowledge the Analytical Imaging Facility of the Albert Einstein College of Medicine for help with electron and confocal microscopy.
REFERENCES
- 1.al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 2.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun D K, Roizman B, Pereira L. Characterization of post-translational products of herpes simplex virus gene 35 proteins binding to the surfaces of full capsids but not empty capsids. J Virol. 1984;49:142–153. doi: 10.1128/jvi.49.1.142-153.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalano C E, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 8.Church G A, Dasgupta A, Wilson D W. Herpes simplex virus DNA packaging without measurable DNA synthesis. J Virol. 1998;72:2745–2751. doi: 10.1128/jvi.72.4.2745-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 11.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 12.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan A, Arbuckle M, McGeoch D J. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 1991;20:97–104. doi: 10.1016/0168-1702(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 14.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 16.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Roizman B. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci USA. 1992;89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Roizman B. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J Virol. 1993;67:1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matusick-Kumar L, Hurlburt W, Weinheimer S P, Newcomb W W, Brown J C, Gao M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J Virol. 1994;68:5384–5394. doi: 10.1128/jvi.68.9.5384-5394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matusick-Kumar L, McCann III P J, Robertson B J, Newcomb W W, Brown J C, Gao M. Release of the catalytic domain No from the herpes simplex virus type 1 protease is required for viral growth. J Virol. 1995;69:7113–7121. doi: 10.1128/jvi.69.11.7113-7121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matusick-Kumar L, Newcomb W W, Brown J C, McCann III P J, Hurlburt W, Weinheimer S P, Gao M. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J Virol. 1995;69:4347–4356. doi: 10.1128/jvi.69.7.4347-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb W W, Brown J C. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J Virol. 1989;63:4697–4702. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 28.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 30.Person S, Laquerre S, Desai P, Hempel J. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. J Gen Virol. 1993;74:2269–2273. doi: 10.1099/0022-1317-74-10-2269. [DOI] [PubMed] [Google Scholar]
- 31.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston V G, Rixon F J, McDougall I M, McGregor M, al Kobaisi M F. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 34.Rixon F J, Cross A M, Addison C, Preston V G. The products of herpes simplex virus type 1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J Gen Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- 35.Robertson B J, McCann P J, Matusick-Kumar L, Newcomb W W, Brown J C, Colonno R J, Gao M. Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids. J Virol. 1996;70:4317–4328. doi: 10.1128/jvi.70.7.4317-4328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmon B, Baines J D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skiba P J, Zha X, Maxfield F R, Schissel S L, Tabas I. The distal pathway of lipoprotein-induced cholesterol esterification, but not sphingomyelinase-induced cholesterol esterification, is energy-dependent. J Biol Chem. 1996;271:13392–13400. doi: 10.1074/jbc.271.23.13392. [DOI] [PubMed] [Google Scholar]
- 39.Spencer J V, Trus B L, Booy F P, Steven A C, Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 40.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 41.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomka M A, Catalano C E. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 45.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 46.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trus B L, Newcomb W W, Booy F P, Brown J C, Steven A C. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrobel I, Collins D. Fusion of cationic liposomes with mammalian cells occurs after endocytosis. Biochim Biophys Acta. 1995;1235:296–304. doi: 10.1016/0005-2736(95)80017-a. [DOI] [PubMed] [Google Scholar]
- 51.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu D, Weller S K. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology. 1998;243:32–44. doi: 10.1006/viro.1998.9041. [DOI] [PubMed] [Google Scholar]
- 53.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]