Summary
Human induced pluripotent stem cell-derived (hiPSC) cardiomyocytes are a promising source for regenerative therapy. To realize this therapy, however, their engraftment potential after their injection into the host heart should be improved. Here, we established an efficient method to analyze the cell cycle activity of hiPSC cardiomyocytes using a fluorescence ubiquitination-based cell cycle indicator (FUCCI) system. In vitro high-throughput screening using FUCCI identified a retinoic acid receptor (RAR) agonist, Am80, as an effective cell cycle activator in hiPSC cardiomyocytes. The transplantation of hiPSC cardiomyocytes treated with Am80 before the injection significantly enhanced the engraftment in damaged mouse heart for 6 months. Finally, we revealed that the activation of endogenous Wnt pathways through both RARA and RARB underlies the Am80-mediated cell cycle activation. Collectively, this study highlights an efficient method to activate cell cycle in hiPSC cardiomyocytes by Am80 as a means to increase the graft size after cell transplantation into a damaged heart.
Keywords: FUCCI, Cell cycle, Cardiac cell transplantation, human induced pluripotent stem cells, cardiomyocyte differentiation, retinoic acid, high throughput screening
Graphical abstract
Highlights
-
•
FUCCI technology can visualize the cell cycle in iPSC cardiomyocytes
-
•
High-throughput screening identified Am80 as a cell cycle activator for cardiomyocytes
-
•
Am80-treated cardiomyocytes showed better engraftment in the damaged mouse heart
-
•
Am80 activates the cell cycle of cardiomyocytes via RARs
We successfully established a high-throughput screening system using FUCCI-expressing iPSC cardiomyocytes and identified Am80, a retinoic acid receptor agonist, as a cell cycle activator of iPSC cardiomyocytes. Am80-treated iPSC cardiomyocytes proliferated via RARA and RARB and created larger grafts when injected into the damaged mouse heart.
Introduction
Human pluripotent stem cell-derived (hPSC) cardiomyocytes are a promising source of cells for several clinical applications, including the in vitro modeling of human heart diseases and related therapies. In the past decade, several investigators have reported the efficacy of cell transplantations using hPSC cardiomyocytes to treat infarcted hearts in animal models (Laflamme et al., 2007; Liu et al., 2018; Shiba et al., 2016). However, the injected cardiomyocytes do not survive or engraft well, resulting in limited graft size and therapeutic effects on the damaged heart (Oikonomopoulos et al., 2018). Overall, better engraftment is required to realize cardiac cell therapies using hPSC cardiomyocytes in the clinical setting.
After cell injection into a damaged heart, the graft size grows rapidly during the first month after transplantation, and the graft continues to grow until 3 months followed by a plateau (Funakoshi et al., 2016). The graft growth is correlated with cell cycle activation of the engrafted cardiomyocytes, which is normally measured by Ki67 or phospho-histone H3 staining (Funakoshi et al., 2016; Romagnuolo et al., 2019). Recently, Zhu et al. (2018) reported that transplanted human induced PSC (hiPSC)-cardiomyocytes that overexpress cyclin D2 (CCND2) show larger graft sizes and cause better improvement of cardiac function than wild-type hiPSC cardiomyocytes. More recently, Bargehr et al. (2019) demonstrated that co-transplantation with hPSC epicardium, which can secrete several proliferative factors and extracellular matrix, promotes proliferation of the engrafted hPSC cardiomyocytes, resulting in a larger graft size than transplanting hPSC cardiomyocytes only. Given that the graft size affects the efficacy of the cell transplantation, enhancing the proliferation of the engrafted cells could be a promising strategy to enlarge the graft size and improve the therapeutic effect of the cardiac cell therapy.
Fluorescence ubiquitination-based cell cycle indicator (FUCCI) is a well-documented and proven technology for monitoring cell cycle activation (Sakaue-Sawano et al., 2008). Briefly, the FUCCI system uses two fluorescent probes, monomeric Kusabira Orange, which is fused to chromosome licensing factor hCdt1 and indicates the G0/G1 phase by emitting red wavelengths, and monomeric Azami Green, which is fused to licensing inhibitor hGeminin and indicates S/G2/M by emitting green wavelengths. This technology can visualize the cell cycle stages from G1 to the late mitosis stage and has been previously studied in the cardiac context. Several reports have demonstrated the utility of cardiac specific FUCCI transgenic organisms to assess cell cycle activities in the developmental process and pathological response (Alvarez et al., 2019; Hashimoto et al., 2014; Uribe et al., 2018). Additionally, FUCCI transgenic zebrafish models, in combination with in vivo high-throughput screening (HTS) systems, have revealed cardiac proliferative factors, such as vitamin D, Hedgehog, insulin-like growth factor, and transforming growth factor β (Choi et al., 2013; Han et al., 2019). However, its application to hPSC cardiomyocytes is not well established.
In this study, we generated a FUCCI-expressing hiPSC line and monitored the cell cycle activities during cardiac differentiation. Using hiPSC cardiomyocytes, we demonstrated differences in gene expression patterns in vitro between cell cycle stages. We also showed that the cell cycle activity before cell transplantation affected proliferation in vivo. Our use of the HTS system with the FUCCI cell line allowed us to identify Am80 as a retinoic acid (RA)-associated proliferative compound for hiPSC cardiomyocytes. We additionally demonstrated that pre-treatment with Am80 enhanced cell engraftment after transplantation into damaged hearts. We further assessed the possible mechanism of RA signaling in the proliferation of hiPSC cardiomyocytes.
Results
Cellular characteristics of the FUCCI-expressing iPSC line
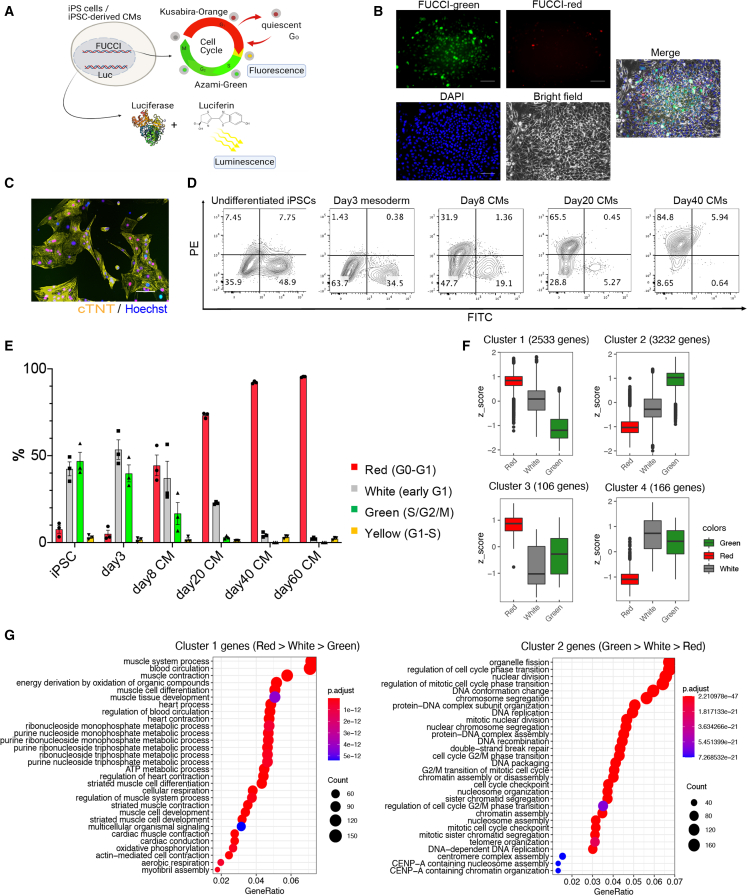
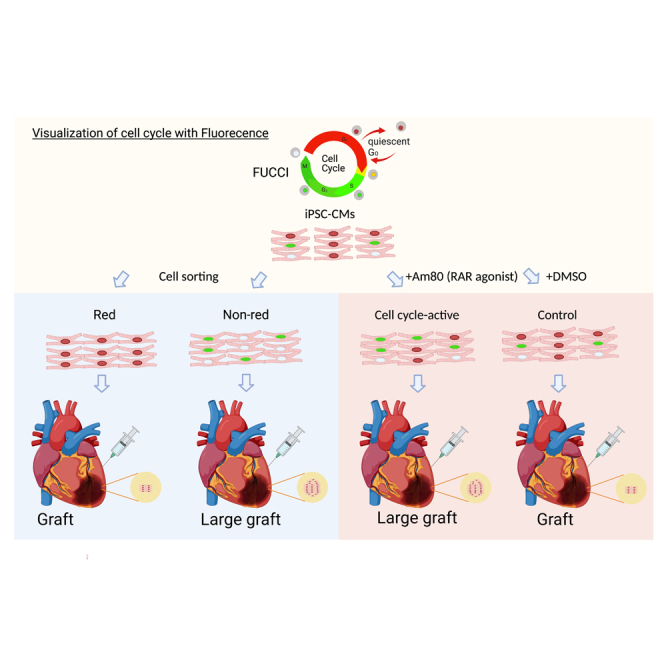
To monitor the cell cycle stages in hPSC cardiomyocytes in vitro and in vivo, we introduced FUCCI probes into an hiPSC line (201B7) continuously expressing luciferase (CAG promoter-driven), in which we can easily monitor engraftment when injected into the heart by in vivo luminescence imaging, as previously described (Figure 1A) (Funakoshi et al., 2016; Miki et al., 2015). Using this FUCCI cell line, we monitored cell cycle activities during cardiac differentiation over a long culture (>40 days). In the undifferentiated iPSC stage, approximately 50% of all cells showed a green fluorescence (S/G2/M) and a smaller percentage showed a FUCCI red fluorescence (G0/G1) (Figure 1B). However, the percentage of green cells gradually decreased during cardiac differentiation, reaching 3%, while the red cells exceeded 70% at day 20 of the differentiation (Figures 1C–1E, S1A, and S1B). White cells (no fluorescence; early G1) were approximately 20% at this time. After the long culture, most cells emitted red fluorescence, suggesting they had become non-proliferative (Figures 1D and 1E).
Figure 1.
Monitoring the cell cycle in iPSC CMs using the FUCCI system
(A) Schema of the luciferase- and FUCCI-expressing iPSC-derived cardiomyocytes (CMs).
(B) Representative fluorescence microscope images of undifferentiated iPSCs (n = 3 independent experiments). Scale bars, 100 μm.
(C) Representative fluorescence microscope image of day-20 iPSC CMs (n = 3 independent experiments). Scale bar, 100 μm.
(D) Representative flow cytometry analysis of FUCCI-expressing iPSC CMs at the indicated times of differentiation (n = 3 independent experiments each).
(E) Percentages of the different FUCCI color intensities (n = 3 independent experiments each). All error bars represent SE.
(F) A cluster analysis showing four clusters of differentially expressed genes among the red, white, and green populations in FUCCI-expressing iPSC CMs. Adjustment p value of <0.01 by likelihood ratio test. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × interquartile range; points, outliers. (n = 3 independent experiments.)
(G) A GO analysis of genes in cluster 1 (red > white > green) and cluster 2 (green > white > red).
Because we previously reported that day-20 hiPSC cardiomyocytes have the greatest engraftment capacity in vivo when injected into infarcted mouse heart (Funakoshi et al., 2016), we focused our efforts on investigating the cellular heterogeneity in cell cycle activity in day-20 differentiated hiPSC cardiomyocytes. We confirmed by live imaging that the FUCCI fluorescence of some of these cells transitioned from red to green, but most remained red, indicating the cells are a mixture of cells in the G0/G1 stage and of cells with an active cell cycle (Video S1). We then sorted the red, white, and green sub-populations (SIRPA positive/lineage negative) and compared the gene expression patterns by RNA sequencing (RNA-seq) (Figure S1C). The differentially expressed genes among the three sub-populations were clustered into 4 groups (Figure 1F). A Gene Ontology (GO) enrichment analysis showed the genes of cluster 2, which consists of cell cycle- and mitosis-related genes, were expressed highest in green cells followed by white cells, indicating that the FUCCI system can monitor accurately the cell cycle stages of hiPSC cardiomyocytes (Figures 1F and 1G). In contrast, genes in cluster 1, which consist of muscle- and contraction-related genes, were expressed most by red cells, suggesting that non-proliferative cardiomyocytes undergo the cellular maturation process earlier than proliferative cardiomyocytes (Figures 1F and 1G). Together, we show that the FUCCI system can detect the cell cycle status and that there exists heterogeneity in the cell maturity and proliferation activity of day-20 hiPSC cardiomyocytes in vitro.
Day-20 cardiomyocytes purified by sorting SIRPA-positive / Lineage-negative cells by flow cytometry and replated onto a fibronectin-coated dish. Video was captured by a live cell imaging analyzer (BioStation CT).
hiPSC cardiomyocytes with activated cell cycle show better engraftment in vivo
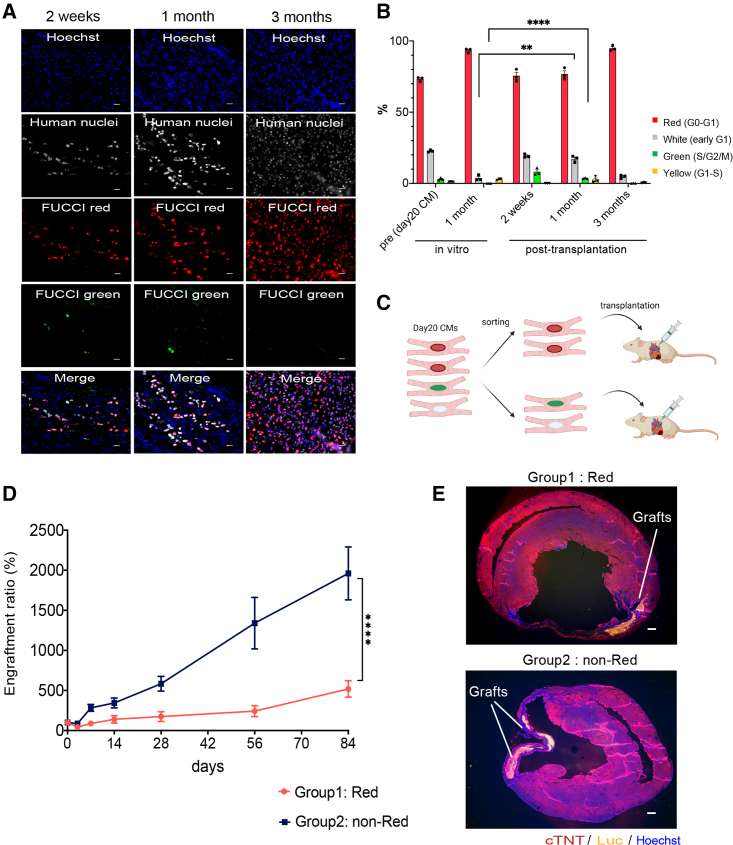
To confirm if our FUCCI system can monitor the cell cycle stages in hiPSC cardiomyocytes in vivo, we injected day-20 hiPSC cardiomyocytes into non-obese diabetic/Shi-SCID IL2Rγnull (NOG) mouse heart with myocardial infarction and monitored the cell cycle stages of the engrafted cells. At 3 months after transplantation, we observed large grafts consisting of human cardiac troponin T (cTNT)-positive cardiomyocytes expressing red fluorescence (Figure S2A). At 2 weeks after transplantation, approximately 10% of the engrafted cells showed green fluorescence and 20% no fluorescence (Figure S2B). The percentage of green cells gradually decreased and was rarely observed at 3 months (Figures 2A and 2B), suggesting that the engrafted cells proliferated during the first few months and became non-proliferative at 3 months. Additionally, when we compared the cell cycle stages at 1 month after transplantation with time-matched day-50 hiPSC cardiomyocytes (1 month after day 20) in vitro, the percentage of day-20 cells with green or no fluorescence signal was significantly higher in vivo than in in vitro day-50 cardiomyocytes (Figure 2B), indicating that the cell cycle of hiPSC cardiomyocytes was maintained actively in vivo during the first month after the cell transplantation.
Figure 2.
Differences in engraftment potential by the cell cycle status of iPSC CMs
(A) FUCCI expression patterns in grafted cells at 2 weeks, 1 month, and 3 months after cardiomyocyte (CM) transplantation (n = 3 independent experiments each). Scale bars, 20 μm.
(B) Percentages of the different FUCCI color intensities (n = 3 independent experiments each). All error bars represent SE. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by two-sided unpaired t test.
(C) Schema of the transplantation of red and non-red (green or white) cells.
(D) Luminescence intensities after transplantation of red or non-red cells (n = 12 independent experiments each). All error bars represent SE. ∗∗∗∗p < 0.0001 by two-way ANOVA test.
(E) Representative fluorescence microscope images of the mouse heart with engrafted group 1 (red) or 2 (non-red) cells (n = 4 independent experiments each). Scale bars, 300 μm.
Because day-20 hiPSC cardiomyocytes have heterogeneous cell cycle stages, we next tested if the engraftment potential is different according to the cell cycle stage before the transplantation. We sorted red from other day-20 hiPSC cardiomyocytes (non-red) and injected 2.5 × 105 cells of one group into infarcted NOG mouse heart (Figure 2C). The luminescence signal intensity was significantly higher in the non-red group than the red group over time, which correlated with the graft size at 3 months (Figures 2D, 2E, and S2C), indicating that other hiPSC cardiomyocytes (S/G2/M/early G1, non-red) before the transplantation engraft and proliferate better than red hiPSC cardiomyocytes (G0/G1). Immunohistology after the transplantation of other hiPSC cardiomyocytes revealed that cTNT-positive cardiomyocytes engrafted and almost all expressed red fluorescence at 3 months, suggesting their cell cycle is G0/G1 at 3 months (Figures S2D and S2E). The sarcomere structure and the expression and distribution of the gap junction protein CX43 in the engrafted cardiomyocytes were comparable between the two groups (Figures S2F and S2G), suggesting that the cell cycle activity before the cell transplantation did not affect the graft quality at 3 months.
Taken together, these results demonstrate that the cell cycle stage in vitro before transplantation impacts proliferation in vivo, with an activated cell cycle stage (S/G2/M/early G1) giving better engraftment potential.
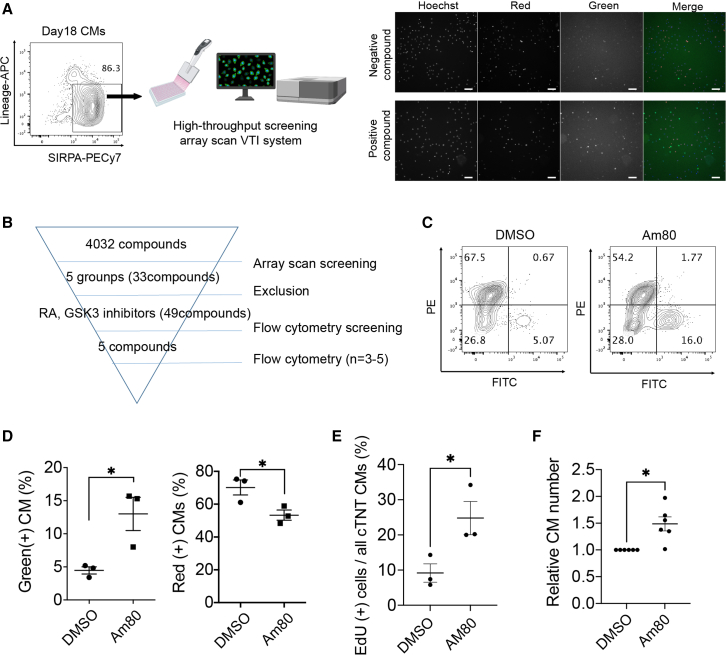
Identification of cardiac proliferation drugs by HTS
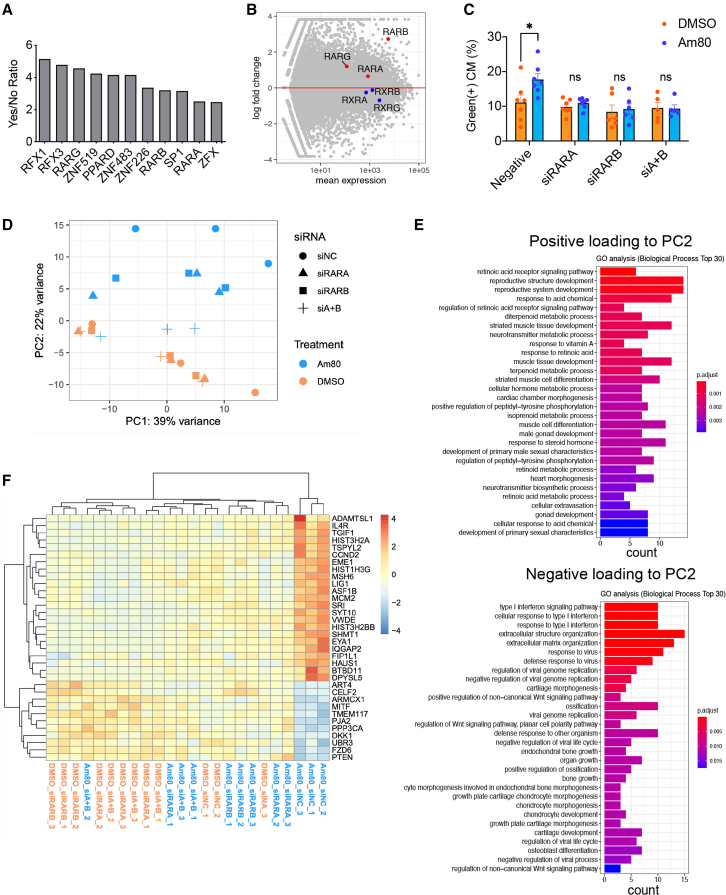
Given that the cell cycle activity affects the engraftment after transplantation, we hypothesized that activating the cell cycle in vitro could enhance the engraftment potential after injection into the heart. We, therefore, tried to discover drugs that proliferate cardiomyocytes. We established HTS using an array scan system (Figure 3A). We searched a drug library consisting of >4,000 compounds and discovered that 33 candidate compounds, including RA-related drugs, GSK3 inhibitors, PI3K inhibitors, isocitrate dehydrogenase inhibitors, and nucleic acid synthesis inhibitors, increased the ratio of green cells after treatment for 2 days (Figure 3B). We excluded anti-cancer drugs, such as nucleic acid synthesis inhibitors, among the compounds and identified RA-related drugs and GSK3 inhibitors as potentially proliferative drugs. We added several RA-related compounds and GSK3 inhibitors that were not contained in the first library and then performed a second screening by flow cytometry among 49 compounds (Figure 3B). Finally, we identified that an RA receptor (RAR) agonist, Am80, resulted in the strongest green fluorescence intensity (Figures S3A–S3C). Treatment with Am80 for 2 days caused a significant increase in the percentage of green cells and a significant decrease in red cells (Figures 3C and 3D). To confirm the proliferative effect, we assessed the incorporation of 5-ethynyl-2′-deoxyuridine (EdU), a marker of DNA synthesis, and observed a significantly higher percentage of EdU-positive hiPSC cardiomyocytes in the Am80-treated cell population than in the non-treated control population and a significant increase in the number of cardiomyocytes within the Am80-treated cells (Figures 3E and 3F). We confirmed a similar proliferative effect in cardiomyocytes derived from another iPSC line (409B2) by the EdU assay (Figures S3D and S3E), suggesting that Am80 is broadly useful as a proliferative drug for hiPSC cardiomyocytes.
Figure 3.
Identification of Am80 by HTS
(A) Left, Schema of the HTS system using FUCCI-expressing day-18 iPSC cardiomyocytes. Right, Example array scan images of negative and positive-hit compounds. Scale bars, 100 μm.
(B) Strategy to identify proliferation compounds. FITC, fluorescein isothiocyanate.
(C) Representative flow cytometry analysis of DMSO-treated (control) and Am80-treated cardiomyocytes (n = 3 independent experiments).
(D) Percentage of green and red cells (n = 3 independent experiments).
(E) Percentage of EdU-positive cells (n = 3 independent experiments).
(F) Number of cardiomyocytes (n = 6 independent experiments). The number was measured 6 days after pre-treatment with Am80 for 2 days. ∗p < 0.05, ∗∗∗p < 0.001 by two-sided unpaired t test (D, E) and by unpaired t test with Welch’s correction (F). All error bars represent SE.
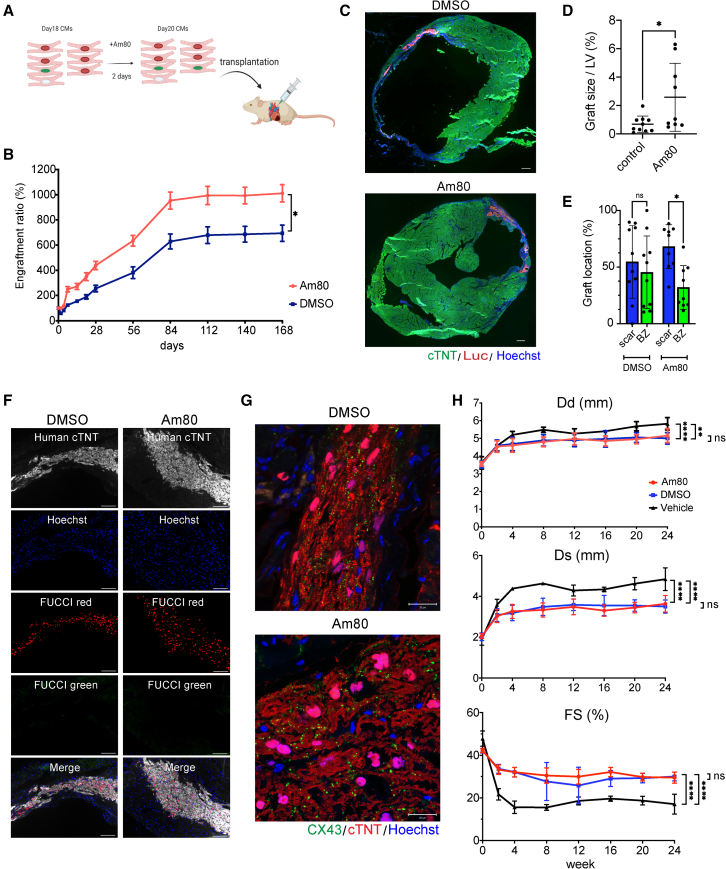
Pre-treatment of Am80 enhances graft size after cell transplantation
To test our hypothesis that pre-treatment with a proliferative drug improves the engraftment potential of hiPSC cardiomyocytes, we injected Am80- or DMSO-treated purified hiPSC cardiomyocytes (1 × 106 cells/mouse) into infarcted NOG mouse heart (Figure 4A) and compared the engraftment by in vivo luminescence imaging. The Am80-treated cardiomyocytes showed significantly greater luminescence during a 6-month follow-up than the control cells (Figure 4B). Especially during the first few weeks, the luminescence signal by the Am80-treated cardiomyocytes increased rapidly, indicating that Am80-treatment strongly induced proliferation just after the cell injection. Consistent with the difference in luminescence, we observed larger grafts in the Am80-treated group than in the control group (Figures 4C and 4D). Regarding graft location, the Am80-treated group is more likely to engraft in the scar lesion compared with the DMSO-treated control group, suggesting that Am80 treatment positively impacts engraftment in the ischemic scar lesion (Figure 4E). Almost all grafted cardiomyocytes in both groups showed FUCCI red signals at 6 months, indicating they were in a non-proliferative state (Figure 4F). With respect to the graft quality, cardiomyocytes from both groups showed well-organized sarcomere structures at 6 months (Figure 4G). Additionally, both groups showed comparable levels and distributions of CX43 expression in the grafts: CX43 in both groups remained distributed throughout the graft, but was not aligned well at the cell-cell junction, indicating that the grafted cells were not fully matured (Figure 4G). Collectively, pre-treatment with Am80 enhances the graft size, but retains a graft quality comparable with untreated cells. Finally, we measured the cardiac function after transplantation by echocardiography. The cell transplantation using both DMSO- and Am80-treated cardiomyocytes showed significantly improved cardiac function including shorter left ventricular diastolic (Dd)/systolic dimension (Ds) and higher fractional shortening (FS) compared with untreated vehicle control, while all the parameters were comparable between DMSO treatment and Am80 treatment 6 months after the cell transplantation (Figure 4H).
Figure 4.
Pre-treatment with Am80 enhances the engraftment of iPSC cardiomyocytes in infarcted mouse heart
(A) Schema of the transplantation of Am80-treated cardiomyocytes.
(B) Luminescence intensity after transplantation of DMSO-treated or Am80-treated cardiomyocytes (n = 6 independent experiments each). All error bars represent SE. ∗p < 0.05, two-way ANOVA test.
(C) Representative fluorescence microscope images of the mouse heart with engrafted DMSO-treated or Am80-treated cardiomyocytes (n = 4 independent experiments each). Scale bars, 300 μm.
(D) Quantification of graft size in the transplantation of DMSO-treated (n = 10 independent experiments) and Am80-treated cardiomyocytes (n = 9 independent experiments). Values are mean ± SD. ∗p < 0.05 by two-sided unpaired t test. LV, left ventricle.
(E) Comparison of graft location in the infarcted mouse heart between the transplantation of DMSO-treated (n = 10 independent experiments) and Am80-treated cardiomyocytes (n = 9 independent experiments). Values are mean ± SD. ∗p < 0.05 by one-way ANOVA test followed by Tukey’s multiple comparison test. BZ, border zone area; ns, not significant; scar, scar area.
(F) Representative FUCCI fluorescence patterns of engrafted DMSO-treated cardiomyocytes or Am80-treated cardiomyocytes 6 months after the transplantation (n = 4 independent experiments each). Scale bars, 100 μm.
(G) Representative CX43 and cTNT expression patterns of engrafted DMSO-treated or Am80-treated cardiomyocytes (n = 4 independent experiments each). Scale bars, 20 μm.
(H) Echocardiographic parameters (Dd, Ds, and FS) during 6-month follow-up (n = 6 independent experiments each). Values are mean ± SD. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by two-way ANOVA test followed by Tukey’s multiple comparison test. Dd: Am80 vs. vehicle, p < 0.01. DMSO vs. vehicle, p < 0.0001. Am80 vs. DMSO, not significant (ns). Ds: Am80 vs. vehicle, p < 0.0001. DMSO vs. vehicle, p < 0.0001. Am80 vs. DMSO, not significant, FS: Am80 vs. vehicle, p < 0.0001. DMSO vs. vehicle, p < 0.0001. Am80 vs. DMSO, not significant.
Am80 enhances cardiomyocyte proliferation through RARs α and β
To elucidate the mechanism underlying the proliferative effect of Am80 treatment, we performed an RNA-seq analysis of Am80-treated day-20 hiPSC cardiomyocytes and compared their gene expression patterns with DMSO-treated day-20 hiPSC cardiomyocytes (Figure S4A). A GO enrichment analysis using differentially expressed genes between these two groups revealed that cell cycle- and mitosis-related genes were enriched in the Am80 group. In contrast, muscle-related genes were enriched in the control cells, confirming that Am80 induces the broad upregulation of cell cycle-related genes (Figures S4B and S4C). Because RA signaling regulates the atrial fate in the developing heart, we confirmed that Am80 treatment did not upregulate several atrial genes, including NR2F2, KCNA5, and KCNJ3. In contrast, several ventricular genes, such as IRX4, MYL2, and GJA1, were highly expressed, consistent with RA signaling for the atrial cell fate being required only in the early phase of cardiac differentiation and that Am80 treatment on day 18 does not affect cell fate specification (Figure S4D). It should be noted that the expression level of some of the ventricular genes gradually decreased, probably because of the proliferative effect of Am80.
To identify the regulatory genes in Am80-treated hiPSC cardiomyocytes, we performed a motif enrichment analysis. We found that several transcription binding motifs were enriched in regions upstream of the highly expressed genes, including RARs alpha, beta, and gamma (RARA, RARB, and RARG) (Figure 5A). The expression level of all three RARs was significantly higher in Am80-treated cells than in control cells. In contrast, the expression level of other RA-related receptors, such as retinoid X receptors (RXRs), was comparable between the two groups (Figure 5B), indicating that RARs play an important role in the proliferative effect of Am80 on cardiomyocytes. We thus performed small interfering RNA (siRNA)-mediated knockdown (KD) of the RARs. We first tested several different siRNAs specific for each RAR and determined the most effective combinations, finding a mixture of three different siRNAs (α1, α2, α3) for RARA (siRARA) and two different siRNAs (β3, β5) for RARB (siRARB), but no efficient siRNAs for RARG, which probably accounts for the lower expression of RARG in cardiomyocytes (Figures 5B and S4E). The inhibition of RARA by siRARA, RARB by siRARB, or their combination (siRARA and siRARB [siA + B]) eliminated the proliferative effect of Am80 (Figure 5C), validating that Am80 shows its proliferative effect through these two receptors. These observations further suggested that Am80 functions on RARA and RARB simultaneously and that there exist common downstream pathways that are essential for the proliferative effect of Am80. To clarify the molecular mechanism in more detail, we performed an RNA-seq analysis of all siRNA-treated cells. Principal-component analysis showed that Am80-treated samples with siA + B were close to DMSO-treated samples in principle component 2 (PC2), but far from Am80-treated samples transfected with a non-targeting siRNA (siNC) (Figure 5D). We thus searched for genes responsible for PC2 by performing a GO analysis. We identified the RAR signaling pathway as enriched in positive loading genes to PC2. In contrast, the interferon (IFN) signaling pathway as well as positive regulation of the non-canonical Wnt signaling pathway were enriched in negative loading genes to PC2, indicating that the KD of both RARA and RARB attenuated the effect of Am80 on the three pathways (Figure 5E, Table S1). We next extracted 485 differentially expressed genes between the DMSO- and Am80-treated samples transfected with siNC. Among those 485 genes, we focused on genes expressed differently between Am80-treated samples transfected with siNC and those transfected with siRARA and/or siRARB, thus identifying 34 genes (Figure S4F). These 34 genes included CCND2, other cell cycle-related genes (MSH6, MCM2), and Wnt/β catenin-related genes (BTBD11, EYA1, TGIF1) (Freyer and Morrow, 2010; Karner et al., 2011; Wang et al., 2017), whose expression levels were upregulated by Am80, as well as several inhibitory genes in the canonical Wnt pathway (DKK1, FZD6, PJA2) (Dawson et al., 2013; Song et al., 2018) and IFN-related genes (CELF2, MITF), whose expression levels were downregulated by Am80 (Figure 5F). All these results suggested that the proliferative effect of Am80 was at least partly caused by activation of the endogenous canonical Wnt pathway through RARA and RARB.
Figure 5.
Molecular mechanism underlying the Am80-induced proliferation of iPSC CMs
(A) Enrichment of upstream regulators among genes highly expressed in Am80-treated cardiomyocytes (CMs) by a motif enrichment analysis. Regulators in which the absolute expression level is >100 normalized counts and the expression level is higher after Am80 treatment than DMSO treatment are included in the graph.
(B) Expression patterns of RARs and RXRs in DMSO-treated and Am80-treated CMs.
(C) Percentage of green cells under the indicated siRNA-mediated KD conditions (N = 5–7 independent experiments).
(D) Principal-component analysis of DMSO- and Am80-treated cardiomyocytes with siNC, siRARA, siRARB, or siA + B (n = 3 independent experiments each).
(E) GO analysis of genes highly responsible for PC2. Results are shown for genes positively and negatively loading to PC2. The top 30 GO terms by adjusted p value are shown.
(F) Heatmap showing the expression level of 34 genes regulated by Am80 through both RARA and RARB. All error bars represent SE. ∗p < 0.05 by two-sided unpaired t test.
Discussion
In this study, using FUCCI-expressing hiPSC cardiomyocytes, we demonstrated heterogeneity in the cell cycle stages in vitro and in vivo. Additionally, we identified a drug, Am80, that promotes the proliferation of cardiomyocytes and demonstrated that pre-treatment with Am80 enhanced the engraftment of the cells in vivo. Furthermore, an RNA-seq analysis of Am80-treated cells indicated that Wnt signaling induces the cardiomyocyte proliferation. Taken together, this study highlights the power of using the FUCCI system to assess the proliferation activity of hPSC cardiomyocytes both in vitro and in vivo.
RA signaling is known to play several essential roles in heart development (Nakajima, 2019; Perl and Waxman, 2019; Xavier-Neto et al., 2015). In the early developing heart, equivalent to embryonic day 6.5–7.5 (E6.5–7.5) in mice, ventricular progenitors, especially left ventricular progenitors, are specified before atrial progenitors (Lescroart et al., 2014). RA signaling is essential for atrial progenitors to specify the atrial fate (Protze et al., 2019). By applying this developmental finding to the cardiac differentiation of hPSCs, it has been reported that atrial and ventricular progenitor cells are derived from different mesoderm populations, and only RALDH2-positive populations (RALDH2 is an RA synthesizing enzyme) can differentiate into atrial cells in vitro (Lee et al., 2017). During the ventricular expansion stage in the embryo, equivalent to E10.5–14.5 in mice, RA synthesized by the RALDH2-positive epicardium induces the endogenous secretion of several proliferative factors, including Wnt, fibroblast growth factors, and insulin-like growth factors, from the epicardium, which induce the proliferation of the outer layer of the ventricle, that is, the compact myocardium (Brade et al., 2011; Lavine et al., 2005; Merki et al., 2005; Zamora et al., 2007). While those indirect effects of RA signaling are well known, the direct effects of RA on cardiomyocytes have not been well documented in this stage of the developing heart. After this stage, the epicardial cells become dormant and lose their RALDH2 expression, resulting in the loss of the source of RA signaling in the heart (Masters and Riley, 2014). However, interestingly, the reactivation of RA signaling after heart injury in the postnatal stage is induced by the re-expression of epicardial RALDH2, which affects cardiac remodeling by modulating the activities of cardiac fibroblasts and immune cells (Kikuchi et al., 2011; Lepilina et al., 2006; Masters and Riley, 2014). Furthermore, it has been reported that the activation of RAR/RXR attenuates left ventricular hypertrophy, fibrosis, apoptosis, and inflammatory responses in the adult heart (Guleria et al., 2013; Pan et al., 2014). It is known that hiPSC cardiomyocytes have a similar molecular signature to in vivo cardiomyocytes in the late-developing heart (Uosaki et al., 2015). Our Am80-treated day-20 cardiomyocytes maintained the ventricular fate and did not acquire the atrial fate, consistent with them having passed the specification stage in the early developing heart. Regarding the endogenous RAR-related pathways, we demonstrated that siRAR treatment inhibited the effect of Am80 treatment. However, it did not decrease the percentage of FUCCI green cells in DMSO-treated cardiomyocytes. These findings suggest that the RAR pathway regulates the cell cycle differently from the pathway that regulates it under baseline conditions in iPSC cardiomyocytes at this stage. Further investigation is necessary to elucidate the mechanisms underlying endogenous cell cycle regulation in iPSC cardiomyocytes. We thus concluded that the phenomena observed in this study reflect how late fetal and perinatal cardiomyocytes react to the exposure of exogenous RA. Consistently, another report using a cardiac-specific beta-major histocompatibility complex (MHC) (fetal MHC)-driven RAR/RXR-overexpression mouse model showed that continuous RA-mediated signaling inhibits the maturation process and maintains the fetal ventricular phenotype (Colbert et al., 1997).
RA signaling has essential roles in regenerative responses of the heart. In zebrafish, whose heart has regenerative capacity, the reactivation of RA signaling through the activated epicardium and endocardium is critical for the regenerative response, and the downregulation of RARA or overexpression of an RA degrading enzyme, CYP26A1, blocks heart regeneration (Kikuchi et al., 2011). However, because the inhibition of RA signaling in that study was systemic, how RA signaling affects cardiomyocyte regeneration directly remains unclear (Cao and Poss, 2018). As for heart regeneration in neonatal mice, the activation of RALDH2 in the epicardium is also induced by injury (Bilbija et al., 2012; van Wijk et al., 2012), suggesting a potential role of RA signaling for mammalian heart regeneration. Because the reactivation of RALDH and other fetal genes in the epicardium has been reported even in adult heart injury (Huang et al., 2012), our finding that Am80 treatment upregulated the expression of RARs and induced the activation of cell cycle genes may be one mechanism underlying the RA-related cardiac repair process in postnatal heart injury.
More specifically, we found that Am80 downregulated the inhibitory genes of canonical Wnt signaling and upregulated Wnt-related genes together with cell cycle genes through RARA and RARB, indicating the relatively activated status of the endogenous canonical Wnt signaling pathway in Am80-treated hiPSC cardiomyocytes. Wnt/β-catenin signaling is activated in response to cardiac injury and is related to the repair process including fibrosis, hypertrophic changes, and regenerative responses (Daskalopoulos and Blankesteijn, 2021; Ozhan and Weidinger, 2015). Although the activation of this pathway includes endothelial cells, fibroblasts, smooth muscle cells, inflammatory cells, and the epicardium, the upregulation of the Wnt pathway in cardiomyocytes has not been established. Our results showing the activation of the endogenous canonical Wnt pathway through RARs suggest that RA secreted from the injured epicardium directly activates an endogenous Wnt pathway in cardiomyocytes during cardiac repair. Furthermore, Wnt signaling promotes the proliferation of hPSC cardiomyocytes in vitro (Buikema et al., 2020). Indeed, our HTS indicated that GSK3 inhibitors, which can enhance Wnt signaling, are also candidate drugs for hiPSC cardiomyocyte proliferation. However, Am80 activated the cell cycle more than any GSK3 inhibitors did, suggesting that there could be other direct or indirect mechanisms to induce cell cycle activation in addition to activation of the endogenous canonical Wnt pathway. We further identified that several genes, including MITF, CELF2, and PTEN, which have been known to interact with the immune system (Ballotti et al., 2020; Diaz-Munoz and Turner, 2018), were downregulated by Am80 through RARs, suggesting that the inflammation levels might be associated with better engraftment. Further investigations are required to confirm this point.
We demonstrated the relationship between cell cycle activity in hiPSC cardiomyocytes and their engraftment potential. Cardiomyocytes in S/G2/M/early G1 phase (FUCCI non-red) had a higher proliferative potential after cell transplantation. However, in a previous study, we demonstrated that day 20-cardiomyocytes derived from hiPSCs had a higher engraftment capacity than day 8-cardiomyocytes (Funakoshi et al., 2016), despite the higher percentage of FUCCI non-red cells in the latter. This suggests that cell cycle activity is not the only factor determining the engraftment capacity in iPSC cardiomyocytes. Other factors, including the expression of cell adhesion molecules and cellular maturation level, could also affect their engraftment potential. Therefore, a specific subpopulation with S/G2/M/early G1 phase in day 20-iPSC cardiomyocytes could be an ideal population for cell engraftment. However, this subpopulation only represents 20%–30% of the total population, resulting in a limited cell numbers. Currently, there are no methods to enrich this subpopulation without using the FUCCI cell line, which could be a significant hurdle for the clinical application of cardiac cell therapy. Thus, we selected a strategy to increase the proportion of FUCCI-green cells in day-20 cardiomyocytes. Further study will be necessary to enrich this subpopulation efficiently. Am80 treatment of this subpopulation might be another effective strategy to increase the graft size if we can generate or efficiently select only this subpopulation in a large-scale culture system without cell sorting.
To conclude, we demonstrated that the pre-treatment of Am80 enhances the engraftment of hiPSC cardiomyocytes in the infarcted heart. In vivo luminescence imaging showed that differences in the signal intensity between Am80-treated and control cells widened mainly during the first few weeks after the transplantation and were maintained for the following 6 months, suggesting that pre-treatment with Am80 enhanced the proliferation of the injected cardiomyocytes during the first few weeks. The luminescence intensity reached a plateau at 3 months, and the Am80-treated grafted cells showed red FUCCI signals at 6 months after transplantation, consistent with the effect of the pre-treatment being limited to the early period and not inducing abnormal growth of the grafts. In this study, we observed the improved cardiac function after cell transplantation, consistent with our previous report (Funakoshi et al., 2016). However, we could not observe any differences in echocardiographic parameters between the transplantation of DMSO-treated cardiomyocytes and that of Am80-treated cardiomyocytes, while the latter demonstrated a larger graft size. In small animal models, the improvement of cardiac function after cell transplantation is mainly caused by the paracrine effect of grafted cells, as hiPSC cardiomyocytes are not able to efficiently couple with the host myocardium because of the rapid heart rate of the host heart (>300 bpm). We assume that the discrepancy between larger graft size and comparable improvement in cardiac function accounts for this issue in the small animal models. In contrast, it has been reported that grafted cardiomyocytes couple with the host heart and the improvement of cardiac function after cell transplantation is correlated with graft size in the large animal model (Shiba et al., 2016). Further investigation using large animal models may enable an evaluation of the impact of larger graft size in Am80-treated hiPSC cardiomyocytes on cardiac function. Additionally, gene expression analysis in Am80-treated cardiomyocytes showed a relatively immature phenotype compared with untreated cardiomyocytes, particularly regarding the expression level of the sarcomere and muscle contraction-related genes. This suggests that Am80-treated cardiomyocytes have some differences in functionality before cell transplantation. However, 3 months after cell transplantation, they exhibited comparable sarcomere structures, CX43 expression, and FUCCI expression patterns, as well as similar improvements in cardiac function measured by echocardiography. This indicates that the relatively immature phenotype of Am80-treated cardiomyocytes before cell transplantation does not negatively impact the efficacy of cell transplantation. Finally, several recent studies reported that graft-related ventricular arrhythmia occurs during the first month after transplantation in large animal models with myocardial infarction (Chong et al., 2014; Romagnuolo et al., 2019; Shiba et al., 2016). How the accelerated proliferation of engrafted cells during this period affects graft-related arrhythmia should be also further investigated in large animal models to elucidate whether pre-treatment with proliferative drugs is feasible in terms of safety.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed and will be fulfilled by the corresponding authors, Yoshinori Yoshida (yoshinor@cira.kyoto-u.ac.jp) and Shunsuke Funakoshi (s.funakoshi@cira.kyoto-u.ac.jp).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
HTS assay by array scan
For the HTS assay, day-20 hiPSC cardiomyocytes were purified by sorting the SIRPA-positive and lineage-negative populations using the Aria Ⅱ and replated onto 384 fibronectin-coated well plates (2,500 cells/well). Two days after the replating, 4,032 compounds (Approved Drug Libraries [ENZO FDA, ENZO ICCB, MICROSOURCE International Drugs, MICROSOURCE US Drugs] and Bioactive Compound Libraries [Selleck Kinase Inhibitor Library, EMD Kinase Inhibitor Library, ENZO Kinase Inhibitor Library, SIGMA LOPAC, and compounds from Tocris Bioscience and Selleck Chemicals]) were administered to the cells. Three days after the treatment, images were captured using an ArrayScan High-Content System (ThermoFisher Scientific). We defined the wells in which the percentage of cells with a green signal higher than the mean signal plus two standard deviations (+2 SD) of all wells as positive wells. Thus, 33 compounds were picked up as possible proliferation compounds in the first screening.
Cell transplantation into mouse myocardial infarction models, in vivo bioluminescence imaging, and echocardiography
All animal experiments were performed in accordance with the Guidelines for Animal Experiments of Kyoto University and the Guide for the Care and Use of Laboratory Animals by the Institute of Animal Resources. Male non-obese diabetic/severe combined immunodeficiency NOG mice (8–12 weeks old) were mechanically ventilated under general anesthesia with inhalation of 2% isoflurane under intubation using a 20G angiocatheter (Terumo Corporation). Mouse hearts were exposed by left anterolateral thoracotomy. Myocardial infarction was generated by the ligation of the left anterior descending artery with an 8-0 Prolene suture (Ethicon, Inc., Johnson & Johnson). hiPSC cardiomyocytes were injected to the ischemic lesion at a single site with a total volume of 20 μL IMDM (Life Technologies). Transplantation procedures were performed using a Hamilton syringe with a 30G needle. To estimate the graft size, bioluminescence imaging was performed using an in vivo bioluminescence imaging system (IVIS, Caplier Life Sciences). D-luciferin (SPI) was administered at a dose of 200 mg/kg intraperitoneally. The engraftment ratio was defined by the signal at each time point divided by the signal immediately after injection on day 0. Both cultured cells and mice were allocated into experimental groups in random for the transplantation experiments. The measurement of the engraftment ratio by IVIS was not performed in a blind manner; the outcome was not dependent on the judgment of the investigators and could not be influenced by prior knowledge of the groups. Cardiac function, including left Dd/Ds and FS, was measured using transthoracic echocardiography (GE, Vivid S, GE) under mild anesthesia with inhaled isoflurane (Abbott Japan). FS was calculated as 100 × (Dd – Ds)/Dd (%). Graft size was measured by calculating the ratio of grafted area divided by the entire left ventricular area by longitudinal analysis from apex to base in the mouse heart.
Quantification and statistical analysis
Statistical significance was determined using the Student’s unpaired t test, unpaired t test with Welch’s correction, or two-way ANOVA test by GraphPad Prism 6 software (GraphPad Software). All statistical parameters are reported in the respective figures and figure legends.
Author contributions
M.K. and S.F. conceived the project, performed experiments, and analyzed data. T.H. and Y.T. performed the animal experiments. C.O. and M.N. performed RNA-seq and analyzed the data. Y.N., M.N., and A.O. performed the HTS experiments. S.F., T.K., and Y.Y. designed the project. M.K., S.F., and Y.Y. wrote the manuscript.
Acknowledgments
We thank Mikako Marx-Mori and Rumi Fujihara for their administrative support and Peter Karagiannis for proofreading the manuscript. Schemas were generated using BioRender. This work was supported by JSPS KAKENHI 17H04176 and 21H02912, by the Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development (AMED) (JP19bm0104001, JP19bm0204003, JP19bm0804008, and JP20bm0804022), the Research on Regulatory Science of Pharmaceuticals and Medical Devices, AMED (JP19mk0104117, JP22mk0101189, 22mk0101241), the Research Project for Practical Applications of Regenerative Medicine, AMED (JP19bk0104095), the Translational Research grant, AMED (JP22ym0126091), the Leducq Foundation (18CVD05), and the iPS Cell Research Fund.
Conflict of interests
Y.Y. is a scientific advisor of Orizuru Therapeutics and received research funding from Takeda Pharmaceutical Co Ltd and Altos Labs, Inc, outside the submitted work. S.F. is a scientific advisor of Orizuru Therapeutics. Kyoto University has filed a patent application (WO2019/156216) relevant to this work. M.K., S.F., T.H., and Y.Y. are the investigators of record listed on the patent application.
Published: July 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.06.006.
Contributor Information
Shunsuke Funakoshi, Email: s.funakoshi@cira.kyoto-u.ac.jp.
Yoshinori Yoshida, Email: yoshinor@cira.kyoto-u.ac.jp.
Supplemental information
Top 100 genes with the highest and lowest loadings that express the direction of the PC2. Values are loading coefficients.
Data and code availability
All data presented in this study are available in the main text, in Supplemental items file, and from the corresponding author upon request. The RNA-seq data in this study has been deposited into the GEO database under the accession code: GSE176303.
References
- Alvarez R., Jr., Wang B.J., Quijada P.J., Avitabile D., Ho T., Shaitrit M., Chavarria M., Firouzi F., Ebeid D., Monsanto M.M., et al. Cardiomyocyte cell cycle dynamics and proliferation revealed through cardiac-specific transgenesis of fluorescent ubiquitinated cell cycle indicator (FUCCI) J. Mol. Cell. Cardiol. 2019;127:154–164. doi: 10.1016/j.yjmcc.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballotti R., Cheli Y., Bertolotto C. The complex relationship between MITF and the immune system: a Melanoma ImmunoTherapy (response) Factor? Mol. Cancer. 2020;19:170. doi: 10.1186/s12943-020-01290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargehr J., Ong L.P., Colzani M., Davaapil H., Hofsteen P., Bhandari S., Gambardella L., Le Novère N., Iyer D., Sampaziotis F., et al. Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nat. Biotechnol. 2019;37:895–906. doi: 10.1038/s41587-019-0197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbija D., Haugen F., Sagave J., Baysa A., Bastani N., Levy F.O., Sirsjö A., Blomhoff R., Valen G. Retinoic acid signalling is activated in the postischemic heart and may influence remodelling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T., Kumar S., Cunningham T.J., Chatzi C., Zhao X., Cavallero S., Li P., Sucov H.M., Ruiz-Lozano P., Duester G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development. 2011;138:139–148. doi: 10.1242/dev.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema J.W., Lee S., Goodyer W.R., Maas R.G., Chirikian O., Li G., Miao Y., Paige S.L., Lee D., Wu H., et al. Wnt Activation and Reduced Cell-Cell Contact Synergistically Induce Massive Expansion of Functional Human iPSC-Derived Cardiomyocytes. Cell Stem Cell. 2020;27:50–63.e5. doi: 10.1016/j.stem.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Poss K.D. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 2018;15:631–647. doi: 10.1038/s41569-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.Y., Gemberling M., Wang J., Holdway J.E., Shen M.C., Karlstrom R.O., Poss K.D. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140:660–666. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J.H., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J., et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert M.C., Hall D.G., Kimball T.R., Witt S.A., Lorenz J.N., Kirby M.L., Hewett T.E., Klevitsky R., Robbins J. Cardiac compartment-specific overexpression of a modified retinoic acid receptor produces dilated cardiomyopathy and congestive heart failure in transgenic mice. J. Clin. Invest. 1997;100:1958–1968. doi: 10.1172/JCI119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalopoulos E.P., Blankesteijn W.M. Effect of interventions in WNT signaling on healing of cardiac injury: a systematic review. Cells. 2021;10 doi: 10.3390/cells10020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson K., Aflaki M., Nattel S. Role of the Wnt-Frizzled system in cardiac pathophysiology: a rapidly developing, poorly understood area with enormous potential. J. Physiol. 2013;591:1409–1432. doi: 10.1113/jphysiol.2012.235382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Muñoz M.D., Turner M. Uncovering the Role of RNA-Binding Proteins in Gene Expression in the Immune System. Front. Immunol. 2018;9:1094. doi: 10.3389/fimmu.2018.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L., Morrow B.E. Canonical Wnt signaling modulates Tbx1, Eya1, and Six1 expression, restricting neurogenesis in the otic vesicle. Dev. Dyn. 2010;239:1708–1722. doi: 10.1002/dvdy.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi S., Miki K., Takaki T., Okubo C., Hatani T., Chonabayashi K., Nishikawa M., Takei I., Oishi A., Narita M., et al. Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci. Rep. 2016;6 doi: 10.1038/srep19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria R.S., Singh A.B., Nizamutdinova I.T., Souslova T., Mohammad A.A., Kendall J.A., Jr., Baker K.M., Pan J. Activation of retinoid receptor-mediated signaling ameliorates diabetes-induced cardiac dysfunction in Zucker diabetic rats. J. Mol. Cell. Cardiol. 2013;57:106–118. doi: 10.1016/j.yjmcc.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Chen A., Umansky K.B., Oonk K.A., Choi W.Y., Dickson A.L., Ou J., Cigliola V., Yifa O., Cao J., et al. Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Dev. Cell. 2019;48:853–863.e5. doi: 10.1016/j.devcel.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Yuasa S., Tabata H., Tohyama S., Hayashiji N., Hattori F., Muraoka N., Egashira T., Okata S., Yae K., et al. Time-lapse imaging of cell cycle dynamics during development in living cardiomyocyte. J. Mol. Cell. Cardiol. 2014;72:241–249. doi: 10.1016/j.yjmcc.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Huang G.N., Thatcher J.E., McAnally J., Kong Y., Qi X., Tan W., DiMaio J.M., Amatruda J.F., Gerard R.D., Hill J.A., et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner C.M., Das A., Ma Z., Self M., Chen C., Lum L., Oliver G., Carroll T.J. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Major R.J., Blum N., Dahn R.D., Begemann G., Poss K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lavine K.J., Yu K., White A.C., Zhang X., Smith C., Partanen J., Ornitz D.M. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell. 2017;21:179–194.e4. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A.N., Kikuchi K., Holdway J.E., Roberts R.W., Burns C.G., Poss K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A., et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014;16:829–840. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.W., Chen B., Yang X., Fugate J.A., Kalucki F.A., Futakuchi-Tsuchida A., Couture L., Vogel K.W., Astley C.A., Baldessari A., et al. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 2018;36:597–605. doi: 10.1038/nbt.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters M., Riley P.R. The epicardium signals the way towards heart regeneration. Stem Cell Res. 2014;13:683–692. doi: 10.1016/j.scr.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E., Zamora M., Raya A., Kawakami Y., Wang J., Zhang X., Burch J., Kubalak S.W., Kaliman P., Izpisua Belmonte J.C., et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. USA. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M., et al. Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell. 2015;16:699–711. doi: 10.1016/j.stem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Nakajima Y. Retinoic acid signaling in heart development. Genesis. 2019;57 doi: 10.1002/dvg.23300. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulos A., Kitani T., Wu J.C. Pluripotent Stem Cell-Derived Cardiomyocytes as a Platform for Cell Therapy Applications: Progress and Hurdles for Clinical Translation. Mol. Ther. 2018;26:1624–1634. doi: 10.1016/j.ymthe.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozhan G., Weidinger G. Wnt/beta-catenin signaling in heart regeneration. Cell Regen. 2015;4:3. doi: 10.1186/s13619-015-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Guleria R.S., Zhu S., Baker K.M. Molecular Mechanisms of Retinoid Receptors in Diabetes-Induced Cardiac Remodeling. J. Clin. Med. 2014;3:566–594. doi: 10.3390/jcm3020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl E., Waxman J.S. Reiterative Mechanisms of Retinoic Acid Signaling during Vertebrate Heart Development. J. Dev. Biol. 2019;7 doi: 10.3390/jdb7020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Lee J.H., Keller G.M. Human Pluripotent Stem Cell-Derived Cardiovascular Cells: From Developmental Biology to Therapeutic Applications. Cell Stem Cell. 2019;25:311–327. doi: 10.1016/j.stem.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Romagnuolo R., Masoudpour H., Porta-Sánchez A., Qiang B., Barry J., Laskary A., Qi X., Massé S., Magtibay K., Kawajiri H., et al. Human Embryonic Stem Cell-Derived Cardiomyocytes Regenerate the Infarcted Pig Heart but Induce Ventricular Tachyarrhythmias. Stem Cell Rep. 2019;12:967–981. doi: 10.1016/j.stemcr.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Shiba Y., Gomibuchi T., Seto T., Wada Y., Ichimura H., Tanaka Y., Ogasawara T., Okada K., Shiba N., Sakamoto K., et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538:388–391. doi: 10.1038/nature19815. [DOI] [PubMed] [Google Scholar]
- Song Y., Lee S., Kim J.R., Jho E.H. Pja2 Inhibits Wnt/beta-catenin Signaling by Reducing the Level of TCF/LEF1. Int. J. Stem Cells. 2018;11:242–247. doi: 10.15283/ijsc18032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H., Cahan P., Lee D.I., Wang S., Miyamoto M., Fernandez L., Kass D.A., Kwon C. Transcriptional Landscape of Cardiomyocyte Maturation. Cell Rep. 2015;13:1705–1716. doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe V., Ramadass R., Dogra D., Rasouli S.J., Gunawan F., Nakajima H., Chiba A., Reischauer S., Mochizuki N., Stainier D.Y.R. In vivo analysis of cardiomyocyte proliferation during trabeculation. Development. 2018;145:dev164194. doi: 10.1242/dev.164194. [DOI] [PubMed] [Google Scholar]
- van Wijk B., Gunst Q.D., Moorman A.F.M., van den Hoff M.J.B. Cardiac regeneration from activated epicardium. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.L., Qi Z., Li Y.H., Zhao H.M., Chen Y.G., Fu W. TGFbeta induced factor homeobox 1 promotes colorectal cancer development through activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:70214–70225. doi: 10.18632/oncotarget.19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier-Neto J., Sousa Costa Â.M., Figueira A.C.M., Caiaffa C.D., Amaral F.N.d., Peres L.M.C., da Silva B.S.P., Santos L.N., Moise A.R., Castillo H.A. Signaling through retinoic acid receptors in cardiac development: Doing the right things at the right times. Biochim. Biophys. Acta. 2015;1849:94–111. doi: 10.1016/j.bbagrm.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora M., Männer J., Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc. Natl. Acad. Sci. USA. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Zhao M., Mattapally S., Chen S., Zhang J. CCND2 Overexpression Enhances the Regenerative Potency of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Remuscularization of Injured Ventricle. Circ. Res. 2018;122:88–96. doi: 10.1161/CIRCRESAHA.117.311504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Day-20 cardiomyocytes purified by sorting SIRPA-positive / Lineage-negative cells by flow cytometry and replated onto a fibronectin-coated dish. Video was captured by a live cell imaging analyzer (BioStation CT).
Top 100 genes with the highest and lowest loadings that express the direction of the PC2. Values are loading coefficients.
Data Availability Statement
All data presented in this study are available in the main text, in Supplemental items file, and from the corresponding author upon request. The RNA-seq data in this study has been deposited into the GEO database under the accession code: GSE176303.