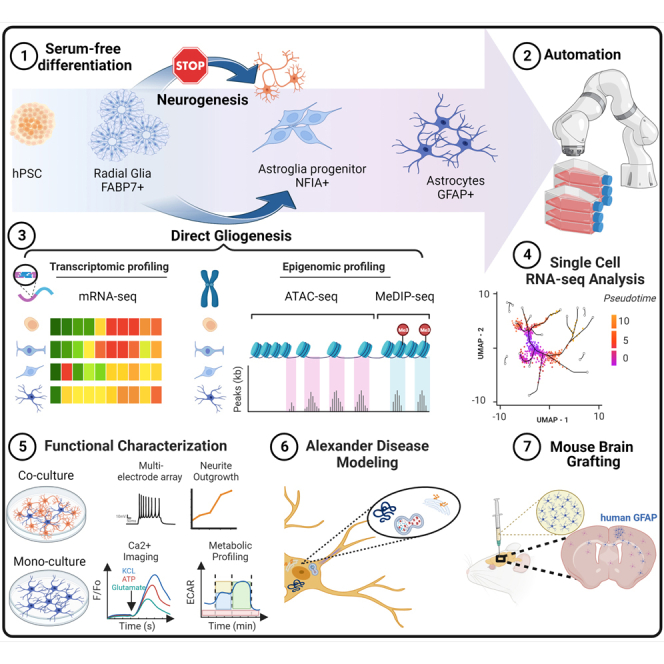
Summary
Human gliogenesis remains poorly understood, and derivation of astrocytes from human pluripotent stem cells (hPSCs) is inefficient and cumbersome. Here, we report controlled glial differentiation from hPSCs that bypasses neurogenesis, which otherwise precedes astrogliogenesis during brain development and in vitro differentiation. hPSCs were first differentiated into radial glial cells (RGCs) resembling resident RGCs of the fetal telencephalon, and modulation of specific cell signaling pathways resulted in direct and stepwise induction of key astroglial markers (NFIA, NFIB, SOX9, CD44, S100B, glial fibrillary acidic protein [GFAP]). Transcriptomic and genome-wide epigenetic mapping and single-cell analysis confirmed RGC-to-astrocyte differentiation, obviating neurogenesis and the gliogenic switch. Detailed molecular and cellular characterization experiments uncovered new mechanisms and markers for human RGCs and astrocytes. In summary, establishment of a glia-exclusive neural lineage progression model serves as a unique serum-free platform of manufacturing large numbers of RGCs and astrocytes for neuroscience, disease modeling (e.g., Alexander disease), and regenerative medicine.
Keywords: iPSCs, neural differentiation, radial glia, astrocytes, gliogenesis, gliogenic switch, serum-free, dual SMAD inhibition, CEPT cocktail, multi-omics, single-cell analysis, robotic cell culture, Alexander disease
Graphical abstract
Highlights
-
•
Efficient serum-free method for radial glia and astrocyte differentiation from hPSCs
-
•
Radial glia can be directly differentiated into astrocytes without preceding neurogenesis
-
•
Extensive molecular and functional characterization of human astrogliogenesis
-
•
Robotic cell culture, mouse brain grafting, and modeling of Alexander disease
Jovanovic, Singeç, and colleagues present a reproducible differentiation method for generation of radial glia and astrocytes from human pluripotent stem cells. Radial glial cells are molecularly distinct from neural precursors generated with dual SMAD inhibition and can be differentiated into astrocytes, obviating neurogenesis and the gliogenic switch. The robustness of the scalable protocol enabled robotic cell culture, detailed multi-omics characterization, functional experiments, identification of new glial markers, and iPSC-based modeling of Alexander disease.
Introduction
Neuroglia were first described by Rudolf Virchow in 1846 as a connective substance, or “brain glue,” that appeared to surround the nervous elements. Ramón y Cajal predicted the importance of astrocytes and dedicated a significant body of work to their morphological characterization (Garcia-Marin et al., 2007). Astrocytes, the most abundant cell type in the central nervous system (CNS), control structural and functional properties of the human brain (e.g., blood-brain barrier, neurotrophic support, synaptogenesis, synaptic pruning, neurotransmitter uptake, ionic milieu control in the extracellular space, innate and adaptive immunity, neural repair, and others) (Liddelow et al., 2017; Rowitch and Kriegstein, 2010; Verkhratsky and Kettenmann, 1996; Wilton et al., 2019). Impairment of these functions can contribute to disorders such as autism, epilepsy, Alzheimer’s disease, amyotrophic lateral sclerosis, and others (Molofsky et al., 2012; Sloan and Barres, 2014; Zuchero and Barres, 2015).
During CNS development, early neuroepithelial cells give rise to RGCs, which generate large numbers of neurons in the cortical plate and provide a glial scaffold for migratory neurons (neurogenic phase) (Noctor et al., 2001; Schmechel and Rakic, 1979). After the progressive waves of neurogenesis, RGCs differentiate into astrocytes and then, later, into oligodendrocytes (gliogenic phase) (Deneen et al., 2006; Hirabayashi et al., 2009; Naka et al., 2008; Rowitch and Kriegstein, 2010). The invariance of this sequence, neurogenesis followed by gliogenesis, is highly conserved throughout vertebrate phylogeny (Deneen et al., 2006). Despite an improved understanding of the spatiotemporal gene-regulatory networks controlling CNS morphogenesis, little is known about the precise cell-intrinsic mechanisms and extracellular signals that coordinate the genesis of human RGCs and their differentiation into astrocytes.
Previous reports generated astrocytes from hPSCs by prolonged culture for 6 months and longer, which seemed necessary to recapitulate the neurogenic-to-gliogenic fate switch (Krencik et al., 2011; Sloan et al., 2017; Velasco et al., 2019). These strategies often required fetal bovine serum (FBS) and cell sorting approaches to enrich for astrocyte-like cells (Leventoux et al., 2020; Sloan et al., 2017; Tcw et al., 2017). Moreover, to accelerate the onset of astrocyte differentiation, recent reports ectopically expressed specific transcription factors in fibroblasts and induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs) (Caiazzo et al., 2015; Canals et al., 2018). Here, we developed a differentiation method that follows the principles of developmental biology that is stepwise and controlled but also scalable and efficient, as required for clinical applications. We first defined conditions for generation of human RGCs, a cell type that is largely understudied. These pure cultures of RGCs were then coaxed exclusively into astrocytes without going through an obligated neurogenic phase. Using this efficient strategy and a robotic cell culture system, we were able to manufacture astrocytes at industrial scale and present a versatile resource to elucidate human glial biology.
Results
Efficient conversion of hPSCs into RGCs
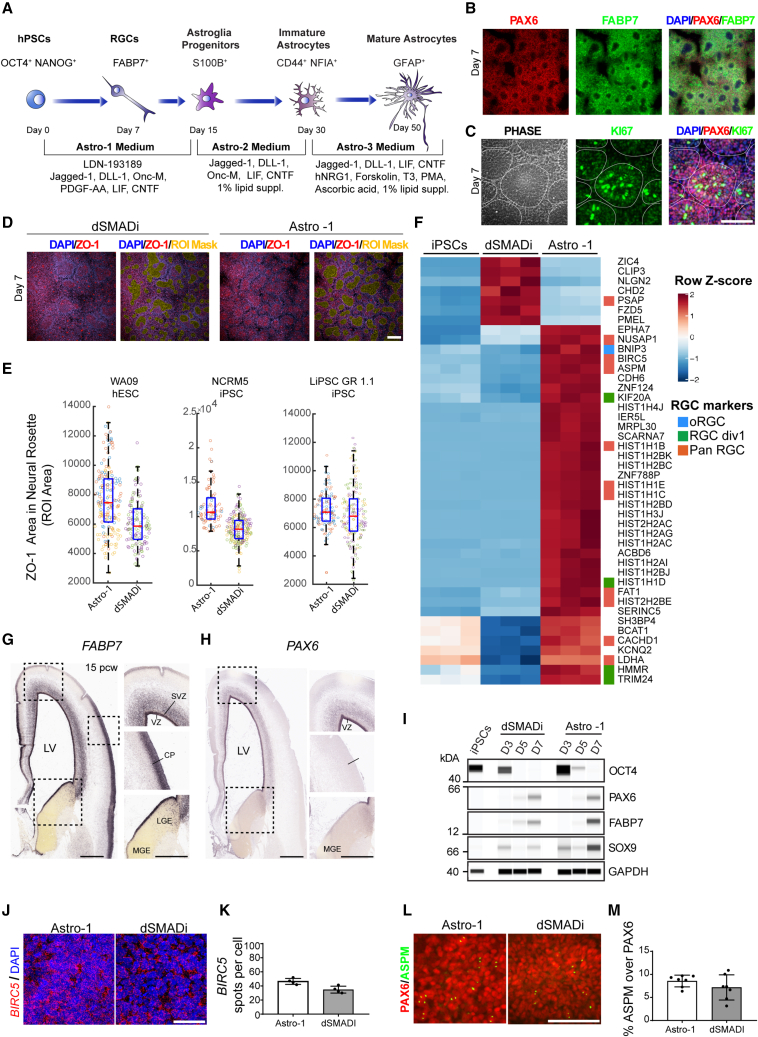
All experiments described here were performed using human embryonic stem cell (hESC) and iPSC lines that were continuously cultured under feeder-free, chemically defined conditions using Essential 8 (E8) medium and vitronectin (VN) as a coating substrate. To improve reproducibility, a defined number of hPSCs (10,000 cells/cm2) was plated on VN-coated dishes in E8 medium supplemented with Chroman 1, Emricasan, Polyamines, and trans-ISRIB (CEPT) small-molecule cocktail for 24 h, which promotes optimal cell survival (Chen et al., 2021). On the following day, the cells were exposed to Astro-1 medium (Figure 1A; experimental procedures), formulated to modulate several cell signaling pathways with importance for RGC differentiation, including NOTCH and JAK/STAT (Deverman and Patterson, 2009). Daily Astro-1 medium changes resulted in efficient neuralization and formation of large numbers of neural rosettes over the course of 7 days (Figures 1B–1D). These cells were strongly immunoreactive for the RGC markers FABP7 (also known as brain lipid-binding protein [BLBP]), PAX6, and the proliferation marker KI-67 (Figures 1B and 1C). Two additional hPSC lines were successfully differentiated into rosette-forming RGCs expressing FABP7 and PAX6 (Figures S1A and S1B). We then compared the neural rosettes generated with our strategy with a widely used neural induction method known as dual SMAD inhibition (dSMADi) (Chambers et al., 2009). We performed unbiased high-content imaging of neuralized cultures (day 7) after performing immunocytochemistry for the tight junction marker ZO-1 to label the lumens of neural rosettes (Elkabetz et al., 2008; Figure 1D). Image analysis of regions of interest (ROIs) showed increased values in three independent cell lines indicating improved neural rosette structures with Astro-1 medium versus dSMADi (Figure 1E). Next, we performed RNA sequencing (RNA-seq) experiments to compare our novel neural induction strategy with dSMADi. Neural cells generated by Astro-1 versus dSMADi were different in their expression of RGC genes (Figure 1F). After normalization to gene expression in pluripotent cells, 44 genes were found to be differentially expressed, with Astro-1 inducing 37 genes and dSMADi inducing 7 genes. Of those 37 genes, 15 genes were reported to be enriched in brain-derived RGCs (Nowakowski et al., 2017): BNIP3, HIST1HD, HMMR, KIF20A, TRIM24, ASPM, BIRC5, CACHD1, FAT1, HIST1H1B, HIST1H1C, HIST1H1E, HIST2H2BE, LDHA, and NUSAP1. In contrast, one RGC gene, PSAP, was induced by dSMADi. The atypical cadherin FAT1 controls RGC proliferation, and its loss leads to abnormal radial glia morphology and neural tube defects (Badouel et al., 2015). We found that FAT1 was strongly expressed by neural rosettes (Figure S1E and S1F). Moreover, the RGC marker FABP7 was expressed at higher levels in cells generated with Astro-1 versus dSMADi, whereas PAX6 was expressed at comparable levels, and OCT4 was undetectable in either condition on day 7 (Figure 1I). Notably, FABP7 is widely expressed by RGCs across the developing brain, whereas PAX6 is restricted to the more anterior regions, such as the dorsal forebrain and diencephalon (Figures 1G and 1H; source: Allen Brain Atlas). SOX9 is an important regulator of glial cell fate (Kang et al., 2012) and was also expressed at higher levels in cells treated with Astro-1 (Figure 1I). Other genes of interest are BIRC5 (SURVIVIN) and ASPM, which control RGC symmetric cell division and maintenance in vivo (Fish et al., 2006; Jiang et al., 2005). Quantitative in situ hybridization (RNA-scope) showed that BIRC5 transcripts were higher in Astro-1 versus dSMADi (Figures 1J and 1K). Similarly, a trend increase was noted for the mitotic spindle assembly protein ASPM in cultures treated with Astro-1 (Figures 1L and 1M). Next, we tested whether these RGCs could be patterned by using developmental cues. Indeed, the telencephalic marker FOXG1 was reduced when CHIR99021 (a WNT activator) was added to Astro-1 medium (Figure S1M), whereas expression of the hindbrain marker HOXA2 was induced (Figure S1O). Adding FGF8 and CHIR99021 to Astro-1 medium reduced FOXG1 expression (Figure S1M) without HOXA2 induction (Figure S1O), while OTX2 expression was detected (Figure S1N), suggesting a midbrain-like identity. These findings were confirmed by using 3 different hPSC lines. Importantly, this experiment also validated FOXG1 protein expression in RGCs (Figure S1M). Collectively, we conclude that Astro-1 medium differentiates hPSCs directly into early RGCs with improved cell type identity compared with dSMADi.
Figure 1.
Controlled RGC differentiation from hPSCs
(A) Schematic depicting RGC and astrocyte differentiation from hPSCs using chemically defined Astro-1, Astro-2, and Astro-3 media (see experimental procedures for details).
(B) Representative immunostaining of RGC cultures (day 7), showing neural rosettes expressing PAX6 and FABP7 derived from iPSCs (NCRM5). See Figure S1 for experimental replicates in hESCs (WA09) and iPSCs (LiPSC GR 1.1).
(C) Representative immunostaining of RGCs (day 7, NCRM5) and neural rosettes expressing the proliferation marker KI-67 and PAX6.
(D) High-content microscopy image analysis of RGC cultures (day 7, WA09), showing neural rosettes expressing ZO-1. ROI is defined as the regions of ZO-1 expression are depicted by a mask in yellow.
(E) Quantification of ROI in one hESC (WA09) and two iPSC lines (NCRM5 and LiPSC GR 1.1). The central area of a well (6 well plate) was imaged using 10× magnification and the Opera Phenix high-content imager (PerkinElmer). Data points in bar graphs represent analyzed images, and different colors of data points represent replicate wells; n ≥ 132 fields of view analyzed from a minimum of 2 replicate wells for each experimental condition. Three independent hPSC lines were tested.
(F) Heatmap showing differentially expressed RGC genes after neural induction with dSMADi and Astro-1. Samples obtained from three independent differentiation experiments are shown in the heatmap (LiPSC GR 1.1).
(G and H) In situ hybridization of the human fetal cortex at post-conceptual week 15, demonstrating expression of FABP7 (G) and PAX6 (H). Note the absence of PAX6 from the cortical plate and the lateral and medial ganglionic eminence (LGE and MGE, respectively). Source of images: Allen Brain Atlas.
(I) Western blot analysis of OCT4, PAX6, FABP7, and SOX9. See the difference in FABP7 and SOX9 between cultures differentiated with Astro-1 versus dSMADi. RGCs (Astro-1) and NPCs (dSMADi) were derived from LiPSC GR 1.1.
(J) BIRC5 expression in RGCs as detected by in situ hybridization (RNA-scope) after differentiation of iPSCs (LiPSC GR 1.1) in Astro-1 medium versus dSMADi (day 7).
(K) Quantification of BIRC5 mRNA transcripts normalized to DAPI+ nuclei; n = 4 replicate wells.
(L) Representative images of cells labeled for ASPM and PAX6 on day 7 (LiPSC GR 1.1).
(M) Quantification of the mitotic spindle protein ASPM normalized to PAX6+ cells after differentiation with Astro-1 versus dSMADi; n = 4 replicate wells (LiPSC GR 1.1).
All data are presented as mean ± SD. Scale bars, 100 μm.
The activated epidermal growth factor (EGF) pathway contributes to RGC-to-astrocyte differentiation at the expense of neurogenesis
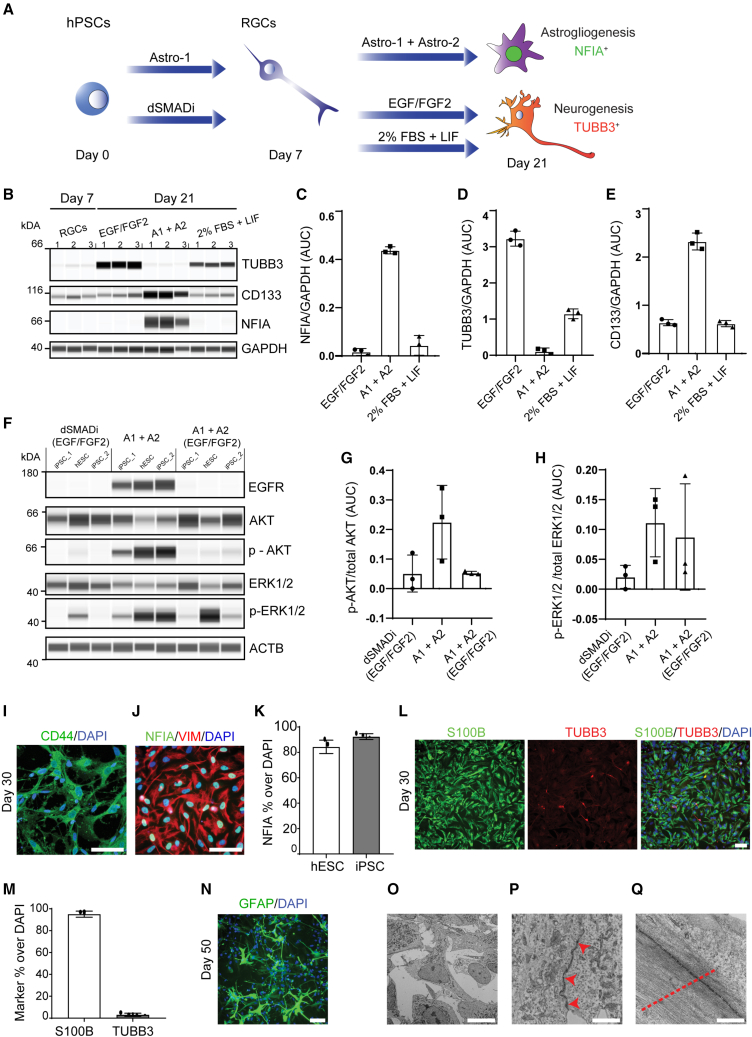
To culture proliferative RGCs in Astro-1 medium, it was important to passage cells at defined time points to avoid overcrowded cultures that can result in cell contact inhibition and compromise directed differentiation. Cells were passaged on days 7, 11, and 14 and plated at 20,000, 30,000, and 40,000 cell/cm2, respectively, to compensate for the decreased proliferation rate over time. Cells were passaged in the presence of the CEPT cocktail (Chen et al., 2021). By day 15, all cells adopted a flat morphology, and approximately 50% of the cells expressed the glial marker S100B (Figures S1G and S1H). At this stage, we removed LDN-193189 and platelet-derived growth factor (PDGF)-AA and added a lipid supplement to provide a more enriched medium (Astro-2 medium), which was necessary when culturing cells in the absence of animal serum (Figure 1A; experimental procedures). Astro-2 medium supplemented with lipids enabled glial differentiation completely under serum-free conditions (experimental procedures). S100B+ cells were less proliferative, and passaging was carried out twice (days 18 and 23). We compared our approach with previous strategies that used either the mitogens EGF and FGF2 or serum-based differentiation (FBS + LIF) to promote the gliogenic switch (Krencik et al., 2011; Tchieu et al., 2019). When RGCs were treated with Astro-1 for 1 week followed by another week of Astro-2 treatment, the critical gliogenic transcription factor nuclear factor Ia (NFIA) (Deneen et al., 2006; Tchieu et al., 2019) was strongly induced. In contrast, NFIA was completely absent when RGCs were treated with EGF/FGF2 or 2% FBS and LIF (Figures 2A–2C). Strikingly, the neuronal marker beta-tubulin 3 (TUBB3) was not induced in cultures differentiated with the Astro-1/Astro-2 combination, whereas the two other tested conditions strongly induced TUBB3 (Figure 2D). We also noted that the glia marker CD133 (prominin-1) was expressed at higher levels after Astro-1/Astro-2 treatment (Figure 2E). To further examine whether the Astro-1/Astro-2 combination is more potent at inducing NFIA, we tested different treatment combinations over the course of 21 days (Figure S2A). Again, by using 3 different hPSC lines, we observed that the strongest average NFIA protein induction was due to Astro-1/Astro-2 treatment (Figures S2B and S2D). Interestingly, dSMADi followed by cell expansion with EGF/FGF2 resulted in the strongest TUBB3 induction (Figures S2B and S2D). Collectively, these experiments not only underscored the importance of Astro-1/Astro-2 media for glia-exclusive differentiation but also demonstrated that RGCs were capable of neuronal fate choice. Next, because Astro-1/Astro-2 media do not include EGF receptor ligands (Figure 1A; experimental procedures) but EGF receptor (EGFR) signaling has been implicated in gliogenesis (Burrows et al., 1997), we further investigated a role of the EGF pathway. To this end, we compared EGFR expression and downstream signaling under three different conditions using three different hPSC lines. Treatment groups included (1) dSMADi followed by EGF/FGF2 treatment, (2) Astro-1/Astro-2, and (3) Astro-1/Astro-2 combined with EGF/FGF2 treatment. Strikingly, strong expression of EGFR was found in all three cell lines after Astro-1/Astro-2 treatment but not when other conditions were used (Figure 2F). This correlated with strong phosphorylation of AKT (Figures 2F and 2G), suggesting that Astro-1/Astro-2 treatment resulted in activated EGF pathway signaling. We also observed that Astro-1/Astro-2 increased phosphorylation of ERK1/2, although variability was found across the different cell lines and treatment conditions (Figure 2H). Unexpectedly, exogenous administration of EGF/FGF2 suppressed EGFR expression and AKT phosphorylation. This finding warrants future investigation, but the experiments described above suggested an important role of EGFR signaling in human gliogenesis even in the absence of recombinant EGF. Next, when culturing cells in Astro-2 until day 30 (Figure 1A), virtually all cells expressed CD44, and more than 85% of total cells expressed NFIA (Figures 2I–2K). At this stage, immunostaining for glial fibrillary acidic protein (GFAP) showed a diffuse signal (Figure S1I), but further maturation in Astro-3 medium (Figure 1A; experimental procedures) produced typical morphologies and GFAP expression (Figures S1D and S1J). Again, this observation was confirmed in several hPSC lines (Figure S1D). To demonstrate the purity of our astrocyte cultures, we performed immunostaining and quantitative analysis, revealing that more than 90% of total cells expressed the glial marker S100B by day 30, with only sporadic occurrence of neurons (3% of total cells) (Figures 2L and 2M). Electron microscopy analysis (days 30 and 50) did not detect neurons, and the derived astrocytes were comparable with control astrocytes (Figure S1J), displaying star-shaped morphologies (Figure 2O) and abundant cytoplasmic intermediate filaments (Figures 2Q and S1L). Astrocytes were rich in organelles such as the endoplasmic reticulum (ER) and Golgi apparatus, indicating secretory activity (Figures S1J and S1K). Well-developed cell junctions were also frequently observed (Figures 2P and S1L).
Figure 2.
Efficient glial differentiation of RGCs with Astro-1 and Astro-2
(A) Experimental design to compare different neural precursors and media.
(B) Western blot analysis of RGCs and neural precursors (day 7) and after treatment for 2 weeks with EGF/FGF2 (25 ng/mL each), Astro-1 and Astro-2 (A1+ A2), or 2% FBS + LIF (10 ng/mL) (NCRM5).
(C–E) Quantification of western blots normalized to GAPDH and performed in triplicates for NFIA (C), TUBB3 (D), and CD133 (E) expression after different treatments. Note the strong expression of NFIA and CD133 after culture in Astro-1/Astro-2 medium and the absence of TUBB3. Three independent experiments were performed (NCRM5).
(F) Western blot analysis of day 21 cultures treated with (1) dSMADi (days 0–7) and EGF/FGF2 (days 7–21), (2) Astro-1/Astro-2, or (3) Astro-1/Astro-2 supplemented with EGF/FGF2 (25 ng/mL each). Replicates represent 3 different hPSC lines (WA09, LiPSC GR 1.1, and NCRM5).
(G and H) Quantification of phosphorylated proteins normalized to total protein for p-AKT (Ser473) expression and p-ERK1/2 (Thr202/Tyr204). Shown are replicates of 3 different hPSC lines (WA09, LiPSC GR 1.1, and NCRM5).
(I and J) Astrocytes expressing CD44, NFIA, and VIMENTIN on day 30 (NCRM5). See Figure S1 for experimental replicates with hESCs (WA09) and iPSCs (LiPSC GR 1.1).
(K) Quantification of NFIA+ nuclei normalized to total number of nuclei (day 30). n = 4 replicate wells for 2 independent cell lines (WA09 and LiPSC GR 1.1).
(L) Immunostaining of cultures (day 30), showing S100B and TUBB3 expression (LiPSC GR 1.1).
(M) Quantification of S100B- and TUBB3-immunoreactive cells normalized to DAPI+ nuclei. n = 4 replicate wells for 2 independent cell lines (WA09 and LiPSC GR 1.1).
(N) Immunostaining showing astrocytes with stellate morphology and GFAP expression (day 50, NCRM5). See Figure S1 for experimental replicates (WA09 and LiPSC GR 1.1).
(O) Electron microscopy confirms typical astrocyte morphologies (day 50).
(P) Depiction of zonula adherens at the ultrastructural level (red arrowheads).
(Q) Presence of abundant intermediate filaments (red dashed line). For ultrastructural analysis, 3 independent samples were included (day 30, day 50 iPSC-astrocytes [NCRM5], and controls [iCell Astrocytes Fujifilm CDI]). See Figure S1 for more details.
All data presented as mean ± SD. Scale bars, 100 μm (I, J, L, and N), 2 μm (O), and 500 nm (P and Q).
To test whether RGCs could produce other neural cell lineages, we generated brain organoids (Figure S1P) following a published protocol (Marton et al., 2019). RGCs formed spheroids (Figure S1Q) and developed neural tube-like structures (Figures S1P and S1R). In different experiments, we asked whether RGCs can be patterned into oligodendrocyte precursors (Lager et al., 2018). When SAG21K, an activator of sonic hedgehog (SHH) signaling, and retinoic acid were added to Astro-1 medium, OLIG2+ cells were generated using two different hPSC lines (Figure S1S). Together, hPSC-derived RGCs displayed appropriate developmental plasticity and produced various neural lineages under different conditions.
Transcriptomic changes confirm RGC and fetal-like astrocyte identity
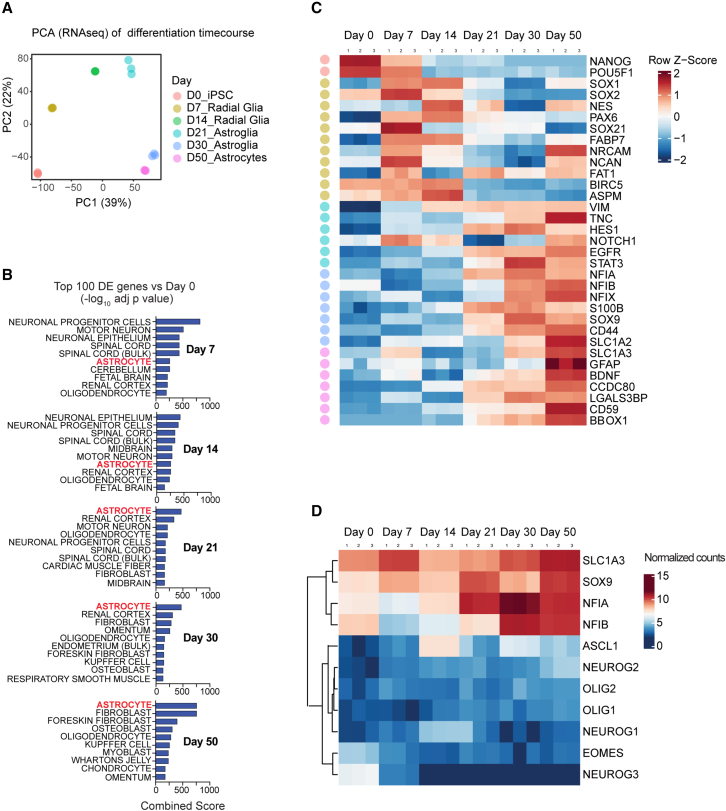
Time-course RNA-seq experiments were performed to characterize human gliogenesis under defined in vitro conditions. Principal-component analysis (PCA) showed distinct clustering of samples across six different time points (Figure 3A). Unbiased comparison of the top 100 differentially upregulated genes using the ARCHS4 tissue database, which contains 84,863 transcriptomes of diverse human samples, and the gene enrichment platform Enrichr (Lachmann et al., 2018), underscored stepwise differentiation, resulting in “astrocyte” as the top category by days 21, 30, and 50 (Figure 3B). Heatmap analysis showed cell type- and stage-specific gene expression and the transcriptional changes, consistent with stepwise and controlled gliogenesis (Figure 3C). Accordingly, the pluripotency-associated genes OCT4 and NANOG were downregulated by day 7, whereas NOTCH1, FABP7, PAX6, NESTIN (NES), NRCAM, and NCAN were strongly induced in RGCs (Figure 3C), as also confirmed by immunocytochemistry and western blot analysis (Figures 1B, 1I, and S1). By days 14 and 21, additional radial glia markers (VIMENTIN [VIM] and TENASCIN C [TNC]) were expressed by RGCs. By day 30, cells expressed canonical astrocyte markers (e.g., NFIA, NFIB, S100B, and CD44) and glutamate transporters (SLC1A2/EAAT2 and SLC1A3/GLAST). Throughout differentiation (days 0–50), we did not detect induction of pro-neuronal transcription factors (NEUROGENIN 1, NEUROGENIN 2, NEUROGENIN 3, and EOMES). In contrast, the gliogenic transcription factors SOX9, NFIA, and NFIB (Canals et al., 2018; Li et al., 2018b) were strongly expressed from day 14 onward (Figure 3D). We also confirmed the absence of oligodendrocyte precursor markers (OLIG1 and OLIG2) throughout the astrocyte differentiation process (Figure 3D). Together, these findings document that RGCs can be directly differentiated into astrocytes without going through a neurogenic phase or artificially inducing the gliogenic switch by forced expression of NFIA in expanded NSCs (Tchieu et al., 2019).
Figure 3.
Transcriptomic analysis of hPSCs differentiating into RGCs and astrocytes
(A) PCA of time-course RNA-seq experiments of iPSCs (day 0) and differentiating cells (days 7, 14, 21, 30, and 50) (NCRM5).
(B) Heatmap depicting cell-type-specific genes at different time points, including pluripotent cells (day 0), RGCs (days 7 and 14), astroglia (days 21 and 30), and astrocytes (day 50).
(C) Gene enrichment analysis (ARCHS4 and Enrichr) based on the top 100 differentially upregulated genes (padj) versus hPSCs (day 0) reveals “astrocytes” as the top hit starting on day 21.
(D) Heatmap with average linkage dendrogram showing induction of astrogliogenic transcription factors (e.g., SOX9, NFIA, and NFIB) and absence of genes representative of neurogenesis (NEUROG 1/2/3 and EOMES) and oligodendrogenesis (OLIG1/2).
n = 3 independent astrocyte differentiation experiments (NCRM5).
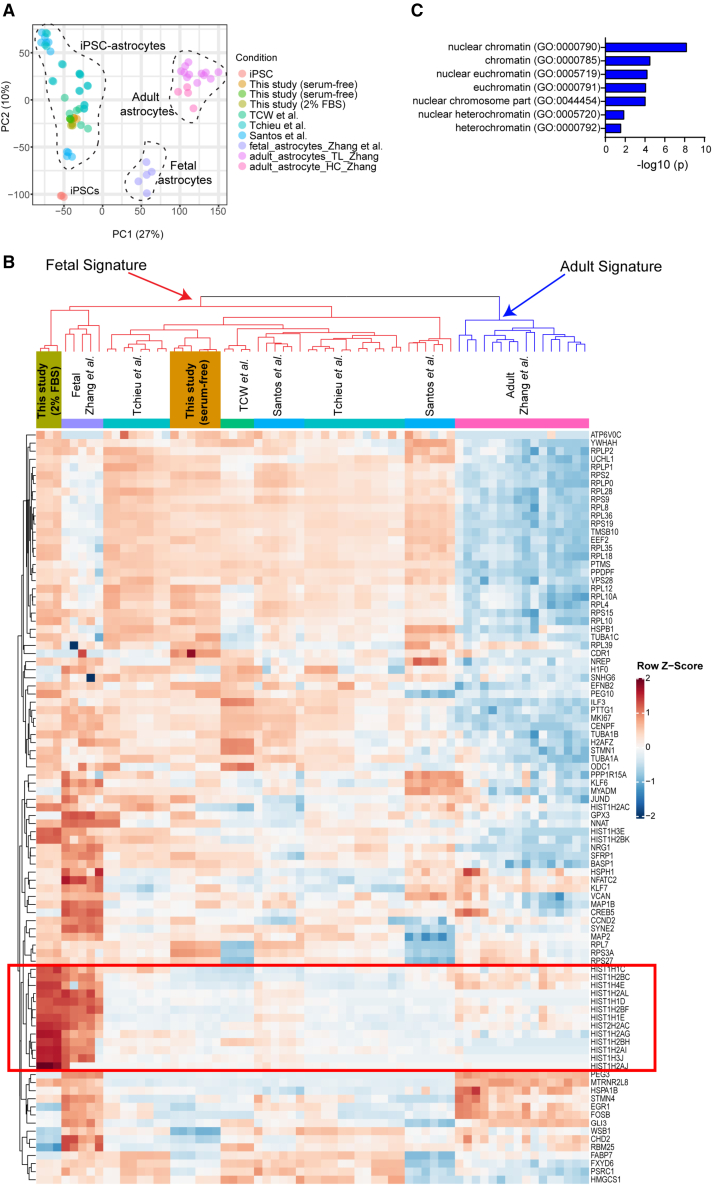
Next, we compared the transcriptomic profiles of astrocytes generated with the present method with previously published reports differentiating hPSCs (Santos et al., 2017; Tchieu et al., 2019; Tcw et al., 2017) and primary astrocytes from fetal and adult human brains (Zhang et al., 2016) (Figure 4A). After batch correction based on expression of 50 “housekeeping” genes, we performed unsupervised hierarchical clustering, focusing on the genes expressed in human fetal astrocytes (Zhang et al., 2016). Our serum-free astrocyte cultures and hPSC-astrocytes of other studies clustered together with samples of primary fetal astrocytes and away from the primary adult astrocytes (Figure 4B). In addition, we treated our astrocyte cultures with 2% FBS, which has been used in other studies (Santos et al., 2017; Tchieu et al., 2019; Tcw et al., 2017). FBS led to a strong induction of genes encoding histone proteins (Figures 4B and 4C), a signature that was also found in primary human fetal astrocytes (Zhang et al., 2016) but not in hPSC-astrocytes reported previously (Santos et al., 2017; Tchieu et al., 2019; Tcw et al., 2017). Next, we addressed whether hPSC-astrocytes showed a distinct regional identity. To perform this analysis in an unbiased fashion, we determined gene sets by focusing on the top 10 genes with the strongest expression across 8 different brain regions during fetal human brain development (CS22) (Eze et al., 2021). Heatmap clustering analysis demonstrated strong expression of cortical markers such as LHX2, TCF4, FEZF2, and FGFR3 in the primary adult cortical astrocytes (Figure S2E). Fetal brain astrocytes expressed regional markers from across different brain regions, such as STMN2 (hindbrain), MEG3 (hypothalamus), and BCL11B (ganglionic eminence), which were not observed in the adult cortical astrocytes (Figure S2E). Despite molecular similarity to fetal astrocytes, hPSC-astrocytes presented an even more diverse expression pattern of regional markers, suggesting an early pre-specification stage (Figure S2E).
Figure 4.
Comparative transcriptomic analysis of hPSC-derived astrocytes
(A) PCA plot of iPSCs and astrocytes generated in the present study (serum free and 2% FBS) and comparison to previous studies after performing batch correction.
(B) Hierarchical clustering based on the top 100 genes expressed by primary human fetal astrocytes according to Zhang et al. (2016). Note that genes regulating histone proteins are strongly induced in iPSC-derived astrocytes (day 30) after adding 2% FBS to Astro-2 medium, making them more similar to fetal brain-derived astrocytes (red boxed area).
(C) Gene enrichment analysis of the prominent histone gene cluster that was induced by 2% FBS (red boxed area in B).
n = 3 independent differentiation experiments (NCRM5) for each of the sample groups included in SRA batch correction from this study (depicted in green and orange in A and B).
Functional characterization of human astrocytes
When co-culturing hPSC-astrocytes and controls with glutamatergic cortical neurons for 8 days, multi-electrode array (MEA) experiments showed enhanced neuronal activity compared with neuron-only cultures. By day 8, neurons co-cultured with astrocytes displayed robust electrical activity (Figure S3A), as indicated by a larger number of spikes (Figure S3B), higher average number of active electrodes (Figure S3C), and robustness of spike intervals across wells (Figure S3D) compared with neuron-only cultures. Next, when astrocytes were co-cultured with glutamatergic neurons for 21 days, the synaptic vesicle protein synaptophysin (SYP) was expressed at higher levels. Similarly, co-culturing resulted in stronger expression of vesicular glutamate transporter 1 (VGlut1) and S100B, which suggests a reciprocal cell maturation effect of both cell types compared with monoculture conditions (Figure S3E). To measure neuronal survival and neurite outgrowth, hPSC-astrocytes were co-cultured with i3 neurons (Fernandopulle et al., 2018), stably expressing green fluorescent protein (GFP) in the nucleus and red fluorescent protein (RFP) in the cytoplasm. To quantify neurite length, continuous live-cell imaging was performed (IncuCyte). i3 neurons co-cultured with astrocytes developed neurites with increased length (Figures S3F and S3G). Periodic acid-Schiff (PAS) staining confirmed intracellular glycogen accumulation (Figure S3H). Similar to their in vivo properties, hPSC-astrocytes were able to remove extracellular glutamate (Figure S3I) and also secreted human complement factor C3 in response to an inflammatory cocktail (interleukin-1b [IL-1b], tumor necrosis factor alpha [TNF-α], and C1Q) (Figure S3J). The use of a fluorescence imaging plate reader (FLIPR) revealed that hPSC-astrocytes (day 30) can elicit calcium transients when stimulated with KCl, ATP, and L-glutamate (Figure S3K). As a normal physiological response, astrocytes can release intracellular calcium through IP3 receptor 2 (IP3R2), expressed on the ER (Sherwood et al., 2021). We tested ATP-induced calcium transients after selective inhibition of IP3R and observed a dose-dependent reduction in the calcium response (Figures S3L and S3M). Astrocyte cultures also displayed slow spontaneous calcium transients spreading across larger cell populations (Video S1), which was comparable with control astrocytes (Video S2).
Video-microscopy of iPSC-derived astrocytes (day 50, LiPSC GR1.1) loaded with calcium dye (FLIPR Calcium 6 kit). Note the spreading of calcium transients across the cell culture.
Video-microscopy of iCell astrocytes (FUJIFILM CDI) loaded with calcium dye (FLIPR Calcium 6 kit). Note that these control astrocytes behave similarly to the iPSC-derived astrocytes shown in Video S1.
Epigenetic regulation of human astrogliogenesis
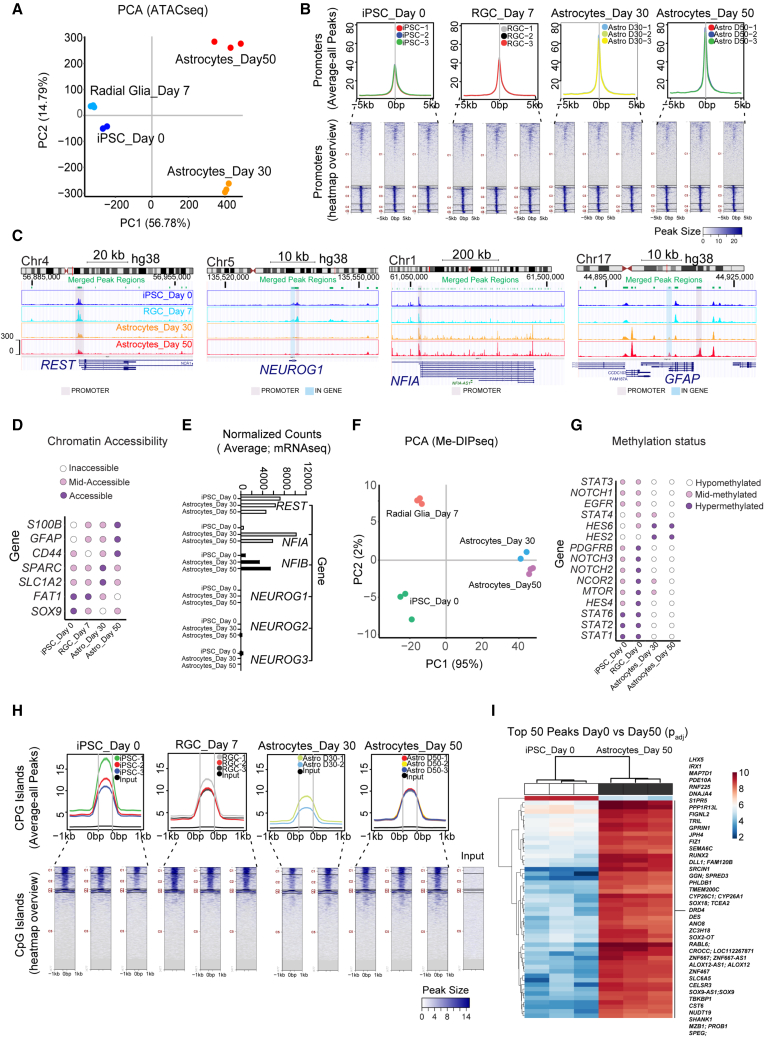
We mapped epigenetic changes by performing genome-wide analyses of chromatin accessibility using the assay for transponase-accessible chromatin with sequencing (ATAC-seq) and DNA methylation by employing methylated DNA imunoprecipitation sequencing (MeDIP-seq) at different time points (days 0, 7, 30, and 50). Distinct chromatin signatures were observed over the time course (Figure 5A). Global promoter accessibility, defined as the average ATAC-seq signal in peaks 5 kb upstream or downstream of the transcription start site (TSS), increased as iPSCs differentiated into RGCs and astrocytes (Figure 5B). The overall number of peaks detected in promoter regions was reduced (Figure 5B). We studied the gene loci of REST (RE1 silencing transcription factor), a master negative regulator of neurogenesis involved in transcriptional repression of neuronal genes in non-neuronal cells (Ballas et al., 2005; Lunyak and Rosenfeld, 2005; Shi et al., 2004). The promoter region of REST remained accessible, and REST transcripts, as also confirmed by RNA-seq, were expressed throughout the glial-specific differentiation trajectory (Figures 5C and 5E). In contrast, the promoter and gene body of pro-neuronal transcription factor NEUROGENIN1 (NEUROG1), known to induce neurogenesis and inhibit gliogenesis (Sun et al., 2001), was inaccessible on days 30 and 50 (Figures 5C and 5E). The NFIA promoter was accessible throughout all time points, whereas the NFIA gene body was inaccessible in iPSCs (day 0) and RGCs (day 7) and then became largely accessible in astrocytes (days 30 and 50) (Figure 5C). Finally, open chromatin at the GFAP promoter was observed only in astrocytes on day 50 (Figure 5C), consistent with the onset of mRNA and protein expression (Figures 2N, 3C, and S1D). Focusing on a set of genes reported previously (Zhang et al., 2016), we identified two clusters (31 and 79 genes) with highly accessible regions (Figure S4A). Gene enrichment analysis revealed that the smaller cluster with the 31 genes annotated processes such as “cell migration” and “cell motility” (Figures S4B–S4E). The larger cluster of 79 genes indicated categories such as “cell cycle and “proliferation” (Figures S4D and S4E). Analysis of open chromatin regions showed highest chromatin accessibility for SOX9 and FAT1 on days 0 and 7, while SLC1A2 and SPARC showed highest accessibility on day 30, and S100B and GFAP were most accessible on day 50 (Figure 5D).
Figure 5.
Genome-wide chromatin accessibility (ATAC-seq) and DNA methylation (MeDIP-seq)
(A) PCA plot of differentiating iPSCs (NCRM5) harvested at different time points (days 0, 7, 30, and 50) to study chromatin accessibility (ATAC-seq).
(B) Top row: average plots for all peaks across target promoter regions defined as TSS ± 5 kb. Bottom row: heatmap representation of all peaks within promoter regions.
(C) UCSC Genome Browser plots showing merged peak regions in REST, NEUROG1, NFIA, and GFAP gene loci. Note that the REST gene promoter remains accessible throughout differentiation. See the global closure of the NEUROG1 region and the global opening of NFIA on days 30 and 50. The GFAP promoter region is open on day 50.
(D) Dot plot of chromatin accessibility, representing peaks of highest variance in genes expressed by RGCs and astrocytes.
(E) Normalized counts (RNA-seq) of gliogenic and neurogenic genes at different time points. Note that REST remains expressed throughout the entire differentiation process.
(F) PCA plot of samples used to study DNA methylation (MeDIP-seq).
(G) Dot plot of DNA methylation, showing peaks with highest variance for genes associated with NOTCH and JAK/STAT signaling: mTOR, NCOR2, and others.
(H) Top row: average plots for all peaks across target promoter regions defined as CpG island ± 1 kb. Bottom row: heatmap representation of all peaks within CpG island regions.
(I) Heatmap of top 50 differentially methylated peaks in iPSCs (day 0) versus astrocytes (day 50). Note that most peaks represent hypermethylated regions within genes (49 of 50) in astrocytes. See Figure S5 for UCSC Genome Browser plots depicting representative gene loci from this heatmap.
n = 3 independent astrocyte differentiation experiments for all datasets presented (NCRM5).
Next, we studied DNA methylation changes over the course of cell differentiation (Figure 5F). Global CpG island hypermethylation, defined as the average MeDIP-seq signal in peaks 1 kb upstream or downstream of the CpG island region, was slightly reduced over the time course of RGC and astrocyte differentiation, while the overall number of peaks increased (Figure 5H). The correlation analysis of peaks (Pearson coefficient) across all samples showed more dramatic changes in chromatin accessibility compared with methylation (Figures S5A and S5B). Of the top 50 most differentially methylated regions between iPSCs and astrocytes, 49 were hypermethylated in astrocytes (Figure 5I). Among these hypermethylated genes were the pro-neuronal transcription factors IRX1 and LHX5 (Cohen et al., 2000; Li et al., 2021) and several genes involved in neuronal function (SHANK1, GPRIN1, and JPH4) (Hogea et al., 2021; Hung et al., 2008)(Figure 5I). Sphingosine-1-phosphate receptor 5 (S1PR5), a gene involved in oligodendrocyte development (Jaillard et al., 2005), and PHLDB1, a gene associated with glioma susceptibility (Baskin et al., 2015), were also hypermethylated in day 50 astrocytes (Figure 5I). Collectively, these analyses substantiated an astrocyte-specific chromatin and methylation landscape (Figures 5I and S5C–S5H).
Next, we asked whether these methylation changes may shed light on the cellular pathways that govern astrogliogenesis. Focusing on the PCA of peaks with the highest variance, we analyzed the promoter regions of JAK/STAT, NOTCH, PDGF, mTOR, and NCOR2 (Figure 5G). Components of JAK/STAT signaling (STAT1, STAT2, STAT3, STAT4, and EGFR), NOTCH (NOTCH1, NOTCH2, NOTCH3, and HES4), and the tyrosine kinase receptor PDGFRB were hypermethylated at the RGC stage and then hypomethylated in astrocytes (Figure 5G). HES6 is an atypical HES gene that inhibits HES1, which is required for activation of NEUROGENIN and NEUROD (Murai et al., 2011). HES6 was hypermethylated in astrocytes (Figure 5I). Last, hypomethylation of mTOR and NCOR2 genes on day 50, but not in immature CD44+ astrocytes (day 30) or RGCs (day 7) (Figure 5I), may indicate potential roles during cell maturation. Indeed, unbiased comparative analysis of the top differentially methylated regions revealed that NCOR2 hypomethylation was the most significant hit (Figure S4F). NCOR2 hypomethylation correlated with increased chromatin accessibility and mRNA expression on day 50. Other differentially hypomethylated genes with potential roles in astrocyte maturation were SYNJ2 and DLG5, which was also confirmed by transcript expression (Figures S4F, S4I, and S4J).
Single-cell transcriptomics identifies new astrocyte markers
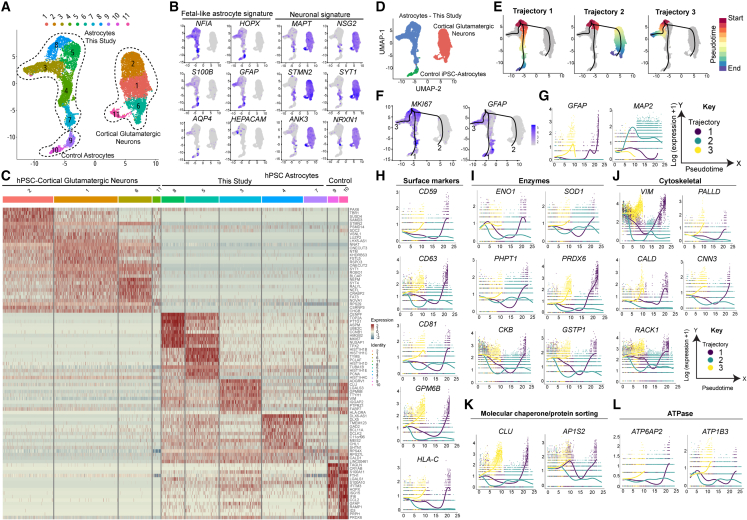
To characterize the molecular underpinnings of RGC-to-astrocyte transition, we performed single-cell RNA-seq (scRNA-seq) of astrocytes (day 50) and compared them with control astrocytes and glutamatergic neurons. We profiled the transcriptomes of 12,771 cells, and unbiased clustering identified 11 transcriptionally distinct cell clusters: 4 representing neurons and 7 representing astrocytes (Figure 6A). Most cells in the astrocyte clusters were characterized by expression of NFIA, HOPX, S100B, and GFAP, while only a few cells expressed AQP4 and HEPACAM, which are thought to be markers of more mature astrocytes (Sloan et al., 2017; Zhang et al., 2016). Neurons showed strong expression of typical markers, including MAPT, NSG2, STMN2, SYT1, ANK3, and NRXN1 (Figure 6B). Heatmap analysis (top 10 genes per cluster) revealed more consistent gene expression across neuronal clusters, whereas astrocyte clusters were more heterogeneous (Figures 6C and S6D–S6G). Next, we performed extensive data integration with single-cell transcriptome data of the developing human telencephalon (Nowakowski et al., 2017). During early development, fetal astrocytes share high transcriptomic similarity with RGCs (Figures S6A–S6C). Data integration revealed 3 distinct clusters: radial glia, astrocytes, and cortical neurons (Figure S6B). The uniform manifold approximation and projection (UMAP) plot revealed bidirectional propagation toward mature astrocytes because cells clustered close to human cortical radial glia of dorsal (Figure S6B, red arrow) and the ventral ganglionic eminence (Figure S6C, blue arrow). This suggests that the first emerging RGCs lack regionalization and do not directly default into a cortical signature, which is consistent with our specification analysis (Figure S2E). To characterize astrocyte differentiation, we performed slingshot analysis (Street et al., 2018) and identified three different trajectories mapping transition of RGCs (pseudotime starting point) into two branches representing astrocytes and one “teleporting” RGCs to excitatory neurons because transitional states were not detected because our differentiation method bypasses neurogenesis (Figures 6D and 6E). The pseudotime starting point was marked by strong MKI67, ASPM, and NUSAP1 expression in clusters 5 and 8, indicating RGC and immature astrocyte identities (Figures 6A, 6C, and 6E). Progression of RGCs into astrocytes did not lead to complete MKI67 downregulation, whereas differentiated neurons were devoid of MKI67, suggesting a postmitotic state (Figures 6F and S6D–S6F). To identify new markers capturing RGC-to-astrocyte differentiation, we correlated gene expression variation with each of the cell types in pseudotime by performing trajectory-based differential expression analysis (tradeSeq) (Van den Berge et al., 2020). Along both astrocyte trajectories (Figure 6G, trajectories 1 and 3, yellow and purple lines), we observed induction of GFAP expression, while the neuronal trajectory displayed MAP2 expression (Figure 6G, trajectory 2, green line). We subcategorized genes differentially upregulated in astrocytes into 5 different categories: surface markers, enzymes, cytoskeletal/scaffolding proteins, membrane-vesicle trafficking, and ATP transporters (Figures 6H–6L). Surface markers that were upregulated over the pseudotime and indicative of astrocyte maturation were CD59, CD63, CD81, GPM6B, and HLA-C (Figure 6H). Astrocytes also showed specific upregulation of enzymes involved in glucose metabolism (ENO1), oxidative metabolism (SOD1 and PRDX6), glutathione biosynthesis (GSTP1), energy metabolism (CKB), and protein dephosphorylation (PHPT1) (Figure 6I). VIM, GFAP, and S100B are well-established astrocyte markers, and our analysis identified PALLADIN (PALLD) and CALDESMON (CALD1) as astrocyte enriched (Figure 6J). CNN3 is a member of the calponin family of actin regulators, highly expressed in the mammalian brain, and involved in regulating the plasticity of dendritic spines (Junghans and Herzog, 2018). The receptor for activated C kinase (RACK1) is important for maintenance of neural progenitors (Zhu et al., 2021). CNN3 and RACK1 were strongly upregulated during astrocyte differentiation (Figure 6J). Similarly, the molecular chaperone CLUSTERIN (CLU) and clathrin-associated adaptor protein complex 1 (AP1S2) (Figure 6K) and specific ATPases (ATP6A2 and ATP1B3) were strongly upregulated during astrocyte differentiation (Figure 6L). Together, these results uncover a set of novel markers that capture the directed differentiation of RGCs into astrocytes. Many of these genes are associated with neurodegenerative diseases (e.g., SOD1, CLU, AP1S2, and ATP1B3) and may serve as biomarkers and elucidate pathology.
Figure 6.
scRNA-seq analysis maps the fetal astrocyte signature and astrocyte maturation markers
(A) UMAP of 12,771 hPSC-derived neural cells showing 11 distinct cell clusters. Clusters are color coded, and dotted lines demarcate different cell types.
(B) Feature plots for genes expressed by fetal and adult astrocytes and neurons.
(C) Heatmap for the top 10 genes expressed across clusters. Rows correspond to genes and columns to cells/clusters. Blue/yellow, low gene expression; red, high expression. Note the clear difference between neuronal and astrocytic signatures.
(D) UMAP of 12,771 hPSC-derived neural cells color-coded by cell type. Blue, astrocytes generated in this study; green, control astrocytes (Fujifilm CDI); red, glutamatergic neurons (Fujifilm CDI).
(E) Slingshot analysis maps 3 different trajectories of cell progression. Red, pseudotime start point; blue, pseudotime endpoint.
(F) Feature plots for the MKI67 and GFAP genes. Note that all pseudotime trajectories start within the clusters with high MKI67 and low GFAP expression and progress to areas with high GFAP expression (1 and 3) or no GFAP expression (2, neurons).
(G) Gene expression dynamics relative to the previously identified trajectories performed using tradeSeq, showing GFAP induction along trajectories 1 and 3 (astrocytes) and MAP2 induction along trajectory 2 (neurons).
(H–L) Genes representing surface markers, enzymes, cytoskeletal/scaffolding proteins, and others with differential induction in maturing astrocytes but not neurons. Data are from 12,771 single cells obtained from experiments using differentiated astrocytes from hESCs (WA09) and commercial products (iAstro and iGluta neurons, Fujifilm CDI). scRNA-seq data were analyzed in the Seurat R package.
Translational utility of hPSC-astrocytes
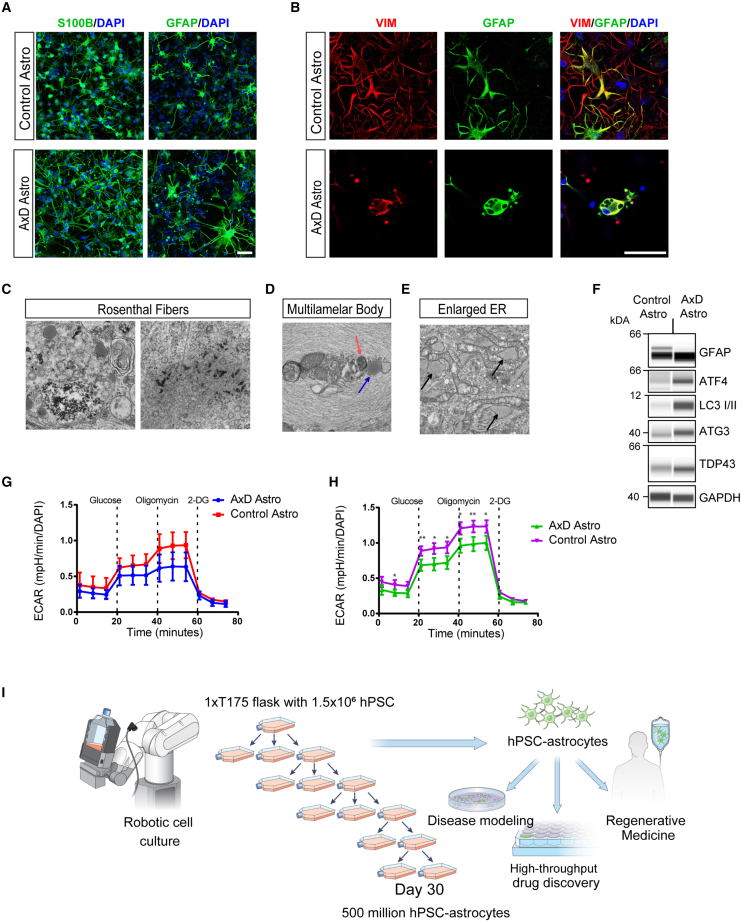
Next, we established a cellular model for Alexander disease (AxD) using an iPSC line from a patient with a mutation in codon 239 of the GFAP gene (729C > T) resulting in arg239-to-cys mutation ARG239CYS (R239C) (Brenner et al., 2001). Clinically, the patient presented with macrocephaly, and MRI scans showed white matter defects and ventricular cysts. The patient experienced first seizures at the age of 3.5 years and died at 6 years of age, with the diagnosis confirmed by autopsy. Using this iPSC line, we generated astrocytes expressing S100B and GFAP (day 50) and compared them with astrocytes from a non-isogenic control line (Figure 7A). Confocal microscopy showed aberrant morphology of AxD astrocytes with prominent GFAP+ aggregates (Figures 7B and S7H), consistent with previous reports (Jones et al., 2018; Li et al., 2018a). Ultrastructural analysis revealed electron-dense material in the cytoplasm of AxD astrocytes, representing pathognomonic protein aggregates, the so-called Rosenthal fibers (Figure 7C). Other pathological changes included multi-lamellar body formation (Figure 7D) and an enlarged ER (Figure 7E). In contrast, these pathological phenotypes were not observed in healthy control astrocytes (Figures S1J–S1L). Furthermore, AxD astrocytes displayed an abnormal unfolded protein response (UPR), as indicated by ATF4/ATG3 protein upregulation. Elevated expression levels of the autophagy marker LC3I/II, GFAP, and total TDP-43, which is associated with several neurodegenerative diseases, suggested the presence of additional pathophysiological mechanisms in AxD astrocytes (Figure 7F). Metabolic profiling (Searhorse XF Analyzer) revealed impairment in glycolysis and glycolytic capacity in AxD astrocytes (Figures 7G and 7H).
Figure 7.
Robotic biomanufacturing of human astrocytes for disease modeling
(A) iPSC-derived astrocytes (S100B+, GFAP+) from an Alexander disease (AxD) patient (GM16825) and an unaffected individual (non-isogenic control, NCRM5).
(B) AxD astrocytes showing typical GFAP+ protein aggregates and aberrant morphology.
(C) Electron microscopy of AxD astrocytes (day 50). Note the dark electron-dense material representing Rosenthal fibers.
(D) Electron micrograph of a multi-lamellar body. Note the multi-lamellar structure surrounding organelles that are being digested. A red arrow indicates mitochondria entering the lysosome (blue arrow).
(E) Black arrows indicate enlarged ER.
(F) Western blot showing elevated expression of markers for ER stress and autophagy. Note that total expression of TDP-43 is higher in AxD astrocytes versus control.
(G) Seahorse glycolysis stress testing glycolytic flux in AxD patient astrocytes and control astrocytes cultured under regular conditions (Astro-3 medium). Glycolytic flux is measured as extracellular acidification rate (ECAR). Data are normalized to total cell numbers per well. n = 6 replicate wells per condition. Control (NCRM5), AxD patient (GM16825).
(H) Glycolysis stress test after 24-h exposure to inflammatory cytokines (TNF-α, C1Q, and IL-1a). Note the decreased glycolysis, measured after glucose administration, and total glycolytic capacity, measured after oligomycin administration to block mitochondrial respiration, compared control astrocytes. n = 6 replicate wells per condition. Control (NCRM5), AxD patient (GM16825).
(I) Schematic of the robotic workflow and scalable astrocyte production using the CompacT SelecT platform (Sartorius) suitable for translational applications.
All data are presented as mean ± SD. Scale bars, 100 μm (A and B) and 500 nm (C–E).
To demonstrate that human astrocytes can be grafted, neonatal and adult mice received iPSC-derived astrocytes via unilateral injections, either intraventricular or cortical, and the brains were analyzed at 4 and 8 week post-transplantation (Figures S7A–S7G). In all experimental groups, immunohistochemical analysis revealed strong immunoreactivity for the human cytoplasm marker (STEM121) at the injection sites as well different brains regions in the ipsi- and contralateral hemispheres. Migratory astrocytes co-expressing human-specific GFAP (hGFAP; STEM123 antibody) and S100B were widely distributed and detectable in the cortex (Figure S7B), corpus callosum (Figure S7C), hippocampus (Figure S7D), thalamus (Figure S7E), and blood vessels in the brain parenchyma (Figure S7F). Quantitative analysis showed that the highest number of transplanted human astrocytes was detected in neonatal animals 8 weeks post grafting (Figure S7G). These experiments demonstrated that hPSC-derived astrocytes engraft, migrate over long distances, and survive in vivo for several weeks.
Last, clinically relevant protocols should enable large-scale biomanufacturing of human cells from hPSCs. We therefore automated our scalable astrocyte differentiation protocol using the CompacT SelecT robotic cell culture platform (Figure 7I). This approach, in combination with the CEPT small-molecule cocktail (Chen et al., 2021), enabled the routine production of up to 500 million astrocytes in a single manufacturing workflow, and astrocytes could be cryopreserved on day 30 and used on demand for various applications.
Discussion
During development, the neural lineage generates the three major cell types in a highly coordinated sequential fashion: neurons, astrocytes, and oligodendrocytes (Rowitch and Kriegstein, 2010). Astrocytes play crucial roles in the normal and diseased human brain, but the precise mechanisms of glial cell fate choice and differentiation remain poorly understood. Here, we established a novel strategy that differentiates hPSCs into RGCs and then directly into astrocytes, obviating neurogenesis and oligodendrogenesis. This stepwise method is based on bone morphogenetic protein (BMP) inhibition and activation of gliogenic pathways (Deverman and Patterson, 2009; Namihira et al., 2009). Our study has several far-reaching implications. First, generation of RGCs with improved capability for neural rosette formation and transcriptomic similarity to resident RGCs of the human fetal telencephalon (Nowakowski et al., 2017) should leverage future studies. RGCs were responsive to rostro-caudal patterning cues and produced neurons and OLIG2-expressing cells under different culture conditions. Therefore, generation of bona fide RGCs could become an alternative to dSMADi (Chambers et al., 2009) and applied to more faithfully recapitulate CNS development in vitro.
Second, if highly controlled RGC differentiation into astrocytes is employed, then strong NFIA expression is obtained on day 21 without long-term expansion of NPCs or genetic manipulation (Tchieu et al., 2019). As one of the contributing mechanisms for gliogenesis, we suggest an activated EGF pathway in the absence of recombinant EGF. Alternative culture conditions, including recombinant EGF/FGF2 administration, promoted neurogenesis and suppressed NFIA expression. Adding FBS to Astro-1/Astro-2 medium induced expression of histone genes, similar to a signature found in human fetal astrocytes. Considering that FBS can contribute to cell maturation or result in reactive astrocytes and that histone genes are involved in various molecular processes (Burda et al., 2022), future work is necessary to elucidate these findings. Our approach confirmed a direct RGC-to-astrocyte differentiation model at the expense of neurogenesis. The chromatin state and expression of SOX9, NFIA, NFIB, and REST and absence of neurogenic genes (e.g., NEUROG1/2/3) across different time points was consistent with exclusive astrogliogenesis. Previous single-cell studies of mouse and human brain development have mapped early cortical neurogenesis and neuronal subtype specification (Di Bella et al., 2021; Eze et al., 2021), whereas differentiation trajectories of human RGCs and early glial specification remain understudied. Hence, our multi-omics datasets represent an invaluable resource for studying human gliogenesis.
Third, derived astrocytes remain immature relative to primary astrocytes from adult brains, which is consistent with previous reports (Canals et al., 2018; Krencik et al., 2011; Li et al., 2018b; Santos et al., 2017; Tchieu et al., 2019; Tcw et al., 2017). Our findings suggest potential important roles for NCOR-2, SYNJ2, and DLG5 in astrocyte maturation. Finally, our demonstration of industrial-scale production of patient- and disease-specific astrocytes may help to deliver on the promise of iPSCs and leverage drug development and personalized therapies for neurological and psychiatric illnesses.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors: Vukasin Jovanovic (vukasin.jovanovic@nih.gov) and Ilyas Singeç (ilyassingec@gmail.com).
Materials availability
All materials and reagents generated in this study will be made available upon request following NIH guidelines.
Data and code availability
Data are presented as the mean ± SD. Statistical analyses (GraphPad Prism) were performed using different tests as appropriate and as described in the figure legends.
Bulk RNA-seq and scRNA-seq, ATAC-seq, and MeDIP-seq FASTQ files have been deposited into the Sequence Read Archive (SRA) under BioProject PRJNA769413. The code for sequencing data analysis is available at https://github.com/cemalley/Jovanovic_methods and https://github.com/jaro-slamecka/Jovanovic_astrocyte_scRNA-seq. Public databases used include Gene Ontology 2019 (http://geneontology.org/), ARCHS4 Tissues (https://maayanlab.cloud/archs4/), and UCSC Cell Browser (https://cells.ucsc.edu/?ds=cortex-dev). Public SRA datasets used were PRJNA412090, PRJNA382448, PRJNA383243, and PRJNA297760.
Cell culture
The hESCs line (WA09), healthy donor iPSC lines (LiPSC-GR1.1, GM25256 [WTC11], and NCRM5 from the NIH Common Fund), and patient-derived AxD iPSCs (GM16825, Coriell) were maintained under feeder-free conditions using E8 medium and VN-coated plates (Thermo Fisher Scientific, A14700). Cells were routinely passaged using 0.5 mM EDTA diluted in phosphate-buffered saline (PBS) without calcium or magnesium (Thermo Fisher Scientific) when culture plates reached about 70%–90% confluency, typically every 3–4 days. For the initial 24 h after cell passaging, E8 medium was supplemented with the CEPT cocktail to optimize cell viability (Chen et al., 2021).
Astrocyte differentiation using Astro-1, Astro-2, and Astro-3 media
Astro-1 medium was composed of DMEM/F12 medium supplemented with N2 (Gibco, 17502048), B27 without vitamin A (Gibco, 12587010), 100 nM LDN-193189 dihydrochloride (Tocris, 6053), and the human recombinant proteins PDGF-AA (R&D Systems, 221-AA), JAGGED-1 (R&D Systems, 1277-JG), DLL-1 (R&D Systems, 1818-DL), ONCOSTATIN M (R&D Systems, 295-OM), LIF (R&D Systems, 7734-LF), and CNTF (R&D Systems, 257-NT) (all at 10 ng/mL concentration).
Astro-2 medium was composed of DMEM/F12 base medium supplemented with N2 (Gibco), B27 complete (Gibco, 17504044), 1% lipid supplement (Gibco, 11905031), and the recombinant proteins JAGGED1, DLL-1, ONCOSTATIN M, LIF, and CNTF (all at 10 ng/mL concentration) (R&D Systems).
Astro-3 medium was composed of DMEM/F12 base medium supplemented with N2, B27 with vitamin A, 1% lipid supplement (Gibco), and JAGGED1, DLL-1, LIF, CNTF (all at 10 ng/mL concentration), hNRG1/EGF domain (20 ng/mL, R&D Systems, 396-HB), 2 μM forskolin (Tocris, 1099), 200 nM phorbol-12 myristate-13 acetate (Tocris, 1201), 40 ng/mL triiodothyronine T3 (Tocris, 6666), and 200 μM ascorbic acid (Tocris, 4055).
Author contributions
V.M.J. and I.S. conceived the project. Experiments, V.M.J., C.S., C.A.T., S.R., P.-H.C., E.B., P.O., J.C.M., S.M., Y.J., J.F.D.S ; data analysis and discussions, V.M.J., J.S., C.W., J.I., M.H., D.G., T.C.V., C.A.T., M.E.W., A.S., and I.S.; manuscript writing, V.M.J. and I.S.
Acknowledgments
We thank H. Hong, P. Francis, T. Deng, J. Freilino, M. Iannotti, C. Pepper Bonney, Y. Gedik, D. Ngan, and H. Baskir for support. We are grateful to A. Hoofring from the NIH Medical Arts Design Section for art designs, K. Nagashima from the Electron Microscopy Laboratory at the National Cancer Institute (NCI) for electron microscopy images, and D. Havas (PsychoGenics) for confocal microscopy and quantification of grafted cells. We also acknowledge funding from the Regenerative Medicine Program (RMP) of the NIH Common Fund and in part by the intramural research program of the National Center for Advancing Translational Sciences (NCATS), NIH. The funders had no role in study design, data collection, and analysis; decision to publish; or preparation of the manuscript.
Conflict of interests
V.M.J., A.S., and I.S. are coinventors on a US Department of Health and Human Services patent covering the described radial glia and astrocyte differentiation method and its use.
Published: July 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.06.007.
Contributor Information
Vukasin M. Jovanovic, Email: vukasin.jovanovic@nih.gov.
Ilyas Singeç, Email: ilyassingec@gmail.com.
Supplemental information
References
- Badouel C., Zander M.A., Liscio N., Bagherie-Lachidan M., Sopko R., Coyaud E., Raught B., Miller F.D., McNeill H. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development. 2015;142:2781–2791. doi: 10.1242/dev.123539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Baskin R., Woods N.T., Mendoza-Fandiño G., Forsyth P., Egan K.M., Monteiro A.N.A. Functional analysis of the 11q23.3 glioma susceptibility locus implicates PHLDB1 and DDX6 in glioma susceptibility. Sci. Rep. 2015;5:17367. doi: 10.1038/srep17367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M., Johnson A.B., Boespflug-Tanguy O., Rodriguez D., Goldman J.E., Messing A. Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat. Genet. 2001;27:117–120. doi: 10.1038/83679. [DOI] [PubMed] [Google Scholar]
- Burda J.E., O'Shea T.M., Ao Y., Suresh K.B., Wang S., Bernstein A.M., Chandra A., Deverasetty S., Kawaguchi R., Kim J.H., et al. Divergent transcriptional regulation of astrocyte reactivity across disorders. Nature. 2022;606:557–564. doi: 10.1038/s41586-022-04739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows R.C., Wancio D., Levitt P., Lillien L. Response diversity and the timing of progenitor cell maturation are regulated by developmental changes in EGFR expression in the cortex. Neuron. 1997;19:251–267. doi: 10.1016/s0896-6273(00)80937-x. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Giannelli S., Valente P., Lignani G., Carissimo A., Sessa A., Colasante G., Bartolomeo R., Massimino L., Ferroni S., et al. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Rep. 2015;4:25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals I., Ginisty A., Quist E., Timmerman R., Fritze J., Miskinyte G., Monni E., Hansen M.G., Hidalgo I., Bryder D., et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Methods. 2018;15:693–696. doi: 10.1038/s41592-018-0103-2. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods. 2021;18:528–541. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D.R., Cheng C.W., Cheng S.H., Hui C.C. Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech. Dev. 2000;91:317–321. doi: 10.1016/s0925-4773(99)00263-4. [DOI] [PubMed] [Google Scholar]
- Deneen B., Ho R., Lukaszewicz A., Hochstim C.J., Gronostajski R.M., Anderson D.J. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Deverman B.E., Patterson P.H. Cytokines and CNS development. Neuron. 2009;64:61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Di Bella D.J., Habibi E., Stickels R.R., Scalia G., Brown J., Yadollahpour P., Yang S.M., Abbate C., Biancalani T., Macosko E.Z., et al. Author Correction: Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 2021;596:E11. doi: 10.1038/s41586-021-03797-5. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze U.C., Bhaduri A., Haeussler M., Nowakowski T.J., Kriegstein A.R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 2021;24:584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., Ward M.E. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 2018;79:e51. doi: 10.1002/cpcb.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J.L., Kosodo Y., Enard W., Pääbo S., Huttner W.B. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Marín V., García-López P., Freire M. Cajal's contributions to glia research. Trends Neurosci. 2007;30:479–487. doi: 10.1016/j.tins.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T.A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hogea A., Shah S., Jones F., Carver C.M., Hao H., Liang C., Huang D., Du X., Gamper N. Junctophilin-4 facilitates inflammatory signalling at plasma membrane-endoplasmic reticulum junctions in sensory neurons. J. Physiol. 2021;599:2103–2123. doi: 10.1113/jp281331. [DOI] [PubMed] [Google Scholar]
- Hung A.Y., Futai K., Sala C., Valtschanoff J.G., Ryu J., Woodworth M.A., Kidd F.L., Sung C.C., Miyakawa T., Bear M.F., et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 2008;28:1697–1708. doi: 10.1523/jneurosci.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard C., Harrison S., Stankoff B., Aigrot M.S., Calver A.R., Duddy G., Walsh F.S., Pangalos M.N., Arimura N., Kaibuchi K., et al. Edg8/S1P5: an oligodendroglial receptor with dual function on process retraction and cell survival. J. Neurosci. 2005;25:1459–1469. doi: 10.1523/jneurosci.4645-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., de Bruin A., Caldas H., Fangusaro J., Hayes J., Conway E.M., Robinson M.L., Altura R.A. Essential role for survivin in early brain development. J. Neurosci. 2005;25:6962–6970. doi: 10.1523/jneurosci.1446-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.R., Kong L., Hanna M.G., 4th, Hoffman B., Krencik R., Bradley R., Hagemann T., Choi J., Doers M., Dubovis M., et al. Mutations in GFAP disrupt the distribution and function of organelles in human astrocytes. Cell Rep. 2018;25:947–958.e4. doi: 10.1016/j.celrep.2018.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans D., Herzog S. Cnn3 regulates neural tube morphogenesis and neuronal stem cell properties. FEBS J. 2018;285:325–338. doi: 10.1111/febs.14338. [DOI] [PubMed] [Google Scholar]
- Kang P., Lee H.K., Glasgow S.M., Finley M., Donti T., Gaber Z.B., Graham B.H., Foster A.E., Novitch B.G., Gronostajski R.M., Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R., Weick J.P., Liu Y., Zhang Z.J., Zhang S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma'ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager A.M., Corradin O.G., Cregg J.M., Elitt M.S., Shick H.E., Clayton B.L.L., Allan K.C., Olsen H.E., Madhavan M., Tesar P.J. Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat. Commun. 2018;9:3708. doi: 10.1038/s41467-018-06102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventoux N., Morimoto S., Imaizumi K., Sato Y., Takahashi S., Mashima K., Ishikawa M., Sonn I., Kondo T., Watanabe H., Okano H. Human astrocytes model derived from induced pluripotent stem cells. Cells. 2020;9:2680. doi: 10.3390/cells9122680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sun L., Peng X.L., Yu X.M., Qi S.J., Lu Z.J., Han J.D.J., Shen Q. Integrative genomic analysis of early neurogenesis reveals a temporal genetic program for differentiation and specification of preplate and Cajal-Retzius neurons. PLoS Genet. 2021;17:e1009355. doi: 10.1371/journal.pgen.1009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tian E., Chen X., Chao J., Klein J., Qu Q., Sun G., Sun G., Huang Y., Warden C.D., et al. GFAP mutations in astrocytes impair oligodendrocyte progenitor proliferation and myelination in an hiPSC model of Alexander disease. Cell Stem Cell. 2018;23:239–251.e6. doi: 10.1016/j.stem.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tao Y., Bradley R., Du Z., Tao Y., Kong L., Dong Y., Jones J., Yan Y., Harder C.R.K., et al. Fast generation of functional subtype astrocytes from human pluripotent stem cells. Stem Cell Rep. 2018;11:998–1008. doi: 10.1016/j.stemcr.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak V.V., Rosenfeld M.G. No rest for REST: REST/NRSF regulation of neurogenesis. Cell. 2005;121:499–501. doi: 10.1016/j.cell.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Marton R.M., Miura Y., Sloan S.A., Li Q., Revah O., Levy R.J., Huguenard J.R., Pașca S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019;22:484–491. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., Krencik R., Ullian E.M., Tsai H.H., Deneen B., Richardson W.D., Barres B.A., Rowitch D.H. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K., Philpott A., Jones P.H. Hes6 is required for the neurogenic activity of neurogenin and NeuroD. PLoS One. 2011;6:e27880. doi: 10.1371/journal.pone.0027880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H., Nakamura S., Shimazaki T., Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat. Neurosci. 2008;11:1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T., Nakashima K. Committed Neuronal Precursors Confer Astrocytic Potential on Residual Neural Precursor Cells. Dev. Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., Kriegstein A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D., et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch D.H., Kriegstein A.R. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Santos R., Vadodaria K.C., Jaeger B.N., Mei A., Lefcochilos-Fogelquist S., Mendes A.P.D., Erikson G., Shokhirev M., Randolph-Moore L., Fredlender C., et al. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Rep. 2017;8:1757–1769. doi: 10.1016/j.stemcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel D.E., Rakic P. Arrested proliferation of radial glial cells during midgestation in rhesus monkey. Nature. 1979;277:303–305. doi: 10.1038/277303a0. [DOI] [PubMed] [Google Scholar]
- Sherwood M.W., Arizono M., Panatier A., Mikoshiba K., Oliet S.H.R. Astrocytic IP(3)Rs: Beyond IP(3)R2. Front. Cell. Neurosci. 2021;15:695817. doi: 10.3389/fncel.2021.695817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Sloan S.A., Barres B.A. Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr. Opin. Neurobiol. 2014;27:75–81. doi: 10.1016/j.conb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Paşca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vicens M., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tchieu J., Calder E.L., Guttikonda S.R., Gutzwiller E.M., Aromolaran K.A., Steinbeck J.A., Goldstein P.A., Studer L. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat. Biotechnol. 2019;37:267–275. doi: 10.1038/s41587-019-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J., Wang M., Pimenova A.A., Bowles K.R., Hartley B.J., Lacin E., Machlovi S.I., Abdelaal R., Karch C.M., Phatnani H., et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 2017;9:600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K., Roux de Bézieux H., Street K., Saelens W., Cannoodt R., Saeys Y., Dudoit S., Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 2020;11:1201. doi: 10.1038/s41467-020-14766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A., Kettenmann H. Calcium signalling in glial cells. Trends Neurosci. 1996;19:346–352. doi: 10.1016/0166-2236(96)10048-5. [DOI] [PubMed] [Google Scholar]
- Wilton D.K., Dissing-Olesen L., Stevens B. Neuron-Glia signaling in synapse elimination. Annu. Rev. Neurosci. 2019;42:107–127. doi: 10.1146/annurev-neuro-070918-050306. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S.B., Li G., et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Chen L., Li Y., Huang M., Shao J., Li S., Cheng J., Yang H., Wu Y., Zhang J., et al. Rack1 is essential for corticogenesis by preventing p21-dependent senescence in neural stem cells. Cell Rep. 2021;36:109639. doi: 10.1016/j.celrep.2021.109639. [DOI] [PubMed] [Google Scholar]
- Zuchero J.B., Barres B.A. Glia in mammalian development and disease. Development. 2015;142:3805–3809. doi: 10.1242/dev.129304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video-microscopy of iPSC-derived astrocytes (day 50, LiPSC GR1.1) loaded with calcium dye (FLIPR Calcium 6 kit). Note the spreading of calcium transients across the cell culture.
Video-microscopy of iCell astrocytes (FUJIFILM CDI) loaded with calcium dye (FLIPR Calcium 6 kit). Note that these control astrocytes behave similarly to the iPSC-derived astrocytes shown in Video S1.
Data Availability Statement
Data are presented as the mean ± SD. Statistical analyses (GraphPad Prism) were performed using different tests as appropriate and as described in the figure legends.
Bulk RNA-seq and scRNA-seq, ATAC-seq, and MeDIP-seq FASTQ files have been deposited into the Sequence Read Archive (SRA) under BioProject PRJNA769413. The code for sequencing data analysis is available at https://github.com/cemalley/Jovanovic_methods and https://github.com/jaro-slamecka/Jovanovic_astrocyte_scRNA-seq. Public databases used include Gene Ontology 2019 (http://geneontology.org/), ARCHS4 Tissues (https://maayanlab.cloud/archs4/), and UCSC Cell Browser (https://cells.ucsc.edu/?ds=cortex-dev). Public SRA datasets used were PRJNA412090, PRJNA382448, PRJNA383243, and PRJNA297760.