Abstract
During the life cycle of hepatitis B virus (HBV), the large envelope protein (L) plays a pivotal role. Indeed, this polypeptide is essential for viral assembly and probably for the infection process. By performing mutagenesis experiments, we have previously excluded a putative involvement of the pre-S2 domain of the L protein in viral infectivity. In the present study, we have evaluated the role of the pre-S1 region in HBV infection. For this purpose, 21 mutants of the L protein were created. The entire pre-S1 domain was covered by contiguous deletions of 5 amino acids. First, after transfection into HepG2 cells, the efficient expression of both glycosylated and unglycosylated L mutant proteins was verified. The secretion rate of envelope proteins was modified positively or negatively by deletions, indicating that the pre-S1 domain contains several regulating sequences able to influence the surface protein secretion. The ability of mutant proteins to support the production of virions was then studied. Only the four C-terminal deletions, covering the 17 amino acids suspected to interact with the cytoplasmic nucleocapsids, inhibited virion release. Finally, the presence of the modified pre-S1 domain at the external side of all secreted virions was confirmed, and their infectivity was assayed on normal human hepatocytes in primary culture. Only a short sequence including amino acids 78 to 87 tolerates internal deletions without affecting viral infectivity. These results confirm the involvement of the L protein in the infection step and demonstrate that the sequence between amino acids 3 and 77 is involved in this process.
Serum from individuals infected by hepatitis B virus (HBV) contains distinct forms of viral particles. Most of them are spherical or filamentous particles of about 22 nm in diameter. These subviral particles consist of a single viral envelope and are therefore not infectious. However, it has been shown that these empty forms enhance the infectivity of duck HBV, a related hepadnavirus (2). The infectious agent is the less abundant form. The virions are spherical 42-nm-diameter particles. The viral DNA is enclosed in an icosahedral structure formed by the association of core proteins. The viral envelope, composed of cellular phospholipids and three virally encoded proteins, the small (S), middle (M), and large (L) polypeptides, surrounds this nucleocapsid. Translation of these hepatitis B surface (HBs) proteins is initiated at three different in-frame start sites within a single open reading frame (ORF) and is ended at a common termination codon (19). Because of this genetic organization, surface proteins are all related to each other by a shared glycosylated or unglycosylated region known as the S domain. The small protein is composed only of this S domain containing 226 amino acids, since it results from initiation at the farthest downstream start site. Initiation at the intermediate start codon leads to the synthesis of the M protein. It includes amino acids of the S protein extended by a 55-amino-acid domain (pre-S2 domain). Utilization of the farthest upstream initiation site generates the L protein, which carries a further 108-amino-acid extension (the pre-S1 domain of the ayw subtype) with respect to the M protein.
All these viral surface proteins are cotranslationally inserted into the lipid bilayer of the endoplasmic reticulum (ER). Apolar segments located in S region mediate their anchorage. The N-terminal extremities of the M and S proteins are immediately translocated in the ER lumen, allowing the carbohydrate modification of the additional glycosylation site located on the fourth amino acid of the pre-S2 domain (11–13). Conversely, a specific internal segment of the pre-S1 domain prevents early translocation of the pre-S region during L protein biogenesis (25). Indeed, in this initial translation product of protein, the pre-S domain remains in the cytosol, as attested by the lack of carbohydrate modification at the two potential sites located in the pre-S1 and pre-S2 regions. Part of the L protein population undergoes translocation only posttranslationally (6, 28, 35), and the other part keeps its initial conformation. Consequently, extracellular viral particles exhibit a mixed population of L protein, with their N-terminal pre-S domains located either inside or outside of the viral structure. This dual topology allows the L protein to potentially play different roles during the viral life cycle. Secretion of complete viral particles requires the isoform of the L protein with a cytoplasmic pre-S region (8). A stretch of amino acids, overlapping the pre-S1/pre-S2 regions, is thought to be involved in the interaction with cytosolic nucleocapsids before the budding event (3, 24, 32). Because of the external exposure of its pre-S domains, the second isoform may participate in the infection process, particularly in the binding to the putative cellular receptor.
Concerning HBV adsorption onto and penetration into the target cells, only few data on the contribution of viral envelope proteins have been reported. The M protein is probably not involved in viral infectivity (14, 24). By contrast, the in vitro infectious ability of HBV requires the presence of the myristate moiety of the L protein (5, 17). In a recent study, we have excluded most of the L protein pre-S2 region from playing a putative role in viral infectivity (24). By using cell-free systems and/or cell lines incompetent for HBV infection, several experiments suggested that the pre-S1 domain was probably important for viral attachment (27, 30, 31, 33).
In this study, we have investigated the involvement of the pre-S1 region in HBV infectivity by using an in vitro model of HBV infection. This model, based on the use of normal human hepatocytes in primary cultures, was developed in previous studies (15, 16). A set of deletions over the pre-S1 domain, was created and the ability of the modified proteins to substitute for the wild-type (WT) form in virion infectivity was evaluated.
MATERIALS AND METHODS
Plasmid engineering.
Plasmid pHBV-ΔEcoRI contains an HBV DNA insert of more than one genome length, starting at position 1232 and ending at position 1984 (17). This viral sequence contains only one copy of the pre-S1–pre-S2–S ORF and is able to support viral replication. Plasmid pHBV L− was derived from pHBV-ΔEcoRI by introduction of a point mutation to prevent L protein expression (24).
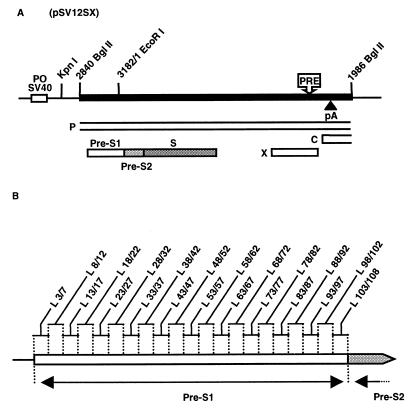
Plasmids have been engineered to express the L surface protein in WT or mutant forms. Plasmid pSV12SX (Fig. 1A) contains the 2,329-bp BglII-BglII fragment of WT HBV DNA, bearing the entire pre-S–S coding regions, cloned downstream of the simian virus 40 early promoter-origin region in plasmid pSV-SPORT 1 (Life Technologies). Short deletions were introduced into pSV12SX by a previously described method (24), and subsequent constructs were sequenced to confirm the expected deletion without other mutation (Fig. 1B).
FIG. 1.
(A) Expression vector of the L protein. The thick line indicates HBV sequence; the thin line indicates plasmid pSV-SPORT 1 sequences with its simian virus 40 early promoter-origin region (PO SV40); boxes indicate ORFs for viral X, P, C, and S proteins. The envelope ORF is divided into pre-S1, pre-S2, and S domains. The approximate locations of the posttranscriptional regulatory element (PRE) (20) and the polyadenylation site (pA) in the HBV sequence are shown. (B) Amino acid deletions in the pre-S1 region of the S gene cloned in different L protein expression plasmids. Deletions (L x/y, where x is the WT position of the N-terminal amino acid flanking the deletion and y is the WT position of the C-terminal amino acid flanking the deletion) are indicated above.
Cell line and transfection.
To produce viral proteins or virions, the permissive HepG2 human hepatoma cell line (1, 38) was transfected with HBV DNA by electroporation. HepG2 cells were cultured in H medium (75% minimum essential medium, 25% medium 199, 5 mg of insulin per liter, 4.5 mg of penicillin per liter, 50 mg of streptomycin per liter) supplemented with 3.5 × 10−7 M hydrocortisone hemisuccinate, 2 mM l-glutamine, and 10% fetal calf serum (FCS).
Virus purification.
HBV particles were isolated from the culture medium of transfected HepG2 cells by precipitation with 6% polyethylene glycol 8000 (PEG 8000; Sigma) for 12 h at 4°C. The precipitates were recovered by centrifugation (10,000 × g for 45 min at 4°C) and concentrated 200-fold in phosphate-buffered saline with 25% FCS.
Primary cell culture and infection.
Fragments of normal adult human liver were obtained from patients undergoing hepatic resection for liver metastases (the fragments were taken at a distance from the metastasis in macroscopically normal liver). Access to this biopsy material was in agreement with French laws and satisfied the requirements of the Ethics Committee of this institution. Hepatocytes were isolated by the procedure of Guguen-Guillouzo and Guillouzo (18) and cultured in H medium supplemented with 3.5 × 10−6 M hydrocortisone hemisuccinate, 2 mM l-glutamine, 50 mg of gentamicin per liter, 2% dimethyl sulfoxide, 5% adult human serum, and 5% FCS. At 3 days after seeding, the cells were infected as described previously (15). The hepatocytes (1.5 × 106 per 10-cm2 petri dish) were covered with 1 ml of serum-free culture medium containing 5% PEG 8000 and 100 μl of inoculum. Infection was performed for 12 h at 37°C. The cells were then washed three times with culture medium and subjected to a further culture.
Intra- and extracellular protein analysis.
Transfected cells and their supernatants were harvested 7 days after transfection. HepG2 cells were lysed and nuclei were removed before the immunoblotting analysis. Released viral particles were precipitated with 6% PEG 8000. Proteins were analyzed by electrophoresis through 12.5% polyacrylamide–sodium dodecyl sulfate gels and transferred onto a nitrocellulose filter (Amersham). Immunoblotting was performed by using enhanced chemiluminescence (Amersham) with primary monoclonal antibody F376 (26) at dilution of 1:5,000 and the secondary anti-mouse antibody linked to horseradish peroxidase at dilution of 1:25,000 (Jackson Immunoresearch Laboratories, Inc).
Assays for HBV-specific proteins.
HBs antigen was detected with a radioimmunoassay kit (Abbott Laboratories, Abbott Park, Ill.) under the conditions recommended by the manufacturer.
DNA extraction and analysis.
Intracellular nucleocapsids were isolated from the cytoplasmic fraction of transfected HepG2 cells as described previously (24). The core particles were immunoprecipitated with an anti-hepatitis B core (HBc) antibody (Dako).
Complete viral particles were immunoprecipitated from the supernatant of transfected HepG2 cells with a polyclonal anti-HBs antibody (Dako) or with a polyclonal antibody raised against a pre-S1 peptide (a generous gift of H. J. Hong).
The covalently closed circular form of HBV DNA (cccDNA) was extracted from human adult hepatocyte cultures by a previously described method (24).
All DNA was analyzed on 1.5% agarose gels. These gels were soaked in 0.25 N HCl for 15 min, and DNA was denatured in situ in 0.4 M NaOH and transferred onto positively charged nylon membranes (Amersham) by the Southern method (36). Hybridization was performed at 65°C with linearized HBV genomic [α-32P]DNA as a probe.
RESULTS
Experimental strategy.
Plasmid pHBV L−, containing an HBV DNA insert of more than one genome length, allowed the transcription of all known viral RNAs under the control of their own promoters. Only the L protein expression was suppressed by introducing an opal mutation in codon 90 of the pre-S1 region. This point mutation remained silent in the overlapping polymerase gene. This defect can be complemented in trans by cotransfecting pHBV L− with a vector expressing the missing WT protein (Fig. 1A) (4). To evaluate the role of the pre-S1 region of the L protein, 21 constructs, each carrying a defined mutation, were used as cotransfecting plasmids and the ability of the mutant proteins to replace the WT form in viral cycle was investigated (Fig. 1B).
Expression and secretion of the mutant proteins.
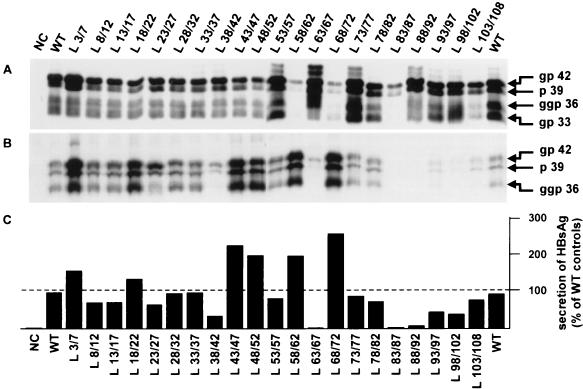
The WT and mutant L proteins were expressed in transiently transfected HepG2 cells. Intra- and extracellular viral proteins were analyzed by Western blotting. The use of a monoclonal anti-pre-S2 antibody revealed the presence of both the L and M proteins. Four main bands were detected in the lysates of cells transfected with WT protein expression vector (Fig. 2A). Consistent with the apparent molecular mass, the two lowest bands correspond to the 33-kDa glycosylated (gp33) and 36-kDa diglycosylated (ggp36) forms of the M protein. A very small amount of unglycosylated form (p30) was revealed when the exposure of the autoradiograph was prolonged. The unglycosylated (p39) and glycosylated (p42) forms of the L protein appeared as a doublet with molecular masses of 39 and 42 kDa, respectively. The identity of these viral surface proteins was confirmed by a deglycosylation experiment (data not shown). Analysis of supernatants of cells transfected with a WT plasmid (Fig. 2B) showed that both unglycosylated and glycosylated forms of the L protein were secreted while the diglycosylated form of the M protein was mainly exported into the medium. Furthermore, secreted HBs antigen (HBsAg) production was measured (Fig. 2C), and it was found to be actively produced by cells transfected with the WT plasmid.
FIG. 2.
Analysis of intra- and extracellular surface viral proteins. HepG2 cells were transfected with 20 μg of different L expression vectors: NC, plasmid without an HBV insert (negative control); WT, expression plasmid driving the synthesis of the WT L protein; L x/y, expression plasmids driving the synthesis of different mutant L proteins. (A and B) Proteins were extracted from cells (A) or precipitated from the culture medium (B) and studied by Western blotting. Samples were analyzed by electrophoresis through a 12.5% polyacrylamide–sodium dodecyl sulfate gel. The primary monoclonal antibody was directed against the pre-S2 region. gp 42, p 39, ggp 36, and gp 33 indicated the migration positions of the glycosylated and unglycosylated L proteins and the diglycosylated and glycosylated M proteins, respectively. (C) HBsAg, secreted by transfected HepG2 cells, was measured by a conventional radioimmunoassay in culture supernatants collected 6 days posttransfection. The data represent the percentage of secreted HBsAg compared to the average of the two WT controls.
All cells transfected with the different mutant plasmids contained L mutant proteins in their two forms (Fig. 2A). However, depending on the deletion, several differences were observed. We distinguished several groups of internal deletions in the pre-S1 region that were able to differently modify the behavior of L proteins and subviral particles for their secretion. While some mutations reduced the retention property of the L protein, other deletions enhanced it. Thus, deletions L3/7, L18/22, L43/47 and L48/52 facilitated the release of viral surface proteins without affecting the intracellular viral protein content. In the L3/7 mutant, the myristylation signal was removed, and the lack of the myristate moiety is known to be sufficient to induce a significant increase of secretion compared with that of WT protein (34). In this mutant, we also observed that the L protein was produced in a larger amount. Although reduced levels of L and M proteins were detected in cells transfected with the L58/62 and L68/72 mutants, their secretions were favored. Then their overall synthesis was probably not affected. Conversely, some deletions (L38/42, L63/67, and L83/87 to L98/102) severely reduced the secretion rate of L protein and the release of HBsAg. A decreased expression level of the M protein was also observed for two of these deletions (L63/67 and L83/87). A Northern blot analysis revealed that it was related to a reduced amount of the major surface antigen transcripts encoding M and S proteins (data not shown). The elimination of the CCAAT element, known as the cis sequence required for S promoter function, could explained this low steady-state RNA level in mutant L83/87 (40). In these two particular cases, impairment of the major surface antigen transcript level could contribute to the drop in secretion of viral surface proteins, since an excess of L with respect to M and S proteins is known to promote their intracellular retention (9, 29, 37). Otherwise, for unknown reasons, cells transfected with the L83/87 mutant contained a reduced amount of L modified protein. This was not due to a low transfection efficiency, since a Northern blot analysis did not show variation in the level of the large surface antigen transcripts (data not shown).
Western blots also revealed that the expression level of the M protein was partially impaired in the L103/108 mutant. The deletion is located just upstream of the start codon of the M protein. A change in the local sequence context of the initiation codon is probably responsible for a negative influence on the M protein translation. However, this mutation did not affect HBsAg production and export.
HBV particle production.
To initiate production of mutant virions, HepG2 cells were transiently cotransfected with 5 μg of the replication-competent L-defective genome together with 200 ng of different L protein expression vectors. During the early steps of virion morphogenesis, pregenomic RNA is encapsidated and serves as template for synthesis of the viral DNA by a reverse transcription mechanism. To verify whether these steps were not influenced in trans by the different deletions introduced in the pre-S1 region of the L protein, cytoplasmic core particles were selectively isolated 6 days after cotransfection and their genetic content was analyzed by the Southern blot procedure (Fig. 3A). Similar patterns were observed in the presence of the WT or mutant L proteins. In all cases, intracellular nucleocapsids preferentially contained a single-stranded viral DNA form. Comparison of the different samples showed no significant variation in the amount of encapsidated DNA. The slightly smaller amount detected for the L58/62 mutant was not observed in repeated experiments. This effect resulted from variation in the transfection efficiency.
FIG. 3.
Southern blot analysis of HBV DNA from intracellular core particles and from extracellular complete viral particles. HepG2 cells were transfected with the L-defective genome complemented with different L expression vectors. NC, plasmid without an HBV insert (negative control); WT, expression plasmid driving the synthesis of the WT L protein; L x/y, expression plasmids driving the synthesis of different mutant L proteins. (A) An anti-HBc antibody was used to immunoprecipitate cytoplasmic core particles from transfected cells. (B) Complete viral particles were immunoprecipitated with a polyclonal anti-HBs antibody from HepG2 cell supernatants collected between days 3 and 6 posttransfection. DNA was extracted from the immunoprecipitates and analyzed on a 1.5% agarose gel. Molecular size markers are indicated in kilobases to the left; the positions of relaxed-circular DNA (RC) and single-stranded DNA (SS) are shown to the right.
After the analysis of the encapsidation step, we evaluated the effect of deletions on viral particle secretion. Supernatants were collected between days 3 and 6 posttransfection and were pooled. Estimation of extracellular HBsAg showed that it was actively secreted by cells transfected with L-defective genome either uncomplemented or complemented with the WT or mutant L protein expression plasmids (data not shown).
We next looked for complete viral particles into the supernatants of transfected cells. For this purpose, a polyclonal anti-HBs antibody was used to immunoprecipitate extracellular viral particles and then viral DNA was visualized by the Southern blot procedure (Fig. 3B). As expected, no virion was secreted in the absence of L protein (negative control), but complementation of the L-defective genome with a WT L protein expression vector restored virion secretion (4). All mutant proteins with an internal deletion between amino acids 3 and 87 were also able to complement the L defect for virion export (mutants L3/7 to L83/87). Conversely, the last four deletions located at the C-terminal extremity of the pre-S1 region totally prevented the secretion of complete viral particles (mutants L88/92 to L103/108).
To verify that the pre-S1 domain was presented at the outer face of virions, an immunoprecipitation was performed with a polyclonal anti-pre-S1 antibody. All mutant virions, which were assembled, were efficiently immunoprecipitated. This proved that they exhibited the pre-S1 domain at their surface (data not shown).
Infectivity of the mutant virions.
After verifying that the original topology of the L mutant proteins was preserved in the mutant viruses, we evaluated their infectious capacity. Mutant virions were concentrated from supernatants of cotransfected cells. Viral infectivity was tested by incubation of human hepatocytes in primary culture with the different inocula. It is important to note that all mutant viruses contain the same replication-competent L-defective genome. Thus, if this modified genome is delivered into hepatocytes by an infectious mutant virus, viral replication will be initiated. Only the assembly process will be inhibited, since no new L protein could be synthesized.
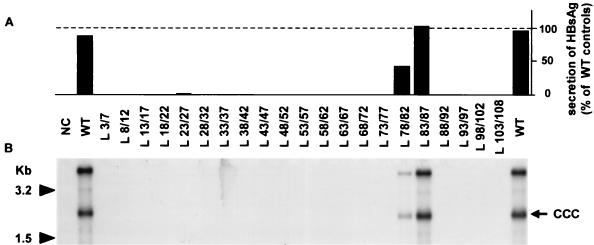
To identify infectious mutant viruses, long-term HBsAg secretion (10 days postinfection) into the culture medium was monitored (Fig. 4A). Like hepatocytes infected with WT virus, those infected with mutant viruses containing either L78/82 or L83/87 proteins secreted HBsAg. No HBsAg secretion was observed for any of the other mutants. These observations suggest that only two types of mutant virus are infectious.
FIG. 4.
Infectivity of complete viral particles containing mutant pre-S1 proteins. Hepatocytes were incubated with concentrated supernatants obtained from HepG2 cells transfected with L-defective genome complemented with different L expression vectors: NC, plasmid without an HBV insert (negative control); WT, expression plasmid driving the synthesis of the WT L protein; L x/y, expression plasmids driving the synthesis of different mutant L proteins. (A) HBsAg, secreted by human hepatocytes following in vitro infection assays, was measured by a conventional radioimmunoassay in hepatocyte primary culture supernatants collected 10 days postinfection. The data represent the percentage of secreted HBsAg compared to the average of the two WT controls. (B) Southern blot analysis of HBV cccDNA in human hepatocytes following in vitro infection assays. Supercoiled viral DNA was selectively extracted from hepatocytes collected 10 days postinfection and analyzed on a 1.5% agarose gel. Molecular size markers are indicated in kilobases to the left; the position of cccDNA is shown to the right (CCC).
After entry into host cell, the relaxed circular DNA contained in infectious virus is converted into supercoiled DNA in the nucleus. Detection of cccDNA in hepatocytes therefore represents reliable proof of viral infectivity. To further confirm the results supplied by HBsAg secretion, we investigated the presence of cccDNA in hepatocytes (Fig. 4B). While this form was found in cells infected by WT virus or L78/82 or L83/87 mutant virions, it was absent in hepatocytes exposed to mutant viral particles with one of the different mutant L proteins, in which deletion was located between amino acids 3 and 77. As expected, both HBsAg secretion and cccDNA detection were negative when cultures were exposed to the supernatants of HepG2 cells containing no virus (L88/92 to L103/108 mutants).
DISCUSSION
In a previous work, we have analyzed the involvement of the pre-S2 region of L protein during the HBV cycle (24). In this region, only the N-terminal 5 amino acids are essential for viral assembly. All the other amino acids are dispensable for both assembly and infectivity. Site-directed mutagenesis was used again in an attempt to study the relevance of the pre-S1 domain to viral infectivity.
The genetic approach we used was based on generation of mutant virions with different contiguous deletions in the L protein pre-S1 domain. This method required several preliminary controls, such as stable expression of the modified proteins. Biosynthesis investigations of the L proteins with deletions showed that all of them were synthesized in their two major forms, unglycosylated and glycosylated. This normal glycosylation state strongly argues for a WT transmembrane topology, even when some deletions (L68/72 to L93/97) overlap the translocation repression signal, which controls the topology of L protein. This was not surprising, since a longer internal deletion in this signal, ranging between amino acids 70 and 94, does not influence the transmembrane topology (25).
We next examined the secretory phenotype of the mutant L proteins. Whereas some internal deletions reinforced the retention property of the protein, others improved their export. Our internal deletions allowed us to identify short domains that were able to positively (amino acids 3 to 7, 18 to 22, 43 to 52, 58 to 62, and 68 to 72) or negatively (amino acids 38 to 42 and 88 to 102) influence the release of surface proteins. Until now, these small amino acid areas with opposite features were not identified, probably because the larger deletions or truncations, used to determine domains governing retention, include domains which regulate the viral surface protein secretion differently. Furthermore, large deletions could affect the crucial transmembrane topology. Although it is likely that small deletions altered the overall folding of the protein less than the largest deletions did, we cannot exclude the possibility that our mutations distantly disturb, in cis, another region(s) regulating the intracellular retention. Two other deletions (L63/67 and L83/87) inhibit the secretion rates of viral surface proteins. In these two cases, this was explained by the decreased amounts of transcripts under the control of the S promoter. However, it is possible that the corresponding amino acids influence the retention property of the L surface protein.
As demonstrated by different mutagenesis experiments, virion morphogenesis required the 17 C-terminal amino acids of the pre-S1 region and the 5 N-terminal amino acids of the pre-S2 domain of the L protein, exposed at the cytosolic face of the ER membrane (3, 7, 8, 24). Four of our mutations overlapped this assembly region and, as expected, were sufficient to prevent the secretion of complete viral particles. Nevertheless, all the other L mutant proteins could substitute for the WT form in extracellular virion release. This assembly ability is another evidence of the normal transmembrane topology of these L mutant proteins.
To potentially ensure contact(s) with the host cell surface, the accessibility of the pre-S1 domain at the surface of mutant virions must be verified. Like WT virus, all assembled mutant virions fulfill this requirement, as demonstrated by their efficient immunoprecipitation with a polyclonal antibody specific for the pre-S1 region. Despite this structural uniformity, most of the modified viruses were unable to initiate viral replication in human hepatocytes after their inoculation. However, two mutant viruses infected cells. Their deletions, located just upstream of the assembly region, are contiguous and delimit a region of 10 amino acids dispensable for the infection process (amino acids 78 to 87). Concerning the deletion of amino acids 3 to 7, the deleterious impact on infection could be ascribed to the destruction of the myristylation signal, since this posttranslational modification of the L protein is absolutely necessary for HBV infectivity (5, 17). The minimal myristylation signal was restricted to the first seven or nine N-terminal residues (21–23, 39); therefore our deletions located beyond these amino acids (L13/17 to L103/108) do not prevent the covalent linkage of myristate. Moreover, in previous studies (17), we have shown that the absence of myristylation changes both the number of secreted mature viral particles, which was increased, and the viral DNA pattern, which preferentially contained low-molecular-weight DNA. These features were found again for the mutant virion with a deletion between amino acids 3 and 7 but not for the mutant virion with an L protein deleted between amino acids 8 and 12 (Fig. 3B, compare lanes L3/7 and L8/12 with lanes WT). It is most likely that the myristylation was not altered by the second deletion. Consequently, the loss of infectivity of the 14 mutant virions (L8/12 to L73/77) could be attributed to the primary structure of the proteinic region. Otherwise, we cannot reject the possibility that amino acids that contribute to viral assembly also play a role during infection.
To date, several studies have provided controversial data about the domain(s) of viral surface proteins implicated in the attachment and penetration of HBV. Some of them suggested that the pre-S1 region is probably crucial for the early events of infection (for a review, see reference 10). However, the experiments were based on the use of either cell-free systems or cell lines refractory to HBV infection. In this study, by using normal human hepatocytes permissive for HBV infection (15, 16), we define a precise region in the pre-S1 domain of the L protein that is crucial for the infection step, probably involved in binding and/or internalization.
ACKNOWLEDGMENTS
This work was supported by INSERM, the Association pour la Recherche contre le Cancer, and the Ligue Nationale contre le Cancer (Comité d’Ille et Vilaine). Jacques Le Seyec and Philippe Chouteau are recipients of fellowships from the Association pour la Recherche contre le Cancer and from the Ligue Nationale contre le Cancer (Comité des Côtes d’Armor), respectively.
We are indebted to Pascal Loyer and Mickaël Rialland for helpful criticism of the manuscript. We gratefully acknowledge Agatha Budkowska for the gift of the F376 antibody and Hyo Jeong Hong for the gift of the pre-S1 peptide.
REFERENCES
- 1.Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruns M, Miska S, Chassot S, Will H. Enhancement of hepatitis B virus infection by noninfectious subviral particles. J Virol. 1998;72:1462–1468. doi: 10.1128/jvi.72.2.1462-1468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss V, Hagelsten J, Gerhardt E, Galle P R. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology. 1996;218:396–399. doi: 10.1006/viro.1996.0209. [DOI] [PubMed] [Google Scholar]
- 6.Bruss V, Lu X Y, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss V, Thomssen R. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J Virol. 1994;68:1643–1650. doi: 10.1128/jvi.68.3.1643-1650.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chisari F V, Filippi P, McLachlan A, Milich D R, Riggs M, Lee S, Palmiter R D, Pinkert C A, Brinster R L. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880–887. doi: 10.1128/jvi.60.3.880-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Meyer S, Gong Z J, Suwandhi W, van Pelt J, Soumillion A, Yap S H. Organ and species specificity of hepatitis B virus (HBV) infection: a review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J Viral Hepat. 1997;4:145–153. doi: 10.1046/j.1365-2893.1997.00126.x. [DOI] [PubMed] [Google Scholar]
- 11.Eble B E, Lingappa V R, Ganem D. Hepatitis B surface antigen: an unusual secreted protein initially synthesized as a transmembrane polypeptide. Mol Cell Biol. 1986;6:1454–1463. doi: 10.1128/mcb.6.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eble B E, Lingappa V R, Ganem D. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol. 1990;64:1414–1419. doi: 10.1128/jvi.64.3.1414-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eble B E, MacRae D R, Lingappa V R, Ganem D. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol Cell Biol. 1987;7:3591–3601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernholz D, Galle P R, Stemler M, Brunetto M, Bonino F, Will H. Infectious hepatitis-B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology. 1993;194:137–148. doi: 10.1006/viro.1993.1243. [DOI] [PubMed] [Google Scholar]
- 15.Gripon P, Diot C, Guguen-Guillouzo C. Reproducible high-level infection of cultured adult human hepatocytes by hepatitis B virus—effect of polyethylene glycol on adsorption and penetration. Virology. 1993;192:534–540. doi: 10.1006/viro.1993.1069. [DOI] [PubMed] [Google Scholar]
- 16.Gripon P, Diot C, Theze N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol. 1988;62:4136–4143. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292–299. doi: 10.1006/viro.1995.0002. [DOI] [PubMed] [Google Scholar]
- 18.Guguen-Guillouzo C, Guillouzo A. Methods for preparation of adult and fetal hepatocytes. In: Guillouzo A, Guguen-Guillouzo C, editors. Isolated and cultured hepatocytes. London, United Kingdom: Les éditions INSERM Paris. John Libbey and Co, Ltd.; 1986. pp. 1–12. [Google Scholar]
- 19.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z M, Yen T S. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamps M P, Buss J E, Sefton B M. Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc Natl Acad Sci USA. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan J M, Mardon G, Bishop J M, Varmus H E. The first seven amino acids encoded by the v-src oncogene act as a myristylation signal: lysine 7 is a critical determinant. Mol Cell Biol. 1988;8:2435–2441. doi: 10.1128/mcb.8.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J Virol. 1998;72:5573–5578. doi: 10.1128/jvi.72.7.5573-5578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffler-Mary H, Werr M, Prange R. Sequence-specific repression of cotranslational translocation of the hepatitis B virus envelope proteins coincides with binding of heat shock protein Hsc70. Virology. 1997;235:144–152. doi: 10.1006/viro.1997.8689. [DOI] [PubMed] [Google Scholar]
- 26.Neurath A R, Adamowicz P, Kent S B, Riottot M M, Strick N, Parker K, Offensperger W, Petit M A, Wahl S, Budkowska A, et al. Characterization of monoclonal antibodies specific for the pre-S2 region of the hepatitis B virus envelope protein. Mol Immunol. 1986;23:991–997. doi: 10.1016/0161-5890(86)90130-6. [DOI] [PubMed] [Google Scholar]
- 27.Neurath A R, Kent S B H, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 28.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persing D H, Varmus H E, Ganem D. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science. 1986;234:1388–1391. doi: 10.1126/science.3787251. [DOI] [PubMed] [Google Scholar]
- 30.Petit M A, Dubanchet S, Capel F, Voet P, Dauguet C, Hauser P. HepG2 cell binding activities of different hepatitis B virus isolates: inhibitory effect of anti-HBs and anti-preS1(21–47) Virology. 1991;180:483–491. doi: 10.1016/0042-6822(91)90062-g. [DOI] [PubMed] [Google Scholar]
- 31.Petit M A, Strick N, Dubanchet S, Capel F, Neurath A R. Inhibitory activity of monoclonal antibody F35.25 on the interaction between hepatocytes (HepG2 cells) and preS1-specific ligands. Mol Immunol. 1991;28:517–521. doi: 10.1016/0161-5890(91)90166-h. [DOI] [PubMed] [Google Scholar]
- 32.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 33.Pontisso P, Ruvoletto M G, Gerlich W H, Heermann K H, Bardini R, Alberti A. Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology. 1989;173:522–530. doi: 10.1016/0042-6822(89)90564-3. [DOI] [PubMed] [Google Scholar]
- 34.Prange R, Clemen A, Streeck R E. Myristylation is involved in intracellular retention of hepatitis B virus envelope proteins. J Virol. 1991;65:3919–3923. doi: 10.1128/jvi.65.7.3919-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 37.Standring D N, Ou J H, Rutter W J. Assembly of viral particles in Xenopus oocytes: pre-surface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc Natl Acad Sci USA. 1986;83:9338–9342. doi: 10.1073/pnas.83.24.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sureau C, Romet-Lemonne J L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 39.Towler D, Gordon J, Adams S, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- 40.Zhou D X, Yen T S. The hepatitis B virus S promoter comprises a CCAAT motif and two initiation regions. J Biol Chem. 1991;266:23416–23421. [PubMed] [Google Scholar]