Abstract
To investigate the mechanism of kinetochore-associated protein 1 (KNTC1) in hepatocellular carcinoma. To query the TCGA database for KNTC1 expression in hepatocellular carcinoma. Detection of protein and mRNA levels of KNTC1 in hepatocellular carcinoma cell lines SK-Hep-1, Huh7, HepG2 and SNU449. Cell proliferation, migration and invasion ability were examined after KNTC1 knockdown in SK-Hep-1 and Huh7. Proteins related to KNTC1 were identified through protein interregulation, and their role in hepatocellular carcinoma was investigated. Our results showed that KNTC1 was significantly upregulated in hepatocellular carcinoma tissues and was associated with poorer prognostic survival. The expression of KNTC1 in hepatocellular carcinoma cell lines SK-Hep-1, Huh7, HepG2 and SNU449 was significantly higher than that in normal hepatocyte line L02. Knockdown of KNTC1 in SK-Hep-1 and Huh7 significantly inhibited cell viability, migration ability and invasion ability. KNTC1 is involved in the regulation of hepatocellular carcinoma through its interaction with cyclin-dependent kinase 1 (CDK1). Knockdown of KNTC1 inhibited CDK1 expression, while CDK1 overexpression was able to rescue the regulation of KNTC1 on the viability, migration and invasive ability of hepatocellular carcinoma cell lines. Knockdown of KNTC1 was found to resulted a cell cycle arrest at the S-phase, potentially through the modulation of CDK1, leading to decreased migration and invasion of hepatocellular carcinoma cells. Moreover, knockdown of KNTC1 in mouse transplanted tumors significantly inhibits tumor growth. Inhibition of high expression of KNTC1 in hepatocellular carcinoma was effective in suppressing the progression of hepatocellular carcinoma cells after knockdown. It may be a potential target for the treatment of hepatocellular carcinoma.
Keywords: KNTC1, CDK1, Liver cancer, Cell cycle arrest
Introduction
Liver cancer is one of the leading causes of cancer-related death (Siegel et al. 2019). Unfortunately, the prognosis for liver cancer is generally poor, with surgical resection being an option for only 5% to 15% of patients in the early stages (Starley et al. 2010). Currently, the main treatments for more advanced stages of liver cancer are trans-arterial chemoembolization or oral sorafenib (El-Serag et al. 2008). However, drug resistance and limited efficacy have made these approaches less effective. Therefore, the exploration of new targets, technologies, and drugs for liver cancer is critical and urgently needed (Anwanwan et al. 2020).
KNTC1 (Kinetochore-associated protein 1) is a subunit of the mitotic protein complex that is highly conserved throughout evolution, and it is essential for spindle assembly and chromosome segregation (Cheeseman 2014). As a key component of the mitotic checkpoint, KNTC1 is involved in multiple mechanisms that ensure proper chromosome segregation during mitosis and prevent premature mitotic arrest (Cheeseman 2014). A variety of proteins that regulate mitosis are overexpressed in human malignancies relative to normal tissue, some of which are oncogenes. In various human malignancies, KNTC1 is differentially expressed or associated with survival. For instance, KNTC1 is highly expressed in esophageal squamous cell carcinoma, and knocking it down suppresses cell viability and induces apoptosis in esophageal squamous cell carcinoma (Liu et al. 2019; Holland and Cleveland 2009). In the bladder, knockdown of KNTC1 inhibits cancer cell proliferation, migration and tumorigenesis in vivo (Huang et al. 2021). Moreover, in colon cancer cells, KNTC1 knockdown dramatically increases apoptosis (Zhengxiang et al. 2021), whereas in cervical cancer cells, KNTC1 knockdown impairs their invasive and migratory capacity (Wang et al. 2022). However, the molecular mechanism of KNTC1 in the growth and metastasis of hepatocellular carcinoma cells is poorly understood. A recent study found that knockdown of KNTC1 inhibited the proliferation and metastasis of hepatocellular carcinoma cells and induced apoptosis through inhibition of the PI3K/Akt pathway (Tong et al. 2023). Nevertheless, the precise role of KNTC1 in the growth and metastasis of hepatocellular carcinoma requires additional research.
Here, we confirmed that KNTC1 expression was upregulated in hepatocellular carcinoma tissues as well as in hepatocellular carcinoma cell lines. Knockdown of KNTC1 induces S-phase cell cycle arrest and thus inhibits tumor growth, primarily through the modulation of CDK1.
Materials and methods
Cell culture
The normal human liver L02 and hepatocellular carcinoma cell lines SK-Hep-1, Huh7, HepG2 and SNU449 were obtained from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. All cells were cultivated in Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) containing 10% FBS (HyClone), 1% penicillin/streptomycin at 37 °C in a 5% CO2 incubator.
Cell transfection
KNTC1 knockdown was achieved by the transfection of shKNTC1 (GenePharma). CDK1 overexpression was achieved by the transfection of the pcDNA3.1-CDK1 plasmid vector (ov-CDK1, GenePharma). Plasmids were transfected with Lipofectamine 3000 (Invitrogen).
Cell viability assay
Once the cells reached the specified culture time, 10 μl CCK-8 (Beyotime Biotechnology, Shanghai, China) was added into each well and cultured in an incubator for 2 h. The absorbance was measured at 450 nm by the Eon™ Microplate Reader (BioTek, Whiting, VT, USA).
Cell proliferation assay
The cultured cells were resuspended in 500 µl of medium and inoculated with 24-well plates (2 × 104 cells/well), and incubated until the cells reached an appropriate growth status. After removing the medium, add medium containing 50 μM EdU to each well and incubate for 2 h. The EdU medium was discarded, and the cells were washed twice with PBS (5 min/ session). After this, the cells were fixed (4% paraformaldehyde) for 30 min, permeabilized (0.5% Triton X-100) for 10 min and then stained (Apollo dye solution) for 30 min. Images were acquired by using the OBSERVER D1/AX10 cam HRC microscope (Zeiss, Oberkochen, Germany) with a charge-coupled device (CCD) camera.
Cell cycle assay
After the plasmid was transfected into SK-Hep-1 and Huh7 cells, digested the cells and resuspended in medium, then incubated cells in 6-well plates for 24 h. And then collected and washed in PBS. After centrifugation, cells were resuspended in PBS (300 μl) containing 5% BSA and fixed for 24 h in 700 μl ethanol, then stained with propidium iodide for 30 min. Cell cycle distribution was analyzed using a flow cytometry on the FACSCalibur system (Becton Dickinson, San Jose, CA, USA).
Wound healing assay
After transfecting SK-Hep-1 and Huh7 cells with the plasmids, resuspension of cells to 1 × 106 cells/ml. Then, 1 ml/well of the cell suspension was added to a 6-mm Petri dish containing 4 ml of serum-free DMEM. After 12 h in culture, make a scratch with a pipette tip. Record the width of the scratch slit. Cells are incubated for 48 h; then, the width of the scratch slit is recorded again and photographed.
Cell invasion assay
After transfecting SK-Hep-1 and Huh7 cells with the appropriate plasmids, they were resuspended to 5 × 104 cells/ml. Then, 200 μl/well of suspension was added to the upper chamber, and 500 μl of DMEM (with 10% FBS) supplemented was added to the lower chamber. After culturing for 24 h, the cells were washed 2 ~ 3 times with PBS and then fixed with 95% alcohol. Then stained with crystalline violet, observed and photographed (Nikon, Tokyo, Japan).
RNA extraction and real-time quantitative PCR
Total RNA was extracted by Trizol regent, (Invitrogen, CA, USA), cDNA was synthesized (HiScript II Reverse Transcriptase, Vazyme, Nanjing, China) after quality control, followed by Real-time quantitative PCR (ChamQ™ SYBR@ qPCR Master Mix, Vazyme, Nanjing, China), and expression was calculated by the 2−ΔΔCT method.
Western blot analysis
Total protein was extracted by strong RIPA (Beyotime Biotechnology, Shanghai, China) lysis, electrophoresed by SDS-PAGE, transferred (Millipore, Massachusetts, USA), blocked, incubated with primary and secondary antibodies, and the grayscale values of the bands were counted after ECL (4A Biotech, Beijing, China) visualization.
In vivo experiments
The BALB/c nude mice (male, 6 weeks old) were obtained from Model Animal Research Center of Nanjing University. Firstly, the knockdown vector against KNTC1 was transfected into Huh7 cells. After 48 h, the transfected Huh7 cells were injected subcutaneously into nude mice. Tumor volumes were measured every 3 days. 28 days after the injection, the tumors were resected from the mice and then weighed. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Central Hospital of Wuhan (P.R. China).
Statistical analysis
All data were presented as mean ± standard deviation (SD). The comparison between two groups was performed by two-tailed paired Student's t test, while the comparison in multiple groups (at least three groups) was conducted by one-way analysis of variance (ANOVA). GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA) was applied to produce statistical graphs. P values < 0.05 were considered statistically significant.
Results
Up-regulation of KNTC1 expression in HCC
To identify changes in KNTC1 expression in HCC patients in the clinical, we queried the CGA-HCC database for KNTC1 levels and found that KNTC1 transcript was significantly increased in HCC compared to non-tumor liver tissue (Fig. 1A). Then, we wondered whether KNTC1 expression affected the survival rate of HCC, and we compared the TCGA-HCC data with high versus low/medium expression of KNTC1 and found that HCC samples with high KNTC1 expression had significantly lower survival rates (Fig. 1B). Considering the high expression of KNTC1 in HCC clinical samples, we speculate that KNTC1 may also be highly expressed in hepatocellular cancer cell lines. We examined protein expression in SK-Hep-1, Huh7, HepG2 and SNU449 hepatocellular cancer cells and found that KNTC1 protein expression was significantly increased compared to human normal hepatocytes L02 (Fig. 1C, left). By qRT-PCR, we found that the mRNA expression level of KNTC1 was also significantly increased in these hepatocellular carcinoma cell lines, with the greatest increase in KNTC1 mRNA in SK-Hep- and Huh7 cells (Fig. 1C, right). These results suggest that KNTC1 is upregulated in the HCC.
Fig. 1.
A TCGA-HCC (Hepatocellular Carcinoma) database for KNTC1 transcript levels in liver cancer tissues (n = 371) and normal tissues (n = 50). B The Kaplan–Meier survival curve of high (n = 91) versus low/medium (n = 274) expression of KNTC1 and in HCC patients. C Increased expression of KNTC1 in hepatocellular carcinoma cells (SK-Hep-1, Huh7, HepG2 and SNU449) compared with human normal liver cells (L02). Left, western blot, right, qRT-PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Knockdown of KNTC1 inhibits the growth of hepatocellular carcinoma cells
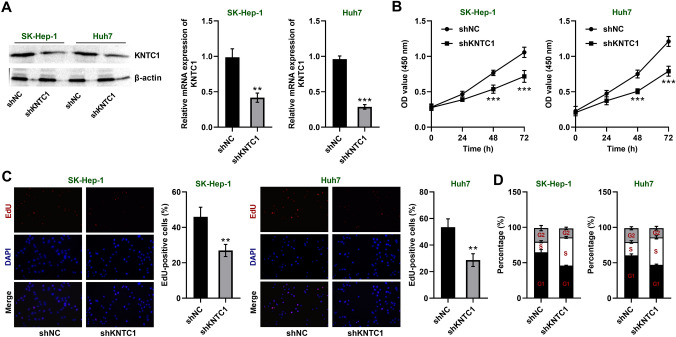
We knocked down KNTC1 in SK-Hep-1 and Huh7 cells to see if it would inhibit the growth of hepatocellular carcinoma cells. We first determined that we had successfully knocked down KNTC1 levels in SK-Hep-1 and Huh7 cells by identifying changes in protein and mRNA (Fig. 2A). We then examined the cell viability by CCK-8 assay and found that knockdown of KNTC1 decreased the proliferative viability of SK-Hep-1 and Huh7 cells (Fig. 2B). We also found that the proliferation ability of SK-Hep-1 and Huh7 cells was significantly reduced after knockdown of KNTC1 by EdU fluorescent labeling (Fig. 2C). In addition, we detected cell cycling by flow cytometry and found that SK-Hep-1 and Huh7 cells were more arrested in S-phase after knockdown of KNTC1 (Fig. 2D). These results suggest that knockdown of KNTC1 inhibited the growth of hepatocellular carcinoma cells.
Fig. 2.
A KNTC1 was knocked down in SK-Hep-1 and Huh7 cells. Left, western blot, right, qRT-PCR. B Knockdown of KNTC1 inhibits cell viability of SK-Hep-1 and Huh7 cells (CCK-8). C Knockdown of KNTC1 inhibits cell proliferation of SK-Hep-1 and Huh7 cells (EdU). D Knockdown of KNTC1 leads to S-phase cell cycle arrest. shNC, Negative control of knock down; shKNTC1, knock down of KNTC1. **, P < 0.01; ***, P < 0.001
Knockdown of KNTC1 inhibits metastasis of hepatocellular carcinoma cells
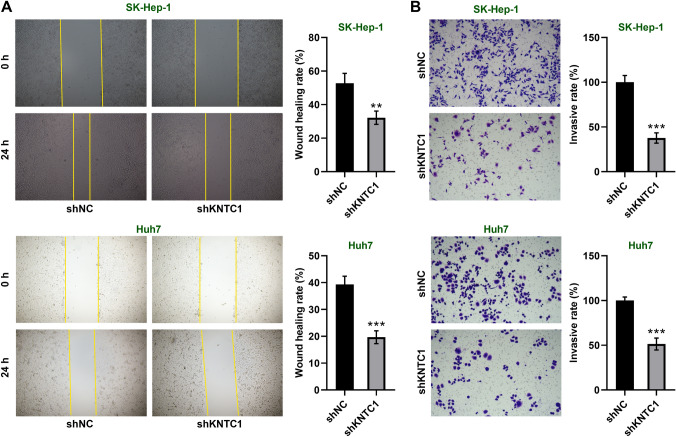
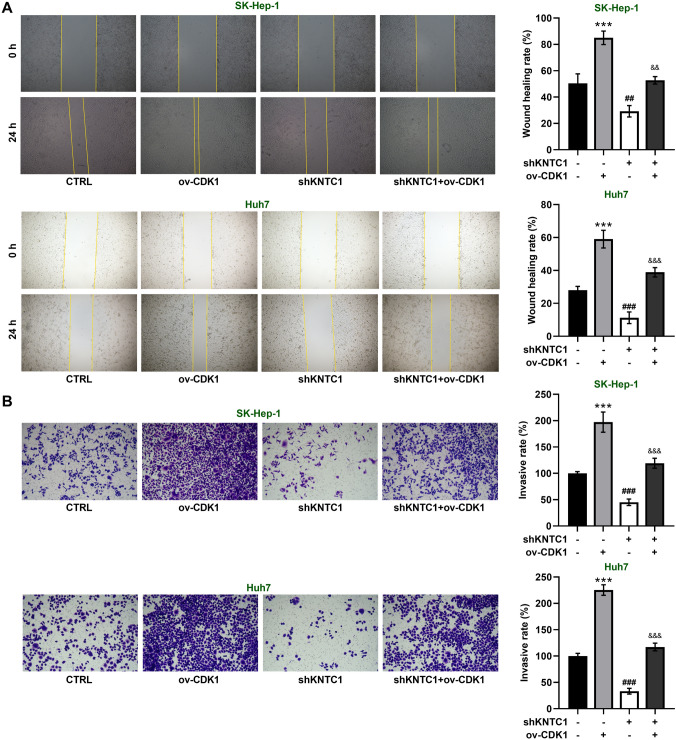
We examined the migration ability of hepatocellular carcinoma cells by scratch assay. We found that knockdown of KNTC1 significantly reduced the migratory ability of SK-Hep-1 and Huh7 cells (Fig. 3A). In addition, we determined the invasive ability of hepatocellular carcinoma cells by transwell assay. The invasive ability of SK-Hep-1 and Huh7 cells was significantly reduced after knockdown of KNTC1 (Fig. 3B). These results indicate that knockdown of KNTC1 inhibits the migration and invasive ability of hepatocellular carcinoma cells.
Fig. 3.
A Knockdown of KNTC1 inhibits cell migration of SK-Hep-1 and Huh7 cells. B Knockdown of KNTC1 inhibits cell invasion of SK-Hep-1 and Huh7 cells. shNC, Negative control of knock down; shKNTC1, knock down of KNTC1. **, P < 0.01; ***, P < 0.001
KNTC1 upregulates CDK1 expression in hepatocellular carcinoma cells
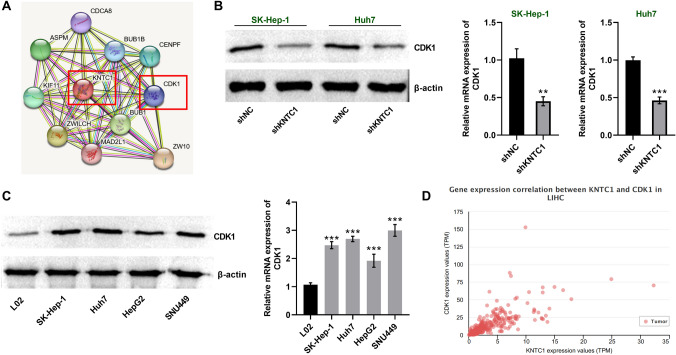
To investigate the mechanisms by which KNTC1 affects hepatocellular carcinoma, we performed predictions of proteins that interrelate with KNTC1 by using the STRING database. The results showed that CDK1 has interaction with KNTC1 (Fig. 4A). We showed that knockdown of KNTC1 in SK-Hep-1 and Huh7 cells significantly reduced the protein as well as mRNA levels of CDK1. This indicates that KNTC1 positive correlation with the expression of CDK1 (Fig. 4B). Next, we examined CDK1 expression in the hepatoma cell lines of SK-Hep-1, Huh7, HepG2 and SNU449 and found that both protein levels and mRNA levels of CDK1 were significantly increased compared to normal hepatocytes (Fig. 4C). Meanwhile, we found that KNTC1 was regulating CDK1 levels in HCC through TCGA database analysis (Fig. 4D). These results suggest that KNTC1 may positively regulate the levels of CDK1 in hepatocellular carcinoma cells, and both proteins are highly expressed.
Fig. 4.
A STRING (https://string-db.org/) predicts the presence of interactions between CDK1 and KNTC1. B Knockdown of KNTC1 repressed CDK1 expression in SK-Hep-1 and Huh7 cells. Left, western blot, right, qRT-PCR. C Elevated expression of CDK1 in hepatocellular carcinoma cells. Left, western blot, right, qRT-PCR. D KNTC1 positive correlation with CDK1 expression in TCGA-HCC. shNC, Negative control of knock down; shKNTC1, knock down of KNTC1. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Inhibition of hepatocellular carcinoma cell growth by CDK1 rescue by KNTC1 knockdown
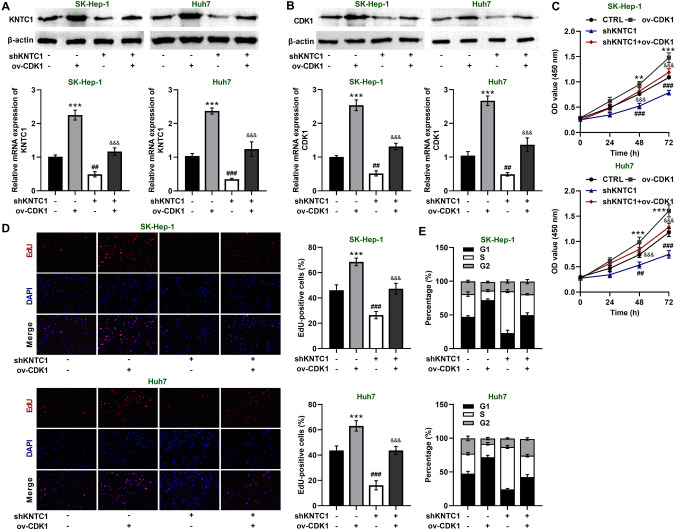
To investigate the role of CDK1 in HCC, we knocked down KNTC1 while overexpressing CDK1 in SK-Hep-1 and Huh7 cells and found that overexpression of CDK1 resulted in significant upregulation of KNTC1 as well as CDK1 protein and mRNA levels (Fig. 5A, B). In addition, we found that CDK1 overexpression rescued the decrease in cell viability as well as proliferative capacity caused by KNTC1 knockdown (Fig. 5C, D). We also identified that CDK1 overexpression rescued the cell cycle arrest caused by KNTC1 knockdown (Fig. 5E). These results imply that CDK1 overexpression can rescue the changes in hepatocellular carcinoma cells caused by KNTC1 knockdown.
Fig. 5.
A Expression of KNTC1 in SK-Hep-1 and Huh7 cells after transfection with shKNTC1 or/and ov-CDK1. B Expression of CDK1 in SK-Hep-1 and Huh7 cells after transfection with shKNTC1 or/and ov-CDK1. C CDK1 overexpression rescues the inhibition of cell viability by silencing KNTC1. D CDK1 overexpression rescues the inhibition of cell proliferation by silencing KNTC1. E KNTC1 knockdown leads to S-phase cell cycle arrest, while CDK1 overexpression promotes cell cycle progression. shKNTC1, knock down of KNTC1. ov, over expression. **P < 0.01, ***P < 0.001, shNC vs. ov-CDK1; ##P < 0.01, ###P < 0.001, snNC vs. shKNTC1; &&& P < 0.001, shKNTC1 vs. shKNTC1 + ov-CDK1
KNTC1 knockdown inhibits hepatocellular carcinoma cell metastasis through downregulation of CDK1
Through scratch assays, we found that CDK1 overexpression resulted in a significant increase in cell migration capacity of SK-Hep-1 and Huh7 cells, with or without inhibition of KNTC1 (Fig. 6A). While KNTC1 knockdown significantly inhibited the invasive ability of SK-Hep-1 and Huh7 cells, CDK1 overexpression increased the invasive ability of SK-Hep-1 and Huh7 cells with or without KNTC1 inhibition (Fig. 6B). These results suggest that knockdown of KNTC1 on the metastasis of hepatocellular carcinoma cells mediated by CDK1.
Fig. 6.
A CDK1 overexpression rescues the inhibition of cell migration by silencing KNTC1. B CDK1 overexpression rescues the inhibition of cell invasion by silencing KNTC1. shKNTC1, knock down of KNTC1. ov, over expression. ***P < 0.001, CTRL vs. ov-CDK1; ##P < 0.01, ###P < 0.001, CTRL vs. shKNTC1; && P < 0.01, &&& P < 0.001, shKNTC1 vs. shKNTC1 + ov-CDK1
Down-regulation of KNTC1 inhibits tumor growth in vivo
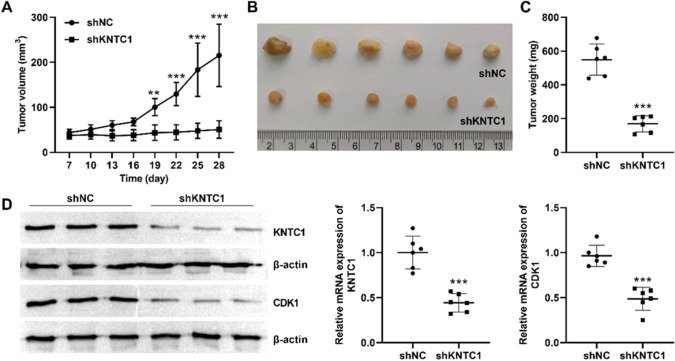
Subsequently, we tested the inhibitory effect of knockdown of KNTC1 on hepatocellular carcinoma cells in mice. We found a significant increase in liver tumor in mice transfected with Huh7, whereas mice transfected with Huh7 after knockdown of KNTC1 had small tumors and did not show a significant increase with observation time (Fig. 7A–C). By comparing the protein levels and mRNA levels in tumor tissues. We found that both protein and mRNA of KNTC1 as well as CDK1 were significantly lower in the KNTC1 knockdown group than in the knockdown low control group (Fig. 7D). These results suggest that KNTC1 knockdown can inhibit the growth of hepatocellular carcinoma cells in vivo.
Fig. 7.
A Inhibition of tumor volume increase in mice by silencing KNTC1. B Image of tumor at day 28 in mouse transplant tumor model in two groups. C Tumor weight at day 28 in mouse transplant tumor model in two groups. D Protein and mRNA levels of KNTC1 and CDK1 in tumor tissues. shNC, Negative control of knock down; shKNTC1, knock down of KNTC1. **P < 0.01, ***P < 0.001
Discussion
Kinetochore-associated proteins are key components of the mitotic checkpoint and are essential for faithful chromosome segregation and spindle assembly during cell division (Katis et al. 2004). Numerous studies have demonstrated that kinetochore-associated proteins are commonly upregulated in various cancer types. For example, mRNA levels of KNTC1 were found to be approximately fivefold higher in pancreatic cancer cell lines, such as SW1990, compared to normal pancreatic ductal epithelial cells (HPDE) (Liu et al. 2022a). Tissue microarray analysis showed that in vivo KNTC1 expression was higher in high-grade squamous intraepithelial lesions (HSILs) than in normal cervix and even higher in cervical cancer (Pan et al. 2023). Similarly, immunohistochemical staining revealed significantly higher levels of KNTC1 in non-small cell lung cancer tumors than in para-carcinoma tissues (Liu et al. 2022b). In addition, significant upregulation of KNTC1 was found in esophageal squamous cell carcinoma, bladder cancer, and colon cancer (Huang et al. 2021; Zhengxiang et al. 2021; Liu et al. 2019). Interestingly, our analysis of the TCGA-HCC database revealed significant upregulation of KNTC1 in hepatocellular carcinoma, compared to normal liver tissue. We confirmed these findings by measuring KNTC1 expression levels in hepatocellular carcinoma cell lines using mRNA and protein assays. Collectively, these results suggest that high levels of KNTC1 expression may have a general association with the occurrence of various cancer types.
Since high KNTC1 expression may be closely related to cancer genesis, down-regulation of KNTC1 could be a potential approach to delaying tumor development. Several studies have identified that knockdown of KNTC1 inhibits the proliferation, invasion and migration ability of tumor cells. KNTC1 knockdown significant reduction in cell growth rates of colon cancer cells RKO (Zhengxiang et al. 2021). Knockdown of KNTC1 suppresses growth and viability of human esophageal squamous cell carcinoma Eca-109 (Liu et al. 2019). Knockdown of KNTC1 inhibits the ability of migration of bladder cancer cells (Huang et al. 2021). In our study, KNTC1 knockdown inhibited the growth, migration and invasion of the hepatocellular carcinoma cell lines SK-Hep-1 and Huh7. Additionally, we observed that KNTC1 knockdown suppressed the volume growth of liver tumors in a mouse transplantation tumor model. Similar results were reported by Hui Tong et al. who found that knockdown of KNTC1 significantly reduced hepatocellular carcinoma cell viability and migration capacity, and tumor tissue growth was inhibited in mouse model (Tong et al. 2023). Collectively, these findings provide compelling evidence that knockdown of KNTC1 can effectively inhibit the progression of hepatocellular carcinoma.
Our study reports the regulatory role of KNTC1 in hepatocellular carcinoma, and our findings indicate that CDK1 mediates this regulation. CDK1 is an essential cell cycle CDK, that can drive cell division in the absence of other interphase CDKs (Santamaría et al. 2007; Asghar et al. 2015), and initiates mitotic initiation. Moreover, CDK1 can phosphorylate various regulatory molecules as well as molecular machines that drive cell-cycle events, making it a key regulator of cell division (Ubersax et al. 2003). There is growing evidence that CDK1 plays a critical role in multiple cancers. For instance, CDK1 can inhibit apoptosis in human colon cancer cells through phosphorylation of p53 (Nantajit et al. 2010), has been associated with poor prognosis in a range of cancers such as colorectal cancer, ovarian cancer, pneumonia, and endometrial cancer (Izadi et al. 2020; Yang et al. 2016; Li et al. 2020; Ying et al. 2021), and can enhance telomerase activity in aggressive cancers through phosphorylation of hTERT (Yasukawa et al. 2020). We confirmed the significant upregulation of CDK1 levels in hepatocellular carcinoma, consistent with previous reports (Zou et al. 2020; Wu et al. 2018; Hao et al. 2021). Furthermore, we found that knockdown of KNTC1 led to a decrease in CDK1 expression and suppressed hepatocellular carcinoma cell growth and migration. This effect was reversed upon re-introduction of CDK1 into cells with KNTC1 knockdown, indicating that CDK1 regulates hepatocellular carcinoma progression downstream of KNTC1. In summary, our study has identified the role of CDK1 in mediating the regulatory effect of KNTC1 in hepatocellular carcinoma, which sheds light on the molecular mechanism of tumor growth and progression in HCC.
Some limitations exist in our study. Although we predicted the interaction of KNTC1 with CDK1 and also clarified that knockdown of KNTC1 inhibited the progression of hepatocellular carcinoma by down-regulating CDK1 levels in vivo and in vitro, we lacked direct evidence of protein interactions between KNTC1 with CDK1. We could not yet explain why overexpression of CDK1 leads to increased levels of KNTC1. In addition, further mechanisms by which KNTC1 knockdown inhibits CDK1 and arrests hepatocellular carcinoma cells in S-phase need to be investigated.
Conclusion
We observed a significant increase in KNTC1 expression in hepatocellular carcinoma tissues. Our experiments also confirmed that KNTC1 knockdown in hepatocellular carcinoma cells led to inhibited cell proliferation and metastasis, due to the downregulation of CDK1 expression and an induced S-phase cell cycle arrest. These findings strongly suggest that KNTC1 plays an important oncogenic role in hepatocellular carcinoma, and thus targeting KNTC1 may be an effective therapeutic strategy for treating this disease.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this manuscript.
Research involving human participants and/or animals
This article does not contain any studies with human participants. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
We affirm that all the authors have seen, prepared, and agreed to the submission of paper and their inclusion of name(s) as co-author(s). We also declare that there are no conflicts of interests for the same.
References
- Anwanwan, D., Singh, S. K., Singh, S., et al., Challenges in liver cancer and possible treatment approaches. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 2020. 1873(1): 188314. [DOI] [PMC free article] [PubMed]
- Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14(2):130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM. The kinetochore. Cold Spring Harb Perspect Biol. 2014;6(7):a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB, Marrero JA, Rudolph L, et al. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134(6):1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- Hao L, Li S, Peng Q, et al. Anti-malarial drug dihydroartemisinin downregulates the expression levels of CDK1 and CCNB1 in liver cancer. Oncol Lett. 2021;22(3):1–14. doi: 10.3892/ol.2021.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Fan, X., Qiao, Y., et al., Knockdown of KNTC1 inhibits the proliferation, migration and tumorigenesis of human bladder cancer cells and induces apoptosis. Critical Reviews™ in Eukaryotic Gene Expression, 2021. 31(1). [DOI] [PubMed]
- Izadi, S., Nikkhoo, A., Hojjat-Farsangi, M., et al., CDK1 in breast cancer: implications for theranostic potential. Anti-Cancer Agents in Medicinal Chemistry (Formerly Current Medicinal Chemistry-Anti-Cancer Agents), 2020. 20(7): 758–767. [DOI] [PubMed]
- Katis VL, Galova M, Rabitsch KP, et al. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14(7):560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Li M, He F, Zhang Z, et al. CDK1 serves as a potential prognostic biomarker and target for lung cancer. J Int Med Res. 2020;48(2):0300060519897508. doi: 10.1177/0300060519897508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CT, Min L, Wang YJ, et al. shRNA-mediated knockdown of KNTC1 suppresses cell viability and induces apoptosis in esophageal squamous cell carcinoma. Int J Oncol. 2019;54(3):1053–1060. doi: 10.3892/ijo.2019.4672. [DOI] [PubMed] [Google Scholar]
- Liu R, Liu R, Guo Z, et al. shRNA-mediated knockdown of KNTC1 inhibits non-small-cell lung cancer through regulating PSMB8. Cell Death Dis. 2022;13(8):685. doi: 10.1038/s41419-022-05140-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Chen, H., Chen, X., et al., KNTC1 as a putative tumor oncogene in pancreatic cancer. J Cancer Res Clin Oncol, 2022a. [DOI] [PMC free article] [PubMed]
- Nantajit D, Fan M, Duru N, et al. Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response. PLoS ONE. 2010;5(8):e12341. doi: 10.1371/journal.pone.0012341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W., Wang, S., Liu, X., et al., KNTC1, regulated by HPV E7, inhibits cervical carcinogenesis partially through Smad2. Experimental Cell Research, 2023. 423(1). [DOI] [PubMed]
- Santamaría D, Barrière C, Cerqueira A, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- Siegel, R. L., Miller, K. D., & Jemal, A., Cancer statistics, 2019. CA: a cancer journal for clinicians, 2019. 69(1): 7–34. [DOI] [PubMed]
- Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51(5):1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- Tong H, Liu X, Peng C, et al. Silencing of KNTC1 inhibits hepatocellular carcinoma cells progression via suppressing PI3K/Akt pathway. Cell Signal. 2023;101:110498. doi: 10.1016/j.cellsig.2022.110498. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425(6960):859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang Y, Liu C, et al. Kinetochore-associated protein 1 promotes the invasion and tumorigenicity of cervical cancer cells via matrix metalloproteinase-2 and matrix metalloproteinase-9. Bioengineered. 2022;13(4):9495–9507. doi: 10.1080/21655979.2022.2061144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CX, Wang XQ, Chok SH, et al. Blocking CDK1/PDK1/β-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics. 2018;8(14):3737. doi: 10.7150/thno.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Cho H, Shin H-Y, et al. Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget. 2016;7(31):49481. doi: 10.18632/oncotarget.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa M, Ando Y, Yamashita T, et al. CDK1 dependent phosphorylation of hTERT contributes to cancer progression. Nat Commun. 2020;11(1):1557. doi: 10.1038/s41467-020-15289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying X, Che X, Wang J, et al. CDK1 serves as a novel therapeutic target for endometrioid endometrial cancer. J Cancer. 2021;12(8):2206. doi: 10.7150/jca.51139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhengxiang, Z., Yunxiang, T., Zhiping, L., et al., KNTC1 knockdown suppresses cell proliferation of colon cancer. 3 Biotech, 2021. 11(6): 262. [DOI] [PMC free article] [PubMed]
- Zou Y, Ruan S, Jin L, et al. CDK1, CCNB1, and CCNB2 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2020;26:e925289–e925281. doi: 10.12659/MSM.925289. [DOI] [PMC free article] [PubMed] [Google Scholar]