Abstract
Background & Aims
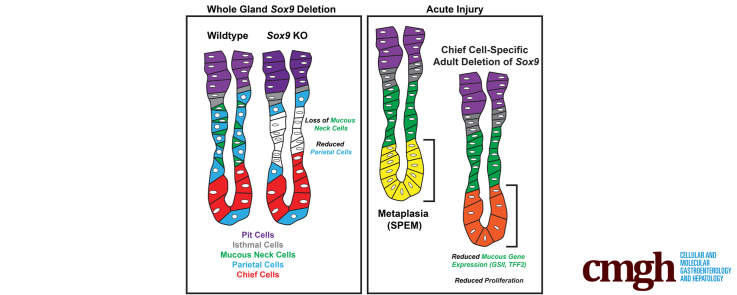
Acute and chronic gastric injury induces alterations in differentiation within the corpus of the stomach called pyloric metaplasia. Pyloric metaplasia is characterized by the death of parietal cells and reprogramming of mitotically quiescent zymogenic chief cells into proliferative, mucin-rich spasmolytic polypeptide–expressing metaplasia (SPEM) cells. Overall, pyloric metaplastic units show increased proliferation and specific expansion of mucous lineages, both by proliferation of normal mucous neck cells and recruitment of SPEM cells. Here, we identify Sox9 as a potential gene of interest in the regulation of mucous neck and SPEM cell identity in the stomach.
Methods
We used immunostaining and electron microscopy to characterize the expression pattern of SRY-box transcription factor 9 (SOX9) during murine gastric development, homeostasis, and injury in homeostasis, after genetic deletion of Sox9 and after targeted genetic misexpression of Sox9 in the gastric epithelium and chief cells.
Results
SOX9 is expressed in all early gastric progenitors and strongly expressed in mature mucous neck cells with minor expression in the other principal gastric lineages during adult homeostasis. After injury, strong SOX9 expression was induced in the neck and base of corpus units in SPEM cells. Adult corpus units derived from Sox9-deficient gastric progenitors lacked normal mucous neck cells. Misexpression of Sox9 during postnatal development and adult homeostasis expanded mucous gene expression throughout corpus units including within the chief cell zone in the base. Sox9 deletion specifically in chief cells blunts their reprogramming into SPEM.
Conclusions
Sox9 is a master regulator of mucous neck cell differentiation during gastric development. Sox9 also is required for chief cells to fully reprogram into SPEM after injury.
Keywords: Stomach, Development, Metaplasia, Paligenosis
Graphical abstract
Summary.
Sox9 is a master regulator of mucous neck cell differentiation during gastric development. Sox9 also is required for chief cells to fully reprogram into spasmolytic polypeptide–expressing metaplasia after injury.
Gastric cancer (GC) is a leading cause of cancer-related death worldwide. GC most commonly arises in the context of chronic inflammation driven by Helicobacter pylori infection. H pylori infection induces altered differentiation patterns within gastric corpus units, termed pyloric or pseudopyloric metaplasia. Pyloric metaplasia is characterized histologically by the loss of mature acid-secreting parietal and digestive enzyme–secreting chief cell lineages and the emergence of highly proliferative mucus-producing cell populations.1,2 The basal-most portion of the expanding mucus-producing cells are spasmolytic polypeptide–expressing metaplasia (SPEM) cells, which are thought to be largely derived from mature chief cells.3, 4, 5, 6, 7, 8, 9 The chronic replacement of normal gastric differentiation with H pylori–associated mucous metaplasia is a major risk factor for progressing toward GC.
The 2 principal mucus-secreting cells in the body of the stomach at homeostasis are called surface (also pit or foveolar cells) and mucous neck cells. Mucous neck cells are sandwiched among parietal cells in the neck region below the foveolar cells. Mucous neck cells are thought to turn over every 2 weeks and are replaced constantly by stem cells located in the isthmus between the neck and foveolar regions.10,11 The molecular pathways that govern the normal development of mucous cells in the stomach are not well understood, and it also is not understood how stem cells give rise to mucous neck cells in the adult. Whether the emergence of mucus-generating SPEM cells involves pathways that recapitulate the development of mucous cells from stem cells also is unknown.10, 11, 12 In mouse models of H pylori–induced metaplasia, markers expressed by homeostatic mucous neck cells, including TFF2 (Trefoil factor 2, spasmolytic polypeptide) and GS-II lectin become expressed in the SPEM population arising at the base of the gastric unit. Mucous neck cell markers also are expressed by the injury-responsive mucous cells in the neck that proliferate after parietal cell death because these cells likely arise from stem cells and existing pre-injury mucous neck cells.13,14
In this study, we show that SRY-box transcription factor 9 (SOX9) is strongly expressed during embryonic development of the stomach and is expressed in nonfoveolar mucous cells: namely mucous neck cells and SPEM and the proliferative neck cells in injury-induced pyloric metaplasia. We determined that without Sox9, mucous neck cells do not arise. Thus, SOX9 is an essential factor in specifying mucous neck cell lineage. Misexpression of Sox9 during either postnatal life or adulthood alters the chief cell lineage such that they lose differentiation markers and acquire markers of mucous cells. Finally, we show that loss of Sox9 in chief cells blunts the induction of metaplastic features.
Results
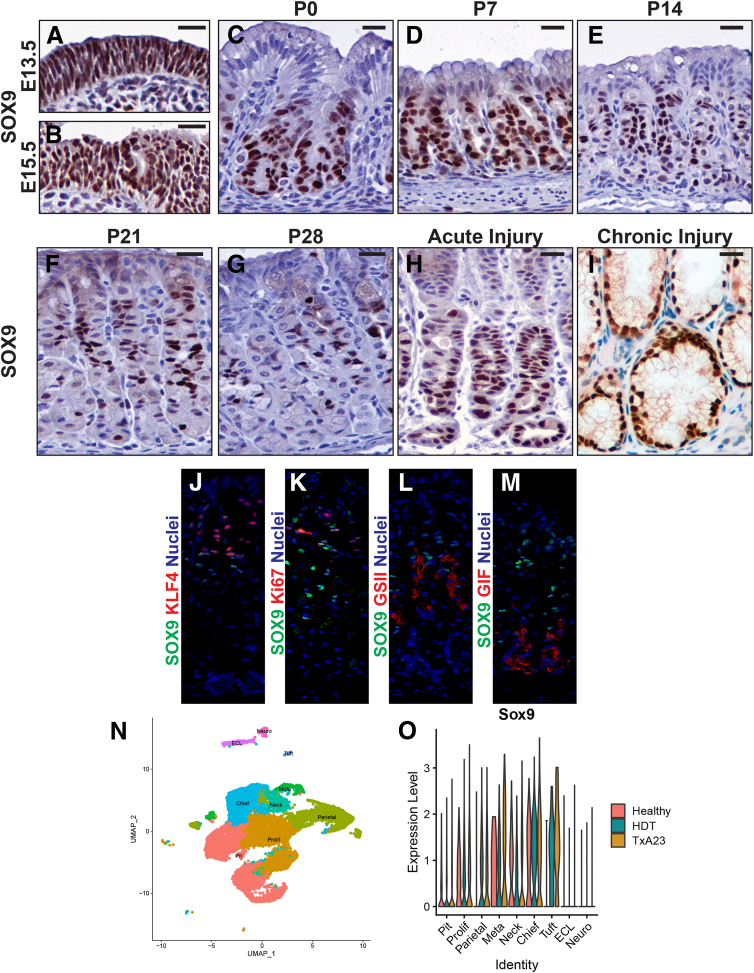
During early gastric development, SOX9 is expressed in all developing gastric epithelial cells (embryonic day [E]13.5 and E15.5) (Figure 1A and B). At the beginning of postnatal life, progenitors at the surface of the epithelium lose SOX9 expression while all nonsurface epithelial (ie, glandular) cells maintain SOX9 expression (Figure 1C). In the first week of postnatal life, SOX9 continues to be expressed throughout all non-surface progenitor cells of the glandular epithelium, including cells at the base of prospective corpus units where gastric chief cells will begin to emerge during the subsequent transition to adulthood (Figure 1D). Beginning in the second week of postnatal life, expression of SOX9 becomes sparser in the base (emerging chief cell zone) of primordial adult corpus units (Figure 1E). By P21 (Figure 1F) and postnatal day (P)28 (Figure 1G), SOX9 expression is most enriched in the neck and subisthmal regions of developing corpus units, with only minor expression in the base of gastric units.
Figure 1.
SOX9 is expressed in progenitor cells and glandular mucous cells during development and injury. During early development, anti-SOX9 immunohistochemistry marks all gastric progenitors: (A) E13.5 and (B) E15.5. In late gestation or early postnatal life, SOX9 is (C) lost from gastric progenitors near the surface (P0), but (D) still expressed throughout all non–surface-associated progenitors in the gland (P7). (E) In the second week of postnatal life, SOX9 expression at the base becomes less intense and ubiquitous at the base (P14). (F) By P21, SOX9 becomes concentrated in the neck region of mature gastric corpus units. (G) In P28 units, the pattern of SOX9 expression resembles that of adult units, with high expression in mucous neck cells. (H) Representative units after high-dose tamoxifen (HDT) injury at peak metaplasia stages (day 3). High SOX9 expression is observed in the neck and base of these injured units. (I) Representative basal metaplasia from the TxA23 autoimmune gastritis model after 12 weeks. SOX9 strongly marks all inflamed metaplastic cells in these stomachs. Immunofluorescence staining of adult gastric tissue stained with SOX9, a general nuclear marker, and representative markers of (J) prepit/pit cells (KLF4), (K) isthmal cells/proliferative zone (Ki67), (L) mucous neck cells (GS-II), and (M) chief cells (GIF). Nuclear expression of SOX9 is detected in some cells of all lineages, with most SOX9 being in isthmus and neck. (N and O) Analysis of previously published scRNA-seq data collected from homeostasis, acute injury, and chronic inflammatory injury conditions. (N) UMAP (Uniform manifold approximation and projection) unbiased clustering of gastric cell suspensions isolated from healthy BALB/c, HDT, and TxA23 mice colored by cluster identity. (O) Violin plots of Sox9 gene expression levels in all identified clusters of the combined healthy and injured scRNA-seq libraries. During homeostasis (except parietal, tuft, enterochromaffin-like [ECL], and endocrine cells) and both injury conditions (except ECL and endocrine cells), Sox9 messenger RNA is detected in all major gastric lineages, with the highest expression in stem and mucous neck cells and in metaplastic cells. All immunohistochemistry sections were counterstained with hematoxylin. Scale bars: 20 μm.
To explore whether SOX9 expression changes during gastric injury, we surveyed the expression pattern of SOX9 after acute chemical injury and chronic inflammatory injury. When delivered at high doses, tamoxifen induces an acute and reversible organ-wide injury that results in the induction of pyloric metaplasia by day 3 of injury.15,16 At these peak metaplasia stages, SOX9 is expressed in the cells in the neck of the corpus unit as well as the SPEM cells in the base (Figure 1H). In the TxA23 model of autoimmune gastritis,17,18 mucus-expressing metaplasia is induced in mice by 12 weeks of age owing to persistent autoimmune destruction of parietal cells and resulting inflammatory signals. In TxA23 animals at 12 weeks, SOX9 also marks all metaplasia, including the developing basal metaplastic (SPEM) cells (Figure 1I). To further explore the expression pattern of SOX9 in adult gastric tissue, we co-stained SOX9 with pit (KLF4, Krüppel-like factor 4), proliferative (Ki67), mucous neck cell (GS-II, Griffonia simplicifolia lectin II), and chief cell markers (GIF, Gastric intrinsic factor) (Figure 1J–M). In adult tissue, we detected nuclear expression of SOX9 in a subset of these cell populations. We previously generated single cell RNA sequencing (scRNA-seq) data sets from corpus tissue during adult homeostasis, after acute injury, or chronic injury.13,14 In all 3 tissue conditions, RNA expression of Sox9 can be detected in most major gastric lineages (Figure 1N and O). Sox9 was not detected in enterochromaffin-like or endocrine cells and was expressed only in parietal and tuft cell lineages after injury. In summary, SOX9 is expressed in early gastric progenitors; in adulthood, it has broad RNA and protein expression in most major gastric lineages, but with strong expression in isthmus and in mucous neck cells. In acute and chronic gastric injury models, SOX9 is strongly expressed in metaplasia, including SPEM.
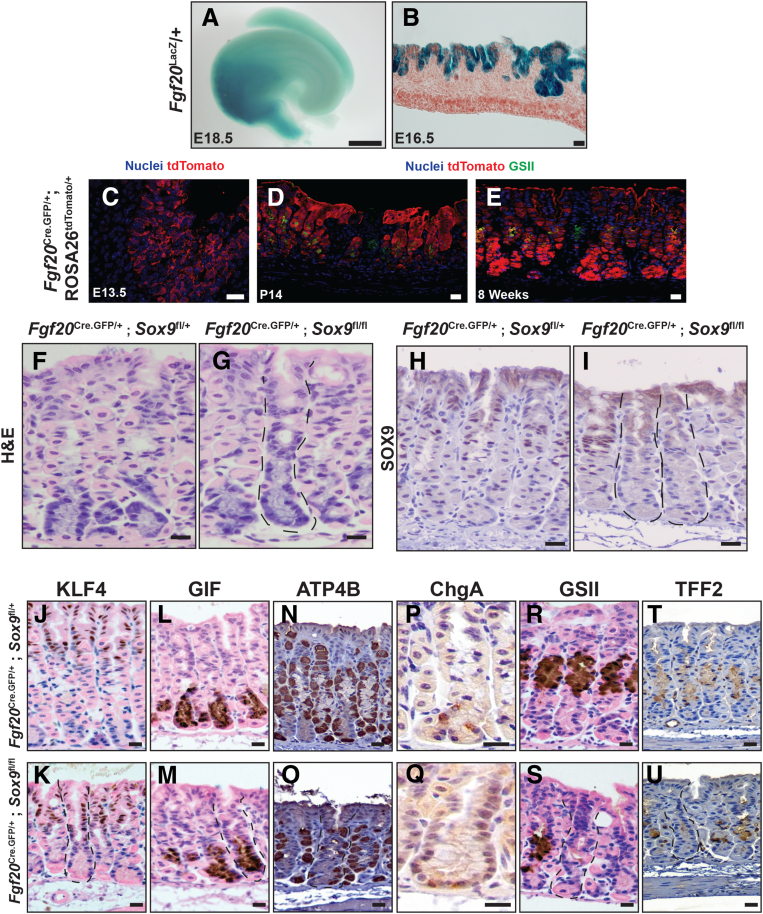
Given the broad expression of SOX9 in primordial gastric progenitors and the expression of SOX9 in mature mucous neck cells, we sought to determine whether Sox9 plays a role in controlling the differentiation of the mucous neck lineage. To remove Sox9 exclusively in the gastric epithelium in early embryogenesis, we characterized a new model for embryonic epithelial conditional gene deletion: Fgf20Cre.GFP.19 Fgf20 expression is specific to the glandular stomach and is not expressed in the squamous epithelium of the forestomach (Fgf20LacZ) (Figure 2A and B).20 To understand Cre recombinase activity under the regulation of the Fgf20 locus, we crossed Fgf20Cre.GFP to a ROSA26 lineage tracing reporter (ROSA26tdTomato) and observed that Fgf20Cre.GFP induces recombination with variable (mosaic) penetrance that is specific to the expanding embryonic progenitor cells during early development (E13.5) (Figure 2C). By late postnatal (P14) (Figure 2D) or adult stages (Figure 2E), Fgf20Cre.GFP/+; ROSA26tdTomato/+ animals showed an all-or-nothing, unit-to-unit recombination pattern: units in the antrum and corpus were either completely labeled or had no recombination at all. Thus, using Fgf20Cre.GFP, we concluded we could induce mosaic recombination of a Sox9 floxed allele (Sox9fl) in early gastric progenitors that would then give rise to Sox9-deficient (ie, Sox9Δ/Δ) adult units along with completely wild-type units that could be used as internal controls.
Figure 2.
Loss of Sox9 prevents mucous neck cell differentiation. (A) X-gal (5-Bromo-4-Chloro-3-Indolyl β-D-Galactopyranoside) staining of whole E18.5 stomach from a Fgf20LacZ/+ animal. X-gal staining was observed specifically within the glandular stomach domain of the stomach. (B) Section-based X-gal staining with nuclear fast red counterstain on E16.5 Fgf20LacZ/+ tissue. X-gal staining is specific to the gastric epithelium. (C) Immunofluorescence staining for nuclei (DRAQ5, blue) and tdTomato (mCherry antibody, red; ROSA26tdTomato). At E13.5 in Fgf20Cre.GFP/+; ROSA26tdTomato/+ animals, recombination of the ROSA26tdTomato was observed in a mosaic, salt-and-pepper pattern specifically within epithelial glandular stomach progenitors. (D and E) Immunofluorescence staining for nuclei (DRAQ5, blue), tdTomato (mCherry antibody, red; ROSA26tdTomatO), and GS-II lectin (mucous neck cell marker, green) at P14 and adult stages. (E) At P14, recombination of the ROSA26tdTomato allele in Fgf20Cre.GFP/+; ROSA26tdTomato/+ follows an all-or-nothing pattern: units have complete recombination throughout the unit from pit to base or no recombination at all. Identical recombination patterns were observed in adult units. (F) Representative histology of control 8-week-old corpus units, hematoxylin and eosin (H&E). (G) Representative Sox9-deficient unit (outlined) from Fgf20Cre.GFP/+; Sox9fl/fl mice. These units have alterations in cell morphology in the neck region, but the pit and base regions appear normal. Representative SOX9 immunohistochemistry from (H) control 8-week-old mice and (I) Fgf20Cre.GFP/+; Sox9fl/fl mice. Outlined units lack SOX9 expression. Representative staining on control animals and Fgf20Cre.GFP/+; Sox9fl/fl for (J and K) KLF4, (L and M) GIF, (N and O) ATP4B, (P and Q) chromogranin A, (R and S) GS-II lectin, and (T and U) TFF2. Control and Fgf20Cre.GFP/+; Sox9fl/fl mice have similar staining patterns for KLF4, GIF, and chromogranin A, indicating the pit, chief, and enteroendocrine lineages are not altered in the absence of Sox9. In contrast, there is a loss of GS-II lectin (S) and TFF2 (U) expression in Sox9-deficient tissue, indicating aberrant mucous neck cell differentiation. Scale bars: (A) 1 mm; (B–S), 20 μm.
Consistent with the pattern of expression of the Fgf20-driven recombination of the ROSA26tdTomato reporter, we observed that Fgf20Cre.GFP/+; Sox9fl/fl animals had a subset of adult corpus units that completely lacked SOX9 (Figure 2F–I). Those units had markedly abnormal neck regions with a paucity of mature parietal cells and the complete absence of normal mucous neck cells. We stained Fgf20Cre.GFP/+; Sox9fl/fl and control tissues with markers for the principal gastric lineages including KLF421,22 (pit cells) (Figure 2J and K), GIF (chief cells) (Figure 2L and M), ATP4B (ATPase H+/K+ transporting subunit beta, parietal cells) (Figure 2N and O), chromogranin A (enteroendocrine cells) (Figure 2P and Q), and TFF2 or GS-II lectin (mucous neck cells) (Figure 2R–U). Within Sox9Δ/Δ units in Fgf20Cre.GFP/+; Sox9fl/fl tissue, the distribution of cells marked with KLF4, GIF, and chromogranin A was similar to that of controls, indicating that the pit, chief, and enteroendocrine lineages were unaffected in the absence of Sox9. However, TFF2 and GS-II lectin staining were absent (GS-II) or nearly absent (TFF2) in Sox9Δ/Δ corpus units in Fgf20Cre.GFP/+; Sox9fl/fl tissue, suggesting there were near-total deficits in mucous neck cell differentiation in the absence of Sox9.
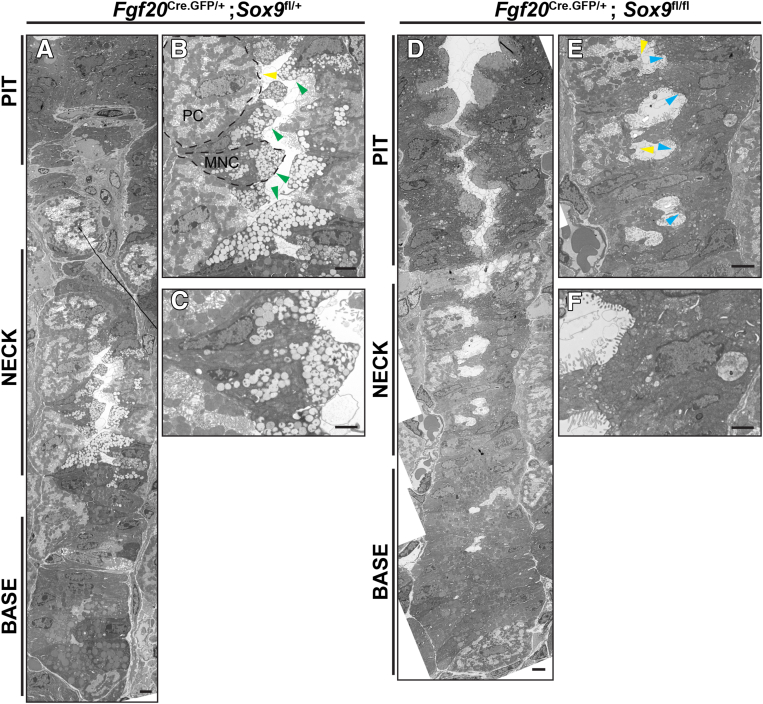
Transmission electron microscopy showed that mucous neck cells in control animals had the expected morphology characterized by largely electron-lucent granules filling a cytoplasm that bulged into the gastric gland lumen11,23 (Figure 3A–C). The mucous neck cells were interspersed, as expected, with parietal cells. In Sox9Δ/Δ units, the foveolar/surface cells in the region superficial to the isthmus (also known as the pit) and the chief cells of the base all appeared similar to the control (Figure 3D–F), consistent with the staining in Figure 2. In the neck, parietal cells were markedly under-represented, but the parietal cells that were present did not appear different from the control, with the usual expanded basal surface, abundant mitochondria, and mixtures of intercanalicular and tubulovesicle structures that harbor the abundant acid-pumping H+-K+-adenosine triphosphatase pumps (Figure 3E). We could not identify any cells with mucous neck morphology in Sox9Δ/Δ units. We noted, however, numerous cells in the neck region that did not have characteristics of any gastric differentiated cell lineage (Figure 3E and F). Overall, the results indicate that, in the absence of Sox9, gastric epithelial embryonic progenitors can generate mature gastric units with most epithelial cell lineages, but they are specifically deficient in their ability to differentiate into mature mucous neck cells. Because most Sox9-deficient gastric units had at least some normal, mature parietal cells, Sox9 does not appear to be directly involved in their differentiation.
Figure 3.
Transmission electron microscopy of adult control and Sox9-deficient units reveals SOX9 is required for cells with mucous neck ultrastructure to develop. (A) Representative adult control corpus unit from pit to base. (B) Higher-magnification images of the neck region of a control unit. Green arrowheads, mucous neck cells; yellow arrowhead, parietal cells. (C) Higher-magnification image of a single neck cell. (D) Representative Sox9-deficient corpus unit, pit to base from an adult Fgf20Cre.GFP/+ ; Sox9fl/fl animal. (E) Higher-magnification images of the neck region of a Sox9-deficient unit. Blue arrowheads, primitive cells lacking ultrastructural features of any differentiated lineage; yellow arrowheads, parietal cells. (F) Higher-magnification image of a single primitive cell in the neck region of a Sox9-deficient unit. Scale bars: (A, B, D, and E) 4 μm; (C and F), 2 μm.
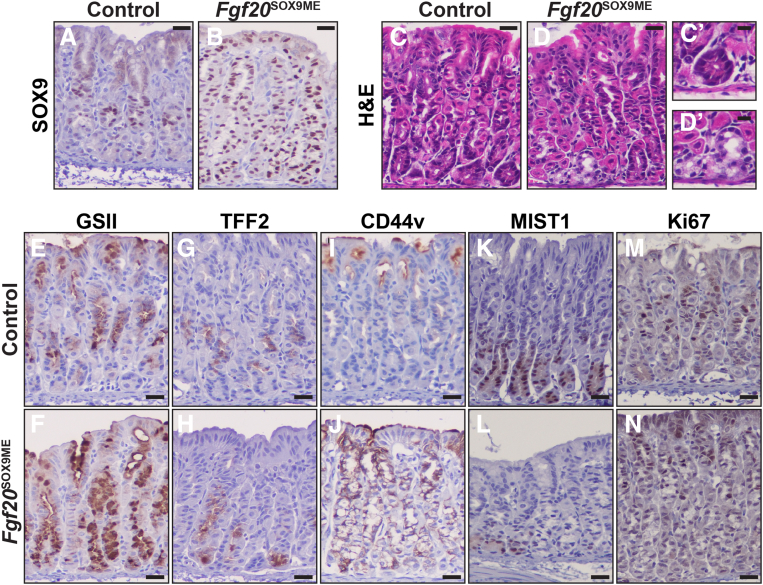
We next sought to determine if Sox9 was sufficient to drive mucous differentiation in the postnatal gastric epithelium. We bred mice carrying alleles for ROSA26rtTA.IRES.EGFP, TetO-Sox9, and Fgf20Cre.GFP. In these mice, cells expressing Cre will induce rtTA from the ROSA26rtTA.IRES.EGFP allele. Thereafter, whenever doxycycline (DOX) is administered, the rtTA will bind and activate the TetO-Sox9 allele to induce the expression of Sox9. Fgf20 is dispensable for gastric development (unpublished data), and Fgf20Cre.GFP/Cre.GFP mice are viable and fertile.20 Thus, we analyzed both Fgf20Cre.GFP/+; ROSA26rtTA.IRES.EGFP/rtTA.IRES.EGFP; TetO-Sox9 pups and Fgf20Cre.GFP/Cre.GFP; ROSA26rtTA.IRES.EGFP/rtTA.IRES.EGFP; TetO-Sox9 pups. Fgf20Cre.GFP/Cre.GFP; ROSA26rtTA.IRES.EGFP/rtTA.IRES.EGFP; TetO-Sox9 pups had increased numbers of units misexpressing Sox9, but were otherwise indistinguishable from Fgf20Cre.GFP/+ ; ROSA26rtTA.IRES.EGFP/rtTA.IRES.EGFP/; TetO-Sox9 pups. For the remainder of this article, we will refer to these triple-allele animals as Fgf20Sox9ME. Controls for all misexpression experiments were biallelic littermates. We administered DOX in the drinking water starting at P0, and maintained DOX administration until P21. At P21, in Fgf20Sox9ME animals, SOX9 was expressed uniformly, pit to base, throughout a subset of corpus units (Figure 4A and B), consistent with the mosaic, unit-to-unit recombination pattern of Fgf20Cre.GFP described earlier. In control mice, most bases were filled, as expected, with nearly mature chief cells characterized by basal–lateral, hematoxylin-stained, rough endoplasmic reticulum and nuclei consistently confined to the bases of cells24 (Figure 4C and C’). In the Sox9-misexpression units, some base regions instead comprised cells with foamy, glassy cytoplasm (Figure 4D and D’), resembling the cytoplasm of mucous neck cells or mucous cells in the bases of the antrum of the stomach. Accordingly, misexpression of SOX9 throughout the gastric unit caused highly increased GS-II lectin reactivity and TFF2 expression beyond the neck zone, with basal cells showing marked ectopic expression of mucous neck cell markers (Figure 4E–H). We also stained for CD44v (ortholog of human CD44v9), which becomes highly expressed in metaplasia after injury.6,25, 26, 27 At P21 in controls, CD44v was, as expected, largely absent from normal corpus units (Figure 4I). In contrast, we observed surprisingly strong expression of CD44v throughout Sox9-misexpression units, from the pit to the base (Figure 4J). Basal cells misexpressing SOX9 also failed to induce the bHLH (basic helix-loop-helix) transcription factor MIST1 (Muscle intestine stomach expression 1; aslo known as BHLHA15), which is essential for terminal maturation of chief cells and normally is induced as fully differentiated chief cells emerge for the first time in development28,29 (Figure 4K and L). Misexpression of SOX9 did not alter cell-cycle behavior in the gastric unit, as indicated by Ki67 expression remaining confined to the isthmus (Figure 4M and N). These findings show that Sox9 is sufficient to drive increased mucous neck cell marker expression in the neck and base, induce metaplasia markers, and suppress the development of mature chief cells.
Figure 4.
Postnatal misexpression of Sox9 leads to the expansion of mucous cells. (A) Anti-SOX9 at P21 in control animals shows SOX9 predominately within the isthmus and neck region of corpus units. (B) SOX9 expression in Fgf20SOX9ME animals. Misexpression units express SOX9 from pit to base. (C) In control animals, H&E shows corpus bases with mature basal (chief cell) phenotype: polarized localization of nuclei, large apical cytoplasms, and elaborate rough endoplasmic reticulum stained with hematoxylin beneath the nucleus. (C’) Higher-magnification image is shown. (D) In Fgf20SOX9ME animals, some base cytoplasm appears clear and foamy, phenocopying corpus mucous neck cells and deep antral mucous cells. (D’) Higher-magnification image is shown. (E–N) Immunohistochemistry to detect representative markers of gastric lineages with hematoxylin counterstain. (E and G) Compared with control animals, Fgf20SOX9ME animals expanded the expression of mucous neck cell markers (F) GS-II lectin and (H) TFF2 into the base region. (I) During homeostasis, CD44v was not expressed in the corpus. (J) However, after Sox9 misexpression, we observed significant expression of CD44v throughout the unit from the pit to the base. (K) At P21, MIST1, a marker of differentiated chief cells, strongly marking all corpus base regions in control animals. (L) In Fgf20SOX9ME animals, several base regions that misexpress Sox9 have completely lost MIST1 expression. (M) By P21, the proliferative center of the unit is located in the isthmus, as observed by the location of Ki67-positive cells in that region. (N) Sox9 misexpression does not alter the proliferative zone in P21 corpus units. Scale bars: (A–N) 20 μm; (C’ and D’) 10 μm.
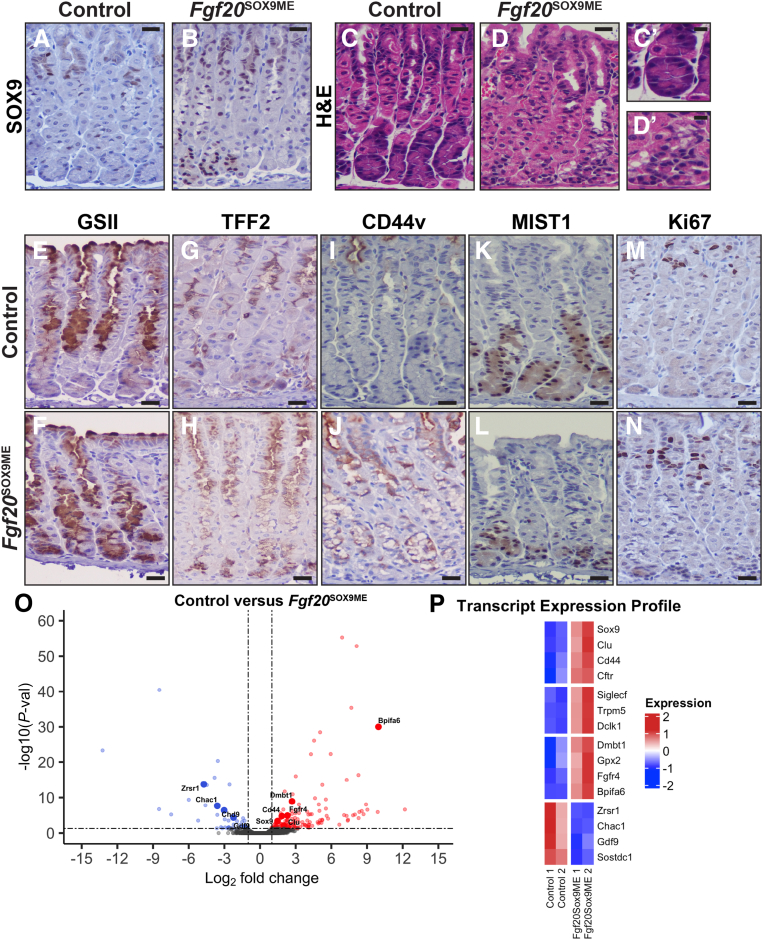
Next, we analyzed the effects of misexpression of Sox9 in adult units. We gave 8-week-old control and Fgf20Sox9ME animals DOX for 2 weeks and examined gastric morphology and differentiation. Again, we observed that a subset of corpus units misexpressed SOX9 from pit to base (Figure 5A and B). Sox9-misexpression units had a normal distribution of gastric lineages; however, chief cells were noticeably smaller, appearing less well differentiated (Figure 5C, D, C’, and D’). In adult Sox9-misexpression units, we observed the expansion of GS-II lectin reactivity and TFF2 expression into base regions (Figure 5E–H). We also observed the expression of CD44v in the neck regions of misexpression units; however, this expression did not extend into base regions (Figure 5I and J). Although we did observe alterations in base morphology, expression of MIST1 was maintained, and we observed no alterations in cell-cycle behavior because Ki67 remained restricted to the isthmal region (Figure 5K–N). To explore transcriptional changes after adult misexpression of Sox9, we isolated RNA from the corpus of Fgf20Sox9ME and control animals and performed RNA-seq (Figure 5O and P). In Fgf20Sox9ME animals, we identified several differentially expressed genes (log2 fold change, >1; false discovery rate, <0.05) (see Supplementary Table 1 for a complete gene list), including Sox9. We detected a metaplasia-associated transcript signature including Cd44, Clu, and Cftr that was up-regulated significantly and a set of tuft cell associated genes (Siglecf, Trpm5, and Dclk1) that were up-regulated after Sox9 misexpression. Sox9 is expressed in tuft cells after acute and chronic injury (scRNA-seq data) (Figure 1O), suggesting Sox9 may play an important role in the tuft cell lineage after gastric injury.
Figure 5.
Relative to postnatal misexpression, adult misexpression of Sox9 causes less-dramatic expansion of mucous cell features. (A) Anti-SOX9 shows that SOX9 is largely restricted to the neck region of adult corpus units. (B) SOX9 expression in Fgf20SOX9ME animals. In a subset of adult units, SOX9 is expressed from pit to base. (C and D) H&E shows that Fgf20SOX9ME units (D, D`) have altered base morphology with reduced cell size compared with control animals (C, C`). (E–N) Immunohistochemistry to detect representative markers of gastric lineages with hematoxylin counterstain. (E and G) Compared with control animals, Fgf20SOX9ME animals have expanded expression of mucous neck cell markers (F) GS-II lectin and (H) TFF2, extended in the base region, although less significantly compared with postnatal misexpression of Sox9. (I) Similar to postnatal tissue, CD44v was not expressed in the corpus during homeostasis. (J) However, after Sox9 misexpression, we observed marked expression of CD44v throughout the pit and neck, but not the base. (K) MIST1 strongly marks all corpus base regions in control animals. (L) After adult Sox9 misexpression, MIST1 expression is maintained in chief cells. (M and N) Adult Sox9 misexpression does not alter the proliferative center of the unit because Ki67-positive cells are still restricted to the isthmus region similar to controls. (O) Analysis of RNA-seq data from whole adult corpus of Fgf20SOX9ME animals treated with doxycycline for 2 weeks compared to control animals. Volcano plot generated from the comparison of bulk RNA-seq from adult Fgf20SOX9ME and control animals: The log2 fold change indicates the mean expression level for each gene. Blue dots, red dots, and black dots represent down-regulated genes, up-regulated genes, and insignificantly differentially expressed genes, respectively. A total of 150 genes were up-regulated, and 46 genes were down-regulated in Fgf20SOX9ME compared with controls, with the thresholds of log2 fold change greater than 1 and false discovery rate less than 0.05. (P) Heatmap (normalized transcripts per million) of 15 differentially expressed genes of interest (log2 fold change, >1; false discovery rate, <0.05) found in our RNA-seq. We detected significant up-regulation of Sox9 and a subset of metaplasia (Cd44, Clu, and Cftr) and tuft-cell–associated (Dclk1, Trpm5, and Siglecf) markers in Fgf20SOX9ME animals. Scale bars: (A–N) 20 μm, (C’ and D’) 10 μm. P-val, P value.
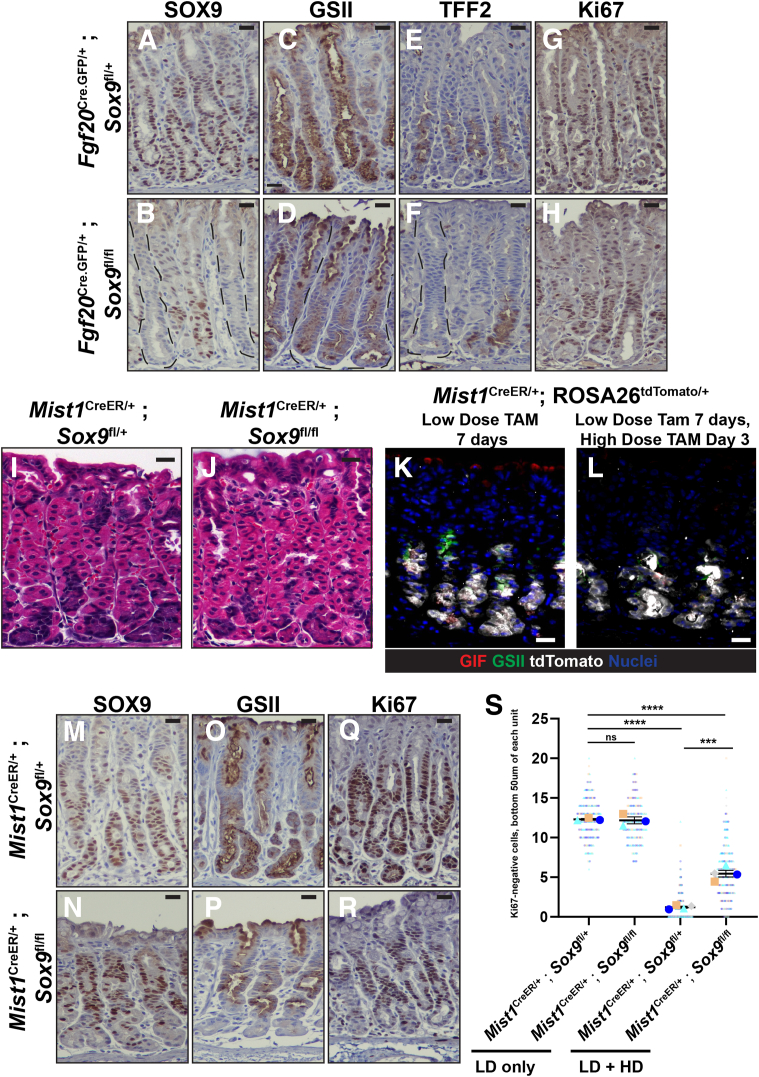
As described in Figure 1, SOX9 is strongly expressed in metaplasia after acute and chronic injury. To determine whether Sox9 plays a role in the induction of metaplasia, we gave control and Fgf20Cre.GFP/+; Sox9fl/fl animals injury-inducing doses of tamoxifen. After injury, we observed that control units had the expected dramatic induction of SOX9 throughout the neck and base of corpus units at peak metaplasia stages. In Fgf20Cre.GFP/+; Sox9fl/fl animals, a subset of units completely lacked SOX9 (Figure 6A and B). Sox9-deficient units had significantly reduced GS-II reactivity and TFF2 expression in both the neck and base after injury (Figure 6C–F). Neck regions of Sox9 knockout corpus units could respond to injury, showing the usual injury-induced proliferation even while lacking mucous neck cell gene expression (Figure 6G and H). Base regions, however, had markedly reduced cell-cycle entry, as observed by the lack of Ki67-positive cells (Figure 6H).
Figure 6.
Loss of Sox9 in gastric chief cells prevents the acquisition of key metaplasia features. (A–H) Eight-week-old control and Fgf20Cre.GFP/+; Sox9fl/fl mice after high-dose tamoxifen treatment stained for markers of SPEM formation. (A) In control animals, SOX9 expression expands to the base regions of corpus units after injury. (C) GS-II lectin and (E) TFF2 expand in the base regions marking SPEM, and the entire gland gets recruited to the cell cycle, (G) as seen by the broad expression of Ki67 after injury. (B) In Fgf20Cre.GFP/+; Sox9fl/fl mice after injury, a subset of units completely lack SOX9. These Sox9Δ/Δ units have significantly reduced expression of (D) GS-II and (F) TFF2 in the neck and base regions. (H) Although the neck region of Sox9Δ/Δ units are recruited to the cell cycle, base regions show reduced Ki67 positivity. Representative histology of (I) Mist1CreER/+; Sox9fl/+ and (J) Mist1CreER/+; Sox9fl/fl corpus tissue stained with H&E after 1 week of LD tamoxifen treatment. LD tamoxifen treatment does not lead to atrophy or metaplasia induction. (K) Immunofluorescence staining for nuclei (DRAQ5, blue), tdTomato (mCherry antibody, white; ROSA26tdTomatO), GS-II lectin (mucous neck cell marker, green), and GIF (chief cell marker, red) in Mist1CreER/+; ROSA26tdTomato/+ mice after LD treatment only. After only low-dose treatment, recombination of the ROSA26tdTomato allele in Mist1CreER/+; ROSA26tdTomato/+ mice was restricted to mature chief cells at the base of corpus units. (L) To induce acute injury after gene deletion, mice were first treated with low-dose tamoxifen for 7 consecutive days followed by a week of rest. Mice then were treated with 2 doses of high-dose tamoxifen to induce injury. Recombination of the ROSA26tdTomato allele in Mist1CreER/+; ROSA26tdTomato/+ mice is still enriched specifically in the base region of injured corpus units. (M-R) Eight-week-old control and Mist1CreER/+; Sox9fl/fl mice after low-dose/high-dose tamoxifen treatment. In control animals, tamoxifen-induced injury causes the expansion of (M) SOX9, (O) GS-II lectin, and (Q) Ki67 in the base region of corpus units. In Mist1CreER/+; Sox9fl/fl mice, after low-dose/high-dose tamoxifen treatment, have corpus bases with reduced (N) SOX9 expression and (P) GS-II lectin reactivity. These bases retain cells with larger, complex architecture and overall have a morphology more similar to control pre-injury bases. (Q) Base regions also have reduced cell-cycle entry, as shown by the lack of Ki67 positivity in corpus base regions. To quantify the reduction in cell-cycle entry, we counted Ki67-negative cells in the bottom 50 um of at least 60 units in control and treated Mist1CreER/+; Sox9fl/fl animals. Mist1CreER/+; Sox9fl/fl animals have significantly increased numbers of Ki67-negative cells compared with control animals. Smaller dots are individual counts (glands) per color-coded mouse replicate. Bold dots are the means of the individual mice, and error bars are the SEM. ∗∗∗P < .001, ∗∗∗∗P < .0001. Scale bars: 20 μm. TAM, Tamoxifen.
To better explore the role of Sox9 in chief cell recruitment after injury, we crossed the Sox9 floxed allele to Mist1CreER. Careful induction of Mist1CreER with a subinjury threshold dose of tamoxifen (low dose [LD]) causes recombination, specifically in mature chief cells,30 allowing for gene deletion without atrophy or metaplasia induction before acute injury (Figure 6I–L).15,16,31 We administered 7 consecutive days of LD tamoxifen to induce Sox9 deletion in homeostatic chief cells. After a week of rest, we administered injury-inducing doses of tamoxifen (high dose [HD]) and examined mice at peak metaplasia stages. In control animals, we observed strong induction of SOX9 throughout the bases of corpus units (Figure 6M). In Mist1CreER/+; Sox9fl/fl animals after LD/HD treatment, we observed a significant reduction of SOX9 expression in gastric bases, consistent with Mist1CreER-mediated gene deletion (Figure 6N). In Mist1CreER/+; Sox9fl/fl mice, base regions harbored chief cells with reduced induction of GS-II reactivity and proliferation (Figure 6O–R). To quantify the changes in cell-cycle recruitment after injury, we stained control and Mist1CreER/+; Sox9fl/fl tissues for Ki67 and counted Ki67-negative epithelial cells in the bottom 50 μm of each unit. After LD tamoxifen treatment only (ie, subinjury dosing to induce Cre recombinase activity), both control and Mist1CreER/+; Sox9fl/fl animals showed essentially no cell proliferation, with both groups having the same census of approximately 12 Ki67-negative cells (ie, normal, mitotically quiescent cells) in the base per corpus unit. After LD/HD treatment, almost all cells in the base were recruited to the cell cycle, so genotype control units had, on average, no more than 1 Ki67-negative cell in the basal 50 μm. In Mist1CreER/+; Sox9fl/fl animals we observed a significant 5-fold increase in Ki67-negative cells in the bottom 50 μm of each unit. In addition, we counted MIST1+ cells in Mist1CreER/+; Sox9fl/+ and Mist1CreER/+; Sox9fl/fl animal after LD/HD treatment. We counted at least 74 units per animal, 4 mice for each condition. Mist1CreER/+; Sox9fl/fl animals had increased levels of MIST1+ cells per unit (3.0 compared with 1.4; P < .05 by unpaired t test). Overall, the injury experiments showed that loss of Sox9 impaired downscaling of differentiated chief cell gene expression, induction of metaplastic gene expression, and cell-cycle entry.
Discussion
Our findings highlight Sox9 as a master regulator of mucous neck cell identity. There is little known about the genes and signaling pathways that control the specification of glandular stomach lineages. During early development, Gata4 is the master regulator of glandular stomach identity. In the absence of Gata4, endoderm in the stomach region adopts a forestomach-like, squamous, rather than glandular, fate; thus, Gata4 controls access to all normal epithelial cell lineages in the corpus and antrum.32 Our findings suggest that Sox9 acts downstream of Gata4, allowing access to the mucous neck cell lineage in corpus units. Spdef is important for deep antral mucous cells, but appears dispensable for mucous neck cells in the corpus.33 Similar regulating genes for the enteroendocrine lineages also have been identified, such as Ngn334,35 and Ascl1.36 There is still no known gene(s) involved in the specification of the pit, parietal, and chief cell lineages. Other transcription factors such as Hnf4a,37 Xbp1,38 Mist1,29 Foxq1,39 and Klf421,22 have been implicated in the maturation of gastric lineages, but do not appear to regulate specification, with the possible exception of Klf4, which does seem to regulate overall fate allocation even if not being entirely required for a specific lineage choice. We also observed consistent loss of parietal cells, to varying degrees, in Sox9-deficient units. It remains unclear, however, how the loss of Sox9 impacts the parietal cell lineage. Future experiments might be able to decipher between: (1) SOX9 being directly involved in differentiation from stem cells through bipotent isthmal progenitors that give rise to mucous neck and parietal cells; or (2) loss of parietal cells occurring indirectly owing to loss of mature mucous neck cells. In any case, it would seem that SOX9 is not involved in the maturation of parietal cells because although they are rare in Sox9-deficient units, they do still arise, and, when they are present, they appear morphologically normal and fully mature (Figure 3).
In tissues such as the stomach and pancreas, the recruitment of terminally differentiated cells to participate in repair is an essential component of tissue homeostasis. Our findings highlight that Sox9 is an essential mediator of chief cell recruitment to proliferating SPEM cells after acute gastric injury. Not only was Sox9 necessary for the morphologic and cell identity changes that occur after injury, but also for the cell-cycle recruitment of chief cells. These observations are consistent with our previous findings that the recruitment of chief cells after acute injury follows an ordered process, which we have termed paligenosis. In those previous findings, we observed that downscaling mature chief cell machinery (through autodegradative pathways) preceded adopting SPEM cell identity (mucinous expression characteristics), which in turn preceded cell-cycle entry.4,40,41 Through treatment with pathway inhibitors and target genetic tools, we determined that blocking early steps in this transformation prevented subsequent behaviors. Given that Sox9-deficient chief cells fail to lose their differentiated architecture, do not adopt the SPEM cell gene expression identity, and do not begin to proliferate, we can conclude that SOX9 must be required at the earliest stages. Perhaps Sox9 can participate in both downscaling (which involves induction of lysosomes and autophagic pathways) and adopting SPEM cell identity, leading to a subsequent robust block in cell-cycle entry. Of note, adult chief cells misexpressing Sox9 were reminiscent of chief cells after acute injury because these chief cells had downscaled cytoplasm and expression of mucous genes. However, SOX9 did not induce cells to enter the cell cycle. We could speculate that SOX9 thus is sufficient to induce the first 2 stages of paligenosis (downscaling and mucinous/SPEM gene expression), but not the final stage (cell-cycle re-entry).
Thus, SOX9 plays a critical role both in normal mucous neck cell differentiation and cell transformation into SPEM. Further investigation will be required to identify Sox9 target genes that are induced after injury to facilitate full downscaling of chief cells and initiation of SPEM cell identity. In addition, it will be important to investigate mechanisms that regulate Sox9 after injury, such as by the immune system or by the tissue environment. Further study could help us identify targetable pathways that can block or reverse mucinous metaplasia in the stomach by interfering with or preventing the function or induction of Sox9.
Material and Methods
Mice and Reagents
Animal experiments followed protocols approved by the Washington University School of Medicine Animal Studies Committee. The mouse allele Sox9-flox (Sox9fl/fl),42 TetO-Sox9,43 ROSA26-rtTA.IRES.EGFP (ROSA26rtTA.IRES.EGFP),44 ROSA26-tdTomato (Ai9; ROSA26tdTomato),45 Mist1-CreER (Mist1CreER),46 Fgf20-CreGFP (Fgf20Cre.GFP),19 Fgf20-LacZ (Fgf20LacZ),20 and TxA2317 were described previously. All mouse experiments were conducted with at least 3 animals per condition unless otherwise noted, with animals from both sexes used. To induce gene deletion but not gastric injury, tamoxifen (1 mg/20 g body weight; Toronto Research Chemicals) was injected intraperitoneally for 7 consecutive days. To cause acute gastric injury, tamoxifen (5 mg/20 g body weight) was injected intraperitoneally for 2 consecutive days.15,31 Tamoxifen was dispersed in 100% ethanol by sonication and then emulsified in sunflower oil (Sigma-Aldrich), 9:1 (oil:ethanol). Doxycycline (Sigma-Aldrich) was administered in drinking water, 2 mg/mL. Table 1 lists the primary antibodies used in this study.
Table 1.
Antibodies and Reagents for Immunohistochemistry and Immunofluorescence
| Antibody | Company | Catalog number | Concentration |
|---|---|---|---|
| SOX9 | Millipore | AB5535 | 1:2000 (IHC) 1:500 (IF) |
| KLF4 | R&D Systems | AF3640 | 1:2000 (IHC) 1:500 (IF) |
| Chromogranin A | DAKO | M0869 | 1:250 (IHC) |
| GIF | Invitrogen | MA5-36032 | 1:1000 (IHC) 1:200 (IF) |
| TFF2 | Abcam | ab49526 | 1:250 (IHC) |
| GS-II | Vector Labs | B-1215-2 | 1:2000 (IHC) 1:500 (IF) |
| mCherry | Abcam | ab167453 | 1:500 (IF) |
| DAPI | Sigma | D9542 | 1:20,000 (IF) |
| DRAQ5 | Cell Signaling | 4084 | 1:20,000 (IF) |
| Ki67 | Abcam | ab15580 | 1:500 (IHC) 1:200 (IF) |
| CD44v | Cosmo Bio | CAC-LKG-M002 | 1:200 (IHC) |
| MIST1 | Cell Signaling | 14896 | 1:1000 (IHC) |
| ATP4B | Abcam | ab176992 | 1:1000 (IHC) |
DAPI, 4′,6-diamidino-2-phenylindole; DRAQ5, 1,5-bis{[2-(di-methylamino) ethyl]amino}-4, 8-dihydroxyanthracene-9,10-dione; IHC, immunohistochemistry.
Imaging
Mouse tissues, immunofluorescence, immunohistochemistry, and X-gal (5-Bromo-4-Chloro-3-Indolyl β-D-Galactopyranoside)staining were prepared as previously described.4,47 Immunofluorescence images were taken on a Nikon Eclipse Ti2 Confocal Microscope. Bright-field images were taken on a Nikon Eclipse 80i. Immunofluorescent and Immunohistochemistry images were acquired and analyzed with the NIS-Element software package suite (Nikon) and Photoshop (Adobe). For ultrastructural analysis, tissue samples were fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Ted Pella) in 100 mmol/L cacodylate buffer, pH 7.2, for 1 hour at room temperature, and then overnight at 4°C. Samples were washed in cacodylate buffer and postfixed in 1% osmium tetroxide (Ted Pella) for 1 hour. Samples then were rinsed extensively in dH2O before en bloc staining with 1% aqueous uranyl acetate (Ted Pella) for 1 hour. After several rinses in dH2O, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX II transmission electron microscope (JEOL USA) equipped with an AMT 8-megapixel digital camera (Advanced Microscopy Techniques). Individual high magnificaiton images were stitched together using Photoshop to generate full-unit images.
scRNA-seq and RNA-seq
Uniform manifold approximation and projection (UMAP) and violin plots were generated using previously published data and methods as described by Bockerstett et al.14 Raw data sets can be downloaded from the National Center for Biotechnology Information BioSample website under the following accession numbers: BALBc SAMN13152839, TAM SAMN13152841, and TXA23 SAMN13152840. Briefly, raw data sets were mapped to a reference mouse genome using the CellRanger 6.0.2 pipeline (10x Genomics), then processed using Seurat to filter low-quality cells, normalize, scale, and integrate data sets. After identifying cluster identities, log-normalized per cell Sox9 expression was measured using the VlnPlot function. Library preparation for bulk RNA-seq was performed with 500 ng to 1 ug of total RNA. Ribosomal RNA was removed by an RNase-H method using RiboErase kits (Kapa Biosystems). Messenger RNA was reverse-transcribed to yield complementary DNA (cDNA) using SuperScript III reverse transcriptase (Life Technologies) and random hexamers. A second-strand reaction was performed to yield ds-cDNA. cDNA was blunt-ended, had an A base added to the 3' ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments then were amplified for 12–15 cycles using primers incorporating unique dual index tags. Fragments were sequenced on an NovaSeq-6000 (Illumina) using paired-end reads extending 150 bases. Quality control of the RNA-seq reads was performed using FastQC v.0.10. RNA-seq reads were mapped to the reference transcriptome and quantified to GRCm38 (Ensembl release 102) transcriptome using Salmon v.1.8.0. Differential gene expression was analyzed using the R package, edgeR.48 Before the analysis, we filtered the data by excluding genes with the lowest count per million based on the default setting of edgeR. The purpose of the filtering was to remove genes from the total data set that have little or no chance of showing significant evidence of differential expression while simultaneously increasing the detection power with a similar false discovery rate.49 Genes with a false discovery rate less than 0.05 and log2-fold change greater than 1 were identified as differentially expressed genes. A heat map of results was generated using ComplexHeatmap 2.12.1. Raw data sets for RNA-seq can be downloaded from the National Center for Biotechnology Information GEO database: GSE230101.
Quantification and Statistics
For Ki67 quantification, the bottom 50 μm of at least 60 individual unit bases in 20× fields of view were counted from each animal. Statistics for Ki67 counts were performed by analysis of variance. Statistically significant differences among various conditions in analysis of variance were determined via the Tukey post hoc tests (for multiple crosswise comparisons of means). For MIST1 quantification, total MIST1+ cells were counted per unit, and statistics were calculated using an unpaired t test.
Acknowledgments
The authors acknowledge the Washington University in St. Louis Molecular Microbiology Imaging Facility for assistance with electron microscopy sample preparation and microscopy and the Washington University in St. Louis Digestive Disease Research Core Center (P30DK052574) for assistance with histology.
CRediT Authorship Contributions
Spencer G Willet, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Nattapon Thanintorn (Data curation: Supporting, Writing – review & editing: Supporting)
Helen McNeill (Supervision: Equal; Writing – review & editing: Supporting)
Sung-Ho Huh (Writing – review & editing: Supporting)
David M. Ornitz (Writing – review & editing: Supporting)
Won Jae Huh (Data curation: Supporting)
Stella G. Hoft (Data curation: Supporting, Writing – review & editing: Supporting)
Richard J. DiPaolo (Supervision: Equal; Writing – review & editing: Supporting)
Jason C. Mills (Conceptualization: Supporting; Funding acquisition: Equal; Supervision: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by National Institutes of Health grant K01 K01DK120800 and a Digestive Diseases Research Core Center (DDRCC) Pilot and Feasibly award (S.G.W.), National Institutes of Health Research Project grants R01CA239645, 5R01DK094989, and R01DK105129 (J.C.M.), and R01HL154747 (D.M.O.), and the Washington University in St. Louis DDRCC grant P30DK052574 (Davidson).
Data Availability RNA-seq data were deposited in the GEO database of the National Center for Biotechnology Information, accession number: GSE230101.
Contributor Information
Spencer G. Willet, Email: willetsg@wustl.edu.
Jason C. Mills, Email: jason.mills@bcm.edu.
Supplementary Material
References
- 1.Goldenring J.R., Nam K.T., Mills J.C. The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the Mist. Exp Cell Res. 2011;317:2759–2764. doi: 10.1016/j.yexcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldenring J.R., Mills J.C. Cellular plasticity, reprogramming, and regeneration: metaplasia in the stomach and beyond. Gastroenterology. 2022;162:415–430. doi: 10.1053/j.gastro.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stange D.E., Koo B.K., Huch M., et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willet S.G., Lewis M.A., Miao Z.F., et al. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 2018;37 doi: 10.15252/embj.201798311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radyk M.D., Burclaff J., Willet S.G., et al. Metaplastic cells in the stomach arise, independently of stem cells, via dedifferentiation or transdifferentiation of chief cells. Gastroenterology. 2018;154:839–843 e2. doi: 10.1053/j.gastro.2017.11.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer A.R., Engevik A.C., Willet S.G., et al. Cystine/glutamate antiporter (xCT) is required for chief cell plasticity after gastric injury. Cell Mol Gastroenterol Hepatol. 2019;8:379–405. doi: 10.1016/j.jcmgh.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leushacke M., Tan S.H., Wong A., et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol. 2017;19:774–786. doi: 10.1038/ncb3541. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.H., Kim S., Han S., et al. p57(Kip2) imposes the reserve stem cell state of gastric chief cells. Cell Stem Cell. 2022;29:826–839 e9. doi: 10.1016/j.stem.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell B., Meyer A.R., Weis J.A., et al. Chief cell plasticity is the origin of metaplasia following acute injury in the stomach mucosa. Gut. 2022;71:1068–1077. doi: 10.1136/gutjnl-2021-325310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burclaff J., Willet S.G., Saenz J.B., et al. Proliferation and differentiation of gastric mucous neck and chief cells during homeostasis and injury-induced metaplasia. Gastroenterology. 2020;158:598–609 e5. doi: 10.1053/j.gastro.2019.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karam S.M., Leblond C.P. Dynamics of epithelial cells in the corpus of the mouse stomach. III. Inward migration of neck cells followed by progressive transformation into zymogenic cells. Anat Rec. 1993;236:297–313. doi: 10.1002/ar.1092360204. [DOI] [PubMed] [Google Scholar]
- 12.Han S., Fink J., Jorg D.J., et al. Defining the identity and dynamics of adult gastric isthmus stem cells. Cell Stem Cell. 2019;25:342–356 e7. doi: 10.1016/j.stem.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bockerstett K.A., Lewis S.A., Noto C.N., et al. Single-cell transcriptional analyses identify lineage-specific epithelial responses to inflammation and metaplastic development in the gastric corpus. Gastroenterology. 2020;159:2116–2129 e4. doi: 10.1053/j.gastro.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bockerstett K.A., Lewis S.A., Wolf K.J., et al. Single-cell transcriptional analyses of spasmolytic polypeptide-expressing metaplasia arising from acute drug injury and chronic inflammation in the stomach. Gut. 2020;69:1027–1038. doi: 10.1136/gutjnl-2019-318930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh W.J., Khurana S.S., Geahlen J.H., et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 2012;142:21–24 e7. doi: 10.1053/j.gastro.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeley T.M., Horita N., Samuelson L.C. Tamoxifen-induced gastric injury: effects of dose and method of administration. Cell Mol Gastroenterol Hepatol. 2019;8:365–367. doi: 10.1016/j.jcmgh.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh R.S., Shevach E.M., Margulies D.H., et al. A T cell receptor transgenic model of severe, spontaneous organ-specific autoimmunity. Eur J Immunol. 2001;31:2094–2103. doi: 10.1002/1521-4141(200107)31:7<2094::aid-immu2094>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T.L., Khurana S.S., Bellone C.J., et al. Autoimmune gastritis mediated by CD4+ T cells promotes the development of gastric cancer. Cancer Res. 2013;73:2117–2126. doi: 10.1158/0008-5472.CAN-12-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh S.H., Warchol M.E., Ornitz D.M. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. Elife. 2015;4 doi: 10.7554/eLife.05921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huh S.H., Jones J., Warchol M.E., et al. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz J.P., Perreault N., Goldstein B.G., et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Miao Z.F., Adkins-Threats M., Burclaff J.R., et al. A metformin-responsive metabolic pathway controls distinct steps in gastric progenitor fate decisions and maturation. Cell Stem Cell. 2020;26:910–925 e6. doi: 10.1016/j.stem.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bredemeyer A.J., Geahlen J.H., Weis V.G., et al. The gastric epithelial progenitor cell niche and differentiation of the zymogenic (chief) cell lineage. Dev Biol. 2009;325:211–224. doi: 10.1016/j.ydbio.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capoccia B.J., Jin R.U., Kong Y.Y., et al. The ubiquitin ligase Mindbomb 1 coordinates gastrointestinal secretory cell maturation. J Clin Invest. 2013;123:1475–1491. doi: 10.1172/JCI65703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khurana S.S., Riehl T.E., Moore B.D., et al. The hyaluronic acid receptor CD44 coordinates normal and metaplastic gastric epithelial progenitor cell proliferation. J Biol Chem. 2013;288:16085–16097. doi: 10.1074/jbc.M112.445551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teal E., Dua-Awereh M., Hirshorn S.T., et al. Role of metaplasia during gastric regeneration. Am J Physiol Cell Physiol. 2020;319:C947–C954. doi: 10.1152/ajpcell.00415.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada T., Ishimoto T., Seishima R., et al. Functional role of CD44v-xCT system in the development of spasmolytic polypeptide-expressing metaplasia. Cancer Sci. 2013;104:1323–1329. doi: 10.1111/cas.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo H.G., Jin R.U., Sibbel G., et al. A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev. 2017;31:154–171. doi: 10.1101/gad.285684.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsey V.G., Doherty J.M., Chen C.C., et al. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 30.Saenz J.B., Vargas N., Cho C.J., et al. Regulation of the double-stranded RNA response through ADAR1 licenses metaplastic reprogramming in gastric epithelium. JCI Insight. 2022;7 doi: 10.1172/jci.insight.153511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saenz J.B., Burclaff J., Mills J.C. Modeling murine gastric metaplasia through tamoxifen-induced acute parietal cell loss. Methods Mol Biol. 2016;1422:329–339. doi: 10.1007/978-1-4939-3603-8_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLaForest A., Kohlnhofer B.M., Franklin O.D., et al. GATA4 controls epithelial morphogenesis in the developing stomach to promote establishment of glandular columnar epithelium. Cell Mol Gastroenterol Hepatol. 2021;12:1391–1413. doi: 10.1016/j.jcmgh.2021.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horst D., Gu X., Bhasin M., et al. Requirement of the epithelium-specific Ets transcription factor Spdef for mucous gland cell function in the gastric antrum. J Biol Chem. 2010;285:35047–35055. doi: 10.1074/jbc.M110.164541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenny M., Uhl C., Roche C., et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C.S., Perreault N., Brestelli J.E., et al. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokubu H., Ohtsuka T., Kageyama R. Mash1 is required for neuroendocrine cell development in the glandular stomach. Genes Cells. 2008;13:41–51. doi: 10.1111/j.1365-2443.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- 37.Moore B.D., Jin R.U., Lo H., et al. Transcriptional regulation of X-Box-binding protein one (XBP1) by hepatocyte nuclear factor 4alpha (HNF4Alpha) is vital to beta-cell function. J Biol Chem. 2016;291:6146–6157. doi: 10.1074/jbc.M115.685750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huh W.J., Esen E., Geahlen J.H., et al. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 2010;139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verzi M.P., Khan A.H., Ito S., et al. Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology. 2008;135:591–600. doi: 10.1053/j.gastro.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radyk M.D., Spatz L.B., Pena B.L., et al. ATF3 induces RAB7 to govern autodegradation in paligenosis, a conserved cell plasticity program. EMBO Rep. 2021;22 doi: 10.15252/embr.202051806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miao Z.F., Lewis M.A., Cho C.J., et al. A dedicated evolutionarily conserved molecular network licenses differentiated cells to return to the cell cycle. Dev Cell. 2020;55:178–194 e7. doi: 10.1016/j.devcel.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akiyama H., Chaboissier M.C., Martin J.F., et al. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., He L., Ma F., et al. SOX9 regulates low density lipoprotein receptor-related protein 6 (LRP6) and T-cell factor 4 (TCF4) expression and Wnt/beta-catenin activation in breast cancer. J Biol Chem. 2013;288:6478–6487. doi: 10.1074/jbc.M112.419184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belteki G., Haigh J., Kabacs N., et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madisen L., Zwingman T.A., Sunkin S.M., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Habbe N., Shi G., Meguid R.A., et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobe C.G., Koop K.E., Kreppner W., et al. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 48.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourgon R., Gentleman R., Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.