Abstract
Liver fibrosis is a common result of liver injury owing to various kinds of chronic liver diseases. A deeper understanding of the pathophysiology of liver fibrosis and identifying potential therapeutic targets of liver fibrosis is important because liver fibrosis may progress to advanced liver diseases, such as cirrhosis and hepatocellular carcinoma. Despite numerous studies, the underlying mechanisms of liver fibrosis remain unclear. Mechanisms of the development and progression of liver fibrosis differ according to etiologies. Therefore, appropriate liver fibrosis models should be selected according to the purpose of the study and the type of underlying disease. Many in vivo animal and in vitro models have been developed to study liver fibrosis. However, there are no perfect preclinical models for liver fibrosis. In this review, we summarize the current in vivo and in vitro models for studying liver fibrosis and highlight emerging in vitro models, including organoids and liver-on-a-chip models. In addition, we discuss the mechanisms and limitations of each model.
Keywords: Liver Fibrosis, In Vivo, In Vitro, Experimental Model
Summary.
Because the development and progression of liver fibrosis differ based on the etiology, it is important to select an appropriate liver fibrosis model according to the purpose of study and type of disease. To study liver fibrosis, many in vivo animal and in vitro models have been developed. This review summarizes and analyzes the various in vivo and in vitro liver fibrosis models and their implications and limitations.
Liver fibrosis is fibrous scar formation by extracellular matrix (ECM) accumulation resulting from chronic liver inflammation caused by conditions including chronic viral hepatitis B and C, autoimmune hepatitis, alcoholic liver disease (ALD), primary biliary cholangitis, primary sclerosing cholangitis (PSC), and nonalcoholic fatty liver disease (NAFLD).1,2 Liver fibrosis may progress to cirrhosis and further to hepatocellular carcinoma (HCC).3 Treating underlying liver disease may ameliorate liver fibrosis, even at the cirrhosis stage.4,5 Although many targets for liver fibrosis were investigated, effective medications have not been developed.6 Therefore, exploring the precise pathophysiologies of liver fibrosis is crucial for better understanding and for discovering new therapeutic targets. Various animal models are used in preclinical studies on liver fibrosis. Because no model is perfect, the selection of relevant animal models to study target fibrotic diseases is crucial. A better understanding of the pathogenesis of each animal model and its implications and limitations is essential. In this review, we summarized the currently available in vivo and in vitro models for studying liver fibrosis and discussed emerging in vitro models.
In Vivo Models of Experimental Liver Fibrosis
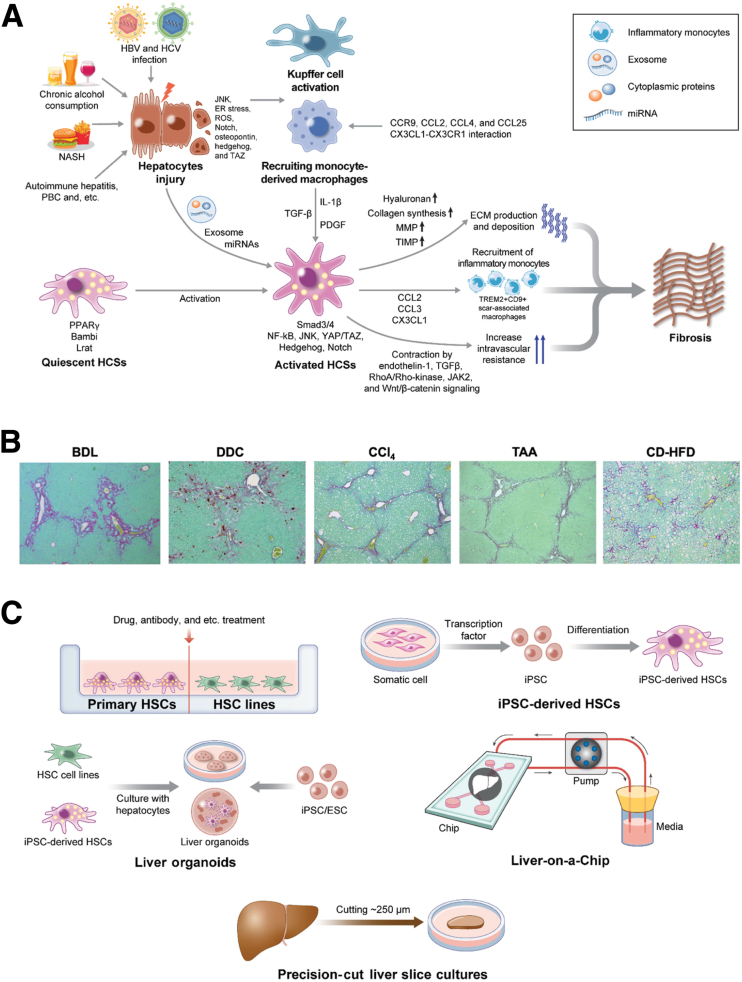
In vivo animal models are the gold standard in studying liver fibrosis. The main effectors that produce ECM in liver fibrosis are activated hepatic stellate cells (HSCs). Various types of liver cells are involved in HSC activation in liver fibrosis. These cells include immune cells (monocyte-derived macrophages, Kupffer cells, B cells, and T cells), cholangiocytes, liver sinusoidal endothelial cells (LSECs), and hepatocytes. These cells produce inflammatory and fibrotic cytokines (transforming growth factor-β [TGF-β], platelet-derived growth factor [PDGF], connective tissue growth factor [CTGF], interleukin 1β, and C-C motif chemokine ligand 2) and mediators (reactive oxygen species and nitric oxide) to affect HSC activation (Figure 1A). Because various cell types are involved in HSC activation and fibrosis, simple in vitro cell culture models cannot entirely recapitulate the disease course of liver fibrosis. To study different etiologies of liver fibrosis, hepatotoxin, cholestasis, and nonalcoholic steatohepatitis (NASH)-induced liver fibrosis models are commonly used (Table 1), and each model shows different patterns of fibrosis (Figure 1B).
Figure 1.
Mechanismof liver fibrosis and in vivo/in vitro models for styding liver fibrosis. (A) Molecular mechanism of liver fibrosis. By chronic liver injury, hepatocytes activate the signaling related to Janus kinase (JNK), Notch, osteopontin, hedgehog, and TAZ,7, 8, 9 and produce exosomes containing microRNAs (miRNAs) for HSC activation.10 Inflammation activates Kupffer cells,11 and recruits monocyte-derived macrophages via C-C motif chemokine receptor (CCR)9, C-C motif chemokine ligand (CCL)2, CCL4, and CCL25.12, 13, 14, 15 The C-X3-C motif chemokine ligand 1 (CX3CL1)–C-X3-C motif chemokine receptor 1 (CX3CR1) interaction regulates macrophage survival, differentiation, and polarization.16, 17, 18 Hepatic macrophages activate HSCs by producing TGF-β, PDGF, and interleukin (IL)1β.19 Quiescent HSCs store vitamin A–containing lipid droplets.20 Activated HSCs produce collagens and other ECM and express inflammatory chemokines CCL2, CCL3, and CX3CL1 to recruit inflammatory monocytes.21,22, 23, 24, 25 HSC-derived matrix metalloproteinase (MMP) and tissue inhibitor of metalloproteinase (TIMP) contribute to ECM maintenance, remodeling, and fibrosis.26 Activated HSCs contribute to contractile force in sinusoids to increase intravascular resistance, leading to portal hypertension, which is influenced by endothelin-1, TGF-β, RhoA/Rho-kinase, JAK2, and Wnt/β-catenin signaling.27, 28, 29 (B) Representative histology of liver fibrosis models. Sirius red staining for bile duct ligation (BDL), DDC, CCl4, TAA, and CD-HFD models. (C) Summary of in vitro model for studying liver fibrosis. Primary HSC. HSC line. iPSC. Liver organoid. Liver-on-a-Chip. Precision-cut liver slice cultures. ER, endoplasmic reticulum; HBV, hepatitis B virus; HCV, hepatitis C virus; HSC, hepatic stellate cell; NF-κB, nuclear factor-κB; PBC, primary biliary cholangitis; ROS, reactive oxygen species.
Table 1.
In Vivo Animal Models of Liver Fibrosis
| Models | Methods | Duration | Advantages | Limitations | References |
|---|---|---|---|---|---|
| CCl4 | IP injection Inhalation |
4–12 wk | Simplicity (IP) High reproducibility, induction of portal hypertension (inhalation) |
High toxicity Risk of peritonitis (IP injection) Requirement of special equipment (inhalation) |
11–13 |
| TAA | IP injection Oral administration |
6–8 wk (IP injection) 2–4 mo (oral) |
Simplicity High reproducibility |
Long time to induce liver fibrosis Highly toxic |
15, 16 |
| DMN | IP injection | 4–8 wk | Simplicity Inducing significant fibrosis |
More suitable to study HCC Risk of carcinogenesis in researcher |
17, 18 |
| HFD | Feeding | 2–6 mo (for fatty liver) 50 wk (for fibrosis) |
Inducing obesity and insulin resistance | Long time to induce liver fibrosis | 27 |
| HFD with glucose/fructose water ± cholesterol | Feeding | 16 wk to 12 mo | Significant feature of metabolic syndrome | Long time to induce liver fibrosis | 29, 30, 32, 33 |
| MCD diet | Feeding | 5–8 wk | Short time to induce fibrosis | Reduces body weight No feature of metabolic syndrome |
38, 39 |
| CDAA diet | Feeding | More than 20 wk | Body weight gain with insulin resistance Useful to study NASH-induced HCC |
Hindrance of studying liver fibrosis by HCC development | 40 |
| CD-HFD | Feeding | 6–24 wk | Human-relevant NASH fibrosis | Long time to induce liver fibrosis | 41, 43, 46, 47 |
| CDA-HFD | Feeding | 12 wk | NASH with severe fibrosis with short duration | No feature of obesity or insulin resistance | 49 |
| BDL | Surgery | 3 wk | Relevant to cholestasis Useful to study relationship between gut microbiome and fibrosis |
High surgery skill level required High mortality rate |
52–56 |
| DDC diet | Feeding | 4–8 wk | Representing chronic cholangiopathy Not requiring surgical expertise |
Not reducing bile flow | 58, 59 |
| Mdr2-/- mice | Genetically modified mice | Age 8–12 wk | Representing chronic cholangiopathy Not requiring surgical expertise |
Developing cholangiocarcinoma at age 4–6 mo Mouse genetic variability in the process of straining C57BL/6 and Balb/c |
61–66 |
| Hepatocyte-specific TAK1-/- mice | Genetically modified mice | 1 month old | Early development of liver fibrosis | Hindrance of studying liver fibrosis by HCC development | 67, 68 |
| ARE-Del-/- mice | Genetically modified mice | 20 weeks of age | Relevant to PBC | Limited data | 69 |
| Alcohol | Mixed with water or liquid diets Intragastric feeding |
10 days (NIAAA) 4–12 wk (Lieber–Decarli) 4 wk to 4 mo (Tsukamoto–French) |
Suitable for studying alcoholic liver disease | Difficult to induce liver fibrosis Need for second hit to induce fibrosis |
73–78, 81, 82 |
ARE, adenylate uridylate-rich element; BDL, bile duct ligation; CDA-HFD, choline-deficient, L-amino acid-defined, high-fat diet; HCC, hepatocellular carcinoma; IP, intraperitoneal; NIAAA, National Institute on Alcohol Abuse and Alcoholism; PBC, primary biliary cholangitis; TAK, transforming growth factor-β–activated kinase.
Hepatotoxin-Induced Liver Fibrosis Models
Carbon tetrachloride
Carbon tetrachloride (CCl4) is the most popular compound used for rodent liver fibrosis models. CCl4 is metabolized to trichloromethyl radical and trichloromethyl peroxide by cytochrome P450 2E1 (CYP2E1), directly injuring hepatocytes and endothelial cells.30, 31, 32, 33 CCl4 activates Kupffer cells and HSCs that induce liver fibrosis. Because CYP2E1 is expressed predominantly in pericentral zone 3 hepatocytes, hepatotoxic CCl4 metabolites are generated and induce hepatocyte injury and ECM deposition in the pericentral area. Liver injury caused by a single injection of CCl4 is recovered quickly. To maintain this injury and ECM deposition, repeated injections are required. The researchers should be aware that CCl4 can cause mortality in mice and that the animal should weigh at least 20 g before initiating CCl4 injection. In CCl4-induced liver fibrosis, serial processes and mechanisms contribute to repeated hepatocyte injury, inflammation, and ECM production, followed by resolution and hepatocyte proliferation, making data interpretation complicated. Although CCl4 is not used currently in daily life, it was used as a solvent in industries and is distributed in nature. Because CCl4 injures hepatocytes, this model may be proposed to study the mechanism of hepatocyte injury–induced liver fibrosis in chronic hepatitis B and C. However, hepatocyte injury in the CCl4 model does not persist, and, instead, pericentral and central–central bridging fibrosis occurs. This finding does not match the pathogenesis of chronic hepatitis B and C, and oral and intraperitoneal administration of CCl4 are common; however, an inhalational model develops cirrhosis and ascites. The inhalation model is useful for studying end-stage liver fibrosis.34 Notably, CCl4 treatment cessation rapidly inactivates HSCs, which are associated with fibrosis regression.35 Activated HSCs express collagen Type I Alpha 1 Chain, α-smooth muscle actin, and tissue inhibitor of metalloproteinase 1. HSCs reexpress their quiescent markers glial fibrillary acidic protein, peroxisome proliferator-activated receptor gamma, and bone morphogenic protein and activin membrane-bound inhibitor after resolution. Gamma-aminobutyric acid type A receptor subunit alpha3 can be used as an inactivated HSC marker.35,36 Thus, the CCl4 model is a useful model for studying the resolution mechanism. Given its high reproducibility, many researchers use this model as a primary model to study liver fibrosis.
Thioacetamide
Thioacetamide (TAA) is the second most commonly used fibrosis-inducing hepatotoxin in rodents.37 TAA itself is nontoxic, but its metabolites, TAA sulfoxide and TAA sulfdioxide, converted by CYP2E1, are hepatotoxic.38 Toxic metabolites induce oxidative stress for centrilobular necrosis and inflammation, thus activating HSCs and inducing fibrosis.39 The TAA model promotes hepatocyte damage in zones 1 and 3, and develop portal–portal and portal–central bridging fibrosis, respectively. Hepatocyte injury is progressive and persistent. Fibrosis regression is lesser after TAA discontinuation than that after CCl4 discontinuation. This model is suitable for studying fibrosis regression by treatment, but inappropriate for studying spontaneous regression. TAA administration through drinking water induces continuous liver damage, which may mimic human chronic hepatitis B and C better than the CCl4 model.
Dimethylnitrosamine
Dimethylnitrosamine (DMN) is a nitrosamine, a known carcinogen, which develops liver fibrosis in rodents and is used commonly in Asian laboratories for preclinical fibrosis studies. DMN induces iron deposition, fat accumulation, centrilobular congestion, and hemorrhagic necrosis.40,41 Fibrosis progression induces porta–portal and portal–central bridging fibrosis by enhancing collagen cross-linking, in which type III collagen is dominant compared with type I.42 The DMN model develops severe fibrosis and displays increased expression of α-smooth muscle actin,43 making it useful in studying advanced fibrosis mechanism.
NASH-Induced Fibrosis
NAFLD is becoming a major cause of chronic liver disease.44 NASH develops in 20%–25% of NAFLD patients. Patients with NASH may develop fibrosis, and some of them progress to cirrhosis.45 Because fibrosis is the most important prognostic factor,46,47 a preclinical NAFLD-fibrosis model is crucial for NAFLD research. Several NAFLD rodent models are available; however, each model has limitations.
A high-fat diet (HFD) containing 40%–60% of fat calories is suitable for studying obesity, insulin resistance, and simple steatosis. Because saturated fat promotes NASH progression better than unsaturated fat,48 a HFD mainly contains saturated fat as the main source.49 Although a HFD increases serum alanine aminotransferase levels and inflammatory gene expression after 2–6 months of feeding, it requires approximately 50 weeks to develop mild fibrosis.50 Modified HFD models can be used. Fructose, which is enriched in sodas and candies, increases hepatic de novo lipogenesis and inhibits fatty acid β-oxidation.51 Administration of a HFD and fructose-/glucose-containing drinking water for 4–6 months induces steatosis, necroinflammation, insulin resistance, and fibrosis.52,53 A HFD supplemented with 0.2%–2% cholesterol also is used because it promotes inflammation and fibrosis.54, 55, 56 The feeding of a HFD supplemented with fructose and cholesterol for 6 months induces NASH with hepatocellular ballooning, progressive fibrosis, and features of the metabolic syndrome.57 A recent systematic review showed that a HFD with a high-fructose model resembles human NAFLD.58 A hybrid model using a HFD and CCl4 is another option to study NASH fibrosis. CCl4 treatment may not be relevant to human NASH unless to study the interaction with environmental toxins. The transcriptomic analysis of this model showed similar gene expression patterns to human NASH.59 Thus, this model is a relevant preclinical model for NASH studies. A Western diet with high sugar water and low-dose CCl4 rapidly developed advanced fibrosis (12 weeks) and HCC (24 weeks).59
Methionine- and choline-deficient (MCD) diets contain high sucrose proportions and moderate amounts of fat, but are methionine and choline deficient. The deficiency of these essential components prevents the export of lipids from hepatocytes, resulting in hepatic lipid accumulation, impaired β-oxidation, and reactive oxygen species production.60 The MCD diet induces NASH in 3 weeks and fibrosis in 5–8 weeks.61 However, this diet does not show systemic metabolic phenotypes, such as body weight gain, dyslipidemia, and insulin resistance.62 Because of its irrelevant systemic metabolic state, the MCD model is less common in the liver research field in the United States. The choline-deficient L-amino acid-defined (CDAA) diet is another choline-deficient, methionine-supplemented NASH diet, which produces low choline amounts, allowing rodents to survive longer. CDAA diet feeding, after 6 months, results in body weight gain and mild insulin resistance with fibrosis.63 A longer CDAA diet feeding (84 weeks) develops HCC; thus, this model is useful to study NASH-induced HCC.63 In contrast, a choline-deficient HFD (CD-HFD) model induces major features of NASH steatosis, inflammation, and fibrosis with systemic metabolic features—body weight gain and insulin resistance. A CD-HFD causes pericellular fibrosis in 6 weeks, bridging fibrosis in 24 weeks, and HCC in 12–15 months.64,65 In patients with the metabolic syndrome, systemic choline levels are decreased as a result of an altered gut microbiome that promotes the conversion of choline to methylamine and reduces plasma levels of phosphatidyl choline.66 Decreased systemic choline levels are associated with NAFLD fibrosis, and dietary choline supplementation improves NAFLD.67, 68, 69, 70, 71 We prefer to use the CD-HFD model for NASH-fibrosis studies and consider it the human-relevant NASH-fibrosis model. Contrarily, some laboratories use a choline-deficient L-amino acid–defined HFD that is a choline-deficient, low-methionine (0.1%)–supplemented diet. This model develops NASH, fibrosis, and HCC more rapidly than the CD-HFD model, but does not show insulin resistance and weight gain.72 This model is distinct from the CD-HFD model and researchers need to select models for their research directions.
Leptin-deficient ob/ob mice develop obesity, insulin resistance, and fatty liver by hyperphagia.73 Because leptin signaling is required for HSC activation, these mice are resistant to liver fibrosis and are not suitable for studying fibrosis.
Biliary Fibrosis Models
Biliary fibrosis is a result of cholestasis, associated with defects in cellular secretion of bile or mechanical obstruction of the bile duct.74 Bile duct ligation is a surgical model that ligates the extrahepatic bile duct.75 After surgery, serum aminotransferase levels are increased for 2–3 weeks76 and bilirubin level is increased for 7 days.77 The biliary obstruction induces ductular reactions by increased biliary epithelial cell proliferation, HSC activation, and profibrotic gene expression.78,79 Periportal fibrosis begins in 10 days. Portal–portal bridging fibrosis develops 3 weeks after bile duct ligation.75 This model induces leaky gut and bacterial translocation and is suitable for studying the gut–liver axis in fibrosis. However, undeveloped surgical skills increase the mortality rate and vitamin K supplementation may increase the survival rate.80
The 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) diet is another model for cholestatic liver fibrosis. DDC feeding induces porphyrin secretion into the bile duct and the formation of porphyrin crystals and plugging in bile ducts, leading to biliary fibrosis.81,82 The DDC diet induces onion skin–like fibrosis in the periductal area after 4–8 weeks of feeding. Although this model represents chronic cholangiopathy, bile flow is not reduced.81,83
The Mdr2-/- mouse model is a genetically modified mouse model that resembles human PSCs. Mdr2 is a homolog of the human ABCB4 gene coding for the canalicular transporter that regulates biliary phospholipid excretion.84,85 Mdr2-/- mice have a defect in phospholipid secretion into bile, leading to periportal fibrosis.86 Mdr2-/- mice show increased profibrogenic gene expression at 2 weeks of age.87 Progressive biliary fibrosis develops at 4–8 weeks of age. HCC develops after 4–6 months of age.88,89 However, its genetic background, FVB/N, differs from that of the popular laboratory mouse strains C57BL/6 and Balb/c.
Another genetically modified liver fibrosis mouse model is a mouse with hepatocyte deletion of the TGF-β–activated kinase 1, a mitogen-activated protein kinase kinase kinase activated by interleukin 1, TGF-β, and Toll-like receptor ligands.90 Hepatocyte-specific TGF-β–activated kinase 1-/- mice spontaneously develop pericellular and periportal liver fibrosis from 1 month of age, followed by HCC formation at 6 months of age.91
Adenylate uridylate-rich element–Del-/- mice can be used as primary biliary cholangitis model, especially in female mice.92 Adenylate uridylate-rich element–Del-/- mice show increased expression of interferon-γ resulting in biliary epithelial injury. Fibrosis is observed at 20 weeks of age and this model is ideal to investigate primary biliary cholangitis.
Alcohol-Induced Fibrosis Models
ALD encompasses diseases from steatosis to severe forms, including alcoholic hepatitis and cirrhosis, resulting from alcohol misuse.93 Chronic alcohol consumption induces steatosis, and 20%–40% of these patients develop fibrosis.93 Although alcohol metabolism and preference are differ between human beings and rodents, partly because of different CYP2E1 activity,30,94 several ALD models are used; however, these models basically do not develop fibrosis.
A chronic ethanol-feeding (Lieber–Decarli) model is a traditional model.95 The feeding of approximately 5% ethanol-containing liquid diet for 4–12 weeks induces steatosis, inflammation, and aminotransferase increase, but no fibrosis.96,97 The intragastric ethanol (Tsukamoto–French) model continuously feeds a liquid ethanol diet to rodents, which then develop severe liver injury and inflammation compared with the Lieber–Decarli model, but fibrosis is mild.98,99 The intragastric model could induce fibrosis in combination with a HFD or Western diet.100,101 Accessibility is a limitation of this model because it requires a special surgical technique and each rodent requires an infusion pump in a separate cage.30 The chronic-binge ethanol feeding (National Institute on Alcohol Abuse and Alcoholism) model is a simple model.102 Mice are fed with 5% ethanol liquid diet for 10 days followed by a single ethanol administration on day 11. This model induces marked aminotransferase increase and steatosis, but no fibrosis.103 The combination of alcohol feeding with low-dose CCl4 treatment induces liver fibrosis.104,105 Collectively, the combination models in rodents induce fibrosis, but alcohol treatment alone does not. The cause of fibrosis, either by ethanol or other supplemental substances, thus is difficult to determine. Researchers should be aware that an adaptation period is needed to shift to feeding high concentrations of ethanol from low concentrations.
In Vitro Experimental Models for Studying Liver Fibrosis
Because various cell types contribute to liver fibrosis, in vivo models are crucial for understanding the disease mechanism. However, complex mechanisms often complicate the interpretation of in vivo models. In contrast, in vitro cell culture models are more simple and are useful for understanding the molecular mechanisms of HSC activation (Table 2 and Figure 1C).
Table 2.
In Vitro Models of Liver Fibrosis
| Models | Characteristics | Advantages | Limitations | References |
|---|---|---|---|---|
| Primary HSCs | Isolation from human and rodent liver | Fresh HSCs such as in vivo state Observation from quiescent HSCs to myofibroblasts |
Complexity of the isolation process Difficulty in isolating activated HSCs Contamination of macrophage Limited life span |
83, 85, 87, 89 |
| HSC lines | Deriving from primary HSCs Immortalization by transformation with SV40T, expression of TERT, subjection to UV light or acquirement during culturing |
Easy access with high scalability Cost effectiveness Efficiency of transfection |
Fully activated state Different response |
91, 92 |
| iPSC-derived HSCs | Induction of HSCs from PSCs using stepwise culture with various factors | Providing enough quiescent HSCs | Including immature HSCs or non-HSC lineages Nonstandardized protocol |
94–96 |
| Liver organoids | Induction from adult or fetal liver tissue, or PSCs | 3D spatial organization Disease modeling Personalized medicine |
Limited cell maturation Expression of fetal markers (PSCs) Limited source of tissue (liver tissue) |
98, 99, 101, 102 |
| Liver-on-a-chip | Culture cells in microfluidic chips with precision control of fluid flow | In vitro culture with physiological liver environment Disease modeling Suitable for testing drug toxicity |
Complex methods to culture cells on a chip Requirement of a perfusion system |
105–109 |
| Precision-cut liver slice cultures | Culture thinly sliced liver tissue | Easy and human relevant model | Short duration of viability | 110–115 |
SV40T, simian virus 40 large T-antigen; TERT, telomerase reverse transcriptase.
Primary HSCs
Primary HSCs are the gold standard in vitro model to investigate liver fibrosis pathogenesis and are isolated from human and rodent livers. In situ collagenase–pronase digestion via hepatic vessels followed by density gradient centrifugation can purify high-quality HSCs.106 Because primary HSCs from the healthy liver are quiescent and their densities are low as a result of lipid droplets,107 density gradient centrifugation effectively separates quiescent HSCs from other liver cells.106,108 Isolated HSCs are cultured on plastic dishes and activated in a time-dependent manner. Day 1 quiescent HSCs are oval-shaped, containing lipid droplets with vitamin A.20 Days 4–5 HSCs change their morphology to star-shaped pseudopodia and lose lipid droplets.109 Fully activated HSCs on day 14 become myofibroblast-like cells without lipid droplets.
Researchers can use HSCs in different activation states. Day 1 HCSs from the normal liver are quiescent and are suitable for studying the function of quiescent HSCs and the mechanism of HSC activation during spontaneous activation or with stimulation with profibrogenic factors, such as TGF-β and PDGF. We can study the function of activated HSCs after 3–7 days of culture. These cells are sensitized and their response to profibrogenic factors is altered compared with that of quiescent HSCs. When activated HSCs are used, we can use culture-activated HSCs or in vivo–activated HSCs isolated from mice with liver fibrosis. Because activated HSCs lose lipid droplets, gradient centrifugation is not effective. HSC isolation from NASH fibrosis has a similar issue because hepatocytes store lipid droplets, which prevents the separation of the HSC fraction from fat-accumulated hepatocytes. Another limitation is the contamination of inflamed liver macrophages. Additional macrophage depletion by magnetic-activated cell sorting, fluorescent-activated cell sorting with genetic labeling (eg, Coll-GFP Tg mice, L-rat Cre-Tomato mice), or by targeting autofluorescence of vitamin A after isolation, or in vivo Kupffer cell depletion by liposomal clodronate will increase the purity of HSCs.109,110 Because in vitro culturing does not reproduce the in vivo HSC activation process, the investigation of in vivo–activated HSCs may provide additional insights into HSC biology. Another consideration is the in vivo ECM conditions, which are crucial for HSC biology. Matrigel and collagen coatings mimic the in vivo ECM scaffold111 and mechanical stiffness, respectively, which helps in understanding the physiological HSC activation process.112 Thus, primary HSCs are the standard in vitro model in the liver fibrosis research field.
HSC Lines
Despite several limitations in using HSC lines that lose the original morphology and function of primary HSCs,113 HSC lines are used as alternatives to primary HSCs for in vitro experiments. HSC lines are easily accessible, highly scalable, and cost effective. Various HSC lines, such as LX-2, HSC-T6, LI90, and GRX, are currently used. HSC lines are myofibroblast-like shaped without lipid droplets, which are considered fully activated.113 LX-2 cells show 98.7% transcriptional similarity with primary HSCs.114 Although the efficiency of gene transfection to primary HSCs is low, it is high for LX-2 cells. Thus, LX-2 cells are useful for studies requiring genetic modifications. Although HSC lines have limitations because of their activated state and different responses to fibrogenic stimuli, their study advances our understanding of HSC biology.
Induced Pluripotent Stem Cell–Derived HSCs
Induced pluripotent stem cells (iPSCs) can be a source for various types of liver cells, including HSCs.115 Several protocols were reported to develop iPSC-HSCs.116, 117, 118 Day 12 iPSC-HSCs have similar gene expression profiles to primary HSCs. Human iPSC-HSCs increase the expression of fibrotic markers in response to TGF-β, lipopolysaccharide, or fetal bovine serum, and are used for studying spontaneous culture activation from the quiescent state.118 iPSC-derived HSCs are also used for high-throughput drug screening to identify therapeutic agents for liver fibrosis.118 However, these cells may include immature undifferentiated cells and some cells that differentiate into non-HSC lineages and may have different characteristics from primary HSCs. Because each protocol has minor differences, it may be necessary to standardize the use of these cells. Nonetheless, iPSC-HSCs are powerful tools for human-based in vitro study.
Liver Organoids
Three-dimensional (3D) organoids are better physiological modalities for disease modeling, drug screening, and regenerative medicine than monolayer culture systems.119,120 Organoids maintain 3D cell-to-cell and cell-to-ECM interactions, which help in studying liver fibrosis in physiological settings. Liver organoids are introduced using adult or fetal liver tissues or stem cells.120,121 A 3D co-culturing system of hepatocytes and HSCs (rat hepatocytes and HSC-T6; HepaRG cells and primary human HSCs) was established.122,123 These organoids are suitable for studying drug toxicity (eg, acetaminophen) and drug-induced fibrosis (eg, methotrexate and alcohol). iPSCs are another approach for generating hepatic organoids. Ouchi et al124 established human iPSC-derived liver organoids containing epithelial and mesenchymal lineage cells, differentiated by treating retinoid and Matrigel scaffolds. This organoid contained hepatocyte-, Kupffer cell–, and HSC-like cells, and developed fatty liver and fibrotic responses after fatty acid challenge. Fibroblast growth factor 19 treatment reduced fat accumulation and fibrosis-like phenotype in the organoid. Liver organoids can be used in the study of genetic disorders. For studying congenital hepatic fibrosis pathogenesis, Guan et al125 developed an organoid model differentiated from human iPSCs with a mutation in autosomal-recessive polycystic kidney disease edited using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology. This model is useful for studying the pathogenesis of congenital hepatic fibrosis. Enlarged bile ducts, ECM deposition, and myofibroblast expansion are observed in the autosomal-recessive polycystic kidney disease–mutant liver organoids. Thus, iPSC-derived liver organoids can be an ex vivo NAFLD-fibrosis model and used for validating drug efficacy and toxicity and for studying genetic fibrotic disorders in a 3D environment.121
Liver-on-a-Chip
The organ-on-a-chip system is an in vitro model for culturing cells in microfluidic chips with control of fluid flow, mimicking the in vivo physiological liver environment.126, 127, 128
The microfluidic chip comprises top and bottom channels separated by an ECM-coated porous membrane, mimicking a hepatic sinusoid-like architecture. The liver-on-a-chip model places hepatocytes in the top channel and LSECs and other liver nonparenchymal cells (eg, HSCs and Kupffer cells) in the bottom channel. The use of human-originated primary cells reproduces human-relevant metabolic states, cell sensitivity to substances, and physiological cell plasticity. Continuous media flow that maintains proper oxygen and nutrient concentrations and washes out metabolites and cell debris and relevant shear stress further mimics the physiological liver microenvironment with longer culture periods of up to 4 weeks. Co-culturing organ- and species-specific endothelial cells help to maintain better hepatocyte metabolism, albumin production capacity, and alcohol and drug metabolism. Fatty acids and ethanol challenges induce lipotoxicity and ethanol-induced liver damage, fat accumulation, and HSC activation, which can be human-relevant ex vivo models for studying NAFLD, ALD, and fibrosis, and for testing drugs for these diseases.129,130 This model may become an indispensable preclinical ex vivo model for validating drug safety and hepatotoxicity.131
Precision-Cut Liver Slice Cultures
Precision-cut liver slices (PCLS) are another 3D culture method that uses thin liver-sliced tissues (∼250 μm) from animal and human specimens.132 To prepare liver-sliced tissues, various instruments and equipment are required, including a tissue slicer, mechanical drill, and incubation cabinet.133 The PCLS model maintains the 3D structure, physiological ECM, and cell–cell interactions. PCLS from normal liver tissues can be stimulated with TGF-β or PDGF to test fibrogenesis. In contrast, PCLS from fibrotic livers can be used to test antifibrotic drugs. Fibrosis in PCLS can regress when treated with effective drugs.134, 135, 136, 137 A limitation is the short duration (24-48 hours) of cell viability resulting from hypoxia and down-regulated hepatocyte functions. The PCLS model is a human-relevant ex vivo model for investigating the mechanism of liver fibrosis and for testing drug efficacies.
Emerging Tools to Study HSC Heterogeneity
Single–cell RNA sequencing (scRNA-seq) is used to examine whole transcriptomics at the single-cell level, determining the heterogeneity of liver cells in normal and fibrotic livers.138 This approach reveals unique liver cell subpopulations, including scar-associated TREM2+CD9+ macrophages, ACKR1+ and PLVAP+ endothelial cells, and PDGFRα+ collagen-producing myofibroblasts. Human HSCs are separated into 2 subpopulations: GPC3+ HSCs and DBH+ HSCs.139 Unique liver subpopulations can be identified through this method.
The general scRNA-seq workflow requires a single-cell suspension from fresh liver tissues. To obtain high-quality data, high-cell viability and yield are crucial and require immediate tissue dissociation and appropriate cell isolation skills. To overcome this limitation, stored frozen tissues can be used for single nuclear RNA sequencing (snRNA-seq), which may show less variation among investigators, but still needs proper optimization of nuclear preparation.140,141 scRNA-seq is good for analyzing nonparenchymal cell populations, especially immune cell populations, but is not suitable for HSCs, LSECs, hepatocytes, and cholangiocytes.142 In contrast, snRNA-seq has the advantage of analyzing HSCs, LSECs, hepatocytes, and cholangiocytes. Although studies have attempted to understand gene profiles in different hepatic zones using zonation markers (zone 1, arginase 1; zone 3, cytochrome P450 family 2 subfamily E member 1), one limitation is that sc/snRNA-seq technology loses spatial information. Recent advancements in spatial transcriptomic and proteomic approaches (eg, NanoString Geo-MX, 10× Visium, CyTOF, and CODEX) could overcome these limitations. Although spatial proteomic analysis can be analyzed at the single-cell level, spatial transcriptomics are still based on regions of interest, not at the single-cell level. However, adding sc/snRNA-seq data can complement single-cell information on spatial analysis. Thus, appropriate uses and combinations of scRNA-seq, snRNA-seq, and spatial transcriptomic/proteomic analyses can enhance further understanding of liver cell heterogeneity in liver fibrosis.
Of Mice and Men: Future Prospective of Preclinical Models
In vitro cell culture models and the validation of results from in vitro results in animal models are the gold standard approaches for elucidating the mechanisms underlying liver fibrosis. This approach can be used to discover and test effective drugs for liver fibrosis treatment. Although many antifibrotic drug candidates have been investigated, most drugs have not been recommended for clinical trials or unsatisfactory results are observed after clinical trials.143,21 There may be a large gap in the pathophysiology of liver fibrosis between rodents and human beings. Enzyme activities crucial for drug and alcohol metabolism (hepatic CYP enzymes) are higher in human beings than those in rodents. The sensitivity of human immune cells and HSCs to fibrotic factors and lipopolysaccharide differ between human beings and rodents (eg, the human body is highly sensitive to lipopolysaccharide). The human gut microbiome is dominant in gram-negative bacteria, whereas the mouse gut microbiome is gram-positive dominant. Genetically, human beings are heterogeneous; however, most laboratory mice are inbred. All of these factors prevent the development of human-relevant liver fibrosis models.144 Therefore, we attempted to identify common mechanisms among various in vitro and in vivo models, while considering that diverse etiologies of human fibrosis have distinct mechanisms that should not be overlooked. One option is to use 3D multicellular culture systems with human-relevant cells to create an experimental platform for mini-human livers.145 Combining human-relevant in vitro cell culture and in vivo animal models will help explore novel mechanisms for liver fibrosis and test novel drugs before moving to clinical trials.
Acknowledgments
CRediT Authorship Contributions
Young-Sun Lee (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Ekihiro Seki, MD PhD (Conceptualization: Lead; Funding acquisition: Lead; Methodology: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Institutes of Health grants R01DK085252 (E.S.) and NRF-2021R1C1C1009445 from the Korean Ministry of Education, Science and Technology (Y.-S.L.).
References
- 1.Friedman S.L. Liver fibrosis–from bench to bedside. J Hepatol. 2003;38(Suppl 1):S38–S53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 2.Bataller R., Brenner D.A. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim Y.S., Kim W.R. The global impact of hepatic fibrosis and end-stage liver disease. Clin Liver Dis. 2008;12:733–746. doi: 10.1016/j.cld.2008.07.007. vii. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M.J. Reversibility of liver fibrosis and cirrhosis following treatment for hepatitis C. Gastroenterology. 2002;122:1525–1528. doi: 10.1053/gast.2002.33367. [DOI] [PubMed] [Google Scholar]
- 5.Chang T.T., Liaw Y.F., Wu S.S., et al. Long-term entecavir therapy results in the reversal of fibrosis/cirrhosis and continued histological improvement in patients with chronic hepatitis B. Hepatology. 2010;52:886–893. doi: 10.1002/hep.23785. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Liu Q., He J., Li Y. Novel therapeutic targets in liver fibrosis. Front Mol Biosci. 2021;8:766855. doi: 10.3389/fmolb.2021.766855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu C., Kim K., Wang X., et al. Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aat0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie G., Karaca G., Swiderska-Syn M., et al. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice. Hepatology. 2013;58:1801–1813. doi: 10.1002/hep.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Zheng Z., Caviglia J.M., et al. Hepatocyte TAZ/WWTR1 promotes inflammation and fibrosis in nonalcoholic steatohepatitis. Cell Metab. 2016;24:848–862. doi: 10.1016/j.cmet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y.S., Kim S.Y., Ko E., et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci Rep. 2017;7:3710. doi: 10.1038/s41598-017-03389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez Perdiguero E., Klapproth K., Schulz C., et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iredale J.P. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–548. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu P.S., Nakamoto N., Ebinuma H., et al. C-C motif chemokine receptor 9 positive macrophages activate hepatic stellate cells and promote liver fibrosis in mice. Hepatology. 2013;58:337–350. doi: 10.1002/hep.26351. [DOI] [PubMed] [Google Scholar]
- 14.Ehling J., Bartneck M., Wei X., et al. CCL2-dependent infiltrating macrophages promote angiogenesis in progressive liver fibrosis. Gut. 2014;63:1960–1971. doi: 10.1136/gutjnl-2013-306294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., You H., Fan X., Jia J. Hepatic macrophages in liver fibrosis: pathogenesis and potential therapeutic targets. BMJ Open Gastroenterol. 2016;3 doi: 10.1136/bmjgast-2016-000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlmark K.R., Zimmermann H.W., Roderburg C., et al. The fractalkine receptor CX(3)CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology. 2010;52:1769–1782. doi: 10.1002/hep.23894. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y.S., Kim M.H., Yi H.S., et al. CX(3)CR1 differentiates F4/80(low) monocytes into pro-inflammatory F4/80(high) macrophages in the liver. Sci Rep. 2018;8:15076. doi: 10.1038/s41598-018-33440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoyama T., Inokuchi S., Brenner D.A., Seki E. CX3CL1-CX3CR1 interaction prevents carbon tetrachloride-induced liver inflammation and fibrosis in mice. Hepatology. 2010;52:1390–1400. doi: 10.1002/hep.23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pradere J.P., Kluwe J., De Minicis S., et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology. 2013;58:1461–1473. doi: 10.1002/hep.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.S., Jeong W.I. Retinoic acids and hepatic stellate cells in liver disease. J Gastroenterol Hepatol. 2012;27(Suppl 2):75–79. doi: 10.1111/j.1440-1746.2011.07007.x. [DOI] [PubMed] [Google Scholar]
- 21.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 22.Seki E., De Minicis S., Osterreicher C.H., et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 23.Marra F., Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014;147:577–594 e571. doi: 10.1053/j.gastro.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 24.Xu F., Liu C., Zhou D., Zhang L. TGF-beta/SMAD pathway and its regulation in hepatic fibrosis. J Histochem Cytochem. 2016;64:157–167. doi: 10.1369/0022155415627681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallowfield J.A., Mizuno M., Kendall T.J., et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. 2007;178:5288–5295. doi: 10.4049/jimmunol.178.8.5288. [DOI] [PubMed] [Google Scholar]
- 26.Benyon R.C., Arthur M.J. Extracellular matrix degradation and the role of hepatic stellate cells. Semin Liver Dis. 2001;21:373–384. doi: 10.1055/s-2001-17552. [DOI] [PubMed] [Google Scholar]
- 27.Rockey D. The cellular pathogenesis of portal hypertension: stellate cell contractility, endothelin, and nitric oxide. Hepatology. 1997;25:2–5. doi: 10.1053/jhep.1997.v25.ajhep0250002. [DOI] [PubMed] [Google Scholar]
- 28.Zhan S., Chan C.C., Serdar B., Rockey D.C. Fibronectin stimulates endothelin-1 synthesis in rat hepatic myofibroblasts via a Src/ERK-regulated signaling pathway. Gastroenterology. 2009;136:2345–2355. doi: 10.1053/j.gastro.2009.01.062. e2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang F., Wang F., He J., et al. Regulation of hepatic stellate cell contraction and cirrhotic portal hypertension by Wnt/beta-catenin signalling via interaction with Gli1. Br J Pharmacol. 2021;178:2246–2265. doi: 10.1111/bph.15289. [DOI] [PubMed] [Google Scholar]
- 30.Yanguas S.C., Cogliati B., Willebrords J., et al. Experimental models of liver fibrosis. Arch Toxicol. 2016;90:1025–1048. doi: 10.1007/s00204-015-1543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber L.W., Boll M., Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33:105–136. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- 32.Boll M., Weber L.W., Becker E., Stampfl A. Mechanism of carbon tetrachloride-induced hepatotoxicity. Hepatocellular damage by reactive carbon tetrachloride metabolites. Z Naturforsch C J Biosci. 2001;56:649–659. doi: 10.1515/znc-2001-7-826. [DOI] [PubMed] [Google Scholar]
- 33.Slater T.F., Cheeseman K.H., Ingold K.U. Carbon tetrachloride toxicity as a model for studying free-radical mediated liver injury. Philos Trans R Soc Lond B Biol Sci. 1985;311:633–645. doi: 10.1098/rstb.1985.0169. [DOI] [PubMed] [Google Scholar]
- 34.Domenicali M., Caraceni P., Giannone F., et al. A novel model of CCl4-induced cirrhosis with ascites in the mouse. J Hepatol. 2009;51:991–999. doi: 10.1016/j.jhep.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Liu X., Xu J., Rosenthal S., et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158:1728–1744 e1714. doi: 10.1053/j.gastro.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal S.B., Liu X., Ganguly S., et al. Heterogeneity of HSCs in a mouse model of NASH. Hepatology. 2021;74:667–685. doi: 10.1002/hep.31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzhugh O.G., Nelson A.A. Liver tumors in rats fed thiourea or thioacetamide. Science. 1948;108:626–628. doi: 10.1126/science.108.2814.626. [DOI] [PubMed] [Google Scholar]
- 38.Hajovsky H., Hu G., Koen Y., et al. Metabolism and toxicity of thioacetamide and thioacetamide S-oxide in rat hepatocytes. Chem Res Toxicol. 2012;25:1955–1963. doi: 10.1021/tx3002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J.S., Wanibuchi H., Morimura K., et al. Role of CYP2E1 in thioacetamide-induced mouse hepatotoxicity. Toxicol Appl Pharmacol. 2008;228:295–300. doi: 10.1016/j.taap.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 40.He J.Y., Ge W.H., Chen Y. Iron deposition and fat accumulation in dimethylnitrosamine-induced liver fibrosis in rat. World J Gastroenterol. 2007;13:2061–2065. doi: 10.3748/wjg.v13.i14.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jezequel A.M., Mancini R., Rinaldesi M.L., et al. A morphological study of the early stages of hepatic fibrosis induced by low doses of dimethylnitrosamine in the rat. J Hepatol. 1987;5:174–181. doi: 10.1016/s0168-8278(87)80570-6. [DOI] [PubMed] [Google Scholar]
- 42.George J., Chandrakasan G. Molecular characteristics of dimethylnitrosamine induced fibrotic liver collagen. Biochim Biophys Acta. 1996;1292:215–222. doi: 10.1016/0167-4838(95)00202-2. [DOI] [PubMed] [Google Scholar]
- 43.Park H.J., Kim H.G., Wang J.H., et al. Comparison of TGF-beta, PDGF, and CTGF in hepatic fibrosis models using DMN, CCl4, and TAA. Drug Chem Toxicol. 2016;39:111–118. doi: 10.3109/01480545.2015.1052143. [DOI] [PubMed] [Google Scholar]
- 44.Riazi K., Azhari H., Charette J.H., et al. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2022;7:851–861. doi: 10.1016/S2468-1253(22)00165-0. [DOI] [PubMed] [Google Scholar]
- 45.Rinella M.E. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 46.Ekstedt M., Hagstrom H., Nasr P., et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–1554. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 47.Lee K.C., Wu P.S., Lin H.C. Pathogenesis and treatment of non-alcoholic steatohepatitis and its fibrosis. Clin Mol Hepatol. 2023;29:77–98. doi: 10.3350/cmh.2022.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosqvist F., Kullberg J., Stahlman M., et al. Overeating saturated fat promotes fatty liver and ceramides compared with polyunsaturated fat: a randomized trial. J Clin Endocrinol Metab. 2019;104:6207–6219. doi: 10.1210/jc.2019-00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eng J.M., Estall J.L. Diet-induced models of non-alcoholic fatty liver disease: food for thought on sugar, fat, and cholesterol. Cells. 2021;10:1805. doi: 10.3390/cells10071805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito M., Suzuki J., Tsujioka S., et al. Longitudinal analysis of murine steatohepatitis model induced by chronic exposure to high-fat diet. Hepatol Res. 2007;37:50–57. doi: 10.1111/j.1872-034X.2007.00008.x. [DOI] [PubMed] [Google Scholar]
- 51.Jensen T., Abdelmalek M.F., Sullivan S., et al. Fructose and sugar: a major mediator of non-alcoholic fatty liver disease. J Hepatol. 2018;68:1063–1075. doi: 10.1016/j.jhep.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohli R., Kirby M., Xanthakos S.A., et al. High-fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology. 2010;52:934–944. doi: 10.1002/hep.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radhakrishnan S., Yeung S.F., Ke J.Y., et al. Considerations when choosing high-fat, high-fructose, and high-cholesterol diets to induce experimental nonalcoholic fatty liver disease in laboratory animal models. Curr Dev Nutr. 2021;5 doi: 10.1093/cdn/nzab138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arguello G., Balboa E., Arrese M., Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim Biophys Acta. 2015;1852:1765–1778. doi: 10.1016/j.bbadis.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 55.Ioannou G.N., Subramanian S., Chait A., et al. Cholesterol crystallization within hepatocyte lipid droplets and its role in murine NASH. J Lipid Res. 2017;58:1067–1079. doi: 10.1194/jlr.M072454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buettner R., Ascher M., Gabele E., et al. Olive oil attenuates the cholesterol-induced development of nonalcoholic steatohepatitis despite increased insulin resistance in a rodent model. Horm Metab Res. 2013;45:795–801. doi: 10.1055/s-0033-1353209. [DOI] [PubMed] [Google Scholar]
- 57.Charlton M., Krishnan A., Viker K., et al. Fast food diet mouse: novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am J Physiol Gastrointest Liver Physiol. 2011;301:G825–G834. doi: 10.1152/ajpgi.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Im Y.R., Hunter H., de Gracia Hahn D., et al. A systematic review of animal models of NAFLD finds high-fat, high-fructose diets most closely resemble human NAFLD. Hepatology. 2021;74:1884–1901. doi: 10.1002/hep.31897. [DOI] [PubMed] [Google Scholar]
- 59.Tsuchida T., Lee Y.A., Fujiwara N., et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 2018;69:385–395. doi: 10.1016/j.jhep.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anstee Q.M., Goldin R.D. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farrell G., Schattenberg J.M., Leclercq I., et al. Mouse models of nonalcoholic steatohepatitis: toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi Y., Soejima Y., Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denda A., Kitayama W., Kishida H., et al. Development of hepatocellular adenomas and carcinomas associated with fibrosis in C57BL/6J male mice given a choline-deficient, L-amino acid-defined diet. Jpn J Cancer Res. 2002;93:125–132. doi: 10.1111/j.1349-7006.2002.tb01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf M.J., Adili A., Piotrowitz K., et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Febbraio M.A., Reibe S., Shalapour S., et al. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnabl B., Brenner D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guerrerio A.L., Colvin R.M., Schwartz A.K., et al. Choline intake in a large cohort of patients with nonalcoholic fatty liver disease. Am J Clin Nutr. 2012;95:892–900. doi: 10.3945/ajcn.111.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu D., Shu X.O., Xiang Y.B., et al. Higher dietary choline intake is associated with lower risk of nonalcoholic fatty liver in normal-weight Chinese women. J Nutr. 2014;144:2034–2040. doi: 10.3945/jn.114.197533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer M.D., Hamp T.J., Reid R.W., et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imajo K., Yoneda M., Fujita K., et al. Oral choline tolerance test as a novel noninvasive method for predicting nonalcoholic steatohepatitis. J Gastroenterol. 2014;49:295–304. doi: 10.1007/s00535-013-0776-3. [DOI] [PubMed] [Google Scholar]
- 71.Sherriff J.L., O'Sullivan T.A., Properzi C., et al. Choline, its potential role in nonalcoholic fatty liver disease, and the case for human and bacterial genes. Adv Nutr. 2016;7:5–13. doi: 10.3945/an.114.007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsumoto M., Hada N., Sakamaki Y., et al. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94:93–103. doi: 10.1111/iep.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikejima K., Takei Y., Honda H., et al. Leptin receptor-mediated signaling regulates hepatic fibrogenesis and remodeling of extracellular matrix in the rat. Gastroenterology. 2002;122:1399–1410. doi: 10.1053/gast.2002.32995. [DOI] [PubMed] [Google Scholar]
- 74.Hirschfield G.M., Heathcote E.J., Gershwin M.E. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology. 2010;139:1481–1496. doi: 10.1053/j.gastro.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 75.Tag C.G., Sauer-Lehnen S., Weiskirchen S., et al. Bile duct ligation in mice: induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J Vis Exp. 2015;96:52438. doi: 10.3791/52438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarcin O., Basaranoglu M., Tahan V., et al. Time course of collagen peak in bile duct-ligated rats. BMC Gastroenterol. 2011;11:45. doi: 10.1186/1471-230X-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huss S., Schmitz J., Goltz D., et al. Development and evaluation of an open source Delphi-based software for morphometric quantification of liver fibrosis. Fibrogenesis Tissue Repair. 2010;3:10. doi: 10.1186/1755-1536-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Georgiev P., Jochum W., Heinrich S., et al. Characterization of time-related changes after experimental bile duct ligation. Br J Surg. 2008;95:646–656. doi: 10.1002/bjs.6050. [DOI] [PubMed] [Google Scholar]
- 79.Iwaisako K., Jiang C., Zhang M., et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111:E3297–E3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beck P.L., Lee S.S. Vitamin K1 improves survival in bile-duct-ligated rats with cirrhosis. J Hepatol. 1995;23:235. doi: 10.1016/0168-8278(95)80345-9. [DOI] [PubMed] [Google Scholar]
- 81.Fickert P., Stoger U., Fuchsbichler A., et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am J Pathol. 2007;171:525–536. doi: 10.2353/ajpath.2007.061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deng X., Zhang X., Li W., et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122 e113. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 83.Erlinger S. What is cholestasis in 1985? J Hepatol. 1985;1:687–693. doi: 10.1016/s0168-8278(85)80012-x. [DOI] [PubMed] [Google Scholar]
- 84.Trauner M., Fickert P., Wagner M. MDR3 (ABCB4) defects: a paradigm for the genetics of adult cholestatic syndromes. Semin Liver Dis. 2007;27:77–98. doi: 10.1055/s-2006-960172. [DOI] [PubMed] [Google Scholar]
- 85.Morita S.Y., Tsuda T., Horikami M., et al. Bile salt-stimulated phospholipid efflux mediated by ABCB4 localized in nonraft membranes. J Lipid Res. 2013;54:1221–1230. doi: 10.1194/jlr.M032425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fickert P., Fuchsbichler A., Wagner M., et al. Regurgitation of bile acids from leaky bile ducts causes sclerosing cholangitis in Mdr2 (Abcb4) knockout mice. Gastroenterology. 2004;127:261–274. doi: 10.1053/j.gastro.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 87.Popov Y., Patsenker E., Fickert P., et al. Mdr2 (Abcb4)-/- mice spontaneously develop severe biliary fibrosis via massive dysregulation of pro- and antifibrogenic genes. J Hepatol. 2005;43:1045–1054. doi: 10.1016/j.jhep.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 88.Mauad T.H., van Nieuwkerk C.M., Dingemans K.P., et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol. 1994;145:1237–1245. [PMC free article] [PubMed] [Google Scholar]
- 89.Ikenaga N., Liu S.B., Sverdlov D.Y., et al. A new Mdr2(-/-) mouse model of sclerosing cholangitis with rapid fibrosis progression, early-onset portal hypertension, and liver cancer. Am J Pathol. 2015;185:325–334. doi: 10.1016/j.ajpath.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 90.Wang W., Gao W., Zhu Q., et al. TAK1: a molecular link between liver inflammation, fibrosis, steatosis, and carcinogenesis. Front Cell Dev Biol. 2021;9:734749. doi: 10.3389/fcell.2021.734749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song I.J., Yang Y.M., Inokuchi-Shimizu S., et al. The contribution of toll-like receptor signaling to the development of liver fibrosis and cancer in hepatocyte-specific TAK1-deleted mice. Int J Cancer. 2018;142:81–91. doi: 10.1002/ijc.31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bae H.R., Leung P.S., Tsuneyama K., et al. Chronic expression of interferon-gamma leads to murine autoimmune cholangitis with a female predominance. Hepatology. 2016;64:1189–1201. doi: 10.1002/hep.28641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crabb D.W., Im G.Y., Szabo G., et al. Diagnosis and treatment of alcohol-associated liver diseases: 2019 practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2020;71:306–333. doi: 10.1002/hep.30866. [DOI] [PubMed] [Google Scholar]
- 94.Holmes R.S., Duley J.A., Algar E.M., et al. Biochemical and genetic studies on enzymes of alcohol metabolism: the mouse as a model organism for human studies. Alcohol Alcohol. 1986;21:41–56. [PubMed] [Google Scholar]
- 95.Lieber C.S., DeCarli L.M., Sorrell M.F. Experimental methods of ethanol administration. Hepatology. 1989;10:501–510. doi: 10.1002/hep.1840100417. [DOI] [PubMed] [Google Scholar]
- 96.Mathews S., Xu M., Wang H., et al. Animals models of gastrointestinal and liver diseases. Animal models of alcohol-induced liver disease: pathophysiology, translational relevance, and challenges. Am J Physiol Gastrointest Liver Physiol. 2014;306:G819–G823. doi: 10.1152/ajpgi.00041.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liang S., Zhong Z., Kim S.Y., et al. Murine macrophage autophagy protects against alcohol-induced liver injury by degrading interferon regulatory factor 1 (IRF1) and removing damaged mitochondria. J Biol Chem. 2019;294:12359–12369. doi: 10.1074/jbc.RA119.007409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsukamoto H., French S.W., Benson N., et al. Severe and progressive steatosis and focal necrosis in rat liver induced by continuous intragastric infusion of ethanol and low fat diet. Hepatology. 1985;5:224–232. doi: 10.1002/hep.1840050212. [DOI] [PubMed] [Google Scholar]
- 99.Ueno A., Lazaro R., Wang P.Y., et al. Mouse intragastric infusion (iG) model. Nat Protoc. 2012;7:771–781. doi: 10.1038/nprot.2012.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.French S.W., Miyamoto K., Tsukamoto H. Ethanol-induced hepatic fibrosis in the rat: role of the amount of dietary fat. Alcohol Clin Exp Res. 1986;10:13S–19S. doi: 10.1111/j.1530-0277.1986.tb05175.x. [DOI] [PubMed] [Google Scholar]
- 101.Kisseleva T., Cong M., Paik Y., et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ki S.H., Park O., Zheng M., et al. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bertola A., Mathews S., Ki S.H., et al. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hall P.D., Plummer J.L., Ilsley A.H., Cousins M.J. Hepatic fibrosis and cirrhosis after chronic administration of alcohol and "low-dose" carbon tetrachloride vapor in the rat. Hepatology. 1991;13:815–819. doi: 10.1016/0270-9139(91)90246-r. [DOI] [PubMed] [Google Scholar]
- 105.Brol M.J., Rosch F., Schierwagen R., et al. Combination of CCl4 with alcoholic and metabolic injuries mimics human liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2019;317:G182–G194. doi: 10.1152/ajpgi.00361.2018. [DOI] [PubMed] [Google Scholar]
- 106.Ramm G.A. Isolation and culture of rat hepatic stellate cells. J Gastroenterol Hepatol. 1998;13:846–851. doi: 10.1111/j.1440-1746.1998.tb00747.x. [DOI] [PubMed] [Google Scholar]
- 107.Blaner W.S., O'Byrne S.M., Wongsiriroj N., et al. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weiskirchen R., Gressner A.M. Isolation and culture of hepatic stellate cells. Methods Mol Med. 2005;117:99–113. doi: 10.1385/1-59259-940-0:099. [DOI] [PubMed] [Google Scholar]
- 109.Chang W., Yang M., Song L., et al. Isolation and culture of hepatic stellate cells from mouse liver. Acta Biochim Biophys Sin (Shanghai) 2014;46:291–298. doi: 10.1093/abbs/gmt143. [DOI] [PubMed] [Google Scholar]
- 110.Mederacke I., Hsu C.C., Troeger J.S., et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gaca M.D., Zhou X., Issa R., et al. Basement membrane-like matrix inhibits proliferation and collagen synthesis by activated rat hepatic stellate cells: evidence for matrix-dependent deactivation of stellate cells. Matrix Biol. 2003;22:229–239. doi: 10.1016/s0945-053x(03)00017-9. [DOI] [PubMed] [Google Scholar]
- 112.Olsen A.L., Bloomer S.A., Chan E.P., et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Herrmann J., Gressner A.M., Weiskirchen R. Immortal hepatic stellate cell lines: useful tools to study hepatic stellate cell biology and function? J Cell Mol Med. 2007;11:704–722. doi: 10.1111/j.1582-4934.2007.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu L., Hui A.Y., Albanis E., et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moradi S., Mahdizadeh H., Saric T., et al. Research and therapy with induced pluripotent stem cells (iPSCs): social, legal, and ethical considerations. Stem Cell Res Ther. 2019;10:341. doi: 10.1186/s13287-019-1455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coll M., Perea L., Boon R., et al. Generation of hepatic stellate cells from human pluripotent stem cells enables in vitro modeling of liver fibrosis. Cell Stem Cell. 2018;23:101–113 e107. doi: 10.1016/j.stem.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 117.Vallverdu J., Martinez Garcia de la Torre R.A., Mannaerts I., et al. Directed differentiation of human induced pluripotent stem cells to hepatic stellate cells. Nat Protoc. 2021;16:2542–2563. doi: 10.1038/s41596-021-00509-1. [DOI] [PubMed] [Google Scholar]
- 118.Koui Y., Himeno M., Mori Y., et al. Development of human iPSC-derived quiescent hepatic stellate cell-like cells for drug discovery and in vitro disease modeling. Stem Cell Reports. 2021;16:3050–3063. doi: 10.1016/j.stemcr.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 120.Bao Y.L., Wang L., Pan H.T., et al. Animal and organoid models of liver fibrosis. Front Physiol. 2021;12:666138. doi: 10.3389/fphys.2021.666138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nuciforo S., Heim M.H. Organoids to model liver disease. JHEP Rep. 2021;3:100198. doi: 10.1016/j.jhepr.2020.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abu-Absi S.F., Hansen L.K., Hu W.S. Three-dimensional co-culture of hepatocytes and stellate cells. Cytotechnology. 2004;45:125–140. doi: 10.1007/s10616-004-7996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leite S.B., Roosens T., El Taghdouini A., et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials. 2016;78:1–10. doi: 10.1016/j.biomaterials.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 124.Ouchi R., Togo S., Kimura M., et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384 e376. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guan Y., Enejder A., Wang M., et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat Commun. 2021;12:6138. doi: 10.1038/s41467-021-26410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hayward K.L., Kouthouridis S., Zhang B. Organ-on-a-Chip systems for modeling pathological tissue morphogenesis associated with fibrosis and cancer. ACS Biomater Sci Eng. 2021;7:2900–2925. doi: 10.1021/acsbiomaterials.0c01089. [DOI] [PubMed] [Google Scholar]
- 127.Jang K.J., Otieno M.A., Ronxhi J., et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aax5516. [DOI] [PubMed] [Google Scholar]
- 128.Nawroth J.C., Petropolis D.B., Manatakis D.V., et al. Modeling alcohol-associated liver disease in a human Liver-Chip. Cell Rep. 2021;36:109393. doi: 10.1016/j.celrep.2021.109393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Freag M.S., Namgung B., Reyna Fernandez M.E., et al. Human nonalcoholic steatohepatitis on a chip. Hepatol Commun. 2021;5:217–233. doi: 10.1002/hep4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Westerouen Van Meeteren M.J., Drenth J.P.H., Tjwa E. Elafibranor: a potential drug for the treatment of nonalcoholic steatohepatitis (NASH) Expert Opin Investig Drugs. 2020;29:117–123. doi: 10.1080/13543784.2020.1668375. [DOI] [PubMed] [Google Scholar]
- 131.Vernetti L.A., Senutovitch N., Boltz R., et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med (Maywood) 2016;241:101–114. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dewyse L., Reynaert H., van Grunsven L.A. Best practices and progress in precision-cut liver slice cultures. Int J Mol Sci. 2021;22:7137. doi: 10.3390/ijms22137137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Graaf I.A., Olinga P., de Jager M.H., et al. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat Protoc. 2010;5:1540–1551. doi: 10.1038/nprot.2010.111. [DOI] [PubMed] [Google Scholar]
- 134.van de Bovenkamp M., Groothuis G.M., Meijer D.K., Olinga P. Precision-cut fibrotic rat liver slices as a new model to test the effects of anti-fibrotic drugs in vitro. J Hepatol. 2006;45:696–703. doi: 10.1016/j.jhep.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 135.Westra I.M., Mutsaers H.A., Luangmonkong T., et al. Human precision-cut liver slices as a model to test antifibrotic drugs in the early onset of liver fibrosis. Toxicol. Vitro. 2016;35:77–85. doi: 10.1016/j.tiv.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 136.Ijssennagger N., Janssen A.W.F., Milona A., et al. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J Hepatol. 2016;64:1158–1166. doi: 10.1016/j.jhep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 137.Dewyse L., De Smet V., Verhulst S., et al. Improved precision-cut liver slice cultures for testing drug-induced liver fibrosis. Front Med (Lausanne) 2022;9:862185. doi: 10.3389/fmed.2022.862185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramachandran P., Dobie R., Wilson-Kanamori J.R., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Payen V.L., Lavergne A., Alevra Sarika N., et al. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021;3:100278. doi: 10.1016/j.jhepr.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arazi A., Rao D.A., Berthier C.C., et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20:902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Strzelecki M., Yin K., Talavera-Lopez C., Martinez-Jimenez C.P. Isolation of nuclei from flash-frozen liver tissue for single-cell multiomics. J Vis Exp. 2022:190. doi: 10.3791/64792. [DOI] [PubMed] [Google Scholar]
- 142.Andrews T.S., Atif J., Liu J.C., et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol Commun. 2022;6:821–840. doi: 10.1002/hep4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Friedman S.L., Pinzani M. Hepatic fibrosis 2022: unmet needs and a blueprint for the future. Hepatology. 2022;75:473–488. doi: 10.1002/hep.32285. [DOI] [PubMed] [Google Scholar]
- 144.Delire B., Starkel P., Leclercq I. Animal models for fibrotic liver diseases: what we have, what we need, and what is under development. J Clin Transl Hepatol. 2015;3:53–66. doi: 10.14218/JCTH.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saxton S.H., Stevens K.R. 2D and 3D liver models. J Hepatol. 2022;78:873–875. doi: 10.1016/j.jhep.2022.06.022. [DOI] [PubMed] [Google Scholar]