Abstract
Few studies have investigated the properties and protein composition of small extracellular vesicles (sEVs) derived from neurons under hypoxic conditions. Presently, the extent of the involvement of these plentiful sEVs in the onset and progression of ischemic stroke remains an unresolved question. Our study systematically identified the characteristics of sEVs derived from neurons under hypoxic conditions (HypEVs) by physical characterization, sEV absorption, proteomics and transcriptomics analysis. The effects of HypEVs on neurites, cell survival, and neuron structure were assessed in vitro and in vivo by neural complexity tests, magnetic resonance imaging (MRI), Golgi staining, and Western blotting of synaptic plasticity-related proteins and apoptotic proteins. Knockdown of Fused in Sarcoma (FUS) small interfering RNA (siRNA) was used to validate FUS-mediated HypEV neuroprotection and mitochondrial mRNA release. Hypoxia promoted the secretion of sEVs, and HypEVs were more easily taken up and utilized by recipient cells. The MRI results illustrated that the cerebral infarction volume was reduced by 45% with the application of HypEVs, in comparison to the non- HypEV treatment group. Mechanistically, the FUS protein is necessary for the uptake and neuroprotection of HypEVs against ischemic stroke as well as carrying a large amount of mitochondrial mRNA in HypEVs. However, FUS knockdown attenuated the neuroprotective rescue capabilities of HypEVs. Our comprehensive dataset clearly illustrates that FUS-mediated HypEVs deliver exceptional neuroprotective effects against ischemic stroke, primarily through the maintenance of neurite integrity and the reduction of mitochondria-associated apoptosis.
Keywords: Hypoxia, FUS, Neuron-derived small extracellular vesicle, Neuroprotection, Mitochondrial mRNA
Graphical abstract
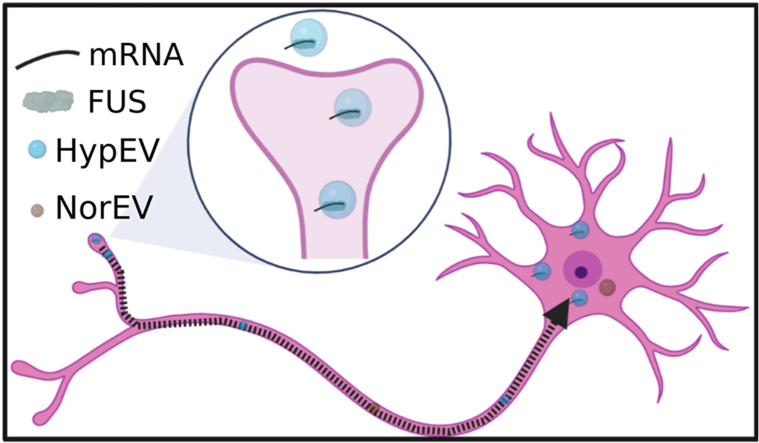
Neurons release more sEVs after being stimulated by hypoxia and promote the entry of FUS into sEVs. Hypoxic sEVs (HypEVs) are more likely to enter receptor neurons. HypEVs containing FUS and mitochondrial mRNA enter neurons through neurite terminals and then move to the cell body to accumulate, triggering neuroprotective effects.
Neurons release more sEVs after being stimulated by hypoxia and promote the entry of FUS into sEVs. Hypoxic sEVs (HypEVs) are more likely to enter receptor neurons. HypEVs containing FUS and mitochondrial mRNA enter neurons through neurite terminals and then move to the cell body to accumulate, triggering neuroprotective effects.
Highlights
-
•
Hypoxia promote the secretion of neuronal sEV.
-
•
Hypoxic sEVs are more easily uptaken by ischemic neurons than normoxic sEVs.
-
•
Primary cortical neuron-derived hypoxic sEV exerts a conspicuous neuroprotection.
-
•
RNA binding protein FUS is essential for hypoxia neuronal sEV's neuroprotection and uptake.
-
•
FUS mediate mitochondrial mRNA transport, reduce mitochondria-associated apoptosis.
1. Introduction
Small extracellular vesicles (sEVs) are structurally and functionally pleiotropic, naturally occurring phospholipid bilayer nanovesicles (approximately 30–150 nm in diameter). They are secreted by almost all cells and perform important functions for intercellular communication by transporting bioactive cargoes and activating signal transduction pathways in target cells [1]. sEVs maintain metabolic homeostasis in the brain by shuttling between neurons, glial cells, and endothelial cells [[2], [3], [4], [5]]. Severe stress, such as hypoxic conditions, can alter the release and contents of sEVs, resulting in either damaging or protective effects [6]. These phenomena have been evidenced in various studies, including those on ischemic stroke [[6], [7], [8], [9]]. In addiction, sEVs derived from hypoxic mesenchymal stromal cells have exhibited potential therapeutic effects in cerebral ischemic injury by promoting angiogenesis [9,10]. However, the role of sEVs derived from damaged neurons within the brain, in either promoting recovery or exacerbating the effects of ischemic stroke, remains uncertain. As such, both neuron-specific sEVs and neuron-targeted sEVs merit further exploration [11,12].
Previous studies have shown that sEV cargo molecules vary considerably depending on the severity of hypoxia and the health state of the donor cells, resulting in extremely different functional outcomes in target cells [13,14]. sEVs can transport RNA which associated with cell damage or provide key nutritional factors for cell survival. In the process of transporting coding and noncoding RNA, RNA-binding proteins (RBPs) are responsible for packaging RNA into sEVs and facilitating RNA transport through RNA-ribonucleoprotein (RNP) complexes [[15], [16], [17], [18], [19]]. Furthermore, RBPs play a vital role in the transport and localization of mRNA at the synapses, which are key processes that allow neurons to quickly respond to stimuli in varying environments by facilitating rapid protein synthesis [20]. In the central nervous system, the disturbance of the RBP equilibrium is responsible for neuropsychiatric and neurodegenerative disorders onset [21]. Identifying the key RBPs that regulate the transportation of RNA via sEVs in hypoxic conditions is necessary to optimize the RNA cargo of sEVs and enhance their potential for drug delivery in future applications.
Herein, we comprehensively describe the properties of sEVs derived from neurons under hypoxic conditions. After identifying the sEV uptake preference of neurons, we evaluated the neuroprotective effects of hypoxia-conditioned sEVs in the cortex of an ischemia mouse model by stereotactic microinjection into the cortex or nasal drops. Our findings, based on mass spectrometry and RNA sequencing, showed a high expression of RNA-binding proteins (RBPs) and mitochondrial mRNA within the sEVs under hypoxic conditions. We also demonstrated that the fused in sarcoma/translocated in liposarcoma protein (FUS/TLS), a RNA-binding protein, plays a significant role in regulation of neuroprotection and mitochondrial mRNA transport (Fig. 1). This study proposes a promising strategy for effective treatment of ischemic stroke and may also guide and enhance our understanding into tissue engineering research, particularly in the research involving cells and sEVs in hypoxic conditions.
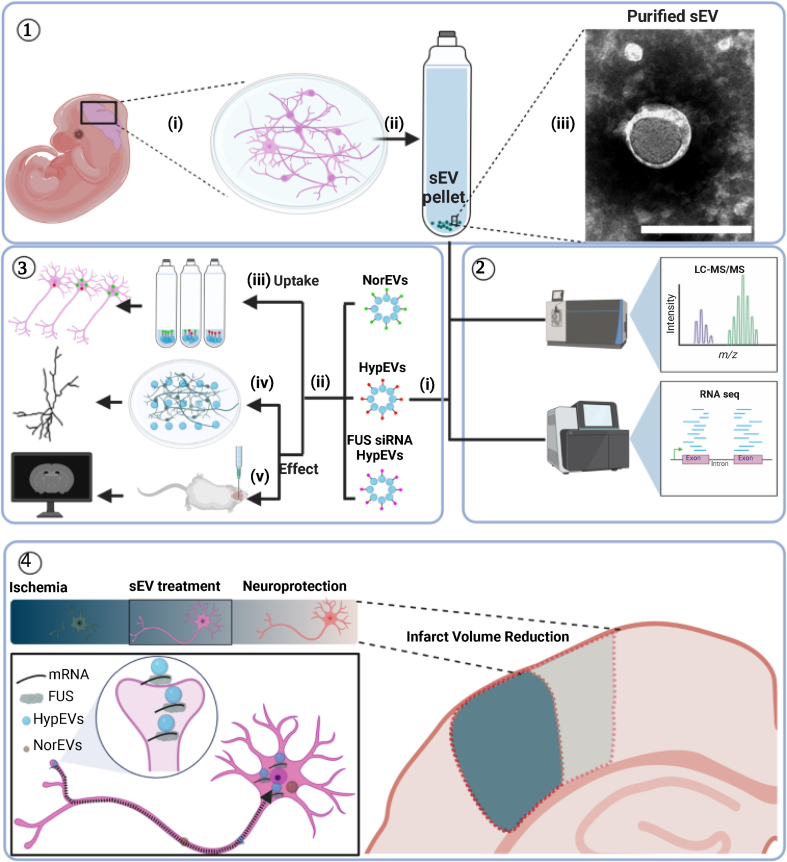
Fig. 1.
Schematic illustration of cortical neuron-derived sEV extraction, characterization, and neuroprotective analysis. ① In vitro culture of primary cortical neurons (i) and extraction of sEVs by ultracentrifugation (ii and iii). ② Proteomic and transcriptomic analysis of neuronal sEVs derived from different culture conditions. ③ Neuronal sEVs were extracted from neurons under normoxic and hypoxic conditions, as well as from FUS siRNA-treated neurons under hypoxic conditions (i), and incubated with neurons or injected into mouse brains (ii). sEV uptake (iii) and neuroprotective effect were analyzed in vitro (iv) and in vivo (v). ④ Compared with normoxic sEVs (NorEVs), hypoxic sEVs (HypEVs) were more likely to enter receptor neurons and exhibit significant neuroprotective effects by maintaining synaptic function and neurite integrity and reducing infarct volume. HypEVs containing FUS and mitochondrial mRNA enter neurons through neurite terminals and then move to the cell body to accumulate, triggering neuroprotective effects.
2. Results
2.1. Hypoxia can stimulate the release and uptake of neural sEVs
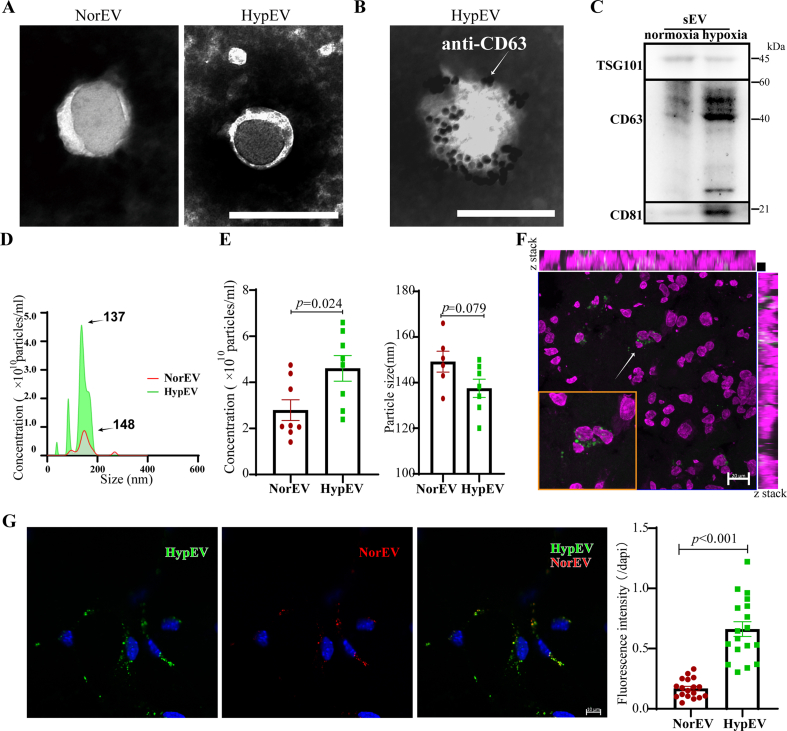
To determine the location of sEVs in the ischemic brain, sEVs derived from neurons under hypoxic conditions (HypEVs) were extracted by ultracentrifugation and labeled with PKH67; subsequently, they were administered intranasally in a mouse ischemia brain model by photothrombosis (PT) or co-incubated with primary cortical neurons under hypoxia stress conditions in vitro (Fig. 1 ③ii). Hypoxia stress was conducted with the oxygen–glucose deprivation/reperfusion (OGD/R) model, and HIF-1α was used to detect hypoxia conditions (Fig. S5B). Initially, we characterized the sEVs by transmission electron microscopy (TEM) and nanoparticle tracking analysis. TEM revealed that the extracted sEVs had a cup-shaped with their membranes expressed CD63 (Fig. 2A and B) and nanoparticle tracking analysis showed diameters ranging from 100 to 200 nm (Fig. 2D). Further investigation via western blotting demonstrated that both sEVs derived from neurons under normoxic conditions (NorEVs) and HypEVs expressed characteristic sEV marker proteins such as CD63, CD81, and TSG101 (Fig. 2C). Importantly, neither type of sEV expressed calnexin, which is a negative marker for sEVs (Fig. S5A). Notably, HypEVs expressed more sEV membrane marker proteins than NorEVs under the identical protein loading conditions. Moreover, we observed that from an equivalent number of neurons, the output production of HypEVs was higher than that of NorEVs, with no difference in particle size (Fig. 2D and E). This finding may be related to the increased quantity of HypEVs compared with NorEV; as the quantity of sEVs increases, the surface area available to express membrane marker proteins present at equivalent concentrations also increases.
Fig. 2.
Investigating the characteristics between sEVs released from primary cortical neurons under normal and hypoxia stress conditions. (A) Transmission electron microscopy (TEM) images of sEVs derived from neurons under normoxic conditions (NorEVs) and hypoxia conditions (HypEVs). Scale bar = 200 nm. (B) Immunogold electron microscopy of CD63-positive HypEV. White arrow indicates 10 nm gold-labeled antibodies. Scale bar = 200 nm. (C) Western blotting analysis of TSG101, CD63, and CD81 expression in NorEV and HypEV. (D) and (E) The size distribution and concentration analysis of NorEVs and HypEVs. Data are expressed as means ± SEM (n = 7). Statistical significance was assessed by Student's t-test; differences between HypEV and NorEV were considered significant when p < 0.05. (F) In vivo fluorescent imaging of PKH67-labeled HypEVs (green) demonstrates its ingestion by peri-infarct neurons (bottom left frame) in PT mice (n = 4). Scale bar = 20 μm. Nissl staining is magenta. (G) Confocal fluorescence images depicting the ratio of sEV uptake by ischemic neurons after co-culturing with a mixture of NorEVs (PKH26, red) and HypEVs (PKH67, green). Scale bar = 10 μm. Nuclei are stained blue (DAPI). Data are expressed as means ± SEM (n = 18 neurons from three experiments). Statistical significance was assessed by Student's t-test; differences between groups were considered significant when p < 0.05.
Subsequently, sEVs were then administered to mice with PT injury by nasal dropping. We then identified the cells that had absorbed the sEVs, which were derived from cultured primary cortical neurons. We found that many of these sEVs entered peri-infarct neurons identified by Nissl staining (Fig. 2F, Fig. S6B), and PKH67-labeled HypEVs were more easily taken up than PKH26-labeled NorEVs (Fig. S6A).
Additionally, we conducted in vitro sEV uptake experiments to evaluate the uptake of NorEVs and HypEVs by neurons. Notably, we observed that neurons more readily absorbed HypEVs compared to NorEVs (Fig. 2G). This pattern was similarly identified with sEVs derived from SH-sy5y cells (Fig. S2B). To substantiate these findings, we undertook fluorescence quantification analysis using flow cytometry. The results obtained through this method were in alignment with those garnered via confocal laser scanning microscopy, further reinforcing our observations (Fig. S10A). Particularly, live-cell imaging experiments revealed that neuronal sEVs entered the neurons via the dendrites and then moved toward the soma after coincubation with recipient neurons, and they finally accumulated with continuous uptake around the soma for at least 24 h (Video 1, Fig. S8).
These findings suggest that HypEVs are involved in brain ischemia/reperfusion pathophysiological processes and trigger some form of reaction in neurons by releasing certain molecular signals. In addition, the results showed that HypEVs are especially prone to entering neurons via contact with neuronal filaments.
2.2. Proteomic profiling reveals upregulation of FUS in neuron-derived HypEVs
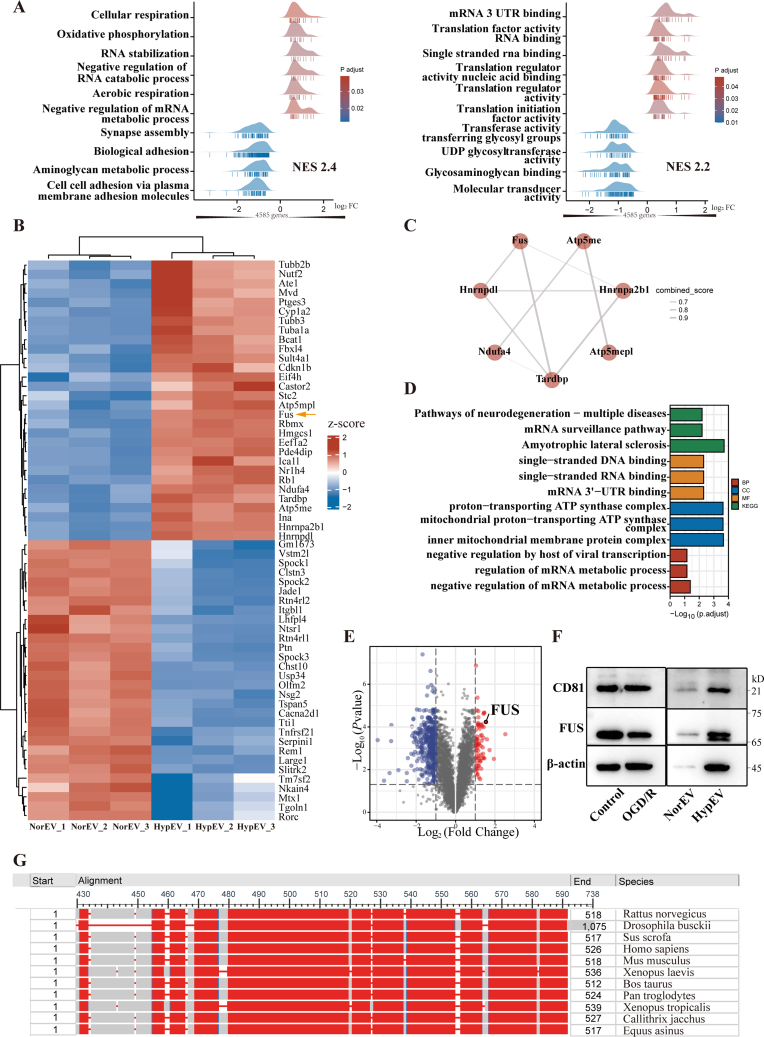
Liquid chromatography/mass spectrometry (LC/MS) was used to identify the protein profiles of neuron-derived sEVs. All identified proteins underwent gene ontology (GO) analysis using DAVID (Table S1.1), which showed that the proteins were related to intercellular exosomes (cellular component, CC), protein transport, mRNA processing (biological process, BP), and RNA binding (molecular function, MF). These results are consistent with the characteristics of sEVs, indicating that our methods of exosome isolation and mass spectrometry were valid (Fig. S1). A total of 4585 proteins were ranked using log 2 fold change (FC) and used for Gene Set Enrichment Analysis (GSEA) (Fig. 3A). The results of the top 10 enriched gene sets of BP suggested that the sEV proteins were related to cellular oxidative phosphorylation, mRNA metabolism, and synapse assembly; the results of CC suggested that proteins were mainly located in the mitochondria, plasma membrane, and synaptic membrane; and the results of MF suggested that the proteins’ functions were related to RNA binding and translational regulation (adjusted p-value <0.05) (Table S1.2–1.4).
Fig. 3.
Proteomics profiles of sEV released from neurons under normoxic and hypoxic conditions. (A) Top 10 enrichment gene sets of all 4585 proteins detected by liquid chromatography/mass spectrometry (LC/MS) using the gene set enrichment analysis (GSEA) database. Left panel: biological process (BP); right panel: molecular function (MF). NES = normalized enrichment score. (B) Heatmap of top 30 upregulated and downregulated proteins (HypEVs vs. NorEVs). FUS is indicated by the yellow arrow. Red: upregulated proteins; blue: downregulated proteins. (C) Protein-protein interaction (PPI) network of 60 proteins (as indicated in the heatmap of Fig. 3B) using STRING. The thicker the line, the stronger the combination. (D) Gene ontology (GO) enrichment analysis of the seven proteins in the PPI network (as shown in C). (E) Volcano plot of twofold differentially expressed proteins (HypEVs vs. NorEVs); FUS is indicated. (F) Representative western blotting of FUS and the sEV marker CD81 for NorEVs, HypEVs, and their parent cells. FUS was significantly upregulated in HypEVs. (G) Conserved sequence analysis of FUS in different species. Sequences in red represent the conserved regions.
We then separately clustered the top 30 differentially expressed proteins (as displayed by a heatmap) by comparing HypEVs with NorEVs and analyzed the protein-protein interactions (PPIs) with STRING software (minimum required interaction score: 0.6) (Fig. 3B and C). The results demonstrated that most upregulated proteins were clustered, revealing a highly interconnected network of RNA-binding proteins (RBPs) (Fig. 3C). However, the clustering of downregulated proteins was difficult to assess. Prominent GO categories of the clustered RBPs were related to RNA binding, RNA metabolism, and neurodegenerative diseases (Fig. 3D, Table S1.5). Notably, RBPs and related proteins were the most abundant, accounting for 14.4% of all identified proteins (Fig. S1). These results demonstrate that sEVs contain many RBPs, implying that they play a role in health and disease; similar results have been reported in the literature [16,22].
In our study, we identified seven RBP-associated proteins (Fig. 3C), and among them, FUS was the most abundant and the consistently enriched protein of the HypEVs with a ratio >2.5. According to the volcano map of the total upregulated proteins in three repeated experiments (Fig. 3E), FUS had the first, fourth, and fifteenth highest expressions. It also showed high interaction scores in PPIs. Western blotting confirmed that FUS was highly expressed in neuronal HypEVs but had low expression in neuron-derived NorEVs (Fig. 3F). FUS interacts with numerous proteins that regulate nuclear processes [23,24], including chromosomal organization, transcription, RNA splicing, RNA processing, RNA transport, translation, and RNA stability. Additionally, conserved sequence analysis revealed that FUS is highly conserved among species (Fig. 3G). These data suggest that FUS and its related RBPs play a key role in mobilizing RNA release from HypEVs.
2.3. HypEVs contribute to the maintenance of neuronal morphology and neurites of ischemia models in vitro and in vivo
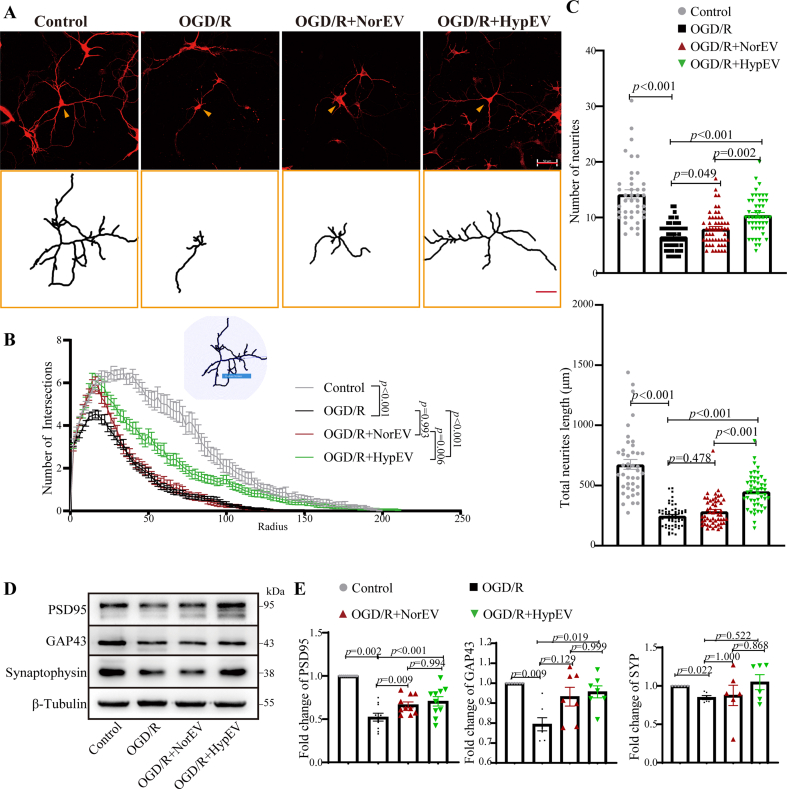
The release of neuron-derived sEVs and their effects on recipient ischemic neurons in vitro or in vivo is still lacking. Given the presence of the FUS protein in neuron-derived sEVs had well-documented critical role in neuronal functions, including its significant involvement in synaptic operations in motor neurons in the context of amyotrophic lateral sclerosis (ALS) [25,26]. Therefore, we evaluated the effects of HypEV and NorEV treatment on neural complexity using microtubule-associated protein 2 (MAP2) labeling by measuring the neuronal branches, dendrite numbers, and total dendrite lengths. HypEV treatment significantly rescued the neural complexity of ischemic neurons more effectively than NorEV treatment (Fig. 4A–C). To some extent, maintaining synaptic structural connections during the acute injury phase facilitated neuron recovery. Furthermore, we examined synaptic plasticity-related proteins such as growth-associated protein 43 (GAP43), a marker of axonal sprouting and plasticity, and synaptophysin and postsynaptic density protein 95 (PSD95), markers of presynaptic and postsynaptic integrity, respectively [27]. Consistently, western blotting showed that HypEVs restored the synaptic integrity and neuronal plasticity markers of ischemic neurons significantly more effectively than NorEVs (Fig. 4D and E). These results indicate that HypEVs exert a beneficial and protective effect on neurons after ischemic injury, and this effect was stronger in HypEVs than NorEVs.
Fig. 4.
The effects of HypEV co-cultured with neurons in vitro. (A) After oxygen-glucose deprivation (OGD) injury for 3 h, the primary cortical neurons were incubated with phosphate-buffered saline (PBS), NorEVs, or HypEVs from primary cortical neurons for an additional 24 h in normal media. Microtubule-associated protein (MAP2) staining was used to characterize neuron morphology in vitro after neurons were incubated with PBS (OGD/reperfusion [R]), NorEV (OGD/R + NorEV), or HypEV (OGD/R + HypEV) (scale bar = 50 μm). The bottom yellow frame outlines single neurons (indicated by yellow arrows) using NeuronJ (scale bar = 50 μm). (B) An illustration showing the quantification of the dendritic complexity by Sholl analysis (upper part of the line chart). Intersections were used to quantify the MAP2-stained neural complexity (as shown in Fig. 4A). Numbers of intersections, total neurite length and neurite number of single neuron (C) are expressed as means ± SEM (n = 42–59 neurons per group from three experiments). Statistical significance was assessed by one-way analysis of variance (ANOVA); differences between groups were considered significant if p < 0.05. The Control, OGD/R and OGD/R + HypEV groups were shared with Fig. 6C. (D) and (E) Representative Western blotting of postsynaptic density 95 (PSD95), growth-associated protein 43 (GAP43), and synaptophysin (a synaptic vesicle protein of the pre-synapse) in each group. Data are presented as means ± SEM, n = 6. Statistical significance was assessed by one-way ANOVA; a value of p < 0.05 was considered significant.
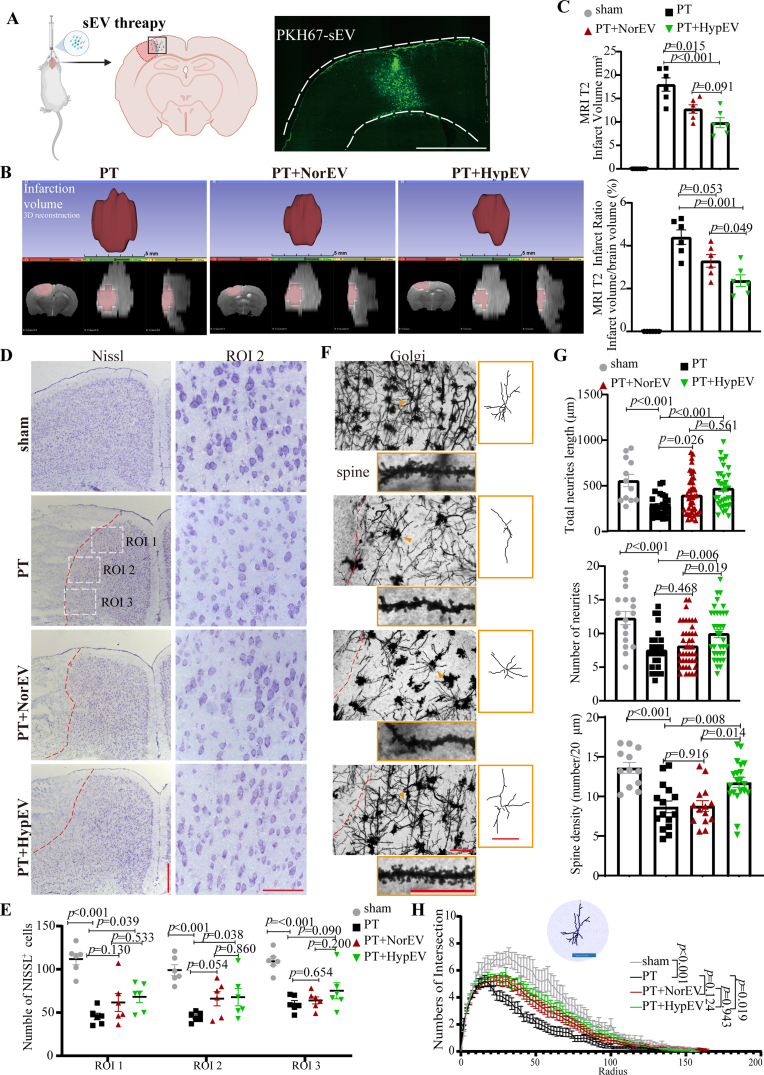
To further confirm the effect of sEVs in vivo, we used PT damage to induce focal brain ischemia in mouse model, followed by stereotactic injection of HypEVs or NorEVs (Fig. 5A). Magnetic resonance imaging (MRI) was performed to determine the volume of the infarction 48 h after sEV administration. T2 weighted images (T2WIs) combined with three-dimensional reconstruction showed that HypEVs reduced the volume of cerebral infarction by 45% when compared with the non- HypEV treatment group (18.0 ± 3.5 mm3 with PT vs. 9.8 ± 2.6 mm3 with HypEV treatment) (Fig. 5B and C). We further analyzed the neural structure in the infarction boundary because most labeled sEVs were distributed around the infarct area (Fig. 5A, D). To detect the number of intact neurons and reflect the protein synthesis capacity of the neurons, we used Nissl staining [28]. After the PT operation, fewer Nissl bodies were observed, and they were smaller and blurrier compared to neurons in the sham group. However, we found that damaged neurons were rescued and the infarct area was completely minimized after PT stroke mice were treated with HypEVs. By contrast, improvement was rare and insignificant after NorEV treatment in PT stroke mice (Fig. 5D and E). Golgi staining was used to detect neural structures. The number, length, and complexity of tracings and the spine number were restored in the PT stroke mice that received HypEV treatment, whereas mice treated with NorEVs showed little difference compared to the PT stroke group (Fig. 5F–H). Overall, these in vivo experiments confirmed the in vitro evidence that HypEVs alleviate damage to ischemic neurons.
Fig. 5.
The effects of HypEVs in vivo. (A) PT mice were injected with PBS, NorEVs, or HypEVs derived from primary cortical neurons into their cortex, and then sacrificed 48 h after PT surgery. Right panel shows the distribution of sEV (green) in the cortex (scale bar = 1500 μm). (B) Representative images of the infarct core (upper) and three-dimensional images of brain infarction (bottom: the view of the coronal section, horizontal section, and sagittal section from left to right) captured by the T2 phase on magnetic resonance imaging (MRI). (C) The infarct volume and infarct volume ratio (percentage of the whole brain) are expressed as means ± SEM (n = 6). Statistical significance was assessed by one-way ANOVA; differences between groups were considered significant if p < 0.05. (D) Nissl staining showing the number of neurons in sham and the peri-infarct region (dotted box) of PT mice treated with PBS, NorEVs, or HypEVs. Low-magnification (left) scale bar = 500 μm; high-magnification (right) scale bar = 100 μm. The red dotted lines indicate the edge between the infarct core and peri-infarct. (E) Data on intact neuron (Nissl staining) number in different dotted boxes are presented as means ± SEM (n = 6). Statistical significance was assessed by one-way ANOVA, and a value of p < 0.05 was considered significant. (F) Golgi-Cox staining was used to distinguish the neural structural changes in the peri-infarct region of PT mice (scale bar = 100 μm). The right yellow frame outlines the single neuron (indicated by triangles using NeuronJ) to show the levels of complexity of dendritic branching of cortical neurons in each group (scale bar = 100 μm). The representative images of the Golgi-Cox-stained spine in each group are shown in the bottom yellow frame (scale bar = 50 μm). (G) Statistical analysis of the total neurite length, neurite number, and spine density of Golgi-Cox-stained neurons (as shown in Fig. 5F); data are expressed as means ± SEM (n = 17–43 neurons per group from 3 to 4 mice). Statistical significance was assessed by one-way ANOVA, and a value of p < 0.05 was considered significant. (H) An illustration showing the quantification of the dendritic complexity by Sholl analysis (upper part of the line chart, bar = 100 μm). Intersections were used to quantify the Golgi-Cox-stained neural complexity (as shown in Fig. 5F). Numbers of intersections are expressed as mean ± SEM (n = 17–43 neurons per group from 3 to 4 mice), and one-way ANOVA was used to assess statistical significance, with a value of p < 0.05 considered significant.
In addition, no significant pathological biotoxicity was observed in the lungs, heart, kidneys, spleen, or liver, and liver and kidney-associated biochemistry parameters were normal after HypEV treatment (Fig. S7).
2.4. Knockdown of FUS diminished HypEV neuroprotection in PT stroke model
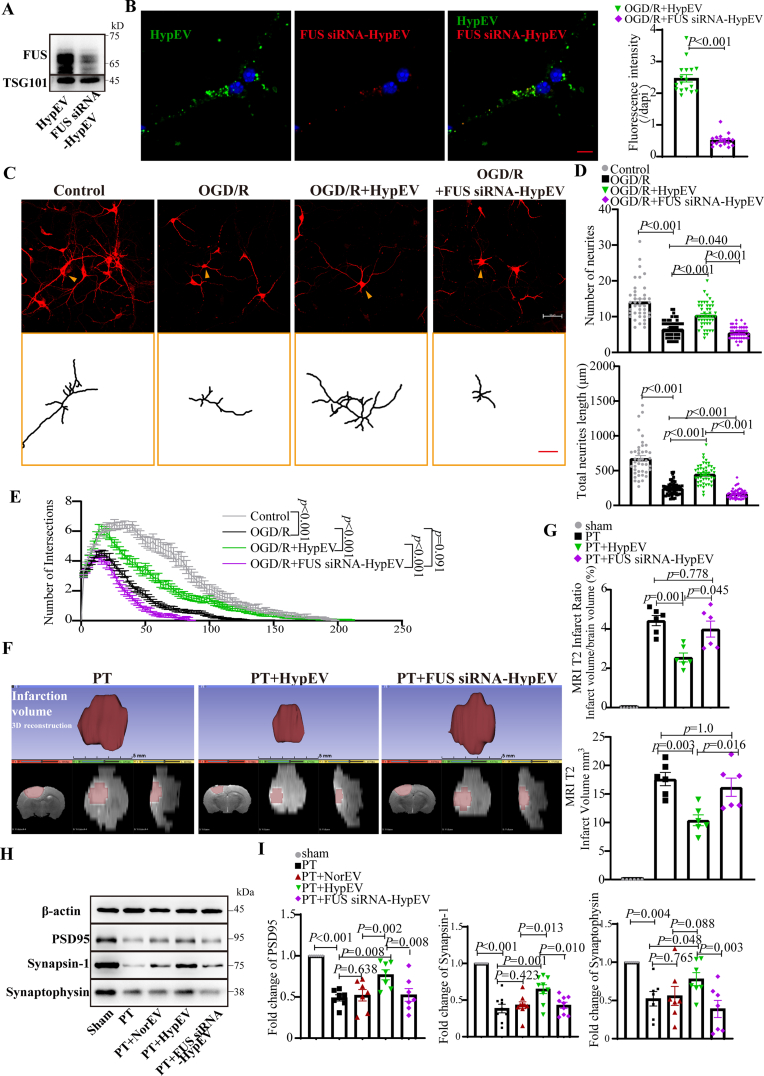
As described previously, a higher number of HypEVs than NorEVs were released from the same number of neurons and taken up by ischemic neurons. Additionally, HypEVs were associated with FUS upregulation, and this finding was also observed in the neuroblastoma cell line SH-sy5y. HypEVs derived from SH-sy5y were preferentially taken up and expressed FUS (Figs. S2A and B). To knock down FUS levels in donor neurons and their sEVs, FUS small interfering RNA (siRNA) was used (Fig. 6A, Fig. S5C). Notably, siRNA-mediated FUS knockdown resulted in diminished FUS expression in HypEVs (FUS siRNA-HypEVs) and reversed the uptake preference for HypEVs in the sEV coincubation experiment (Fig. 6B). To further confirm the different efficiencies of internalization and exclude color bias, equal quantities of NorEVs, HypEVs, and FUS siRNA-HypEVs were stained with PKH26 and co-cultured with recipient neurons for long-term living cell observation (Fig. S8). The results indicated that HypEVs uptake was rapid and abundant, whereas NorEVs and FUS siRNA-HypEVs were inferior to HypEVs. To put it differently, HypEVs are more easily taken up by neurons than NorEVs.
Fig. 6.
Knockdown FUS reduced neuroprotective effects of HypEVs in vitro and in vivo. (A) FUS expression in HypEV was significantly reduced after specific siRNA knockdown in parent cells. (B) Confocal fluorescence images depict sEV uptake by ischemic neurons after co-culturing FUS siRNA-treated HypEV (FUS siRNA-HypEV; PKH26, red) and HypEV (PKH67, green) from primary cortical neurons for 24 h. Scale bar = 20 μm. Nuclei are stained blue (DAPI). Data are expressed as mean ± SEM (n = 17 neurons), and statistical significance was assessed by Student's t-test with differences between groups were considered significant if p < 0.05. (C) MAP2 staining was used to characterize neuron morphology in vitro for neurons incubated with PBS (OGD/R), FUS siRNA–HypEVs (OGD/R + FUS siRNA–HypEV), or HypEVs (OGD/R + HypEV) (scale bar = 50 μm). The bottom yellow frame outlines single neurons (indicated by yellow arrows using NeuronJ) to show the dendritic branching complexity of neurons with reperfusion (24 h) after OGD (3 h) and under normoxic conditions (control) (scale bar = 50 μm). (E) The dendritic complexity was quantified by Sholl analysis, with the numbers of intersections (as shown in Fig. 6C), total neurite length, and neurite number of single neurons (D) expressed as means ± SEM (n = 42–59 neurons per group from three experiments). Statistical significance was assessed by one-way ANOVA with a value of p < 0.05 was considered significant. The Control, OGD/R and OGD/R + HypEV groups were shared with Fig. 4A. (F) Representative images of the infarct core (upper) and three-dimensional images of brain infarction (bottom: the view of the coronal section, horizontal section, and sagittal section from left to right) captured by MRI (T2 phase) in PT mice treated with PBS, HypEVs, or FUS siRNA-HypEVs. (G) The infarct volume and the corresponding ratio (percentage of whole brain) were expressed as means ± SEM (n = 6). Statistical significance was assessed by one-way ANOVA, and differences between groups were considered significant when p < 0.05. (H) and (I) Western blotting analysis of PSD95, synapsin-1 and synaptophysin (synaptic vesicle proteins of the pre-synapse). Data are presented as means ± SEM, n = 7 from 3 mice after MRI analysis. Statistical significance was assessed by one-way ANOVA with a value of p < 0.05 was considered significant.
To compare the effect of FUS on HypEV uptake directly, we performed overexpression (FUSKO + OE) with stable FUS knockout (FUSKO) SH-sy5y strains. We confirmed the difference between the presence and absence of FUS in HypEVs(Figs. S9A–D). In vitro, the uptake of HypEVs from FUSKO cells was sharply decreased compared to those derived from wild-type cells. However, the uptake efficiency of HypEVs was rescued following FUS supplementation, as evidenced by the increased uptake of FUSKO + OE HypEVs. These results suggest that FUS is necessary for the superior internalization of HypEVs.
Furthermore, we investigated whether FUS is necessary for the neuroprotective effect of HypEVs. Neural complexity analysis using MAP2 labeling in vitro revealed that HypEVs significantly rescue the length, number, and complexity of neuronal branches under hypoxic conditions. However, these benefits were significantly diminished by FUS siRNA treatment in HypEVs (Fig. 6C–E). Similar neuroprotective effects were observed when sEVs were derived from hypoxic SH-sy5y cells. HypEVs derived from SH-sy5y cells showed a neuroprotective effect in OGD/R neurons compared with non- sEVs treatment group. However, the benefit was sharply decreased after treating OGD/R neurons corresponding to HypEVs with HypEVs from FUSKO cells. Nevertheless, it was restored after treatment with HypEVs from FUSKO + OE cells (Figs. S9E–G). These results suggest that FUS is necessary for the neuroprotective effects of HypEVs.
The volume of cerebral infarction was significantly reduced by 45% after HypEV treatment in vivo, but this effect was significantly diminished after FUS knockdown in HypEVs (9.4 ± 2.4 mm3 with HypEV treatment vs. 14.6 ± 3.2 mm3 with FUS siRNA-HypEV treatment) (Fig. 6F and G, Fig. S3). Additionally, HypEVs were found to significantly restore the expression of synaptic integrity and neuronal plasticity markers of ischemic neurons, such as PSD95, synaptophysin, and synapsin-1, in the area surrounding the infarction boundary compared to NorEVs (Fig. 6H and I). However, this upregulation of synaptic proteins was eliminated when FUS knockdown was performed in HypEVs. These results are consistent with the in vitro findings discussed earlier, except for the results regarding the expression of GAP43 (Fig. S4E). Interestingly, high levels of GAP43 were found in the infarction boundary area, and no significant differences were observed between the sEV treatment group and the PBS group, likely due to its increased expression when neurons are damaged or stimulated [29], which is different from the observations in developing neurons cultured in vitro. Taken together, these results indicate that FUS plays a crucial role in the uptake and neuroprotective effects of sEVs under ischemic conditions.
However, the mechanism by which FUS affects uptake remains unclear. It is known that the entry of sEVs into recipient cells relies on the interaction between sEV surface proteins and cellular receptors, such as tetraspanins, integrins, lipids, lectins, extracellular matrix (ECM) components, and intercellular adhesion molecules [30]. Although TEM and ELISA rarely detected FUS on the surface of sEVs (Fig. S2C). An increase in FUS inside sEVs was observed after sEV membranelysis (Fig. S2D). This indicated that sEVs contained FUS. Moreover, blocking FUS on the surface of sEVs did not reduce sEV uptake (Fig. S2E). Thus, the effect of FUS knockdown on sEV uptake may be a result of its impact on sEV assembly and RNA sorting. Further investigation is necessary to fully elucidate the role of FUS in sEV-mediated neuroprotection.
2.5. FUS-mediated mitochondrial mRNA transport via HypEVs
FUS, is a highly expressed protein in HypEVs, and it plays a crucial role in various physiological processes, such as DNA repair, RNA processing and transport, and RNA stabilization [24]. Its capacity is attributed to binding, sorting, and carrying RNA into sEVs and then transporting RNA to recipient cells [17]. FUS binds to mRNA and is involved in synaptic mRNA metabolism [25], making it a key protein responsible for the neuroprotective effects of sEVs.
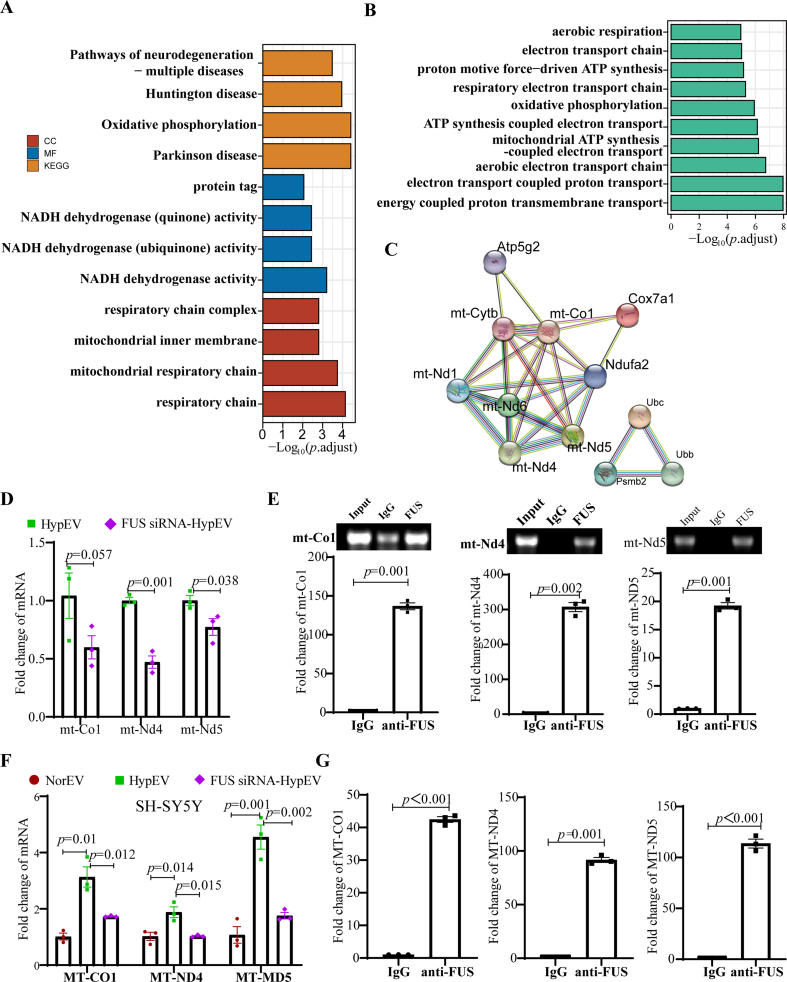
To investigate whether FUS interacts with mRNA transported by sEVs, we performed RNA sequence analysis and identified 209 upregulated mRNAs (log2FC ≥ 1) in HypEVs. GO enrichment of upregulated mRNA revealed that it was related to oxidative metabolism, the respiratory chain, ATP production, Parkinson's disease, and oxidative phosphorylation pathways (Fig. 7A and B) (Table S2.1–2.2). The enrichment results of BP and Kyoto Encyclopedia of Genes and Genomes (KEGG) were all related to several mitochondrial mRNAs, such as cytochrome c oxidase subunit 7A1 (Cox7a1), mitochondrially encoded NADH dehydrogenase 6 (mt-Nd6), mitochondrially encoded NADH dehydrogenase 4 (mt-Nd4), mitochondrially encoded cytochrome c oxidase I (mt-Co1), and mitochondrially encoded NADH dehydrogenase 5 (mt-Nd5) (Table S2.3). The PPI network of these differentially expressed gene-coded proteins was analyzed by STRING software, which showed that they are exclusively mitochondrial genes related to respiration and ATP synthesis (Fig. 7C). The quantitative reverse transcription polymerase chain reaction (RT-qPCR) results confirmed that the expression of these mitochondrial mRNAs was higher in HypEVs from neurons than in NorEVs (Fig. S4G). The binding between FUS and these mRNAs was calculated by different prediction algorithms (catRAPID and RBPsuite) (Table S2.4). Two RNA-binding domains were identified in FUS, including the conserved protein domain family RRM_1 (PF00076), and the most frequently detected RNA-binding motif was GGUG (Fig. S4F). To validate the prediction, RBP immunoprecipitation (RIP) and FUS knockdown experiments were conducted. First, the RIP assay performed on OGD/R primary cortical neurons confirmed that FUS bind to mt-Co1 mRNA, mt-Nd4 mRNA, and mt-Nd5 mRNA (Fig. S4H, Fig. 7E); we then conducted FUS knockdown experiments to further validate our findings. We found that FUS knockdown reduced the abundance of these mRNAs in FUS siRNA-HypEVs compared with that in HypEVs (Fig. 7D) but did not regulate Cox7a1 or mt-Nd6 (Figs. S4I and J). These results were also validated in SH-sy5y because SH-sy5y-derived HypEVs have high FUS expression (Fig. 7F and G). On this basis, we attempted to carry out RIP in SH-sy5y-derived HypEVs, and found that FUS can bind to MT-CO1 and MT-ND4 mRNA (Fig. S10B). These results suggest that FUS is involved in extracellular sorting and transporting mitochondria-associated mRNA via HypEVs.
Fig. 7.
FUS mediated mitochondrial mRNA transport via HypEVs. (A) GO enrichment analysis of upregulated mRNA, log2FC>1 (HypEV vs. NorEV). Enrichment results of cellular component (CC), molecular function (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG). (B) Enrichment results of biological process (BP). (C) The interacting network of the enrichment gene-coding proteins (as shown in Table S2.3). (D) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) results of mRNA in HypEVs or FUS siRNA- HypEVs from primary cortical neurons. Data are expressed as means ± SEM (n = 3). Statistical significance was assessed by Student's t-test; differences between groups were considered significant if p < 0.05. (E) RT-qPCR results of FUS-pulled down mRNA in OGD/R primary cortical neurons via RBP immunoprecipitation (RIP) (bottom). Data are expressed as means ± SEM (n = 3). Statistical significance was assessed by Student's t-test; differences between groups were considered significant when p < 0.05. Top: PCR products on agarose gel. (F) RT-qRCR results of mRNA in NorEVs, HypEVs or FUS siRNA- HypEVs from SH-sy5y. Data are expressed as means ± SEM (n = 3). Statistical significance was assessed by one-way ANOVA; a value of p < 0.05 was considered significant. (G) RT-qPCR results of FUS-pulled down mRNA in OGD/R SH-sy5y using RIP. Data are expressed as means ± SEM (n = 3). Statistical significance was assessed by Student's t-test; differences between groups were considered significant when p < 0.05.
Mt-Nd4 and mt-Nd5 are subunits of the mitochondrial membrane respiratory complex I, and mt-Co1 is a subunit of respiratory chain complex IV; these subunits are located in the inner mitochondrial membrane. To test whether the mRNA transported by FUS compensates for these mitochondrial proteins, actinomycin D (ACTD) was used to inhibit mRNA transcription in hypoxia receipt neurons. We found that the addition of sEVs promoted the expression of mitochondrial proteins in the recipient neurons, compared with ACTD-treated OGD/R neurons, even if they were treated with ACTD (Fig. S5E).
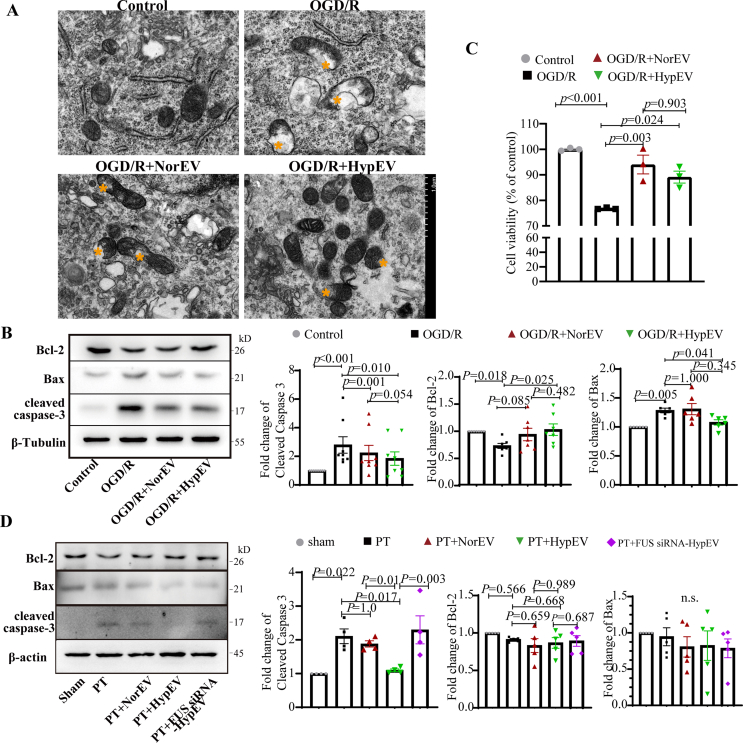
Given the role of FUS in carrying mitochondria-associated mRNAs and facilitating their transport via sEVs, we used TEM to observe the integrity of mitochondrial morphology. This observation was conducted neurons that after the co-culturing of primary cortical neurons derived sEVs. Our results highlighted that the mitochondrial ridges in neurons treated with HypEVs were distinct and well-organized compared to those in neurons untreated with OGD/R or treated with NorEVs (Fig. 8A). For a more comprehensive comparison, we quantified the degree of mitochondrial damage through the proportion of mitochondrial vacuolization, which is a key indicator of mitochondrial damage [31]. Notably, neurons treated with HypEVs showed a significant decrease in mitochondrial vacuolization compared to those treated with NorEVs, accompanied by an increase in mitochondrial quantity (Fig. S5F). Additionally, we observed a vesicle-releasing process that was similar to that of mitochondria-derived vesicles (MDVs) [32] (Fig. S4A), indicating that HypEVs may contain mitochondrial material that can be released from parent cells [33] (Fig. S4B). Furthermore, the expression of mitochondrial apoptotic proteins, including the anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) and the pro-apoptotic proteins Bcl2-associated X protein (Bax) and cleaved caspase-3, was evaluated by western blotting. The results showed that both HypEVs and NorEVs significantly reduced OGD/R-induced cleaved caspase-3 formation, but only HypEVs promoted Bcl-2 expression and reduced Bax expression (Fig. 8B). The Cell Counting Kit-8 assay demonstrated that both NorEVs and HypEVs were beneficial for cell survival, but little difference was observed between NorEV- and HypEV-treated OGD/R neurons (Fig. 8C). HypEV treatment in vivo also significantly reduced the levels of cleaved caspase-3; other pro-apoptotic proteins, including Bak and cleaved BID (Fig. 8D, Figs. S4C–D); although the change in Bcl-2 levels was small. HypEVs exhibited a stronger anti-apoptotic ability than NorEVs in terms of their ability to resist cleaved caspase-3 formation, both in vitro and in vivo. However, FUS knockdown in HypEVs eliminated the anti-apoptosis effect of HypEVs, suggesting that FUS is critical for the function of HypEVs.
Fig. 8.
HypEVs reduce mitochondria-associated apoptosis in neurons in vitro and in vivo
(A) Electron microscopic images of primary neurons treated with cultured primary neuron derived sEVs in vitro in each group. The morphology of the mitochondria was clearly altered. Yellow * represents mitochondrial vacuolization. (B) and (D) Representative western blotting of B-cell lymphoma 2 (Bcl-2), Bcl2-associated X protein (Bax), and cleaved caspase-3 expression levels in each group in vitro and in vivo. Data are presented as means ± SEM, n = 6 in vitro; n = 5 from 3 mice after MRI analysis in vivo. Statistical significance was assessed by one-way ANOVA; a value of p < 0.05 was considered significant. The “n.s.” stands for “no significant difference” among groups. (C) Cell viability was determined by Cell Counting Kit-8 (CCK-8) assay. Data are presented as means ± SEM, n = 3. Statistical significance was assessed by one-way ANOVA; differences between groups were considered significant when p < 0.05.
To eliminate the possibility of apoptotic-associated proteins in sEVs (including Bcl-2, Bax, and cleaved caspase-3) contributing to the observed effects, we assessed their expression levels in sEVs. We found that Bcl-2 and Bax were expressed at low levels in sEVs, and although cleaved caspase-3 was present, its levels were lower in HypEVs than in NorEVs or the FUS knockdown groups (Fig. S5D). Moreover, the high expression of cleaved caspase-3 in NorEVs did not induce apoptosis (Fig. 8B and C). Therefore, we conclude that the observed anti-apoptotic effect is likely due to FUS rather than apoptotic-associated proteins carried by sEVs.
These results strongly suggest that FUS, which is present in HypEVs, plays a key role in transporting mitochondrial mRNA and reducing apoptosis in recipient neurons. Based on these findings, we hypothesize that FUS and the mitochondrial mRNA it carries work together to protect neurons against mitochondrial dysfunction and apoptosis.
The results of these experiments provide a comprehensive understanding of the features of neuron-derived sEVs during hypoxic conditions. The proteomic and transcriptomic analysis indicated that FUS is a key molecule in the neuroprotective function of HypEVs. Furthermore, these findings demonstrate that FUS facilitates the transport of mitochondrial mRNA through HypEVs and plays a vital role in mitigating mitochondria-associated apoptosis.
3. Discussion
The prevalence of ischemic stroke is on the rise, leading to significant disability and mortality rates. Given the production of multiple hypoxic neurons during ischemic stroke, it is imperative to explore the potential contribution of hypoxia-induced sEVs to the pathophysiological mechanisms of ischemic stroke. To this end, we conducted a study in which we simulated the release of sEVs from hypoxic neurons in the mouse brain by generating sEVs from primary neuronal cultures exposed to hypoxic conditions, and systematically assessed their impact on ischemic brain injury. What's more, sEVs exhibit unique characteristic of tissue specificity and possess cell type-specific proteins presented on the parent cell membrane enabling delivery to disease affected tissues or organs [34,35]. We use them to investigate the homing effect of neuronal sEVs and pave the way for the development of targeted nanomaterials for neurons.
This study highlights the differences in the release, absorption, and function of sEVs derived from neurons under normoxic and hypoxic conditions. The findings indicate that sEVs derived from neurons under hypoxic conditions may offer more neuroprotection during neuroregeneration than those derived from neurons under normoxic conditions. To simulate ischemia in mice, the PT stroke model was selected as it is repeatable and can imitate the process of intravascular thrombosis [36]. Our results showed that most neuron-derived sEVs were taken up by neurons in vivo and HypEVs were more readily taken up and utilized in vitro than NorEVs. Furthermore, the neuroprotective effects of HypEVs involve RBPs, which are abundant in neuron-derived sEVs. Among these RBPs, FUS plays a crucial role in protecting neurites and promoting neuroplasticity. Mechanistically, FUS participates in sorting and transporting mitochondrial mRNA and reducing mitochondria-related apoptosis.
RBPs, including FUS, can selectively bind to specific RNAs and transport them into sEVs [16,19]. FUS, a highly conserved protein, was initially identified as a fusion oncogene in human liposarcomas [37] and has been shown to play a critical role in normal central nervous system development and neuron survival [38,39]. FUS mutations have been linked to neurodegenerative disorders, such as ALS and frontotemporal dementia (FTD) [40,41], making FUS a key target for therapeutic intervention. FUS reportedly mediates nucleocytoplasmic shuttling, splicing, transcription, microRNA processing, RNA transport and translation, and stress granule formation [42]. In addition to inducing diverse changes in mRNA [43], FUS is also involved in transporting mRNA to dendrites to maintain neuronal plasticity and dendritic integrity. Notably, many RNAs involved in synaptic functions are known targets of FUS, supporting its critical role in synaptic development, maintenance, and plasticity [25,26]. Our data suggest that FUS-mediated RNA sorting and transport into sEVs is essential for maintaining neuronal plasticity and dendritic integrity, as these critical RNAs are released into the extracellular space before neurons are damaged.
Although FUS has been extensively studied in cells, little is known about the transportation of FUS and FUS-binding mRNA. Therefore, to investigate this, we performed RNA sequencing, which revealed that hypoxia significantly upregulated mitochondrial mRNA in sEVs. However, synaptic mRNA was relatively rare in differentially expressed genes. Notably, we also found that FUS bind to mitochondrial mRNAs and affect the entry of these mRNA into sEVs. FUS is known to interact with mitochondrial proteins and impair mitochondrial quality [44,45], as well as sequester respiratory chain mRNA and induce mitochondrial dysfunction [46]. Mitochondrial homeostasis and mitochondrial energy supply are critical for dendritic integrity and function in the central nervous system [47,48]. Because of the long distances covered by axons [20,49,50], axon mRNA is transported to specific subcellular sites and locally translated in the neurons of the healthy, mature central nervous system to sustain mitochondrial and synaptic function [[51], [52], [53], [54], [55]] with the assistance of RBPs and endosomes.
Mitochondria components can be transported via sEVs to target cells and trigger a damage-associated molecular pattern release to cause cell damage [56] or trigger self-adaption, such as ischemic preconditioning (IPC) [33], to protect cells. Research has confirmed that MDVs containing mitochondrial material can enter multivesicular bodies (MVBs) and be secreted from cells via exosomes [33]. Our mass spectrometry and RNA sequencing results also showed that both mitochondria-associated proteins and RNA were transported into sEVs, which may be related to the entry of MDVs into MVBs (exosome precursors) (Figs. S4A and B). On the basis of our data, we hypothesized that mitochondrial mRNA was rapidly transported to the cytoplasm via the dendrites by HypEVs containing FUS (Video 1) and that subsequently, followed by translation on endosomes to compensate for the absence of mitochondrial proteins. Therefore, we found that sEVs contained FUS, and the FUS is contributed to maintain morphology and function of mitochondrial under the condition of ischemic injury. However, the mechanism by which mRNA-encoded proteins protect mitochondria and neurons must be systematically verified in further studies.
The preferential targeting of neurons by sEVs remains unclear. Mesenchymal stem cell exosomes (MSC-Exos) were visualized in vivo by noninvasive computed tomography (CT), which showed that MSC-Exos selectively targeted brain areas and neurons affected by pathology in mouse models of ischemic stroke, Parkinson's disease, Alzheimer's disease, and autism disorder [11]. This revealed a homing phenomenon of MSC-Exos in vivo, which was attributed to inflammation. In an in vitro primary neuron-astrocyte co-culture model, sEVs somewhat targeted neurons but not target astrocytes [57]. The homing phenomenon is a major advantage of targeted therapy using sEVs [58], which is more common in tumor cells. Tumor-derived sEVs from lung or liver cancer, when injected into normal mice via intravenous injection, can lead to the localization of sEVs in their corresponding organs [35]. Thus, we speculate that neuronal sEVs may have a specific affinity for neurons, and used them as a research object. The time point at which we collected sEVs was on the 7th day of in vitro neuronal culture. After 7 days in vitro (DIV7) culture, neurons can be considered mature because they exhibit the morphology, cellular metabolism, and neuronal polarity characteristics of mature neurons, including the appearance of dendritic spines and abundant synaptic connections [59,60]. We hope this can simulate the hypoxic injury encountered by mature neurons within the skull.
Increasing studies have indicated that hypoxia promotes sEV secretion [6,13,14], but it is unclear why HypEVs are taken up preferentially. Our findings showed that FUS was not located on the surface of sEVs, suggesting that the reduction in HypEV uptake resulting from FUS knockdown may be a consequence of FUS reduction rather than FUS itself. A review [61] highlighted the molecular mechanisms of the recognition and interaction of recipient cells, which depend on the interaction of sEV surface proteins and cellular receptors, including tetraspanins, integrins, lipids, lectins, extracellular matrix (ECM) components, and intercellular adhesion molecules [30]. Although, immunoblotting of sEVs showed that HypEVs expressed higher levels of CD63 and CD81 than NorEVs, which could explain the preferential uptake of HypEVs, the molecules mentioned above (e.g., integrins, lectins, and adhesion molecules) were expressed at low levels in HypEVs in the results of proteomics. In addition, the uptake of sEVs is affected not only by their surface proteins but also by their pH value [13]. We think the uptake of sEVs may be primarily influenced by FUS-related proteins and their pH value, with RBPs potentially located on the surface of sEVs [62]. Nonetheless, the precise cellular and molecular mechanisms that govern the efficient targeting of neurons by sEVs are currently unknown, necessitating further investigation.
Behavioral recovery studies provide valuable insights into the functional outcomes of therapeutic interventions. The absence of behavioral recovery studies in our current research could be viewed as a limitation. Our study's primary focus was to elucidate the underlying mechanisms exerted by hypoxia-conditioned sEVs in an ischemic stroke context. Our future research can build upon our findings by incorporating behavioral recovery assessments to explore the clinical relevance and therapeutic efficacy of our approach.
4. Conclusion
This study provides new insights into the regulation of ischemic stroke by sEVs released from neurons under hypoxia conditions. Specifically, we have shown that FUS and its carried mitochondrial mRNA are released into the extracellular space, along with other RBPs, via sEVs when neurons experience such an injury. Our findings provide important insights into the mechanisms underlying HypEVs-mediated neuroprotection and highlight the potential therapeutic applications of HypEVs-based interventions for ischemic injury.
5. Materials and methods
5.1. Study approval
All animal procedures were approved by the Institutional Animal Care and Use Committee of Jinan University (approval ID: 20,201,028–03).
5.2. Animals
A total of 81 adult male and female Balb/c mice weighing between 22.0 and 25.5 g and aged between 5 and 7 weeks [63], with half of them being males and the other half females, were purchased from the Institute of Laboratory Animal Science of the Chinese Academy of Medical Sciences (Guangzhou, China). Mice were assigned to specific experimental groups without bias using a random number generator (https://www.calculator.net/random-number-generator.html), and no mice were excluded from the study. The mice were housed in a strictly controlled environment with constant temperature and humidity. Food and water were available all day and night. The study was carried out in accordance with the recommendations of the NIH Guide (NIH Publications No. 8023, revised 1978) for the Care and Use of Laboratory Animals. All experiments were carefully conducted in accordance with the guidelines for Animal Experimentation of Jinan University. Endeavors were made to reduce the total number of animals used as well as their potential pain and suffering.
The PT stroke model has good stability and high repeatability for research. Focal cortical ischemia was induced in mice by photothrombosis of cortical microvessels as previously described [64]. Briefly, the mice were anesthetized with isoflurane gas (4% induction anesthesia, 1.5% maintenance anesthesia; RWD Life Sciences, Shenzhen, China) and placed in a stereotaxic device. After the skull was exposed through a midline incision, the optic fiber was positioned to the coordinates (ML 2.0 mm, AP -0.5 mm) from bregma, as close as possible to skull. Five minutes after administration of Rose Bengal (40 mg/kg, Sigma), the brain was illuminated for 15 min by fiber cable of 110 mW intensity (GL532TA-100 F C, Shanghai Laser & Optics Century). Finally, light exposure was stopped, and the mice were treated with sEVs or PBS. Four mice were used for in vivo uptake experiment.
For the detection of the neuroprotection of HypEVs in vivo, 40 mice were divided into four groups: sham (10), PT + PBS (10), PT + NorEV (10), and PT + HypEV (10). After the MRI scan (n = 6), brains were collected for Nissl staining (n = 6) or Golgi staining (n = 4).
To detect the inhibitory effect of FUS siRNA on the neuroprotection of HypEVs in vivo, 24 mice were divided into 5 groups: sham (3), PT + PBS (6), PT + NorEV (3), PT + HypEV (6), PT + FUS siRNA-HypEV (6). Eighteen mice from PT + PBS, PT + HypEV and PT + FUS siRNA-HypEV were used to detect the infarct volume by MRI scanning (n = 6). After MRI scanning, 3 samples per group and 6 another samples (3 mice for SHAM, 3 mice for PT + NorEV) were used for western bloting.
5.3. Cell cultures
For primary cortical neuron culture, primary cortical neurons were obtained from the cerebral cortex of Balb/c mouse embryos (E18-E19) purchased from the Institute of Laboratory Animal Science of the Chinese Academy of Medical Sciences (Guangzhou, China) [63,65]. After separation, cortical neurons were cultured in DMEM/F12 with 10% FBS for 4 h, and then changed to neurobasal medium (Cat No. 21103049, Gibco, USA) with 2% B27 (Cat No. 177504044, Gibco, USA) for further cultivation in a 37 °C, 5% CO2 incubator. After 5 days in vitro (DIV5), cells were used for the following experiments. To initiate OGD, cell culture media were switched to Glucose-free DMEM after washing twice with PBS, and incubated in a hypoxia incubator (95% N2 5% CO2, 37 °C) for 3 h. For OGD/R injury, the glucose-free DMEM medium was replaced by neurobasal medium in 37 °C, 5% CO2 incubator for further culture 24 h as reperfusion period. The cells without any treatment were utilized as a control. Cells were divided into 4 groups: Control, OGD/R + PBS, OGD/R + HypEV, OGD/R + NorEV (or OGD/R + Fus siRNA_HypEV). PBS or sEVs (1:5000, total 60 μg) were added at the beginning of reperfusion.
SH-sy5y cells were obtained from American Type Culture Collection and cultured in high-glucose DMEM containing 10% FBS in a humidified incubator (37 °C, 5% CO2). The OGD/R model was established as described above.
5.4. Transfection
FUS siRNA (Ribobio, China) was synthesized to knockdown FUS in sEVs using riboFECT™ CP transfection kits (Cat No. C10511-05, Ribobio, China). FUS siRNA target sequence: CAAGCAGATTGGTATTATT (Ribobio, stB0001791 A). According to transfection reagent instructions, DIV3 neurons supernatant were replaced with fresh complete medium (neurobasal medium + 2% B27) contained FUS siRNA/liposome complexes with the siRNA concentration of 50 nM for 48 h. Then the fresh media replaced transfection medium for the following experiments, such as OGD/R model establishing and HypEV production.
5.5. sEVs isolation
Usually, total 100 ml primary neuron culture supernatant from ten 10-cm cell culture dishes (8 × 106 cells per dish) are needed to extract sEVs per group. sEVs were purified from primary neuron cell culture supernatant under two conditions using ultrahigh speed centrifugation techniques: After 5 days in vitro (DIV5), neurons were cultured with neurobasal (without B27) medium for 48 h in 37 °C, 5% CO2 incubator for NorEV production; or neurons were cultured with Glucose-free DMEM for 3 h in hypoxia incubator (95% N2 5% CO2) and replaced by neurobasal medium (without B27) for further culture 45 h in 37 °C, 5% CO2 incubator for HypEV production. The supernatant was harvested and centrifuged at 300 g for 10 min to remove cells, at 2000 g for 20 min to remove cell debris and apoptotic bodies, and at 12,000 g for 40 min to remove large particles and organelles. Next, the supernatant was passed through a 0.22-μm filter, and ultracentrifuged at 120,000 g for 70 min. Finally, supernatant was discarded, and sEV precipitation was washed with PBS and ultracentrifuged again, followed by resuspended with 20 μl PBS. The sEV were identified by electron microscopy, western blotting and nanoparticle tracking analysis (NTA) using a Nanosight NS300 (Malvern, England).
sEVs were extracted from SH-sy5y as above. To extract sEVs from SH-sy5y and its derivative strains, a total of 60–80 ml of culture supernatant from 6 to 8 dishes is required. Cells seeded in 10-cm dishes were treated with two conditions after they reach 90% confluence: high-glucose DMEM (without FBS) for 24 h in 37 °C, 5% CO2 incubator (NorEV source); or Glucose-free DMEM for 3 h in hypoxia incubator (95% N2 5% CO2) and replaced by high-glucose DMEM (without FBS) for further culture 21 h in 37 °C, 5% CO2 incubator (HypEV source). The supernatant was harvested and processed as above to obtain sEV precipitation.
5.6. sEV uptake experiment
To verify the uptake of neuronal sEVs in vitro, we stained sEVs with PKH67/PKH26 (Cat No. Mini 67, Sigma-Aldrich, USA) as our previously published article [63,66]. For each sEV staining, we used 20 μl of sEVs (2 μg/μl). First, we add 500 μl Diluent C to the sEV suspension and prepare the staining solution by adding 2 μl dye to 500 μl Diluent C. Next, PKH67/PKH26 (2 μl dye/500 μl Diluent C) were added to the sEV suspension to label sEVs by pipetting for 5 min at room temperature, followed by 10 ml DMEM complete medium (DMEM+ 10% exosome-free FBS) adding to stop the staining. Finally, the sEV were retained using ultrahigh speed centrifugation and resuspended by 10 μl of PBS.
In vitro, 5 μl dye-stained sEVs was added to primary cortical neurons in each experiment. After 24 h incubation, the cells were fixed with 4% PFA and stained with DAPI for fluorescent quantitative analysis by confocal laser scanning microscopy (CLSM) using a Zeiss LSM888 (Zeiss, Oberkochen, Germany). The staining of SH-sy5y-derived sEVs is similar to that described above.
To detect whether FUS stayed at the sEV membrane, anti-FUS antibody (20 μg, Cat No. Sc47711, Santa Cruz) were added to 500 μl of sEV suspension with rotation at 4 °C overnight to block sEV surface antigens. sEVs were restored by ultrahigh speed centrifugation and labeled with PKH67 the next day, followed by co-cultured with SH-sy5y.
For the detection of the localization of sEVs in vivo, 4 PT stroke mice were immediately applied for nose-dripping therapy (nasal drop) with 10 μl sEV suspension after PT surgery. Dye-stained sEVs were administered intranasally as drops with a small pipette every 1 min into alternating sides of the nasal cavity for a total of 20 min, delivering a total volume of 10 μl into the nasal cavity. After 24 h, brains were harvested after cardiac perfusion with 0.9% saline and PFA followed by PFA fixation and dehydration. Finally, the brain was cut into 10 μm frozen sections for immunofluorescence.
One normal mouse was applied for visualizing the sites of sEV using stereotaxic injection. 3 μl of stained sEVs were stereotaxically injected into the same location in the cortex.
5.7. Flow cytometry
In order to enhance the accuracy of fluorescence quantification, cell flow cytometry analysis was performed using SH-sy5y co-cultured with PKH26-labeled sEVs. SH-sy5y-derived sEVs were labeled with PKH26 and subsequently co-cultured with SH-sy5y cells for 24 h. After treatment, cells from each group were enzymatically detached using EDTA-free trypsin (Cat No. 15050057, Gibco, USA), washed with PBS, and subjected to analysis using a BC Gallios flow cytometer (Beckman Coulter, USA) within 1 h. FlowJo software (FlowJo LLC) was utilized for data analysis.
5.8. ELISA
The ELISA assay was carried out in accordance with instruction of Human FUS ELISA Kit (Cat No. EH14546, FineTest). sEV samples were isolated from conditioned medium of SH-sy5y and divided into two equal portions. One portion was directly added to ELISA plates, and the other portion was added after lysed with lysate buffer.
5.9. sEV characterization
Ten μl fresh sEV suspension was dripped on the surface of a carbon-coated copper grid for 10 min to dry, followed by a fixation in 2.5% glutaraldehyde for 5 min. Next, the fixed grid was washed with PBS, followed by negatively stained with 3% phosphotungstic acid (Cat No. G1871, solarbio) for 2 min. After air-dry, sEV micrographs were captured through Transmission Electron Microscopy (TEM) (Leica USA) with voltage setting at 120 Kv.
Nanoparticle tracking analysis (NTA; Nanosight NS300, Malvern, England) was performed to observe the concentration and size distribution of sEVs. Western blotting was performed to verify sEV's classical marker [67,68], CD63, CD81, TSG101, and calnexin as a negative control.
5.10. Immunoelectron microscopy
To visualize sEV's protein expression using immunoelectron microscopy, the fixed grid was blocked with 5% BSA/PBS for 10 min, and incubated with primary antibodies (CD63, 1:20, Cat No. Ab193349, abcam; FUS, 1:20, Cat No. Sc47711, Santa Cruz) diluted in 0.1% (v/v) BSA/PBS for 30 min followed by six washes of 0.1% BSA/PBS. Then the grid was incubated with gold-conjugated secondary antibody (10 nm colloidal gold antibody, 1:20; Cat No. Bs-0296G Bioss) diluted in 0.1% (v/v) BSA/PBS for 30 min followed by six washes of 0.1% BSA/PBS. Finally, the grid was fixed with 1% glutaraldehyde for 2 min, followed by washed and negatively stained with 3% phosphotungstic acid for 2 min. After air-dry, sEV micrographs were captured through Transmission Electron Microscopy (TEM) (Leica USA) with voltage setting at 120 Kv.
5.11. Proteomics
TMT quantitative proteomics was completed with assistance of PTM Biolab. sEVs were collected and total protein was extracted. After tryptic digestion of proteins, the peptides were dissolved in 0.5 M TEAB, and were tagged with TMT kit (Thermo) according to the manufacturer's instructions. The peptides were fractionated by high-pH reverse HPLC, using an Agilent 300 Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). Then the peptides analysis was conducted using liquid chromatography (EASY-Nlc 1200) coupled with mass spectrometry (Q ExactiveTM Plus). Intact peptide and their secondary fragments were detected and analyzed in the Orbitrap at a resolution of 17,500. The resulting mass spectral data were retrieved using MaxQuant v.1.5.2.8 (http://www.maxquant.org/).
5.12. RNA sequencing
Three biological samples in each group were used for RNAseq. Sequencing libraries were prepared, and the raw read data of each group were tested and filtered using fastp software (https://github.com/OpenGene/fastp). Raw read Mapping were performed by Hisat2 (http://ccb.jhu.edu/software/hisat2/index.shtml). HTSeq were applied to calculate the counts of the Reads mapped the genome [69]. FPKM(Fragments Per Kilo base Million Reads)was used to standardize the expression data.
5.13. Bioinformatics analysis
Tandem mass spectra were searched against the UniProt database (http://www.ebi.ac.uk/GOA/). Protein gene ontology (GO) annotation information was primarily obtained from the UniProt Gene Ontology Annotation (GOA) database (http://www.ebi.ac.uk/GOA/). For any identified proteins that were not annotated in the GOA database, sequence alignment in InterProScan was used to de-annotate the GO classification of the proteins. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation information was obtained from the KEGG database, and protein pathways were annotated by the KEGG Automatic Annotation Server (KAAS), a KEGG online service tool. The STRING database (https://cn.string-db.org/) was used for protein interaction network analysis. Enrichment analysis was performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/home.jsp) [70,71], and the top 30 upregulated and downregulated proteins were highlighted. The R package clusterProfiler (https://guangchuangyu.github.io/software/clusterProfiler) was used to process the gene set enrichment analysis (GSEA, https://www.gsea-msigdb.org/gsea/msigdb/index.jsp), and the results were visualized and plotted in R (version 3.6.3). We utilize the online algorithms RBPsuite [72] and catRAPID [73] to forecast the binding of the FUS protein to mitochondrial mRNAs.
5.14. Living-cell imaging
To capture the process of sEV uptake, DIV5 primary cortical neurons cultured in glass-like polymer coverslip (Cat No. D35-20–1.5 P, Cellvis) were subjected to OGD/R injury, and then were added with stained sEVs. The cells were placed in a living cells workstation (37 °C, 5% CO2) and imaged by time lapse living-cell confocal imaging using a Zeiss LSM888 for 2 h. Pictures were taken every 3 min to capture uptake process. To detect the efficiency of sEVs uptake, pictures were taken at 1, 3, 6, 18, 24 h for long-term living cell observation.
5.15. Stable cell lines
The FUS knockout (FUSKO) stable SH-sy5y cell line was constructed by FUS/TLS CRISPR KO Plasmids (Cat No. Sc-400612-NIC-2, Santa Cruz). Cells were transfected plasmids once they reached 70–80% confluency using FuGENE HD transfection reagent (Cat No. E2311, Promega) at a 3:2 FuGENE® HD Transfection Reagent: DNA ratio. After 48 h incubation, successful transfection of CRISPR/Cas9 KO Plasmid was visually confirmed by cell fluorescence. The lowest concentration of puromycin that killed 100% of non-transfected cells in 3 days from the start of puromycin selection was determined. Finally, the cells were selected with Puromycin (Cat No. 60209ES10, Yeasen) at concentration of 2.5 μg/ml for 7 days, followed by puromycin maintenance.
pLV [Exp]-mCherry/Neo-EF1A > FLAG/hFUS [NM_004960.4] vectors (VectorBuilder) was transfected to FUSKO cell line to generate co-transfection of FUS knockout and FUS overexpression (FUSKO + OE) SH-sy5y cell line. Finally, the cells were selected with G418 (Cat No. 108321-42-2, Aladdin) at a concentration of 600 μg/ml for 14 days, followed by G418 maintenance. The successful construction of FUSKO + OE and FUSKO stable SH-sy5y strains was confirmed by western blotting.
5.16. mRNA transcription inhibition
Actinomycin D (ACTD; Cat No. 1036–50, Biovision), a transcriptional inhibitor, was used to inhibit new mRNA synthesis. After OGD stimulation as describe above, DIV5 neurons were treated with 1.6 μM ACTD with or without HypEVs at the beginning of reperfusion. Each sample was harvested 24 h after treatment with actinomycin D to test the protein expression of Nd4, Nd5 and mt-Co1.
5.17. sEV administration in vivo
After the PT stroke model, Balb/c mice were microinjected with different sEVs (3 μg/μl, 3 μl, diluted in PBS) or PBS (3 μl) into the left cortex closed to the infarction boundary (coordinates relative to bregma: ML 2.0 mm, AP +0.2 mm, and DV -1.5 mm) by stereotactic injections [74].
5.18. MRI for mice and three-dimension reconstruction
MRI for mice was conducted using a 9.4 T small animal MRI scanner (Bruker PharmaScan) as before [63]. Mice were anesthetized using 2% isoflurane through a nose cone, and the body temperature and respiratory rate were monitored. T2-weighted imaging (T2WI) imaging was conducted at 48 h after the PT model using the following scanning parameters: 2D fast-spin echo sequence (3500/33 ms of repetition time/echo time, 2 average), field of view (FOV) = 20 × 20 mm, 17 axial slices with a slice thickness of 1 mm, a matrix of 256 × 256. It was positioned over the brain, excluding the olfactory bulb. Under the same scale and brain slices of PT mouse images, T2WI imaging was scanned and quantified using 3D slicer software [75] (https://www.slicer.org/). Three-dimension images were reconstructed by 3D slicer according T2WI images, then the infarct and non-infarct regions were identified by threshold adjustment. Finally, the volumes of infarct determined by software by an investigator blinded to the groups.
5.19. Histology
48 h after the PT operation, the mice were perfused with PBS and fixated with 4% paraformaldehyde (PFA). The brains were incubated in PFA for 24 h, followed by 30% sucrose for 48 h twice and stored at 4 °C. Finally, the brain was cut into 10 μm frozen sections.
For Nissl staining, brain frozen sections from randomly selected mice after MRI analysis were fixed with 4% PFA, washed with PBS, and stained by Nissl staining solution (Cat No. C0117, Beyotime, China) for 30 min at RT. Next, sections were washed with PBS, followed by transparent for 5 min with xylene twice. Finally, the sections were sealed with resinene, and observed under an optical microscope.
To test the HypEVs are safe and nontoxic on live animals, mice were microinjected with 3 μl of (i) PBS, (ii) sEVs (3 μg/μl) (n = 4, including female and male, each gender 2 mice). Two days later, blood was bled to examine serum biochemistry parameters, and sacrificed for tissue histological analysis by an independent pathology service (servicebio, Guangzhou, China). Susceptible tissues (heart, spleen, liver, lung, and kidney) were harvested from the above three groups (sham, PT + PBS and PT + HypEV), paraffin-embedded, sectioned, and stained with HE.
5.20. Golgi staining
Golgi staining was performed using modified Golgi-Cox impregnation method. Briefly, 15 mice were perfused with 4% paraformaldehyde and brains were removed and post fixed with 4% paraformaldehyde over 24 h. The brains were cut into tissue pieces with a thickness of 2∼3 mm, then the pieces were immersed in Golgi dye (Servicebio, China) in the shade and avoided light for 14 days. The Golgi dye was changed every 3 days. Next, tissue pieces were dehydrated with sucrose and soften with 75% glacial acetic acid, followed by cutting into brain slices 100 μm and air-dry overnight. Finally, the slices were washed with distilled water, dyed in concentrated ammonia water for 10 min, washed with distilled water, sealed with glycerin gelatin, and photographed.
5.21. Immunofluorescence and neuronal structure complexity
The cells or frozen brain sections were fixed with 4% PFA for 15 min and permeabilized with PBS containing 0.3% Triton X-100 for 15 min, followed by 5% bovine serum albumin (BSA) blocking for 30 min at room temperature. Without washing, the cells were incubated with the primary antibodies anti-MAP2 (Cat No. Ab32454, abcam) overnight, followed by incubation with a mixture of fluorescent secondary antibodies for 1 h at room temperature. The images were acquired using a confocal laser microscopy Zeiss LSM888. For Fluorescent Nissl Stains, frozen brain sections were incubated with Nissl staining solution (Cat No. N-21483, NeuroTrace Fluorescent Nissl Stains, Thermo) for 30 min after permeabilized treatment.
MAP2 labeling neurons and Golgi staining morphology were traced by NeuronJ, a plugin of ImageJ [76]. And the number and length of tracings can be calculated. To quantify neuronal complexity, the tracings were used for sholl analysis [77].
5.22. Electron microscope for mitochondria
Cells were collected and fixed by 2.5% glutaraldehyde solution. The fixed cells were washed with 0.1 M phosphate buffer (PBS, pH7.4), and embedded into agarose, subsequently, the cells were post-fixed with 1% OsO4 in 0.1 M PB (pH 7.4) for 2 h. Then the agarose blocks were washed with PB and dehydrated using ethanol gradient and acetone in sequence at room temperature. After that, cells were embedded in EMbed 812 resin (SPI, 90,529-77-4), and sectioned onto the 150 meshes cuprum grids with formvar film at a thickness of 60–80 nm. Finally, cuprum grids were stained with 2% uranyl acetate in saturated alcohol for 8 min, followed by 2.6% lead citrate for 8 min. The cuprum grids were dried overnight at room temperature are observed under TEM (hitachi, HT7800). The number of mitochondrial vacuolization were repeatedly recorded in the three different cells by a skilled analyst. The proportion of mitochondrial vacuolization was calculated as the number of vacuoles divided by the total number of mitochondria. Mitochondrial vacuolization rate of control was used for normalization.
5.23. Western blotting
Cells or tissues were collected and total protein was extracted as in a previous study [66]. Protein concentration was determined using BCA assay kit (Thermo Fisher, USA). Five to fifteen μg of protein was separated by SDS-PAGE gel, and then transferred to NC membrane. After the membrane was blocked with 5% milk, the membranes were incubated with the primary antibodies overnight at 4 °C. After washing 5 times with TBST for 10 min, the membranes were incubated with secondary antibodies for 1 h at room temperature. After washing, the density of the protein bands was visualized and analyzed using a Tanon 2500 gel imaging system (Tanon, Shanghai, China). The antibodies used for western blotting (WB, 1:1000) are as follows: anti-β tubulin (CST, #2128), anti-β actin (CST, #4970), anti-PSD95 (CST, #3450), anti-synapsin-1 (CST, ab52903), anti-GAP43 (CST, #8945), anti-synaptophysin (abcam, ab32127), anti-Bcl 2 (wanlei, WL01556), anti-Bax (prointech, 50,599), anti-cleaved caspase 3 (CST, #9664), anti-Bak (Wanlei, WL0129a), anti-cleaved BID (Wanlei, WL01129), anti-FUS(Santa Cruz, sc47711).
5.24. RNA extraction and RT-qPCR
Total RNA was extracted from cell lysates using Trizol Reagent (Cat No. 15596–026, Thermo Fisher Scientific, Waltham, MA, USA). The relative values of mitochondrial mRNA were determined by comparing them with those in NorEVs. Quantitative RT-PCR (RT-qPCR) was carried out using Prime Script RT Master Mix (Cat No. RR047A, Takara, Japan) for reverse-transcription and LightCycler® 480 SYBR Green I Master (Cat No. 04887352001, Roche, United States) for real-time PCR, respectively. The primer sequences are shown in Supplementary Table S2.5. The relative target mRNA levels were determined using the 2−ΔΔCt method, and 18s was used as an internal reference for normalization.
5.25. RBP immunoprecipitation (RIP)
SH-sy5y (8 × 106 cells per plate) or primary cortical neuron (6 × 107 cells per plate) were seeded on 10 cm plates. After various treatments, RIP was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Cat No. MAGNARIP01, Millipore, Bedford, MA) according to the manufacturer's instructions. Lysates were incubated with 12 μg of control mouse IgG or FUS antibody conjugated-beads with rotation at 4 °C overnight. Finally, the immunoprecipitated RNAs were extracted using Trizol Reagent and analyzed by RT-qPCR. PCR products were separated by agarose gel electrophoresis. For the HypEVs RIP experiment, a total of 160 ml of culture supernatant from 16 dishes is required for HypEVs extraction. After precipitation and lysis of sEVs, they were incubated with FUS antibody or IgG using the same method as described above.
5.26. Statistics analysis
All statistical analyses were performed with SPSS (Windows version 27.0; SPSS Inc., Chicago, IL, USA) and visualized with GraphPad Prism 8.01 software (GraphPad Software, Inc., La Jolla, CA, USA). Student's two-sample t-test was performed to compare values between two groups. One-way ANOVA tests were used to examine the statistical significance of differences between the data of three or more groups. Data are expressed as means ± SEM. P-values of 0.05 or less were considered statistically significant. All representative images were selected without bias and had characteristics typical of the data or overall trend.
CRediT author statement
Y.W: Conceptualization, Validation, Formal analysis, and Writing – original draft, X.H: Conceptualization, Validation, Formal analysis, and Writing – original draft, Z.T: Conceptualization, Validation, Formal analysis, and Writing – original draft. D.L: Conceptualization, Writing – review & editing, Visualization, and Supervision, A.X: Conceptualization, Writing – review & editing, Visualization, and Supervision, H.M.: Conceptualization, Writing – review & editing, Visualization, and Supervision. J.Z: Formal analysis and Writing – original draft, M.P: Formal analysis and Writing – original draft, N.H: Formal analysis and Writing – original draft, T.Z: Formal analysis and Writing – original draft. The manuscript is attributed to contributions from all authors. All authors have approved of the final version of the manuscript.
Ethics approval
All animal procedures were approved by the Institutional Animal Care and Use Committee of Jinan University (approval ID: 20,201,028–03). The study was carried out in accordance with the recommendations of the NIH Guide (NIH Publications No. 8023, revised 1978) for the Care and Use of Laboratory Animals. All experiments were carefully conducted in accordance with the guidelines for Animal Experimentation of Jinan University.
Declaration of competing interest
The authors declared that they have no conflicts of interest to this work.
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82271304, 81801150, 81971121, 82171316 and 81671167), the Science and Technology Planning Project of Guangdong Province, China (2017A020215049, 2019A050513005), Natural Science Foundation of Guangdong Province (2018A0303130182, 2020A1515010279 and 2022A1515012311), the Fundamental Research Funds for the Central Universities (21621102),Science and Technology Projects in Guangzhou, China (2014Y2-00505, 202002020003, 202201010127, and SL2023A03J01214), Science and Technology Program of Guangzhou: Key Lab of Guangzhou Basic and Translational Research of Pan-vascular Diseases(202201020042), Young Talent Support Project of Guangzhou Association for Science and Technology (QT-2023-024), Guangdong Basic and Applied Basic Research Foundation (2021A1515111226), China Postdoctoral Science Foundation (2022M710058). The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.07.009.
Contributor Information
Hongcheng Mai, Email: hongchengmai@163.com, hongcheng.mai@helmholtz-munich.de.
Anding Xu, Email: tlil@jnu.edu.cn.
Dan Lu, Email: ludan@jnu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
The following is the Supplementary data to this article:Gene ontology analysis results of proteins identified by mass spectrometry in Fig. S1; assessment of FUS on the surface of sEV in Fig. S2; additional results of sEV protection and mitochondrial associated mRNA prediction in Figs. S3 and S4; additional results of neuronal sEV's composition, uptake and safety in Figs. S5–8; additional results of SH-sy5y-released sEV's composition, uptake and neuroprotection in Figs. S9–10; detailed results of mass spectrometric analysis in Table S1; detailed results of transcriptomes in Table S2.
References
- 1.Hong S.-B., Yang H., Manaenko A., et al. Potential of exosomes for the treatment of stroke. Cell Transplant. 2019;28(6):662–670. doi: 10.1177/0963689718816990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauré J., Lachenal G., Court M., et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 2006;31(4):642–648. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Zagrean A.-M., Hermann D.M., Opris I., et al. Multicellular crosstalk between exosomes and the neurovascular unit after cerebral ischemia. Therapeutic implications. Front. Neurosci. 2018;12:811. doi: 10.3389/fnins.2018.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B., Zhang Y., Du X.F., et al. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27(7):882–897. doi: 10.1038/cr.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Men Y., Yelick J., Jin S., et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019;10(1):4136. doi: 10.1038/s41467-019-11534-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bister N., Pistono C., Huremagic B., et al. Hypoxia and extracellular vesicles: a review on methods, vesicular cargo and functions. J. Extracell. Vesicles. 2020;10(1) doi: 10.1002/jev2.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang C.-S., Fu S.-J., Hsu C.-L., et al. Neuronal exosomes secreted under oxygen-glucose deprivation/reperfusion presenting differentially expressed miRNAs and affecting neuronal survival and neurite outgrowth. NeuroMolecular Med. 2021;23(3):404–415. doi: 10.1007/s12017-020-08641-z. [DOI] [PubMed] [Google Scholar]
- 8.Deng M., Xiao H., Peng H., et al. Preservation of neuronal functions by exosomes derived from different human neural cell types under ischemic conditions. Eur. J. Neurosci. 2018;47(2):150–157. doi: 10.1111/ejn.13784. [DOI] [PubMed] [Google Scholar]
- 9.Gregorius J., Wang C., Stambouli O., et al. Small extracellular vesicles obtained from hypoxic mesenchymal stromal cells have unique characteristics that promote cerebral angiogenesis, brain remodeling and neurological recovery after focal cerebral ischemia in mice. Basic Res. Cardiol. 2021;116(1):40. doi: 10.1007/s00395-021-00881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Börger V., Sardari M., et al. Mesenchymal stromal cell-derived small extracellular vesicles induce ischemic neuroprotection by modulating leukocytes and specifically neutrophils. Stroke. 2020;51(6):1825–1834. doi: 10.1161/STROKEAHA.119.028012. [DOI] [PubMed] [Google Scholar]
- 11.Perets N., Betzer O., Shapira R., et al. Golden exosomes selectively target brain pathologies in neurodegenerative and neurodevelopmental disorders. Nano Lett. 2019;19(6):3422–3431. doi: 10.1021/acs.nanolett.8b04148. [DOI] [PubMed] [Google Scholar]
- 12.Betzer O., Perets N., Angel A., et al. In vivo neuroimaging of exosomes using gold nanoparticles. ACS Nano. 2017;11(11):10883–10893. doi: 10.1021/acsnano.7b04495. [DOI] [PubMed] [Google Scholar]
- 13.He G., Peng X., Wei S., et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol. Cancer. 2022;21(1):19. doi: 10.1186/s12943-021-01440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H., Zhao H., Zhang M., et al. Hypoxia induced changes of exosome cargo and subsequent biological effects. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.824188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groot M., Lee H. Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells. 2020;9(4) doi: 10.3390/cells9041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Statello L., Maugeri M., Garre E., et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei H., Chen Q., Lin L., et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021;17(1):163–177. doi: 10.7150/ijbs.53671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabbiano F., Corsi J., Gurrieri E., et al. RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J. Extracell. Vesicles. 2020;10(2) doi: 10.1002/jev2.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalla Costa I., Buchanan C.N., Zdradzinski M.D., et al. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2021;22(2):77–91. doi: 10.1038/s41583-020-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schieweck R., Ninkovic J., Kiebler M.A. RNA-binding proteins balance brain function in health and disease. Physiol. Rev. 2021;101(3):1309–1370. doi: 10.1152/physrev.00047.2019. [DOI] [PubMed] [Google Scholar]
- 22.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135(8):851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamelgarn M., Chen J., Kuang L., et al. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim. Biophys. Acta. 2016;1862(10):2004–2014. doi: 10.1016/j.bbadis.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Kodavati M., Britz G.W., et al. DNA damage and repair deficiency in ALS/FTD-Associated neurodegeneration: from molecular mechanisms to therapeutic implication. Front. Mol. Neurosci. 2021;14 doi: 10.3389/fnmol.2021.784361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling S.-C. Synaptic paths to neurodegeneration: the emerging role of TDP-43 and FUS in synaptic functions. Neural Plast. 2018;2018 doi: 10.1155/2018/8413496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahadevan S., Hembach K.M., Tantardini E., et al. Synaptic FUS accumulation triggers early misregulation of synaptic RNAs in a mouse model of ALS. Nat. Commun. 2021;12(1):3027. doi: 10.1038/s41467-021-23188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Ding X., Zhou Z., et al. Sirtuin 1 alleviates diabetic neuropathic pain by regulating synaptic plasticity of spinal dorsal horn neurons. Pain. 2019;160(5):1082–1092. doi: 10.1097/j.pain.0000000000001489. [DOI] [PubMed] [Google Scholar]
- 28.Hanz S., Fainzilber M. Retrograde signaling in injured nerve-the axon reaction revisited. J. Neurochem. 2006;99(1):13–19. doi: 10.1111/j.1471-4159.2006.04089.x. [DOI] [PubMed] [Google Scholar]
- 29.Guo L., Huang Z., Huang L., et al. Surface-modified engineered exosomes attenuated cerebral ischemia/reperfusion injury by targeting the delivery of quercetin towards impaired neurons. J. Nanobiotechnol. 2021;19(1):141. doi: 10.1186/s12951-021-00879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKelvey K.J., Powell K.L., Ashton A.W., et al. Exosomes: mechanisms of uptake. J Circ Biomark. 2015;4:7. doi: 10.5772/61186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahu A., Mamiya H., Shinde S.N., et al. Age-related declines in α-Klotho drive progenitor cell mitochondrial dysfunction and impaired muscle regeneration. Nat. Commun. 2018;9(1):4859. doi: 10.1038/s41467-018-07253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiura A., McLelland G.-L., Fon E.A., et al. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33(19):2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crewe C., Funcke J.-B., Li S., et al. Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metabol. 2021;33(9) doi: 10.1016/j.cmet.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L., Fan M., Huang D., et al. Clodronate-loaded liposomal and fibroblast-derived exosomal hybrid system for enhanced drug delivery to pulmonary fibrosis. Biomaterials. 2021;271 doi: 10.1016/j.biomaterials.2021.120761. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino A., Costa-Silva B., Shen T.-L., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uzdensky A.B. Photothrombotic stroke as a model of ischemic stroke. Transl Stroke Res. 2018;9(5):437–451. doi: 10.1007/s12975-017-0593-8. [DOI] [PubMed] [Google Scholar]
- 37.Rabbitts T.H., Forster A., Larson R., et al. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4(2):175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 38.Sharma A., Lyashchenko A.K., Lu L., et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 2016;7 doi: 10.1038/ncomms10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjuan-Ruiz I., Govea-Perez N., McAlonis-Downes M., et al. Wild-type FUS corrects ALS-like disease induced by cytoplasmic mutant FUS through autoregulation. Mol. Neurodegener. 2021;16(1):61. doi: 10.1186/s13024-021-00477-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng H., Gao K., Jankovic J. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 2014;10(6):337–348. doi: 10.1038/nrneurol.2014.78. [DOI] [PubMed] [Google Scholar]
- 41.Guerrero E.N., Wang H., Mitra J., et al. TDP-43/FUS in motor neuron disease: complexity and challenges. Prog. Neurobiol. 2016;145–146:78–97. doi: 10.1016/j.pneurobio.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda A., Takeda J.-I., Ohno K. FUS-mediated regulation of alternative RNA processing in neurons: insights from global transcriptome analysis. Wiley Interdiscip Rev RNA. 2016;7(3):330–340. doi: 10.1002/wrna.1338. [DOI] [PubMed] [Google Scholar]
- 44.Deng J., Yang M., Chen Y., et al. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 2015;11(9) doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng J., Wang P., Chen X., et al. FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc. Natl. Acad. Sci. U. S. A. 2018;115(41):E9678–E9686. doi: 10.1073/pnas.1806655115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai Y.-L., Coady T.H., Lu L., et al. ALS/FTD-associated protein FUS induces mitochondrial dysfunction by preferentially sequestering respiratory chain complex mRNAs. Genes Dev. 2020;34(11–12):785–805. doi: 10.1101/gad.335836.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devine M.J., Kittler J.T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 2018;19(2):63–80. doi: 10.1038/nrn.2017.170. [DOI] [PubMed] [Google Scholar]
- 48.Rossi M.J., Pekkurnaz G. Powerhouse of the mind: mitochondrial plasticity at the synapse. Curr. Opin. Neurobiol. 2019;57:149–155. doi: 10.1016/j.conb.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel P., Buchanan C.N., Zdradzinski M.D., et al. Nucleic Acids Res; 2022. Intra-axonal Translation of Khsrp mRNA Slows Axon Regeneration by Destabilizing Localized mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo S., van Niekerk E.A., Merianda T.T., et al. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp. Neurol. 2010;223(1):19–27. doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim E., Jung H. Local mRNA translation in long-term maintenance of axon health and function. Curr. Opin. Neurobiol. 2020;63:15–22. doi: 10.1016/j.conb.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Müntjes K., Devan S.K., Reichert A.S., et al. Linking transport and translation of mRNAs with endosomes and mitochondria. EMBO Rep. 2021;22(10) doi: 10.15252/embr.202152445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cioni J.-M., Lin J.Q., Holtermann A.V., et al. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176(1–2) doi: 10.1016/j.cell.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossoll W., Bassell G.J. Crosstalk of local translation and mitochondria: powering plasticity in axons and dendrites. Neuron. 2019;101(2):204–206. doi: 10.1016/j.neuron.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 55.Dennerlein S., Wang C., Rehling P. Plasticity of mitochondrial translation. Trends Cell Biol. 2017;27(10):712–721. doi: 10.1016/j.tcb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Todkar K., Chikhi L., Desjardins V., et al. Selective packaging of mitochondrial proteins into extracellular vesicles prevents the release of mitochondrial DAMPs. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-21984-w. 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venturini A., Passalacqua M., Pelassa S., et al. Exosomes from astrocyte processes: signaling to neurons. Front. Pharmacol. 2019;10:1452. doi: 10.3389/fphar.2019.01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Batrakova E.V., Kim M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Contr. Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agostini M., Romeo F., Inoue S., et al. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23(9):1502–1514. doi: 10.1038/cdd.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arimura N., Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 2007;8(3):194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 61.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu R., Greening D.W., Chen M., et al. Surfaceome of exosomes secreted from the colorectal cancer cell line SW480: peripheral and integral membrane proteins analyzed by proteolysis and TX114. Proteomics. 2019;19(8) doi: 10.1002/pmic.201700453. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Li Y., Zang J., et al. CircOGDH is a penumbra biomarker and therapeutic target in acute ischemic stroke. Circ. Res. 2022;130(6):907–924. doi: 10.1161/CIRCRESAHA.121.319412. [DOI] [PubMed] [Google Scholar]
- 64.Labat-gest V., Tomasi S. Photothrombotic ischemia: a minimally invasive and reproducible photochemical cortical lesion model for mouse stroke studies. J. Vis. Exp. 2013;76 doi: 10.3791/50370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fath T., Ke Y.D., Gunning P., et al. Primary support cultures of hippocampal and substantia nigra neurons. Nat. Protoc. 2009;4(1):78–85. doi: 10.1038/nprot.2008.199. [DOI] [PubMed] [Google Scholar]
- 66.Zang J., Wu Y., Su X., et al. Inhibition of PDE1-B by vinpocetine regulates microglial exosomes and polarization through enhancing autophagic flux for neuroprotection against ischemic stroke. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.616590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomes A.R., Sangani N.B., Fernandes T.G., et al. Extracellular vesicles in CNS developmental disorders. Int. J. Mol. Sci. 2020;21(24) doi: 10.3390/ijms21249428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 71.Sherman B.T., Hao M., Qiu J., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic. Acid. Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan X., Fang Y., Li X., et al. RBPsuite: RNA-protein binding sites prediction suite based on deep learning. BMC Genom. 2020;21(1):884. doi: 10.1186/s12864-020-07291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armaos A., Colantoni A., Proietti G., et al. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49(W1):W72–W79. doi: 10.1093/nar/gkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchthal B., Weiss U., Bading H. Post-injury nose-to-brain delivery of activin A and SerpinB2 reduces brain damage in a mouse stroke model. Mol. Ther. 2018;26(10):2357–2365. doi: 10.1016/j.ymthe.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fedorov A., Beichel R., Kalpathy-Cramer J., et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schindelin J., Arganda-Carreras I., Frise E., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferreira T.A., Blackman A.V., Oyrer J., et al. Neuronal morphometry directly from bitmap images. Nat. Methods. 2014;11(10):982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.