Abstract
Augmentation of regenerative ability is a powerful strategy being pursued for the biomedical management of traumatic injury, cancer, and degeneration. While considerable attention has been focused on embryonic stem cells, it is clear that much remains to be learned about how somatic cells may be controlled in the adult organism. The tadpole of the frog Xenopus laevis is a powerful model system within which fundamental mechanisms of regeneration are being addressed. The tadpole tail contains spinal cord, muscle, vasculature, and other terminally differentiated cell types and can fully regenerate itself through tissue renewal—a process that is most relevant to mammalian healing. Recent insight into this process has uncovered fascinating molecular details of how a complex appendage senses injury and rapidly repairs the necessary morphology. Here, we review what is known about the chemical and bioelectric signals underlying this process and draw analogies to evolutionarily conserved pathways in other patterning systems. The understanding of this process is not only of fundamental interest for the evolutionary and cell biology of morphogenesis, but will also generate information that is crucial to the development of regenerative therapies for human tissues and organs.
Keywords: Xenopus, frog, tadpole, tail, regeneration, growth factor, bioelectricity, ion pump
INTRODUCTION
The restoration of lost or damaged body parts has been a long-standing topic of interest in biology. In the mid-seventeenth century, Abraham Trembley showed that hydra can regrow into complete animals after being cut into several pieces (Dinsmore, 1991). In the early 20th century, T.H. Morgan systematically studied the ability of planaria to regenerate even after being cut into over 100 sections (Morgan, 1901). Subsequent studies have continued to focus interest on animals with high regenerative abilities (Birnbaum and Sánchez Alvarado, 2008).
Urodele amphibians, such as newts and axolotls, are noted for their capacity to regenerate multiple tissues, including limbs, tail, heart, lens, spinal cord, brain, jaw, and retina. Studies have revealed that urodele regeneration involves a high degree of cellular plasticity (for review, see Brockes and Kumar, 2002). Existing cells at an injury site can undergo de-differentiation to become proliferating precursor cells and later re-differentiate into multiple cell types (also known as metaplasia) (Lo et al., 1993; Kumar et al., 2000). Although these models have traditionally been difficult to manipulate genetically, this situation is changing with the development of molecular techniques in these systems (Echeverri and Tanaka, 2002, 2003; Atkinson et al., 2006; Kumar et al., 2007).
More recently, work has extended to zebrafish and planaria, where loss-of-function genetic screens using mutagenesis (Poss et al., 2003) or RNAi interference (Reddien et al., 2005), respectively, have been successful in the identification of genes involved in the regenerative process. In contrast to the animals mentioned above, mammals have very limited regenerative ability (Heber-Katz et al., 2004; Price et al., 2005). Thus, in these systems, the major focus has been on identifying, isolating, and characterizing stem cells, with the goal of using them to generate multiple cell types that can then be transplanted into animals to replace the lost or damaged tissues. However, most of these studies have been performed in vitro. Because of the three-dimensional complexity of real organs, in vivo studies are more likely to give a complete understanding of the interactions required during the regenerative process.
The anuran frog, Xenopus laevis, is a well-studied model for the understanding of developmental events. The majority of work has focused on early embryogenesis, taking advantage of the relatively large size of the embryos, which can be obtained with ease and cultured to allow access to any stage of development. Interestingly, later-stage Xenopus larva, or tadpoles, are also capable of regeneration. Through most of development, larval tails can regenerate fully after injury, making this a potentially fruitful model for the study of regeneration (Deuchar, 1975). There are several important advantages. First, embryos quickly develop into tadpoles with tails within 4 days after fertilization; the tail is highly accessible and can be simply and reproducibly amputated with a scalpel blade. Second, the tail is transparent and easily amenable to most microscopy techniques, including fluorescent marker tracking, physiological reporter dyes, FRET, photo-bleaching, and laser ablation. Third, the embryos do not require circulating water for growth, facilitating culture of large numbers of animals. Most importantly, many genetic techniques are available for the Xenopus system, allowing for direct molecular studies (Sive et al., 2000). Together, the fast development time, ease of culture, accessibility of the tail, and the availability of molecular approaches make the vertebrate Xenopus tadpole an excellent model in which to examine and develop our understanding of regeneration as a process in which a complex appendage recognizes damage and performs the necessary repairs.
OVERVIEW OF Xenopus TAIL REGENERATION
The developing Xenopus larval tail is a complex tissue consisting of muscle, notochord, the spinal cord, and vasculature. In the tail, the spinal cord is located on top of the notochord, and the two central structures are flanked by muscle bilaterally, with dorsal and ventral fins at the midline. The tail is a developmental structure that enables tadpoles to move, gather food, and, in nature, escape predators. During thyroid-hormone-driven metamorphosis, the tail is gradually resorbed and disappears as the tadpole transforms into a frog (Weber, 1965).
A scalpel blade can be used to amputate four-day-old larval tails at the midpoint between the posterior end of the hindgut and the caudal tip of the tail (Fig. 1). Regeneration is rapid, and by 7 days post-amputation (dpa), the previously injured tadpole is mostly indistinguishable from its uncut siblings, although molecular differences have been described between the primary and regenerated tissues (Mochii et al., 2007). Remarkably, tadpoles continuously grow in size during development, and the process of tail replacement is significantly faster than the normal tail growth rate, allowing regenerates to catch up in size to uncut siblings.
Figure 1.
Example of Xenopus tail regeneration. (A) A schematic of the tail tip amputated in the tadpole. (B) A tadpole at 7 days of development. (C) A tadpole that has regenerated its tail (green arrow); several time-points during regeneration are shown in C′ (24 hpa), C″ (48 hpa), and C‴ (96 hpa). Blue stain in panel C″ indicates new tissue produced in the core of the tail regenerate. (D) A tadpole at 7 days that was treated with a V-ATPase inhibitor. The wound healed, but the tail did not regenerate (red arrowhead; purple line shows plane of amputation).
Throughout its larval life, the tail of the Xenopus tadpole is able to regenerate fully when injured, with the exception of a short period of time, at approximately the developmental stages when tadpoles begin to feed (although the precise timing is somewhat variable). During this “refractory” period, tadpoles with amputated tails fail to replace the lost tissues and instead, produce a thick “skin-like” epidermis covering (Beck et al., 2003). After this phase, tadpoles again regain tail-regenerative ability. It is unknown what, if any, advantage this might confer on the tadpole. However, the existence of this refractory period provides an important endogenous context in which to compare and contrast regenerative and non-regenerative states. In particular, this may be an important model where the known age-dependent reductions of human regenerative ability may be explored (Douglas, 1972; Illingworth, 1974).
In Urodeles, such as newts, limb amputation leads to the formation of a blastema that contains de-differentiated (or potentially multipotent) cells, some of which have the ability to generate multiple cell types (Echeverri and Tanaka, 2002). Cell lineage studies during tail regeneration have sought to determine whether similar mechanisms exist in the anuran Xenopus, or whether precursor cells are lineage-restricted.
Lineage Analysis of Cell Types Regenerated in the Tail
Individual cell lineages in the regenerating tail were examined following the development of specifically marked tissues by tissue grafting or electroporation of marker DNA into selected cells. To determine the origin of the regenerated notochord and spinal cord, investigators used transgenic embryos expressing green fluorescent protein (GFP) under the control of cytomegalovirus early promoter (CMV) as a donor source for transplantation studies (Gargioli and Slack, 2004). The CMV promoter directs ubiquitous expression of GFP in Xenopus embryos; thus, grafts of selected tissues marked with GFP can be obtained and integrated into normal, unmarked embryos so that their fates can be tracked. Grafts of GFP-labeled notochord cells specifically populated the notochord of the GFP-negative recipient embryos. After amputation of the tail, only the regenerated notochord showed GFP expression, indicating that notochord cells are limited to generating their specific cell population (Gargioli and Slack, 2004). The same result was obtained with spinal cord tissue grafts.
To examine the source of the regenerated muscle tissue, investigators examined muscle cells marked with GFP either by the Cre-Lox recombinase method (Ryffel et al., 2003; Gargioli and Slack, 2004) or by tissue grafts (Gargioli and Slack, 2004), after tail amputation. The regenerated non-muscle cells did not express GFP, suggesting that transdifferentiation is unlikely to occur in myocytes. Interestingly, the labeled differentiated muscle cells did not contribute to the regenerating myocyte population (Gargioli and Slack, 2004; Mochii et al., 2007). Instead, Pax7-expressing muscle satellite cells were identified as the progenitor source for regenerating muscle tissue (Gargioli and Slack, 2004; Chen et al., 2006). Muscle satellite cells are mononucleate cells that are found in the basal layer of muscle fibers (Gros et al., 2005; Kassar-Duchossoy et al., 2005; Relaix et al., 2005). They are resident progenitor cells that express the transcription factor Pax7, and are able to proliferate and contribute to the regeneration of muscle fibers upon injury or damage (Charge and Rudnicki, 2004). After tail amputation, the number of Pax7-expressing cells is increased. Inducing the expression of a dominant-negative Pax7 transgene in Xenopus larvae reduced the number of satellite cells in regenerating tails (Chen et al., 2006), which subsequently contained little or no muscle tissue, even though the notochord and the spinal cord were comparable with the wild-type regenerate.
To date, studies have provided no evidence of transdifferentiation during this process, in contrast to newt regeneration (Tsonis et al., 1995; Echeverri and Tanaka, 2002), or of a dependence on a large adult stem cell population, as occurs in planaria (Reddien and Sánchez Alvarado, 2004). Thus, Xenopus appendage regeneration is likely to be highly similar to the process of mammalian cell renewal, in which lineage-restricted progenitors proliferate to generate cells of one specific type that subsequently differentiate to replace lost tissue. Although the tadpole is still a developing organism, the ability of differentiated neurons to proliferate bodes well for regenerative approaches based on the modulation of somatic cells. Indeed, consistent with the role of bioelectric parameters in regenerative control (discussed below), it is now known that mature CNS neurons from amniotes can be induced to re-enter the cell cycle (begin mitosis) by sustained depolarization (Stillwell et al., 1973; Cone and Cone, 1976).
Molecular Tools for the Study of Xenopus Regeneration
The study of Xenopus tail regeneration has been facilitated by the development of chemical and molecular biology technologies that can be used to manipulate molecular pathways. A well-established method of gene transfer in Xenopus is transgenesis. The generation of Xenopus transgenics involves the isolation of sperm nuclei that are treated to allow the integration of a DNA vector containing the gene of interest into the haploid genome (Kroll and Amaya, 1996). The treated sperm are then injected into unfertilized eggs to produce embryos that can mature to become transgenic frog lines. More recently, another approach, meganuclease-aided transgenesis, has been developed for the creation of transgenic frogs (Ogino et al., 2006; Pan et al., 2006). This method bypasses sperm treatment. Instead, the desired transgene is cloned into a vector that contains flanking 18-base-pair recognition sites for the meganuclease. The construct, along with the I-SceI meganuclease enzyme, is then directly injected into fertilized eggs. Because the Xenopus laevis genome is pseudotetraploid, experiments that can induce gain-of-function activity through over-expression or loss-of-function through the use of dominant-negative transgenes are likely to result in tractable phenotypes, whereas RNAi interference or similar methods targeting inactivation of individual genes may not have similar effects.
Both methods allow for the use of promoters to create embryos with localized and specific transgene expression, and well-characterized mammalian promoters (such as the ubiquitous CMV, and the nerve-specific β-tubulin) have been used successfully in Xenopus to drive gene expression. However, one problem is that gene expression driven by some promoters during embryogenesis causes developmental defects or lethality. In this case, a heat shock (Beck et al., 2006) or a chemically inducible promoter (Das and Brown, 2004) can be used for temporal control of expression in the tadpole. An alternative method of gene transfer into Xenopus tadpoles is electroporation (Lin et al., 2007; Mochii et al., 2007), which can allow for the transfection of a small, targeted, population of cells (Eide et al., 2000; Haas et al., 2001, 2002; Sasagawa et al., 2002).
Another important and highly useful approach to the manipulation of gene expression in Xenopus does not require any direct modification of DNA or mRNA. Chemical genetics is the use of small molecules to modulate protein function (Yeh and Crews, 2003; den Hertog, 2005). With the ever-increasing number of available compounds with known specificity (Smith and Remsen, 2006), loss-of-function analysis by drug inhibitor screening has become an excellent tool for the rapid and inexpensive identification of genes and gene families that participate in specific biological events. A powerful strategy first uses compounds with broad targeting, and then proceeds to more specific reagents when positive results are obtained in the chosen assay from a compound targeting a particular protein set. This strategy (Adams and Levin, 2006) relies on a hierarchical grouping of many different molecular pathways, which can thus be probed by an efficient binary tree approach. While any pharmacological result must ultimately be validated by molecular-genetic analysis, there are several advantages for the use of chemical genetics in Xenopus embryos as a screen to identify proteins functioning in specific aspects of regenerative response. First, it bypasses the requirement to generate transgenic frogs to control gene function. Second, animals can be treated with small molecules at specific timepoints. This feature is highly useful in the determination of the temporal requirement for a gene function. Third, the availability of chemical libraries and hierarchical databases can facilitate rapid in vivo screening to identify agents that disrupt a molecular pathway or biological process of interest. Furthermore, because tadpoles can be reared in an area as small as a single well of a six-well tissue culture plate, they are particularly well-suited for vertebrate studies involving small-molecule probes (Adams and Levin, 2006).
In Xenopus, the use of chemical genetics has resulted in the successful identification of several important pathways that regulate tail regeneration, including Notch (Beck et al., 2003), ion flow (Adams et al., 2007), apoptosis (Tseng et al., 2007), TGF-β (Ho and Whitman, 2008), and FGF (Lin and Slack, 2008). Because the specificity of each inhibitor can vary, results from studies using chemical genetics must ultimately be validated with molecular, gene-specific techniques.
In contrast to studies that focus on the effects of specific molecular targets, genome-wide approaches present a global perspective on the changes in gene expression that occur in the tail, and help to identify the pathways that participate in a biological event such as tail regeneration. A cDNA macroarray containing EST clones covering 17,000 Xenopus genes was examined for differential expression patterns between 0, 1.5, and 3 days post-amputation (dpa) (Tazaki et al., 2005). Further spatiotemporal analysis of the selected genes by RNA in situ enabled these targets to be categorized into several groups. Importantly, these genes were specifically expressed in the regenerating tail, providing fertile ground for further functional analysis.
The early-response genes identified from the macroarray are largely involved in inflammation response, wound healing, cell proliferation, and signaling, processes that are known to be required after injury. The late-response group includes genes that participate mostly in cell proliferation and differentiation. In addition, genes were identified that were novel or had no known function. For identification of the mechanisms that are regeneration-specific, it will be necessary to characterize the functions of the genes that do not participate in primary tail development, but are expressed during tail regeneration.
PATHWAYS THAT REGULATE TAIL REGENERATION
Overview of Tail Regeneration
Xenopus tail regeneration consists of three main steps: wound healing, formation of the regeneration bud, and subsequent outgrowth and patterning. After tail amputation, a basal layer of p63-positive cells covers the wound by 2 hrs post-amputation (hpa) (Ho and Whitman, 2008). Wound healing is rapidly completed within 6–12 hpa as the apical wound epithelium is formed. By 24 hpa, an outgrowth called the regeneration bud is visible at the amputation site. The bud consists mainly of undifferentiated mesenchymal cells, the neural ampulla (resulting from the closing of the cut spinal cord), the terminated notochord with a group of precursor cells directly gathered at its distal end, and degenerating muscle fibers (Gargioli and Slack, 2004). Tail outgrowth follows as cells in the bud undergo proliferation and patterning. A fully functional tail can be reformed 7–21 days after amputation, depending on the age of the larvae.
Within the last 10 yrs, studies of tail regeneration have identified several molecular pathways that regulate this process, and ongoing work is attempting to integrate these into a detailed stepwise synthesis of the regenerative process.
Early Requirements
Wound Healing
It has long been known that the wound epithelium is a key factor in the regeneration and patterning of new tissue (Tassava and Mescher, 1975; Borgens, 1984; Rajnicek et al., 1988; Yoshii et al., 2005; Armstrong et al., 2006; Campbell and Crews, 2007; Raja et al., 2007). Recently, it was observed that TGF-β signaling plays a role in wound healing of the injured tail after amputation. The TGF-β pathway functions through signaling by serine-threonine kinase receptors that phosphorylate downstream targets which then translocate into the nucleus and interact with transcriptional components (Massagué, 1998). It is involved in multiple processes during development and repair (Gordon and Blobe, 2008; Rhett et al., 2008). Expression of 2 ligands, xTGFβ-2 and xTGFβ-5, can be seen in the early tail regeneration bud by 4 hpa (Ho and Whitman, 2008). Treatment of amputated tails with SB-431542, a specific TGF-β inhibitor, prevented regeneration. In the presence of SB-431542, blot clotting after amputation appears to be normal. However, the injury fails to heal since the wound epithelium does not form and cellular material continues to leak from the site (Ho and Whitman, 2008). In contrast, normal closure of an untreated cut is completed by 6–8 hpa. Subsequent withdrawal of SB-431542 at 8 hpa appears to rescue the healing defect, since a wound epidermis is observed by 2 dpa. Phosphorylated Smad2, a transducer of TGF-β activity, is present at the wild-type tail amputation edge in a single layer by 1 hpa and remains there until 8 hpa. This expression is lost in SB-431542-treated tail wound sites. Interestingly, p-Smad2 expression in untreated wounds is seen in the basal layer of the wound epidermis, suggesting that closure of the apical layer is TGF-β–independent.
An examination of amputated tails treated with SB-431542 from 0–8 hpa showed that inhibition of TGF-β signaling during wound healing also causes defects in the regeneration bud. At 2 dpa, SB-treated tail buds show reduced proliferation, as well as a disorganized neural ampulla, and a reduction in numbers of notochord precursor and muscle satellite cells (Ho and Whitman, 2008), resulting in a regenerative failure. Together, these results indicate that TGF-β is required for proper formation of the wound epidermis after injury in Xenopus tails.
V-ATPase and Bioelectric Events
An extensive body of classic results suggests that endogenous bioelectric events are involved in the control of regeneration (Borgens, 1983; Levin, 2007). Analysis of recent data has begun to reveal molecular details by which ion flows control the early steps of tail regeneration (Adams et al., 2007). The V-ATPase is a pump that exports H+ through membranes at the expense of ATP; while many V-ATPases occur on intracellular vesicles, the regeneration-relevant subtype functions on the cell surface to pump H+ ions out of the cytoplasm, inducing hyperpolarization of the transmembrane potential (Wieczorek et al., 1999). V-ATPase is up-regulated by 6 hpa at the plasma membrane of cells on the surface of the regeneration bud, and functional studies indicate that its activity is required in the first 24 hpa. Loss of V-ATPase function in Xenopus, either through treatment with a specific inhibitor, concanamycin, or through expression of a dominant-negative form of this pump complex, specifically abolished tail regeneration (although this pump is not required for wound healing or primary tail development). Importantly, it is the H+ flux activity per se that is required for regeneration, and not ion-independent roles of the V-ATPase, since the pharmacological-dependent defect can be rescued by expression of a concanamycin-insensitive yeast proton pump, PMA1 (Masuda and Montero-Lomeli, 2000). It was further revealed (Adams et al., 2007) that the control of regeneration by V-ATPase activity is mediated by V-ATPase’s regulation of cell proliferation in the bud as well as of axonal patterning (a morphogenetic cue).
Examination of the physiology of the regeneration bud showed that, after tail amputation, the wild-type bud is depolarized by 6 hpa, but becomes repolarized by 24 hpa. In contrast, buds lacking V-ATPase function remain depolarized at 24 hpa. Similarly, refractory-stage buds also remain depolarized at 24 hpa. Functional experiments have indicated that the physiology of bud cells is a major determining factor in tail regeneration. Under permissive conditions, the bud is polarized and regeneration proceeds, while under non-permissive conditions, the bud remains depolarized and growth is inhibited. Analysis of the data indicated that direct manipulation of membrane potential was sufficient to modulate regeneration of the tail, suggesting that a primary role of V-ATPase in this context is to regulate membrane voltage. While this is consistent with the role of membrane potential in the regulation of cell proliferation and migration (McCaig et al., 2005; Levin, 2007), it is still not known whether localized pH gradients (intracellular or extracellular), as well as long-range electric fields also generated by V-ATPase, are involved in this process.
Apoptosis
Another pathway that appears to play an important early role in determining the regenerative ability of the tail is apoptosis (Elmore, 2007). In uncut larvae, a small number of randomly distributed apoptotic cells can be observed throughout the tail. However, immunostaining with an activated Caspase-3 antibody, an indicator of programmed cell death, identified a specific endogenous group of apoptotic cells that exists in the regeneration bud by 12 hpa and extends even to 48 hpa (Tseng et al., 2007). Treatment of amputated tails with 2 distinct chemical apoptosis inhibitors, M50054 (Tsuda et al., 2001) and NS3694 (Lademann et al., 2003), induced a 20% decrease in apoptotic activity and specifically inhibited regeneration (Tseng et al., 2007), suggesting—counter-intuitively—that cell death is required for the process of rebuilding a tail.
In the Xenopus tail, the requirement for apoptosis in regeneration occurs in the first 24 hrs after amputation, since treatment with inhibitors starting at 24 hpa or later did not affect normal regenerative ability. Furthermore, apoptosis-inhibited tail regeneration buds showed decreased proliferation and disrupted neuronal patterning. Unexpectedly, refractory-stage tail amputation resulted in an increase in the apoptotic population in the bud as compared with the regenerative stages, suggesting that a tight control of a specific level of apoptosis is required for proper regeneration. The epistasis between apoptosis and membrane voltage control mechanisms in the process of regeneration is currently under investigation in our lab, because of the known links between transmembrane potential and programmed cell death (Gilbert et al., 1996; Wang, 2004).
Tail Outgrowth and Patterning
BMP Signaling
After the establishment of the regeneration bud by 24 hpa, the initiation and maintenance of regenerative growth and patterning are regulated by several pathways. A main component is Bone Morphogenetic Protein (BMP) signaling. BMPs are growth factors that belong to the TGF-β superfamily of proteins and have important roles in embryogenesis (von Bubnoff and Cho, 2001). In the amputated tail, BMP-2 is expressed at the edge of the regenerating fin by 24 hpa. Its antagonist, Noggin, is expressed at the tip of the spinal cord (Beck et al., 2006). Three hours prior to tail amputation, inhibition of BMP by Noggin expression did not affect regenerative ability (Beck et al., 2006). However, Noggin induction at either 24 or 48 hpa reduced regeneration significantly, demonstrating a requirement for BMP signaling in this process. Loss of BMP activity caused a reduction of cells labeled by BrdU in the spinal cord and the notochord tissues of the regeneration bud (Beck et al., 2006). TUNEL staining showed that there was no increase in apoptosis at the amputation site. Together, these results indicate that the inhibition of BMP signaling leads to reduced proliferation in the tail. Importantly, promoting BMP signaling by expression of a constitutively active BMP receptor, Alk3, can induce tail regeneration during the refractory stage (Beck et al., 2003).
It has been proposed that since BMP loss reduces proliferation in notochord and spinal cord cells, but activation of BMP signaling rescues regenerative growth of all tissues, the signal for regeneration is non-cell-autonomous (Beck et al., 2003). Furthermore, perhaps proliferation of neural and notochord cells is the main component that drives regeneration. However, it is important to note that the experiments in which activated BMP expression rescued regeneration during the refractory period utilized a constitutively active Alk3 transgene that was under the control of the Hsp70 promoter, which drives expression ubiquitously in the embryo. Thus, it is possible that BMP may also drive growth outside of its native expression regions. This hypothesis could be more rigorously tested by specifically driving expression of the Alk3 transgene in the notochord and neural ampulla of tails amputated during the non-regenerative state.
Msx1
Msx1 is a transcription factor that has been implicated in the regeneration of several tissues (Odelberg et al., 2000; Kumar et al., 2004), and BMP activity can activate Msx1 expression (Suzuki et al., 1997). In amputated tails, the inhibition of BMP by Noggin abolishes Msx1 expression in the neural ampulla of the regeneration bud, suggesting that, in this event, Msx1 also acts downstream of BMP (Beck et al., 2006). To determine whether Msx1 can recapitulate BMP function, investigators induced the expression of a hyper-activated Msx1 transgene in tails cut during the refractory period (Beck et al., 2003). Activated Msx1 was able to promote normal regeneration, indicating that its activity can fully substitute for BMP in tail outgrowth, and that it is an important regulator of Xenopus regeneration.
Notch Pathway
The Notch pathway is involved in multiple processes, including proliferation and cell fate specification (Lai, 2004; Bolos et al., 2007; Fiuza and Arias, 2007). Genes involved in the Notch pathway are expressed in the regeneration bud by 24 hpa. Treatment of amputated tails with MG132, an inhibitor that blocks Notch activity, prevents regeneration without affecting overall development (Beck et al., 2003). Furthermore, the presence of MG132 blocks the ability of the activated Alk3 transgene, when expressed, to rescue tail regeneration during the refractory period, showing that Notch likely functions downstream of BMP signaling. By itself, expression of a transgene-containing activated Notch induces incomplete tails during the refractory period, with only notochord and spinal cord present, but missing muscle and fin tissue. Interestingly, Msx1 expression was also not induced. These observations suggest that Notch signaling has a more specific role in driving tail outgrowth downstream of BMP and, by itself, is not sufficient to recapitulate complete regeneration.
Additional Pathways
Recently, 2 additional pathways regulated by BMP signaling have been identified that also are required for regenerative tail outgrowth (Lin and Slack, 2008). Inhibition of FGF signaling either with a chemical inhibitor, SU5402, or by expression of a dominant-negative FGF receptor transgene abolished tail regeneration. Similarly, expression of a transgene, Dkk1, which inhibits Wnt/β-catenin activity, also prevented tail regeneration. Detailed epistasis analyses with chemical inhibitors and induction of transgene expression showed that Wnt activity can regulate FGF expression (Lin and Slack, 2008). However, both of these components appear to act downstream of BMP activity. BMP4 expression is unaltered in SU5402-treated or Dkk1-expressing larvae. However, ectopic expression of the BMP antagonist, Noggin, in amputated tails abolished expression of several members of the FGF and Wnt families.
Last, there is a second role for TGF-β signaling in promoting regenerative outgrowth. Although TGF-β is well-known for its ability to inhibit proliferation, it can drive proliferation in several cell types and conditions (Ruscetti et al., 2005; Seoane, 2006), and activin (a TGF-β ligand) has recently been shown to promote zebrafish fin regeneration (Jazwinska et al., 2007). At 48 hpa in Xenopus tails, p-Smad2 was expressed throughout the regeneration bud and blastema, but not in apical epidermis. Treatment of amputated tails at 2 dpa with SB-431542 stopped regenerative growth and resulted in tail stumps (Ho and Whitman, 2008). Quantification of the number of phospho-Histone H3-positive cells, a marker of mitosis, showed a significantly reduced cell proliferation population in the regeneration bud at 3 dpa. However, proliferation rates in the non-regenerating tissues surrounding the regeneration bud remained constant, suggesting that this is a regeneration-specific phenomenon. Inhibition of TGF-β activity by SB-431542 appears to inhibit cell division directly, since treatment with cell-cycle-specific inhibitors resulted in a similar phenotype. Thus, TGF-β signaling can regulate cell proliferation during regenerative outgrowth.
TAIL REGENERATION: RECAPITULATION OF DEVELOPMENT OR NOVEL EVENT?
Do the genes that function during tail regeneration recapitulate developmental events, or should regeneration be considered a distinct process? Several of the pathways discussed in the previous section, including BMP and Notch, are important in tail development, and many genes that are involved during tail bud growth are also similarly expressed in the tail regenerate (Beck et al., 2003; Christen et al., 2003; Tazaki et al., 2005), suggesting that regeneration utilizes at least some of the developmental pathways to drive appendage repair. However, a few genes showed differential expression patterns in both processes (Sugiura et al., 2004), and thus may have different functions. Furthermore, some processes, such as apoptosis and V-ATPase activity, do not appear to play a role in normal tail development. However, it is logical that regeneration should integrate some of the existing tail developmental pathways, especially those that are involved in outgrowth and patterning. The differences are likely to occur during the early phase after tail injury (first 24 hpa), when the animal makes the decision to undergo regeneration or not. Such events include, but are not limited to, wound healing and the establishment of the regeneration bud. Interestingly, the pathways that participate in these events have characteristics that also relate them to other developmental processes, and we next consider the components of tail regeneration more broadly, as they relate to limb development and other instances of physiological healing.
Epithelial Wound Healing: Drosophila, Xenopus, and Mammals
Wound healing after larval tail amputation requires the movement of epithelial cells to cover the injury. How widely conserved are the mechanics of this process? One normal developmental process that is highly similar to wound healing is Drosophila dorsal closure during embryogenesis, which has served as an excellent model for the study of tissue repair in animals (for review, see Martin and Parkhurst, 2004; Redd et al., 2004). Near the end of gastrulation, after germband movements, an elliptical hole remains on the dorsal surface of the embryo. Dorsal closure occurs when the epithelial edges are brought together to close the hole; this process involves an actin filament cable at the leading edge, transient filopodia, and permanent adherens junctions.
There are several strong similarities between this morphogenetic event and Xenopus wound healing after tail injury. Both processes involve epithelial cell movement, do not require cell proliferation, and do not generate a scar. In addition, wound healing in Xenopus larvae also appears to begin at the fin edges of the amputation site (Ho and Whitman, 2008), similar to the ‘zipper effect’ of dorsal closure. While there is no inflammation response during fly embryo dorsal closure, it is not yet known whether larval tail wound healing induces an inflammation response after injury, although several inflammation response genes have been shown to be up-regulated during tail regeneration (Tazaki et al., 2005).
A gene family that has been widely implicated in wound healing is the matrix metalloproteinases (MMPs), which are involved in tissue remodeling through the degradation of extracellular matrix. Members of the MMP family have been implicated in epithelial wound healing of the cornea, and the skin (Azar et al., 1996; Fini et al., 1998; Ye and Azar, 1998), and in the regeneration of tissues including mouse muscle (Kherif et al., 1999), zebrafish heart (Lien et al., 2006), and urodele appendages (Yang and Bryant, 1994; Miyazaki et al., 1996; Yang et al., 1999). In particular, Xenopus MMP-9 is expressed after epithelial wound healing. During the first 2 days after amputation, MMP-9 is specifically up-regulated in both the ectodermal and mesodermal cells in the limb stump (Carinato et al., 2000), suggesting that genes that participate in wound healing are also likely to be important for the proper formation of Xenopus wound epidermis and subsequent repair. Furthermore, cutaneous immunity genes that are implicated in mammalian skin repair and regeneration are also expressed during wound healing of amputated Xenopus limbs (Mescher et al., 2007). However, the roles of these immunity and wound-healing genes in tail regeneration have not yet been characterized. Although analysis of the expression data indicates their potential importance in Xenopus regeneration, it remains to be determined whether these mechanisms are functionally conserved.
Epigenetic Control in Appendage Development
The ability of ion flows to regulate cellular events is an important and widely conserved mechanism of epigenetic control over wound healing and subsequent regeneration (McCaig et al., 2005; Zhao et al., 2006; Levin, 2007). Bioelectric events have been shown to have roles in wide-ranging biological processes, including limb development (Borgens et al., 1983; Borgens, 1984), embryonic patterning (Metcalf et al., 1994; Shi and Borgens, 1995), and establishment of left-right asymmetry (Levin and Mercola, 1998, 1999; Levin et al., 2002; Adams et al., 2006; Hibino et al., 2006; Shimeld and Levin, 2006). During development, the presence of a current leakage due to breakdown in tight junctions between epithelial cells results in an outward ion flow that predicts the location of limb bud emergence in chick, Xenopus laevis, and mouse embryos by several days (Borgens et al., 1983; Robinson, 1983; Altizer et al., 2001). Subsequently, the limb bud is populated by a combination of neural crest and mesodermal cells that accumulate through migration (Balinsky, 1981). It has been hypothesized that the leaky ion flow at the site of presumptive limb bud directs the migration of mesenchymal cells to the site (Borgens, 1984) by means of galvanotaxis (Nuccitelli and Erickson, 1983; Nuccitelli et al., 1993). Furthermore, modification of the presumptive limb ion flow by the imposition of a reverse current interfered with proper limb outgrowth (Altizer et al., 2001), suggesting that this is a causal effect.
The role for ion flows during limb development may be helpful in understanding its role in Xenopus tail regeneration. Amputation of the tail generates a surface that strongly expels H+ ions, resulting in a net outflow that correlates to the one seen prior to limb bud emergence in several species. Moreover, evidence for ion flux in guiding cell migration is also present during tail regeneration: The activity of V-ATPase H+ pump in the regeneration bud provides a morphological cue for cell migration, since its inhibition results in the mis-patterning of axons at the amputation site (Adams et al., 2007). Together, the evidence indicates that the mechanism underlying appendage regeneration in Xenopus may be highly similar to that of limb development.
A recent study has shown that another form of epigenetic control is also important for regeneration in the Xenopus limb. Changes in DNA methylation of promoter and enhancer regions can be used by cells to regulate gene expression at the transcriptional level without altering the coding sequence (Bird, 2002). DNA methylation regulates a variety of biological processes, including genomic imprinting and X-chromosome inactivation. Recent work has also shown that control of Sonic Hedgehog (Shh) expression in Xenopus regeneration is regulated by DNA methylation (Yakushiji et al., 2007).
Shh plays an important in role patterning in many systems. After Xenopus tadpole limb amputation, Shh is expressed at 4 dpa in the posterior-distal region in the regenerate. In contrast, after amputation in the non-regenerative froglet limb, Shh expression was not detected. Examination of the status of the major limb-specific Shh enhancer, known as MFCS1, after amputation, showed that it was hypomethylated in the regenerating tadpole limb, but hypermethylated in the non-regenerating froglet limb. In addition, urodeles able to regenerate limbs during adulthood show constitutive hypomethylation at the MFCS1 site. Thus, there is a strong correlation between differences in DNA methylation states and Shh expression during limb regeneration. Moreover, DNA methylation may represent an additional mechanism that can contribute to the determination of regenerative capability. A highly methylated state may prevent the expression of genes necessary for regeneration. To support this hypothesis, potential targets of methylation control during tail regeneration would have to be identified, and their modification states determined. Nevertheless, it is apparent that epigenetic pathways, either through promoter methylation to mediate gene expression, or through modulation of ion flows by pumps, are key regulators of regeneration in Xenopus.
The Role of Apoptosis in Development
It is well-known that cell death is required for the sculpturing of avian and mammalian digits during limb development (Saunders and Gasseling, 1962). In contrast, amphibian digit formation depends on differential proliferative growth, but apoptosis is largely absent (Vlaskalin et al., 2004). Thus, the early requirement for endogenous apoptosis in tail regeneration is surprising (Tseng et al., 2007). After all, regenerative growth requires extensive proliferation to replace the lost tissue. We have hypothesized that the endogenous group of apoptotic cells in the tail regeneration bud carries an inhibitory signal that needs to be abolished before regenerative growth can proceed (Tseng et al., 2007). However, the mechanism that underlies this process remains to be identified. Interestingly, a detailed examination of the role of apoptosis during development shows that it exists endogenously during Xenopus limb development (Suzuki et al., 2005).
Early-stage Xenopus larval limbs are also able to regenerate after amputation. Four days after limb amputation, the normal blastema contains apoptotic cells at the distal periphery. In contrast, in denervated blastemas that subsequently fail to regenerate properly, there is a great increase in the number of apoptotic cells located throughout the region (Mescher et al., 2000; Suzuki et al., 2005). This is analogous to the situation in the tail, where, during the refractory period, there is a significantly expanded region of apoptosis in the regeneration bud. Notably in all cases, apoptosis is also seen in the wound epidermis (Mescher et al., 2000; Suzuki et al., 2005; Tseng et al., 2007). The increased apoptosis during non-regenerative states may hinder the ability of the animal to replace the lost tissue. However, it is unclear why a specific amount of apoptosis is required to drive regenerative growth.
SUMMARY AND CONCLUSION
The current focus of studies on Xenopus tail regeneration has generated an increasingly detailed understanding of how this process works in a vertebrate model, both morphologically and molecularly (Fig. 2). Immediately after amputation, wound healing is observed as epidermal cells undergo migration to cover the injury. This process is rapid, occurring within 6 hpa, and requires TGF-β activity, which is active by 2 hpa. Inhibition of TGF-β signaling prevents wound closure. By 6 hpa, the V-ATPase H+ proton pump is expressed in the presumptive regeneration bud, which is depolarized. Within 24 hpa, a regeneration bud is established at the amputation site, containing mainly undifferentiated mesenchymal cells and precursor cells of the spinal cord and notochord. The first 24 hpa appears to be the critical time in which regenerative ability is determined. During this period, two events are known to be required. First, V-ATPase activity acts to re-polarize the bud. Second, programmed cell death removes a specific cell population.
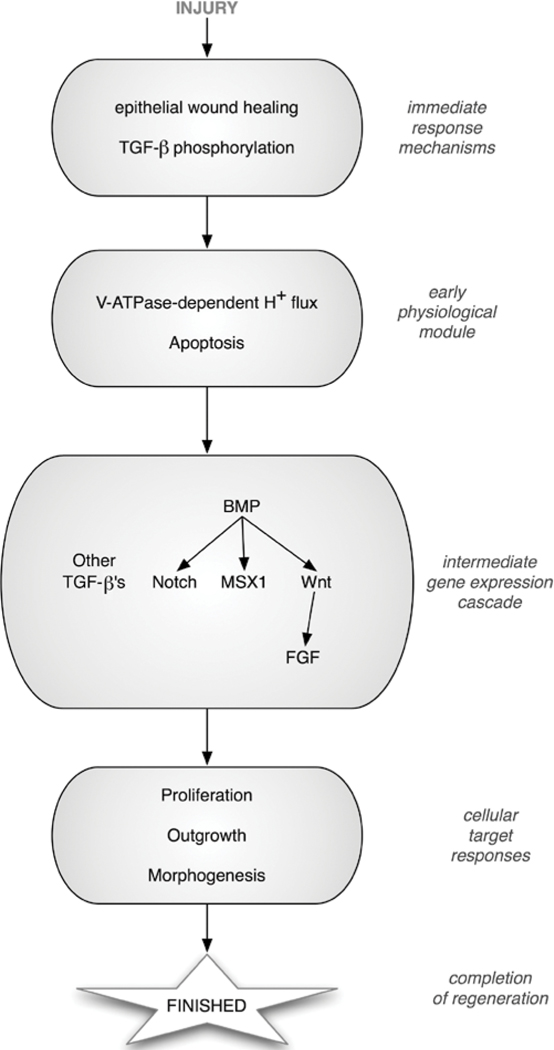
Figure 2.
A schematic of functional modules in tail regeneration. Injury triggers an immediate early response via still-unknown mechanisms. The early response includes the generation of a wound epithelium and rapid protein-level events. Next follows a set of physiological responses, including up-regulation of specific ion transporters (and the resulting bioelectric events) and programmed cell death of a specific cell group. Downstream lies a cascade of gene expression changes, resulting in the secretion of factors that modulate subsequent cell behaviors, such as mitotic rates and migration. The process completes when the system determines (via an unknown mechanism) that it has caught up to the correct tail size.
By 24 hpa, the tail regeneration bud initiates appendage outgrowth by induction of cell proliferation and subsequent patterning. The main component is the BMP signaling pathway, which is required for extension, although the early function of the V-ATPase is required for proper axonal outgrowth in subsequent days. TGF-β activity regulates downstream targets, including the Notch pathway, the transcriptional repressor, Msx1, and the FGF and Wnt/β-catenin pathways, ultimately regulating cell proliferation rates as required for regeneration.
What Are the Key Determining Factors for Initiating Tail Regeneration?
The definitive event initiating regeneration (mechanism of recognition of damage) remains unknown. Under non-regenerative conditions, tail amputation results in wound healing that contains a thickened epithelium as compared with regenerative healing. Is the thickened wound epidermis a consequence of the failure to regenerate, or is it actually a mechanism to prevent regeneration? Interestingly, the observation that the refractory period exists only for a short duration indicates that there is a temporal-specific control present in the tadpole. It would be an important insight to uncover the mechanisms involved in determining regenerative ability, to be able to induce regeneration under non-permissive conditions.
Currently, the role of TGF-β in wound healing is the earliest molecular event identified in tail regeneration. Continuous inhibition of TGF-β activity after tail injury prevents the formation of a wound epidermis, but inhibition from 0–8 hpa results in the formation of a thickened epithelium without regeneration (Ho and Whitman, 2008). However, the expression and function of TGF-β in non-regenerative conditions would need to be characterized to determine whether it may act as a key initiator of regeneration.
How Do the Molecular Pathways Interact to Drive Regeneration?
Multiple pathways have now been identified that participate in Xenopus tail regeneration. Moreover, epistatic relationships have been established for the well-known signaling pathways that are important in driving regeneration tail outgrowth, including BMP, Notch, FGF, and Wnt (Beck et al., 2003, 2006; Lin and Slack, 2008). However, it is not understood whether the early processes (TGF-β, ion flux, and apoptosis) interact with each other, or how they relate to the downstream pathways. It is likely that additional unidentified molecular pathways are involved in this process. One hypothesis is that very early TGF-β signaling may induce V-ATPase expression. But how does apoptosis of cells in the regeneration bud fit into this model? And how are ion flux signals transduced to activate downstream pathways such as BMP or Notch?
Recently, the protein Ci-VSP was identified (Murata et al., 2005). Notably, it contains a voltage-sensing transmembrane domain and a cytoplasmic domain similar to phosphate and tensin homologue (PTEN). PTEN genes are well-known regulators of growth and cell behavior in cancer and stem cells (Dong et al., 2003; Li et al., 2003), and are specifically implicated in electrically mediated corneal wound healing (Zhao et al., 2006). The discovery of Ci-VSP raises the possibility that it may transduce the V-ATPase-driven voltage changes to downstream pathways in regeneration. Characterization of such transducer proteins that can participate in the regenerative processes would represent a major step in the understanding of how changes in ion flux are interpreted by the regeneration bud cells. Thus, VSP is an also interesting candidate to act as an ion flux sensor in other systems.
CONCLUSION
The Xenopus tadpole is a well-characterized and genetically tractable vertebrate model with high regenerative ability. The easy accessibility of its tail for manipulation and visualization, coupled with the availability of established molecular techniques, makes it an ideal system for the study of regeneration. Notably, tissues undergoing regeneration in the tail are formed from their specific cell types, and no transdifferentiation is observed, suggesting that tail regeneration correlates well with mammalian tissue renewal.
Tail regeneration occurs at a significantly faster rate than normal growth, since it must replace a lost structure even as the larvae continues to develop, grow, and mature. Crucially, this process also knows when to stop; it is unknown how the regenerating system knows when it is done, and the required target morphology has been rebuilt. This aspect of somatic cell regulation is important, because utilizing these pathways in regenerative medicine requires that we know how to induce growth that is patterned and does not result in runaway tumor induction, as has been observed in stem cell transplantation approaches (Wakitani et al., 2003; Arnhold et al., 2004).
Processes such as wound healing and establishment of the regeneration bud may integrate components that participate in related endogenous events, such as dorsal closure and limb bud formation, to generate a response to tissue injury. Furthermore, a recurring theme in studies of Xenopus and other models is that the signaling pathways discussed here are used reiteratively in widely varying regenerating systems (Stoick-Cooper et al., 2007). For example, in addition to the Xenopus tail, BMPs have roles in the regeneration of zebrafish fin, mouse digit tips, and chick retina (Quint et al., 2002; Smith et al., 2006; Haynes et al., 2007), among others. Interestingly, modulation of BMP activity has been utilized in potential regenerative therapies for craniofacial defects (Suzuki et al., 2002; Jin et al., 2003). Thus, with the rapidly accumulating knowledge relative to the molecular pathways that participate in Xenopus tail regeneration, it is likely that a detailed understanding of vertebrate appendage regeneration can be achieved and leveraged into the development of therapeutics for augmenting human regenerative repair.
ACKNOWLEDGMENTS
We thank the members of the FCRDB for many useful discussions. The authors gratefully acknowledge support from the NSF (IBN#0347295), NHTSA (DTNH22-06-G-00001), DARPA (W911NF-07-1-0572), and NIH (HD055850). This review was prepared in a Forsyth Institute facility renovated with support from Research Facilities Improvement Grant Number CO6RR11244 from the National Center for Research Resources, NIH.
REFERENCES
- Adams DS, Levin M (2006). Inverse drug screens: a rapid and inexpensive method for implicating molecular targets. Genesis 44:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, et al. (2006). Early, H+−V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development 133:1657–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Masi A, Levin M (2007). H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development 134:1323–1335. [DOI] [PubMed] [Google Scholar]
- Altizer AM, Moriarty LJ, Bell SM, Schreiner CM, Scott WJ, Borgens RB (2001). Endogenous electric current is associated with normal development of the vertebrate limb. Dev Dyn 221:391–401. [DOI] [PubMed] [Google Scholar]
- Armstrong MT, Turlo K, Elges CJ, Dayton SM, Lee J, Armstrong PB (2006). A novel form of epithelial wound healing of the embryonic epidermis. Exp Cell Res 312:2415–2423. [DOI] [PubMed] [Google Scholar]
- Arnhold S, Klein H, Semkova I, Addicks K, Schraermeyer U (2004). Neurally selected embryonic stem cells induce tumor formation after long-term survival following engraftment into the subretinal space. Invest Ophthalmol Vis Sci 45:4251–4255. [DOI] [PubMed] [Google Scholar]
- Atkinson DL, Stevenson TJ, Park EJ, Riedy MD, Milash B, Odelberg SJ (2006). Cellular electroporation induces dedifferentiation in intact newt limbs. Dev Biol 299:257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azar DT, Hahn TW, Jain S, Yeh YC, Stetler-Stevensen WG (1996). Matrix metalloproteinases are expressed during wound healing after excimer laser keratectomy. Cornea 15:18–24. [PubMed] [Google Scholar]
- Balinsky BI (1981). An introduction to embryology. Philadelphia: Saunders College Publishing. [Google Scholar]
- Beck CW, Christen B, Slack JM (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell 5:429–439. [DOI] [PubMed] [Google Scholar]
- Beck CW, Christen B, Barker D, Slack JM (2006). Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech Dev 123:674–688. [DOI] [PubMed] [Google Scholar]
- Bird A (2002). DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21. [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sánchez Alvarado A (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL (2007). Notch signaling in development and cancer. Endocr Rev 28:339–363. [DOI] [PubMed] [Google Scholar]
- Borgens RB (1983). The role of ionic current in the regeneration and development of the amphibian limb. Prog Clin Biol Res 110:597–608. [PubMed] [Google Scholar]
- Borgens RB (1984). Are limb development and limb regeneration both initiated by an integumentary wounding? A hypothesis. Differentiation 28:87–93. [DOI] [PubMed] [Google Scholar]
- Borgens RB, Rouleau MF, DeLanney LE (1983). A steady efflux of ionic current predicts hind limb development in the axolotl. J Exp Zool 228:491–503. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A (2002). Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat Rev Mol Cell Biol 3:566–574. [DOI] [PubMed] [Google Scholar]
- Campbell LJ, Crews CM (2007). Wound epidermis formation and function in urodele amphibian limb regeneration. Cell Mol Life Sci 65:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carinato ME, Walter BE, Henry JJ (2000). Xenopus laevis gelatinase B (Xmmp-9): development, regeneration, and wound healing. Dev Dyn 217:377–387. [DOI] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA (2004). Cellular and molecular regulation of muscle regeneration. Physiol Rev 84:209–238. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin G, Slack JM (2006). Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development 133:2303–2313. [DOI] [PubMed] [Google Scholar]
- Christen B, Beck CW, Lombardo A, Slack JM (2003). Regeneration-specific expression pattern of three posterior Hox genes. Dev Dyn 226:349–355. [DOI] [PubMed] [Google Scholar]
- Cone CD Jr, Cone CM (1976). Induction of mitosis in mature neurons in central nervous system by sustained depolarization. Science 192:155–158. [DOI] [PubMed] [Google Scholar]
- Das B, Brown DD (2004). Controlling transgene expression to study Xenopus laevis metamorphosis. Proc Natl Acad Sci USA 101:4839–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J (2005). Chemical genetics: drug screens in Zebrafish. Biosci Rep 25:289–297. [DOI] [PubMed] [Google Scholar]
- Deuchar EM (1975). Regeneration of the tail bud in Xenopus embryos. J Exp Zool 192:381–390. [DOI] [PubMed] [Google Scholar]
- Dinsmore CE (1991). A history of regeneration research: milestones in the evolution of a science. Cambridge: Cambridge University Press. [Google Scholar]
- Dong XY, Su YR, Qian XP, Yang XA, Pang XW, Wu HY, et al. (2003). Identification of two novel CT antigens and their capacity to elicit antibody response in hepatocellular carcinoma patients. Br J Cancer 89:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas BS (1972). Conservative management of guillotine amputation of the finger in children. Aust Paediatr J 8:86–89. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM (2002). Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science 298:1993–1996. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM (2003). Electroporation as a tool to study in vivo spinal cord regeneration. Dev Dyn 226:418–425. [DOI] [PubMed] [Google Scholar]
- Eide FF, Eisenberg SR, Sanders TA (2000). Electroporation-mediated gene transfer in free-swimming embryonic Xenopus laevis. FEBS Lett 486:29–32. [DOI] [PubMed] [Google Scholar]
- Elmore S (2007). Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fini ME, Cook JR, Mohan R (1998). Proteolytic mechanisms in corneal ulceration and repair. Arch Dermatol Res 290:12–23. [DOI] [PubMed] [Google Scholar]
- Fiuza UM, Arias AM (2007). Cell and molecular biology of Notch. J Endocrinol 194:459–474. [DOI] [PubMed] [Google Scholar]
- Gargioli C, Slack JM (2004). Cell lineage tracing during Xenopus tail regeneration. Development 131:2669–2679. [DOI] [PubMed] [Google Scholar]
- Gilbert MS, Saad AH, Rupnow BA, Knox SJ (1996). Association of BCL-2 with membrane hyperpolarization and radioresistance. J Cell Physiol 168:114–122. [DOI] [PubMed] [Google Scholar]
- Gordon KJ, Blobe GC (2008). Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta 1782:197–228. [DOI] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C (2005). A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435:954–958. [DOI] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, Cline HT (2001). Single-cell electroporation for gene transfer in vivo. Neuron 29:583–591. [DOI] [PubMed] [Google Scholar]
- Haas K, Jensen K, Sin WC, Foa L, Cline HT (2002). Targeted electroporation in Xenopus tadpoles in vivo—from single cells to the entire brain. Differentiation 70:148–154. [DOI] [PubMed] [Google Scholar]
- Haynes T, Gutierrez C, Aycinena JC, Tsonis PA, Del Rio-Tsonis K (2007). BMP signaling mediates stem/progenitor cell-induced retina regeneration. Proc Natl Acad Sci USA 104:20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E, Leferovich J, Bedelbaeva K, Gourevitch D, Clark L (2004). The scarless heart and the MRL mouse. Philos Trans R Soc Lond B Biol Sci 359:785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Ishii Y, Levin M, Nishino A (2006). Ion flow regulates left-right asymmetry in sea urchin development. Dev Genes Evol 216:265–276. [DOI] [PubMed] [Google Scholar]
- Ho DM, Whitman M (2008). TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev Biol 315:203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth CM (1974). Trapped fingers and amputated finger tips in children. J Pediatr Surg 9:853–858. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Badakov R, Keating MT (2007). Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol 17:1390–1395. [DOI] [PubMed] [Google Scholar]
- Jin QM, Anusaksathien O, Webb SA, Rutherford RB, Giannobile WV (2003). Gene therapy of bone morphogenetic protein for periodontal tissue engineering. J Periodontol 74:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S (2005). Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev 19:1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherif S, Lafuma C, Dehaupas M, Lachkar S, Fournier JG, Verdiere-Sahuque M, et al. (1999). Expression of matrix metalloproteinases 2 and 9 in regenerating skeletal muscle: a study in experimentally injured and mdx muscles. Dev Biol 205:158–170. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E (1996). Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122:3173–3183. [DOI] [PubMed] [Google Scholar]
- Kumar A, Velloso CP, Imokawa Y, Brockes JP (2000). Plasticity of retrovirus-labelled myotubes in the newt limb regeneration blastema. Dev Biol 218:125–136. [DOI] [PubMed] [Google Scholar]
- Kumar A, Velloso CP, Imokawa Y, Brockes JP (2004). The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol 2(8):E218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318:772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lademann U, Cain K, Gyrd-Hansen M, Brown D, Peters D, Jaattela M (2003). Diarylurea compounds inhibit caspase activation by preventing the formation of the active 700-kilodalton apoptosome complex. Mol Cell Biol 23:7829–7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC (2004). Notch signaling: control of cell communication and cell fate. Development 131:965–973. [DOI] [PubMed] [Google Scholar]
- Levin M (2007). Large-scale biophysics: ion flows and regeneration. Trends Cell Biol 17:261–270. [DOI] [PubMed] [Google Scholar]
- Levin M, Mercola M (1998). Gap junctions are involved in the early generation of left right asymmetry. Dev Biol 203:90–105. [DOI] [PubMed] [Google Scholar]
- Levin M, Mercola M (1999). Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node. Development 126:4703–4714. [DOI] [PubMed] [Google Scholar]
- Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M (2002). Asymmetries in H+/K+−ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell 111:77–89. [DOI] [PubMed] [Google Scholar]
- Li L, Liu F, Ross AH (2003). PTEN regulation of neural development and CNS stem cells. J Cell Biochem 88:24–28. [DOI] [PubMed] [Google Scholar]
- Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT (2006). Gene expression analysis of zebrafish heart regeneration. PLoS Biol 4(8):e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Slack JM (2008). Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol 316:323–335. [DOI] [PubMed] [Google Scholar]
- Lin G, Chen Y, Slack JM (2007). Regeneration of neural crest derivatives in the Xenopus tadpole tail. BMC Dev Biol 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP (1993). Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci USA 90:7230–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Parkhurst SM (2004). Parallels between tissue repair and embryo morphogenesis. Development 131:3021–3034. [DOI] [PubMed] [Google Scholar]
- Massagué J (1998). TGF-beta signal transduction. Annu Rev Biochem 67:753–791. [DOI] [PubMed] [Google Scholar]
- Masuda CA, Montero-Lomeli M (2000). An NH2-terminal deleted plasma membrane H+−ATPase is a dominant negative mutant and is sequestered in endoplasmic reticulum derived structures. Biochem Cell Biol 78:51–58. [PubMed] [Google Scholar]
- McCaig CD, Rajnicek AM, Song B, Zhao M (2005). Controlling cell behavior electrically: current views and future potential. Physiol Rev 85:943–978. [DOI] [PubMed] [Google Scholar]
- Mescher AL, White GW, Brokaw JJ (2000). Apoptosis in regenerating and denervated, nonregenerating urodele forelimbs. Wound Repair Regen 8:110–116. [DOI] [PubMed] [Google Scholar]
- Mescher AL, Wolf WL, Moseman EA, Hartman B, Harrison C, Nguyen E, et al. (2007). Cells of cutaneous immunity in Xenopus: studies during larval development and limb regeneration. Dev Comp Immunol 31:383–393. [DOI] [PubMed] [Google Scholar]
- Metcalf MEM, Shi R, Borgens RB (1994). Endogenous ionic currents and voltages in amphibian embryos. J Exp Zool 268:307–322. [Google Scholar]
- Miyazaki K, Uchiyama K, Imokawa Y, Yoshizato K (1996). Cloning and characterization of cDNAs for matrix metalloproteinases of regenerating newt limbs. Proc Natl Acad Sci USA 93:6819–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochii M, Taniguchi Y, Shikata I (2007). Tail regeneration in the Xenopus tadpole. Dev Growth Differ 49:155–161. [DOI] [PubMed] [Google Scholar]
- Morgan T (1901). Regeneration. New York: Macmillan. [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y (2005). Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435:1239–1243. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, Erickson CA (1983). Embryonic cell motility can be guided by physiological electric fields. Exp Cell Res 147:195–201. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, Smart T, Ferguson J (1993). Protein-kinases are required for embryonic neural crest cell galvanotaxis. Cell Motil Cytoskeleton 24:54–66. [DOI] [PubMed] [Google Scholar]
- Odelberg SJ, Kollhoff A, Keating MT (2000). Dedifferentiation of mammalian myotubes induced by msx1. Cell 103:1099–1109. [DOI] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM (2006). Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev 123:103–113. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T (2006). I-SceI mega-nuclease-mediated transgenesis in Xenopus. Dev Dyn 235:247–252. [DOI] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A (2003). Tales of regeneration in zebrafish. Dev Dyn 226:202–210. [DOI] [PubMed] [Google Scholar]
- Price J, Faucheux C, Allen S (2005). Deer antlers as a model of mammalian regeneration. Curr Top Dev Biol 67:1–48. [DOI] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, et al. (2002). Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci USA 99:8713–8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja KS, Garcia MS, Isseroff RR (2007). Wound re-epithelialization: modulating keratinocyte migration in wound healing. Front Biosci 12:2849–2868. [DOI] [PubMed] [Google Scholar]
- Rajnicek AM, Stump RF, Robinson KR (1988). An endogenous sodium current may mediate wound healing in Xenopus neurulae. Dev Biol 128:290–299. [DOI] [PubMed] [Google Scholar]
- Redd MJ, Cooper L, Wood W, Stramer B, Martin P (2004). Wound healing and inflammation: embryos reveal the way to perfect repair. Philos Trans R Soc Lond B Biol Sci 359:777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A (2004). Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20:725–757. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435:948–953. [DOI] [PubMed] [Google Scholar]
- Rhett JM, Ghatnekar GS, Palatinus JA, O’Quinn M, Yost MJ, Gourdie RG (2008). Novel therapies for scar reduction and regenerative healing of skin wounds. Trends Biotechnol 26:173–180. [DOI] [PubMed] [Google Scholar]
- Robinson KR (1983). Endogenous electrical current leaves the limb and prelimb region of the Xenopus embryo. Dev Biol 97:203–211. [DOI] [PubMed] [Google Scholar]
- Ruscetti FW, Akel S, Bartelmez SH (2005). Autocrine transforming growth factor-beta regulation of hematopoiesis: many outcomes that depend on the context. Oncogene 24:5751–5763. [DOI] [PubMed] [Google Scholar]
- Ryffel GU, Werdien D, Turan G, Gerhards A, Goosses S, Senkel S (2003). Tagging muscle cell lineages in development and tail regeneration using Cre recombinase in transgenic Xenopus. Nucleic Acids Res 31:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa S, Takabatake T, Takabatake Y, Muramatsu T, Takeshima K (2002). Improved mRNA electroporation method for Xenopus neurula embryos. Genesis 33:81–85. [DOI] [PubMed] [Google Scholar]
- Saunders JW Jr, Gasseling MT (1962). Cellular death in morphogenesis of the avian wing. Dev Biol 5:147–178. [DOI] [PubMed] [Google Scholar]
- Seoane J (2006). Escaping from the TGFbeta anti-proliferative control. Carcinogenesis 27:2148–2156. [DOI] [PubMed] [Google Scholar]
- Shi R, Borgens RB (1995). Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev Dyn 202:101–114. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, Levin M (2006). Evidence for the regulation of left-right asymmetry in Ciona intestinalis by ion flux. Dev Dyn 235:1543–1553. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM (2000). Early development of Xenopus laevis. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA (2006). Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblasts differentiation and function. Dev Biol 299:438–454. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Remsen D (2006). Using Pharmabase to perform pharmacological analyses of cell function. Curr Protoc Bioinformatics Chapter 14:Unit 14.2. [DOI] [PubMed] [Google Scholar]
- Stillwell EF, Cone CM, Cone CD (1973). Stimulation of DNA synthesis in CNS neurones by sustained depolarisation. Nat New Biol 246:110–111. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Moon RT, Weidinger G (2007). Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev 21:1292–1315. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Taniguchi Y, Tazaki A, Ueno N, Watanabe K, Mochii M (2004). Differential gene expression between the embryonic tail bud and regenerating larval tail in Xenopus laevis. Dev Growth Differ 46:97–105. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ueno N, Hemmati-Brivanlou A (1997). Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development 124:3037–3044. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Satoh A, Ide H, Tamura K (2005). Nerve-dependent and -independent events in blastema formation during Xenopus froglet limb regeneration. Dev Biol 286:361–375. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Bessho K, Fujimura K, Okubo Y, Segami N, Iizuka T (2002). Regeneration of defects in the articular cartilage in rabbit temporomandibular joints by bone morphogenetic protein-2. Br J Oral Maxillofac Surg 40:201–206. [DOI] [PubMed] [Google Scholar]
- Tassava RA, Mescher AL (1975). The roles of injury, nerves, and the wound epidermis during the initiation of amphibian limb regeneration. Differentiation 4:23–24. [DOI] [PubMed] [Google Scholar]
- Tazaki A, Kitayama A, Terasaka C, Watanabe K, Ueno N, Mochii M (2005). Macroarray-based analysis of tail regeneration in Xenopus laevis larvae. Dev Dyn 233:1394–1404. [DOI] [PubMed] [Google Scholar]
- Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M (2007). Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol 301:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsonis PA, Washabaugh CH, Del Rio-Tsonis K (1995). Transdifferentiation as a basis for amphibian limb regeneration. Sem Cell Biol 6:127–135. [DOI] [PubMed] [Google Scholar]
- Tsuda T, Ohmori Y, Muramatsu H, Hosaka Y, Takiguchi K, Saitoh F, et al. (2001). Inhibitory effect of M50054, a novel inhibitor of apoptosis, on anti-Fas-antibody-induced hepatitis and chemotherapy-induced alopecia. Eur J Pharmacol 433:37–45. [DOI] [PubMed] [Google Scholar]
- Vlaskalin T, Wong CJ, Tsilfidis C (2004). Growth and apoptosis during larval forelimb development and adult forelimb regeneration in the newt (Notophthalmus viridescens). Dev Genes Evol 214:423–431. [DOI] [PubMed] [Google Scholar]
- von Bubnoff A, Cho KW (2001). Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol 239:1–14. [DOI] [PubMed] [Google Scholar]
- Wakitani S, Takaoka K, Hattori T, Miyazawa N, Iwanaga T, Takeda S, et al. (2003). Embryonic stem cells injected into the mouse knee joint form teratomas and subsequently destroy the joint. Rheumatology (Oxford) 42:162–165. [DOI] [PubMed] [Google Scholar]
- Wang Z (2004). Roles of K+ channels in regulating tumour cell proliferation and apoptosis. Pflugers Arch 448:274–286. [DOI] [PubMed] [Google Scholar]
- Weber R (1965). Inhibitory effect of actinomycin D on tail atrophy in Xenopus larvae at metamorphosis. Experientia 21:665–666. [DOI] [PubMed] [Google Scholar]
- Wieczorek H, Brown D, Grinstein S, Ehrenfeld J, Harvey WR (1999). Animal plasma membrane energization by proton-motive V-ATPases. Bioessays 21:637–648. [DOI] [PubMed] [Google Scholar]
- Yakushiji N, Suzuki M, Satoh A, Sagai T, Shiroishi T, Kobayashi H, et al. (2007). Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev Biol 312:171–182. [DOI] [PubMed] [Google Scholar]
- Yang EV, Bryant SV (1994). Developmental regulation of a matrix metalloproteinase during regeneration of axolotl appendages. Dev Biol 166:696–703. [DOI] [PubMed] [Google Scholar]
- Yang EV, Gardiner DM, Carlson MR, Nugas CA, Bryant SV (1999). Expression of Mmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev Dyn 216:2–9. [DOI] [PubMed] [Google Scholar]
- Ye HQ, Azar DT (1998). Expression of gelatinases A and B, and TIMPs 1 and 2 during corneal wound healing. Invest Ophthalmol Vis Sci 39:913–921. [PubMed] [Google Scholar]
- Yeh JR, Crews CM (2003). Chemical genetics: adding to the developmental biology toolbox. Dev Cell 5:11–19. [DOI] [PubMed] [Google Scholar]
- Yoshii Y, Matsuzaki T, Ishida H, Ihara S (2005). Wound healing ability of Xenopus laevis embryos. II. Morphological analysis of wound marginal epidermis. Dev Growth Differ 47:563–572. [DOI] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, et al. (2006). Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature 442:457–460. [DOI] [PubMed] [Google Scholar]