Abstract
There is an increasing appreciation for the heterogeneous nature of extracellular vesicles (EVs). In addition, two non-vesicular extracellular nanoparticles (NVEPs), exomeres and supermeres, have been discovered recently that are enriched in many cargo previously ascribed to EVs. The EV field has largely focused on EV isolation and characterization, while studies on NVEPs are limited. At this juncture, it is critically important to have robust and reliable methods to separate distinct populations of EVs and NVEPs to assign cargo to their correct carrier. Here, we provide a comprehensive step-by-step protocol for sequential isolation of large and small EVs, non-vesicular fractions, exomeres and supermeres from the same starting material. We describe in detail the use of differential ultracentrifugation, filtration, concentration, and high-resolution density-gradient fractionation to obtain purified fractions of distinct populations of Evs and NVEPs. This protocol allows assignment and enrichment of a biolmolecule of interest to its specific extracellular compartment. Compared to other isolation methods our protocol has unique advantages including high purity and reproducibility with minimal expertise required. The protocol can be applied to purification of EVs and NVEPs from cell culture media and human plasma and requires about 72 h to complete. Adoption of this protocol will help translational investigators identify potential circulating biomarkers and therapeutic targets for a host of human diseases, and allow basic scientists to better understand EV and NVEP biogenesis and function. Overall, this protocol will allow those interested in isolating extracellular vesicles and particles to advance scientific inquiry to answer outstanding questions in the field.
Keywords: Extracellular vesicles, exomeres, supermeres, exosomes, nanoparticles, plasma, albumin
Editor Summary
This protocol isolates large and small EVs, and non-vesicular nanoparticles known as exomeres and supermeres from cell-conditioned medium or human plasma, by differential ultracentrifugation, filtration, concentration, and high-resolution density-gradient fractionation.
Tweet
Isolation of large and small extracellular vesicles, exomeres and supermeres from cell-conditioned medium or human plasma using differential ultracentrifugation, filtration, concentration, and high-resolution density-gradient fractionation.
Introduction
Extracellular vesicles (EVs) and non-vesicular extracellular nanoparticles (NVEPs) play pivotal roles in both physiological and pathological conditions1–4. However, a major challenge in the field of EVs and NVEPs is their heterogeneity and the methods used to isolate and purify distinct populations3,5–12. Furthermore, the field has largely focused on studies related to EVs while studies on extracellular amembranous NVEPs, including the recently discovered exomeres5,10 and supermeres13–15, are limited. However, it is becoming increasingly clear that different classes of EVs may contain specific cargo, and, equally important, that NVEPs, such as exomeres and supermeres, contain many of the biomolecules, including proteins, RNA and DNA, that have previously been ascribed to exosomes3,9,15,16. EVs range in size from small (s)EVs (< 200 nm), including exosomes generated from endosomal compartments, to large (l)EVs (> 200 nm), including microvesicles and large oncosomes, which are shed from the plasma membrane3,6,7,9,11. NVEPs include a wide range of size of particles including lipoproteins, exomeres and supermeres5,15,17–19. Fluid-phase atomic force microscopy (AFM) has revealed that supermeres have distinct morphological features in comparison to both sEVs and exomeres15. To understand the roles of EVs and NVEPs in basic cell biology, as well as to realize their full clinical potential, robust and reliable methods are needed to separate distinct populations of these particles. Diverse EV isolation methods have been extensively described, including differential ultracentrifugation, size-exclusion chromatography (SEC), ultrafiltration, immunocapture and microfluidics9,20–24. Ultracentrifugation is the gold standard for isolation of EV and NVEPs from cells, tissues, and plasma9,10,15 and high resolution density-gradient purification has been shown to further separate non-vesicular material from purified vesicles9,15,18,25–28. Centrifugation-based isolation schemes have the advantage of being relatively high yield, with ultracentrifuges being widely available to basic and clinical research labs. In the protocol herein, we provide a detailed description of how to reproducibly obtain highly purified lEVs, sEVs, exomeres and supermeres from human cell lines and human plasma9,10,15. Additionally, we describe how to improve the purification of exomeres and supermeres from plasma by the addition of an albumin-depletion step. Methods for isolation of lipoproteins from plasma are not described here; however, methods for their isolation have been described and reviewed elsewhere17,19,27.
Development and overview of the protocol
Major challenges in the field involve the heterogeneity of EVs and NVEPs and the variety of methods used to isolate and purify distinct populations5–11. It is increasingly clear that traditionally isolated “exosome” or “EV” samples contain a heterogeneous mixture of EVs and non-vesicular (NV) components9. Progress in the EV field has been hampered by the lack of methods to separate the various secreted vesicles from non-vesicular components. Furthermore, the focus has been on EVs and lipoproteins6,9,17–19,26,27,29–34 while studies of the recently discovered exomeres and supermeres are very limited5,10,15,35. Isolation of EVs and NVEPs from plasma and other body fluids is challenging and most EV studies have been focused on isolation of EVs from cell-conditioned medium, while reports for EV and lipoprotein isolation from plasma and other body fluids are more limited but a number of protocols using combinations of different techniques are available18–20,26,27,30,33,34,36–39.
Our comprehensive protocol is based on methods introduced in three manuscripts by our group9,10,15. These protocols entail a series of sequential centrifugation, concentration, filtration, and high-resolution density-gradient centrifugation steps to sequentially isolate lEVs, sEVs, exomeres and supermeres from cell-conditioned media (Figs. 1–2), and human plasma (Fig. 3). Some of the major steps and modifications are listed below:
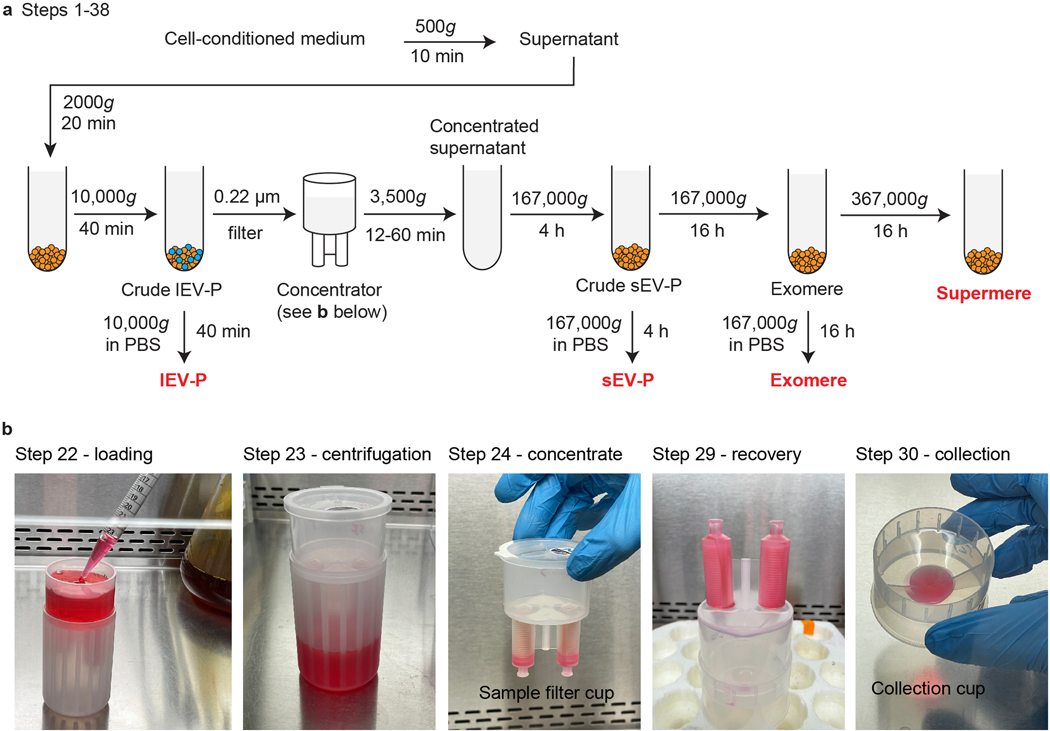
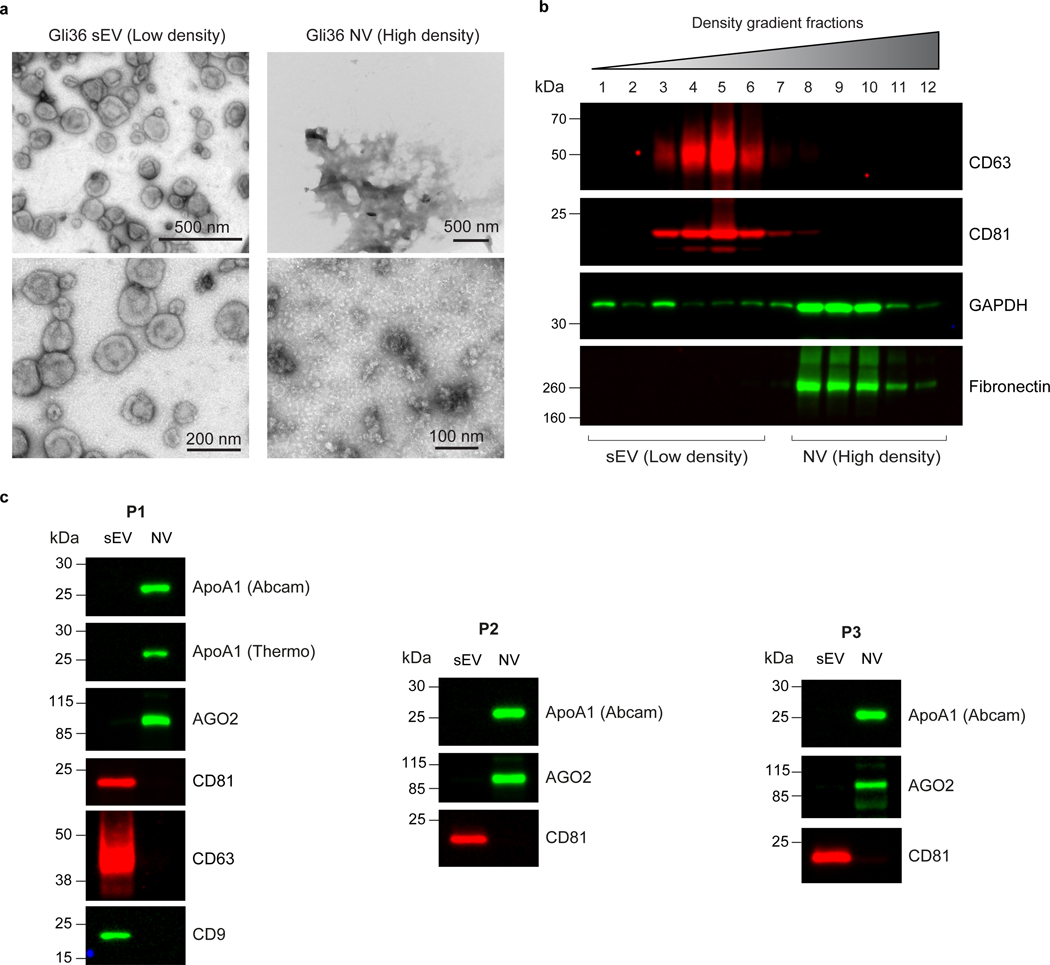
Fig. 1. Overview of steps for isolation of EVs and NVEPs from cell-conditioned medium.
a, Schematic for isolation of large EV pellets (lEV-Ps), small EV pellets (sEV-Ps), exomeres and supermeres. Serum-free conditioned medium is centrifuged (500g and 2000g) to remove dead cells, cellular debris and apoptotic bodies. The lEV-P is obtained after centrifugation of the supernatant at 10,000g centrifugation for 40 min. The leftover supernatant is first concentrated and then subjected to ultracentrifugation at 167,000g for 4 h to obtain the sEV-P (washed one time in PBS by ultracentrifugation at 167,000g for 4 h). The supernatant from the previous step is centrifuged at 167,000g for 16 h to isolate the exomeres (washed one time in PBS by ultracentrifugation at 167,000g for 16 h). The supernatant from the previous step is centrifuged at 367,000g for 16 h to isolate supermeres. b, Representative photographs of the most important steps during the concentrator procedure from a.
Fig. 2. Overview of high-resolution density gradient fractionation of EVs.

a, Schematic of the generation of lEVs or sEVs and non-vesicular (NV) fractions by high-resolution iodixanol density-gradient fractionation (12–36%). Crude pellets of lEV-Ps or sEV-Ps were resuspended in ice-cold PBS and mixed with ice-cold iodixanol (OptiPrep)/PBS for a final 36% iodixanol solution. The suspension was added to the bottom of a centrifugation tube and solutions of descending concentrations of iodixanol (30%, 24%, 18%, 12%) in PBS were carefully layered on top yielding the complete gradient. The bottom-loaded 12%–36% gradient was subjected to ultracentrifugation at 120,000g for 15 h. Twelve individual fractions of 1 ml were collected from the top of the gradient. The first six fractions are pooled in a tube, and the last five fractions are pooled in a second tube. The tubes are filled with PBS and mixed. Following ultracentrifugation at 120,000g for 4 h, the two pellets represent purified EVs and NVs, respectively. The images in panel a are modified from Zhang et al.15 and Jeppesen et al.9. b, Representative photographs of the most important steps during the high-resolution gradient fractionation procedure from a.
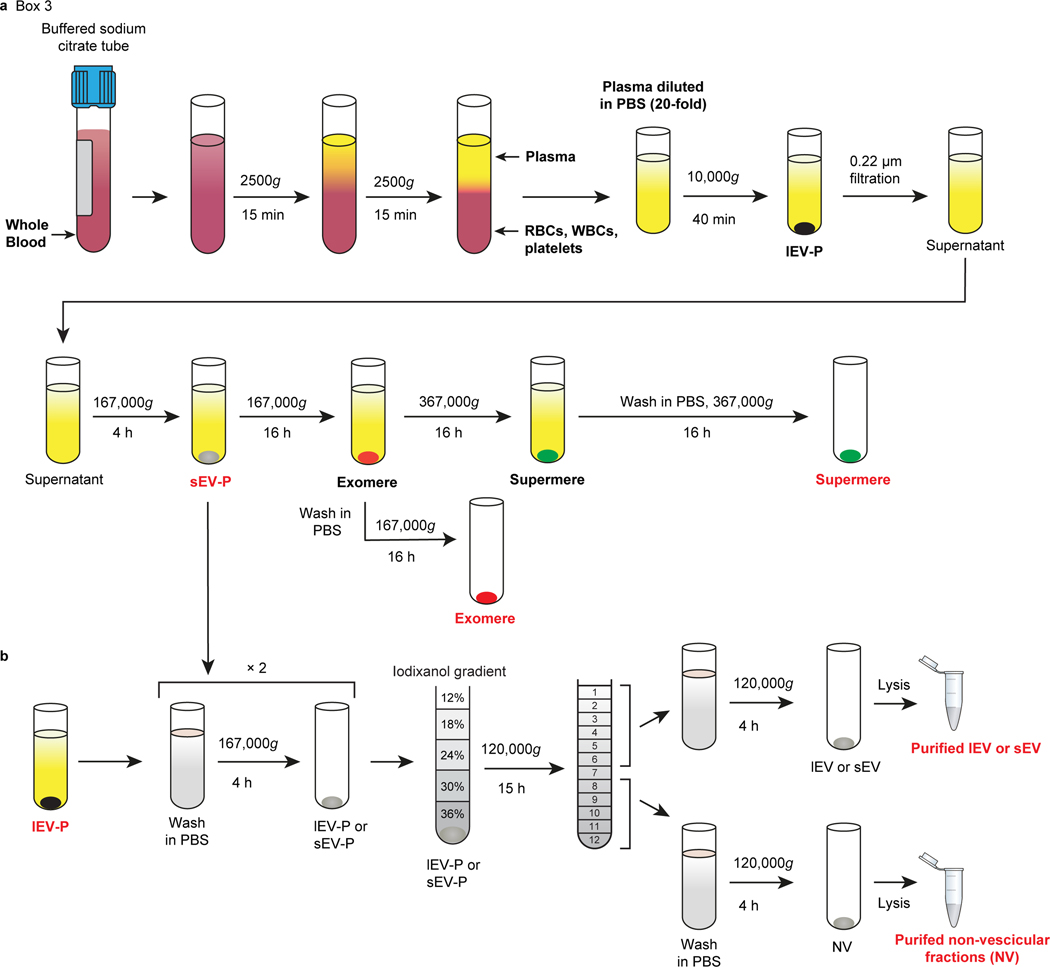
Fig. 3. Schematic of the isolation procedure for lEV, sEV, exomeres and supermeres from human plasma.
a, Schematic for isolation of different fractions from plasma. Plasma is generated by centrifugation of the blood at 2500g for 15 min twice at room temperature (RT). The resulting plasma samples are immediately diluted ∼1:10–20 in ice-cold PBS-H and centrifuged at 10,000g for 40 min to pellet lEV-Ps. The supernatant is filtered through a 0.22 μm pore PES filter and the resulting supernatants are subjected to sequential ultracentrifugation at 167,000g for 4 h, and 16 h, and then at 367,000g for 16 h to isolate sEV-Ps, exomeres and supermeres, respectively. b, Schematic of the generation of purified plasma lEVs, sEVs and NV fractions by high-resolution iodixanol density-gradient fractionation (12–36%). Crude pellets of lEV-Ps or sEV-Ps Crude pellets of lEV-Ps or sEV-Ps are processed as described in Fig. 2. The images are modified from Zhang et al.15 and Jeppesen et al.9. The research conducted as part of this protocol complies with all the relevant ethical regulations. The use of the human samples was approved by the Vanderbilt University Medical Center Institutional Review Board (IRB; IRB nos 161529 and 151721).
Removal of cells and cell debris from cell culture media and human plasma by a series of centrifugation steps. We also describe an albumin-depletion step (Fig. 4) that increases the purity of exomeres and supermeres from human plasma.
Isolation of lEVs by a combination of ultracentrifugation and high-resolution 12–36% iodixanol density-gradient fractionation (Figs. 1–3). Bottom loading of the high-resolution gradient is important to our method as it removes contaminating NV fractions from EV samples.
A filtration step to ensure that any remaining lEVs are removed so that only sEVs, exomeres and supermeres remain for subsequent steps.
A dual-purpose concentration step. A 100,000 molecular-weight cutoff concentrator is employed to concentrate the sample from a large to small volume and to facilitate the removal of free proteins. This step is omitted from most published EV protocols but is crucial to maximize yield of exomere and supermere fractions15 and to remove free proteins that would otherwise contaminate exomere and supermere samples.
Isolation of sEV samples by a combination of high-speed ultracentrifugation and high-resolution 12–36% iodixanol density-gradient fractionation9,15 (Figs. 1–3). The bottom-loaded high-resolution gradient has been demonstrated to remove contaminating non-vesicular (NV) fractions from sEV samples, including contaminating vault structures and nucleosomes9. Bottom loading is necessary as the centrifugation time (15 h) and speed (120,000g) is insufficient for non-vesicular components to reach their buoyant densities if samples are top-loaded.
Isolation of exomeres by 167,000g ultracentrifugation10,28. This simple step is an alternative to asymmetric flow field-flow fractionation (AF4)5,35, which is more costly and results in a lower yield (Figs. 1, 3).
Isolation of supermeres by 367,000g high-speed ultracentrifugation15. This method of purification was used in our recent Nature Cell Biology manuscript that described the discovery of supermeres and is the only published method for their isolation (Figs. 1, 3).
Albumin-depletion using a commercially available kit. When isolating sEVs, exomeres and supermeres from plasma, the biggest challenge for downstream analysis is albumin contamination. The albumin-depletion step greatly improves the purity of exomere and supermere samples, thereby aiding downstream analysis and characterization.
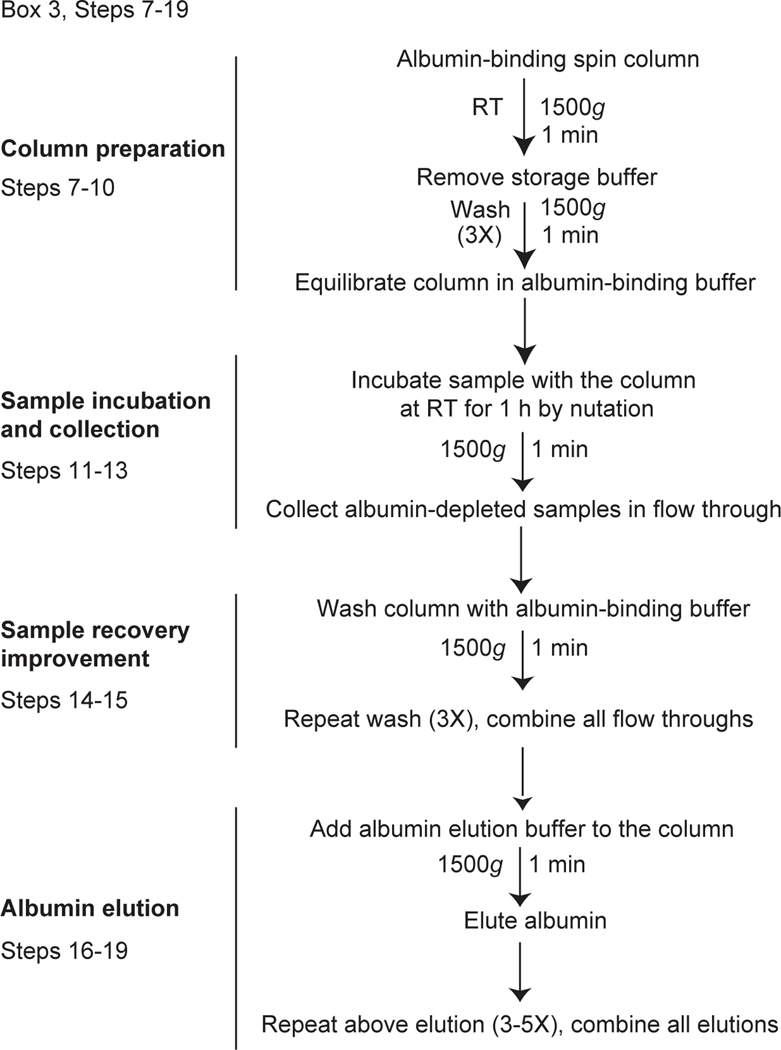
Fig. 4. Flowchart of albumin depletion steps from human plasma-derived sEV-Ps, exomeres and supermeres.
Albumin spin columns are equilibrated in albumin binding buffer by centrifugation at 1,500g for 1 min at RT. The samples are then applied to the albumin-binding columns and incubated for 1 h at RT with rotation. The columns were centrifuged, and the albumin-depleted samples are collected in the flow- through. Albumin is eluted by centrifugation using albumin elution buffer. This protocol was modified from the Albumin Depletion Kit procedure (Abcam). The research conducted as part of this protocol complies with all the relevant ethical regulations. The use of the human samples was approved by the Vanderbilt University Medical Center Institutional Review Board (IRB; IRB nos 161529 and 151721).
The protocol we previously described15 allows reasonable quantities of supermeres to be isolated from cell conditioned media and plasma. We detail the necessary steps, including crucial filtration and concentration steps, to maximize yield (Figs. 1, 3 and Steps 1–38).
A further development of the EV purification schema is the use of a high-resolution density gradient9,15 to increase the resolving power of separating both lEVs and sEVs from non-vesicular contaminants such as nucleosomes, vault structures and nanoparticles (Fig. 2 and Steps 39–51). For purification of EVs from human plasma, the gradient allows separation of EVs and high-density lipoprotein (HDL) particles9. We provide a detailed description of the steps in our 12–36% iodixanol gradient protocol with the rationale for both the concentrations we use and for bottom loading of the sample in contrast to the widely practiced top-loading method.
New to our protocol is the addition of an albumin-depletion step for purification of sEVs, exomeres and supermeres from plasma that contain large amounts of albumin (Fig. 4 and Box 3). The removal of contaminating albumin greatly improves downstream analysis, such as immunoblotting, thereby allowing detection of low-abundance target proteins as potential biomarkers and/or therapeutic targets.
Box 3. Isolation of lEV-Ps, sEV-Ps, exomeres and supermeres from human plasma, and albumin depletion Timing: (3 d, 4.5 h).
EVs, exomeres and supermeres can also be purified from human plasma using the procedure described in this protocol. Rather than collect cell-conditioned media, blood is drawn, plasma prepared and sample diluted as outlined in the procedure below. After isolation of EVs, exomeres and supermeres, albumin depletion is performed to increase purity of the samples.
For this study, blood was collected from individuals at VUMC. A group of 13 colorectal cancer patients ranged in age from 30 – 68 years old with an average age of 52.2 years old. Three normal healthy individuals ranged in age from 44 – 71 years old with an average age of 56 years old. All procedures on human peripheral blood specimens were approved and performed in accordance with the VUMC Institutional Review Board (IRB#161529 and #151721). All human participants provided informed consent (clinical trial registration number: NCT03263429). Participants did not receive compensation. Consent to publish this information was provided.
Procedure
Draw the blood into BD Vacutainer (9NC) blood collection tubes (BD Bioscience) containing buffered sodium citrate as an anticoagulant. The first tube collected should be discarded.
! CRITICAL One tube of 4.5 ml of blood normally yields about 2.2 ml of plasma, from which we can isolate enough EVs and NVEPs for downstream analysis including immunoblotting and flow cytometry, especially for exomeres and supermeres.
Transfer the blood to a new 15 ml centrifuge tube (CellStar) with a 5 ml pipette and centrifuge the blood at 2500g for 15 min at room temperature (RT) using a swinging-bucket centrifuge (Thermo Fisher Scientific, Sorvall LYNX 4000) to pellet red cells, white blood cells and platelets.
CRITICAL STEP Tubes are kept at RT and should be processed within 2 h after collection. If a blood sample shows any significant hemolysis after the initial pellet, it will be difficult to remove contaminating hemoglobin.
Transfer the plasma from Step 2 to a new 15 ml centrifuge tube (CellStar) with a 5 ml or 10 ml pipette and centrifuge the plasma at 2500g for 15 min at RT using a swinging bucket centrifuge (Thermo Fisher Scientific, Sorvall LYNX 4000) to isolate platelet-poor plasma.
CAUTION Be careful when pipetting off the plasma to avoid red and white blood cell pellet. Leave a small amount of supernatant on top of the pellet to avoid contamination. The type of pipette used depends on the initial blood volume collected.
PAUSE POINT The platelet-poor plasma can be frozen at −80 °C (long-term storage).
Pass the plasma collected in Step 3 through a 0.22 μm syringe filter (Millipore) connected to a syringe (BD) to remove large vesicles, cell debris and other contaminants.
Dilute the resulting plasma samples immediately at least 1:10 in ice cold PBS-H. Mix well by pipetting up and down.
PAUSE POINT The diluted plasma can be kept at either 4 °C (short-term storage for 1–2 weeks) or used immediately.
Isolate lEV-Ps, sEV-Ps, exomeres and supermeres from plasma as described in Steps 11–18, 31–38 for EV and NVEP isolation from cell conditioned media.
CRITICAL STEP: Optionally, further isolate plasma lEVs and sEVs on high-resolution density gradients as described in Steps 39–51 if highly purified EVs are required.
? TROUBLESHOOTING
Albumin depletion from sEV-Ps, exomeres and supermeres
CRITICAL Samples of sEV-Ps, exomeres and supermeres isolated from human plasma have high levels of albumin that can interfere with downstream analyses such as immunoblotting. Therefore, depletion of albumin and other highly abundant proteins from plasma-derived exomeres and supermeres will improve downstream analysis. We performed albumin-depletion from isolated exomeres and supermeres using an Albumin Depletion kit (Abcam) according to the manufacturer’s instructions with some modifications as follows.
Warm up the binding buffer, elution buffer and the column to RT before use.
Conduct a 1:5 dilution of plasma-derived exomeres and supermeres isolated from Box 3 by mixing 40 μl of sample with 160 μl of albumin-binding buffer.
CRITICAL STEP Although the kit recommends diluting unprocessed plasma samples 10-fold with the maximum sample loading volume being 100 μl, we have found that dilution of the isolated sample 5-fold with a maximum sample loading volume of 200 μl can efficiently remove albumin contamination. We found that this kit also works for unprocessed plasma samples by diluting the sample 10-fold. The decision to use unprocessed plasma, or the isolated extracellular samples, as the starting material for albumin depletion will depend on the downstream sample analysis.
! CAUTION The maximum sample protein concentration is 5 – 7 mg/ml, so the sample concentration needs to be adjusted to within the loading limit.
Remove the bottom closure of the column by hand and slightly loosen the column cap. Put the column in a 1.5 ml sterile microcentrifuge tube (Fisher Scientific) and centrifuge at 1500g for 1 min at RT on a benchtop microcentrifuge (Thermo Scientific, Sorvall Legend Micro 17R) to remove the storage buffer. Discard the storage buffer.
! CAUTION Be careful to not let the column touch the liquid on the side of the tube.
Remove the column carefully and place it in a new 1.5 ml sterile microcentrifuge tube, add 200 μl of albumin-binding buffer and centrifuge as above to wash the column. Discard the flow through and repeat the wash step for a total of three times.
Add 200 μl of diluted sample from Step 8 onto the spin column. Close the cap tightly to avoid losing sample from the bottom of the column. Invert the column 2 – 3 times.
Incubate the column for 1 h on a sample mixer at RT (Invitrogen) by 360° end-over-end rotation with vortexing at 10 rpm periodically.
CRITICAL STEP We have observed that a 200 μl volume of sample can cover the column better than 100 μl, allowing more albumin to bind the column. If the volume is limited, then the sample will not contact the column well. Also, longer incubation time (1 h) helps albumin bind better with the column than the 30 min recommended by the manufacturer.
! CAUTION At each step, make sure there is no residual solution at the bottom of the column before placing the column in new microcentrifuge tube.
Transfer the column to a new microcentrifuge tube, loosen the cap and centrifuge at 1500g for 1 min at RT. Keep flow through (FT) (this is the albumin-depleted sample).
Optionally, to recover more albumin-depleted sample, add 200 μl of albumin-binding buffer to the column that was transferred to a new microcentrifuge tube to wash the column, then repeat step 13. Repeat this wash several times based on the protein concentrations in the flow through.
Combine the initial flow through with the flow through from the washes and concentrate using Amicon Ultra-15 50k (Millipore) or 30k (Millipore) or other available concentration methods.
Add 200 μl of albumin elution buffer to the column and spin down at 1500g for 1 min at RT to elute the bound albumin from the column.
Optionally, to elute more bound albumin from the column, repeat the elution step 3 or 4 times by transferring the column to a new microcentrifuge tube each time. Combine the initial elution with the elution from the other repeats and concentrate using Amicon Ultra-15 or other available concentration methods.
CRITICAL STEP The number of washes (Step 14) or elutions (Step 17) need to be optimized based on the downstream analyses.
Measure the OD280 of the albumin-depleted exomere and supermere samples, using Nano Drop One (Thermo Scientific). Also quantify the proteins in the samples using a Direct Detect spectrometer, or alternatively, by the BCA or Bradford methods.
Run 30 μl of pre-albumin depletion exomere and supermere samples together with post-albumin depletion exomere and supermere samples on a NuPAGE 4–12% Bis-Tris gel (Invitrogen). Stain with SimplyBlue (Invitrogen) to confirm the albumin depletion efficiency or perform immunoblotting to confirm the enrichment of specific cargo in exomere and supermere samples after albumin depletion.
? TROUBLESHOOTING
Applications of the method
It is of utmost importance for the future therapeutic potential and design of treatment interventions to correctly identify the compartment and mechanisms by which specific DNA, RNA, and proteins are secreted in human disease9,15,40. A strength of this protocol is that it allows investigators to use the same starting material to identify the compartment(s) in which specific secreted cargo reside. The methods can be applied to isolate lEVs, sEVs, exomeres and supermeres from conditioned medium of cells in culture and from plasma.
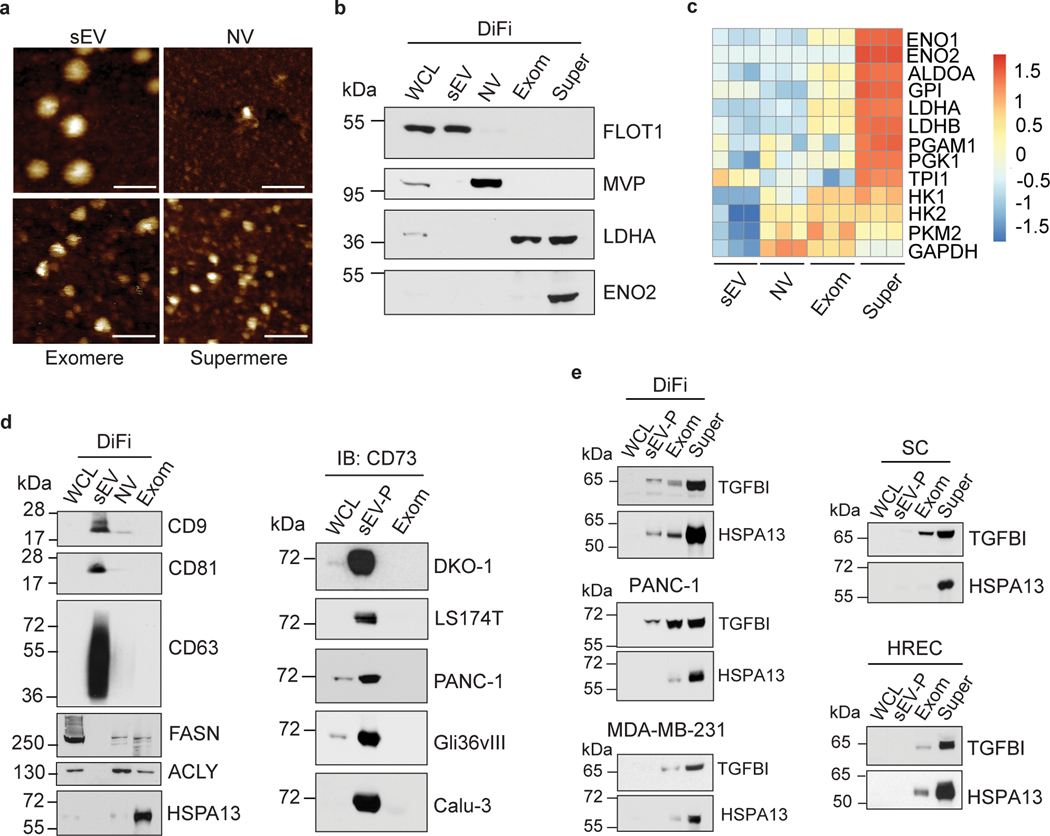
We recently developed a high-speed ultracentrifugation procedure to isolate and characterize a novel small extracellular NVEP that we have termed supermere (supernatant of exomeres)15. Supermeres display a markedly greater uptake in a variety of organs in vivo compared with sEVs and exomeres. Supermeres are highly enriched in cargo involved in multiple disease states, including cancer, Alzheimer’s disease, and cardiovascular disease. Cancer-derived supermeres play important biological functions, including transfer of cetuximab resistance15. Thus, supermeres are a distinct functional new NVEP replete with potential circulating biomarkers and therapeutic targets for a host of human diseases.
In our recent publication on exomeres10,28, we described a high-yield ultracentrifugation-based method for exomere isolation and demonstrated that exomeres contain and transfer functional cargo. These findings should accelerate advances in determining the composition and biological functions of exomeres along with their potential use in biomarker discovery and therapeutic applications. In our third key supporting research manuscript9, we have employed differential ultracentrifugation together with a unique high-resolution density-gradient fractionation to precisely characterize the RNA, DNA, and protein constituents of sEVs and non-vesicular material. We show that extracellular RNA, RNA-binding proteins, and other cellular proteins are differentially expressed in sEVs and non-vesicular compartments. This study provides a framework for a clearer understanding of EV heterogeneity.
Isolation of crude EV pellets (EV-Ps), exomeres and supermeres has been performed in multiple human cancer cell lines and normal human kidney epithelial cells15. The EV and NVEP isolation protocol has also been performed on plasma from both normal and colorectal cancer patients. The final yield, purity, and characteristics of these EVs and NVEPs have been examined in diverse sets of cell lines and patient conditions. Optimization is necessary based on cell type and growth conditions, patient conditions and plasma volumes.
These refined isolation methods for parsing of lEVs, sEVs (a subset of which are exosomes), exomeres and supermeres are vital to the advancement of basic science investigation, as well as clinical translation, for many disorders such as cancer, Alzheimer’s disease, cardiovascular disease, and infectious diseases like COVID-19. Overall, these techniques will improve biomarker discovery and identification of therapeutic targets. Protocols for isolation of exomeres and supermeres from other biofluids, including urine and milk, have not yet been developed, but will likely be detailed in the near future due to the importance of these nanoparticles in health and disease.
We cannot exclude that other subsets of EVs and NVEPs will be identified as methods are refined and new technologies are developed. However, we have provided a comprehensive and detailed protocol for isolation of lEVs, sEVs, exomeres and supermeres from the same starting material. The protocols for isolation of the different extracellular compartments require the use of high-speed ultracentrifuges and rotors capable of reaching the required high centrifugal forces. This complete protocol requires approximately 3 days to complete.
Comparison with other methods
Our high-resolution iodixanol density-gradient fractionation of lEV and sEV pellets reliably separates EVs from contaminating non-vesicular fractions9. A key feature of this method is bottom loading of a 12–36% gradient, which allows EVs to float upwards while higher density components will remain at the bottom portion of the gradient. Previous studies have used different percentages of iodixanol density gradients (for example, 5–40%) and top-loaded iodixanol density gradients to isolate EVs from NV fractions. However, these methods cannot be assumed to produce the same results. Our method of fractionation reliably separates lEV or sEVs from non-vesicular fractions to ensure that cargo can be assigned to their correct compartments9,15.
Our ultracentrifugation method for isolation of exomeres has several advantages over the alternative AF4 protocol that has been used by other groups5,35. Besides the centrifugation method to isolate exomeres10,28 detailed in this protocol, AF4 is the only other published method for isolation of exomeres5,35. AF4 represents a step forward by fractionating exomeres from exosome subpopulations based on their size and hydrodynamic properties5,35; however, the technique relies on a specialized instrument8, and the input and yield are limited. The loading capacity of AF4 is limited and typically involves ultracentrifugation to concentrate EVs and NVEPS before AF4 isolation, making comprehensive characterization and functional analysis challenging41. The exomere yield using ultracentrifugation can be comparatively high (Table 1). AF4 also requires costly instrumentation and considerable technical expertise that may limit its widespread adoption8.
Table 1. Protein concentrations and ratios of the sEV-P, exomeres and supermeres produced from cell lines in equal volumes.
Note that the size of the sample preparations (number of cell culture plates) is not equal between different cell lines. This table is modified from Zhang et al.15.
| Concentration (μg/μl) | Ratio | ||||||
|---|---|---|---|---|---|---|---|
| Cell line | Cancer | sEV-P | Exomere | Supermere | sEV-P | Exomere | Supermere |
| DiFi | Colorectal | 8.7 | 4.5 | 13.8 | 1 | 0.52 | 1.59 |
| LIM1215 | Colorectal | 14.1 | 8.9 | 36.4 | 1 | 0.63 | 2.58 |
| LS174T | Colorectal | 10.1 | 7.5 | 20.8 | 1 | 0.74 | 2.06 |
| CC | Colorectal | 4.5 | 2.2 | 15.4 | 1 | 0.49 | 3.42 |
| CC-CR | Colorectal | 7.2 | 3.1 | 21.5 | 1 | 0.43 | 2.99 |
| SC | Colorectal | 7.3 | 3.7 | 21.1 | 1 | 0.51 | 2.89 |
| MDA-MB-231 | Breast | 16.0 | 9.6 | 36.0 | 1 | 0.60 | 2.25 |
| PANC-1 | Pancreas | 17.7 | 12.8 | 33.8 | 1 | 0.72 | 1.91 |
| Calu-3 | Lung | 1.3 | 1.2 | 2.1 | 1 | 0.92 | 1.62 |
| HREC | Normal kidney | 3.2 | 1.8 | 6.7 | 1 | 0.56 | 2.09 |
Although differential ultracentrifugation has been widely used for isolation of EVs based on size and density, it has some disadvantages. For example, the EVs and NVEPs isolated using ultracentrifugation may be heterogenous, including subpopulations of different sizes. Also, some soluble proteins and nucleic acids may be co-isolated with each fraction. Other disadvantages include the cost of high-speed ultracentrifuges, along with the time and labor involved in processing the samples. However, to date, high-speed ultracentrifugation is the only method described to isolate biologically functional superemeres15.
Expertise needed to implement the protocol
Previous experience with cell culture and high-speed ultracentrifugation using different rotors is needed to perform the basic isolation protocol. First-hand isolation of EVs, exomeres and supermeres from plasma requires experience in handling and collecting human blood samples.
Limitations
In the isolation of exomeres and supermeres from human plasma, the albumin-depletion step may not remove all albumin. Also, during the depletion process, some albumin-binding proteins may be lost and thus will not be detected during downstream analysis. Therefore, the compatibility of the albumin-depletion step with a specific target protein in mind should be empirically validated to ensure that the target is not unintentionally depleted along with the albumin.
This protocol allows separation of plasma EVs from HDL particles9; however, other lipoprotein particles, such as low-density lipoprotein (LDL), very-low-density lipoprotein (VLDL), intermediate-density lipoproteins (IDL) and chylomicrons present in human plasma are not removed. Therefore, if minimizing the presence of these other lipoprotein particles is a priority, other published protocols may be more suitable18,19,26,27,33. Also, we cannot exclude the possibility that other types of NVEPs, including lipoproteins, may be co-isolated with exomeres and supermeres from human plasma. Other biofluids, including urine, milk, and stool, may present unique challenges (such as the highly abundant Tamm-Horsfall protein in urine30) for isolation of EVs and NVEPs, and, in those instances, other published protocols may represent better alternatives18,30,31,34.
Experimental design
Cell line choice
Our lab has used a human colorectal cancer cell line, DiFi, as a standard for isolation and characterization of EVs and NVEPs10,15. The Extracellular RNA Community Consortium (ERCC)2 is using secreted fractions from this line for its benchmarking studies42. A typical preparation from DiFi cells consists of 80 culture dishes (15 cm) with approximately 1.34 × 108 cells per dish at the time of harvest. The typical protein yield is approximately 4 mg for sEV-P, 2.5 mg for exomeres and 7 mg for supermeres. Depending on the downstream application, our methods can be applied to any cell type; we have applied this method to human cancer cell lines (colorectal, breast, lung, pancreas, and glioblastoma) and human primary renal proximal tubule epithelial cells (Table 1). The protocol is suitable for both cell culture media from conventional two-dimensional (2D) cultures, as well as from bioreactor (3D) cultures32,43,44. However, the yield and purity of different extracellular fractions may be influenced by cell type, cell growth conditions, culturing methods, serum-free or exosome-depleted serum-containing medium, cell density, cell stress, and drug exposures. Additional factors, including the number of culture dishes and the serum-starvation times, will also influence yield.
Isolation of lEV-Ps, sEV-Ps, exomeres and supermeres from cell-conditioned medium (Steps 1–38) or plasma (Box 3)
From the same starting material, we present an optimized protocol that sequentially isolates crude lEV pellets (lEV-Ps), crude sEV-Ps, exomeres and supermeres, which are released from cells, using sequential centrifugation, concentration and filtration (Fig. 1a and Steps 1–38).
In our published protocol, serum-free conditioned medium was centrifuged for 10 min at 500g followed by 2000g for 20 min at 4 °C to remove cellular debris and apoptotic bodies. The resulting supernatant was then subjected to 10,000g for 40 min to isolate lEV-Ps. Then the supernatant was filtered through a 0.22 μm polyethersulfone filter (Millipore) to reduce potential lEV contamination of the later sEV samples (Steps 1–18).
The supernatant filtrate was concentrated using a centrifugal concentrator with a 100,000 molecular-weight cutoff (Millipore) (Fig. 1b and Steps 19–30). A 100,000 molecular-weight cutoff concentrator can concentrate the sample from a large (several hundreds of milliliters to several liters depending on the number of cultured dishes) to a smaller volume (less than 37 ml depending on the preparation size). The concentration procedure serves two purposes: to facilitate the next centrifugation steps by reducing the sample volume and to reduce the amount of potentially contaminating free soluble proteins by allowing them to pass through the concentrator cores. The extent to which the conditioned medium is concentrated (fold-concentration) will depend both on the volume of the media collected and on the producer cells. Some cell types will produce more EVs and NVEPs so the media will need to be concentrated less as the starting concentration is higher. Conversely, cells that release lesser amounts of EVs and NVEPs will require a greater degree of concentration as the starting conditioned media is more dilute. The procedure is flexible in terms of the fold-concentration and can be determined by the investigator to suit their individual needs and concerns.
The concentrate, containing sEV-Ps, exomeres and supermeres, is then subjected to high-speed ultracentrifugation at 167,000g for 4 h at 4 °C in a SW32 Ti swinging-bucket rotor (k factor of 133, Beckman Coulter). The resulting pellet is resuspended in PBS-HEPES (25 mM) (PBS-H) and washed by centrifuging again at 167,000g for 4 h. The washed pellet contains sEVs and non-vesicular components and is designated sEV-P. In many studies, sEV-Ps are isolated by ultracentrifugation for shorter times, including 1 h, 70 min or 2 h, although some studies have also used extended overnight centrifugation. Short centrifugation times may not completely pellet all the sEVs32. Longer centrifugation times will allow more complete pelleting of sEVs but at the cost of co-sedimenting increasing amounts of non-vesicular material, including exomeres. Likewise, the choice of centrifugation g force (and the k factor of the chosen rotor type) will influence the efficiency of sEV sedimentation, with higher g forces leading to more effective pelleting of sEVs but at the potential cost of increased levels of co-sedimented non-vesicular material29,32. We have previously used centrifugation g forces ranging from 120,000g - 167,000g for 4 - 6 h with good yields of intact sEVs9,10,15,45. Yield and purity of sEV-Ps are two important factors to consider during this step of isolation, but the protocol is flexible enough to allow g forces from 100,000g - 167,000g and spin times from 70 min to 16 h (overnight), depending on the needs of the investigator.
We have developed and optimized a simple but high-yield method of separating exomeres from sEV-Ps. To isolate exomeres, the supernatant collected from the 4 h ultracentrifugation, which is normally discarded, is ultracentrifuged at 167,000g for 16 h at 4 °C in a SW32 Ti swinging-bucket rotor (k factor of 133, Beckman Coulter). The resulting pellet is resuspended in PBS-H and washed by centrifuging again at 167,000g for 16 h. The washed pellet represents biologically active exomeres10.
To isolate supermeres, the supernatant from the pelleting of exomeres is subjected to ultracentrifugation at 367,000g using a Beckman Coulter SW55 Ti rotor (k factor of 48, Beckman Coulter) for 16 h at 4 °C. The resulting pellet is resuspended in PBS-H and is designated supermeres.
An optional step in the protocol (Box 2) is to pass pelleted samples sequentially through increasingly narrow pore size syringes when monodispersion of the sample is required (Steps 35, 37 and 38).
Box 2. Optional protocol modification to further homogenize sEV-Ps, exomeres and supermeres by sequential syringing.
For many types of downstream analyses, the samples in PBS-H can be resuspended and adequately dispersed by pipetting up and down using a P1000 manual pipette. But for some downstream analyses, such as flow cytometry or functional assays, having the samples fully monodispersed may be required or deemed beneficial.
Procedure
To fully homogenize the sample solution containing sEV-Ps, exomeres or supermeres, pass the pelleted material in PBS-H through sequential needles of decreasing pore size: 22G x 1 ½ (0.7 mm x 40 mm), 27G x 1 ½ (0.4 mm x 30 mm), and 30G x 1 ½ (0.3 mm x 13 mm) attached to a 3 ml syringe (BD Plastipak). The syringing is performed at least 7 times/needle in order to obtain a more homogenous, monodispersed particle preparation.
High-resolution density gradient fractionation of lEVs and sEVs from non-vesicular fractions (Steps 39–51)
In our protocol, we demonstrate separation of lEVs and sEVs from NV components by a combination of high-speed ultracentrifugation and bottom-loaded high-resolution iodixanol density-gradient fractionation9 (Fig. 2 and Steps 39–51). The bottom-loaded high-resolution gradient (12–36%) allows EVs to float upwards in the gradient and reach their buoyant densities in the top portion of the gradient while the NV components remain in the bottom portion of the gradient due their higher density. This approach has been shown to remove contaminating NV fractions, including vault structures, nucleosomes, and non-EV RNA binding proteins9. Bottom loading of iodixanol gradients has also been shown to be preferable for isolation of EVs from urine30,31. The bottom-loaded high-resolution (12–36%) gradient can be used to separate HDL from EVs due to the relatively higher density of HDL particles9. However, due to their relatively lighter density, other lipoprotein particles such as LDL, VLDL, IDL and chylomicrons, will have overlapping buoyant densities with EVs. For plasma samples where these lipoproteins are abundant, a top-loading gradient approach may be preferable if minimizing the presence of these lower density lipoprotein particles is a priority18,26,27. The majority of circulating Argonaute 2 (AGO2)-miRNA complexes are non-vesicular and can be separated from EVs in plasma by bottom-loading the samples for the high-resolution gradient9,46.
Isolation of sEV-Ps, exomeres and supermeres from human plasma and a step to deplete albumin (Box 3)
In addition to EVs, exomeres and supermeres, plasma contains other types of particles (including lipoproteins) and proteins (including albumin, globulins, and clotting factors such as fibrinogen). Other published protocols have implemented various combinations of centrifugation, size-exclusion chromatography and density-gradient fractionation to improve isolation of EVs from plasma 19,26,27,33. However, in this protocol, we focus on isolation of EVs and the newly identified exomeres and supermeres from plasma using ultracentrifugation and density-gradient methods9,10,15. Unlike cell-conditioned media, plasma samples are not concentrated, but rather diluted at least 10-fold prior to isolation of exomeres and supermeres due to the inherent high viscosity of plasma. Albumin is the most abundant protein in plasma, accounting for roughly 60% of the total protein. We describe a method to isolate lEV-Ps, sEV-Ps, exomeres and supermeres from plasma by sequential ultracentrifugation15 (Fig. 3 and Box 3). However, due to the abundance of albumin in exomeres and supermeres, it is difficult to perform downstream analysis of these NVEPs using proteomics or immunoblotting. We describe a method to deplete albumin from isolated exomeres and supermeres using an albumin depletion kit (Abcam) (Fig. 4 and Box 3). We can also deplete albumin from plasma before EV and NVEP isolation, but plasma should be diluted at least 10-fold before performing the albumin depletion step; however, due to the low plasma input volume on the column (each column can hold 100 μl), and the high volume of diluted plasma, it may be too costly to routinely deplete the plasma before isolation of EVs and NVEPs. The sEV-P isolated from plasma can be further fractioned into sEV and non-vesicular fractions9 using high-resolution 12–36% iodixanol gradient fractionation (Steps 39–51) to further increase the purity of sEVs for downstream analysis. Our comprehensive isolation of EVs, exomeres and supermeres from plasma, including albumin depletion, can enhance biomarker discovery.
Characterization of isolated EVs and NVEPs
After isolation, we recommend characterization of EVs and NVEPs in compliance with guidelines outlined in MISEV201847 prior to further analyses to validate the quality of isolated fractions. Protocols are available to assess the size, morphology, and ultrastructure by AFM48 and electron microscopy49 while standard protocols for immunoblotting or ELISA can be used to assess the presence of expected marker proteins for lEVs, sEVS, exomeres and supermeres6,9,10,15,47. NTA can be used to count total particle numbers and estimate size distribution based on the scattering of light50, with the addition of detergents improving the discrimination between EVs and NVEP9. However, it should be noted that both exomeres and supermeres are likely to be below the size that can reliably be detected with NTA and caution should therefore be taken when normalizing by particle number for downstream analysis or functional assays. Normalization by protein quantification is the preferred option at this time.
Controls
For depletion of albumin, the plasma before albumin depletion is used as a control to compare with the plasma obtained after albumin depletion to determine if the depletion has been successful.
Materials
Biological Materials
Human plasma samples from normal and colorectal cancer patients.
We have isolated EV and NVEPs from human plasma from both normal and colorectal cancer patients at Vanderbilt University Medical Center.
! CAUTION The research conducted as part of this protocol complies with all the relevant ethical regulations. The use of the human samples was approved by the Vanderbilt University Medical Center Institutional Review Board (IRB; IRB nos 161529 and 151721).
The DiFi (RRID:CVCL_6895) colorectal cancer cell line was developed and characterized by the Robert Coffey laboratory.
The DKO-1 (RRID:CVCL_9798) colorectal cancer cell line was obtained from T. Sasazuki at Kyushu University.
The Gli36 (RRID:CVCL_RL88) human glioblastoma cell line was obtained from Xandra Breakefield at Harvard Medical School.
The B16-F1 (RRID:CVCL_0158) mouse melanoma cell line was obtained from the American Type Culture Collection. https://scicrunch.org/resolver/RRID:CVCL_0158.
General laboratory reagents
Phosphate-buffered saline (PBS) without calcium and magnesium, pH 7.4 (Gibco, cat. no. 14190–144)
HEPES (Sigma, cat. no. H0887)
DMEM (Corning, cat. no.10–017-CV)
Non-essential amino acid solutions (100X, Cytiva HyClone™, Fisher Scientific, cat. no. SH3023801)
Penicillin Streptomycin (100X, Cytiva HyClone™, Fisher Scientific, cat. no. SV30010)
L-Glutamine (Cytiva SH3003401)
Bovine Calf Serum (BCS) (Cytiva HyClone™, Fisher Scientific, cat. no. SH3054103)
Distilled H2O (Corning, cat. no. 46–300-cl)
Ethyl alcohol 200 proof (Pharmco, cat. no. 111000200) ! CAUTION Ethanol is highly flammable liquid and vapor. Causes serious eye irritation. Keep container tightly closed and store in a well-ventilated place.
Iodixanol (OptiPrep) (Sigma-Aldrich, cat. no. D1556)
Albumin depletion kit (Abcam, cat. no. ab241023)
SimplyBlue SafeStain (Invitrogen, cat. no. LC6060)
Trypan Blue Solution 0.4% (w/v) (Cellgro, cat. no. 25–900-cl)
TGFBI (RRID:AB_2202311) antibody (Proteintech, cat. no.10188–1-AP) https://scicrunch.org/resolver/RRID:AB_2202311
EQUIPMENT
General equipment
Microcentrifuge (Thermo Scientific Sorvall Legend Micro 17R, cat. no. 75–002-543)
Centrifuge (Thermo Fisher Scientific, Sorvall LYNX 4000, superspeed, swing bucket, cat. no. 75006580)
Benchtop swinging-bucket centrifuge (Thermo Fisher Scientific, Sorvall Legend XTR, cat. no.75–217-420)
Optima XPN-100 Ultracentrifuge (Beckman Coulter, cat. no. A94469)
Optima XE-100 Ultracentrifuge (Beckman Coulter, cat. no. A94516)
SW 41 Ti swinging-bucket rotor, titanium (Beckman Coulter, cat. no. 331362), The k factor of this rotor is 124, the maximum speed is 41,000 rpm
SW 32 Ti swinging-bucket rotor, titanium (Beckman Coulter, cat. no. 369694), The k factor of this rotor is 133, the maximum speed is 32,000 rpm
SW 55 Ti swinging-bucket rotor, titanium (Beckman Coulter, cat. no. 342194), The k factor of this rotor is 48, the maximum speed is 55,000 rpm
Direct Detect Spectrometer (Millipore, cat. no. DDHW00010-WW)
Direct Detect cards (Millipore, cat. no. DDAC00010-GR)
Balance (Ohaus, Explorer Pro Precision, cat. no. 80108933)
Cabinet (Class II, Type A2 biological safety cabinet, Nuaire, cat. no. NU-425–400)
Nutating mixer (Barnstead International, cat. no. M26125)
Electronic pipettor (Eppendorf Repeater E3x - Electronic Multi-Dispenser Pipette, cat. no. 4987000410)
Sample mixer (Invitrogen, HulaMixer, cat. no. 15920D)
NanoDrop One (Thermo Scientific, cat. no. 13–400-518)
T-175 cell culture flask (175 cm2, Greiner CELLSTAR, cat. no. 660–175)
T-75 cell culture flask (Falcon 75cm² Rectangular Straight Neck Cell Culture Flask with Vented Cap, Corning, cat. no. 353–110)
145/20 mm cell culture dish (Greiner CELLSTAR, cat. no. 639160)
60/15 mm cell culture dish (Falcon, Corning, cat. no. 353002)
50 ml conical centrifuge tube (Corning, cat. no. 430829)
15 ml conical centrifuge tube (CellStar, cat. no. 188261)
1.5 ml centrifuge tube (Fisher Scientific, cat. no. 05–408-129)
10 ml pipette (Greiner CELLSTAR serological pipette, cat. no. GN607180)
5 ml pipette (Greiner CELLSTAR serological pipette, cat. no. GN606180)
25 ml pipette (Greiner CELLSTAR serological pipette, cat. no. GN760180)
22G x 1 ½ needles (0.7 mm x 40 mm, Precision Glide Needle, BD, cat. no. 305156)
27G x 1 ½ needles (0.4 mm x 30 mm, Precision Glide Needle, BD, cat. no. 305136)
30G x 1 ½ needles (0.3 mm x 13 mm, Precision Glide Needle, BD, cat. no. 305106)
3 ml syringe (BD Plastipak, cat. no. 309657)
5 ml syringe (BD Plastipak, cat. no. 309646)
Centricon Plus-70 (100,000 MV cut off, Millipore, cat. no. UFC710008)
SETON open top polyclear centrifuge tubes 1X3 ½ inch (25X89mm) (SETON, cat. no. 7052)
Ultra-Clear centrifuge tubes (13×51mm) (Beckman Coulter, cat. no. C14295, for supermere isolation)
Ultra-Clear centrifuge tubes (25×89 mm) (38.5 ml, sterile, Open-Top Thin Wall, Beckman Coulter, cat. no. C14292, for sEV-P and exomere isolation)
Ultra-Clear centrifuge tubes (14×89 mm) (Beckman Coulter, cat. no. C14293, for gradient purification of EVs)
Stericup quick release (Millipore express Plus 0.22uM PES, cat. no. S2GPU11RE)
Syringe filter (Millex-GP Syringe Filter Unit, 0.22 μm, polyethersulfone, 33 mm, gamma sterilized, Millipore, cat. no. SLGP033RS)
Blood Collection Tubes (BD Bioscience, cat. no. 369714)
Amicon Ultra-15 centrifugal filters (Millipore, 50k, cat. no. UFC905024)
Amicon Ultra-15 centrifugal filters (Millipore, 30k, UFC903024)
NuPAGE Bis-Tris Gel (Invitrogen, 4–12%, NP0322BOX)
Reagent setup
70% (vol/vol) sterile ethanol
To prepare 1 liter of 70% ethanol, mix 700 ml of ethyl alcohol 200 proof with 300 ml of Milli-Q H2O, pass through 0.22 μm filter and store at room temperature (RT; 22 °C) for up to 6 months.
Complete DMEM medium (10% BCS) (vol/vol)
In a tissue culture hood, to a bottle of 450 ml of DMEM medium, add 50 ml of bovine growth serum, 5 ml of non-essential amino acid solutions (100X), 5 ml of L-glutamine (100X, 200 mM) and 5 ml of penicillin streptomycin (100X, 10,000 U/ml), mix well and keep at 4 °C up to one week.
Serum-free DMEM medium
In a tissue culture hood, to a bottle of 500 ml of DMEM medium, add 5 ml of non-essential amino acid solutions (100X), 5 ml of L-glutamine (100X) and 5 ml of penicillin streptomycin (100X), mix well and keep at 4 °C for up to one week.
PBS-HEPES (PBS-H) (25 mM) (vol/vol)
Add 12.8 ml of 1 M sterile HEPES (pH 7.0–7.6) to a bottle of 500 ml sterile PBS, mix well and keep at 4 °C for up to one month.
Iodixanol (OptiPrep) 12–36% (wt/vol)
To make the density solutions, mixing 60% (wt/vol) iodixanol with sterile PBS based on the volumes listed in Table 2. Mix well. Solutions should be made fresh before the experiment and kept at 4 °C for up to a few days.
Table 2.
Iodixanol(OptiPrep) solutions
| Final iodixanol solution concentration | Volume of 60% iodixanol stock (ml) | PBS Volume (ml) |
|---|---|---|
| 50% (wt/vol) | 20 | 4 |
| 36% (wt/vol) | 14.4 | 9.6 |
| 30% (wt/vol) | 12 | 12 |
| 24% (wt/vol) | 9.6 | 14.4 |
| 18% (wt/vol) | 7.2 | 16.8 |
| 12% (wt/vol) | 4.8 | 19.2 |
Procedure
Isolation of lEV-Ps, sEV-Ps, exomeres and supermeres from conditioned medium of cell culture. Timing: (variable, depending on the growth rate of the cells and size of the preparation)
CRITICAL The DiFi colorectal cancer cell line has been used as a standard in this protocol. This protocol is applicable to a variety of cell lines from different organs and origins. (see Experimental Design for further discussion).
CRITICAL: The procedure below is for cell conditioned media but the isolation of lEV-Ps, sEV-Ps, exomeres and supermeres can also be performed for human plasma (see Box 3). For plasma samples, skip the media collection (Steps 1–10) and media concentration (Steps 19–30) procedures below and instead perform the steps outlined in Box 3.
Medium collection
CRITICAL Perform Steps 1–6 in a class II, type A2 biological safety cabinet (Nuaire).
-
Seed cells in 175T flasks in 35 ml of warm (37 °C ) DMEM complete medium (listed in the reagent setup) in the tissue culture hood. The number of flasks of cells needed depends on the size of the preparation needed for downstream applications.
CRITICAL: The number of cells per flask would depend on the specific cell line as they vary in size. A visual estimation that the cells cover about 70–80% of the flask is sufficiently precise.
! CAUTION Cells should be routinely tested for mycoplasma contamination. We routinely assess cell viability using trypan blue exclusion (Cellgro). Only medium collected from cells with > 95% viability is used for isolation of EVs and NVEPs.
Grow cells in the tissue culture incubator with 5% CO2 at 37 °C.
-
Seed cells into 15 cm dishes in 18 ml of DMEM complete medium (or T175 culture flask in 25–30 ml of media) when the cells reach about 70–80% confluency and grow in the tissue culture incubator with 5% CO2 at 37 °C until the cells reach 70 – 80% confluency. Normally it takes about 48 h to reach 70 – 80% confluency depending on the growth rate and the seeding density of the cells.
CRITICAL STEP The number of dishes (or T175 flasks) needed depends on the size of the preparation and the downstream application. A standard preparation for comprehensive profiling and functional in vivo assays normally uses 80 – 120 dishes but smaller setups can be used for biochemical and molecular characterization.
-
Wash the cells twice with at least 12 ml of pure DMEM medium without any supplements.
! CRITICAL STEP Tilt the plate for 1 min after aspirating the medium to be sure all the medium is completely removed.
-
Starve the cells in 18 ml of serum-free DMEM medium (without BCS but include all the other supplements listed in the reagent setup) for 48 h in the tissue culture incubator with 5% CO2 at 37 °C.
! CRITICAL STEP The EVs and NVEPs are isolated from serum-free medium to avoid any bovine serum-derived EV contamination. If bovine growth serum is used, EV-depleted fetal bovine serum should be prepared (see Box 1).
BOX 1. Optional protocol modification for using media containing EV-depleted bovine serum.
As an alternative to using serum-free media for harvesting EV- and NVEP-containing cell-conditioned media, media containing EV-depleted serum can be used. It is important to note that while bovine EVs can be mostly eliminated, some contaminating EVs may remain52, and bovine NVEPs and bovine RNA-protein complexes53 will also still be present. Additionally, growth factors present in the serum that support cellular viability may be removed by the procedure to eliminate bovine EVs53. Of course, the use of serum-free media will also influence cellular growth and stress with possible consequences for subsequent isolation and content of EV and NVEPs. Thus, no perfect solution is currently available, and investigators will have to choose serum-free or EV-depleted serum based on the most important downstream analyses they wish to perform.
Procedure
Dilute bovine calf serum or fetal calf serum 1:5 fold in DMEM.
Centrifuge the diluted serum-DMEM at 100,000g for 16–18 h at 4 °C in a SW 32 Ti swinging-bucket rotor (Beckman Coulter).
After centrifugation, the EV-depleted media is contained in the supernatant whereas the bovine serum EVs will form the pellets at the bottom of the tubes. Remove the EV-depleted media by decanting or pipetting. Discard the pellets.
Pass the EV-depleted media through a 0.22 μm polyethersulfone filter (Millipore). Add the filtered EV-depleted media (20% serum) to DMEM to obtain the desired final concentration of serum in the media.
-
Collect the serum-free conditioned medium into 50 ml conical tubes.
! CAUTION Tilt the plate for 1 min after aspirating the medium to be sure the medium is completely removed.
Large Extracellular Vesicle-Pellet (lEV-P) Isolation (2 h)
Centrifuge the conditioned media at 500g for 10 min at 4 °C to pellet cells and cell debris.
-
Transfer the supernatant to a new 50 ml conical tube by decanting or pipetting the supernatant in the tissue culture hood. Discard the pellet.
CRITICAL STEP Perform this step as soon as the centrifugation is completed to avoid dispersion of the pellet. If transferring by pipetting, leave 0.5 ml of the supernatant above the pellet to avoid possible contamination.
Centrifuge the supernatant at 2000g for 20 min at 4 °C to remove cell debris and apoptotic bodies.
-
Transfer the supernatant to a new 50 ml conical tube by decanting or pipetting the supernatant in the tissue culture hood. Discard the pellet.
CRITICAL STEP Perform this step as soon as the centrifugation is completed to avoid dispersion of the pellet. If transfer by pipetting, leave 0.5 ml of the supernatant above the pellet to avoid possible contamination.
-
Centrifuge the supernatant at 10,000g for 40 min at 4 °C to collect lEV-P.
CRITICAL STEP Depending on the experimental design, if only the sEV-Ps, exomeres and supermeres are to be isolated but not lEV-Ps, then skip Steps 11–17, continuing at Step 18 to filter the media supernatant collected in Step 10 through a 0.22 μm pore PES filter.
-
Transfer the supernatant to a T175 flask by pouring off carefully or pipetting the supernatant. Tilt the tube, carefully transfer the rest of the supernatant out to avoid disturbing the pellet. This pellet is the lEV-P.
CRITICAL STEP Perform this step as soon as the centrifugation is completed to avoid dispersion of the pellet. If transferring by pipetting, leave 0.5 ml of the supernatant above the pellet to avoid possible contamination.
Repeat Steps 11–12 until there is no more medium left. CRITICAL STEP: The number of times this needs to be repeated will depend on the total volume of conditioned media collected. Higher volume of media will take longer time to process.
Keep all the supernatant in the T175 cm flasks at 4 °C for downstream isolation of sEVs, exomeres and supermeres, starting from Step 19.
Wash the lEV-P pellet with PBS-H. Add 1ml of PBS-H to each pellet in the 50 ml tubes and re-suspend the pellet thoroughly by pipetting to wash away free proteins and other contaminants trapped in the lEV-P. Combine resuspended pellets to a new 50 ml centrifuge tube by pipetting. Rinse each tube by adding 1 ml more of PBS-H and transfer the liquid to the 50 ml tube containing the resuspended lEV-P. Fill this tube with PBS-H till it reaches 50 ml.
Centrifuge the re-suspended lEV-P at 10,000g for 40 min at 4 °C to collect washed lEV-P. Discard the supernatant by decanting or pipetting. Tilt the tube, and carefully transfer the rest of the supernatant out using a 1 ml pipette to avoid disturbing the lEV pellet.
-
Resuspend the pellet in PBS-H and transfer the lEV solution to a 1.5 ml microcentrifuge tube using a P1000 manual pipette. Take out 10 μl of the lEV-P material and quantify protein concentration using a Direct Detect Spectrometer as described by the manufacturer’s protocol (Millipore), or alternative assays for protein quantification such as BCA or Bradford51.
? TROUBLESHOOTING
PAUSE POINT Depending on the downstream application purpose, the lEV-Ps can be kept at either 4 °C (short-term storage) for characterization including immunoblotting, electron microscopy, nanoparticles tracking analysis (NTA), as well as functional studies, −80 °C (long-term storage) or used immediately.
CRITICAL STEP The amount of buffer added depends on the size of the preparation, the host cell line and the downstream applications. For example, if the preparation size is eighty 15-cm plates of DiFi cells, we resuspend the pellet in 500 μl of PBS-H or lysis buffer.
-
Pass the media supernatant from Step 14 through a 0.22 μm pore PES filter (Stericup 1000ml, Millipore express Plus 0.22 μm PES) to remove any remaining lEVs. Keep the supernatant in the filter bottle at 4 °C for up to a few days or continue immediately to the next step for sEV-P isolation.
PAUSE POINT The filtered media supernatant can be kept at 4 °C for a short time (one or two days) before sEV-P isolation.
Cell-conditioned media supernatant concentration using concentrator (time variable depends on the total volume of media and the host cells),
-
Rinse the centrifugal filter device by adding 15 ml of 70% ethanol to the sample filter cup and shake the sample filter cup several times upside down to sanitize the device (Centricon Plus-70, Millipore, 100,000 NMWL).
CRITICAL STEP 70% ethanol needs to be filtered through a 0.22 μm pore PES filter (Stericup 1000 ml, Millipore express Plus 0.22 μm PES).
-
Centrifuge the filter device at 3500g for 5 min at 4 °C using a benchtop swing bucket centrifuge (Thermo Fisher Scientific, Sorvall Legend XTR). Shake the bottom filtrate collection cup with 70% ethanol to sanitize the device and pour off the ethanol.
! CAUTION The maximum centrifugal force is 3500g for a concentration spin using the filter device. Do not use a fixed angle rotor.
-
Rinse the filter device by adding 15 ml sterile PBS-H into the top sample filter cup. Shake it well. Spin the filter device at 3500g for 5 min at 4 °C. Shake the bottom filtrate collection cup several times and pour off the PBS-H. The device is ready for sample concentration.
CRITICAL STEP Keep the membrane wet once it is rinsed with 70% ethanol and PBS-H. Leave the fluid on the membrane until it is used.
-
Shake the bottle with media from Step 18 and load 60 ml to the sample filter cup. Carefully put on the cap.
! CAUTION The maximum volume capacity of the collection cup is 70 ml. Add no more than 70 ml media to the collection cup, each time.
-
Centrifuge the filter device at 3500g at 4 °C until no media is above the top of the filter cores. Proteins with a molecular weight smaller than 100 kD will go through the filter.
! CAUTION Balance the centrifuge with a second filter device and an equal volume of PBS or medium.
CRITICAL STEP The centrifugation time varies from 12–60 min depending on the host cell line and media protein concentration.
Remove the filter device and separate the sample filter cup from filtrate collection cup. Carefully pour the media out of the filtrate collection cup and discard it.
-
Repeat Steps 22–24 until no media is remaining in the top of the sample filter cup.
! CAUTION If the media flow rate slows to less than1 ml/ min, then a new filter device should be used. Typically, a filter device can filter 1 liter of medium, but this depends on the number of EVs and NVEPs released into the condition medium. The final volume of the concentrate depends on the fold concentration desired.
Add 60 mL of PBS-H to the sample filter cup and centrifuge at 3500g for 10 min at 4 °C to wash the concentrate. Pour off the flow through in the filtrate collection cup and discard it.
Concentrate Recovery (5–10 min)
Rinse the concentrate collection cups (see Fig. 1) with 70% ethanol, dry in the tissue culture hood with air for at least 30 min, and then expose the cups under the UV in the tissue culture hood for at least 30 min to sterilize.
Turn the concentrate collection cup upside down and place it on top of the sample filter cup.
-
Invert the cups carefully and place them in a benchtop centrifuge, then centrifuge at 1000g for 2 min at 4 °C.
! CAUTION The maximum centrifugal force for the recovery spin is 1000g. Sample may be lost if higher than a 1000g spin force is used.
CRITICAL STEP Wipe off any excess fluid on the side of the filter cup.
-
Remove the concentrate collection cup containing the concentrated sample from the sample filter cup. Transfer the sample with a 1 ml pipette to a 50 ml conical tube. Rinse the collection cup with PBS-H several times and then combine into the 50 ml tube.
PAUSE POINT The concentrated samples can be kept at 4 °C for up to 16 h or can be immediately subjected to downstream sEV-P isolation.
Sequential high-speed ultracentrifugation to isolate sEV-Ps, exomeres and supermeres (3 d)
-
Load the concentrate from Step 30 to Ultra-Clear Centrifuge Tubes (25 × 89 mm, 38.5 ml, sterile, Beckman Coulter). Add PBS-H to reach a total volume around 37–37.5 ml. Mix by pipetting up and down.
! CAUTION Each tube can hold a maximum of 38.5 ml of solution, but is normally filled to 37–37.5 ml. If the solution volume is less than that, the tubes may be crushed due to the high-speed ultracentrifugation power. The tubes should be filled to the recommended volume with PBS-H.
CRITICAL STEP If the centrifuge tubes are not sterile, for example, SETON open top Polyclear centrifuge tubes 1X3 ½ inch (25X89 mm), they need to be sanitized. Rinse the tubes with 70% 0.22 μm -filtered ethanol first, pour off the ethanol, air dry the tubes in the tissue culture hood, and then expose the tubes to UV to sanitize. These centrifuge tubes are used for sEV-P and exomere isolation.
CRITICAL STEP The centrifuge tubes must be carefully balanced by weight. Tubes numbered 1 and 4, 2 and 5, 3 and 6 should be balanced in pairs.
! CAUTION Incorrect use of an ultracentrifuge may result in equipment failure and damage, and cause loss of the sample. Always ensure that tubes are filled nearly to the top, that rotor buckets are correctly closed and attached to the rotor, and that opposing buckets with filled tubes have the same weight to ensure balance.
-
Carefully load the tubes on the rotor and centrifuge the concentrate at 167,000g for 4 h at 4 °C in a SW 32 Ti swinging-bucket rotor (Beckman Coulter).
! CAUTION The tube number must match the number on the rotor. For example, put tube 1 onto the rotor where it is labeled 1. Be careful to be sure the tubes are secured on the rotor.
CRITICAL STEP The rotor and holders should be kept at 4 °C when not in use.
-
Transfer the supernatant to a new sterile ultracentrifuge tube by decanting carefully or pipetting off the supernatant and keep at 4 °C for exomere and supermere isolation. If pouring off, tilt the tube and remove the extra liquid above the pellet by pipetting using a 1 ml manual pipette to avoid disturbing the pellet and then discard the supernatant. If transferring the supernatant by pipetting, use a 10 or 25 ml sterile disposable pipette and leave about 1 ml of the supernatant over the pellet. Transfer the residual volume using a 1 ml pipette.
CRITICAL STEP Perform this step as soon as the centrifugation is completed to avoid dispersion of the pellet.
To wash the pellet, add 1 ml PBS-H and pipette up and down several times using a 1 ml pipette. Transfer the resuspension to a new sterile ultracentrifuge tube and add 36.5 ml PBS-H to the residual pellet to transfer the rest of the pellet to the tube. Then repeat Steps 31 and 32.
-
Discard the supernatant (PBS-H) by decanting or pipetting as soon as the centrifugation is completed. Tilt the tube, carefully transfer the rest of the supernatant off using a 1ml pipette to avoid dispersing the pellet. The resulting washed crude pellet is designated the sEV-P. Resuspend the sEV-P in PBS-H or lysis buffer depending on downstream analysis and transfer the solution to a 1.5 ml microcentrifuge tube using a 1 ml pipette. Optionally, the crude sEV-P can be further homogenized by sequential passage through syringes (Box 2). Pipette out a small amount (2 μl per measurement) of the sEV-P material and quantify protein concentration by Direct Detect Spectrometer as described by the manufacturer’s protocol (Millipore), or alternative assays for protein quantification such as BCA or Bradford.
? TROUBLESHOOTING
PAUSE POINT Depending on the downstream applications, the sEV-P sample can be kept at either 4 °C (short-term storage), −80 °C (long-term storage) or used immediately.
CRITICAL STEP The amount of buffer added depends on the size of the preparation/pellet and the donor cell line. For example, if the preparation size is 80 15-cm dishes of DiFi cells, resuspend the pellet in 1 ml of PBS-H or lysis buffer.
! CAUTION Perform this step as soon as the centrifugation is completed to avoid dispersion of the pellet.
To isolate exomeres, ultracentrifuge the supernatant collected from the 4 h ultracentrifugation in Step 33 at 167,000g for 16 h at 4 °C in a SW 32 Ti swinging bucket rotor (Beckman Coulter). After the first centrifugation is completed, keep the supernatant at 4 °C for later supermere isolation.
-
Fully resuspend and wash the pellet in 37.5 ml PBS-H and centrifuge at 167,000g for 16 h at 4 °C in a SW 32 Ti swinging-bucket rotor. The resulting washed pellet is designated exomeres. Resuspend the exomere pellet in 0.5–1.0 ml PBS-H or lysis buffer depending on downstream analysis and transfer the solution to a 1.5 ml microcentrifuge tube using 1 ml pipette. Optionally, the exomeres can be further homogenized by sequential passage through syringes (Box 2). Take out a small amount of the exomere material (2 μl per measurement) and quantify protein concentration by Direct Detect Spectrometer (Millipore), or alternative assays for protein quantification such as BCA or Bradford.
? TROUBLESHOOTING
PAUSE POINT Depending on the downstream applications, the exomere solutions can be kept at either 4 °C (short-term storage) or −80 °C (long-term storage) for future use or used immediately.
-
To isolate supermeres, load the supernatant collected from the pelleting of exomeres (Step 36) to Ultra-Clear centrifuge tubes (13 × 51mm, Beckman Coulter). Subject the supernatant to ultracentrifugation at 367,000g using a Beckman Coulter SW55 Ti rotor (Beckman Coulter) for 16 h at 4 °C. Each ultracentrifuge tube (13×51mm, Beckman Coulter) can hold 5 ml of supernatant and the rotor can hold 6 tubes, resulting in a total of 30 ml of the supernatant that can be used for isolation of the supermeres. The tubes need to be balanced and loaded on the rotor as stated in Step 31. Discard the supernatant. The resulting pellet is designated as supermeres. Optionally, wash the supermere in PBS-H by an additional round of 367,000g centrifugation for 16 h at 4 °C. Resuspend the supermere pellet in 0.5–1.0 ml PBS-H or lysis buffer depending on downstream analysis, and transfer the supermere solution to a 1.5 ml microcentrifuge tube using 1 ml pipette. Optionally, the supermeres can be further homogenized by sequential passage through syringes (Box 2). Take out a small amount of the supermere material (2 μl per measurement) and quantify protein concentration by Direct Detect Spectrometer (Millipore), or alternative assays for protein quantification such as BCA or Bradford.
? TROUBLESHOOTING
PAUSE POINT Depending on the downstream applications, the supermere sample can be kept at either 4 °C (short-term storage) or −80 °C (long-term storage) for future use or used immediately.
High-resolution (12%–36%) iodixanol density-gradient fractionation of crude EV samples (1 d)
The crude lEV-Ps and sEV-Ps samples isolated in Steps 18 and 35 above are heterogenous, containing both EVs and non-vesicular (NV) material in separate fractions. To obtain highly purified EVs that have been separated from contaminating NV fractions, a bottom-loaded high-resolution (12% – 36%) iodixanol density gradient fractionation procedure is performed. Note that the individual fractions can either be processed individually for downstream analyses or pooled (highlighted as optional in the steps below) to represent purified EVs and NVs, respectively (see also Fig. 2).
-
Prepare the iodixanol (OptiPrep) (Sigma-Aldrich) density solutions as outlined in Table 2 in ice-cold PBS immediately, or shortly before use. The amounts listed are enough for 4–5 gradients.
! CAUTION The iodixanol density solutions should be made fresh each time before use, or shortly before use and kept at 4 °C.
-
Carefully mix 700 μl of a PBS-suspension containing crude lEV-Ps and/or sEV-Ps, isolated in Steps 18 and 35, with 1700 μl ice-cold 50% (wt/vol) iodixanol in a 15 ml tube to obtain a final volume of 2.4 ml of 36% (wt/vol) iodixanol EV-P solution.
CRITICAL STEP To ensure complete mixing of the 50% iodixanol solution with the crude EVs suspension in PBS to generate the 36% iodixanol EV-P solution, slowly pipette (using a P1000 manual pipette) up and down in the 15 ml tube until the mixture looks completely homogeneous.
Use a P1000 manual pipette to carefully transfer the 2.4 ml of the 36% (wt/vol) iodixanol EV-P suspension from the 15 ml tube to the bottom of a 14 × 89 mm Ultra-Clear centrifuge tube (Beckman Coulter) taking care not to deposit material on the sides of the tube.
Tilt the centrifugation tube sideways to an angle of 30–45° and carefully dispense 2.4 ml of the 30% (wt/vol) iodixanol solution on top of the previous layer using an automatic pipettor (Eppendorf Repeater E3x - Electronic Multi-Dispenser Pipette) set to the lowest dispensing speed with a Combitips advanced 2.5 ml pippete tip. Alternatively, manually dispense the solution using a P1000 manual pipette, taking care not to perturb the previous layer.
Repeat Step 42 three more times in sequence 2.4 ml of 24% (wt/vol) then 2.4 ml of 18% (wt/vol), then 2.4 ml of 12% (wt/vol) iodixanol solution each on top of the previous layer to complete the gradient.
Weigh the loaded centrifugation tube containing 12 ml of 12–36% iodixanol gradient on a balance (Ohaus, Explorer Pro Precision), to make sure it matches exactly the second tube that will run opposite in the ultracentrifugation rotor. The second tube can be another gradient containing an EV sample, or it can be an empty 12–36% gradient that can be used for later determination of fraction densities by refractometry. If an empty (no EV-P sample) gradient is used, instead of loading the 2.4 ml 36% iodixanol EV-P sample at the bottom of the centrifugation tube dispense 2.4 ml of 36% iodixanol solution to the bottom of the tube (Table 2) and proceed to add the lower density steps as detailed. In order for the tube weights to match exactly, carefully add a few drops of 12% (wt/vol) iodixanol solution on top of either tube to make sure the weight of both tubes match exactly.
-
Place the two tubes opposite of each other in cooled (4 °C) buckets of a SW41 TI swinging-bucket rotor (k factor of 124, Beckman Coulter) and centrifuge at 120,000g for 15 h at 4 °C.
! CAUTION Incorrect use of an ultracentrifuge may result in equipment failure and damage, and cause loss of the sample. Always make sure that tubes are filled nearly to the top, that rotor buckets are correctly closed and attached to the rotor and that opposing buckets with filled tubes have the same weight to ensure balance.
-
Remove the tubes from the bucket, and slowly and carefully collect twelve individual fractions of 1 ml each from the top of the gradient using a P1000 pipette taking care not to disturb the underlying fractions. Dispense each individual 1 ml fraction in a labeled 14 × 89 mm Ultra-Clear centrifuge tube. In total, there are 12 fractions.
CRITICAL STEP: Optionally, instead of isolating each of the 12 individual fractions, the first six fractions (fractions 1–6 from the top) representing EVs, can be pooled in a Ultra-Clear centrifuge tube (13 × 51mm, Beckman Coulter). The seventh fraction is discarded, and the last five fractions (fractions 8–12 from the top) representing NV, can then be pooled in a second tube (see Fig. 2).
-
Add 11 ml of PBS to each of the twelve tubes containing a 1 ml fraction and mix (by pipetting or tube inversion) the total volume of 12 ml until the solution appears homogenous. Adjust with PBS to make sure the total volume in each tube is equal and to make sure the weight of the tubes matches exactly.
CRITICAL STEP: Optionally, if fractions have been pooled in two tubes, add 30 ml of PBS to each of the two tubes and mix. Adjust with PBS to make sure the total volume in each tube is equal and the weight of both tubes match exactly.
-
Wash the first six fractions (fractions 1–6, taken from the top of the gradient) in a cooled (4°C) SW41 TI swinging-bucket rotor (k factor of 124, Beckman Coulter) by centrifugation at 120,000g for at least 4 h at 4 °C.
CRITICAL STEP: Optionally, if fractions have been pooled in two tubes, wash the two pools in a cooled (4°C) SW32 TI swinging-bucket rotor by centrifugation at 120,000g for at least 4 h at 4 °C.
-
Discard the supernatant by decanting or pipetting and resuspend the individual pellets of highly purified lEVs or sEVs that are at the bottom of each of the first six centrifugation tubes in a small volume (30 to 100 μl) of PBS and transfer to a microcentrifuge tube.
CRITICAL STEP: Optionally, if fractions have been pooled in two tubes, discard the supernatant and resuspend the two pellets, representing the final purified EVs and NVs, respectively, in a small volume (30 to 100 μl) of PBS. Transfer samples to microcentrifuge tubes. For pooled samples, this concludes the procedure and Steps 50–51 are not applicable.
? TROUBLESHOOTING
Wash the second six fractions (fractions 7–12, taken from the top of the gradient) in a cooled (4 °C) SW41 TI swinging-bucket rotor (k factor of 124, Beckman Coulter) by centrifugation at 120,000g for at least 4 h at 4 °C.
-
Discard the supernatant by decanting or pipetting and resuspend the individual pellets of NV material that are at the bottom of each of the second six centrifugation tubes in a small volume (30 to 100 μl) of PBS. Transfer samples to microcentrifuge tubes.
PAUSE POINT The gradient-purified lEV (fractions 1–6) or sEV (fractions 1–6) and NV (fractions 7–12) samples can be used immediately or stored at 4°C for a short duration. If the samples are extracted in lysis buffer, they can be stored long term at −80 °C.
Troubleshooting
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 17 | Low lEV-P yield | Low cell density when the conditioned medium was collected The lEV-P was lost after 10,000g centrifugation |
Starve cells when 70– 80% confluent Carefully collect and transfer the supernatant once the spin is complete The pellet may be firmly attached to the tube so it may be necessary to dislodge and solubilize it using the pipette tip. Simply rinsing the tube may not be sufficient |
| 35, 37, 38 | Low yields of sEV-P, exomere and supermere | Low cell density when the conditioned medium was collected The sEV-P and NVEPs were lost during the concentration step due to filter membrane leakage The sEV-P and NVEPs were lost during the concentrate recovery step due to high-speed centrifugation The sEV-P, exomere and supermere were lost after ultracentrifugation at 167,000g for 4 h and 16 h for sEV-Ps and exomeres and 367,000g for 16 h for supermeres |
Starve cells when 70– 80% confluent Save the flow through and keep it at 4 °C and re-concentrate it with a new filter device Spin the concentrate in the collection cup at 1000g Carefully collect and transfer the supernatant once the spin is complete The pellet may be firmly attached to the tube so it may be necessary to dislodge and solubilize it using the pipette tip. Simply rinsing the tube may not be sufficient |
| 37, 38 | Very high yield of exomeres and supermeres from cell culture | The cell-conditioned media harvested from cells contain bovine serum | Shift to serum-free media 48 h before harvest of cell-conditioned media and wash supermeres in PBS by 367,000g centrifugation |
| 49 | Low lEV- or sEV yield | The initial input of lEV-Ps or sEV-Ps is too low as the sample will be dispersed across several fractions Insufficient dilution of the harvested iodixanol-containing fractions in PBS may cause a decrease in the recovered material after the wash step |
Increase the amount of input lEV-P or sEV-P material Increase the volume of PBS used to dilute the fractions for the wash step |
|
Box 3 Step 6 |
Low yield of EV and NVEP derived from human plasma | Blood was kept for too long at room temperature or 4 °C The plasma was not diluted enough before EV and NVEP isolation making the plasma sample too viscous and dense thus preventing efficient pelleting The sEV-P, exomere and supermere were lost after ultracentrifugation at 167,000g for 4 h and 16 h for sEV-Ps and exomeres and 367,000g for 16 h for supermeres |
Human blood needs to be processed at room temperature within 2 h after collection Dilute the plasma at least 10- fold before isolation of EV and NVEPs Carefully collect and transfer the supernatant once the spin is complete The pellet may be firmly attached to the tube so it may be necessary to dislodge and solubilize it using the pipette tip Simply rinsing the tube may not be sufficient |
|
Box 3 Step 19 |
Albumin was not sufficiently depleted Low yield of albumin-depleted samples |
Too much sample was placed on the column. The column was saturated Residual albumin bound to the column after elution The column was not washed enough with albumin binding buffer, some of the albumin depleted samples still bind the column |
Reduce the input load, each column can only bind about 2 to 3 mg of albumin Wash the column several more times with albumin binding buffer to elute residual albumin Wash the column several times with albumin binding buffer to release more samples |
Timing
Step 1–6, cell growth and medium collection, variable (depending on the number of dishes with cells that will be used for medium collection)
Steps 7–17, lEV-P Isolation, 2 h (depending on the size of the preparation)
Step 18, media filtration 5–10 min
Steps 19–26, media concentration using concentrator, variable (depending on the volume of the media collected and the host cells)
Steps 27–30 media concentrate recovery,5 – 10 min
Steps 31–35, isolation of sEV-Ps from cell-conditioned media, 8 – 9 h
Steps 36–37, isolation of exomeres from cell-conditioned media, 33 – 34 h
Step 38, isolation of supermeres from cell-conditioned media, 16 – 17 h
Steps 39–51, High resolution iodixanol gradient fractionation of crude EV samples, 1 d
Box 3: isolation sEV-Ps, exomeres and supermeres from human plasma, and albumin depletion, 3 d, 4.5 h
Anticipated results
We have provided a comprehensive protocol for the sequential isolation of lEVs, sEVs, NV fractions, exomeres and supermeres from cells and human plasma. We describe in step-by-step detail the use of differential ultracentrifugation, filtration, concentration, and high-resolution density-gradient fractionation to obtain pure EVs, exomeres and supermeres. Representative AFM images of these distinct fractions derived from DiFi cells are shown in Fig. 5a. Similar samples isolated from other cell lines, and plasma are expected to have broadly similar morphology as those from DiFi cells. Proteomic analyses have revealed that lEVs, sEVs, NV fractions, exomeres and supermeres have distinct proteome profiles9,10,15. Representative immunoblots of select proteins in the distinct fractions are shown in Fig. 5b. Characteristically, the EV protein flotillin-1 is restricted to the gradient-purified sEVs and the vault protein MVP is restricted to the NV fractions. The expression of the metabolic enzyme LDHA is high in both exomeres and supermeres while the metabolic enzyme ENO2 is highly enriched in the supermeres (Fig. 5b). Metabolic enzymes are a category of proteins enriched in exomeres and supermeres (Fig. 5c). Characteristically, the specific EV marker proteins including CD9, CD81, CD63 and CD73 will also be enriched in the sEVs (Fig. 5d) while Transforming Growth Factor Beta Induced (TGFBI) and HSPA13 are highly expressed in exomeres and supermeres (Fig. 5e).
Fig. 5. Anticipated results.
a, Representative fluid-phase atomic force microscopy (AFM) topographic images of all the fractions derived from DiFi cells. sEVs (top left), NV fractions (top right), exomeres (bottom left) and supermeres (bottom right). Twenty microliters of isolated sEVs, NV fractions, exomeres and supermeres are diluted 1:1 with PBS and then incubated over (3-aminopropyl) triethoxysilane (AP)-modified mica substrates (Ted Pella Inc.) for 3 min. The substrates were washed twice with 50 μl of PBS to remove unbound particles and imaged in PBS at RT. Scale bars, 100 nm. b, Representative immunoblots of the proteins FLOT1, MVP, LDHA and ENO2 in distinct fractions obtained from DiFi cells. Equal amounts of proteins from each fraction are used. Exom, exomere: NV, non-vesicular; Super, supermere; WCL, whole cell lysate. c, Heatmap of normalized spectral counts for select proteins and enzymes involved in glycolysis in sEVs, exomeres and supermeres from DiFi cells. sEVs, NV fractions, exomeres and supermeres are isolated and fractionated as described in the protocol (steps 1–51). Equal amounts of proteins from each fraction are used for proteomic analysis and selected metabolic proteins and enzymes were further analyzed. d, Representative immunoblots of CD9, CD81, CD63, FASN, ACLY and HSPA13 in distinct fractions obtained from DiFi cells (left), and immunoblots for CD73 in fractions obtained from indicated cell lines (right). Equal amounts of proteins from each fraction are used. e, Representative immunoblots of TGFBI and HSPA13 in distinct fractions obtained from indicated cell lines. Equal amounts of proteins from each fraction are used. The images (panels a-e) are modified from Zhang et al.15.
In our modified protocol, we first isolate the crude lEV-P, sEV-P, exomeres and supermeres sequentially. However, crude lEV-Ps and sEV-Ps are heterogenous and include both EVs and contaminating NV material9. Performing additional high-resolution 12–36% iodixanol density gradient fractionation of crude EV-P samples with bottom loading of samples successfully separates the EVs from NV components as assessed by electron microscopy and immunoblotting ( Figs. 5 - 6). Fractions from the gradient can be either pooled to represent purified EVs and NV fractions, respectively (Fig. 6a,c) or each fraction can be analyzed individually (Fig. 6b). For crude plasma sEV-P samples, performing the high-resolution density gradient fractionation partitions HDL particles and AGO2 complexes in the NV fractions away from the purified sEVs (Fig. 6c). By using this optimized protocol, cargo that has previously been claimed to be in exosomes or EVs can be assigned to their correct extracellular compartment9,15.
Fig. 6. High-resolution density gradient fractionation separates small extracellular vesicles from non-vesicular components.
a, Negative stain transmission electron microscopy (TEM) of pooled sEV (low density) and NV (high density) fractions derived from Gli36 cells obtained from high-resolution density gradients. The purified sEVs display the expected cup-shaped morphology for EVs while the NV fractions display few distinct structures. b, Density-gradient fractionation of crude small EV pellet (sEV-Ps) derived from DKO-1 cells. After flotation of the sample in high-resolution iodixanol gradients (12 – 36%), equal volumes of each fraction are loaded on SDS-PAGE gels, and membranes were immunoblotted with indicated antibodies. NV, non-vesicular; sEV, small EV. c, High-resolution density gradient fractionation of crude human plasma sEV-Ps to obtain purified sEV and NV fractions. Immunoblots of plasma samples from three normal human individuals (P1–3) following fractionation. ApoA1 (marker for HDL particles) and AGO2 are enriched in the NV fraction while CD81, CD63 and CD9 (markers for sEVs) are enriched in the sEV fraction. The images (panels a-c) are modified from Jeppesen et al.9.
DiFi cells have been used as a standard for our isolation method. A standard production lot of DiFi consists of 80 culture dishes (15 cm) with approximately 1.34 × 108 cells per dish at time of harvest. The typical protein yield is approximately 4 mg for sEV-P, 2.5 mg for exomeres and 7 mg for supermeres. Yields and ratios of different fractions derived from other type of cells are listed (Table 1)15. The yields for sEV-Ps, exomeres and supermeres can vary based on host cells, growth conditions, number of culture dishes, etc. The yields presented in Table 1 are from cells that are switched to, and incubated in, serum-free media for the purpose of harvesting cell-conditioned media for isolation of EVs and NVEPs. If serum-containing media is used, even if this media has been depleted for bovine EVs, the presence of bovine serum proteins is a likely contaminant in exomere and supermere samples. Washing the supermeres in PBS by including an extra 367,000g step may improve the result, but the measured protein yield will likely be an overestimation of supermere content due to the presence of bovine serum proteins.
EV and NVEP isolation from human plasma is complicated due to the inherent complexity of blood. The yield of sEV-P samples from plasma is lower relative to exomeres and supermeres when compared to the yield from cells in culture. One of the reasons for this is the high abundance of albumin and other soluble blood proteins present in plasma samples that will contaminate EV and NVEP samples. With the albumin-depletion step, the protein yield of exomeres dropped to about 1/3 while the yield of supermeres dropped to about ¼ (Fig. 7a–c). Albumin depletion dramatically enriched low abundance proteins in exomeres and supermeres, which would be otherwise masked in plasma. The albumin-depletion step may aid in biomarker discovery for plasma-derived nanoparticles such as exomeres and supermeres. As an example, TGFBI is the most abundant proteins in supermeres derived from DiFi cells and it can also be detected in plasma-derived NVEPs by ELISA15. However, it was not possible to detect TGFBI by immunoblotting in supermeres derived from CRC patient plasma without albumin depletion, likely due to high levels of albumin contamination in the supermere samples. After albumin depletion, TGFBI was clearly detectable in supermeres by immunoblotting (Fig. 7d), confirming that removal of albumin can help reveal proteins that may serve as potential biomarkers for a variety of diseases.
Fig. 7. Depletion of albumin from sEV-Ps, exomeres and supermeres isolated from CRC patient plasma.
The extracellular samples are isolated from a CRC patient plasma as described in the protocol (Box 3) with concentrations of 4.8, 78.3 and 83.3 μg/μl proteins for sEV-Ps, exomeres and supermeres respectively. Equal volumes (40 μl) of each fraction are diluted 5-fold in albumin binding buffer and applied to the column for albumin depletion using the Albumin Depletion Kit (Abcam) as described in the protocol (Box 3). Equal volume (12 μl) of input (pre-albumin depletion), albumin depleted samples (two flow-throughs, FT1 and FT2) and eluted albumin samples (two elutions, E1 and E2, mainly containing albumin) are analyzed by 4–12% SDS-PAGE and stained by Coomassie blue. a, albumin depletion from sEV-Ps. b, albumin depletion from exomeres. c, albumin depletion from supermeres. d, Immunoblot analysis of TGFBI levels in albumin-depleted supermeres derived from colorectal cancer patient plasma. Albumin was depleted in supermeres as described above, and analyzed by immunoblotting for TGFBI expression. Equal amount (30 μg) of pre- and after albumin-depleted supermeres are used for immunoblot analysis with the antibodies indicated. Research conducted as part of this protocol complies with all the relevant ethical regulations. The use of the human samples was approved by the Vanderbilt University Medical Center Institutional Review Board (IRB; IRB nos 161529 and 151721). NS, non-specific background.
Supplementary Material
Acknowledgements
The work was supported by NCI R35 CA197570, UG3 241685, P01 CA229123 and P50 236733 to R.J.C. The authors acknowledge the generous support of the Nicholas Tierney GI Cancer Memorial Fund.
Footnotes
Related Links
Key references using this protocol
Jeppesen, DK. et al. Cell 177, 428–445 (2019): https://doi.org/10.1016/j.cell.2019.02.029
Zhang, Q. et al. Cell Reports 27, 940–954 (2019): https://doi.org/10.1016/j.celrep.2019.01.009
Zhang, Q. et al. Nature Cell Biology 23, 1240–1254 (2021): https://doi.org/10.1038/s41556–021-00805–8
Competing interests
The authors declare no competing interests.
Data availability
All data used to generate protein expression heatmaps are provided in the supporting primary research article by Zhang et al15.
References
- 1.Yates AG, et al. In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo: Part II: Pathology: Part II: Pathology. J Extracell Vesicles 11, e12190 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates AG, et al. In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo: Part I: Health and Normal Physiology: Part I: Health and Normal Physiology. J Extracell Vesicles 11, e12151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Niel G, et al. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat Rev Mol Cell Biol 23, 369–382 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Roefs MT, Sluijter JPG & Vader P. Extracellular Vesicle-Associated Proteins in Tissue Repair. Trends Cell Biol 30, 990–1013 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 20, 332–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113, E968–977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Niel G, D’Angelo G. & Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Willms E, Cabanas C, Mager I, Wood MJA & Vader P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol 9, 738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeppesen DK, et al. Reassessment of Exosome Composition. Cell 177, 428–445 e418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, et al. Transfer of Functional Cargo in Exomeres. Cell Rep 27, 940–954 e946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu M, Martin-Jaular L, Lavieu G. & Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Cheng L. & Hill AF Therapeutically harnessing extracellular vesicles. Nat Rev Drug Discov (2022). [DOI] [PubMed] [Google Scholar]
- 13.Jeppesen DK, Zhang Q, Franklin JL & Coffey RJ Are Supermeres a Distinct Nanoparticle? J Extracell Biol 1(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosar JP, Cayota A. & Witwer K. Exomeres and Supermeres: monolithic or diverse? J Extracell Biol 1(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nature Cell Biology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tosar JP, Witwer K. & Cayota A. Revisiting Extracellular RNA Release, Processing, and Function. Trends Biochem Sci 46, 438–445 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K, Wong DK, Luk FS, Kim RY & Raffai RL Isolation of Plasma Lipoproteins as a Source of Extracellular RNA. Methods Mol Biol 1740, 139–153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tulkens J, De Wever O. & Hendrix A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc 15, 40–67 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Borg EGF, Liaci AM, Vos HR & Stoorvogel W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles 9, 1791450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, et al. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat Biomed Eng 5, 678–689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boing AN, et al. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 3(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahoo S, et al. Therapeutic and Diagnostic Translation of Extracellular Vesicles in Cardiovascular Diseases: Roadmap to the Clinic. Circulation 143, 1426–1449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheruvanky A, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol 292, F1657–1661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clos-Sansalvador M, Monguio-Tortajada M, Roura S, Franquesa M. & Borras FE Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur J Cell Biol 101, 151227 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Hinger SA, et al. Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Sci Rep 10, 15804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onodi Z, et al. Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography From Blood Plasma. Front Physiol 9, 1479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vergauwen G, et al. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J Extracell Vesicles 10, e12122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, et al. Angiotensin-converting Enzyme 2-containing Small Extracellular Vesicles and Exomeres Bind the Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein. Gastroenterology 160, 958–961 e953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cvjetkovic A, Lotvall J. & Lasser C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 3(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhondt B, et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J Extracell Vesicles 9, 1736935 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhondt B, Lumen N, De Wever O. & Hendrix A. Preparation of Multi-omics Grade Extracellular Vesicles by Density-Based Fractionation of Urine. STAR Protoc 1, 100073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeppesen DK, et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles 3, 25011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimi N, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci 75, 2873–2886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zonneveld MI, et al. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J Extracell Vesicles 3(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H. & Lyden D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat Protoc (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojmar L, et al. Extracellular vesicle and particle isolation from human and murine cell lines, tissues, and bodily fluids. STAR Protoc 2, 100225 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshino A, et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 182, 1044–1061 e1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller L, Hong CS, Stolz DB, Watkins SC & Whiteside TL Isolation of biologically-active exosomes from human plasma. J Immunol Methods 411, 55–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostenfeld MS, et al. miRNA profiling of circulating EpCAM(+) extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles 5, 31488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clancy JW, Boomgarden AC & D’Souza-Schorey C. Profiling and promise of supermeres. Nat Cell Biol 23, 1217–1219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucotti S, Kenific CM, Zhang H. & Lyden D. Extracellular vesicles and particles impact the systemic landscape of cancer. EMBO J 41, e109288 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateescu B, et al. Phase 2 of extracellular RNA communication consortium charts next-generation approaches for extracellular RNA research. iScience 25, 104653 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeppesen DK, et al. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics 14, 699–712 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Mitchell JP, Court J, Mason MD, Tabi Z. & Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J Immunol Methods 335, 98–105 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Higginbotham JN, et al. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles 5, 29254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thery C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skliar M. & Chernyshev VS Imaging of Extracellular Vesicles by Atomic Force Microscopy. J Vis Exp (2019). [DOI] [PubMed] [Google Scholar]
- 49.Rikkert LG, Nieuwland R, Terstappen L. & Coumans FAW Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles 8, 1555419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachurski D, et al. Extracellular vesicle measurements with nanoparticle tracking analysis - An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J Extracell Vesicles 8, 1596016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olson BJ & Markwell J. Assays for determination of protein concentration. Curr Protoc Protein Sci Chapter 3, Unit 3 4 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Pham CV, et al. Bovine extracellular vesicles contaminate human extracellular vesicles produced in cell culture conditioned medium when ‘exosome-depleted serum’ is utilised. Arch Biochem Biophys 708, 108963 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Lehrich BM, Liang Y. & Fiandaca MS Foetal bovine serum influence on in vitro extracellular vesicle analyses. J Extracell Vesicles 10, e12061 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used to generate protein expression heatmaps are provided in the supporting primary research article by Zhang et al15.