Abstract
The gut-associated lymphoid tissue (GALT) faces a considerable challenge. It encounters antigens derived from an estimated 1014 commensal microbes and greater than 30 kg of food proteins yearly. It must distinguish these harmless antigens from potential pathogens and mount the appropriate host immune response. Local and systemic hyporesponsiveness to dietary antigens, classically referred to as oral tolerance, comprises a distinct complement of adaptive cellular and humoral immune responses. It is increasingly evident that a functional epithelial barrier engaged in intimate interplay with innate immune cells and the resident microbiota is critical to establishing and maintaining oral tolerance. Moreover, innate immune cells serve as a bridge between the microbiota, epithelium, and the adaptive immune system, parlaying tonic microbial stimulation into signals critical for mucosal homeostasis. Dysregulation of gut homeostasis and the subsequent disruption of tolerance therefore has clinically significant consequences for the development of food allergy.
Keywords: oral tolerance, microbiome, food allergy, dysbiosis
INTRODUCTION
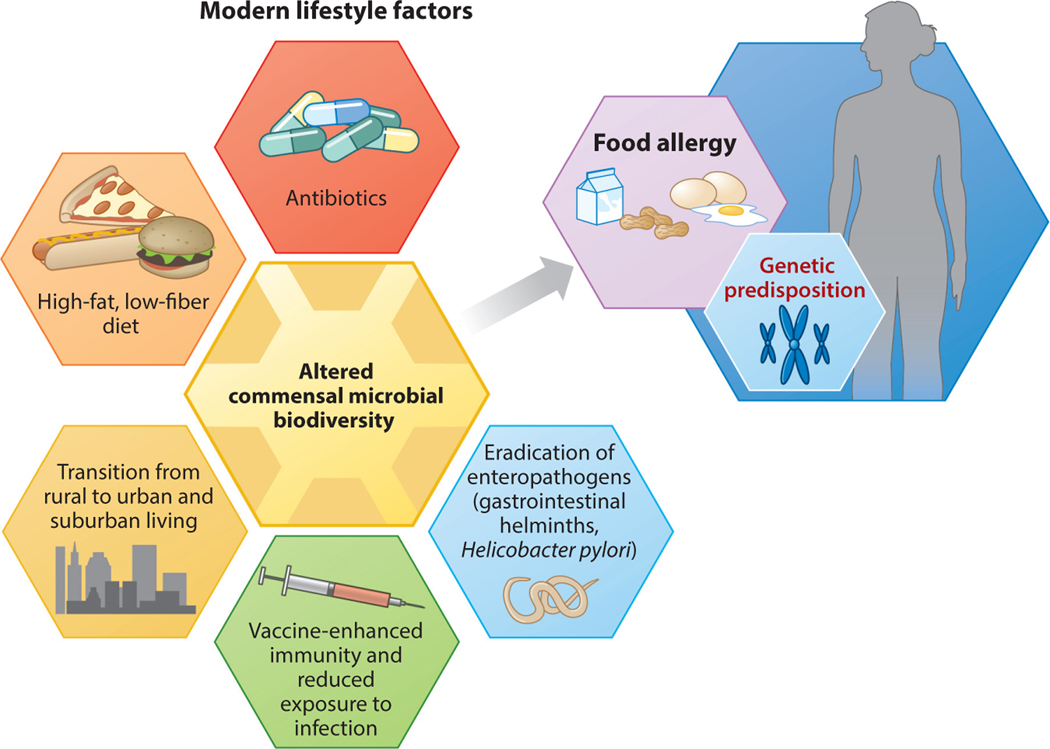
Food allergies are a major public health concern and represent an unmet clinical need (1, 2). An individual can become allergic to any food, at any time, but eight foods cause most reactions: milk, eggs, peanuts, tree nuts, soy, wheat, fish, and shellfish. Reactions can range from swelling and urticaria to life-threatening anaphylactic shock. No treatment other than strict avoidance is currently available for the 15 million Americans who suffer from food allergies. These numbers represent a marked generational increase in disease prevalence that has been noted in industrialized societies worldwide. The rising rate of food allergies parallels increases in what have been called the diseases of Western society, including obesity, diabetes, asthma, autism, and inflammatory bowel disease (among others). How do we account for this kind of generational change? Food allergies often present as part of a constellation of allergic diseases, referred to as the atopic (or allergic) march (3). Atopic dermatitis appears first in infancy, followed by food allergies between ages 2 and 5 years. Asthma and allergic rhinitis typically arise at school age. The increased prevalence of food allergy has followed an earlier rise in asthma and other allergic diseases in heavily industrialized nations (4, 5). A number of theories have been proposed to explain the rising prevalence of allergic disease (Table 1). The hygiene hypothesis initially linked the environment to allergic disease (6). It posited that improved cleanliness and household conditions reduced exposure to infectious disease and increased susceptibility to allergy. In particular, infection in early childhood due to “unhygienic contact” with older siblings or transmitted prenatally from mothers infected by their older children protected against the development of hay fever (6). We and others first suggested a critical role for immunoregulatory signals from commensal bacteria in the regulation of allergic hyperreactivity (7, 8). This led to a reformulation of the hygiene hypothesis as the “old friends” or the “biodiversity” hypotheses of allergy, which propose that changes in the environment, diet, and lifestyle associated with Westernized, industrialized countries have altered the diversity of the gut and skin microbiomes (9–11). Industrialized populations are exposed to both prescribed antibiotics, which dramatically alter the microbiome (12), and residual antibiotics used in agribusiness to enhance livestock growth (reviewed in 13). Modern Western diets contain large quantities of highly processed, high-fat, low-fiber foods which also cause shifts in microbial communities (14, 15). Epidemiologic studies point to a decreased prevalence of food allergies among populations where parasitic helminth infection is endemic compared to populations where helminth infection is rare (16). Increased microbial exposure in rural (particularly farming) populations suggests that higher biodiversity contributes to protection against disease (17). The available evidence therefore indicates that the loss of beneficial symbiotic relationships between humans, parasites, bacteria, and other microbes acquired throughout human evolution has increased the risk for developing atopic diseases (Figure 1). The timing and route of first exposure to food allergens also seem to play a role. The dual allergen hypothesis suggests that sensitization is promoted by allergen contact with skin and prevented by ingestion of food allergens early in life (18). We have proposed that tolerance to dietary antigen (and prevention of food allergy) requires both a food-antigen-specific regulatory response and a commensal-bacteria-induced intestinal-epithelial-barrier-protective response (19). In this review, we explore the many ways in which the microbiome regulates the response to dietary antigens.
Table 1.
Theories to explain the development and rising prevalence of food allergy
| Purpose of theory | Hypothesis | Reference | Summary |
|---|---|---|---|
| To explain the rising prevalence of food allergy | Hygiene hypothesis | 6 | In large families, infection in early childhood due to “unhygienic contact” with family members prevents the development of atopic disease. Shrinking family size and decreased infection exposure leads to increases in atopic disease development. |
| Balanced microbiota and microbial stimulation hypotheses | 7, 8 | Antibiotic use and dietary differences in industrialized countries have upset normal commensal gastrointestinal microbial communities, disrupting tonic microbial signaling through microbial pattern recognition receptors and preventing the development of tolerogenic mucosal immune networks that protect against allergic hyperreactivity. | |
| Old friends and biodiversity hypotheses | 9–11 | Changes in living environment, diet, and lifestyle associated with industrialized, Westernized countries impact commensal microbial diversity in gut and on skin, disrupting the immunoregulatory function of the microbiota at these sites, thus predisposing to allergic sensitization. | |
| To explain the failure of oral tolerance and development of allergic sensitization | Dual-allergen exposure hypothesis | 18 | Low-dose cutaneous exposures to food predispose to allergic sensitization while early ingestion of higher doses of food proteins leads to oral tolerance |
| Barrier regulation hypothesis of allergic sensitization | 19 | Twenty-first-century lifestyle factors have depleted allergy-protective bacterial populations in the intestinal mucosa that are required to maintain epithelial barrier integrity and limit allergen access to the systemic circulation. |
Figure 1.
Elements of a modern, industrialized lifestyle trigger shifts in the commensal microbiota, predisposing to the development of food allergy in genetically susceptible individuals. Adapted from Reference 20 with permission from Springer Nature.
COMMENSAL BACTERIA PROTECT AGAINST FOOD ALLERGY
Commensal Bacteria Regulate Antigen Presentation in the Gut
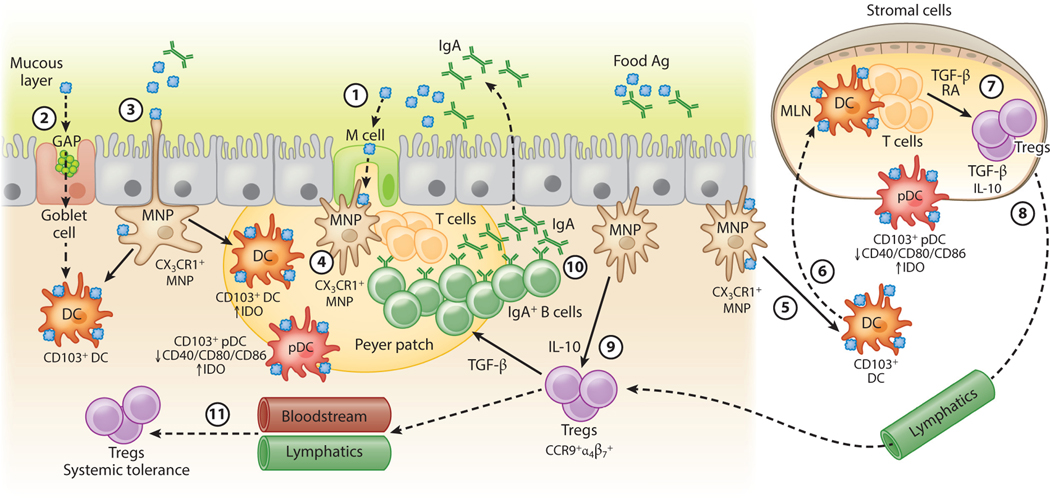
The paradigmatic view has been that the primary mechanism regulating tolerance to dietary antigen is the induction of food-antigen-specific regulatory T cells (Tregs) (21). Tolerogenic responses to luminal antigens depend on the translocation of these antigens across the gut epithelial barrier. M (microfold) cells, specialized intestinal epithelial cells located above collections of small intestinal submucosal lymphoid follicles called Peyer patches (PPs), are critical for the transcytosis of particulate antigens and macromolecules from the gut lumen into the submucosa (22, 23). Recent work has shown that goblet cell–associated antigen passages (GAPs) also play an important role in transferring luminal antigens to antigen-presenting cells (APCs) (24, 25). Multiple, functionally distinct subsets of APCs, including dendritic cells (DCs) and mononuclear phagocytes (MNPs) capture these transported antigens in the subepithelial dome (SED) of the follicle-associated epithelium (26). Antigen-loaded DCs migrate from the SED to the mesenteric lymph nodes (MLNs), which drain the small intestine. Presentation of antigen to naive T cells in the presence of the vitamin A metabolite retinoic acid (RA) favors TGF-β−dependent conversion to Foxp3+ Tregs (27–29). Concomitant upregulation of the gut-homing receptors CCR9 and α4β7 (30) allows these committed Tregs to home back to the lamina propria (LP) and expand under the influence of IL-10 produced by resident MNPs (31). Some Tregs exit the mucosa via the lymph or bloodstream to promote systemic tolerance (Figure 2).
Figure 2.
Tolerance to dietary antigen: the food-antigen-specific Treg paradigm. Luminal antigens are translocated across the gut epithelial barrier via M cells (①) or GAPs (②). CX3CR1+ lamina propria–resident MNPs sample luminal antigens; some form transepithelial dendrites that penetrate gut epithelia (③). Different populations of antigen-presenting cells, including tolerogenic IDO-expressing CD103+ DCs and pDCs reside in the subepithelial dome below M cells (④). CX3CR1+ MNPs transfer luminal antigens to CD103+ DCs and pDCs (⑤) that traffic to the draining MLNs (⑥) and display antigen to naive T cells in the presence of TGF-β and RA, favoring the generation of food-antigen-specific Tregs (⑦) that traffic through lymphatics (⑧) back to the lamina propria in a CCR9- and α4β7-dependent manner. MNP-derived IL-10 promotes Treg proliferation (⑨). TGF-β produced by Tregs regulates B cell antibody class switching to IgA; IgA is transported across the epithelial barrier via the polymeric immunoglobulin receptor (pIgR, not shown) into the intestinal lumen, where it acts to exclude luminal food antigens (⑩). Proliferating Tregs eventually exit the gut and enter the circulation to promote systemic tolerance to dietary antigen (⑪). Dashed lines represent the movement/trafficking of cells from one area to another. Solid lines represent the production of factors by a cell; the transfer of materials from one cell to another; or how chemical factors or cells influence the behavior of other cells. Abbreviations: Ag, antigen; DC, dendritic cell; GAP, goblet cell–associated antigen passage; IDO, indoleamine 2,3-dioxygenase; M, microfold; MLN, mesenteric lymph node; MNP, mononuclear phagocyte; pDC, plasmacytoid DC; RA, retinoic acid; Treg, regulatory T cell.
What is driving allergic, rather than tolerogenic, responses to food antigens? The role of the microbiota in regulating both antigen uptake and antigen presentation is beginning to be unraveled. In the small intestine, where nutrients (and food antigens) are absorbed, MNPs resident in the LP express the chemokine receptor CX3CR1; some form transepithelial dendrites that penetrate gut epithelia and sample luminal antigens (32, 33). Microbial stimulation of intestinal epithelial cell Toll-like receptors (TLRs) and signaling through the adaptor molecule MyD88 upregulate the numbers of these DC extensions in the small intestine (34). Both antigen presentation and IL-10 production by CX3CR1+ MNPs are required for oral tolerance (35); genetic ablation of either MHC-II or IL-10 in CX3CR1+ MNPs abrogates the induction of tolerance, although it is not yet clear whether the same cell must perform both functions. In the absence of MHC-II expression on CX3CR1+ MNPs, the induction of dietary-antigen-specific Tregs is hampered (35). Interestingly, the commensal microbiota regulates the tolerance-inducing capabilities of this MNP subset. CX3CR1+ MNPs from antibiotic-treated mice lose the ability to produce IL-10 (35). The enteric microbiota therefore appears to be critical to the antigen presentation function of CX3CR1+ APCs in the gut.
Notably, rather than migrating directly to lymphoid tissues, CX3CR1+ MNPs transfer their antigenic cargo to CD11c+CD103+ DCs in a manner dependent on gap junction proteins like Connexin-43 (36). Gut epithelial cells that produce TGF-β and RA create a microenvironment that drives DCs to express CD103 (37). These CD103+ DCs then migrate from the LP through afferent lymphatic vessels in a CCR7-dependent manner to present antigen to naive T cells in the MLNs. MLN stromal cells are conditioned by the intestinal microbiota to express higher levels of the enzyme critical for RA production, RALDH, compared to peripheral lymph nodes; RA derived from these cells triggers expression of the gut-homing molecules α4β7 and CCR9 on activated T cells (38, 39). Mice deficient in CCR9, or the integrins α4β7 or MAdCAM-1, fail to develop tolerance to orally delivered antigen (31, 40). Multiple DC subsets have been described in the MLNs. Elegant lineage-depletion studies confirm that CD103+ DCs are critical for oral tolerance (41). CD103+CD11b− DCs expressing IRF8 seem to be the most potent producers of TGF-β and RA (41).
TGF-β is critical to the regulation of both cellular and humoral immunity in the gut (42, 43). It is produced by multiple gut-associated immune cells, including DCs and Tregs, as well as by intestinal epithelial cells (44). Interestingly, inflammation can induce expression of the integrin αvβ8 by Foxp3+ Tregs (45). This integrin, in addition to other metalloproteinases and integrins, cleaves the latency-associated peptide that binds the inactive form of TGF-β, converting it to an active form that binds to TGFβRII in the TGF-βR complex and regulates host immunity (44). LP CD103+ DCs express high levels of αvβ8 integrin, which can activate TGF-β to generate Foxp3+ Tregs. Expression of αvβ8 integrin depends on RA and TGF-β as well as bacterial signaling through the TLR adaptor protein MyD88. Boucard-Jourdin and colleagues (46) showed that mice deficient in TGF-β signaling or fed a vitamin A–deficient diet had reduced β8 integrin expression. β8 integrin expression was similarly reduced in mice treated with antibiotics and in MyD88-deficient mice. In a chemical-injury-based colitis model, the gut microbiota drives proinflammatory cytokine production to modulate retinaldehyde dehydrogenases, which are critical to the synthesis of RA (47). Although this has yet to be shown in an oral tolerance or food allergy model, this finding raises the intriguing possibility that microbes influence colonic LP Treg numbers by modulating RA and TGF-β levels in the enteric microenvironment.
Other work has shown that CD103+ DCs expressing indoleamine 2,3-dioxygenase (IDO), an enzyme involved in tryptophan metabolism, can push CD4+ T cells toward a Foxp3+ regulatory phenotype. Blocking IDO expression in vivo hinders the development of antigen-specific Tregs and impairs oral tolerance induction (48). IDO digests tryptophan into the metabolite kynurenine, and the kynurenine:tryptophan ratio can be used as a marker of IDO activity. In subjects with food allergy, the serum kynurenine:tryptophan ratio is significantly reduced compared to that in healthy controls (49), suggesting that dysregulated IDO activity may hinder the development of tolerance to dietary antigens. Tryptophan is also a substrate for trillions of gut microbes that synthesize serotonin and other metabolically active compounds. Multiple studies have demonstrated reductions in tryptophan metabolism and diminished diversity of tryptophan and indole metabolites in germfree and antibiotic-treated mice compared to specific-pathogen-free (SPF) controls (50–52). One could postulate that gut dysbiosis and the resultant alterations in host serum concentrations of tryptophan and indole-containing compounds impact IDO expression in DCs, which in turn may hinder DCs’ ability to induce antigen-specific Foxp3+ Tregs. Another DC subset, plasmacytoid DCs (pDCs), has also been shown to express IDO (50) and varying levels of gut-homing receptor molecules like α4β7, CCR9, and CD103 whether they reside in the GALT or in systemic tissues like spleen and peripheral blood (53). In contrast to CD103+ DCs, pDCs express low levels of costimulatory molecules like CD40, CD80, and CD86, hampering their ability to stimulate antigen-specific CD4+ T cell proliferation and suggesting a tolerance-promoting role for pDCs in the gut (53). Using a mouse model, Uto and colleagues (53) showed that MLN pDCs induced antigen-specific Foxp3+ Tregs in the presence of TGF-β and upregulated expression of the Aldh1a2 gene that encodes RALDH, an enzyme needed to generate RA. Foxp3+ Tregs that have homed to the intestines can differentiate into Foxp3− T follicular helper cells in the PPs and promote the generation of IgA-producing B cells (54). The role of IgA antibodies in facilitating oral tolerance is not well defined, but feeding dietary antigen does generate food-specific IgA antibodies that may promote oral tolerance through immune exclusion (55). Interestingly, recent work has shown that much of the commensal microbiota is coated with IgA, particularly in the small intestine, where IgA is predominantly produced (56, 57). Whether and how IgA coating of commensal bacteria contributes to tolerance to dietary antigens has not yet been determined.
Commensal microbes also influence Treg development directly through microbial fermentation products, like butyrate, a short-chain fatty acid (SCFA) produced from dietary fiber predominantly by Clostridia, a class of mucosa-associated Firmicutes (58, 59). Comparative nuclear magnetic resonance–based metabolome analysis showed a positive correlation between colonic Treg cell numbers and luminal concentrations of SCFAs (59). In particular, butyrate stimulated Treg differentiation under a number of conditions, which were associated with histone H3 acetylation at the Foxp3 promoter and conserved, noncoding sequences (58, 59). These findings suggest that a class of commensal bacteria can promote Treg development through epigenetic regulation of the signature Treg transcription factor Foxp3. Moreover, colonic-microbiota-induced Foxp3+ Tregs also express RORγt, the transcription factor that canonically controls the Th17 pathway (60, 61). Mice deficient in RORγt+Foxp3+ Tregs show increased Th2 responses in both oxazolone colitis and helminth infection models, suggesting that these type 3 Tregs are required for the regulation of Th2 immunity (61). What, however, is the specificity of these Tregs, and how do they regulate tolerance to food antigens? Bacteria-induced Tregs with bacteria-specific T cell receptors have been described in the colonic LP (62, 63). Although Foxp3+ Tregs are depleted in the colons of germfree mice, they are found in numbers comparable to SPF mice in the small intestines. Elegant studies with germfree mice fed an elemental (antigen-free) diet demonstrated that Foxp3+Tregs with specificity for dietary antigens dominate in the small intestine (64). Interestingly, most of these small intestine Tregs develop after weaning to solid food and are essential to suppress immunity to dietary antigens. Taken together the data suggest that both bacteria- and food-antigen-induced Foxp3+ Tregs cooperate to prevent allergic responses to food.
Allergic Effector Cells Are Responsive to Bacterial Stimuli
Classical food allergy (and atopic disease in general) is characterized by the generation of antigen-specific IgE antibody responses. Allergen activation at epithelial barrier surfaces elicits the production of epithelial alarmins including TSLP, IL-33, and IL-25, which stimulate type 2 innate lymphoid cells (ILC2s) to produce Th2 cytokines and prime DCs to elicit allergen-specific immunity (65). B cells are licensed to class switch to IgE in a Th2 cytokine microenvironment rich in IL-4, IL-13, and IL-5 (19). Class switching to allergen-specific IgE leads to sensitization and is a requirement for the development of IgE-mediated food allergy. Subsequent exposure to allergen cross-links IgE bound to the FcεRI on allergic effector cells, like mast cells and basophils, that become decorated with IgE generated during sensitization. The symptoms associated with an acute allergic reaction arise following this cross-linking and subsequent degranulation, with the release of inflammatory mediators like histamine, other vasoactive amines, lipid mediators, and cytokines. This response encompasses fast-acting chemical mediators of the immediate hypersensitivity response and the synthesis of additional cytokines and chemokines that contribute to later phases of the allergic inflammatory response (66).
Commensal bacteria regulate allergic effector cell numbers at sites of allergic inflammation. In an allergic airway inflammation model, increased numbers of basophils accumulated in the airways of germfree mice compared to SPF mice. Germfree mice also demonstrated increased airway hyperresponsiveness following intranasal ovalbumin challenge compared to their SPF counterparts (67). Antibiotic-treated and germfree mice have higher resting serum IgE levels and an increased frequency of circulating basophils (68). Moreover, Hill and colleagues (69) have shown that the absence of MyD88 in B cells triggers a rise in IgE and circulating basophils. Human subjects with hyper-IgE syndrome due to loss-of-function mutations in the gene encoding dedicator of cytokinesis 8 (DOCK8) experience frequent infections, increased risk of atopic dermatitis, on average tenfold increases in serum IgE levels, and higher frequencies of circulating basophils compared to the general population (69). A correlation between elevated serum IgE and elevated circulating basophil numbers in the setting of impaired commensal microbial signaling has led some to postulate the existence of a commensal bacteria–IgE–basophil axis of allergic inflammation, particularly since IgE has been shown to shape elements of granulocyte homeostasis (70). Consistent with this model, Hill et al. (69) demonstrated that in anti-IgE-treated mice, antibiotic treatment did not increase circulating basophil numbers as it did in control animals. In addition, commensal microbial signals influenced basophil development by limiting the proliferation of bone marrow–derived basophil precursor populations in a manner dependent on the IL-3 receptor (IL-3R) and IgE. Thus, commensal microbial signaling, in part through IgE, modulates the development of allergic effector cells like basophils and the generation of the Th2 cytokine environment.
A Bacteria-Induced Barrier-Protective Response is Required to Prevent Allergic Responses to Food
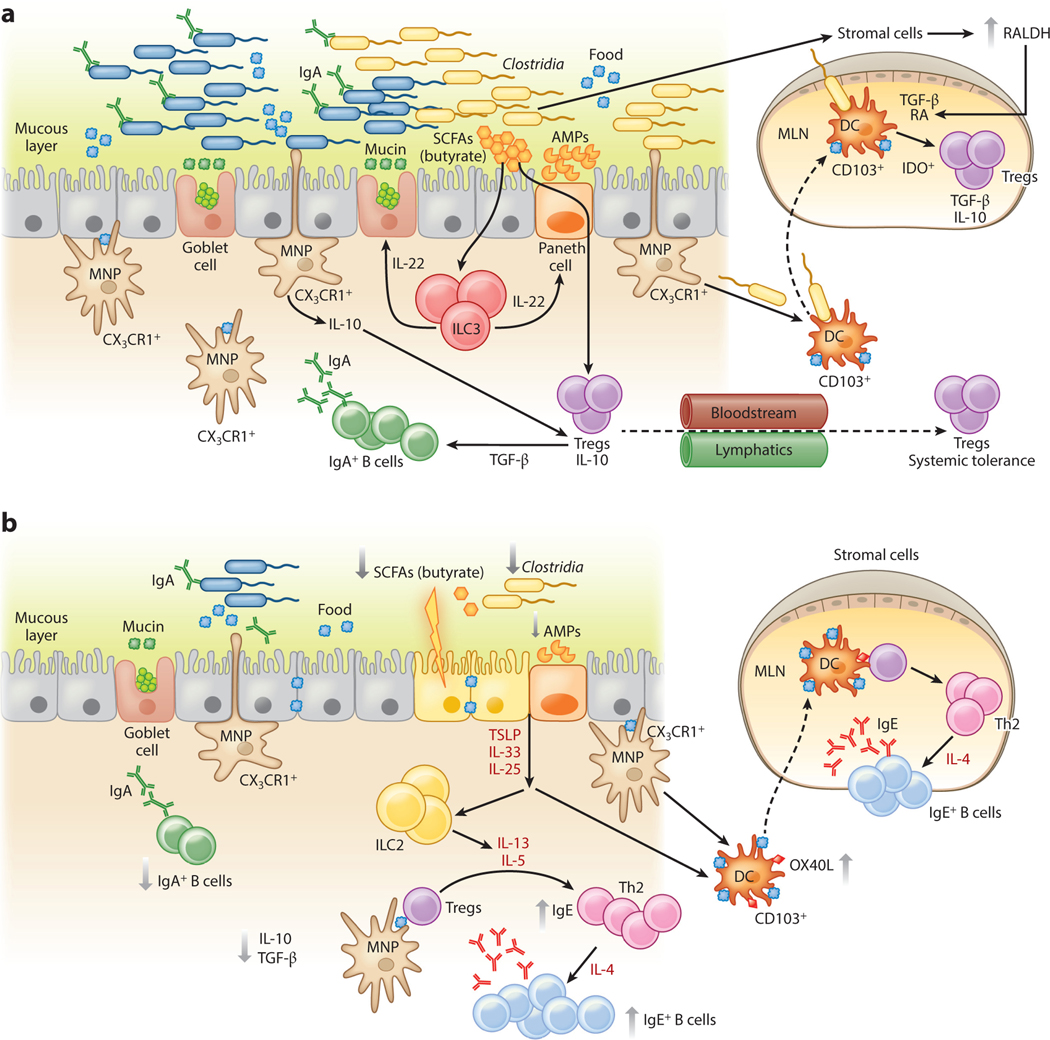
Physiologically, tolerance to dietary antigens results in the development of neutralizing, noninflammatory IgG or IgA humoral immune responses. IgA, the most abundant isotype at mucosal surfaces, is critical to the maintenance of homeostasis. Its functions include regulating bacterial adherence or translocation, sampling luminal antigens, immune exclusion, and influencing the composition of the intestinal microbiota (56). Early studies in germfree mice suggested that intestinal bacteria or microbial products like lipopolysaccharide (LPS) were important for the generation of tolerance to dietary antigens (71–73). In particular, Kiyono et al. (72) showed that oral tolerance could not be induced in LPS-hyporesponsive C3H/HeJ mice. After Sampson and colleagues described a model of systemic anaphylaxis to peanut in C3H/HeJ mice (74), we showed that this strain’s susceptibility to food allergy was linked to its inability to signal via TLR4 (75). We hypothesized that the TLR4 ligand originated from commensal bacteria and demonstrated that reducing the commensal bacterial load in TLR4-sufficient neonatal mice using a cocktail of broad-spectrum antibiotics induced an allergic response equivalent to that seen in TLR4-mutant mice (75). Subsequent murine model studies from our laboratory have shown that both germfree mice and mice treated with a cocktail of antibiotics, beginning preweaning, are highly susceptible to allergic sensitization to food (76). To identify allergy-protective bacterial populations, we selectively colonized germfree mice with representatives of bacterial orders (Bacteroidales and Clostridiales) numerically predominant in the murine gut. All mice were intragastrically sensitized with peanut plus the mucosal adjuvant cholera toxin. Using this approach, we identified mucosa-associated sporeforming Firmicutes in the Clostridia class as the taxa responsible for protection against allergic sensitization (76). Sensitization to a food allergen was blocked in antibiotic-treated mice that received a Clostridia-containing microbiota (76). Microarray analysis of intestinal epithelial cells isolated from the colonized gnotobiotic mice identified a novel innate mechanism by which Clostridia protect against sensitization to dietary antigens. We found that Clostridia colonization stimulated ILC3s in the colonic LP to produce the barrier-protective cytokine IL-22. IL-22 fortified epithelial barrier function by regulating the secretion of mucus by goblet cells and the production of Paneth cell antimicrobial peptides (76, 77). These IL-22-dependent responses reduced the access of orally administered dietary antigen to the systemic circulation and protected against allergic sensitization (76). In mice lacking a Clostridia-induced barrier-protective response, the immunodominant peanut allergens Ara h 2 and Ara h 6 were readily detectable in serum by ELISA postgavage. Their detection by ELISA indicates that these food proteins resisted proteolytic degradation in the gut; this may be a feature common to the dominant food allergens (78). In the barrier regulation hypothesis of allergic sensitization to food we proposed that a bacteria-induced barrier-protective response is required to reduce allergen access to the systemic circulation and prevent allergic responses to food (19) (Figure 3). Clostridia, however, do not signal directly via TLR4 since they do not bear LPS. What, then, does LPS have to do with it (79)? Interestingly, other work has identified a role for LPS variants in protection against autoimmune and allergic disease (80). Finnish and Estonian children have a higher incidence of autoimmune and allergic disease than children with similar genetic ancestry in Russian Karelia. Examination of fecal samples from each group from birth until three years of age showed that the microbiota of the Russian children was significantly less diverse than that of the Finnish or Estonian children during the first year of life. Escherichia coli, with its hexa-acylated form of lipid A (one component of LPS) dominated in the healthy Russian children and promoted endotoxin tolerance (80). By contrast, the allergy-prone Finnish and Estonian children had a higher abundance of Bacteroides, which carry a penta- or tetra-acylated form of lipid A and are unable to effectively mediate endotoxin tolerance, promoting inflammatory responses in later life (80). These data suggest that multiple distinct bacteria-induced immunostimulatory pathways are involved in the prevention of allergic responses to food.
Figure 3.
An epithelial barrier in equilibrium with commensal bacteria is required for protection against allergic sensitization. (a) In healthy individuals, Clostridia (and possibly other allergy-protective commensal bacteria) maintain epithelial barrier integrity by stimulating ILC3s to produce IL-22. IL-22 promotes the production of mucus from goblet cells and antimicrobial peptides from Paneth cells and reduces the ability of food allergens to gain access to the systemic circulation. Clostridial metabolites, including SCFAs, directly induce Treg development. Treg-derived TGF-β favors local IgA antibody production, and circulating Tregs promote systemic tolerance. (b) Twenty-first-century lifestyle factors like antibiotics and high-fat, low-fiber diets promote microbial dysbiosis, predisposing to allergic sensitization. Depletion of Clostridia leads to loss of IL-22-dependent barrier functions and decreased concentrations of SCFAs, impairing barrier integrity and increasing allergen access. The stressed epithelium produces alarmins like TSLP, IL-33, and IL-25, which promote the generation of a Th2 immune response in part by stimulating ILC2s to produce Th2 cytokines. CD103+ DCs in a TSLP-rich microenvironment upregulate OX40L expression and traffic to MLNs or interact with lamina propria–resident T cells, where they stimulate the generation of antigen-specific Th2 cells producing IL-4 that triggers antibody class switching to IgE. Adapted from Reference 19 with permission from Cell Press. Abbreviations: AMP, antimicrobial peptide; DC, dendritic cell; ILC3, type 3 innate lymphoid cell; MLN, mesenteric lymph node; MNP, mononuclear phagocyte; RA, retinoic acid; SCFA, short-chain fatty acid; Treg, regulatory T cell.
Healthy Infants Harbor Intestinal Bacteria That Protect Against Food Allergy
Humans have coevolved with their microbiota for millennia. Colonization with a founder microbiota occurs at birth; lactobacilli from the mother’s vaginal tract are the dominant founder bacteria (81, 82). A highly ordered ecological succession then follows, with taxa emerging later dependent on the metabolites and environment created by the founder bacteria (83). Cesarean delivery disrupts this process and introduces founder bacteria derived largely from the skin (81); some data suggest that this effect may be short-lived (84). It is clear, however, that the maternally acquired microbiota profoundly affects innate immune system development after birth (85, 86). Other work has hinted at a role for gut dysbiosis in the pathogenesis of food allergy (87–89). To begin to understand the influence of intestinal bacteria on the development of food allergies, we compared the composition and diversity of fecal samples collected from 4- to 5-month-old infants at the time of diagnosis with IgE-mediated cow’s milk allergy (CMA) with that of samples collected from age- and gender-matched healthy infants attending a vaccination clinic (90). All infants belonged to the same Neapolitan cohort. The microbiota of the healthy infants was dominated by taxa from the orders Lactobacillales, Bifidobacteriales, and Enterobacteriales (90), as shown in other reports on the infant microbiome (84). Lactobacilli and bifidobacteria are readily culturable bacteria that have been highly studied for potential probiotic activity (91). To our surprise the CMA infants had an adult-type microbiota, dominated by Bacteroidales and Clostridiales, as if the maturation of their microbiota had occurred at an accelerated pace (90). Dietary management with an extensively hydrolyzed casein formula (EHCF) containing the probiotic Lactobacillus rhamnosus GG (LGG) resulted in a higher rate of tolerance acquisition in infants with CMA than other formulas studied (90, 92, 93). When we examined the influence of this dietary intervention on the composition of the gut microbiota, we found that it did not result in an increased abundance of lactobacilli detectable in the feces of the treated infants. Instead, treatment with EHCF plus LGG, but not EHCF alone, was associated with changes in microbial community structure that included the expansion of particular subsets of butyrate-producing Clostridia (90). CMA infants treated with EHCF plus LGG also had significantly higher levels of butyrate detectable in their feces and an enhanced acquisition of tolerance to cow’s milk (90). While the small sample size examined is a limitation of this study, our findings have been corroborated in a much larger cohort (94). The Consortium of Food Allergy Research (CoFAR) collected fecal samples from 226 children enrolled in an observational study of milk allergy (94). They reported that certain taxa within Clostridia were enriched between ages 3 and 6 months in children with CMA whose disease had resolved by 8 years of age when compared to those with persistent disease.
To examine whether commensal bacteria play a causal role in protection against food allergy, we used human feces from four healthy and four IgE-mediated-CMA infant donors matched for age, gender, and mode of birth to colonize germfree mice. Stable colonization of germfree mice with human feces depends, in part, on the animals’ diet (95). The colonized mice were therefore fed the same formulas consumed by their human infant donors to maintain the donor-derived bacterial food source (in addition to plant-based mouse chow). All of the mice were then sensitized to the cow’s milk allergen β-lactoglobulin (BLG). In agreement with earlier studies (68, 76), germfree mice were highly susceptible to food-induced anaphylaxis. Following colonization, germfree mice that received bacteria from healthy infants were protected against sensitization to BLG (96). By contrast, germfree mice colonized with bacteria from CMA infants had significantly higher serum levels of BLG-specific IgE and experienced anaphylaxis after BLG challenge. Healthy and CMA-colonized mice exhibited differences in bacterial composition and unique transcriptome signatures in ileal intestinal epithelial cells (96). Correlation of ileal bacteria with differentially expressed genes in the ileum of healthy-colonized mice allowed us to identify a butyrate-producing clostridial species, Anaerostipes caccae, as a candidate allergy-protective bacterial taxa (96). Strikingly, monocolonization of germfree mice with this single species mimicked the effect of the healthy microbiota and was sufficient to protect against food allergy. Our findings demonstrate a causal role for the intestinal microbiota for protection against allergic responses to dietary antigens and indicate that targeted modulation of specific bacterial communities may represent a viable therapeutic strategy for food allergy (see below).
THE DUAL ALLERGEN EXPOSURE HYPOTHESIS AND ALLERGIC SENSITIZATION TO FOOD
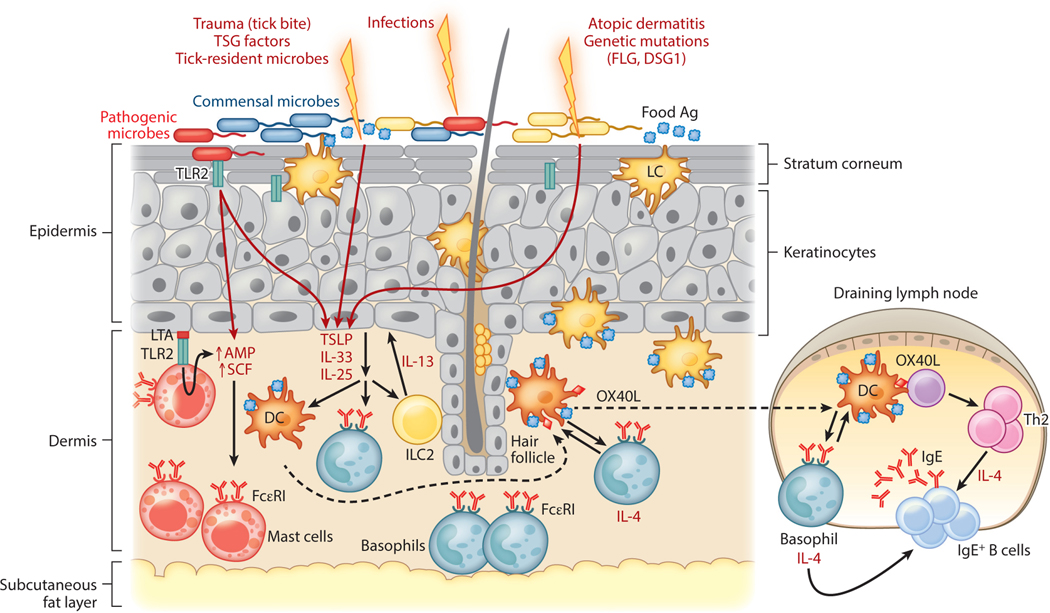
Atopic children frequently present with evidence of sensitization to a food antigen (typically peanut or tree nuts) with no previous history of ingestion of the food in question. This observation suggests that these sensitized children are exposed to food antigens through a route independent of oral ingestion. In the case of peanut allergy, sources for epicutaneous exposure include peanut allergen on tabletops, on hands after spreading peanut butter (even after rinsing hands with water) (97) and as a component in skin oils, and in household dust (98). As exposure to environmental sources of food allergen appears to be a risk factor, this has led some to hypothesize that while oral ingestion of food promotes immune tolerance to food antigens, epicutaneous exposure to food allergens promotes allergic sensitization (18) (Table 1). This hypothesis has been referred to as the dual allergen exposure hypothesis (18) (Figure 4).
Figure 4.
A compromised skin barrier elicits allergic sensitization to foods. A population of skin dendritic cells called Langerhans cells has the ability to sample high-molecular-weight antigens applied to the skin. Defects in skin barrier function due to trauma (e.g., tick bite, TSG factors), infection-mediated alterations in skin commensal microbiota, atopic dermatitis, and genetic defects in barrier proteins like FLG and DSG1 trigger the release of TSLP and other alarmins (IL-25, IL-33) by the skin epithelium. In the epidermis, LTA-mediated activation of the pattern recognition receptor TLR2 on keratinocytes induces secretion of SCF and TSLP and also upregulates AMP production by both mast cells and keratinocytes. These cytokines promote Th2 immune responses by stimulating Th2 cytokine production by ILC2s, upregulation of OX40L by antigen-laden DCs, and IL-4 production by basophils. IL-4 promotes antibody class switching to IgE and allergic sensitization once IgE binds Fcε receptors (FcεRI) on basophils and mast cells. Abbreviations: Ag, antigen; AMP, antimicrobial peptide; DC, dendritic cell; ILC2, type 2 innate lymphoid cell; LC, Langerhans cell; LTA, lipoteichoic acid; SCF, stem cell factor; TLR2, Toll-like receptor 2; TSG, tick salivary gland.
As with the intestinal epithelial barrier, it appears that allergen exposure through an intact skin epithelial barrier promotes tolerogenic responses to the allergen (99). Dioszeghy et al. (100) found that when ovalbumin (OVA) was applied to intact skin of mice it did not passively cross the skin epithelial barrier and was not systemically detectable. However, prolonged application to intact skin led to the internalization and transport of OVA to the draining lymph nodes by DCs in the stratum corneum and the induction of antigen-specific Tregs (100), providing conceptual support for the use of epicutaneous skin patches as an immunotherapeutic approach to allergen-specific desensitization (see below). Hair serves as a critical component of the skin’s physical barrier, and the hair follicle is home to dense populations of specialized APCs, including CD14+CX3CR1+ Langerhans DCs, with the ability to sample high-molecular-weight antigens applied to the skin (101). The trafficking of these cells is controlled by chemokines generated by hair follicles (102). Epicutaneous antigen exposure to intact skin generates Foxp3+ Tregs that traffic between the site of immunization and the draining skin lymph nodes (103, 104). In mouse models, these Tregs have been shown to suppress inflammatory hypersensitivity responses to epicutaneously applied protein antigens (100, 104). Skin barrier compromise leads to dysregulated immune responses to antigens introduced epicutaneously (99). Sensitization to food allergens like egg and peanut is far more common in children with eczematous skin and atopic dermatitis (105, 106). Up to 50% of individuals with atopic dermatitis have loss-of-function mutations in filaggrin (FLG), a protein that modulates the integrity of the stratum corneum and regulates the skin’s permeability to water and antigens (107). Brough et al. (108) found an increased risk of peanut sensitization and allergy in children with FLG gene mutations, while Venkataraman and colleagues (109) also showed an association between FLG mutations and food allergy in older children due to eczema and food allergen sensitization in early childhood. In mice orally sensitized with peanut and the adjuvant cholera toxin, epicutaneous peanut antigen delivered to intact skin muted Th2 cytokine production and the IgE response to oral challenge with peanut, whereas epicutaneous peanut antigen delivered through injured, tape-stripped skin reinforced systemic Th2 immune responses to peanut (110). When mice had hair removed with depilatory cream followed by epicutaneous exposure to peanut extract, they also developed Th2-biased IgE and IgG1 responses and anaphylaxis following intraperitoneal challenge with peanut (111). Skin barrier impairment is associated with increased expression of TSLP, the cytokine that links innate immune responses at epithelial barriers to Th2-skewed adaptive immune responses (112, 113). Epicutaneous food allergen exposure in the setting of increased TSLP production by inflamed skin is associated with an increase in serum allergen-specific IgE and an expansion of basophils in the skin that amplify Th2 allergen-specific cytokine responses (114). Recent findings by Hussain and colleagues (115) suggest that TSLP-expanded basophils work in concert with DCs through direct cell contact to promote a Th2 polarized microenvironment. This interaction enhances OX40L expression on DCs and stimulates IL-4 production by basophils critical to generate IgE-mediated hypersensitivity responses (115). Moreover, compared to wild-type mice, clinical signs of food allergy are significantly reduced after epicutaneous sensitization in mice whose basophils cannot produce IL-4. This underscores the critical role for IL-4 derived from TSLP-expanded basophils in the development of food allergy (115).
Commensal Bacteria Maintain a Tolerogenic Environment in the Skin
Analogous to their role in the maintenance of homeostasis at mucosal surfaces, commensal bacteria are critical for creating and maintaining a noninflammatory, tolerogenic environment in normal, intact skin (116). Skin bacteria in humans are found in distinct niches, like the hair follicles (117) that are also home to skin-resident DCs, putting skin bacteria proximal to key sentinels of the skin’s immunologic barrier. In normal human skin, microbial DNA can be amplified from several different cutaneous compartments including epidermis, hair follicles, dermis, and subcutaneous adipose tissue (118). The skin microbiome is dominated by members of the phylum Proteobacteria with some representation from Actinobacteria, Firmicutes, and Bacteroidetes (118). In a model system mapping out the antigen-specific response to Staphylococcus epidermidis, Scharschmidt et al. (116) found that microbial colonization of neonatal, but not adult, mice with this skin commensal within a two-week window was required to establish a healthy host-microbe interface regulated by highly activated skin Tregs. Colonization with S. epidermidis can also induce IL-17A+CD8+ T cells, in a manner dependent on CD103+ skin-resident DCs, that migrate to the epidermis to fortify skin barrier immunity against invasive pathogens (119). Infection with pathogenic microbes like Staphylococcus aureus, a common complication in individuals with severe atopic dermatitis, can cause significant skin barrier dysfunction (120). S. aureus produces exotoxins, proteases, and lipases that promote skin breakdown. Nakamura et al. (121) provided mechanistic insight into the relationship between S. aureus colonization and allergic skin disease with the demonstration that the S. aureus δ toxin activates mast cells and induces their degranulation. In children with atopic dermatitis, Jones et al. (120) found a link between S. aureus colonization and allergic responses to peanut, egg white, and cow’s milk. These studies suggest that through direct insult to the skin epithelium, S. aureus disrupts the tolerogenic presentation of food antigens epicutaneously and promotes the generation of Th2 immune responses to the antigen that lead to sensitization. Interestingly, an increase in the presence of S. aureus (and reduced diversity of other species) in the skin microbiome has been associated with flares of atopic dermatitis and worsening disease activity (122). Similarly, FLG-deficient skin has significantly less microbial diversity than FLG-sufficient skin, with underrepresentation of bacterial taxa that use histidine. Keratinocytes exhibit different cytokine and antimicrobial peptide responses depending on the composition of the bacteria to which they are exposed (123). In culture, human keratinocytes can be stimulated with TLR ligands and S. aureus membranes via TLR2 to produce TSLP, bridging innate immune responses at epithelial barriers to Th2-skewed adaptive immunity (124). In addition, compared with conventional mice, germfree mice have keratinocytes expressing abnormally low levels of stem cell factor (SCF), a factor critical for mast cell differentiation, and higher numbers of immature dermal mast cells with impaired ability to degranulate. Dermal mast cell maturation and degranulation can be rescued when staphylococcal lipoteichoic acid (LTA), absent in the epidermis and hair follicles of germfree mice, is injected into the skin. The microbe-derived LTA stimulates keratinocytes to produce SCF through a TLR2-dependent pathway (125). Skin mast cells can also express TLR2 and bind LTA directly, which can increase their activity against vaccinia viruses and enhance their production of antimicrobial peptides (126). Thus, alterations to microbial skin diversity in atopic dermatitis patients, by changing the cytokine and antimicrobial peptide makeup at the epithelial surface, may serve as another factor pushing the skin epithelial barrier to promote Th2-skewed responses to epicutaneously delivered antigens.
EOSINOPHILIC ESOPHAGITIS—AN EMERGING FOOD-RELATED DISEASE
Classic IgE-mediated food allergy is not the only food intolerance associated with dysbiosis. Eosinophilic esophagitis (EoE) is a chronic inflammatory disease characterized by eosinophilic inflammation of the esophagus with a peak count of ≥15 eosinophils per high-power field of esophageal biopsy tissue and an associated Th2 inflammatory response (127). On endoscopy, linear furrows with little to no vascularity, mucosal rings and exudates, strictures, and a narrow esophageal lumen can be found in both adult and pediatric patients with this condition, although up to one-third of pediatric patients may have a normal endoscopy despite clinically active EoE (128). Clinically, individuals present with symptoms related to esophageal dysfunction. Although no clear role for IgE has been described in the pathogenesis of EoE (127, 129), there are significant associations between genetic loci specific for EoE and loci for general atopic disease, which may act synergistically (130). IgE-mediated sensitization to ingested, inhaled, or epicutaneously introduced allergens is often present in subjects with EoE, even those with no history of anaphylaxis to food (127, 131, 132). Esophageal exposure to food, and in some individuals, to aeroallergens, triggers the disease. As with IgE-mediated food allergy, population-based studies point to a rise in the incidence of EoE starting in the early 2000s in Westernized societies (133, 134). The sharp rise in the incidence of EoE in the United States corresponds with widespread introduction of antibiotics in the mid-twentieth century and a decline in the prevalence of Helicobacter pylori infection (131, 135–137). Recent work shows that, like food allergy, early-life environmental factors (prenatal, intrapartum, and postnatal) influence susceptibility to EoE; positive risk factors include antibiotic treatment, cesarean delivery, maternal fever, preterm labor, and acid suppressant use in infancy (138). Postnatal dog exposure is a negative risk factor (138). Topical glucocorticoids and dietary therapy, including the six-food elimination diet (in which dairy, wheat, egg, peanuts/tree nuts, fish/shellfish, and soy, the six most common US allergens are removed from the diet) and elemental diets, are the current mainstays of treatment (127). It is tempting to speculate that the efficacy of elemental diets is due, in part, to diet-induced alterations in the esophageal microbiome.
Several challenges surround research into the esophageal microbiome and the relationship with EoE, including the current invasiveness and cost of obtaining endoscopic brushings or esophageal biopsies (131). While endoscopic techniques have been reported to be most effective at securing bacterial samples within the esophageal microbiome (139), there are ongoing attempts to make sampling and analysis of the esophageal microbiome less invasive using nonendoscopic techniques like the Esophageal String Test (140), a mesh capsule or cytosponge (141), and inflatable balloons to sample the upper gastrointestinal tract (131). However, contamination of samples obtained by these methods with the oral microbiota is common (131). 16S rRNA–targeted sequencing of esophageal biopsies (142) and esophageal mucosa and mucosal secretions collected with the Esophageal String Test (143) from patients with or without a diagnosis of EoE have identified over 300 bacterial species among the esophageal microbiota. The most common genera detected were Streptococcus, Prevotella, and Veillonella, with an increased abundance of members of the Haemophilus genus in patients with untreated EoE (143).
Barrier Dysfunction Underlies the Pathogenesis of Eosinophilic Esophagitis
Similar to IgE-mediated food allergy, a growing body of research has implicated a dysfunctional epithelial barrier in the pathophysiology of EoE. The esophagus, like the skin, comprises stratified squamous epithelium that protects underlying tissues in areas at high risk for mechanical or chemical injury (144). Subjects with mutations in skin barrier proteins like filaggrin or desmoglein-1 (DSG1) develop atopic dermatitis. DSG1 is an adherens junction protein and a desmosomal component linking the cell surface to the keratin cytoskeleton. In the absence of functional DSG1, allergic dermatitis is particularly severe (145). Interestingly, DSG1 is reduced in esophageal biopsies from EoE patients and was shown to be critical for esophageal epithelial integrity (146). Davis et al. (128) showed that IL-13, present in inflamed esophageal tissue in EoE, triggered the loss of DSG1 expression by enhancing production of CAPN14, an intracellular calcium-activated protease in the calpain family of proteases, whose overexpression in the esophageal epithelium reduced DSG1 levels, impairing epithelial barrier function. Recent work has identified a possible role for the serine protease inhibitor SPINK7, constitutively generated by differentiated esophageal squamous epithelium, in the pathogenesis of EoE (147). The absence of SPINK7 promoted proinflammatory responses in the esophagus that compromised barrier integrity, including dilated intercellular spaces and increased epithelial permeability, dysfunctional DSG1, and absent FLG expression. It also triggered a transcriptome signature consistent with allergic inflammation and stimulated TSLP release in esophageal cell lines stimulated via TLR3 (147). TLR-mediated TSLP production in the absence of SPINK7 suggests that esophageal microbial signaling in genetically susceptible individuals may influence esophageal epithelial barrier integrity. Future studies using germfree and gnotobiotic mouse models of EoE could begin to address the relationship between the esophageal microbiome and esophageal barrier function.
Human studies and mouse models suggest that invariant natural killer T (iNKT) cells are also involved in eosinophil recruitment to the esophagus in EoE. Lexmond and colleagues (148) observed significant upregulation of the human iNKT cell–associated invariant TCR Vα24, the MHC-I-like molecule CD1d, and the chemokine CXCL16, which has been associated with the recruitment of iNKT cells to mucosal surfaces, in esophageal biopsies from EoE patients. Studies in germfree mice have shown enhanced accumulations of iNKT cells in the colonic LP and lung and heightened morbidity in models of inflammatory bowel disease and allergic asthma in comparison to SPF mice (149). Moreover, in a murine model, treatment of neonatal mice with glycosphingolipids derived from the commensal bacterium Bacteroides fragilis protected these mice against oxazolone-induced colitis in adulthood (150). Colonization of germfree neonatal mice with a conventional microbiota induces epigenetic regulation of the CXCL16 promoter, decreasing hypermethylation and reducing CXCL16 production and iNKT cell recruitment (149, 150). These findings in the lower intestines raise the possibility that commensal microbiota in the upper gastrointestinal tract and esophagus may also regulate iNKT cell levels in the submucosa, which in turn can modulate eosinophil recruitment to this organ.
Whether the change in esophageal microbiota is predisposing individuals to EoE or whether the influx of eosinophils triggers alterations in the esophageal microbiota requires additional study. Treatments used to address dental disease and other gastrointestinal diseases like gastroesophageal reflux disease (GERD) may impact the esophageal microbiota and predispose patients to EoE. Alternatively, it is possible that current pharmacologic treatments used to control EoE, such as proton pump inhibitors and oral corticosteroids (127), also contribute to the quelling of the disease by altering the esophageal microbiota. Additional larger-scale studies in the vein of Benitez et al. (142) and Harris et al. (143) will be useful to expand our understanding of how treatments like antibiotics, proton pump inhibitors, and oral corticosteroids change the microbiota in those with EoE and those without. Such studies may also identify a microbial class (similar to Clostridia in food allergy) critical for the maintenance of a normal esophageal epithelial barrier and with the potential to prevent or treat EoE.
TICK MICROBIOTA AND MEAT ALLERGY
Alpha-gal mammalian meat allergy is characterized by a delayed allergic response to the ingestion of mammalian meat in subjects with circulating IgE specific for the carbohydrate galactose-α−1,3-galactose (alpha-gal) (151). In contrast to conventional food allergy responses, allergic reactions to alpha-gal in ingested mammalian meat are usually delayed at least 3 h and up to 8 h after ingestion. Reactions to alpha-gal may not occur with every allergen exposure. The ingestion of lipid-rich meats is associated with more consistent, severe reactions (152). The mechanisms underlying sensitization in alpha-gal allergy remain unclear. Alpha-gal-specific IgE rises in humans and mice following tick bites from the lone star tick, Amblyomma americanum, in the United States and bites from several other tick species across the globe (153, 154). Alpha-gal motifs are found in all nonprimate mammalian tissues (2) and in microbes, including commensal gut bacteria (155). One group has detected alpha-gal in the tick midgut (156), likely present following a blood meal, which may explain the mechanism of sensitization in humans. Yet, larval ticks with no prior blood meal can also induce alpha-gal-specific IgE (153).
High titers of T cell–independent natural human serum IgG antibodies against alpha-gal were first described by Galili and colleagues (157). Subsequent studies reported circulating levels of anti-alpha-gal-specific IgM and IgA as well (155). Interestingly non-IgE antibody isotypes directed against alpha-gal motifs play a role in acute organ rejection (2) and in protection against malaria driven by Plasmodium, which possesses alpha-gal epitopes (158). The high titers of alpha-gal-specific IgG, IgM, and IgA in the human circulation are likely driven by chronic antigenic stimulation by alpha-gal-containing commensal gut microbes (155, 158). It is unclear, however, how tick bites license alpha-gal-specific B cells to class switch and begin producing alpha-gal-specific IgE antibodies that predispose to red meat allergy.
One intriguing possibility, drawing on the dual allergen exposure hypothesis, is that tick bites introduce alpha-gal to the host epicutaneously while imposing multiple hits to the skin epithelial barrier. Tick mouthparts include chelicerae, which are barbed, telescoping structures effective at penetrating beyond the epidermis and anchoring into host skin, and a hypostome, which serves as a blood-drawing tube (159). During a blood meal, these tick mouthparts induce trauma to the skin. Another tick-mediated breach of skin barrier integrity may involve the disruption of the skin microbiota with the addition of bacterial orders like Rickettsiales, present in tick gut and saliva (160–162). Currently, there are no specific tick microbiota identified as critical for breaking or inducing tolerance to meat antigens. Future studies comparing the salivary microbiomes of the different tick species associated with alpha-gal sensitization may reveal a common microbe necessary and/or sufficient to induce IgE to alpha-gal. Profiling the microbial composition of biopsies of healthy skin and comparing it to tick-bitten skin from alpha-gal-allergic and nonallergic volunteers may allow for the identification of a microbial signature in the skin commonly associated with alpha-gal allergy.
Insults to the skin epithelial barrier may drive the production of TSLP, IL-33, and IL-25, predisposing skin DCs to induce Th2-skewed immune responses to tick-borne alpha-gal, leading to IgE-mediated sensitization (163). Upon reingestion of alpha-gal-containing mammalian meat, the sensitized host then mounts an allergic response. This model does not fully explain why the alpha-gal-sensitized host does not subsequently mount persistent IgE-mediated allergic responses to gut commensal microbial alpha-gal but selectively responds to mammalian-derived alpha-gal-containing glycoproteins and glycolipids. One explanation is that gut commensal alpha-gal remains sequestered in the local mucosal immune microenvironment, off-limits to the systemic immune system. By contrast, food proteins like alpha-gal-containing glycolipids and glycoproteins from mammalian meat can enter the bloodstream through the mesenteric and portal venous circulation, or in the case of dietary lipid, transit as chylomicrons in the lymphatics, eventually emptying into the systemic circulation (164).
Allergic sensitization to alpha-gal can be induced in wild-type and alpha-gal-deficient SPF C57Bl/6J mice injected intradermally with tick salivary gland extract (TSGE) from adult A. americanum (Aa) ticks. Alpha-gal-specific IgE levels rise following each intradermal TSGE injection. Both wild-type and alpha-gal-deficient mice generate an allergic response to oral challenge with pork sausage characterized by pruritus and drop in core body temperature. Peak allergic response is often seen more than 2 h after oral challenge, with allergic symptoms more exaggerated in alpha-gal-deficient mice than in wild-type mice (165). Pretreating these animals with broad-spectrum antibiotics prior to TSGE intradermal injection is a blunt-force approach to exploring the role of the intestinal and skin microbiota on the induction of alpha-gal-specific IgE. Recreating the alpha-gal sensitization model in a gnotobiotic mouse facility would allow for a more targeted approach to the question of whether the presence of particular bacteria hinders or enhances the development of alpha-gal-IgE and allergic reactions to mammalian meat.
HOPE ON THE HORIZON
The standard of care in treating food allergy is strict avoidance of known food allergens and, in the event of accidental exposure, epinephrine injection and oral or intravenous antihistamines (2). There are currently no curative or disease-modifying treatments for food allergy approved by national or international drug regulatory agencies. However, the significant public health and economic burden of food allergy has spurred considerable scientific and financial investment in developing new therapeutics for this condition. As the prevalence of food allergy began to rise in the late 1990s, the American Academy of Pediatrics (166) recommended that mothers of children at risk for allergy avoid consumption of allergenic foods during pregnancy and breast-feeding and exclude allergenic foods from their infants’ diets. More than 15 years later, a landmark study from Lack and colleagues led to the opposite recommendation; they showed that oral introduction of peanuts early in life significantly reduced the development of peanut allergy in high-risk infants (167). In addition to revising the recommendations regarding introduction of allergenic foods in infancy (168), these findings have heightened interest in allergen-specific immunotherapy. Immunotherapy strives to reset the aberrant immune response to food, abrogating Th2-skewed responses and promoting regulatory, tolerogenic responses (169). During allergen immunotherapy, escalating doses of target allergen are delivered to a subject until a maintenance dose is achieved. Immunotherapy can be oral (OIT), sublingual (SLIT), epicutaneous (EPIT), or subcutaneous. Currently, all food allergy therapeutics are investigational. However, OIT has shown the greatest efficacy, with rates of desensitization from 36% to 100% of subjects (compared to 10–70% for SLIT and 46–48% for EPIT) (169). With the results of phase 3 trials now reported (170), peanut OIT is closest to coming to market. Yet concerns surrounding the tolerability of OIT for all food allergy patients persist. Subjects on OIT typically experience mild adverse allergic reactions like mouth itch or abdominal discomfort, but systemic reactions or anaphylaxis do occur (171) and EoE may develop in up to 2.7% of patients following OIT initiation (172). Thus, emerging treatments for food allergy increasingly include adjuvant therapies designed to enhance or accelerate the tolerogenic immune response and/or reduce adverse allergic events during OIT (169). Anti-IgE therapy has been shown to increase the threshold of peanut sensitivity during oral peanut challenge nearly 18-fold (173). Multiple studies are exploring a role for monoclonal anti-IgE antibody omalizumab in rush and multiple-food OIT protocols (174–176). Early signals suggest that omalizumab enhances the efficacy of multiple-food OIT, allowing for safe, accelerated desensitization (177). Given the critical role of IL-4 production in the development of food allergy, there may be a role for dupilumab, a monoclonal antibody against IL-4Rα recently approved to treat moderate to severe atopic dermatitis, as a supplement to food immunotherapy. Some adjuncts to food immunotherapy, like the naturally occurring TLR4 agonist monophosphoryl lipid A and its synthetic version glucopyranosyl lipid A and TLR9 agonist CpG oligonucleotides (169), target microbial pattern recognition receptors, possibly harnessing tolerogenic mucosal immune networks to protect against allergic reactions (see microbial stimulation hypothesis in Table 1).
As emphasized in this review, the microbiota regulates all aspects of the development of tolerance to dietary antigen. There is now clear evidence for a causal protective role for the microbiota in food allergy (96). The identification of protective bacterial taxa, and their metabolites, in both asthma and food allergy models has spurred interest in the development of microbiome-modulating therapeutics to treat allergic disease (96, 178, 179). The first 1,000 days of life may be most crucial for prevention strategies, but adjunctive therapies pairing microbiome-modulating therapeutics with OIT or biologics (as discussed above) are also under consideration. The new group of live biotherapeutic bacteria described herein are quite different from the largely ineffective (but readily culturable) probiotics of the past. Therapeutic administration of obligate, oxygen-sensitive anaerobes, like Clostridia, will present many challenges for oral delivery. Combination approaches that include live bacteria (bugs as drugs), microbial metabolites, and prebiotic formulations may be most effective (180). For example, Clostridia ferment butyrate largely from insoluble dietary fiber. Mackay and colleagues have demonstrated that administration of dietary fiber can protect against food allergen sensitization in murine models (181). Butyrate is of particular interest because of its broad range of immunomodulatory activities. It modulates Treg function through histone deacetylase (HDAC) inhibition (58, 59); part of its efficacy is also related to its ability to signal via G protein–coupled receptors, particularly GPR43 and GPR109A (181, 182). Butyrate is also a critical source of metabolic energy for intestinal epithelial cells (183). Recent work has shown that butyrate induces colonocytes to consume oxygen through β-oxidation, generating and sustaining a locally hypoxic niche for butyrate-producing obligate anaerobes like Clostridia (184). These emerging data suggest that it may be possible to combine dietary fiber, butyrate-producing taxa, and orally administrable forms of butyrate to create feed-forward pathways that restore these lost allergy-protective bacteria to their former niche.
ACKNOWLEDGMENTS
We thank Drs. Taylor Feehley and Erin Steinbach for critical review of this manuscript. The Nagler and Iweala laboratories are funded by the National Institute of Allergy and Infectious Diseases. Additional support was received from the Sunshine Foundation (C.R.N.) and the UNC Food Allergy Initiative (O.I.I.).
DISCLOSURE STATEMENT
C.R.N. is President and Co-Founder of ClostraBio, Inc., a company developing microbiome-modulating therapeutics to prevent or treat food allergy.
LITERATURE CITED
- 1.Sicherer SH, Allen K, Lack G, Taylor SL, Donovan SM, Oria M. 2017. Critical issues in food allergy: a National Academies consensus report. Pediatrics 140:e20170194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iweala OI, Burks AW. 2016. Food allergy: our evolving understanding of its pathogenesis, prevention, and treatment. Curr. Allergy Asthma Rep 16:37. [DOI] [PubMed] [Google Scholar]
- 3.Spergel JM, Paller AS. 2003. Atopic dermatitis and the atopic march. J. Allergy Clin. Immunol 112:S118–27 [DOI] [PubMed] [Google Scholar]
- 4.Tordesillas L, Berin MC, Sampson HA. 2017. Immunology of food allergy. Immunity 47:32–50 [DOI] [PubMed] [Google Scholar]
- 5.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. 2016. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol 137:984–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strachan DP. 1989. Hay fever, hygiene, and household size. Br. Med. J 299:1259–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prioult G, Nagler-Anderson C. 2005. Mucosal immunity and allergic responses: lack of regulation and/or lack of microbial stimulation? Immunol. Rev 206:204–18 [DOI] [PubMed] [Google Scholar]
- 8.Noverr MC, Huffnagle GB. 2005. The ‘microflora hypothesis’ of allergic diseases. Clin. Exp. Allergy 35:1511–20 [DOI] [PubMed] [Google Scholar]
- 9.Rook GA. 2010. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin. Exp. Immunol 160:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rook GA, Lowry CA, Raison CL. 2013. Microbial ‘Old Friends’, immunoregulation and stress resilience. Evol. Med. Public Health 2013:46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, et al. 2012. Environmental biodiversity, human microbiota, and allergy are interrelated. PNAS 109:8334–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wypych TP, Marsland BJ. 2018. Antibiotics as instigators of microbial dysbiosis: implications for asthma and allergy. Trends Immunol. 39:697–711 [DOI] [PubMed] [Google Scholar]
- 13.Lambrecht BN, Hammad H. 2017. The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol 18:1076–83 [DOI] [PubMed] [Google Scholar]
- 14.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, et al. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS 107:14691–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maizels RM, McSorley HJ. 2016. Regulation of the host immune system by helminth parasites. J. Allergy Clin. Immunol 138:666–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ober C, Sperling AI, von Mutius E, Vercelli D. 2017. Immune development and environment: lessons from Amish and Hutterite children. Curr. Opin. Immunol 48:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lack G.2008. Epidemiologic risks for food allergy. J. Allergy Clin. Immunol 121:1331–36 [DOI] [PubMed] [Google Scholar]
- 19.Wesemann DR, Nagler CR. 2016. The microbiome, timing, and barrier function in the context of allergic disease. Immunity 44:728–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feehley T, Stefka AT, Cao S, Nagler CR. 2012. Microbial regulation of allergic responses to food. Semin. Immunopathol 34:671–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabst O, Mowat AM. 2012. Oral tolerance to food protein. Mucosal Immunol. 5:232–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraehenbuhl J-P, Neutra MR. 2000. Epithelial M cells: differentiation and function. Ann. Rev. Cell. Dev. Biol 16:301–32 [DOI] [PubMed] [Google Scholar]
- 23.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. 2013. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 6:666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, et al. 2012. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 483:345–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. 2015. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8:198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnardel J, Da Silva C, Wagner C, Bonifay R, Chasson L, et al. 2017. Distribution, location, and transcriptional profile of Peyer’s patch conventional DC subsets at steady state and under TLR7 ligand stimulation. Mucosal Immunol. 10:1412–30 [DOI] [PubMed] [Google Scholar]
- 27.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, et al. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med 204:1775–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. 2007. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med 204:1765–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, et al. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med 204:1757–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson-Lindbom B, Svensson M, Wurbel MA, Malissen B, Marquez G, Agace W. 2003. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J. Exp. Med 198:963–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, et al. 2011. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34:237–46 [DOI] [PubMed] [Google Scholar]
- 32.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, et al. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol 2:361–67 [DOI] [PubMed] [Google Scholar]
- 33.Niess JH, Brand S, Gu X, Landsman L, Jung S, et al. 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254–58 [DOI] [PubMed] [Google Scholar]
- 34.Chieppa M, Rescigno M, Huang AY, Germain RN. 2006. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med 203:2841–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M, Galan C, Hill AA, Wu WJ, Fehlner-Peach H, et al. 2018. Critical role for the microbiota in CX3CR1+ intestinal mononuclear phagocyte regulation of intestinal T cell responses. Immunity 49:151–63.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzini E, Massimiliano L, Penna G, Rescigno M. 2014. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity 40:248–61 [DOI] [PubMed] [Google Scholar]
- 37.Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. 2009. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. 2:340–50 [DOI] [PubMed] [Google Scholar]
- 38.Molenaar R, Greuter M, van der Marel AP, Roozendaal R, Martin SF, et al. 2009. Lymph node stromal cells support dendritic cell-induced gut-homing of T cells. J. Immunol 183:6395–402 [DOI] [PubMed] [Google Scholar]
- 39.Cording S, Wahl B, Kulkarni D, Chopra H, Pezoldt J, et al. 2014. The intestinal micro-environment imprints stromal cells to promote efficient Treg induction in gut-draining lymph nodes. Mucosal Immunol. 7:359–68 [DOI] [PubMed] [Google Scholar]
- 40.Cassani B, Villablanca EJ, Quintana FJ, Love PE, Lacy-Hulbert A, et al. 2011. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 141:2109–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esterhazy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. 2016. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral Treg cells and tolerance. Nat. Immunol 17:545–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagler-Anderson C.2001. Man the barrier! Strategic defenses in the intestinal mucosa. Nat. Rev. Immunol 1:59–67 [DOI] [PubMed] [Google Scholar]
- 43.Agace WW, McCoy KD. 2017. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46:532–48 [DOI] [PubMed] [Google Scholar]
- 44.Bauche D, Marie JC. 2017. Transforming growth factor beta: a master regulator of the gut microbiota and immune cell interactions. Clin. Transl. Immunol 6:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worthington JJ, Kelly A, Smedley C, Bauche D, Campbell S, et al. 2015. Integrin αvβ8-mediated TGF-β activation by effector regulatory T cells is essential for suppression of T-cell-mediated inflammation. Immunity 42:903–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boucard-Jourdin M, Kugler D, Endale Ahanda ML, This S, De Calisto J, et al. 2016. β8 integrin expression and activation of TGF-β by intestinal dendritic cells are determined by both tissue microenvironment and cell lineage. J. Immunol 197:1968–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, et al. 2016. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8+ T cell-mediated immunity in colorectal cancer. Immunity 45:641–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, et al. 2010. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 59:595–604 [DOI] [PubMed] [Google Scholar]
- 49.Buyuktiryaki B, Sahiner UM, Girgin G, Birben E, Soyer OU, et al. 2016. Low indoleamine 2,3-dioxygenase activity in persistent food allergy in children. Allergy 71:258–66 [DOI] [PubMed] [Google Scholar]
- 50.Van der Leek AP, Yanishevsky Y, Kozyrskyj AL. 2017. The kynurenine pathway as a novel link between allergy and the gut microbiome. Front. Immunol 8:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, et al. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. PNAS 106:3698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desbonnet L, Clarke G, Traplin A, O’Sullivan O, Crispie F, et al. 2015. Gut microbiota depletion from early adolescence in mice: implications for brain and behaviour. Brain Behav. Immun 48:165–73 [DOI] [PubMed] [Google Scholar]
- 53.Uto T, Takagi H, Fukaya T, Nasu J, Fukui T, et al. 2018. Critical role of plasmacytoid dendritic cells in induction of oral tolerance. J. Allergy Clin. Immunol 141:2156–67.e9 [DOI] [PubMed] [Google Scholar]
- 54.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, et al. 2009. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science 323:1488–92 [DOI] [PubMed] [Google Scholar]
- 55.Tordesillas L, Berin MC. 2018. Mechanisms of oral tolerance. Clin. Rev. Allergy Immunol 55:107–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC. 2018. IgA function in relation to the intestinal microbiota. Annu. Rev. Immunol 36:359–81 [DOI] [PubMed] [Google Scholar]
- 57.Bunker JJ, Bendelac A. 2018. IgA responses to microbiota. Immunity 49:211–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, et al. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature 504:451–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, et al. 2013. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature 504:446–50 [DOI] [PubMed] [Google Scholar]
- 60.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, et al. 2015. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349:993–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, et al. 2015. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349:989–93 [DOI] [PubMed] [Google Scholar]
- 62.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, et al. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478:250–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nutsch K, Chai JN, Ai TL, Russler-Germain E, Feehley T, et al. 2016. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep. 17:206–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim KS, Hong SW, Han D, Yi J, Jung J, et al. 2016. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351:858–63 [DOI] [PubMed] [Google Scholar]
- 65.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 14:141–53 [DOI] [PubMed] [Google Scholar]
- 66.Strait R, Morrist SC, Finkelman FD. 2004. Cytokine enhancement of anaphylaxis. Novartis Found. Symp 257:80–91 [PubMed] [Google Scholar]
- 67.Herbst T, Sichelstiel A, Schar C, Yadava K, Burki K, et al. 2011. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med 184:198–205 [DOI] [PubMed] [Google Scholar]
- 68.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. 2013. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 14:559–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, et al. 2012. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med 18:538–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hill DA, Artis D. 2013. The influence of commensal bacteria-derived signals on basophil-associated allergic inflammation. Gut Microbes 4:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sudo N, Sawamura S-A, Tanaka K, Aiba Y, Kubo C, Koga Y. 1997. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol 159:1739–45 [PubMed] [Google Scholar]
- 72.Kiyono H, McGhee JR, Wannemuehler MJ, Michalek SM. 1982. Lack of oral tolerance in C3H/HeJ mice. J. Exp. Med 155:605–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wannemuehler MJ, Kiyono H, Babb JL, Michalek SM, McGhee JR. 1982. Lipopolysaccharide (LPS) regulation of the immune response: LPS converts germfree mice to sensitivity to oral tolerance induction. J. Immunol 129:959–65 [PubMed] [Google Scholar]
- 74.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, et al. 2000. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J. Allergy Clin. Immunol 106:150–58 [DOI] [PubMed] [Google Scholar]
- 75.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. 2004. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J. Immunol 172:6978–87 [DOI] [PubMed] [Google Scholar]
- 76.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, et al. 2014. Commensal bacteria protect against food allergen sensitization. PNAS 111:13145–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sabat R, Ouyang W, Wolk K. 2014. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat. Rev. Drug Discov 13:21–38 [DOI] [PubMed] [Google Scholar]
- 78.Valenta R, Hochwallner H, Linhart B, Pahr S. 2015. Food allergies: the basics. Gastroenterology 148:1120–31.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feehley T, Belda-Ferre P, Nagler CR. 2016. What’s LPS got to do with it? A role for gut LPS variants in driving autoimmune and allergic disease. Cell Host Microbe 19:572–74 [DOI] [PubMed] [Google Scholar]
- 80.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, et al. 2016. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165:842–53. Erratum. 2016. Cell 165:1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, et al. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS 107:11971–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. 2011. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 140:1713–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, et al. 2011. Succession of microbial consortia in the developing infant gut microbiome. PNAS 108(Suppl. 1):4578–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. 2017. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med 23:314–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. 2015. The infant microbiome development: mom matters. Trends Mol. Med 21:109–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, et al. 2016. The maternal microbiota drives early postnatal innate immune development. Science 351:1296–302 [DOI] [PubMed] [Google Scholar]
- 87.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, et al. 2013. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol 131:201–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, et al. 2014. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol 80:2546–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hua X, Goedert JJ, Pu A, Yu G, Shi J. 2016. Allergy associations with the adult fecal microbiota: analysis of the American Gut Project. EBioMedicine 3:172–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, et al. 2016. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 10:742–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, et al. 2014. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol 11:506–14 [DOI] [PubMed] [Google Scholar]
- 92.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, et al. 2012. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J. Allergy Clin. Immunol 129:580–82 [DOI] [PubMed] [Google Scholar]
- 93.Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, et al. 2013. Formula selection for managment of children with cow milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. J. Pediatr 163:771–77 [DOI] [PubMed] [Google Scholar]
- 94.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, et al. 2016. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol 138:1122–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, et al. 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351:aad3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, et al. 2019. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perry TT, Conover-Walker MK, Pomes A, Chapman MD, Wood RA. 2004. Distribution of peanut allergen in the environment. J. Allergy Clin. Immunol 113:973–76 [DOI] [PubMed] [Google Scholar]
- 98.Brough HA, Santos AF, Makinson K, Penagos M, Stephens AC, et al. 2013. Peanut protein in household dust is related to household peanut consumption and is biologically active. J. Allergy Clin. Immunol 132:630–38 [DOI] [PubMed] [Google Scholar]
- 99.Matsumoto K, Saito H. 2013. Epicutaneous immunity and onset of allergic diseases—per-“eczema”tous sensitization drives the allergy march. Allergol. Int 62:291–96 [DOI] [PubMed] [Google Scholar]
- 100.Dioszeghy V, Mondoulet L, Dhelft V, Ligouis M, Puteaux E, et al. 2011. Epicutaneous immunotherapy results in rapid allergen uptake by dendritic cells through intact skin and downregulates the allergen-specific response in sensitized mice. J. Immunol 186:5629–37 [DOI] [PubMed] [Google Scholar]
- 101.Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, et al. 2008. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity 29:497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, et al. 2012. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol 13:744–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tomura M, Honda T, Tanizaki H, Otsuka A, Egawa G, et al. 2010. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J. Clin. Investig 120:883–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Majewska-Szczepanik M, Zemelka-Wiacek M, Ptak W, Wen L, Szczepanik M. 2012. Epicutaneous immunization with DNP-BSA induces CD4+ CD25+ Treg cells that inhibit Tc1-mediated CS. Immunol. Cell Biol 90:784–95 [DOI] [PubMed] [Google Scholar]
- 105.Hill DJ, Sporik R, Thorburn J, Hosking CS. 2000. The association of atopic dermatitis in infancy with immunoglobulin E food sensitization. J. Pediatr 137:475–79 [DOI] [PubMed] [Google Scholar]
- 106.Lack G, Fox D, Northstone K, Golding J, Avon Longitud. Study of Parents Child. Study Team. 2003. Factors associated with the development of peanut allergy in childhood. N. Engl. J. Med 348:977–85 [DOI] [PubMed] [Google Scholar]
- 107.O’Regan GM, Sandilands A, McLean WHI, Irvine AD. 2008. Filaggrin in atopic dermatitis. J. Allergy Clin. Immunol 122:689–93 [DOI] [PubMed] [Google Scholar]
- 108.Brough HA, Simpson A, Makinson K, Hankinson J, Brown S, et al. 2014. Peanut allergy: effect of environmental peanut exposure in children with filaggrin loss-of-function mutations. J. Allergy Clin. Immunol 134:867–75.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Venkataraman D, Soto-Ramirez N, Kurukulaaratchy RJ, Holloway JW, Karmaus W, et al. 2014. Filaggrin loss-of-function mutations are associated with food allergy in childhood and adolescence. J. Allergy Clin. Immunol 134:876–82.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mondoulet L, Dioszeghy V, Puteaux E, Ligouis M, Dhelft V, et al. 2012. Intact skin and not stripped skin is crucial for the safety and efficacy of peanut epicutaneous immunotherapy (EPIT) in mice. Clin. Transl. Allergy 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tordesillas L, Goswami R, Benede S, Grishina G, Dunkin D, et al. 2014. Skin exposure promotes a Th2-dependent sensitization to peanut allergens. J. Clin. Investig 124:4965–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leyva-Castillo JM, Hener P, Jiang H, Li M. 2013. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J. Investig. Dermatol 133:154–63 [DOI] [PubMed] [Google Scholar]
- 113.Leyva-Castillo JM, Hener P, Michea P, Karasuyama H, Chan S, et al. 2013. Skin thymic stromal lymphopoietin initiates Th2 responses through an orchestrated immune cascade. Nat. Commun 4:2847. [DOI] [PubMed] [Google Scholar]
- 114.Noti M, Kim BS, Siracusa MC, Rak GD, Kubo M, et al. 2014. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietin-basophil axis. J. Allergy Clin. Immunol 133:1390–99.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hussain M, Borcard L, Walsh KP, Pena Rodriguez M, Mueller C, et al. 2018. Basophil-derived IL-4 promotes epicutaneous antigen sensitization concomitant with the development of food allergy. J. Allergy Clin. Immunol 141:223–34.e5 [DOI] [PubMed] [Google Scholar]
- 116.Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, et al. 2015. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43:1011–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337:1115–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. 2013. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun 4:1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, et al. 2015. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520:104–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones AL, Curran-Everett D, Leung DYM. 2016. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J. Allergy Clin. Immunol 137:1247–48.e3 [DOI] [PubMed] [Google Scholar]
- 121.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, et al. 2013. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503:397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kong HH, Oh J, Deming C, Conlan S, Grice EA, et al. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22:850–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zeeuwen PL, Ederveen TH, van der Krieken DA, Niehues H, Boekhorst J, et al. 2017. Gram-positive anaerobe cocci are underrepresented in the microbiome of filaggrin-deficient human skin. J. Allergy Clin. Immunol 139:1368–71 [DOI] [PubMed] [Google Scholar]
- 124.Takai T, Chen X, Xie Y, Vu AT, Le TA, et al. 2014. TSLP expression induced via Toll-like receptor pathways in human keratinocytes. Methods Enzymol. 535:371–87 [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Lack G. 2017. Food allergy: unmet needs and new perspectives. J. Allergy Clin. Immunol. Pract 5:295. [DOI] [PubMed] [Google Scholar]
- 126.Wang Z, MacLeod DT, Di Nardo A. 2012. Commensal bacteria lipoteichoic acid increases skin mast cell antimicrobial activity against vaccinia viruses. J. Immunol 189:1551–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.O’Shea KM, Aceves SS, Dellon ES, Gupta SK, Spergel JM, et al. 2018. Pathophysiology of eosinophilic esophagitis. Gastroenterology 154:333–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, et al. 2016. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight 1:e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aceves SS. 2014. Food allergy testing in eosinophilic esophagitis: what the gastroenterologist needs to know. Clin. Gastroenterol. Hepatol 12:1216–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin LJ, He H, Collins MH, Abonia JP, Biagini Myers JM, et al. 2018. Eosinophilic esophagitis (EoE) genetic susceptibility is mediated by synergistic interactions between EoE-specific and general atopic disease loci. J. Allergy Clin. Immunol 141:1690–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.May M, Abrams JA. 2018. Emerging insights into the esophageal microbiome. Curr. Treat. Options Gastroenterol 16:72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roy-Ghanta S, Larosa DF, Katzka DA. 2008. Atopic characteristics of adult patients with eosinophilic esophagitis. Clin. Gastroenterol. Hepatol 6:531–35 [DOI] [PubMed] [Google Scholar]
- 133.Hruz P, Straumann A, Bussmann C, Heer P, Simon HU, et al. 2011. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J. Allergy Clin. Immunol 128:1349–50.e5 [DOI] [PubMed] [Google Scholar]
- 134.Prasad GA, Alexander JA, Schleck CD, Zinsmeister AR, Smyrk TC, et al. 2009. Epidemiology of eosinophilic esophagitis over three decades in Olmsted County, Minnesota. Clin. Gastroenterol. Hepatol 7:1055–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dellon ES, Peery AF, Shaheen NJ, Morgan DR, Hurrell JM, et al. 2011. Inverse association of esophageal eosinophilia with Helicobacter pylori based on analysis of a US pathology database. Gastroenterology 141:1586–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, et al. 2013. The oesophageal string test: A novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut 62:1395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spergel JM, Book WM, Mays E, Song L, Shah SS, et al. 2011. Variation in prevalence, diagnostic criteria, and initial management options for eosinophilic gastrointestinal diseases in the United States. J. Pediatr. Gastroenterol. Nutr 52:300–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jensen ET, Kuhl JT, Martin LJ, Rothenberg ME, Dellon ES. 2018. Prenatal, intrapartum and postnatal factors are associated with pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol 141:214–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gall A, Fero J, McCoy C, Claywell BC, Sanchez CA, et al. 2015. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLOS ONE 10:e0129055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fillon SA, Harris JK, Wagner BD, Kelly CJ, Stevens MJ, et al. 2012. Novel device to sample the esophageal microbiome—the esophageal string test. PLOS ONE 7:e42938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Elliott DRF, Walker AW, O’Donovan M, Parkhill J, Fitzgerald RC. 2017. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol. Hepatol 2:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, et al. 2015. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, et al. 2015. Esophageal microbiome in eosinophilic esophagitis. PLOS ONE 10:e0128346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maghsoudlou P, Eaton S, De Coppi P. 2014. Tissue engineering of the esophagus. Semin. Pediatr. Surg 23:127–34 [DOI] [PubMed] [Google Scholar]
- 145.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, et al. 2013. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat. Genet 45:1244–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, et al. 2014. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 7:718–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, et al. 2018. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci. Transl. Med 10:eaap9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, et al. 2014. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am. J. Gastroenterol 109:646–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Olszak T, An D, Zeissig S, Vera MP, Richter J, et al. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.An D, Oh SF, Olszak T, Neves JF, Avci FY, et al. 2014. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156:123–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Commins SP, Satinover SM, Hosen J, Mozena J, Borish L, et al. 2009. Delayed anaphylaxis, angioedema, or urticaria after consumption of red meat in patients with IgE antibodies specific for galactose-alpha-1,3-galactose. J. Allergy Clin. Immunol 123:426–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Steinke JW, Pochan SL, James HR, Platts-Mills TAE, Commins SP. 2016. Altered metabolic profile in patients with IgE to galactose-alpha-1,3-galactose following in vivo food challenge. J. Allergy Clin. Immunol 138:1465–67.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Commins SP, Platts-Mills TA. 2013. Tick bites and red meat allergy. Curr. Opin. Allergy Clin. Immunol 13:354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hashizume H, Fujiyama T, Umayahara T, Kageyama R, Walls AF, Satoh T. 2018. Repeated Amblyomma testudinarium tick bites are associated with increased galactose-alpha-1,3-galactose carbohydrate IgE antibody levels: a retrospective cohort study in a single institution. J. Am. Acad. Dermatol 78:1135–41.e3 [DOI] [PubMed] [Google Scholar]
- 155.Mangold A, Hercher D, Hlavin G, Liepert J, Zimmermann M, et al. 2012. Anti-alpha-Gal antibody titres remain unaffected by the consumption of fermented milk containing Lactobacillus casei in healthy adults. Int. J. Food Sci. Nutr 63:278–82 [DOI] [PubMed] [Google Scholar]
- 156.Hamsten C, Tran TAT, Starkhammar M, Brauner A, Commins SP, et al. 2013. Red meat allergy in Sweden: association with tick sensitization and B-negative blood groups. J. Allergy Clin. Immunol 132:1431–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Galili U, Rachmilewitz EA, Peleg A, Flechner I. 1984. A unique natural human IgG antibody with anti-alpha-galactosyl specificity. J. Exp. Med 160:1519–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, et al. 2014. Gut microbiota elicits a protective immune response against malaria transmission. Cell 159:1277–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Richter D, Matuschka FR, Spielman A, Mahadevan L. 2013. How ticks get under your skin: insertion mechanics of the feeding apparatus of Ixodes ricinus ticks. Proc. Biol. Sci 280:20131758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Qiu Y, Nakao R, Ohnuma A, Kawamori F, Sugimoto C. 2014. Microbial population analysis of the salivary glands of ticks: a possible strategy for the surveillance of bacterial pathogens. PLOS ONE 9:e103961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ponnusamy L, Gonzalez A, Van Treuren W, Weiss S, Parobek CM, et al. 2014. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl. Environ. Microbiol 80:354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, et al. 2015. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl. Environ. Microbiol 81:6200–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wilson JM, Schuyler AJ, Schroeder N, Platts-Mills TA. 2017. Galactose-alpha-1,3-galactose: atypical food allergen or model IgE hypersensitivity? Curr. Allergy Asthma Rep 17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang ZK, Yang YS. 2013. Upper gastrointestinal microbiota and digestive diseases. World J. Gastroenterol 19:1541–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Commins SP, Karim S. 2017. Development of a novel murine model of alpha-gal meat allergy. J. Allergy Clin. Immunol 139:AB193 [Google Scholar]
- 166.Am. Acad. Pediatr. Comm. Nutr. 2000. Hypoallergenic infant formulas. Pediatrics 106:346–49 [PubMed] [Google Scholar]
- 167.Du Toit G, Roberts G, Sayre PH, Bahson HT, Radulovic S, et al. 2015. Randomized trial for peanut consumption in infants at risk for peanut allergy. N. Engl. J. Med 372:803–13. Erratum. 2016. N. Engl. J. Med. 375:398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Togias A, Cooper SF, Acebal JD, Assa’ad A, Baker JR Jr., et al. 2017. Addendum guidelines for the prevention of peanut allergy in the United States: Report of the National Institute of Allergy and Infectious Disease—sponsored expert panel. Ann. Allergy Asthma Immunol 118:166–73 [DOI] [PubMed] [Google Scholar]
- 169.Virkud YV, Wang J, Shreffler WG. 2018. Enhancing the safety and efficacy of food allergy immunotherapy: a review of adjunctive therapies. Clin. Rev. Allergy Immunol 55:172–89 [DOI] [PubMed] [Google Scholar]
- 170.PALISADE Group Clin. Investig., Vickery BP, Vereda A, Casale TB, Beyer K, et al. 2018. AR101 oral immunotherapy for peanut allergy. N Engl. J. Med 379:1991–2001 [DOI] [PubMed] [Google Scholar]
- 171.Varshney P, Steele PH, Vickery BP, Bird JA, Thyagarajan A, et al. 2009. Adverse reactions during peanut oral immunotherapy home dosing. J. Allergy Clin. Immunol 124:1351–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lucendo AJ, Arias A, Tenias JM. 2014. Relation between eosinophilic esophagitis and oral immunotherapy for food allergy: a systematic review with meta-analysis. Ann. Allergy Asthma Immunol 113:624–29 [DOI] [PubMed] [Google Scholar]
- 173.Leung DY, Sampson HA, Yunginger JW, Burks AW Jr., Schneider LC, et al. 2003. Effect of anti-IgE therapy in patients with peanut allergy. N. Engl. J. Med 348:986–93 [DOI] [PubMed] [Google Scholar]
- 174.Nadeau KC. 2015. Multi Immunotherapy to Test Tolerance and Xolair (M-TAX). Study Rec. NCT02626611. https://clinicaltrials.gov/ct2/show/NCT02626611 [Google Scholar]
- 175.Li X-M. 2016. E-B-FAHF-2, Multi OIT and Xolair (Omalizumab) for Food Allergy. Study Rec. NCT02879006. https://clinicaltrials.gov/ct2/show/NCT02879006 [Google Scholar]
- 176.Nadeau KC. 2015. Study Using Xolair in Rush Multi Oral Immunotherapy in Multi Food Allergic Patients (MAP-X). Study Rec. NCT02643862. https://clinicaltrials.gov/ct2/show/NCT02643862 [Google Scholar]
- 177.Andorf S, Purington N, Block WM, Long AJ, Tupa D, et al. 2018. Anti-IgE treatment with oral immunotherapy in multifood allergic participants: a double-blind, randomised, controlled trial. Lancet Gastroenterol. Hepatol. 3:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, et al. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med 7:307ra152 [DOI] [PubMed] [Google Scholar]
- 179.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, et al. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med 22:1187–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olle B.2013. Medicines from microbiota. Nat. Biotechnol 31:309–15 [DOI] [PubMed] [Google Scholar]
- 181.Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, et al. 2016. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep. 15:2809–24 [DOI] [PubMed] [Google Scholar]
- 182.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. 2014. The role of short-chain fatty acids in health and disease. Adv. Immunol 121:91–119 [DOI] [PubMed] [Google Scholar]
- 183.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, et al. 2011. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13:517–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Byndloss MX, Olsan EE, Rivera-Chavez F, Tiffany CR, Cevallos SA, et al. 2017. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–75 [DOI] [PMC free article] [PubMed] [Google Scholar]