Abstract
Background
Although immune checkpoint blockade (ICB) therapy has shown remarkable benefits in cancers, a subset of patients with cancer exhibits unresponsiveness or develop acquired resistance due to the existence of abundant immunosuppressive cells. Tumor-associated macrophages (TAMs), as the dominant immunosuppressive population, impede the antitumor immune response; however, the underlying mechanisms have not been fully elucidated yet.
Methods
Single-cell RNA sequencing analysis was performed to portray macrophage landscape and revealed the underlying mechanism of component 1q (C1q)+ TAMs. Malignant pleural effusion (MPE) of human and mouse was used to explore the phenotypes and functions of C1q+ TAMs.
Results
C1q+ TAMs highly expressed multiple inhibitory molecules and their high infiltration was significantly correlated with poor prognosis. C1q+ TAMs promote MPE immunosuppression through impairing the antitumor effects of CD8+ T cells. Mechanistically, C1q+ TAMs enhance fatty acid binding protein 5 (FABP5)-mediated fatty acid metabolism, which activate transcription factor peroxisome proliferator-activated receptor-gamma, increasing the gene expression of inhibitory molecules. A high-fat diet increases the expression of inhibitory molecules in C1q+ TAMs and the immunosuppression of MPE microenvironment, whereas a low-fat diet ameliorates these effects. Moreover, FABP5 inhibition represses the expression of inhibitory molecules in TAMs and tumor progression, while enhancing the efficacy of ICB therapy in MPE and lung cancer.
Conclusions
C1q+ TAMs impede antitumor effects of CD8+ T cells promoting MPE immunosuppression. Targeting C1q+ TAMs effectively alleviates the immunosuppression and enhances the efficacy of ICB therapy. C1q+ TAMs subset has great potential to be a therapeutic target for cancer immunotherapy.
Keywords: immunomodulation; immunotherapy; macrophages; tumor microenvironment; lymphocytes, tumor-infiltrating
WHAT IS ALREADY KNOWN ON THIS TOPIC
Several studies based on single-cell RNA sequencing have identified the component 1q (C1q)+ macrophage subset in various types of cancer. Most of the studies have revealed the potential immunosuppressive function of C1q+ macrophages. However, there is a lack of experimental evidence regarding the function of C1q+macrophages and their action mechanisms remain elusive.
WHAT THIS STUDY ADDS
C1q+ macrophages highly express multiple inhibitory molecules and suppress the antitumor activities of CD8+ T cells in malignant pleural effusion. C1q+ macrophages exert an immunosuppressive function through fatty acid binding protein 5 (FABP5)-mediated reprogramming of fatty acid metabolism. FABP5 inhibition significantly alleviates the immunosuppressive tumor microenvironment and enhances the efficacy of immune checkpoint blockade therapy.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The functions and mechanisms of C1q+ macrophages in various tumor types warrant further investigation. C1q has a potential to be a target for cancer immunotherapy.
Background
Recent advances in tumor immunotherapies, especially immune checkpoint blockade (ICB), aiming to reinvigorate tumor-infiltrating T cells have demonstrated unprecedented benefits in advanced malignant tumors, including lung cancer, melanoma, renal cancer, and Hodgkin’s diseases.1–3 However, a significant proportion of patients with cancer remain unresponsive to ICB therapy or develop acquired resistance.4 ICB resistance primarily arises from aberrant antigen presentation and signaling pathways, inhibition by the tumor microenvironment (TME), as well as activation of inhibitory immune checkpoints.5 Notably, myeloid cells play a critical role in promoting immunosuppression within the TME, including facilitating tumor immune escape and tumor-infiltrating T-cell anergy.6–8 Tumor-associated macrophages (TAMs) as abundant components of myeloid cells within the TME are recognized as the dominant population suppressing the function of tumor-killing T cells.8 TAMs highly express immune inhibitory molecules, including programmed cell death-ligand 1 (PD-L1), recognized by inhibitory receptors on T cells, thus inhibiting T-cell activation and inducing their exhaustion.9 TAMs also overexpress inhibitory receptors, such as programmed cell death-1 (PD-1), activated by PD-L1 on tumor cells, curbing the ability of antigen presentation and phagocytosis of macrophages.10 In addition, TAMs can secrete immunomodulatory cytokines including interleukin-10 (IL-10) and transforming growth factor-β (TGF-β) to restrain T-cell function.11 Therefore, depleting or reprogramming the immunosuppressive TAMs is a promising strategy to ameliorate drug resistance in tumor immunotherapy.6 8 Nevertheless, the phenotypic and functional characteristics of TAM subsets remain elusive, and the underlying mechanisms by which TAMs exert immunosuppressive effects are not completely understood. It has provided impetus to search for novel co-inhibitory molecules on TAMs, which could be exploited to enhance the response of patients with cancer to immunotherapeutic agents.
It is widely recognized that TAMs are considerably plastic and heterogeneous, encompassing both immunostimulatory and immunosuppressive subsets which can mutually transform under specific conditions within the TME.6 12 For a long time, macrophages have been divided into pro-inflammatory M1 or tumor-promoting M2 subsets. However, growing evidence indicates that the dichotomy is too simplistic and it is more appropriate to classify macrophages into distinct subpopulations based on specific molecular markers.6 The emergence of single-cell RNA sequencing (scRNA-seq) technologies has enabled the identification of efficient markers that define different cell subsets and the exploration of potential functions of cell subpopulations within the TME.13 Recently, increasing studies based on scRNA-seq analysis have identified a distinct subset of TAMs that expresses complement component 1q (C1q) in various types of cancer, including colon cancers, clear cell renal carcinoma, hepatocellular carcinoma and pancreatic ductal adenocarcinoma, correlating with worse prognosis and enabling to become a potential target for immunotherapy.14–18 C1q as a tumor-promoting factor has been recently observed in the TME, but a more comprehensive understanding of the function and action mechanisms for C1q+ TAM is still lacking.19 20
Malignant pleural effusion (MPE) is a prevalent and debilitating complication of advanced malignancies, heralding a fatal prognosis and a limited life expectancy.21 22 As a prototypical “cold tumor”, MPE is characterized by the enrichment of immunosuppressive cell populations, including TAMs, myeloid derived suppressor cells (MDSC), and T-regulatory cells (Tregs).21 Making “cold” MPE “hot” exhibits great potential as an effective strategy for MPE immunotherapy. TAMs are one of the most abundant components in MPE, contributing to immunomodulation and immunotherapy resistance.23 Targeting protumor TAMs is expected to alleviate immunosuppression and improve the outcomes of MPE patients. Here, we implemented scRNA-seq to identify a distinct macrophage subset in MPE, C1q+ macrophage subset, which highly expresses multiple immune inhibitory molecules. In MPE, a high degree of infiltration by C1q+ TAMs correlated with tumor progression and poor prognosis. We found that C1q+ TAMs impaired the tumor-killing function of CD8+ T cells. C1q+ TAMs exhibited enhanced fatty acid metabolism through upregulating the expression of fatty acid binding protein 5 (FABP5), which subsequently activated peroxisome proliferator-activated receptor-gamma (PPAR-γ). PPAR-γ increased the gene transcription of inhibitory molecules in C1q+ TAMs, thereby promoting MPE immunosuppression. In addition, FABP5 inhibition suppressed C1q+ TAMs-mediated immunosuppression and tumor progression, while enhancing anti-PD-1 therapy efficacy in MPE and lung cancer. These results suggest that C1q+ TAMs subset is a potential therapeutic target for cancer immunotherapy.
Material and methods
Cell lines
LLC (CRL-1642) is a Lewis lung carcinoma cell line from American Type Culture Collection (ATCC). LLC-LUC (ATCC, CRL-1642-LUC2) is a luciferase-expressing LLC cell line. Cells were cultured in Dulbecco’s Modified Eagle’s Medium containing 10% heat-inactivated fetal bovine serum (FBS). Cells were grown at 37°C in 5% CO2.
Patients and MPE collection
Sixty MPE specimens were obtained from patients who were diagnosed as lung cancer at the Department of Respiratory and Critical Care Medicine of the Union Hospital of Tong Medical College (Wuhan, China) between March 2019 and March 2021. The untreated MPE specimens were collected and detected immediately. Cytological and pathological diagnoses were confirmed. All specimens were obtained with appropriate informed consent from the patients and approved by Wuhan Union Hospital ethics committee. Clinicopathological information is presented in table 1. All of the patients were regularly followed-up, and the overall survival time was determined from the date of diagnosis until either death or a period of 2 years.
Table 1.
Clinicopathological features of 60 MPE patients and the infiltration levels of C1q+ TAMs
| Parameters | Group | Cases | C1q+ TAMs low | C1q+ TAMs high | P value |
| Gender | Male | 30 | 14 | 16 | 1 |
| Female | 30 | 14 | 16 | ||
| Age | <60 | 24 | 14 | 10 | 0.139 |
| ≥60 | 36 | 14 | 22 | ||
| Tumor type | Adenocarcinoma | 50 | 23 | 27 | 0.766 |
| Squamous carcinoma | 7 | 3 | 4 | ||
| Small cell lung cancer | 3 | 2 | 1 | ||
| Smoke | Yes | 13 | 6 | 7 | 0.967 |
| No | 47 | 22 | 25 | ||
| MPE volume | Low | 6 | 4 | 2 | 0.495 |
| Medium | 18 | 9 | 9 | ||
| High | 36 | 15 | 21 | ||
| Pathological stage | IVa | 35 | 18 | 17 | 0.382 |
| IVb | 25 | 10 | 15 | ||
| Tumor burden | Low | 39 | 26 | 13 | <0.0001 |
| High | 21 | 2 | 19 | ||
| Total | 60 | 28 | 32 |
P<0.05 represents statistical significance; χ2 test.
C1q, component 1q; MPE, malignant pleural effusion; TAM, tumor-associated macrophage.
Moue models and treatments
C57BL/6J mice were purchased from the Beijing Vital River Experimental Animal Technology (Beijing, China). To establish MPE model, sex-matched C57BL/6 mice (4–5 weeks old, 16–18 g) were injected intrapleurally with 2×105 LLC-LUC in 100 mL of phosphate buffer saline (PBS) after anesthetized by 0.5% pentobarbital sodium. Five days after inoculation with LLC-LUC cells, mice were observed by bioluminescence imaging to ensure that MPE models were established successfully and uniformly. Mice were then randomized to four groups (10 mice/group), including control, SBFI-26, anti-PD-1 monoclonal antibody (mAb), and SBFI-26 and anti-PD-1 mAb groups. Mice were treated by intraperitoneal injection with anti-PD-1 mAb (10 mg/kg) and/or SBFI-26 (1 mg/kg) three times at 2-day intervals. For diet treatment, MPE mice were randomized to five groups, including RD (mice with regular diet), RD+SBFI-26 (mice with regular diet and SBFI-26), HFD (mice with high-fat diet, 45% fat), HFD+SBFI-26 (mice with high-fat diet and SBFI-26) and HFDR, (mice with high-fat diet and low-fat diet recovery after 5 days, 4.3% fat). The survival time of MPE mice without treatment is approximately 14 days. Therefore, MPE specimens were collected and the volume was measured at 14th day. The survival status of MPE mice was observed daily until a fatal outcome for all animals
To establish the subcutaneous xenograft LLC model, 1×106 LLC cells in 100 mL of PBS were subcutaneously injected in the right flank of each mouse. Seven days after LLC inoculation, mice were randomized to four groups, including control, SBFI-26, anti-PD-1 mAb, and SBFI-26 and anti-PD-1 mAb groups. Mice were treated by intraperitoneal injection with anti-PD-1 mAb (10 mg/kg) and/or SBFI-26 (1 mg/kg) four times at 2-day intervals. The length (L) and width (W) of each tumor were measured using a vernier caliper every 2 days for 20 days, and the tumor size (V) was assessed with the formula V=(L×W2)/2. The survival status of mice was observed daily, and mice were sacrificed when the tumor volume reached 1500 mm3.
Bioluminescence imaging
MPE mice were intraperitoneally injected with D-luciferin (150 mg/kg in PBS) on being anesthetized with 0.5% pentobarbital sodium. After 15 min, mice were imaged by the Bruker In Vivo FX PRO Imager. The luminescent images were acquired with 3 min exposure time, and the reflectance images were acquired with 0.175 s exposure time. Bruker MI SE software was used to analyze total photon flux.
Isolation of immune cells from human and mouse specimens
The cell suspensions of human and mouse MPE and blood were collected by centrifugation and subsequent filtration using a 70 mm nylon cell strainer. Immune cells were separated by Lymphoprep. Mouse tumor tissue was cut into small pieces and then digested with collagenase IV (1 mg/mL) and DNase I (0.25 mg/mL) at 37°C for 60 min. Splenic cell suspensions were prepared by grinding the mouse spleens using 40 mm nylon cell strainer. After being separated by Lymphoprep, immune cells were collected.
Flow cytometry and cell sorting
Immune cells were resuspended in a staining buffer and incubated with Fc block (BioLegend) for 10 min. Then surface antibodies were added and stained for 30 min at 4°C in the dark. Cells were permeabilized by Fixation/Permeabilization Concentrate and Fixation/Perm Diluent (eBioscience) at a ratio of 1:3 for 30 min at 4°C in the dark, and then washed twice with diluted 1×Permeabilization Buffer. Subsequently, intracellular antibodies were added and stained for 30 min at 4°C in the dark. Finally, cells were fixed by Fixation Buffer and analyzed by flow cytometer (BD). To detect cytokines secreted by CD8+ T cells, cells were stimulated with cell activation cocktail (BioLegend) for 4–6 hours and then were performed the above staining.
For magnetic-activated cell sorting (MACS) of CD8+ T cells, CD8+ T cells were purified using mouse or human CD8+ T-cell isolation kit (STEMCELL). TAMs of human MPE were isolated using CD14 positive selection kit. C1q+ TAMs (major histocompatibility complex class II (MHC-II)/human leukocyte antigen-DR (HLA-DR)+CX3CR1+) and C1q– TAMs (MHC-II/HLA-DR–CX3CR1–) were isolated from MPE using fluorescence-activated cell sorting (FACS) by BD FACSAria II Cell Sorter. The panels were listed in online supplemental table S1, all antibodies and reagents were itemized in online supplemental table S2.
jitc-2023-007441supp010.pdf (3.3MB, pdf)
jitc-2023-007441supp011.pdf (113.1KB, pdf)
jitc-2023-007441supp001.pdf (3.6MB, pdf)
Co-culture assay for macrophages and T cells
Human and mouse CD8+ T cells were isolated by MACS and were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% FBS and activated by anti-CD3 (2 mg/mL) and anti-CD28 antibodies (5 mg/mL) for 72 hours. Human and mouse C1q+ and C1q– TAMs were sorted by FACS, and were cultured with activated CD8+ T cells (TAMs: CD8+ T ratio=2:1) for 72 hours. In the end, the cytokines secreted by CD8+ T cells and exhausted molecules were detected by flow cytometry.
Proliferation assay
CD8+ T cells were isolated from peripheral blood of healthy donors and were preactivated by anti-CD3 (2 mg/mL) and anti-CD28 antibodies (5 mg/mL). For the proliferation assay, CD8+ T cells were labeled with carboxyfluorescein succinimidyl ester (CFSE, BD) in a dilution of 1:1000 according to the manufacturer’s instructions. Subsequently, the cells were co-cultured with C1q+ TAMs, C1q– TAMs, or SBFI-26 treated C1q+ TAMs. After 4 days of co-culture, CD8+ T cells were collected and the expression of CFSE was measured by flow cytometry.
Ex vivo pHrodo green Escherichia coli phagocytosis assay
Isolated immune cells of MPE were incubated with 100 µL pHrodo green Escherichia coli bioparticles (Invitrogen) in 1 mL medium for 2 hour at 37°C. Then cells were washed twice with PBS and stained with macrophage antibodies. The phagocytosis assay was analyzed by flow cytometry. TAMs that were fluorescein Iiothiocyanate (FITC)-positive were considered to be phagocytosing.
Lipid content measurement
BODIPY 493/503 stain was used to measure cellular lipid content. After corresponding treatments, cells were stained for 15 min with BODIPY 493/503 at room temperature in PBS, and examined promptly using flow cytometry or confocal microscopy (Nikon).
Immunofluorescence
For immunofluorescence staining, human and mouse TAMs were sorted from MPE and were seeded in 24-well culture plates with cell climbing slices. Then the cells were fixed with 4% paraformaldehyde solution for 15 min. After washing with PBS, cells were permeabilized with 2 mL 0.2–0.5% Triton X-100 (in PBS) for 10 min. Subsequently, blocking cells with 2% bovine serum albumin (BSA) for 30 min, cells were washed three times with PBS and added with primary antibodies at room temperature for 1 hour (or overnight at 4°C). After washing three times with PBS, cells were added with secondary antibodies and incubated at room temperature for 30 min. Finally, cell climbing slices were added with an antifade solution with 4,6-diamino-2-phenyl indole (DAPI) and observed using confocal microscopy (Nikon).
Chromatin immunoprecipitation
1×107 TAMs were isolated from human MPE by MACS. Chromatin immunoprecipitation (ChIP) assays were performed with an enzymatic ChIP Kit (Cell Signaling Technology) according to the manufacturer’s protocol. Anti-PPAR-γ (1:500, ProteinTech) antibody and normal IgG were used for ChIP. Genes that contain PPAR-γ-binding elements were subsequently confirmed by quantitative PCR (qPCR). The sites of PPAR-γ binds to promoters of inhibitory molecules (triggering receptor expressed by myeloid cells-2 (TREM2), T-cell immunoglobulin-3 (Tim-3), signal regulatory protein α (SIRPα), PD-1, PD-L1) were predicted by JASPAR (http://jaspar.genereg.net). The primers were synthesized by Tsingke Biotech (Beijing, China) and the sequences were listed in online supplemental table S3.
jitc-2023-007441supp012.pdf (153.4KB, pdf)
Luciferase reporter assay
Luciferase activity was measured with the Dual Luciferase Reporter Assay Kit (Vazyme, Nanjing, China) according to the manufacturer’s protocols. DNA fragments of inhibitory molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1) messenger RNA were cloned into the pMIR-REPORT vector to construct firefly luciferase reporters. TAMs were isolated from human MPE by MACS. The firefly luciferase reporter plasmids and renilla luciferase reporter vectors were co-transfected to TAMs to detect luciferase activities. Subsequently, the firefly luciferase reporter plasmids and PPAR-γ-overexpression plasmids were co-transfected to TAMs using Lipofectamine 3000. Dual-Luciferase Reporter Assay system (Promega, Madison, Wisconsin, USA) was used to measure the luciferase activities.
Western blotting
TAMs were isolated and then lysed in a radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors and phosphatase inhibitor cocktails on ice for 30 min. The liquid was centrifuged and the supernatant (TAM protein) was collected. Protein was quantified using the BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). Equal amount of protein was mixed with a 1× loading buffer and placed in a metal bath at 100°C for 10 min. Subsequently, they were separated in a sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane on ice. The membrane was blocked with 5% skim milk in tris-buffered saline with 0.05% Tween 20 (tris buffered saline with tween, TBST) at room temperature for 1 hour and incubated with primary antibody overnight at 4°C. Next, they were washed three times with TBST at room temperature for 15 min each time and incubated with secondary antibody at room temperature for 1 hour. After washing with TBST, chemiluminescent substrate solution (Vazyme, Nanjing, China) was added, and the images were captured using UVP chemiluminescence imaging system (Upland, California, USA).
Real-time quantitative PCR (RT-qRCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), and RNA concentration was determined using NanoDrop (NanoDrop Technologies, USA). Purified RNA was reverse-transcribed into complementary DNA using HiScript qRT-PCR SuperMix (Vazyme, Nanjing, China) in accordance with the manufacturer’s instructions. The RT-qPCR reaction was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) and the ABI 7500 real-time PCR system were used to measure the expression of target genes. Relative gene expression levels were calculated using the 2–ΔΔct method. The primers were synthesized by Tsingke Biotech (Beijing, China) and the sequences are listed in online supplemental table S4.
jitc-2023-007441supp014.pdf (27.2KB, pdf)
Immunohistochemistry
Mouse tumor tissues were fixed and paraffin embedded. On that day, they were sectioned, dewaxed and rehydrated. After being blocked with 1% BSA, the slides were incubated with primary antibody overnight at 4°C. They were then incubated with secondary antibody for 1 hour at room temperature. After washing with PBS, the slides were incubated with Streptomyces antibiotin-peroxidase solution for 15 min at room temperature and were then added with diaminobenzidine for 3 min at room temperature.
Single-cell RNA-seq analysis
scRNA-seq analysis of C1q+ TAMs was based on our previous MPE scRNA-seq data (PRJNA970083). Seven MPE specimens of patients with untreated lung cancer were collected, and CD45+ immune cells were subsequently isolated using MACS for scRNA-seq. The scRNA-seq data were preprocessed to filter the low-quality cells according to the previous study.15 16 24 The primary analyses were accomplished with the Seurat R package (V.4.2.0). The data was normalized using a scaling factor of 10,000 and the top 2000 variable genes were identified through the FindVariableFeatures function for subsequent principal component analysis. The first 30 principal components were used in the FindNeighbors algorithm, while the clusters were determined by the FindClusters function. The identified clusters were visualized using the t-distributed stochastic neighbor embedding method. Differentially expressed genes (DEGs) of the clusters were identified by the FindAllMarkers function. Enrichment analysis was performed using the clusterProfiler package (V.4.2.0) and gene set variation analysis (GSVA) package (V.1.42.0) based on the gene sets in the MSigDB database. CellPhoneDB (V.2.1.5) was used to analyze cell–cell interactions between different cell types.
Publicly available scRNA-seq data of non-small cell lung cancer (NSCLC), kidney renal clear cell carcinoma (KIRC), and colorectal cancer (CRC) were separately downloaded from Gene Expression Omnibus (GEO) database (GSE127465, GSE207493, GSE146771). The Cancer Genome Atlas (TCGA) RNAseq data and clinical data were downloaded by the TCGAbiolinks R package (V.2.25.3). Based on the average expression levels of C1q+ TAM signatures (C1QA, C1QB, C1QC, GPNMB, PLTP, MACRO, TREM2, APOE, APOC1, SPP1, FABP5) normalized with the expression of PTPRC, the abundance of C1q+ TAM subset was inferred and patients were classified into low and high groups. Survival (V.3.4.0) and survminer (V.0.4.9) packages were used for analysis and visualization.
Statistical analysis
Statistical analyses were performed using Prism V.8 (GraphPad Software). Statistical significance was determined using two-tailed Student’s t-test, one-way analysis of variance, or Pearson correlation coefficient. Survival time was calculated using the Kaplan-Meier method and analyzed using the log-rank test. Data were presented as the mean±SD. P value<0.05 was used to indicate significance. P values are indicated as follows: *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. ns., no significance.
Results
C1q+ macrophage subset detected by scRNA-seq correlates with poor prognosis of MPE patients
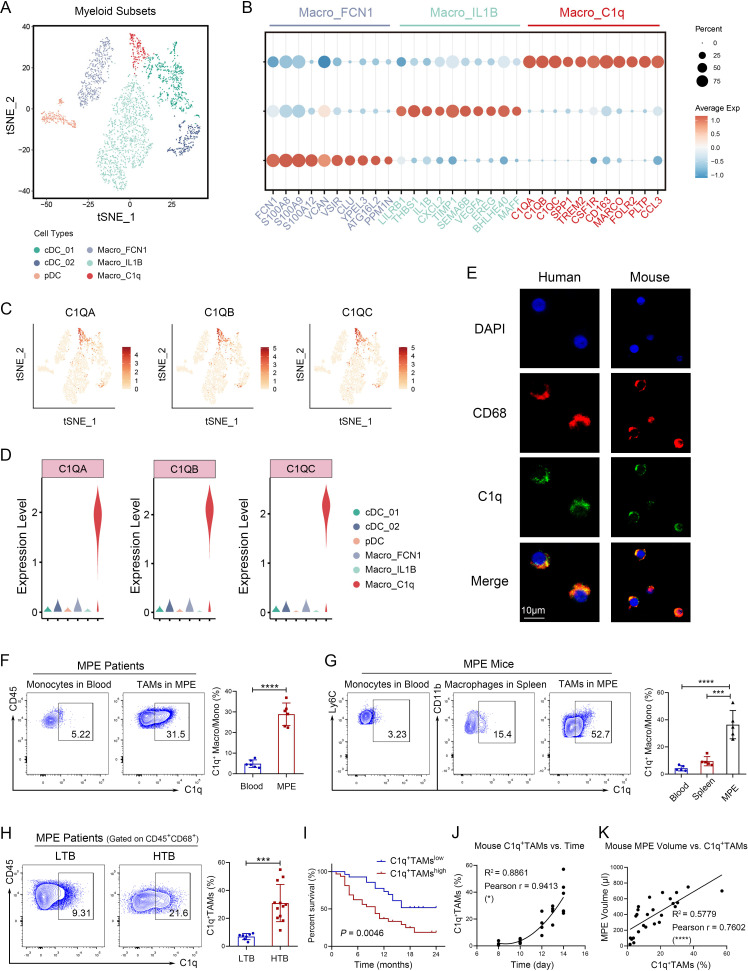
Considering the phenotypic heterogeneity of macrophages, scRNA-seq analysis was performed to portray macrophage landscape in MPE. CD45+ immune cells from seven MPE specimens of patients with untreated lung cancer were isolated using MACS for scRNA-seq. Based on gene signatures, three subsets of macrophages and dendritic cells were identified among myeloid cells, including C1q+ macrophages with high expression of C1QA, C1QB, and C1QC (figure 1A,B and online supplemental table S5). C1q is a component of the complement C1 complex, which serves as a promoter for the classical activation pathway of the complement system in an innate immune system. In the TME, C1q+ macrophage has recently been identified as a protumor subset without in-depth study. According to scRNA-seq analysis, C1q was predominantly expressed on macrophages in human MPE (figure 1C,D). We demonstrated that C1q was highly expressed on macrophages in human and mouse MPE rather than peripheral blood or spleen (figure 1E–G). Similarly, C1q+ TAMs were also observed in the other types of cancers, such as NSCLC, KIRC, and CRC based on scRNA-seq (online supplemental figure S1A). As revealed by Single-cell Analysis from the Tumor Immune Single-cell Hub database, C1q is mainly expressed on TAMs in lung cancer, particularly the M2 type (online supplemental figure S1B). The expression of C1q on CD68+ macrophages was further confirmed in human lung cancer tissues (online supplemental figure S1C). Moreover, the percentage of C1q+ TAMs in MPE was higher in patients with high tumor burden (tumor cells >30%) (figure 1H), leading to a poor prognosis (figure 1I). Apart from MPE, C1q+ TAMs can also serve as a prognostic marker for multiple types of cancer. A high infiltration of C1q+ TAMs was found to be significantly associated with worse prognosis of patients in lung adenocarcinoma, KIRC, and rectum adenocarcinoma (online supplemental figure S1D). In mice, the proportion of C1q+ macrophages/monocytes in MPE increased over time, while it decreased in spleen and peripheral blood (figures 1J, 2A and B). Furthermore, MPE volume of mice increased with the proportion of C1q+ TAMs (figure 1K). These results indicate that a high level of C1q+ TAMs infiltration in the TME is associated with poor prognosis and can serve as a prognostic indicator for patients with cancer.
Figure 1.
C1q+ macrophage subset detected by scRNA-seq correlates with poor prognosis of MPE patients. (A–D) Seven MPE specimens of patients with untreated lung cancer were collected, and CD45+ immune cells were isolated using magnetic-activated cell sorting for scRNA-seq. A t-distributed stochastic neighbor embedding (t-SNE) plot based on scRNA-seq revealed six clusters of myeloid cells, comprising three subsets of macrophages and three subsets of dendritic cells (A). Heat map of gene signatures for macrophage subsets (B). Expression levels of C1QA, C1QB, and C1QC genes in t-SNE space (C). Expression levels of C1QA, C1QB, and C1QC genes in distinct myeloid subsets (D). (E) Immunofluorescence staining revealed co-localization of C1q and CD68+ macrophages in both human and mouse MPE. Red, CD68; green, C1q; and blue, DAPI for nucleus. Scale bar 10 µm. (F) The percentages of C1q+ TAMs (CD45+ CD68+ C1q+) in human MPE and C1q+ monocytes (CD45+ CD14+ C1q+) in corresponding peripheral blood were quantified by flow cytometry (n=6). (G) The percentages of C1q+ macrophages (CD45+ F4/80+ CD11b+ C1q+) in mouse MPE and corresponding spleen, as well as C1q+ monocytes (CD45+ Ly6C+ C1q+) in corresponding peripheral blood, were quantified by flow cytometry (n=5). (H) MPE patients were stratified into high tumor burden group (HTB, tumor cells >30%, n=12) and low tumor burden group (LTB, tumor cells <30%, n=6). The percentages of C1q+ TAMs in both groups were determined by flow cytometry analysis. (I) Correlation between C1q+ TAMs and survival in MPE was analyzed using Kaplan-Meier analysis (n=28–32). The median survival time of C1q+ TAMslow was 19 months, while the median survival time of C1q+ TAMshigh was 10 months. (J and K) The MPE mouse model was established using LLC cells. Correlation analyses were conducted to investigate the relationship between the proportion of C1q+ TAMs and time (I, n=18), as well as the proportion of C1q+ TAMs and MPE volume (J, n=26). Data shown in (E–K) are representative of at least three independent experiments (mean±SD). Statistical analysis was performed using paired two-tailed Student’s t-test (F), unpaired two-tailed Student’s t-test (G, H), log-rank test (I), or Pearson correlation coefficient (J, K). *p<0.05, ***p<0.001, ****p<0.0001. C1q, component 1q; cDC, conventional dendritic cells; DAPI, 2- (4-Amidinophenyl)-6-indolecarbamidine dihydrochloride; IL, interleukin; LLC, Lewis lung cancer cells; MPE, malignant pleural effusion; pDC, plasmacytoid dendritic cells; scRNA-seq, single-cell RNA sequencing; TAM, tumor-associated macrophage.
Figure 2.
C1q+ TAMs are characterized by the higher expression levels of immune inhibitory molecules. (A and B) Differentially expressed genes (A, volcano plots) were identified between C1q+ and C1q– macrophages, including macrophage markers, inhibitory markers and metabolism-related genes (B, heatmap). (C and D) Tumor-infiltrating immune cells were isolated from human MPE, and the macrophage markers of C1q+ TAMs were determined by flow cytometry analysis, including the expression levels of HLA-DR, CSF1R and MS4A4A (n=4–9). (E–H) Tumor-infiltrating immune cells were isolated from human MPE; M2-like markers (CD163, CD206, CX3CR1) and inhibitory molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1) were detected by flow cytometry (n=5–10). (I and J) Tumor-infiltrating immune cells were isolated from mouse MPE; M2-like markers (MHC-II, CD206, CX3CR1; H) and inhibitory molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1; I) were detected by flow cytometry (n=8–13). Data shown in (C–J) are representative of at least three independent experiments (mean±SD). Statistical analysis was performed using paired two-tailed Student’s t-test (D, F, H–J). *p<0.05, ***p<0.001, ****p<0.0001. CSF1R, colony stimulating factor 1 receptor; CX3CR1, C-X3-C motif chemokine receptor 1; C1q, component 1q; IL, interleukin; HLA, human leukocyte antigen; MHC, major histocompatibility complex; MPE, malignant pleural effusion; PD-1, programmed cell death-1; PD-L1, programmed cell death-ligand 1; SIRPα, signal regulatory protein α; TAM, tumor-associated macrophage; Tim-3, T-cell immunoglobulin-3; TREM2, triggering receptor expressed by myeloid cells-2.
jitc-2023-007441supp015.xlsx (260.9KB, xlsx)
jitc-2023-007441supp002.pdf (3.4MB, pdf)
C1q+ TAMs are characterized by high expression levels of immune inhibitory molecules
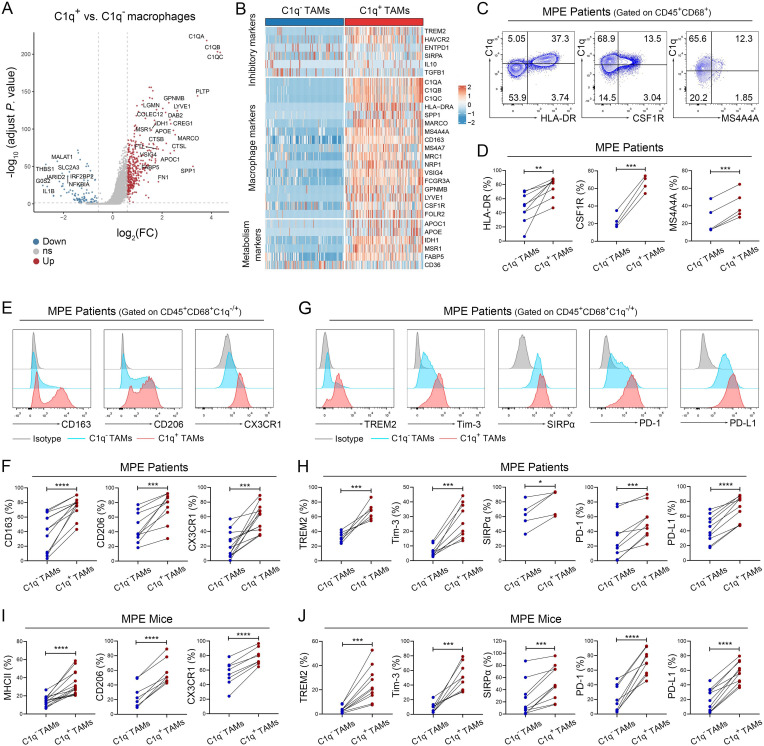
To determine the phenotype of C1q+ TAMs, scRNA-seq was performed to identify the DEGs between C1q+ TAMs and C1q– TAMs. Due to the presence of multiple molecules related to immunosuppression and metabolism in the DEGs, we classified them into three clusters: macrophage markers, inhibitory markers, and metabolism-related markers (figure 2A,B). We demonstrated that C1q+ TAMs from human MPE exhibited higher expression levels of macrophage markers, including HLA-DR (an MHC-II molecule in human), colony stimulating factor 1 receptor (CSF1R), and MS4A4A; as well as tumor-promoting molecules CD163, CD206 (also named as MRC1 or mannose receptor C-type 1), C-X3-C motif chemokine receptor 1 (CX3CR1); and also immunosuppressive molecules TREM2, Tim-3, SIRPα, PD-1, and PD-L1 compared with C1q– TAMs using flow cytometry (figure 2C–H). In addition, C1q in TAMs co-expressed with CD163 and CD206 (online supplemental figures S2G,H). In accordance with human MPE, C1q+ TAMs in mouse MPE also exhibited higher expression of MHC-II, CD206, CX3CR1, as well as immunosuppressive molecules TREM2, Tim-3, SIRPα, PD-1, and PD-L1 (figure 2I, J, E and F) compared with C1q− TAMs. Moreover, the co-expression of TREM2 and Tim-3 with C1q was observed on TAMs in mice (online supplemental figure S2I). Similar with previous studies,14 15 25 these results provide further evidence that C1q+ TAMs highly express multiple tumor-promoting and immunosuppressive molecules in MPE.
jitc-2023-007441supp003.pdf (3MB, pdf)
C1q+ TAMs impair the antitumor responses of CD8+ T cells
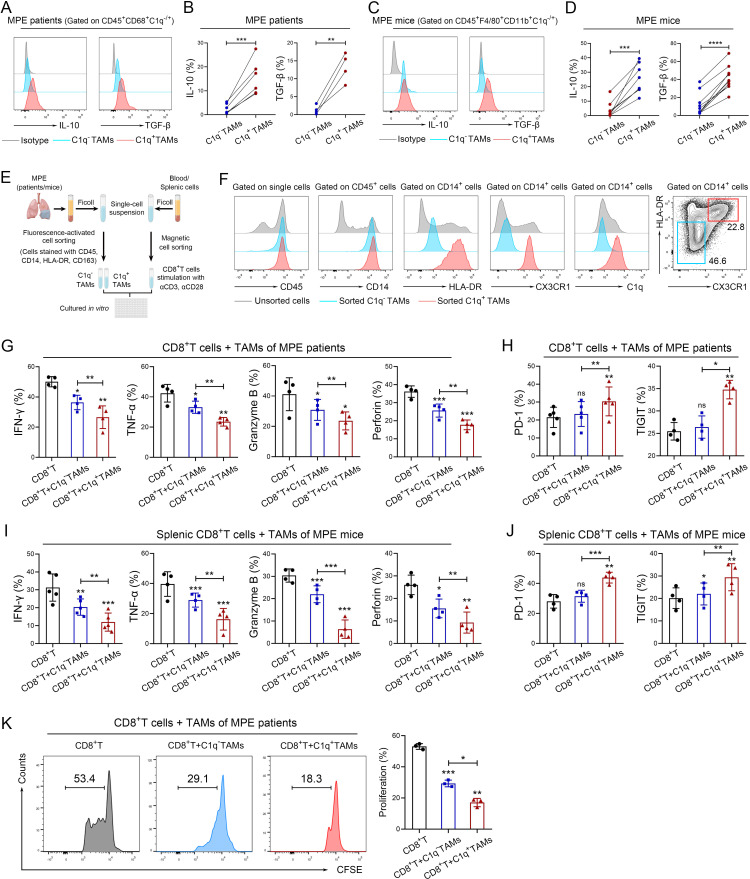
Previous studies have indicated the immunomodulatory phenotype and predicted the potential function of C1q+ TAMs; however, limited experimental evidence is currently available to substantiate their roles in the TME. To elucidate the functions of C1q+ TAMs in MPE, we assessed their capacity to secrete immunosuppressive cytokines and phagocytose, as well as their impact on effector cells. As shown in figure 3A–D, C1q+ TAMs expressed higher levels of IL-10 and TGF-β in both human and mouse MPE compared with C1q– TAMs. It has been reported that TAMs can induce immunosuppression in the TME by secreting IL-10 and TGF-β, which leads to the suppression of CD8+ T-cell cytotoxicity.26–28 In addition, Dong et al have revealed that the interplay between C1q+ TAMs and CD8+ T cells is mediated by the CXCL10-CXCR3 axis based on scRNA-seq.25 We also demonstrated potential interaction between C1q+ TAMs and CD8+ effector T cells by scRNA-seq of MPE (online supplemental figure S3). Therefore, we established a co-culture system of C1q+/C1q– TAMs and CD8+ T cells to investigate the impact of C1q+ TAMs on CD8+ T-cell function (figure 3E). A recent study has demonstrated that MHC-II can be serve as a signature distinctive gene to discriminate mouse C1q+ and C1q– TAMs, and using both MCH-II and CX3CR1 enables to separate the two subsets.25 Thus, the two subsets of TAMs were sorted by FACS based on their surface markers (HLA-DR/MHC-II and CX3CR1), distinguishing C1q+ TAMs (HLA-DR/MHC-II+CX3CR1+) from C1q– TAMs (HLA-DR/MHC-II–CX3CR1–) (HLA-DR for human, MHC-II for mouse) using TAMs from MPE (online supplemental figures S4A and B). Flow cytometry analysis confirmed that HLA-DR/MHC-II+ CX3CR1+ TAMs (C1q+ TAMs) expressed much higher levels of C1q than HLA-DR/MHC-II– CX3CR1– TAMs (C1q– TAMs) (figures 3F and 4C). Human CD8+ T cells were isolated from peripheral blood of healthy donors, while mouse CD8+ T cells were isolated from spleen tissues. Co-culturing with C1q+ TAMs suppressed the expression of interferon (IFN)-γ, tumor necrosis factor-α, granzyme B, perforin and the proliferation of CD8+ T cells, while increasing the expression of exhaustion molecules PD-1 and T cell immunoglobulin and ITIM domains (TIGIT) compared with co-culture with C1q– TAMs (figures 3G–K and 4D,E). Furthermore, an ex vivo phagocytosis assay was performed using pHrodo-labeled E. coli bioparticles, which revealed that C1q+ TAMs in both human and mouse MPE exhibited a heightened degree of phagocytic activity towards the E. coli bioparticles compared with C1q− TAMs (online supplemental figures S5A–D). Collectively, our results underpin that C1q+ TAMs impair the tumor-killing ability of CD8+ T cells and induce their functional exhaustion.
Figure 3.
C1q+ TAMs impair the antitumor responses of CD8+ T cells. (A–D) Tumor-infiltrating immune cells were isolated from human MPE (A–B) and mouse MPE (C–D); The expression of IL-10 and TGF-β in TAMs were analyzed using flow cytometry (n=4–8). (E) Schematic diagram of a co-culture system involving macrophages and CD8+ T cells (macrophages: CD8+ T cells=2:1) was drawn by FigDraw (www.figdraw.com; ID: IYYPP2af53). (F) The efficiency of fluorescence-activated cell sorting (FACS) for C1q+ TAMs in human MPE was confirmed by the expression of C1q. (G and H) C1q+ and C1q− TAMs were isolated from human MPE by FACS, CD8+ T cells were isolated from peripheral blood of healthy donors by magnetic-activated cell sorting (MACS). The tumor-killing activities (IFN-γ, TNF-α, granzyme B and perforin) and exhaustion‐related molecules (PD-1, TIGIT) of CD8+ T cells were analyzed by flow cytometry after co-culturing with TAMs for 72 hours (n=4–5). (I and J) C1q+ and C1q− TAMs were isolated from MPE mice by FACS, and CD8+ T cells were isolated from mouse spleen by MACS; tumor-killing activities (IFN-γ, TNF-α, granzyme B and perforin) and exhaustion‐related molecules (PD-1, TIGIT) of CD8+ T cells were determined by flow cytometry after co-cultured with TAMs for 72 hours (n=4–5). (K) C1q+ and C1q− TAMs were isolated from human MPE by FACS, CD8+ T cells were isolated from peripheral blood of healthy donors by MACS. CFSE assay was performed to detect the proliferation of CD8+T cells after co-cultured with TAMs (n=3). Data shown in (A–D), (F–K) are representative of at least three independent experiments (mean±SD). Statistical analysis was performed using paired two-tailed Student’s t-test (B, D, G–K). *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns: not statistically significant. C1q, component 1q; CFSE,carboxyfluorescein succinimidyl ester; HLA, human leukocyte antigen; IFN, interferon; IL, interleukin; MPE, malignant pleural effusion; PD-1, programmed cell death-1; TAM, tumor-associated macrophage; TGF-β, transforming growth factor-β; TIGIT, T cell immunoglobulin and ITIM domains; TNF, tumor necrosis factor.
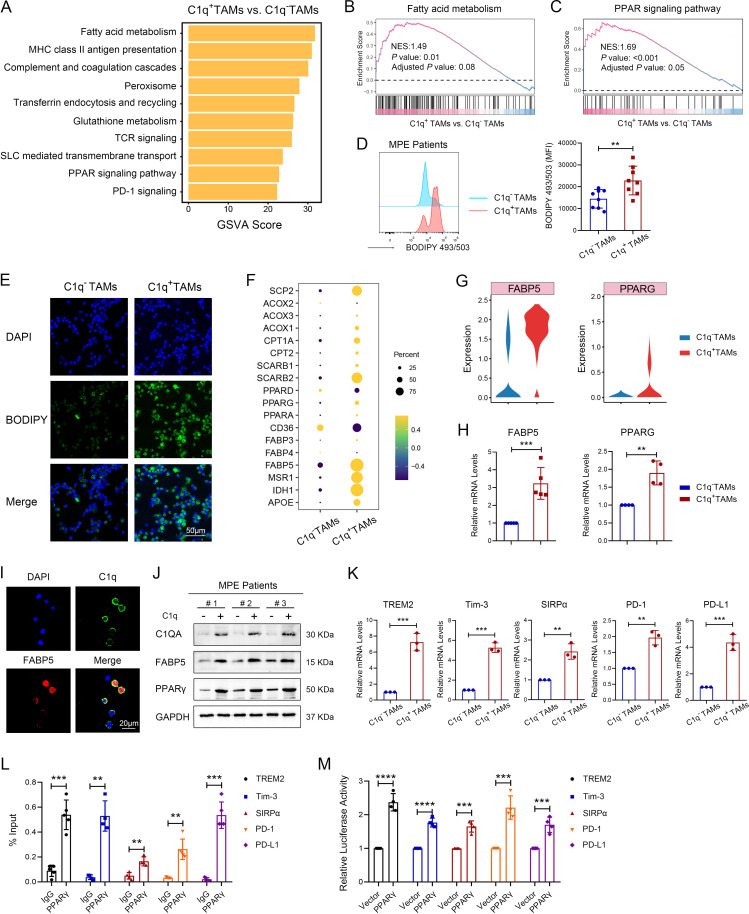
Figure 4.
C1q+ TAMs determine the immunosuppressive phenotypes by programming lipid metabolism. (A) Gene set variation analysis (GSVA) identified the differentially expressed signaling pathways enriched in C1q+ TAMs compared with C1q– TAMs. (B and C) Gene-set enrichment analysis was performed on gene sets of fatty acid metabolism (B) and PPAR signaling pathway (C). NES, Normalized Enrichment Score. Positive NES indicate higher expression in C1q+ TAMs. (D) Fatty acid accumulation was assessed by BODIPY 493/503 stain using flow cytometry (n=8). (E) Representative images of BODIPY staining in TAMs of human MPE, scale bar 50 µm. (F) Dot plot of differentially expressed genes related to lipid metabolism. (G) The expression of FABP5 and PPARG (PPAR-γ) extracted from the single-cell sequencing data. (H) The relative expression levels of FABP5 and PPARG genes were measured by qRCR in sorted C1q+ TAMs and C1q– TAMs isolated from human MPE (n=4–5). (I) Representative images of C1q and FABP5 staining in TAMs of human MPE. Red: FABP5; green: C1q; and blue: DAPI for nucleus. Scale bar 20 µm. (J) The protein expression levels of C1q, FABP5, and PPAR-γ in sorted C1q+ TAMs and C1q– TAMs isolated from human MPE by FACS were measured by western blotting. (K) The relative expression levels of TREM2, Tim-3, SIRPα, PD-1 and PD-L1 genes in sorted C1q+ TAMs and C1q– TAMs isolated from human MPE by FACS were measured by qRCR (n=3). (L) The binding of PPAR-γ to the promoter regions of TREM2, Tim-3, SIRPα, PD-1 and PD-L1 was determined by chromatin immunoprecipitation-PCR in TAMs from human MPE (n=3–5). (M) Effects of PPAR-γ on the transcriptional regulation of TREM2, Tim-3, SIRPα, PD-1 and PD-L1 were measured by luciferase assays in TAMs from human MPE (n=4). Data shown in (D−E), (H−M) are representative of at least three independent experiments (mean±SD). Statistical analysis was performed using paired two-tailed Student’s t-test (E) or unpaired two-tailed Student’s t-test (H, K, L, M). **p<0.01, ***p<0.001, ****p<0.0001. C1q, component 1q; DAPI, 4,6-diamino-2-phenyl indole; FABP5, fatty acid binding protein 5; FACS, fluorescence-activated cell sorting; MHC, major histo compatibility complex; MPE, malignant pleural effusion; mRNA, messenger RNA; PD-1, programmed cell death-1; PD-L1, programmed cell death-ligand 1; PPAR-γ, peroxisome proliferator-activated receptor-γ; qPCR, quantitative PCR; SIRPα, signal regulatory protein α; SLC,solute carriers; TAM, tumor-associated macrophage; TCR, T cell receptor; Tim-3, T-cell immunoglobulin-3; TREM2, triggering receptor expressed by myeloid cells-2.
jitc-2023-007441supp004.pdf (1.6MB, pdf)
jitc-2023-007441supp005.pdf (4.4MB, pdf)
jitc-2023-007441supp006.pdf (791.5KB, pdf)
C1q+ TAMs determine the immunosuppressive phenotypes by programming fatty acid metabolism
To explain the immunosuppressive function of C1q+ TAMs, we used GSVA to enrich the differentially expressed signaling pathways between C1q+ TAMs and C1q– TAMs. The result showed a significant difference in the fatty acid metabolism pathway and PPAR signaling pathway (figure 4A), which were predicted to be activated in C1q+ TAMs through gene-set enrichment analysis (figure 4B and C). Consistently, fatty acid metabolism pathway and PPAR signaling pathway were also upregulated in NSCLC, KIRC, and CRC (online supplemental figures S6A–C). To confirm the activation of fatty acid metabolism pathway, a BODIPY 493/503-labeled fatty acid probe was used to detect fatty acid accumulation using flow cytometry and immunofluorescence; our findings were consistent with the scRNA-seq analysis (figure 4D,E, online supplemental figure S7A). To further determine the molecular mechanisms regulating the fatty acid metabolism pathway, DEGs involved in this pathway were identified by scRNA-seq, including significantly upregulated FABP5 and PPARG (PPAR-γ) in C1q+ TAMs (figure 4F). FABP5, a lipid chaperone protein expressed in macrophages, binds with long-chain fatty acids and other hydrophobic ligands to regulate the uptake, transport, and metabolism of fatty acid.29 30 PPAR-γ induces fatty acid oxidation, thereby mediating M2-type macrophage polarization and promoting their immunosuppression function.29 31 A study has demonstrated that FABP5 can bind to PPAR and subsequently activate the downstream genes of PPAR family.30 Therefore, we speculated that FABP5 and PPAR-γ might be involved in regulating the immunosuppressive effects of C1q+ TAMs. Consistently, scRNA-seq analysis showed significantly high expression of FABP5 and PPARG genes in C1q+ TAMs and these were also confirmed by qPCR (figure 4G,H, online supplemental figure S7B, C). Moreover, FABP5 co-expressed with C1q in TAMs of human MPE (figure 4I). In vitro results confirmed that C1q+ TAMs expressed higher levels of FABP5, PPAR-γ, and immune inhibitory molecule genes (TREM2, Tim-3, SIRPα, PD-1, PD-L1) compared with C1q– TAMs (figure 4J,K, online supplemental figure S7D and E). To verify the interaction between transcription factor PPAR-γ and immune inhibitory molecules, ChIP and luciferase reporter assay were performed; results demonstrated that PPAR-γ was capable of binding to the promoter regions of TREM2, Tim-3, SIRPα, PD-1, PD-L1 (figure 4L,M). Together, these data suggest that C1q+ TAMs enhance fatty acid metabolism by upregulating FABP5, which activates PPAR-γ and promotes the transcription of immune inhibitory molecules.
jitc-2023-007441supp007.pdf (1,005.1KB, pdf)
jitc-2023-007441supp008.pdf (1.5MB, pdf)
FABP5 regulates the immunosuppressive activities of C1q+ TAMs
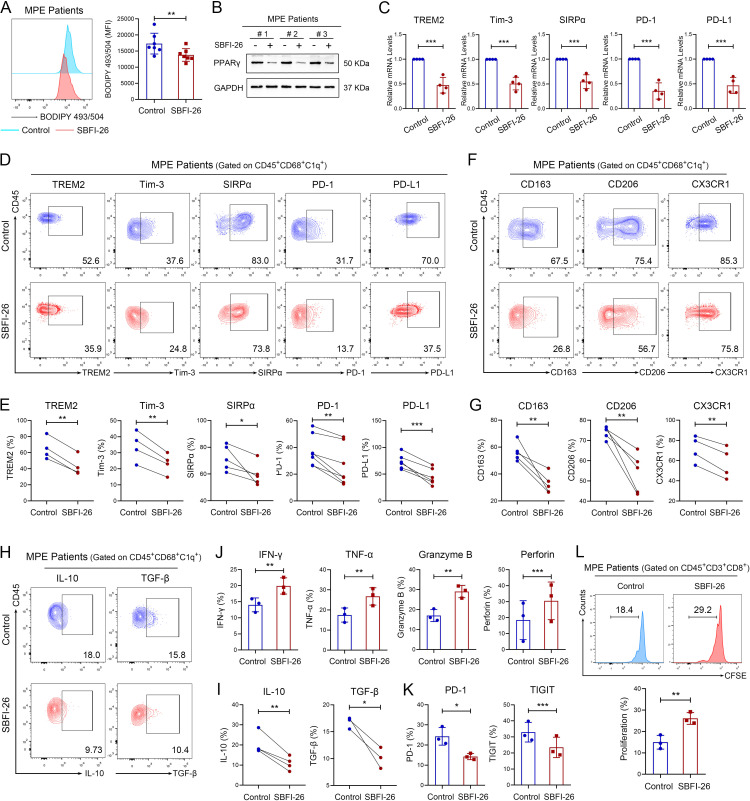
To confirm the regulatory role of FABP5 in the immunosuppressive function of C1q+ TAMs, we treated TAMs with SBFI-26, an FABP5 inhibitor.32 SBFI-26 reduced fatty acid accumulation and downregulated the expression levels of PPAR-γ as well as immune inhibitory molecule genes (TREM2, Tim-3, SIRPα, PD-1, PD-L1) in human and mouse TAMs (figure 5A–C, online supplemental figure S7F–H). In addition, FABP5 inhibition significantly downregulated the expression of immune inhibitory molecules, tumor-promoting molecules (CD163, CD206, CX3CR1), and immunomodulatory cytokines (IL-10, TGF-β) in C1q+ TAMs, but slightly or not decreased their expression in C1q– TAMs (figure 5D–I, online supplemental figure S8A–C). SBFI-26 also inhibited the impaired tumor-killing capacity, functional exhaustion, and proliferative inhibition of CD8+ T cells induced by C1q+ TAMs (figure 5J–L). These results indicate that FABP5 regulates the immunosuppressive function of C1q+ TAMs by upregulating multiple immune inhibitory molecules and immunomodulatory cytokines, thereby contributing to the CD8+ T-cell dysfunction.
Figure 5.
FABP5 regulates the immunosuppressive activities of C1q+ TAMs. TAMs were sorted from human MPE and were treated with 100 µM SBFI-26 (an FABP5 inhibitor) or 0.1% DMSO for 72 hours. (A) Fatty acid accumulation was assessed by BODIPY 493/503 stain using flow cytometry (n=7). (B) The protein expression level of PPAR-γ in TAMs from human MPE was measured by western blotting. (C) The relative expression levels of TREM2, Tim-3, SIRPα, PD-1 and PD-L1 genes in TAMs from human MPE were measured by qRCR (n=4). (D and E) Immune inhibitory molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1) in C1q+ TAMs were detected by flow cytometry (n=4–7). (F and G) M2-like markers (CD163, CD206, CX3CR1) in C1q+ TAMs were detected by flow cytometry (n=4–5). (H and I) The expression of IL-10 and TGF-β were detected by flow cytometry (n=3–4). (J−L) CD8+ T cells were isolated from peripheral blood of healthy donors and co-cultured with C1q+ TAMs of human MPE treated with SBFI-26 or DMSO for 72 hours. The tumor-killing activities (J), exhaustion‐related molecules (K), and proliferation (L) of CD8+ T cells were assessed by flow cytometry (n=3). Data shown in (A−L) are representative of at least three independent experiments (mean±SD). Statistical analysis was performed using unpaired two-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. C1q, component 1q; CX3CR1, C-X3-C motif chemokine receptor 1; DMSO, Dimethyl sulfoxide; IFN, interferon; IL, interleukin; FABP5, fatty acid binding protein 5; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; MFI, mean fluorescent intensity; MPE, malignant pleural effusion; mRNA, messenger RNA; PD-1, programmed cell death-1; PD-L1, programmed cell death-ligand 1; PPAR-γ, peroxisome proliferator-activated receptor-γ; qPCR, quantitative PCR; SIRPα, signal regulatory protein α; TAM, tumor-associated macrophage; TGF-β, transforming growth factor β; TIGIT, T cell immunoglobulin and ITIM domains; Tim-3, T-cell immunoglobulin-3; TNF, tumor necrosis factor; TREM2, triggering receptor expressed by myeloid cells-2.
jitc-2023-007441supp009.pdf (1.1MB, pdf)
High-fat diet facilitates C1q+ TAMs-mediated immunosuppression in MPE
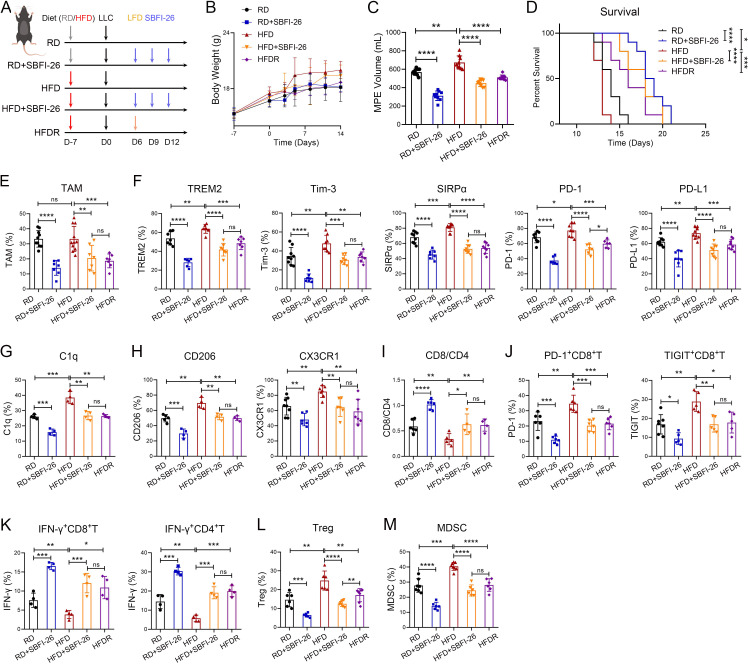
As FABP5-mediated fatty acid metabolism induced C1q+ TAMs-mediated immunosuppression, we proceeded to investigate the impact of high/low-fat diet and FABP5 inhibition on TAMs and MPE immunosuppression. We found that MPE mice fed with a high-fat diet exhibited an increase in body weight, while those on a low-fat diet showed recovery of their body weight (figure 6A,B). High-fat diet (HFD) resulted in an increase in MPE volume and a decrease in survival time of MPE mice; while SBFI-26 treatment and low-fat diet recovery after HFD (HFDR) led to a reduction in MPE volume and prolonged the survival time (RD: 14 days, RD+SBFI-26: 18.5 days, HFD: 13 days, HFD+SBFI-26: 18 days, HFDR: 16 days) (figure 6C,D). Moreover, the proportion of TAMs, immunosuppressive molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1), C1q, and tumor-promoting molecules (CD206, CX3CR1) in TAMs was found to increase in HFD group but decrease in SBFI-26 and HFDR groups (figure 6E–H). To reveal the impact of dietary intervention and FABP5 inhibition on MPE microenvironment, we detected the proportion of CD8+ T lymphocytes, effector CD8+ T cells (CD8+IFN-γ+), Th1 cells (CD4+IFN-γ+), exhausted CD8+ T cells, Tregs (CD4+CD25+CD127−), and MDSC (CD11b+Gr1+) infiltration in mouse MPE. The results indicated that an HFD resulted in a reduction of the CD8+/CD4+ T-cell ratio as well as the percentages of effector CD8+ T cells and Th1 cells, an increase in the percentages of PD-1+CD8+ T cells, TIGIT+CD8+ T cells, Tregs and MDSC; on the contrary, the opposite results were observed in SBFI-26 and HFDR groups (figure 6I–M). Collectively, HFD facilitates C1q+ TAMs and MPE microenvironment induced immunosuppression, while a low-fat diet and FABP5 inhibition partially alleviate MPE immunosuppression, offering potential therapeutic strategies for MPE treatment.
Figure 6.
High-fat diet facilitates C1q+ TAMs-mediated immunosuppression in MPE. (A–D) Schematic diagram of MPE mouse model was drawn by FigDraw (www.figdraw.com; ID: IYYPP2af53). The MPE mice were randomly assigned to five groups. RD, mice with a regular diet; RD+SBFI-26, mice with a regular diet and SBFI-26; HFD, mice with a high-fat diet (45% fat); HFD+SBFI-26, mice with a high-fat diet and SBFI-26; HFDR, mice with a high-fat diet and then low-fat diet (4.3% fat) recovery (A). Body weight (n=6) (B), MPE volume (n=9) (C) and Kaplan-Meier survival plot (n=10) (D) were observed. (E−M) Tumor-infiltrating immune cells were isolated from mouse MPE. The percentages of TAMs in immune cells (E), TREM2, Tim-3, SIRPα, PD-1, PD-L1 (F), C1q (G), CD206 and CX3CR1 (H) in TAMs, CD8/CD4 (I), PD-1+ and TIGIT+ CD8+ T cells (J), IFN-γ+ CD8+ and IFN-γ+ CD4+ T cells (K), Treg (CD45+CD3+CD4+CD25+CD127−, (L) and MDSC (CD45+CD11b+Gr1+, (L) were analyzed by flow cytometry (n=4–10). Data shown in (B−M) are representative of at least three mice (mean±SD). Statistical analysis was performed using one-way analysis of variance followed by Tukey’s post hoc test (C, E−L) or log-rank test (D). *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns: not statistically significant. C1q, component 1q; CX3CR1, C-X3-C motif chemokine receptor 1; HFD, high-fat diet; HFDR, high-fat diet recovery; IFN, interferon; LLC, Lewis lung cancer cells; MDSC, myeloid derived suppressor cells; MPE, malignant pleural effusion; PD-1, programmed cell death-1; PD-L1, programmed cell death-ligand 1; RD, regular diet; SIRPα, signal regulatory protein α; TAM, tumor-associated macrophage; TIGIT, T cell immunoglobulin and ITIM domains; Tim-3, T-cell immunoglobulin-3; Tregs, T-regulatory cells;TREM2, triggering receptor expressed by myeloid cells-2.
FABP5 inhibition alleviates immunosuppression and potentiates response to ICB therapy
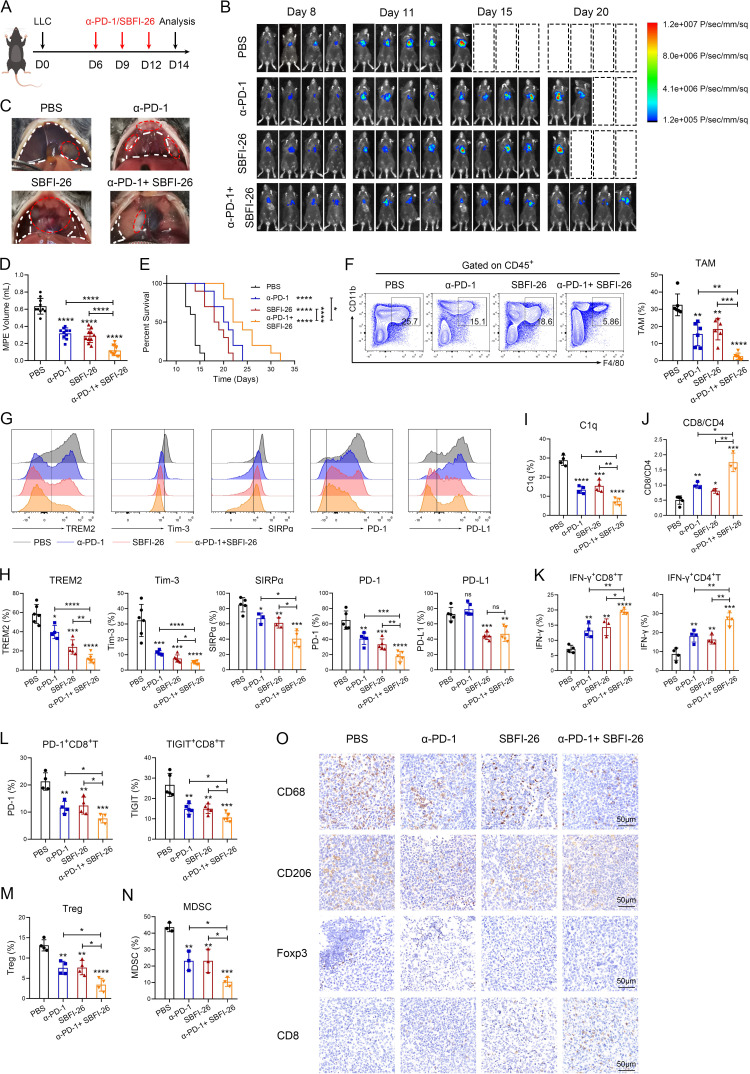
Overwhelming evidence suggests that combination of TAMs-targeted therapy and ICB therapy enhances response to ICB and produces synergetic effects in cancer treatment.6 8 Therefore, our focus lies in exploring the potential of C1q+ TAMs-targeted therapy in combination with ICB for MPE and lung cancer immunotherapy. We found that both SBFI-26 and anti-PD-1 treatments significantly delayed MPE progression, reduced MPE volume and pleural tumor size, and prolonged survival time (PBS: 14 days, α-PD-1: 20.5 days, SBFI-26: 18.5 days, α-PD-1+ SBFI-26: 23 days) in MPE mice; the combined therapy exhibited a synergetic effect (figure 7A–E). In addition, SBFI-26 and anti-PD-1 treatments inhibited the expression of immunosuppressive molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1) and C1q in TAMs; decreased the infiltration of TAMs, exhausted CD8+ T cells (PD-1+CD8+ T, TIGIT+CD8+ T), Tregs and MDSC; and increased the proportion of CD8+/CD4+ T cells, effector CD8+ T cells (IFN-γ+ CD8+T), and Th1 cells (IFN-γ+ CD4+ T); the combined therapy also exhibited superior efficacy in MPE (figure 7F–N). In pleural tumors of MPE mice, SBFI-26 and anti-PD-1 therapies reduced the infiltration of TAMs, M2-type TAMs and Tregs, while increasing the infiltration of CD8+ T cells (figure 7O).
Figure 7.
Fatty acid binding protein 5 inhibition alleviates immunosuppression and potentiates response to immune checkpoint blockade therapy. (A–E) Schematic diagram of MPE mouse model was drawn by FigDraw (www.figdraw.com; ID: IYYPP2af53). The MPE mice were randomly assigned to four groups: mice with PBS, mice with anti-PD-1 antibody (α-PD-1), mice with SBFI-26, mice with α-PD-1 and SBFI-26 (A). (B) Representative in vivo bioluminescence images of the growth of mice MPE (n=4). (C) Representative images of mouse MPE and pleural cavity tumors in different groups. MPE volume (D) and Kaplan-Meier survival plot of MPE mice (E) were observed (n=10). (F–N) Tumor-infiltrating immune cells were isolated from mouse MPE. The percentages of TAMs in immune cells (F), TREM2, Tim-3, SIRPα, PD-1, PD-L1 (G and H, and C1q (I) in TAMs, CD8/CD4 (J), IFN-γ+CD8+ and IFN-γ+CD4+ T cells (K), PD-1+ and TIGIT+ CD8+ T cells (L), Treg (M) and MDSC (N) were analyzed by flow cytometry (n=3–6). (O) Paraffin-embedded sections of pleural cavity tumors were analyzed by CD68, CD206, Foxp3 and CD8 staining. Data shown in (B−O) are representative of at least three mice (mean±SD). Statistical analysis was performed using one-way analysis of variance followed by Tukey’s post hoc test (C, E–K) or log-rank test (D, F, H–M). *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns: not statistically significant. C1q, component 1q; IFN, interferon; LLC, Lewis lung cancer cells; MDSC, myeloid derived suppressor cells; MPE, malignant pleural effusion; PBS, phosphate buffer saline; PD-1, programmed cell death-1; PD-L1, programmed cell death-ligand 1; SIRPα, signal regulatory protein α; TAM, tumor-associated macrophage; TIGIT, T cell immunoglobulin and ITIM domains; Tim-3, T-cell immunoglobulin-3; Tregs, T-regulatory cells; TREM2, triggering receptor expressed by myeloid cells-2.
Consistent with MPE, C1q+ TAMs-targeted therapy demonstrated favorable outcomes in mouse subcutaneous model with lung cancer. SBFI-26 and anti-PD-1 treatments significantly reduced tumor size and weight, as well as delayed tumor progression; the combined therapy exhibited superior antitumor effects (online supplemental figures S9A–D). Moreover, both treatments downregulated the expression levels of immunosuppressive molecules and C1q in TAMs, reduced the proportion of TAMs, Tregs and MDSC, increased the proportion of CD8+/CD4+ T cells, effector CD8+ T cells, and Th1 cells in lung cancer. Consistently, the combined therapy also exhibited a synergetic effect (online supplemental figures S9E–L). Overall, these data indicate that inhibiting FABP5 can inhibit TAMs infiltration, retard the progression of MPE and lung cancer, partially rescue immunosuppression in the TME, and improve the efficacy of ICB.
Discussion
The function and action mechanism of TAM subsets, particularly C1q+ TAMs, have remained elusive. In this study, we discovered that C1q+ TAMs were enriched in MPE with high expression levels of immunosuppressive molecules and were associated with poor prognosis in MPE patients. Furthermore, C1q+ TAMs impaired the tumor-killing capacity of CD8+ T cells by promoting FABP5-mediated fatty acid metabolism, which contributed to the upregulation of immunosuppressive molecules.
Based on scRNA-seq analysis of MPE, three subsets of macrophages have been identified among myeloid cells, including FCN1+, IL-1B+, and C1q+ macrophages. FCN1+ macrophages highly express genes associated with pro-inflammatory monocytes and are recognized as the precursors of TAM development.15 18 IL-1β is a well-known marker of M1 macrophages and the IL-1β-producing macrophages exert a pro-inflammatory role.33 While C1q+ macrophages have been identified as a marker of poor prognosis for patients with cancer, correlating with TME immunosuppression.34 Therefore, our study focused on immunosuppressive C1q+ macrophages to reveal their function and action mechanism, with the aim of exploring their potential as a novel target for cancer immunotherapy. In our study, C1q+ TAMs constituted a proportion of 0–40% within the total population of TAMs in MPE. The percentage of C1q+ TAMs in total TAMs was positively associated with tumor burden in MPE. As a result, the limited infiltration of tumor cells in MPE specimens for scRNA-seq led to a low count of C1q+ TAMs.
Although various studies have employed different definitions for C1q+ TAMs based on scRNA-seq, the majority of these studies revealed the co-expression of C1Q, TREM2 and apolipoprotein E (APOE).14 16 35 Zhang et al have reported the co-expression of C1Q, HLA-DR, and MARCO in colon cancer15; Zilionis et al have indicated the co-expression of C1Q and MRC1 in lung cancer24; Dong et al have revealed the high expression of CX3CR1 in C1q+ TAMs.25 In our study, we found that C1q+ TAMs highly expressed several macrophage markers (SPP1, MARCO, MS4A4A, CD163, MRC1, CX3CR1, CSF1R, FOLR2 and APOE) and inhibitory molecules (TREM2, Tim-3, SIRPα, PD-1, PD-L1), implying an immunosuppressive effect in MPE. TREM2, a protumorigenic marker expressed on the surface of tumor infiltrating macrophage, is associated with non-response to ICB therapy via unknown mechanisms.36 37 Tim-3 is expressed in T cells and myeloid cells, such as dendritic cells and macrophages, and is expected to be a target for tumor therapy. Tim-3 inhibits antitumor immunity by mediating T-cell depletion, and blocking Tim-3 pathway promotes the production of IFN-γ by T cells and enhances the antitumor function.38 CD47, the ligand of SIRPα, is expressed in normal cells and functions as a “don’t eat me” signal to prevent the host cells from being phagocytic and cleared by SIRPα-expressing macrophages. In numerous types of cancer, tumor cells evade immune surveillance by upregulating CD47 expression to avoid phagocytosis by macrophages. Inhibition of CD47-SIRPα axis can enhance phagocytic activity and antigen uptake of macrophage.39 Thus, C1q+ TAMs exhibit a potent immunosuppressive phenotype, suggesting a tumor-promoting role in MPE.
However, several studies have identified C1q+ TAMs with distinct functions. Most studies support that C1q+ TAMs correlate with CD8+ T-cell dysfunction and tumor progression; only one study indicates that C1q+ TAMs may recruit or activate T cells based on scRNA-seq analysis of colon cancer.15–17 25 Consistent with most studies, we found that C1q+ TAMs restrained the antitumor effects of CD8+ T cells by upregulating the expression levels of inhibitory molecules, resulting in MPE progression and poor prognosis. This functional heterogeneity may be attributed to distinct TME; further experiments are warranted to be conducted to determine the function in other malignancies. Consistent with the research of Dong et al, MCH-II and CX3CR1 molecules were used as markers to segregate C1q+ and C1q– TAMs.25 CX3CR1, as the marker of tissue resident macrophages, distinguishes them from bone marrow-derived macrophages. Studies have indicated that CX3CR1+ macrophages exhibit M2-like phenotypes in TME thus promoting tumorigenesis.40 41 Both scRNA-seq analysis of human MPE and mouse lung cancer have revealed the high expression of CX3CR1 and HLA-DR (MHC-II in mice) in C1q+ TAMs. Therefore, we selected these two molecules to separate C1q+ and C1q– TAMs after experimental validation. Moreover, we observed that C1q+ TAMs expressed higher levels of IL-10 and TGF-β in MPE. IL-10 and TGF-β have been documented to inhibit CD8+ T-cell cytotoxicity, and the blockade of IL-10 and TGF-β can potentiate antitumor immune response of T cells.26–28 In this study, there is a limit that C1q+ TAMs may modulate T-cell function dependent on immunosuppressive cytokines IL-10 and TGF-β, which will be further explored. In addition, C1q+ TAMs can affect the function of CD4+ T cells. In CRC, C1q+ TAMs interact with T-cell subsets by CXCL10-CXCR3 axis, suggesting the potential role in the recruitment of CD4+ T cells and activation of the Th1 response.15 The impact of C1q+ TAMs on CD4+ T cells warrants further investigation.
A study has reported that the expression of PD-1 on TAMs inhibits their phagocytosis against colon cancer cells.10 Despite high expression of PD-1, C1q+ TAMs showed more potent phagocytic capacity in MPE. Considering that macrophages uptake antigens through phagocytosis, pinocytosis, or opsonization and subsequently present them to T cells,12 and given the high expression of HLA-DR in C1q+ TAMs, we speculated that C1q+ TAMs could enhance their phagocytic ability to promote antigen presentation to CD8+ T cells. This would result in persistent immune activation leading to cellular exhaustion. Consistently, our study demonstrated that C1q+ TAMs indeed inhibited the antitumor effects and induced functional exhaustion of CD8+ T cells in MPE. Nevertheless, besides upregulating immune inhibitory molecules, it remains unclear whether and how the antigen presentation of C1q+ TAMs contributes to CD8+ T-cell dysfunction, necessitating further investigation in the future.
To reveal the underlying mechanism by which C1q+ TAMs with high expression of inhibitory molecules impede antitumor response of CD8+ T cells, we conducted scRNA-seq analysis to identify DEGs and enriched signaling pathways between C1q+ TAMs and C1q– TAMs. Notably, the fatty acid metabolism pathway with an increased expression level of FABP5 was significantly upregulated in C1q+ TAMs. Considering that tumor-promoting TAMs preferentially use mitochondria-dependent fatty acid oxidation as their energy supply,42 we hypothesize that C1q+ TAMs may exert immunosuppressive effects by FABP5-mediated fatty acid metabolic reprogramming. In addition, the PPAR signaling pathway was also upregulated in C1q+ TAMs with high expression level of PPAR-γ, which can be activated by FABP5.30 It is well-established that fatty acid oxidation upregulation in TAMs depends on the transcription factor PPAR-γ, which upregulates the expression of its target genes and is vital for protumor TAMs polarization.43 Studies have reported that PPAR-γ can upregulate the expression of PD-L1 in CRC cells,44 and M2 markers (Arg1, Ym1, and Fizz1) in macrophages45 by binding to and activating DNA-motifs similar to cognate PPAR-γ responsive elements located in the promoter regions of PD-L1, Arg1, Ym1, and Fizz1 genes, suggesting that high expression of inhibitory molecules in C1q+ TAMs may attribute to PPAR-γ upregulation. Therefore, we proposed that C1q+ TAMs could enhance fatty acid metabolism to promote MPE immunosuppression through FABP5 upregulation, which activates PPAR-γ and increases the expression of inhibitory molecules in C1q+ TAMs. Encouragingly, the in vitro results were consistent with the hypothesis. Whereas, it appears that C1q does not directly regulate the expression of FABP5. Our study in vitro observed a significant increase in C1q expression levels on lactate treatment, while FABP5 expression remained unchanged. Further investigation will be conducted to explore the interaction between C1q and FABP5, as well as the factors that induce the upregulation of FABP5 in C1q+ TAMs.
The upregulation of fatty acid metabolism provides survival advantages for the tumor to resist antitumor treatments, and contributes to an immunosuppressive TME which promotes immune escaping of tumor cells.46 Similarly, fatty acid metabolism of C1q+ TAMs was upregulated to promote MPE immunosuppression. Due to the protumor effect of fatty acid in several cancer types,47 48 we wonder whether diets with varying fat content can impact TAM phenotype and then MPE microenvironment. Our results show that an HFD enhances TAMs-mediated immunosuppression in MPE and reduces the efficacy of FABP5 inhibition; while a low-fat diet partially ameliorates the immunosuppression induced by an HFD. These findings imply that an HFD exacerbates MPE immunosuppression and is detrimental to MPE treatment. Moreover, FABP5 inhibition can enhance the efficacy of ICB treatment in MPE and lung cancer, indicating C1q+ TAMs can be harnessed as a therapeutic target in combination with ICB therapy for antitumor immunotherapy.
Nonetheless, it remains elusive whether the secretory protein C1q exerts a direct impact on the TME. C1q, mainly produced by macrophages, is associated with poor prognosis in multiple cancers.34 As the initiating protein of the complement cascade, C1q promotes tumor progression instead of inducing tumor cell death in some cancer types.34 It could be attributed to immunosuppressive TME and T-cell exhaustion triggered by C5-mediated chronic inflammation, which is caused by low expression of terminal pathway components in tumor and the limited formation of membrane attack complex.16 17 20 Besides promoting tumor progression via the complement cascade, C1q may inhibit the response of tumor-infiltration CD8+ T cells to antigen stimulation, inducing their functional exhaustion.34 Our study unveils the mechanism that C1q+ TAMs upregulate the expression of inhibitory molecules to promote immunosuppression in MPE, but the direct effects of C1q on the TME remain unknown and need to be further investigated. Additionally, the mechanisms by which C1q-triggered activation of complement cascade facilitates tumor progression remain incompletely understood and deserve in-depth exploration.
jitc-2023-007441supp013.pdf (21.4KB, pdf)
jitc-2023-007441supp016.pdf (123KB, pdf)
Footnotes
SZ, WP, HW and XX contributed equally.
Contributors: SZ, WP and QZ designed the project. SZ and HW performed most of the experiments. XX, LY, XW, ZW and QX helped to perform a part of experiments. SZ analyzed data obtained from the experiments. WP performed single-cell RNA sequencing data analysis. SZ wrote the manuscript and prepared figures. QZ, as the guarantor for the overall content, approved and supervised the project. All authors reviewed the final manuscript.
Funding: This work is funded by the National Natural Science Foundation of China (No. 82170105, No. 81973990, No. 82100111, No. 82200113).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. The human malignant pleural effusion data set is available at the Sequence Read Archive with BioProject accession no. PRJNA970083. The single-cell RNA sequencing data sets from non-small cell lung cancer (Zilionis et al, 2019), kidney renal clear cell carcinoma (Yu et al, 2023) and colorectal cancer (Zhang et al, 2020) were available from GEO: GSE127465, GEO: GSE207493, and GEO: GSE146771, respectively. Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All animals were maintained in the specific pathogen-free barrier facility in the Animal Center of Huazhong University of Science and Technology. All animal experiments were approved by the Committee on Ethics of Animal Experiments at the Tongji Medical College of Huazhong University of Science and Technology (IACUC Number: 2973), and were performed in accordance with the Guide for the Care and Use of Laboratory Animals.
References
- 1. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 2. Antonia SJ, Villegas A, Daniel D, et al. Overall survival with Durvalumab after Chemoradiotherapy in stage III NSCLC. N Engl J Med 2018;379:2342–50. 10.1056/NEJMoa1809697 [DOI] [PubMed] [Google Scholar]
- 3. Wei SC, Levine JH, Cogdill AP, et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 Checkpoint blockade. Cell 2017;170:1120–33. 10.1016/j.cell.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Higgins KA, Puri S, Gray JE. Systemic and radiation therapy approaches for locally advanced non-small-cell lung cancer. J Clin Oncol 2022;40:576–85. 10.1200/JCO.21.01707 [DOI] [PubMed] [Google Scholar]
- 5. Vesely MD, Zhang T, Chen L. Resistance mechanisms to anti-PD cancer Immunotherapy. Annu Rev Immunol 2022;40:45–74. 10.1146/annurev-immunol-070621-030155 [DOI] [PubMed] [Google Scholar]
- 6. Zhang SY, Song XY, Li Y, et al. Tumor-associated Macrophages: A promising target for a cancer Immunotherapeutic strategy. Pharmacol Res 2020;161:S1043-6618(20)31419-5. 10.1016/j.phrs.2020.105111 [DOI] [PubMed] [Google Scholar]
- 7. Wang ZH, Peng WB, Zhang P, et al. Lactate in the tumour Microenvironment: from immune modulation to therapy. EBioMedicine 2021;73:103627. 10.1016/j.ebiom.2021.103627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mantovani A, Allavena P, Marchesi F, et al. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov 2022;21:799–820. 10.1038/s41573-022-00520-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang H, Liang Y, Anders RA, et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest 2018;128:580–8. 10.1172/JCI96061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated Macrophages inhibits Phagocytosis and tumour immunity. Nature 2017;545:495–9. 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and Immunotherapy. Nat Rev Immunol 2019;19:369–82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christofides A, Strauss L, Yeo A, et al. The complex role of tumor-infiltrating Macrophages. Nat Immunol 2022;23:1148–56. 10.1038/s41590-022-01267-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren X, Zhang L, Zhang Y, et al. Insights gained from single-cell analysis of immune cells in the tumor Microenvironment. Annu Rev Immunol 2021;39:583–609. 10.1146/annurev-immunol-110519-071134 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Q, He Y, Luo N, et al. Landscape and Dynamics of single immune cells in hepatocellular carcinoma. Cell 2019;179:829–45. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 15. Zhang L, Li Z, Skrzypczynska KM, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 2020;181:442–59. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 16. Obradovic A, Chowdhury N, Haake SM, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor Macrophages. Cell 2021;184:2988–3005. 10.1016/j.cell.2021.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J, Lin P, Yang M, et al. Integrated Genomic and Transcriptomic analysis reveals unique characteristics of hepatic metastases and pro-metastatic role of complement C1Q in Pancreatic Ductal adenocarcinoma. Genome Biol 2021;22:4. 10.1186/s13059-020-02222-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Q, Zhang H, Wei T, et al. Single-cell RNA sequencing reveals the heterogeneity of tumor-associated macrophage in non-small cell lung cancer and differences between sexes. Front Immunol 2021;12:756722. 10.3389/fimmu.2021.756722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bulla R, Tripodo C, Rami D, et al. C1Q acts in the tumour Microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun 2016;7:10346. 10.1038/ncomms10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roumenina LT, Daugan MV, Noé R, et al. Tumor cells hijack macrophage-produced complement C1Q to promote tumor growth. Cancer Immunol Res 2019;7:1091–105. 10.1158/2326-6066.CIR-18-0891 [DOI] [PubMed] [Google Scholar]
- 21. Murthy P, Ekeke CN, Russell KL, et al. Making cold malignant pleural effusions hot: driving novel Immunotherapies. Oncoimmunology 2019;8:e1554969. 10.1080/2162402X.2018.1554969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye L-L, Peng W-B, Niu Y-R, et al. Accumulation of Tnfr2-expressing regulatory T cells in malignant pleural effusion of lung cancer patients is associated with poor prognosis. Ann Transl Med 2020;8:1647. 10.21037/atm-20-7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu M-F, Lin C-A, Yuan T-H, et al. The M1/M2 spectrum and plasticity of malignant pleural effusion-macrophage in advanced lung cancer. Cancer Immunol Immunother 2021;70:1435–50. 10.1007/s00262-020-02781-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zilionis R, Engblom C, Pfirschke C, et al. Single-cell Transcriptomics of human and Mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 2019;50:1317–34. 10.1016/j.immuni.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong L, Chen C, Zhang Y, et al. The loss of RNA N(6)-Adenosine methyltransferase Mettl14 in tumor-associated Macrophages promotes Cd8(+) T cell dysfunction and tumor growth. Cancer Cell 2021;39:945–957. 10.1016/j.ccell.2021.04.016 [DOI] [PubMed] [Google Scholar]
- 26. Zhang B, Vogelzang A, Miyajima M, et al. B cell-derived GABA elicits IL-10(+) Macrophages to limit anti-tumour immunity. Nature 2021;599:471–6. 10.1038/s41586-021-04082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sullivan KM, Jiang X, Guha P, et al. Blockade of interleukin 10 potentiates Antitumour immune function in human colorectal cancer liver metastases. Gut 2023;72:325–37. 10.1136/gutjnl-2021-325808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Batlle E, Massagué J. Transforming growth factor-Β signaling in immunity and cancer. Immunity 2019;50:924–40. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El Kharbili M, Aviszus K, Sasse SK, et al. Macrophage programming is regulated by a cooperative interaction between fatty acid binding protein 5 And peroxisome Proliferator-activated receptor gamma. FASEB J 2022;36:e22300. 10.1096/fj.202200128R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo Y, Liu Y, Zhao S, et al. Oxidative stress-induced Fabp5 S-Glutathionylation protects against acute lung injury by suppressing inflammation in Macrophages. Nat Commun 2021;12:7094. 10.1038/s41467-021-27428-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishtiaq SM, Arshad MI, Khan JA. PPARγ signaling in Hepatocarcinogenesis: mechanistic insights for cellular Reprogramming and therapeutic implications. Pharmacol Ther 2022;240:S0163-7258(22)00192-9. 10.1016/j.pharmthera.2022.108298 [DOI] [PubMed] [Google Scholar]
- 32. Lei Q, Yu Z, Li H, et al. Fatty acid-binding protein 5 aggravates pulmonary artery fibrosis in pulmonary hypertension secondary to left heart disease via activating WNT/Β-Catenin pathway. J Adv Res 2022;40:197–206. 10.1016/j.jare.2021.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu J, Han Z, Wu Z, et al. GABA regulates IL-1Beta production in Macrophages. Cell Rep 2022;41:S2211-1247(22)01653-9. 10.1016/j.celrep.2022.111770 [DOI] [PubMed] [Google Scholar]
- 34. Revel M, Sautès-Fridman C, Fridman W-H, et al. C1Q+ Macrophages: passengers or drivers of cancer progression. Trends Cancer 2022;8:517–26. 10.1016/j.trecan.2022.02.006 [DOI] [PubMed] [Google Scholar]
- 35. Huang Z-Y, Shao M-M, Zhang J-C, et al. Single-cell analysis of diverse immune phenotypes in malignant pleural effusion. Nat Commun 2021;12:6690. 10.1038/s41467-021-27026-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molgora M, Esaulova E, Vermi W, et al. Trem2 modulation Remodels the tumor myeloid landscape enhancing anti-PD-1 Immunotherapy. Cell 2020;182:886–900. 10.1016/j.cell.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H, Liu Z, Wen H, et al. Immunosuppressive Trem2(+) Macrophages are associated with undesirable prognosis and responses to anti-PD-1 Immunotherapy in non-small cell lung cancer. Cancer Immunol Immunother 2022;71:2511–22. 10.1007/s00262-022-03173-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev 2017;276:97–111. 10.1111/imr.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ribas A, Wolchok JD. Cancer Immunotherapy using Checkpoint blockade. Science 2018;359:1350–5. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishida Y, Kuninaka Y, Yamamoto Y, et al. Pivotal involvement of the Cx3Cl1-Cx3Cr1 axis for the recruitment of M2 tumor-associated Macrophages in skin carcinogenesis. J Invest Dermatol 2020;140:1951–61. 10.1016/j.jid.2020.02.023 [DOI] [PubMed] [Google Scholar]
- 41. Rivas-Fuentes S, Salgado-Aguayo A, Arratia-Quijada J, et al. Regulation and biological functions of the Cx3Cl1-Cx3Cr1 axis and its relevance in solid cancer: A mini-review. J Cancer 2021;12:571–83. 10.7150/jca.47022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nomura M, Liu J, Rovira II, et al. Fatty acid oxidation in macrophage polarization. Nat Immunol 2016;17:216–7. 10.1038/ni.3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu S, Zhang H, Li Y, et al. S100A4 enhances Protumor macrophage polarization by control of PPAR-gamma-dependent induction of fatty acid oxidation. J Immunother Cancer 2021;9:e002548. 10.1136/jitc-2021-002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gutting T, Hauber V, Pahl J, et al. Ppargamma induces PD-L1 expression in MSS+ colorectal cancer cells. Oncoimmunology 2021;10:1906500. 10.1080/2162402X.2021.1906500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tian Y, Yang C, Yao Q, et al. Procyanidin B2 activates Ppargamma to induce M2 polarization in Mouse Macrophages. Front Immunol 2019;10:1895. 10.3389/fimmu.2019.01895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corn KC, Windham MA, Rafat M. Lipids in the tumor Microenvironment: from cancer progression to treatment. Prog Lipid Res 2020;80:S0163-7827(20)30035-7. 10.1016/j.plipres.2020.101055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miska J, Chandel NS. Targeting fatty acid metabolism in glioblastoma. J Clin Invest 2023;133:e163448. 10.1172/JCI163448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sena LA, Denmeade SR. Fatty acid synthesis in prostate cancer: vulnerability or Epiphenomenon Cancer Res 2021;81:4385–93. 10.1158/0008-5472.CAN-21-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007441supp010.pdf (3.3MB, pdf)
jitc-2023-007441supp011.pdf (113.1KB, pdf)
jitc-2023-007441supp001.pdf (3.6MB, pdf)
jitc-2023-007441supp012.pdf (153.4KB, pdf)
jitc-2023-007441supp014.pdf (27.2KB, pdf)
jitc-2023-007441supp015.xlsx (260.9KB, xlsx)
jitc-2023-007441supp002.pdf (3.4MB, pdf)
jitc-2023-007441supp003.pdf (3MB, pdf)
jitc-2023-007441supp004.pdf (1.6MB, pdf)
jitc-2023-007441supp005.pdf (4.4MB, pdf)
jitc-2023-007441supp006.pdf (791.5KB, pdf)
jitc-2023-007441supp007.pdf (1,005.1KB, pdf)
jitc-2023-007441supp008.pdf (1.5MB, pdf)
jitc-2023-007441supp009.pdf (1.1MB, pdf)
jitc-2023-007441supp013.pdf (21.4KB, pdf)
jitc-2023-007441supp016.pdf (123KB, pdf)
Data Availability Statement
Data are available upon reasonable request. The human malignant pleural effusion data set is available at the Sequence Read Archive with BioProject accession no. PRJNA970083. The single-cell RNA sequencing data sets from non-small cell lung cancer (Zilionis et al, 2019), kidney renal clear cell carcinoma (Yu et al, 2023) and colorectal cancer (Zhang et al, 2020) were available from GEO: GSE127465, GEO: GSE207493, and GEO: GSE146771, respectively. Data are available upon reasonable request.