Abstract
Human cytomegalovirus (HCMV)-specific CD8+ cytotoxic T lymphocytes (CTL) appear to play an important role in the control of virus replication and in protection against HCMV-related disease. We have previously reported high frequencies of memory CTL precursors (CTLp) specific to the HCMV tegument protein pp65 in the peripheral blood of healthy virus carriers. In some individuals, the CTL response to this protein is focused on only a single epitope, whereas in other virus carriers CTL recognized multiple epitopes which we identified by using synthetic peptides. We have analyzed the clonal composition of the memory CTL response to four of these pp65 epitopes by sequencing the T-cell receptors (TCR) of multiple independently derived epitope-specific CTL clones, which were derived by formal single-cell cloning or from clonal CTL microcultures. In all cases, we have observed a high degree of clonal focusing: the majority of CTL clones specific to a defined pp65 peptide from any one virus carrier use only one or two different TCRs at the level of the nucleotide sequence. Among virus carriers who have the same major histocompatibility complex (MHC) class I allele, we observed that CTL from different donors that recognize the same peptide-MHC complex often used the same Vβ segment, although other TCR gene segments and CDR3 length were not in general conserved. We have also examined the clonal composition of CTL specific to pp65 peptides in asymptomatic human immunodeficiency virus-infected individuals. We have observed a similarly focused peptide-specific CTL response. Thus, the large population of circulating HCMV peptide-specific memory CTLp in virus carriers in fact contains individual CTL clones that have undergone extensive clonal expansion in vivo.
CD8+ cytotoxic T lymphocytes (CTL) recognize virus-infected cells via the T-cell receptor (TCR), an αβ heterodimer that has specificity for the peptide antigen presented by major histocompatibility complex (MHC) class I molecules. During T-cell development in the thymus, the TCR β-chain is constructed by rearrangement of variable (V), diversity (D), and joining (J) gene segments, and the α-chain by rearrangement of V and J segments. Additional diversity is generated by imperfect joining of these segments, exonucleotide nibbling at the joins, and addition of non-germ line-encoded N-region nucleotides (25). The regions spanning the V-D-J and V-J joins constitute the hypervariable CDR3 regions which are thought to interact with the middle of the bound peptide and to account for approximately 50% of the TCR’s interaction with peptide (14, 15, 20). The α- and β-chain complementarity determining regions CDR1, which reside within the TCR V segments, are thought to interact with the N and C termini of a peptide that is bound to MHC. By contrast, Vα and Vβ CDR2s are thought to interact predominantly with the MHC itself (14, 15).
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that infects between 60 and 90% of individuals, depending on the population studied. After primary HCMV infection, the virus persists lifelong in a latent state in cells of the myeloid lineage and under the control of the immune system (5). HCMV reactivation can, however, cause serious disease in immunocompromised individuals, such as patients with advanced human immunodeficiency virus (HIV) infection (30) and patients who have undergone bone marrow transplantation (33). Evidence from animal models (32) and from studies of immunosuppressed humans (39) indicates that virus-specific CD8+ CTL have a role in protection against CMV disease.
We previously studied in detail the HCMV-specific CTL response in healthy virus carriers. All seropositive donors had high frequencies of MHC-restricted HCMV-specific memory CTL precursors in peripheral blood and strongly recognized one of the viral tegument proteins, pp65. In some donors, the CTL response to this protein was highly focused, recognizing only a single epitope within pp65, whereas in others the CTL recognized multiple pp65 peptides (41 and unpublished data).
The aim of this study was to examine the clonal composition of the memory CTL response to HCMV pp65 by determining how many different CTL clones are involved in the recognition of a given pp65 peptide. In order to do this, we analyzed the TCR α- and β-chain usage of multiple independently derived peptide-specific CTL clones from healthy virus carriers.
Previous studies have examined the heterogeneity of the CTL response to other human virus infections within single subjects (2, 8, 11, 18, 19, 22, 38) or between different donors (2, 6, 8, 11, 23, 38). In the most extreme cases, a very high degree of TCR focusing has been seen: in a study of one HIV-positive individual’s CTL response to an HLA-B14-restricted HIV env peptide, the same TCR was used by 9 of 10 peptide-specific CTL clones, each derived at different time points over the course of 36 months (22). Similarly, multiple independent CTL clones specific to an HLA-B8-restricted Epstein-Barr virus (EBV) peptide derived from one virus carrier at one time point all used the same TCR (2). The CTL response to different human T-lymphotropic virus type 1 (HTLV-1) peptides has been observed to be oligoclonal within individual donors (38). However, in a variety of other human and mouse viral infections within a given individual, the repertoire of CTL specific for a given peptide has been highly heterogeneous (8, 11, 18, 19).
The TCRs of CTL obtained from different donors that recognize the same peptide-MHC complex often show some conservation of gene segment usage, although they differ in hypervariable sequence. For example, Vβ segments and certain β-chain CDR3 motifs were conserved between TCR that recognized an HLA-A2-restricted influenza virus peptide in CTL clones derived from different donors (23); the same phenomenon has been seen for an HLA-B27 restricted influenza virus peptide (6) and an HLA-A11-restricted EBV peptide (8). A much higher degree of TCR conservation has also been seen; the same TCR α- and β-chain protein sequences were used by CTL clones from four of five unrelated donors that recognized an HLA-B8 restricted EBV peptide (2). In the case of HTLV-1, CTL from different donors that were specific to the same peptide used largely unrelated TCR (38).
For all of the human viruses so far studied, the clonal composition of virus-specific CTL has only been examined for a very few viral peptide-MHC combinations, sometimes in only one donor or at only one time point. In this study, we have therefore examined multiple CTL clones specific to a total of four pp65 peptides, all restricted by three different HLA alleles. We have derived these clones from six healthy virus carriers at one to four time points up to 18 months apart. To identify CTL clonotypes for longitudinal studies and to determine whether HIV infection modifies the clonal composition of HCMV-specific CTL, we have also examined pp65-specific memory CTL in two asymptomatic HIV-infected subjects who are HCMV seropositive. For any given individual, whether HIV seropositive or seronegative, our results indicate that the memory CTL response to individual HCMV pp65 epitopes is highly focused and contains CTL clones that have undergone extensive expansion in vivo.
MATERIALS AND METHODS
Donors.
Six healthy HIV-seronegative laboratory donors and two asymptomatic HIV-seropositive donors attending the HIV clinic at Addenbrooke’s Hospital, Cambridge, United Kingdom, were studied. All donors were HCMV seropositive as determined by an immunoglobulin G enzyme-linked immunosorbent assay (IgG ELISA; Captia HCMV IgG immunoassay; Centocor Inc., Malvern, Pa.). MHC class I tissue types were established for each of the donors by serological typing (Lymphotype ABC-120; Biotest, AG, Dreieich, Germany) and are shown in Table 1.
TABLE 1.
MHC class I tissue types of donors
| Donor | HCMV IgG | HIV | HLA type |
|---|---|---|---|
| 001 | + | − | A2 B12(44) B27 |
| 002 | + | − | A9(24) A19(33) B7 B12(44) |
| 009 | + | − | A2 A3 B7 B37 |
| 011 | + | − | A2 A9(23) B15(62) B12(44) |
| 015 | + | − | A2 A3 B7 B7 |
| 016 | + | − | A3 A3 B7 B7 |
| H0009 | + | + | A2 A9(24) B15(62) B27 |
| H0018 | + | + | A2 A3 B7 B12(44) |
Viruses and cell lines.
HCMV AD169 (ATCC VR-538) was grown in GMO5387 fibroblasts (Coriell Cell Repositories) infected at a multiplicity of infection (MOI) of 0.01. Whole infected cultures were harvested 5 days after a 100% cytopathic effect was evident and were spun at 8,000 rpm. Pellets were pooled, resuspended in 5 ml of RPMI 1640 medium, and sonicated in an ice-cooled water sonication bath. The resulting mixture was divided into 200-μl samples and frozen at −70°C. Titers of stocks were determined on 12-well plates of GMO5387s by using 10-fold dilutions of virus and a 10-day incubation before reading. Recombinant vaccinia viruses expressing the HCMV protein pp65 (Vacpp65; a gift of S. Riddell, Fred Hutchinson Cancer Research Center, Seattle, Wash.) and a negative control expressing an irrelevant protein bacteriophage RNA polymerase T7 (VacT7) were grown in BHK cells infected at an MOI of 0.1. After 48 to 72 h the infected cells were harvested and subjected to three rounds of freeze-thawing followed by sonication. The cell debris was removed by centrifugation, and supernatant containing virus was divided into aliquots and stored at −70°C. The titer of one aliquot was subsequently determined on Vero cells; aliquots normally contained between 1.5 × 107 to 10 × 107 PFU/ml. B-lymphoblastoid cell lines (LCL) were established from peripheral blood mononuclear cells (PBMC) by EBV transformation (B95.8) and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 105 IU of penicillin per liter, and 100 mg of streptomycin per liter.
Production of peptides from the matrix protein pp65.
A panel of 15-amino-acid (15mer) peptides overlapping by 9 amino acids were constructed as described previously (41). These peptides are numbered 1 to 80; peptide 1 starts at amino acid 85 of pp65, and peptide 80 ends at the C terminus. Minimal 9- or 10-amino-acid peptides corresponding to proposed MHC-restricted epitopes were synthesized and purified (>95%) by high-performance liquid chromatography (Affiniti Research Products, Ltd., Exeter, United Kingdom). All peptides were dissolved in RPMI 1640 at 200 μg/ml and frozen in small aliquots at −70°C.
Generation of HCMV-specific CTL in LDA.
The methodology of limiting dilution analysis (LDA) as used for this study has previously been described in detail (9). Briefly, PBMC were prepared from fresh heparinized venous blood samples by Ficoll-Hypaque (Lymphoprep; Nyegaard, Oslo, Norway) density gradient centrifugation. In order to remove natural killer (NK) cells, 1.5 × 107 PBMC were incubated with 20 μl of anti-CD16 IgM monoclonal antibody (Leu11b; Becton-Dickinson) for 30 min at 4°C. After one wash in phosphate-buffered saline (Oxoid), the antibody-labeled PBMC were mixed with a 1:2 dilution of baby rabbit complement in RPMI 1640 and incubated for 45 min at 37°C. After a washing in RPMI 1640, the NK-cell-depleted PBMC were used as responder cells in LDA. Replicate microcultures (n = 18 to 27) were set up in 96-well round-bottom plates in which the initial number of responder cells per well was progressively reduced over an appropriate range of dilutions in RPMI 1640 supplemented with 10% human HCMV-seronegative AB serum (Blood Transfusion Service, Addenbrookes Hospital), 2 mM l-glutamine, 105 IU of penicillin per liter, and 100 mg of streptomycin per liter (from now on referred to as RPMI-HuAB). As antigen-presenting cells, autologous PBMC were pulsed for 1 h with either HCMV at an MOI 0.01 or with defined pp65 peptides irradiated (2,400 rad) and added at 5 × 104 per well. The medium was further supplemented with human recombinant interleukin-2 (IL-2) to give a final concentration of 5 IU/ml. The cultures were incubated at 37°C in 5% CO2 and refed with RPMI-HuAB supplemented with 5 IU of IL-2 per ml on days 5 and 10. On day 14, the cells in each individual well were resuspended and divided into five aliquots that were assayed for cytotoxicity against radiolabeled target cells expressing different HCMV antigens in 4-h 51Cr release cytotoxicity assays. Target cells comprised autologous or MHC-mismatched LCL (4 × 103 cells/well) that had been infected for 18 h with Vacpp65 or VacT7 (MOI = 10) and radiolabeled with 51Cr (Amersham) for 45 min at 37°C.
Preparation of target cells expressing peptide.
pp65 peptide-pulsed target cells were prepared by first labeling LCL with 51Cr (Amersham) for 45 min at 37°C and then pulsing them with 50 μl of peptide at a concentration of 40 μg/ml for an additional hour. Target cells were washed three times in 5 ml of RPMI 1640–10% FCS and counted for use in the chromium release assays.
Analysis of LDA.
The method of analysis has been described previously (9, 41). For each target cell type, the percentage of specific lysis was calculated for each well by using the following formula: [(test release − spontaneous release)/(maximum release − spontaneous release)] × 100. A well was considered, by split-well analysis, to be positive for MHC-restricted cytotoxicity if the percentage of specific lysis against the autologous target was 10% greater than the percentage of specific lysis against the MHC-mismatched control target. Limiting dilution plots for MHC-restricted killing were produced by plotting the proportion of negative wells against the initial responder cell number per well on a semilogarithmic plot. From the single-hit Poisson model, the frequency of antigen-specific CTL precursors (CTLp) was estimated from the initial responder cell number at which 37% of the wells were negative for cytotoxicity (9). All calculations were performed by using a series of Excel Macros written by one of the authors (M.P.W.).
Monoclonal antibodies and surface phenotyping.
The cell surface phenotype of CD16-depleted PBMC and responder cell populations (either the progeny of individual LDA microcultures or cells pooled from multiple microcultures after 14 days of stimulation) was determined by flow cytometry. Results were analyzed on a FACSort flow cytometer (Becton Dickinson). TCR Vβ-chain usage was determined by three-color immunofluorescence with a panel of TCR-specific fluorescein isothiocyanate-conjugated monoclonal antibodies (the panel included Vβ 1, 2, 3, 5.1, 5.2, 6.7, 7, 8.1, 11, 12, 13.6, 14, 16, 17, 19, 20, 21.3, and 22 (Coulter); Vβ 13.1/13.3; and Vα2 [Serotech]) with phycoerythrin (PE)-conjugated anti-CD8 and PerCP-conjugated CD3 (Becton Dickinson).
Derivation of formal single-cell clones and expansion of low-dilution CTL microcultures.
Formal single-cell clones were obtained by subculture of selected LDA microcultures by dilution at 0.5 cells/well in 96-well U-bottom plates. At day 0, each well received 5 × 104 irradiated (2,400 rad) allogeneic PBMC in RPMI-HuAB plus 10% FCS (Myoclone; Gibco) plus human recombinant IL-2 (50 IU/ml) plus phytohemagglutinin at a final concentration of 2 μg/ml. Clones were incubated at 37°C in 5% CO2, refed with RPMI-HuAB supplemented with 10% Myoclone and 50 IU of IL-2 per ml twice weekly, and supplemented with 5 × 104 irradiated allogeneic PBMC once weekly. After 2 to 3 weeks, the cells in each individual well were resuspended, and two aliquots were removed to assay for cytotoxicity against 51Cr-labeled target cells pulsed with the relevant peptide or left unpulsed. Clones were selected on the basis of strong killing of the peptide-pulsed target and minimal killing of the unpulsed target. Clonally derived LDA microcultures (selected from master plate dilutions at which fewer than 50% of the microcultures showed peptide-specific cytotoxicity) were expanded by refeeding in a twice-weekly manner as described above, until large cell pellets were visible. These cultures were retested, and those showing peptide-specific cytotoxicity were selected. We have demonstrated separately that individual low-dilution LDA microcultures contain single CTL clones (unpublished observations).
mRNA extraction and cDNA synthesis.
T-cell clones and lines were cultured without feeder cells for at least 10 days prior to RNA preparation. Total RNA was extracted from 1 × 105 to 10 × 105 cells with an RNA extraction kit (Qiagen, West Sussex, United Kingdom). First-strand cDNA was reverse transcribed with an oligo(dT) primer and avian myeloblastosis virus reverse transcriptase by using a reverse transcription kit (Promega, Madison, Wis.) according to the manufacturer’s instructions.
PCR amplification to determine β- or α-chain V region usage.
PCR was performed by using either a panel of TCR Vβ primers based on those published by Loveridge et al. (24) and Rosenberg et al. (35) or a panel of TCR Vα primers based on those published by Kalams et al. (22). In order to ensure that every possible TCR Vβ and Vα subfamily could be recognized by these panels, we designed new subfamily-specific primers. We made a comparison between the sequence of the published Vβ or Vα primer (22, 24, 35) and the sequence of the corresponding region to which the primer should bind for each of the subfamilies (most recently published by Arden et al. [1]). Where it was predicted that a family-specific primer would be unable to bind effectively to a subfamily gene segment due to a sequence difference, a new subfamily-specific primer was designed (for example, our Vβ11 primer had 5′-to-3′ sequence AACAGTCTCCAGAATAAGGACG. The corresponding region in the Vβ11.2 gene segment differed at the last two nucleotides, making efficient priming improbable. In order to be able to detect any Vβ11.2+ TCRs, we therefore designed a new subfamily-specific primer, sequence AACAGTCTCCAGAATAAGGATA. In total, nine new Vβ subfamily-specific TCR primers were designed: Vβ5.5, CATGACTGTTGCTCTGAGATG; Vβ6.3, GGCCTGAGGGATCCATCTC; Vβ6.4, AAGGGATCTTTCTCCACCT; Vβ8.4, TGCAGGGACTGGAATTGCTG; Vβ9.2, ACTCTCCAGACAAAGTTCAT; Vβ11.2, AACAGTCTCCAGAATAAGGATA; Vβ12.3, AGATAAAGGAGAAGTCCCCGAT; Vβ13.4, CACTGACAAAGGAGAAGTCCC; and Vβ13.6, GATAAAGGAGAAGTCCCGAAT. Also, three new Vα subfamily-specific TCR primers were designed: Vα1.2, TTACCCTGGGAGGAACCAGAG; Vα1.4, TTTTTCCAGGAACTGCCAGAG; and Vα8.2, GGAGAGAGTGTGGGGCTGCATC. In addition, corresponding C-region specific primers were used: Cβ, AGGCGGCTGCTCAGGCAGTATCTGGAGTCA (from Loveridge et al. [24]) and Cα, GCTGTTGTTGAAGGCGTTTGCACATGCAAA (from Baer et al. [3]) (synthesized by Genosys Biotechnologies, Inc., Cambridge, United Kingdom). Each reaction was carried out in a total volume of 50 μl containing a 1 mM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim), 3.75 mM MgCl2, each primer at a final concentration of 1 μM, and 1 U of Taq polymerase (Promega) in buffer supplied by the manufacturer. A total of 2 μl of cDNA was used for PCR amplification in each case. The reaction was overlaid with mineral oil, and amplification was performed for 40 cycles of PCR. Conditions were 1 min of denaturation at 94°C, 30 s of annealing at 60°C, and 30 s of extension at 72°C on a DNA thermal cycler (Cetus 9600 Instrument; Perkin-Elmer, Norwalk, Conn.). Next, 45 μl of each PCR product was separated on a 1.3% agarose gel. Expression of Vα or Vβ genes was considered positive when an approximately 300- to 400-nucleotide rearranged band could be visualized with ethidium bromide staining.
TCR α- and β-chain sequencing.
Individual PCR reactions were performed as described above with a selected V-region-specific primer labeled with a 5′ biotin tag (Genosys Biotechnologies). Amplification was confirmed by separating 5 μl of each PCR product on a 1.3% agarose gel and visualizing it by ethidium bromide staining. A negative reagent control was included in each case; no PCR product was ever visible in the negative control lanes. The amplified biotinylated fragments were isolated by using M280-streptavadin-conjugated Dynabeads according to the manufacturer’s instructions (Dynal AS, Oslo, Norway). The biotinylated strands were resuspended in 7 μl of H2O and sequenced with a Cβ or Cα primer positioned 5′ within the C region compared to the corresponding primer used for PCR amplification (Cβseq, AGATCTCTGCTTCTGATG; Cαseq, ATAGGCAGACAGACTTGT). Sequencing was performed with a Sequenase 2.0 kit, and labeling was carried out with [35S]dATP according to the manufacturer’s instructions (Amersham). Completed sequencing reactions were incubated at 80°C for 5 min. Dynabeads were sedimented, and 4 μl of each reaction was separated on a 6% polyacrylamide sequencing gel.
Sequence data.
All TCR sequence data are available from EMBL-GenBank-DDBJ under accession numbers AJ010874-877, AJ010878-883, A010884-886, AJ010887-895, and AJ010896-900.
RESULTS
We previously identified four CTL epitopes restricted by three different HLA alleles from pp65, as shown in Table 2 (reference 41 and unpublished data).
TABLE 2.
pp65 peptides used in this study and their class I MHC restriction elements
| Peptide no.a | Amino acid sequence | Amino acid position in pp65 | Restricting HLA allele |
|---|---|---|---|
| 31 | RPHERNGFTV | 265–274 | B7 |
| 56 | TPRVTGGGAM | 417–426 | B7 |
| 69 | NLVPMVATV | 495–503 | A2 |
| 72 | EFFWDANDIY | 511–525 | B12(44) |
Numbering is as reported by Wills et al. (41).
Characterization of the clonal composition of the memory CTL response to pp65 peptides. (i) Peptide 69.
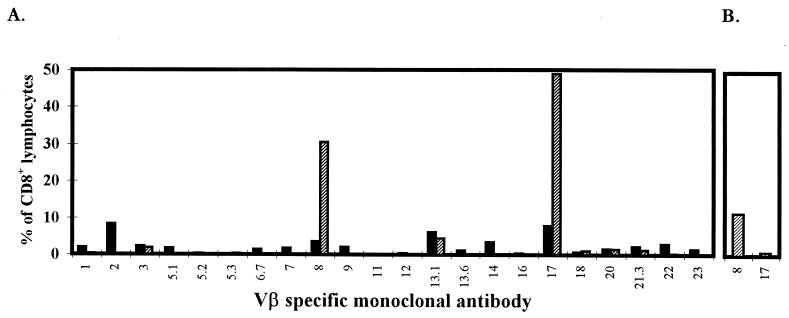
In order to address the question of how many different CTL clones recognize a given peptide-MHC complex, we first determined how many different TCR Vβ gene segments were used by CTL specific to this peptide. Peptide-specific CTL lines were obtained by pooling the residual cells from LDA microcultures that showed strong peptide-specific killing, and their composition was studied by using 2-color immunofluorescence with monoclonal antibodies specific for defined Vβ gene segments. Figure 1A illustrates the percentage of CD8+ lymphocytes from donor 011 that express each Vβ segment. Upon stimulation with peptide 69, Vβ8+ cells are expanded to approximately 31% of the CD8+ population and Vβ17+ cells are expanded to approximately 49%. These Vβ expansions were specific for CTL stimulated with peptide 69; CTL lines generated by stimulation with a different pp65 peptide did not show expansions of Vβ8+ or Vβ17+ cells (Fig. 1B).
FIG. 1.
(A) Analysis of Vβ gene usage by CD8+ T cells in unstimulated PBMC (■) and peptide 69-specific CTL after in vitro stimulation with peptide in LDA (▨) for donor 011. (B) Control analysis of Vβ8 and Vβ17 gene usage by peptide 72-specific CTL derived from LDA in the same experiment. Results are expressed as the percentage of CD8+ T cells that are Vβ-positive.
We next addressed the question of how many different individual CTL clones contribute to these Vβ8+ and Vβ17+ peptide 69-specific CTL populations. At limiting dilution, where <50% of replicate microcultures showed peptide-specific killing, we identified three individual microcultures that showed strong peptide-specific killing; residual cells from each well were subcultured at 0.5 cells per well to generate formal single-cell CTL clones. These clones were retested for peptide-specific cytotoxicity, and the TCRs of clones that showed peptide-specific killing were sequenced. Subclones from two of the three independent limiting-dilution cultures were Vβ8+ and had identical sequences; the third clone was Vβ17+ (Table 3). In order to determine whether these clones were representative of the peptide 69-specific CTL population, we amplified material from multiple independent clonally derived CTL cultures at the same low dilution at a second time point and determined their sequences (Table 3). We found that for Vβ8+ peptide 69-specific CTL, the same three clonotypes could be isolated at a third time point a year later.
TABLE 3.
Amino acid and nucleotide sequences of the TCR β-chains of peptide 69-specific CTL clones from donor 011
| Date of blood sample | Formal clone or CTL culture | % Specific lysis
|
TCR β-chain sequence
|
||||
|---|---|---|---|---|---|---|---|
| With peptide | Control | Vβ | CDR3 | Jβ | Cβ | ||
| 11/96a | Clone 2 | 89 | 4 | Vβ17.1 | C A S M G N S A G A N V L T F G | Jβ2.6 | Cβ2 |
| tgt gcc agt atg ggt aat tca gct ggg gcc aac gtc ctg act ttc ggg | |||||||
| 5/97b | Culture 2 | 56 | 0 | Vβ17.1 | C A S M G N S A G A N V L T F G | Jβ2.6 | Cβ2 |
| tgt gcc agt atg ggt aat tca gct ggg gcc aac gtc ctg act ttc ggg | |||||||
| Culture 3 | 21 | 0 | Vβ17.1 | Jβ2.6 | Cβ2 | ||
| Culture 4 | 22 | 0 | Vβ17.1 | Jβ2.6 | Cβ2 | ||
| Culture 6 | 28 | 0 | Vβ17.1 | Jβ2.6 | Cβ2 | ||
| Culture 8 | 39 | 0 | Vβ17.1 | Jβ2.6 | Cβ2 | ||
| 11/96c | Clone 1 | 88 | 3 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 |
| tgt gcc agc agt tca gct aac tat ggc tac acc ttc ggt | |||||||
| Clone 3 | 87 | 8 | Vβ8 | Jβ1.2 | Cβ1 | ||
| 5/97d | Culture 1 | 54 | 2 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 |
| tgt gcc agc agt tca gct aac tat ggc tac acc ttc ggt | |||||||
| Culture 7 | 27 | 0 | Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | |
| tgt gcc agc agt tct gtc tct gaa gct ttc ttt gga | |||||||
| Culture 9 | 39 | 0 | Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | |
| tgt gcc agc agt tct gtc tct gaa gct ttc ttt gga | |||||||
| Culture 10 | 34 | 0 | Vβ8 | C A S S S V N E Q F F G | Jβ2.1 | Cβ2 | |
| tgt gcc agc agt tca gtt aat gag cag ttc ttc ggg | |||||||
| 4/98e | Culture 3 | 53 | 0 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 |
| Culture 4 | 75 | 0 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 | |
| Culture 7 | 41 | 1 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 | |
| Culture 8 | 76 | 2 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 | |
| Culture 2 | 62 | 0 | Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | |
| Culture 5 | 41 | 1 | Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | |
| Culture 6 | 72 | 0 | Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | |
| Culture 1 | 55 | 1 | Vβ8 | C A S S S V N E Q F F G | Jβ2.1 | Cβ2 | |
Sequence of one Vβ17+ formal clone. The CDR3 region is as defined by Kabat and Wu (21) as starting at position 95 of Vβ and finishing at the phenylalanine of the motif FGXG within Jβ.
Sequences of five independent Vβ17+ CTL cultures. (All five sequences were identical.)
Sequences of two independent Vβ8+ formal clones. (The two sequences were identical.)
Sequences of four independent Vβ8+ CTL cultures.
Sequences of eight independent Vβ8+ CTL cultures.
Peptide 69-specific CTL were analyzed in other virus carriers in a similar manner, by first deriving two to four formal peptide-specific single-cell clones and then sequencing their TCRs. Subsequently, we identified LDA microcultures that showed peptide-specific killing; we then expanded the residual cells and sequenced the TCR. Results of this analysis are shown in Table 4. In all of the donors studied, the memory CTL responses to peptide 69 were similarly focused: for any given donor one predominant β-chain sequence was expressed by multiple independently derived peptide-specific formal clones and also by clonally derived CTL cultures. Similar results were observed for HIV-seropositive donors H0009 and H0018.
TABLE 4.
TCR β-chain sequences derived from eight donors showing the proportion of peptide-specific CTL that use a particular TCR β-chain sequence
| pp65 peptide | Donor | TCR β-chain sequence
|
No. of clones/culture containing this β-chain | |||
|---|---|---|---|---|---|---|
| Vβ | Sequence | Jβ | Cβ | |||
| 69 | 011 | Vβ17.1 | C A S M G N S A G A N V L T F G | Jβ2.6 | Cβ2 | 6/6 |
| Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 | 7/14 | ||
| Vβ8 | C A S S S V S E A F F G | Jβ1.1 | Cβ1 | 5/14 | ||
| Vβ8 | C A S S S V N E Q F F G | Jβ2.1 | Cβ2 | 2/14 | ||
| 015 | Vβ8 | C A S S S A N Y G Y T F G | Jβ1.2 | Cβ1 | 3/3a | |
| 001 | Vβ13.1 | C A S S Y Q T G T G N Y G Y T F G | Jβ1.2 | Cβ1 | 3/4 | |
| Vβ13.1 | C A S S Y S G H V Y E Q Y F G | Jβ2.7 | Cβ1 | 1/4 | ||
| H0009 | Vβ13.1 | C A S R G Q G F S Y E Q Y F G | Jβ2.7 | Cβ2 | 4/4b | |
| Vβ6 | C A S S F L G L N E Q F F G | Jβ2.1 | Cβ2 | 2/2c | ||
| H0018 | Vβ6 | C A S S L D I P S Y N E Q F F G | Jβ2.1 | Cβ2 | 5/5d | |
| 72 | 011 | Vβ6.4 | C A S S L G G N L Y E Q Y F G | Jβ2.7 | Cβ2 | 9/9 |
| 56 | 002 | Vβ6.4 | C A S S S H D T G G Y N S P L H F G | Jβ1.6 | Cβ1 | 2/5 |
| Vβ6.4 | C A S S F R D Y G N Y E Q Y F G | Jβ2.7 | Cβ2 | 3/5 | ||
| 016 | Vβ6.4 | C A S S L I G V S S Y N E Q F F G | Jβ2.1 | Cβ2 | 3/3 | |
| H0018 | Vβ6.4 | C A S S S H D R Q G A S S P L H F G | Jβ1.6 | Cβ1 | 1/1e | |
| 015 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | 8/8 | |
| Vβ14.1 | C A S K V G A G G L Y E Q Y F G | Jβ2.7 | Cβ2 | 1/1f | ||
| 009 | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | 5/5 | |
| 31(10mer) | 015 | Vβ7.1 | C A S S Q A A L A G F G Y E Q Y F G | Jβ2.7 | Cβ2 | 3/3 |
| 016 | Vβ7.2 | C A S S P A R N T E A F F G | Jβ1.1 | Cβ1 | 1/1e | |
| H0018 | Vβ7.2 | C A S S P S R N T E A F F G | Jβ1.1 | Cβ1 | 1/5 | |
| Vβ7.2 | C A S S P H R N T E A F F G | Jβ1.1 | Cβ1 | 4/5 | ||
Two clones and one bulk CTL line used identical Vα8.2 CAE IHNNARLM FG Jα12.
Four cultures on 4/97.
One clone on 2/96; one clone on 9/96.
Three clones on 6/96; two cultures on 1/98.
Only one clone was available for analysis.
One clone of nine analyzed was Vβ14+; the remaining eight were Vβ6.4+.
(ii) Peptide 72.
In peptide 72-specific CTL lines derived from donor 011, we were unable to detect any Vβ expansions with a monoclonal antibody panel that recognizes 50 to 70% of all Vβ segments. We therefore determined the TCR Vβ usage of these CTL lines by extracting mRNA, reverse transcribing it, and then performing PCR with a panel of Vβ-specific primers to amplify the VDJ region of the TCR β-chain.
We amplified material from multiple independently derived formal clones and CTL lines specific for peptide 72 derived at three time points. We found that, in every case, each formal clone or CTL line expressed both Vβ6.4 and Vβ9.2. We sequenced both TCR β-chains (Table 5) and found that the Vβ9.2+ β-chain nucleotide sequence contained a stop codon in the middle of the hypervariable region. It therefore appeared, in this case, that during T-cell development an error had occurred in the rearrangement of the DNA encoding the Vβ9.2+ β-chain of this clone causing it to rearrange another one, the Vβ6.4+ β-chain. We further sequenced the complementary strand of the Vβ9.2+ PCR product and confirmed the presence of the stop codon. We also performed individual PCR reactions with a panel of Vα-specific primers on two of the peptide 72-specific Vβ6.4+-Vβ9.2+ formal clones. In both cases, Vα4 was the only Vα segment expressed by these clones. We sequenced the Vα4+ α-chains of all peptide 72-specific clones and CTL cultures and found in each case that every clone or culture expressed the same α-chain sequence (Table 5).
TABLE 5.
TCR α- and β-chain usage by a pp65 peptide 72-specific CTL clone from donor 011a
| Date of blood sample | Formal clone or CTL culture | % Specific lysis
|
TCR β-chain usage
|
||
|---|---|---|---|---|---|
| With peptide | Control | Vβb | CDR3 | ||
| 11/96 | Clone 1 | 69 | 2 | Vβ6.4 | C A S S L G G N L Y E Q Y |
| tgt gcc agc agc tta ggg gga aac ctt tac gag cag tac | |||||
| Clone 2 | 59 | 0 | Vβ9.2 | C A S S Q E E G V Z | |
| R D P D Q R | |||||
| tgt gcc agc agc caa gaa gaa ggg gta taa gg gac cca gat cag cgc | |||||
| 5/97 | Culture 1 | 22 | 5 | ||
| Culture 2 | 48 | 2 | |||
| Culture 3 | 21 | 1 | |||
| Culture 4 | 27 | 0 | |||
| 1/98 | Clone 1D | 70 | 1 | ||
| Clone 2B | 73 | 0 | |||
| Clone 4B | 69 | 3 | |||
| TCR β-chain usage
|
TCR α-chain usage
|
||||
|---|---|---|---|---|---|
| CDR3 | Jβ | Cβ | Vα | CDR3 | Jα |
| F G | Jβ2.7 | Cβ2 | Vα4.1 | C I L R G G G A D G L T F G | Jα45 |
| ttc ggg | Jβ2.7 | Cβ2 | |||
| Jβ2.7 | Cβ2 | ||||
| Y E Q Y F G | |||||
| tac gag cag tac ttc ggg | |||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
| Jβ2.7 | Cβ2 | ||||
The usage and sequence data presented are identical for all nine clones and cultures listed here.
This clone uses a productively rearranged Vβ6.4+ β-Chain and nonproductively rearranged Vβ9.2+ β-chain. The Vβ9.2+ TCR β-chain sequence is shown in reading frames 1 and 3. Amino acid sequences of the TCR α- and β-chains of nine independent peptide 72-specific CTL clones and microcultures derived from donor 011 over the course of 14 months are shown. All nucleotide sequences are identical and show a very high degree of focusing of this donor’s peptide 72-specific CTL.
(iii) Peptide 56.
For five donors, we amplified material from multiple independently derived formal clones and LDA microcultures that recognized peptide 56 (Table 6). We again observed a high degree of focusing. This dominant clonal response was also seen in bulk CTL lines. For donors 015 and 016, we performed an entire Vβ PCR screen on peptide 56-specific CTL lines. In each case dominant bands were seen corresponding to Vβ6.4 and Vβ14. We sequenced both of the Vβ6.4+ PCR products. Either pure sequence was seen, or sequence contaminated with very faint bands derived from other TCRs was seen. The predominant sequence was always identical to the sequence derived from the independent peptide-specific Vβ6.4+ formal clones, indicating that this population of peptide-specific CTL was highly focused. For donors 015 and 016 we also analyzed the α-chain usage of two peptide 56-specific CTL clones and the bulk peptide 56-specific CTL lines. We confirmed that a given β-chain was always associated with the same α-chain.
TABLE 6.
Clonal focusing of CTL specific to HLA-B7-restricted pp65 peptide 56
| Donora | Date of blood sample | Formal clone or CTL culture | TCR β-chain sequence
|
TCR α-chain sequence
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Vβ | CDR3 | Jβ | Cβ | Vα | CDR3 | Jα | |||
| 002 | 5/98 | Clone A | Vβ6.4 | C A S S S H D T G G Y N S P L H F G | Jβ1.6 | Cβ1 | |||
| Culture 5 | Vβ6.4 | C A S S S H D T G G Y N S P L H F G | Jβ1.6 | Cβ1 | |||||
| Clone B | Vβ6.4 | C A S S F R D Y G N Y E Q Y F G | Jβ2.7 | Cβ2 | |||||
| Clone C | Vβ6.4 | C A S S F R D Y G N Y E Q Y F G | Jβ2.7 | Cβ2 | |||||
| Culture 7 | Vβ6.4 | C A S S F R D Y G N Y E Q Y F G | Jβ2.7 | Cβ2 | |||||
| 009 | 10/97 | Clone B56 | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | |||
| Clone C56 | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | |||||
| 2/98 | Clone 1a | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | ||||
| Clone 1b | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | |||||
| Clone 2a | Vβ14.1 | C A S S I G P A L N T E A F F G | Jβ1.1 | Cβ1 | |||||
| 015 | 1/97 | Clone 5 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | Vα6.1 | C A M R E G M D S S Y K L I F G | Jα31 |
| Clone 7 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | Vα6.1 | C A M R E G M D S S Y K L I F G | Jα31 | ||
| Bulk CTL line | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | Vα6.1 | C A M R E G M D S S Y K L I F G | Jα31 | ||
| 7/97 | Clone 3 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | ||||
| 1/98 | Clone 5 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | ||||
| 3/98 | Clone 2 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | ||||
| Clone 5 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | |||||
| Clone 6 | Vβ6.4 | C A S S L H D R G S R T E A F F G | Jβ1.1 | Cβ1 | |||||
| Clone 3 | Vβ14.1 | C A S K V G A G G L Y E Q Y F G | Jβ2.7 | Cβ2 | |||||
| 016 | 7/97 | Clone 8 | Vβ6.4 | C A S S L I G V S S Y N E Q F F G | Jβ2.1 | Cβ2 | Vα3.1 | C A T V L R M D S S Y K L I F G | Jα21 |
| 9/97 | Clone 4 | Vβ6.4 | C A S S L I G V S S Y N E Q F F G | Jβ2.1 | Cβ2 | Vα3.1 | C A T V L R M D S S Y K L I F G | Jα31 | |
| Bulk CTL line | Vβ6.4 | C A S S L I G V S S Y N E Q F F Q | Jβ2.1 | Cβ2 | Vα3.1 | C A T V L R M D S S Y K L I F G | Jα31 | ||
| H0018 | 1/98 | Clone 1B | Vβ6.4 | C A S S S H D R Q G A S S P L H F G | Jβ1.6 | Cβ1 | |||
TCR α- and β-chain amino acid sequences are shown from CTL clones derived from five donors. These TCRs exclusively use Vβ6.4 or Vβ14. CTL from donors 015 and 016 also both use Jα31.
For donor 002, we sequenced the TCRs of peptide 56-specific formal CTL clones and LDA microcultures that had been stimulated in two different ways: either with PBMC pulsed with peptide 56 (cultures 5 and 7) or with PBMC pulsed with HCMV (clones A, B, and C) (Table 6). Clone A and culture 5 had the same Vβ6.4+ β-chain nucleotide sequence. Clones B and C and culture 7 shared a single Vβ6.4+ β-chain sequence which was different from the β-chain sequence of clone A. This demonstrates that both methods of stimulation in vitro activate the same CTL clonotypes.
(iv) Peptide 31(10mer).
All HLA-B7-positive donors in our study also recognized a 10mer epitope of peptide 31. We studied the clonal composition of CTL specific to this epitope (Table 4).
A summary of all TCR β-chain sequences derived from the eight subjects studied is given in Table 4. For each donor, the proportion of CTL clones to the cultures analyzed that share identical β-chain sequences is also shown.
DISCUSSION
We have shown a high degree of clonal focusing among the memory CTL specific for a given HCMV pp65 peptide-MHC complex within each of the eight donors studied here. In general, for any given donor, CTL specific to a particular peptide used only one or two Vβ segments; in most cases only one or two predominant β-chain clonotype sequences were detected in multiple independently derived peptide-specific formal clones and clonally derived CTL cultures that we analyzed. Whenever we examined α-chain usage of some of these independent CTL clones and microcultures, we always found that a given β-chain was associated with the same α-chain.
It is important to consider how closely the HCMV-specific TCR repertoire analyzed in vitro corresponds to that present in vivo. The CTL clones that we studied were selected both for growth in vitro and for peptide-specific CTL activity (range, 21 to 89% specific lysis). It is possible that the composition of peptide-specific CTL clones in vivo may be more heterogeneous than was suggested by our in vitro analysis because some peptide-specific clones may show reduced in vitro growth and/or have lower TCR affinity for a specific peptide-MHC. To address whether the method of stimulation in vitro influences the observed degree of clonal focusing, we studied in donor 002 peptide 56-specific CTL derived from limiting-dilution cultures that had been stimulated in two different ways: either with PBMC pulsed with peptide or with PBMC pulsed with HCMV. For the multiple independent cultures analyzed, the same two Vβ6.4+ clonotypes were seen regardless of the stimulation protocol used (Table 6); the degree of focusing we have observed was thus independent of the method of CTL generation in vitro. One approach to studying clonal composition without selection for in vitro growth has been to use staining with MHC-peptide tetramers to purify peptide-specific CD8+ T cells directly ex vivo (42). When the TCR of purified HIV gag peptide-specific CD8+ cells were sequenced, 10 of 18 clones were one clonotype and 8 of 18 clones were a second clonotype. These results indicate that this HIV peptide-specific CTL response was highly focused in vivo, although MHC-peptide tetramers may preferentially select high-affinity CTL clonotypes rather than low-affinity CTL clones (if any are present in vivo).
Some donors used more than one Vβ segment to recognize the same peptide-MHC. For example, donor 011 used Vβ8 and Vβ17 to recognize peptide 69. Vβ17+ CTL recognizing peptide 69 had highly focused TCR usage: six independently derived peptide-specific clones or clonal cultures generated at two time points 6 months apart all had identical TCR β-chain sequences. By contrast, Vβ8+ CTL recognizing peptide 69 were more diverse; two independent formal clones derived at the first time point had identical β-chain sequences, whereas only one of four clonal microcultures that were subsequently analyzed expressed this TCR. Three of these four microcultures expressed different TCRs, although all showed some conservation at the level of the hypervariable region motif (either SA or SV). The CDR3 lengths of these TCR β-chains also differed, at either 7 or 8 amino acids, which may be because each TCR CDR3 can interact with different regions of peptide 69. In order to determine how diverse the Vβ8+ response was, eight peptide 69-specific CTL microcultures were analyzed at a third time point a year later. Only the three clonotypes that had been identified at the previous time point were detected, and the clonotype originally identified in the first analysis of two formal clones accounted for four of eight of these cultures.
In order to study donor 011’s peptide 72-specific CTL, we derived nine formal peptide-specific clones and CTL cultures at three time points over the course of 14 months. For every clone or culture, the same Vα4+ α-chain was detected, along with a productively rearranged Vβ6.4+ β-chain and a nonproductively rearranged Vβ9.2+ β-chain. It is very unlikely that during thymic development several different T-cell clones independently acquired an identical nonproductively rearranged Vβ9.2+ β-chain. This Vβ9.2+ β-chain therefore acts as a distinctive genetic marker for all progeny of the single CTL clone in which the defective rearrangement took place. Since all nine peptide 72-specific clones or cultures we analyzed were positive for this marker, this confirms that the vast majority of 011’s peptide 72-specific CTL originate from the expansion of a single precursor cell in vivo.
We observed that CTL from different donors that recognize the same peptide-MHC complex often used the same Vβ gene segment. For example, three Vβ regions were predominantly used by peptide 69-specific CTL derived from the five A2-positive donors we studied: Vβ regions 6, 8, and 13.1. We have now observed Vβ13.1 usage by peptide 69-specific CTL for a total of 4 of 10 HLA-A2-positive HCMV-seropositive donors examined. Interestingly, the same TCR β-chain protein sequence, although not the same nucleotide sequence, was used by Vβ8+ peptide 69-specific CTL from donors 011 and 015 (Table 4), implying that this TCR may be able to bind this peptide efficiently.
In all five HLA-B7-positive donors studied, CTL that recognized peptide 56 used either Vβ6.4 or Vβ14; some donors had both Vβ6.4+ and Vβ14+ peptide 56-specific CTL. Among Vβ6.4+ peptide 56-specific clones from four donors, CDR3 length varied between 11 and 13 amino acids. The CDR3 region from three donors contained the motif HD; two clones from different donors used Jβ1.6. The CTL from donors 015 and 016 that were specific to peptide 56 also used Jα31 but with different Vα segments (Table 6). All HLA-B7-positive donors recognized peptide 31 (10mer); peptide-specific CTL from the three donors studied exclusively used Vβ7 family (Table 4). In addition, TCR β-chain sequences of peptide 31-specific CTL from donors 016 and H0018 were highly similar; three different CTL clones from these two donors used Vβ7.1 and Jβ1.1 and had CDR3 amino acid sequences of the same length that differed at only one position.
We studied the clonal composition of pp65 peptide-specific CTL in two subjects with asymptomatic HIV infection. A similar pattern of clonal focusing was seen in the CTL of these donors.
In the experiments where we analyzed the clonal composition of multiple LDA microcultures, we simultaneously quantified the frequencies of peptide-specific CTLp by using LDA (unpublished data). It is therefore possible to estimate the number of identical functional peptide-specific clones present in vivo. Donor 009’s peptide 56-specific CTLp were present at a frequency of 14,000 per 106 CD8+ T cells (1 of 71 CD8+ T cells). As a conservative estimate, it can be assumed that at least 50% of the observed peptide-specific response in LDA in this donor is mediated by a single CTL clone (since five of five clones or cultures had identical Vβ14+ sequences). If any given individual has 5 × 1010 CD8+ lymphocytes (13), this would imply that donor 009 had approximately 108 identical peptide 56-specific CTL. The lowest peptide-specific CTLp frequency measured was for donor 011’s peptide 72-specific CTL response of 143 peptide-specific CTLp per 106 CD8+ T cells. With the same assumptions, the corresponding clonal size would be 106 cells. If the cloning efficiency of LDA is less than 100%, the actual clonal sizes in vivo may be even larger. Although there may be other CTL clonotypes in the HCMV peptide-specific repertoire in vivo in addition to those that we have characterized here, it is clear that these in vitro-derived clones must have undergone extensive expansion in vivo.
Our results indicate that the large population of circulating memory CTL specific for a given pp65 peptide contains individual CTL clones that have undergone extensive clonal expansion. This finding may provide an explanation for many previous observations of oligoclonal expansions within CD8+ memory T cells in normal human subjects for which no function has hitherto been identified (4, 12, 16, 17, 26, 28, 29, 31, 36, 40). Such expansions may be due to clones of CD8+ CTL specific to HCMV or other common persistent viruses.
There are a number of possible reasons for the high degree of clonal focusing of pp65 peptide specific memory CTL in a healthy virus carrier. The sequence of the peptide eliciting the CTL response in vivo may be a factor: small changes in the amino acid composition of one peptide can change a response from being focused into being more polyclonal (10). Overall, the MHC class I haplotype may have an effect: this has been reported to be an important determinant of whether a heterogeneous or homogeneous CTL response is mounted in response to a B*4402-restricted EBV epitope (7). As HCMV infection is persistent and may reactivate intermittently, the focused pp65-specific CTL response we have observed in HCMV carriers may be the result of repeated exposure to viral antigen with selection of CTL that express certain high-affinity TCRs during maturation into long-term memory (37). Repeated stimulation by persistent viral antigen may also explain the very large clonal sizes we have observed. Supportive evidence comes from studies of other persistent virus infections which have also been associated with large clonal expansions of peptide-specific CTL: EBV (2), HIV-1 (22), and HTLV-1 (38). In contrast to HCMV, the magnitude of the CTL response to a nonpersistent virus, influenza virus, is 10- to 100-fold lower (23) and diminishes after resolution of acute infection (27). It has, however, been proposed that the conserved TCR usage seen by CTL specific to A2-restricted influenza virus peptide M58-66 may be due to exposure to repeatedly encountered antigen during asymptomatic reinfection (23). It will therefore be important to examine the clonal composition of CTL generated in response to primary infection by HCMV and to determine how the composition of this response changes over time.
It has previously been reported that HCMV-specific CTL immunity can be reconstituted by adoptive transfer of cloned CTL (34). The data presented here may explain the relative ease by which clones can be obtained in vitro and show that reconstitution with clones of CTL may in fact provide immunity similar to that generated after natural infection.
In summary, there are very high frequencies of CTLp specific to a variety of pp65 peptides restricted by different class I MHC alleles. There is a high degree of clonal focusing among CTL specific to each of these peptides in the donors studied, and the same clone or clones were repeatedly isolated at multiple time points up to a year apart. The memory CTL response to individual HCMV pp65 epitopes thus contains individual clones that have undergone extensive expansion in vivo.
ACKNOWLEDGMENTS
This work was supported by a program grant from the Medical Research Council. M.P.W. is supported by a Wellcome Trust Prize Studentship, and A.J.C. is a Lister Institute Research Fellow.
REFERENCES
- 1.Arden B, Clark S P, Kabelitz D, Mak T W. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 2.Argaet V P, Schmidt C W, Burrows S R, Silins S L, Kurilla M G, Doolan D L, Suhrbier A, Moss D J, Kieff E, Sculley T B, Misko I S. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J Exp Med. 1994;180:2335–2340. doi: 10.1084/jem.180.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer R, Lefranc M P, Minowada J, Forster A, Stinson M A, Rabbitts T H. Organization of the T-cell receptor alpha-chain gene and rearrangement in human T-cell leukaemias. Mol Biol Med. 1986;3:265–277. [PubMed] [Google Scholar]
- 4.Batliwalla F, Monteiro J, Serrano D, Gregersen P K. Oligoclonality of CD8+ T cells in health and disease: aging, infection, or immune regulation? Hum Immunol. 1996;48:68–76. doi: 10.1016/0198-8859(96)00077-8. [DOI] [PubMed] [Google Scholar]
- 5.Borysiewicz L K, Graham S, Hickling J K, Mason P D, Sissons J G. Human cytomegalovirus-specific cytotoxic T cells: their precursor frequency and stage specificity. Eur J Immunol. 1988;18:269–275. doi: 10.1002/eji.1830180214. [DOI] [PubMed] [Google Scholar]
- 6.Bowness P, Moss P A, Rowland Jones S, Bell J I, McMichael A J. Conservation of T cell receptor usage by HLA B27-restricted influenza-specific cytotoxic T lymphocytes suggests a general pattern for antigen-specific major histocompatibility complex class I-restricted responses. Eur J Immunol. 1993;23:1417–1421. doi: 10.1002/eji.1830230702. [DOI] [PubMed] [Google Scholar]
- 7.Burrows S R, Khanna R, Burrows J M, Moss D J. An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTL) cross-reactive with a single Epstein-Barr virus CTL epitope: implications for graft-versus-host disease. J Exp Med. 1994;179:1155–1161. doi: 10.1084/jem.179.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos-Lima P O, Levitsky V, Imreh M P, Gavioli R, Masucci M G. Epitope-dependent selection of highly restricted or diverse T cell receptor repertoires in response to persistent infection by Epstein-Barr virus. J Exp Med. 1997;186:83–89. doi: 10.1084/jem.186.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmichael A, Jin X, Sissons P, Borysiewicz L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1)-specific cytotoxic T lymphocyte (CTL) response at different stages of HIV-1 infection: differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien Y H, Davis M M. How alpha beta T-cell receptors “see” peptide/MHC complexes. Immunol Today. 1993;14:597–602. doi: 10.1016/0167-5699(93)90199-u. [DOI] [PubMed] [Google Scholar]
- 11.Cole G A, Hogg T L, Woodland D L. The MHC class I-restricted T cell response to Sendai virus infection in C57BL/6 mice: a single immunodominant epitope elicits an extremely diverse repertoire of T cells. Int Immunol. 1994;6:1767–1775. doi: 10.1093/intimm/6.11.1767. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald J E, Ricalton N S, Meyer A C, West S G, Kaplan H, Behrendt C, Kotzin B L. Analysis of clonal CD8+ T cell expansions in normal individuals and patients with rheumatoid arthritis. J Immunol. 1995;154:3538–3547. [PubMed] [Google Scholar]
- 13.Fleury S, De Boer R J, Rizzardi G P, Wolthers K D, Otto S A, Welbon C C, Graziosi C, Knabenhans C, Soudeyns H, Bart P, Gallant S, Corpataux J, Gillet M, Meylan P, Schnyder P, Meuwly J, Spreen W, Glauser M P, Miedema F, Pantaleo G. Limited CD4+ T-cell renewal in early HIV-1 infection: effect of highly active antiretroviral therapy. Nat Med. 1998;4:794–801. doi: 10.1038/nm0798-794. [DOI] [PubMed] [Google Scholar]
- 14.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 15.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 16.Grunewald J, Jeddi Tehrani M, Dersimonian H, Andersson R, Wigzell H. A persistent T cell expansion in the peripheral blood of a normal adult male: a new clinical entity? Clin Exp Immunol. 1992;89:279–284. doi: 10.1111/j.1365-2249.1992.tb06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hingorani R, Choi I H, Akolkar P, Gulwani-Akolkar B, Pergolizzi R, Silver J, Gregersen P K. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J Immunol. 1993;151:5762–5769. [PubMed] [Google Scholar]
- 18.Horwitz M S, Yanagi Y, Oldstone M B. T-cell receptors from virus-specific cytotoxic T lymphocytes recognizing a single immunodominant nine-amino-acid viral epitope show marked diversity. J Virol. 1994;68:352–357. doi: 10.1128/jvi.68.1.352-357.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imarai M, Goyarts E C, van Bleek G M, Nathenson S G. Diversity of T cell receptors specific for the VSV antigenic peptide (N52-59) bound by the H-2Kb class I molecule. Cell Immunol. 1995;160:33–42. doi: 10.1016/0008-8749(95)80006-5. . (Erratum, 162:340.) [DOI] [PubMed] [Google Scholar]
- 20.Jorgensen J L, Esser U, de St Groth B F, Reay P A, Davis M M. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 21.Kabat E A, Wu T T. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 22.Kalams S A, Johnson R P, Trocha A K, Dynan M J, Ngo H S, D’Aquila R T, Kurnick J T, Walker B D. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J Exp Med. 1994;179:1261–1271. doi: 10.1084/jem.179.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehner P J, Wang E C, Moss P A, Williams S, Platt K, Friedman S M, Bell J I, Borysiewicz L K. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loveridge J A, Rosenberg W M, Kirkwood T B, Bell J I. The genetic contribution to human T-cell receptor repertoire. Immunology. 1991;74:246–250. [PMC free article] [PubMed] [Google Scholar]
- 25.Marrack P, Kappler J. The T cell receptor. Science. 1987;238:1073–1079. doi: 10.1126/science.3317824. [DOI] [PubMed] [Google Scholar]
- 26.Maryanski J L, Jongeneel C V, Bucher P, Casanova J L, Walker P R. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 27.McMichael A J, Gotch F M, Dongworth D W, Clark A, Potter C W. Declining T-cell immunity to influenza, 1977–82. Lancet. 1983;i:762–764. doi: 10.1016/s0140-6736(83)92297-3. [DOI] [PubMed] [Google Scholar]
- 28.Monteiro J, Hingorani R, Choi I H, Silver J, Pergolizzi R, Gregersen P K. Oligoclonality in the human CD8+ T cell repertoire in normal subjects and monozygotic twins: implications for studies of infectious and autoimmune diseases. Mol Med. 1995;1:614–624. [PMC free article] [PubMed] [Google Scholar]
- 29.Morley J K, Batliwalla F M, Hingorani R, Gregersen P K. Oligoclonal CD8+ T cells are preferentially expanded in the CD57+ subset. J Immunol. 1995;154:6182–6190. [PubMed] [Google Scholar]
- 30.Peters B S, Beck E J, Anderson S, Coleman D, Coker R, Main J, Migdal C, Harris J R, Pinching A J. Cytomegalovirus infection in AIDS. Patterns of disease, response to therapy and trends in survival. J Infect. 1991;23:129–137. doi: 10.1016/0163-4453(91)91987-9. [DOI] [PubMed] [Google Scholar]
- 31.Posnett D N, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.”. J Exp Med. 1994;179:609–618. doi: 10.1084/jem.179.2.609. . (Erratum, 179:1077.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinnan G V, Jr, Burns W H, Kirmani N, Rook A H, Manischewitz J, Jackson L, Santos G W, Saral R. HLA-restricted cytotoxic T lymphocytes are an early immune response and important defense mechanism in cytomegalovirus infections. Rev Infect Dis. 1984;6:156–163. doi: 10.1093/clinids/6.2.156. [DOI] [PubMed] [Google Scholar]
- 33.Reusser P, Riddell S R, Meyers J D, Greenberg P D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 34.Riddell S R, Watanabe K S, Goodrich J M, Li C R, Agha M E, Greenberg P D. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg W M, Moss P A, Bell J I. Variation in human T cell receptor V beta and J beta repertoire: analysis using anchor polymerase chain reaction. Eur J Immunol. 1992;22:541–549. doi: 10.1002/eji.1830220237. [DOI] [PubMed] [Google Scholar]
- 36.Schwab R, Szabo P, Manavalan J S, Weksler M E, Posnett D N, Pannetier C, Kourilsky P, Even J. Expanded CD4+ and CD8+ T cell clones in elderly humans. J Immunol. 1997;158:4493–4499. [PubMed] [Google Scholar]
- 37.Silins S L, Cross S M, Elliott S L, Pye S J, Burrows S R, Burrows J M, Moss D J, Argaet V P, Misko I S. Development of Epstein-Barr virus-specific memory T cell receptor clonotypes in acute infectious mononucleosis. J Exp Med. 1996;184:1815–1824. doi: 10.1084/jem.184.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utz U, Banks D, Jacobson S, Biddison W E. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter E A, Greenberg P D, Gilbert M J, Finch R J, Watanabe K S, Thomas E D, Riddell S R. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 40.Wang E C, Moss P A, Frodsham P, Lehner P J, Bell J I, Borysiewicz L K. CD8highCD57+ T lymphocytes in normal, healthy individuals are oligoclonal and respond to human cytomegalovirus. J Immunol. 1995;155:5046–5056. [PubMed] [Google Scholar]
- 41.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson J D K, Ogg G S, Allen R L, Goulder P J R, Kelleher A, Sewell A K, O’Callaghan C A, Rowland-Jones S L, Callan M F C, McMichael A J. Oligoclonal expansions of CD8+ T cells in chronic HIV infection are antigen specific. J Exp Med. 1998;188:785–790. doi: 10.1084/jem.188.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]