Highlights
-
•
Consistent upregulation of cyclin H in various human cell types by strains of HCMV.
-
•
Reduced or increased vCDK/pUL97 kinase activity as a consequence of cyclin H knock-down or exogenous addition, respectively.
-
•
Significant trans-stimulation of human CDK7 activity by pUL97 in vitro.
-
•
Impaired HCMV replication efficiency under conditions of cyclin H knock-down.
-
•
Codetermination of viral replication efficiency by pUL97, cyclin H and CDK7 and novel targeting options for antiviral drugs.
Keywords: Human cytomegalovirus, Viral cyclin-dependent kinase (CDK) ortholog, vCDK/pUL97–cyclin binding, Functional importance of cyclin H, Modulation of vCDK/pUL97 activity in vitro, Cyclin H knock-down, pUL97-mediated CDK7 trans-stimulation, Codetermination of viral replication, Options for antiviral drug targeting
Abstract
Human cytomegalovirus (HCMV) infection is shaped by a tightly regulated interplay between viral and cellular proteins. Distinct kinase activities, such as the viral cyclin-dependent kinase ortholog (vCDK) pUL97 and cellular CDK7 are both crucial for efficient viral replication. Previously, we reported that both kinases, vCDK/pUL97 and CDK7, interact with cyclin H, thereby achieving an enhanced level of kinase activity and overall functionality in viral replication. Here we provide a variety of novel results, as generated on a methodologically extended basis, and present a concept for the codetermination of viral replication efficiency through these kinase activities: (i) cyclin H expression, in various human cell types, is substantially upregulated by strains of HCMV including the clinically relevant HCMV Merlin; (ii) vCDK/pUL97 interacts with human cyclin H in both HCMV-infected and plasmid-transfected cell systems; (iii) a doxycycline-inducible shRNA-dependent knock-down (KD) of cyclin H significantly reduces pUL97 activity (qSox in vitro kinase assay); (iv) accordingly, pUL97 in vitro kinase activity is seen significantly increased upon addition of recombinant cyclin H; (v) as a point of specific importance, human CDK7 activity shows an increase by vCDK/pUL97-mediated trans-stimulation (whereas pUL97 is not stimulated by CDK7); (vi) phosphosite-specific antibodies indicate an upregulated CDK7 phosphorylation upon HCMV infection, as mediated through a pUL97-specific modulatory effect (i.e. shown by pUL97 inhibitor treatment or pUL97-deficient viral mutant); (vii) finally, an efficient KD of cyclin H in primary fibroblasts generally results in an impaired HCMV replication efficiency as measured on protein and genomic levels. These results show evidence for the codetermination of viral replication by vCDK/pUL97, cyclin H and CDK7, thus supporting the specific importance of cyclin H as a central regulatory factor, and suggesting novel targeting options for antiviral drugs.
1. Introduction
Human cytomegalovirus (HCMV) is a worldwide distributed herpesvirus (β-subfamily member) that causes life-long latency in the infected human host. Facing several unresolved medical problems of viral pathogenesis, HCMV is considered a major human pathogen. In immunocompetent individuals, HCMV may remain asymptomatic, but severe symptoms and even life-threatening viral pathogenesis can occur in the immunosuppressed (i.e. transplant recipients, tumor and AIDS patients) or the immunonaïve host (i.e. the unborn and neonates) (Boehmer and Nimonkar, 2003; Goodrum et al., 2021; Gugliesi et al., 2020). Most seriously, congenital HCMV infection (cCMV) during pregnancy, associated with an exceptionally high rate of mother-to-child transmission, represents the most serious risk to obtain developmental defects or cytomegalovirus inclusion disease (Njue et al., 2020; Revello and Gerna, 2002; Tsutsui, 2009). In general, HCMV-related pathogenesis is closely linked to the efficiency of viral replication in individual host cell types and tissues. In this context, the role of host factors, especially the interaction through virus–host protein complexes including their regulatory interplay, possess main importance in the course of virus infection and cell-directed modulatory mechanisms (Marschall et al., 2017; Marschall et al., 2020; Milbradt et al., 2014; Schütz et al., 2022; Schütz et al., 2021).
Notably, HCMV replication drastically interferes with cell cycle regulation, a process, in which the HCMV-encoded protein kinase pUL97 directly interacts with cellular components of the CDK–cyclin machinery (Graf et al., 2013; Schütz et al., 2021; Steingruber et al., 2019). Notably, the HCMV-encoded serine-threonine protein kinase pUL97 represents a viral CDK ortholog (vCDK) as it combines typical structural and functional features of host CDKs (Hume et al., 2008; Iwahori et al., 2017; Schütz et al., 2021). CDKs are generally regulated by (i) phosphorylation of the T-loop and (ii) interaction with their respective cyclins. Previously, we characterized the specific association of pUL97 with at least three types of human cyclin, namely in our investigations most abundantly with T1, H, and B1 (Graf et al., 2016; Graf et al., 2013; Steingruber et al., 2019; Steingruber et al., 2016; Steingruber et al., 2015). Based on these observations, pUL97 is considered a multiple cyclin-binding kinase, and most recent findings pointed to the particular relevance of cyclin H for HCMV replication (Schütz et al., 2022). Cyclins comprise a family of 29 proteins in humans, structurally defined by the presence of the so-called cyclin box, a domain of approximately 100 amino acid residues that form α-helices and provides an interface for binding to CDKs. The transient abundance of individual cyclins underlies cyclical oscillations, i.e. the specific fluctuations in cyclin gene expression, to give rise, as regulatory cofactors, to the stimulated activity of either cell cycle- or transcription-associated CDKs (Malumbres, 2014; Martínez-Alonso and Malumbres, 2020). Of note, the cell cycle-related CDKs are typically characterized by multiple cyclin binding, whereas transcriptional CDKs are mostly single cyclin binding (Malumbres, 2014). As a characteristic feature, the CDK–cyclin complex formation results in activation of the CDK kinase domain, and complete activation requires distinct regulatory steps of site-specific phosphorylation.
In the case of viral vCDK/pUL97, also a multiple cyclin-binding phenotype has been identified so that the current understanding considers the multifunctional nature of this viral kinase as a consequence of these combined regulatory cyclin interactions. Interestingly, the role of cyclin types H and T1 has first been assigned to a bridging function of pUL97–pUL97 dimerization/oligomerization (Graf et al., 2013; Schütz et al., 2021; Steingruber and Marschall, 2020). Very recently, we provided evidence for the molecular details of pUL97–cyclin T1 interaction, which strongly supported the formation of a quaternary complex (Schütz et al., 2021). In this regard, it is important that the minimal binding regions responsible for pUL97–cyclin T1 interaction and pUL97–pUL97 oligomerization showed a complete overlap in the N-terminal amino acid region 231-280. Viral genomic deletion mutants in this region of ORF-UL97 pointed to the fact that it determines not only cyclin T1 interaction, but also cyclin H interaction, and that these interactions are functionally required for efficient viral replication (Schütz et al., 2021). Functionally most relevant was the finding that an impairment of cyclin H binding to pUL97 (e.g. achieved through an experimental setting of transient cyclin H knock-out) resulted in a marked reduction of viral replication (Schütz et al., 2022). In this context, it should be mentioned that the detailed 3D structure of HCMV pUL97 has not been deciphered yet, but molecular modelling, based on available CDK crystal structures, provided highly valuable information (Hutterer et al., 2016; Romaker et al., 2006; Steingruber et al., 2019; Steingruber et al., 2015; Webel et al., 2011; Webel et al., 2012; present study). Notably, pUL97 consists of a widely unstructured N-terminal portion, including two classical nuclear localization signals (NLS1 and NLS2), and a large globular domain in its C-terminal half (Steingruber and Marschall, 2020). Our computational approach suggested that cyclin H and T1 may bind to parts of the pUL97 N-lobe, while B1 binding may involve both N- and C-lobes (Steingruber et al., 2016). More specifically, we identified in our previous investigations two main candidate regions in pUL97 that represent determinants of cyclin interaction, i.e. amino acids 231-280, as a linear binding motif contained within the unstructured N-terminal pUL97 region, termed interface 2 (IF2, amino acids 231-280; Schütz et al., 2021), and a larger contact area within the C-terminal globular kinase region termed interface (IF1). In the case of cyclin T1 binding, this larger interface IF1 was not able to confer a stable pUL97–cyclin interaction by itself but was dependent on the presence of the dominant IF2 (Schütz et al., 2021). Importantly, the deletion of IF2 (Δ231-280) led to the loss of pUL97 interaction with cyclins T1 and H (but not B1), thus emphasizing its binding-determining, dominant role within the pUL97 N-terminal region.
With the present study, we provide important novel data, as generated on the basis of various experimental approaches, that jointly illustrate the binding vCDK/pUL97 to cyclin H, the functional importance of this interaction, and the cyclin H-dependent modulation of vCDK/pUL97 activity in vitro. Novel aspects are a refined bioinformatics-based modelling of the pUL97–cyclin H–CDK7, a first successful demonstration of the mutual interplay in activity between these two kinases, and a new antiherpesviral drug targeting concept based on covalently binding PPI-blocking small molecules. Specifically, a trans-stimulation of CDK7 by vCDK/pUL97 was identified, in the context of a codetermination of the efficiency of viral replication through the activity of these two cyclin H-associated kinases. Thus, the findings provide a distinct confirmation of our previous statements on the functional relevance of HCMV-specific interaction with these components of the CDK–cyclin machinery. Regarding newly recognized opportunities to pharmacologically interfere with this type of virus–host interaction, these novel and application-oriented options for antiviral drug targeting are discussed.
2. Materials and methods
2.1. Cell culture and virus infection
Primary human foreskin fibroblasts (HFFs, C0045C, Thermo Fisher Scientific, Waltham, MA, USA) and 293T cells were cultured at 37°C, 5% CO2, and 80% humidity. Culture media for HFF (MEM, 21090055, Thermo Fisher Scientific, Waltham, MA, USA) and 293T (DMEM, 11960044, Thermo Fisher Scientific, Waltham, MA, USA) were supplemented with 10% FCS, 1x GlutaMAXTM (35050038, Thermo Fisher Scientific, Waltham, MA, USA), and 10 g/mL gentamicin (22185.03, SERVA, Heidelberg, Germany). Stably transduced HFF and 293T were selected by adding 2.5 µg/ml puromycin to the media. Tetracycline-negative FCS (FCS-TET-12A, Capricorn Scientific, Ebsdorfergrund, Germany) was used for the cultivation of cyclin H knock-down cells. Expression of cyclin H-specific shRNA was induced by the addition of 500 ng/ml doxycycline every other day (dox, D9891, Sigma-Aldrich, St. Louis, MO, USA). HCMV strains AD169 or Merlin were used for infection experiments at a multiplicity of infection (MOI) of 0.5 or less, as indicated for each experiment. For reasons of the high replication efficiency of HCMV in the established model of primary fibroblasts (HFFs), we were primarily using the laboratory strain AD169. Since this viral strain, due to its adaptation to cultured cells, acquired certain genomic deletions, we additionally used the genomically intact, clinically relevant strain Merlin as a confirmation in specific aspects of virus–cell interaction (see cyclin H upregulation upon HCMV infection and conditional cellular cyclin H KD in chapters 3.1 and 3.4, respectively). Virus inocula were replaced with fresh growth medium after incubation at 37°C for 90 min. For a more efficient infection with HCMV Merlin, a 30-min centrifugation step of 2,500 rpm (2,300 x g) at room temperature was included in the 90 min inoculation time. As a routine procedure, we monitored infection rates by costainings of viral protein (Wb, IVKA, CoIP with samples from HCMV-infected cells) or viral reporter expression (HCMV GFP- or YFP-based replication assays). For the transfection of 293T cells, polyethyleneimine (PEI) was used as described before using PBS instead of HBS (Schregel et al., 2007).
2.2. Coimmunoprecipitation (CoIP), SDS-PAGE and Western bot (Wb) analysis, non-radioactive in vitro kinase assay (IVKA)
5 × 106 293T cells were seeded in 10 cm dishes, transfected, and harvested two days post-transfection (d p.t.) and CoIP was performed as described previously (Sonntag et al., 2016). Input controls, immunoprecipitation (IP), and CoIP samples were subjected to standard SDS-PAGE and Wb analysis (Hutterer et al., 2013; Thomas et al., 2009). The antibodies used for IP and Wb are listed in Section 2.3. For densitometric determination of Wb signal intensities, two independent Wbs were analyzed using the AIDA Image Analyzer (V4.23) software. Non-radioactive in vitro kinase assay (IVKA) was performed as described previously (Schütz et al., 2021). A coimmunoprecipitate of kinase and substrate was incubated for 60 min at 30°C in 30 µl kinase assay buffer (20 mM Tris/HCl at pH 7.5, 0.5 mM MnCl2, 1 µM ATP, and 200 ng ATPγS). Samples incubated in assay buffer without ATPγS served as a negative control. ATPγS served as a donor to thiophosphorylate kinase substrates. After 60 min, 3 µl of 25 mM p-nitrobenzyl mesylate (PNBM, Abcam) was added to the reaction to alkylate thiophosphates for 30 min at 25°C. After a denaturation step at 95°C for 10 min, samples were subjected to standard SDS-PAGE and Wb analysis, and thiophosphate esters (TPE) were detected using specific antibodies.
2.3. Antibodies and small molecules
The following antibodies were used in this study: mAb-UL97.01, mAb-UL53, mAb-UL50, and mAb-UL56 (kindly provided by T. Lenac and S. Jonick, University of Rijeka, Croatia), mAb-IE1 (kindly provided by William Britt, Birmingham, AL, USA), mAb-UL44 and mAb-MCP (kindly provided by Bodo Plachter, University of Mainz, Mainz, Germany), mAb-pp65 and mAb-UL69 (kindly provided by T. Stamminger, Ulm, Germany), mAb-β-actin (A5441, Sigma Aldrich), pAb-cyclin H (LS-C331195, LS Bio), pAb-CDK7 pSer164 (PA5-105583, Sigma-Aldrich), pAb-CDK7 pThr170 (ab155976, Abcam), pAb-CDK9 pThr186 (2549, Cell signaling), mAb-CDK7 (sc-7344, Santa Cruz Biotechnology), pAb-RNAPII CTD pSer2 (NB100-1805, Novus Biologicals), mAb-Flag (F1804, Sigma-Aldrich). Kinase inhibitors were provided by Lead Discovery Center GmbH, Dortmund, Germany, as a series of compounds of specific interest for current developmental purposes. All these compounds are small molecules possessing the indicated molecular masses (MW range 257–557 Da) and are derived from various chemical classes, with scaffolds typically representing ATP-competitive kinase inhibitors (i.e. the chemical class of quinazolines applies to LDC599, the chemical class of diaminopyrimidines applies to LDC745, and the class of tihiazolo-pyrimidinones applies to LDC492). Importantly, as a common feature of the series of LDC compounds analyzed in chapter 3.5, they possess covalent binding properties, in most cases mediated through an acceptor of the Michael addition reaction (Johansson, 2012; Péczka et al., 2022), which links the warhead to a distinct amino acid, i.e. a cysteine residue in most cases of the target protein(s). As non-covalently binding reference kinase inhibitors, LDC4297 and MBV were used for the specific activity assessments of the two kinases of interest: LDC4297 is a selective developmental CDK7 inhibitor (for a human kinome selectivity panel, i.e. in vitro SelPan, see Hutterer et al. 2015), and MBV is a clinically approved and selective vCDK/pUL97 inhibitor (for HCMV infected cell-based in vivo selectivity, see Wild et al. 2022). Stock aliquots of all compounds were prepared in DMSO at a concentration of 10 mM.
2.4. Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)
HFFs or TEV-1 were infected in 12-well plates with HCMV Merlin at MOI of 0.5. Cells were harvested at the indicated time point and total RNA was extracted using the Quick-RNA MiniPrep Plus Kit (R1057, Zymo Research). Relative cyclin H mRNA levels were determined by a one-step RT-qPCR using the LunaScript RT SuperMix Kit (E3010, NEB) and cyclin H-specific primers and probe, as follows:
cyclin H forward 5’ CAACGGGAAGGTTCTTCCGA;
cyclin H reverse 5’ TACAAGCCGTACCCACAACA;
probe 5’ CATGAAGAAATGACACTCTGCAAATACTAT modified with 5’ FAM and 3’ TAMRA. The values obtained were normalized to the mock-infected controls.
2.5. Quantitative polymerase chain reaction (qPCR)
Viral genome copy numbers were determined as described before (Lorz et al., 2006). In brief, 2.25 × 105 HFFs were seeded in 12-well plates and infected the next day with HCMV AD169 or Merlin at MOI of 0.001 for 90 min at 37°C. Viral supernatant was taken at indicated time points and replication kinetics were analyzed via qPCR using a FAM/TAMRA-labelled probe directed against the HCMV MIE region exon 4. Regarding replication kinetics of HCMV Merlin, qPCR was performed using a Roche 480 LightCycler with Kapa Sybr Fast qPCR master mix (Merck) as described elsewhere (Hamilton et al., 2020).
2.6. Generation of cyclin H knock-down (KD) cell populations
shRNAs were either selected based on important features associated with microRNA processing (Fellmann et al., 2011) or using the novel splash algorithm (Pelossof et al., 2017). The improved mirE backbone was used to ensure optimal pri-mRNA recognition and processing by Drosha and Dicer RNA endonucleases (Fellmann et al., 2013). shRNA cloning was performed according to Fellmann et al. (2013). 97-mer oligonucleotides harboring the selected shRNA sequence were PCR amplified. Final vectors were obtained by XhoI/EcoRI cloning of the PCR product into the mirE vector. Lentiviral particles were produced by transfecting 5 × 106 293T cells seeded in 10 cm dishes with 4 µg of the desired shRNA mirE vector together with 4 µg of pCMVdeltaR8.9 and 2 µg of pVSV-G using Lipofectamine 2000 (Thermo Fisher Scientific). All the following experimental steps regarding transduction and selection of HFF and 293T cells were performed exactly as described before (Schütz et al., 2022). shRNA was induced by the addition of 500 ng/ml dox every other day for a total of five days before the start of the experiment.
2.7. Quantitative Sox peptide-based in vitro kinase assay (qSox-IVKA)
A recently developed qSox-IVKA was used for sensitive and quantitative measurement of in vitro kinase activity of pUL97 (Schütz et al., 2022; kinase-specific sensor peptide AQT0258, AssayQuant Inc.) and CDK7 (AQT0459). For this purpose, 5 × 106 293T cells were seeded and transfected as described in Section 2.1. Cells were harvested 2 d p.t. and CDK7 and/or pUL97 were immunoprecipitated using specific antibodies as described in 2.3. A mouse Fc fragment, instead of IP Ab, served as a negative control. Washed IP samples were eluted in 100-200 µl enzyme buffer (20 mM HEPES, pH 7.5, 0.1% Brij-35, 5% glycerol, 1 mg/ml bovine serum albumin, BSA). Volumes of 33 µl of the eluted kinase were added to 132 µl of the kinase reaction mix (final mix: 54 mM HEPES, pH 7.5, 1.0 mM DTT, 1.0 mM ATP, 0.012% Brij-35, 0.52 mM EGTA, 1% glycerol, 0.2 mg/ml BSA, 10 mM MgCl2, 15 µM PhophoSenS® cysteine-Sox kinase sensor peptide AQT0258/AQT0459) and kinase activity was measured in triplicates with 50 µl each. Reactions were performed in Corning NBS 96-well half area plates (3992, Corning) at 30°C for approx. 80 min in a kinetic manner (measuring every 30 sec to 1 min) using a Victor 1420 multilabel counter (Ex355/Em480, Perkin-Elmer). To address the impact of increased levels of cyclin H, recombinant human cyclin H, commercially available as a full-length product, carrying GST-tag and expressed in insect cells (ABIN6699744, antibodies-online.com), was exogenously added to the reactions at the indicated concentrations. To achieve a quantitatively adjusted basis, 25 µl of IP samples were used for Wb normalization in duplicates to ensure equal kinase reactivities in the qSox-IVKA reaction. Control samples that were derived from empty vector transfection and Fc fragment pseudo-IP were subtracted from the kinase signal to obtain a corrected output of the kinase-specific activity. To measure the trans-stimulating potential of pUL97 onto CDK7, the in vitro kinase activity of pUL97 was measured in settings of single immunoprecipitation (pUL97 only) and then subtracted from the value of the combined kinase reaction (both CDK7 plus pUL97).
2.8. NanoBiT® protein–protein interaction system (Promega)
The NanoBiT (Promega) system was used to perform a first-time analysis of putative inhibitors of the pUL97−cyclin H interaction. To this end, 5 × 105 293T cells in 6-well plates were transfected with 2 µg of the corresponding expression plasmid (single transfection combined with empty vector, or double transfection with LgBiT plus SmBiT constructs) using PEI as described in Section 2.1. Biological triplicates of cells were transferred to a 96-well plate at 2 d p.t. and inhibitors were added using the given concentrations 4 h later. Luciferase assay was performed immediately after the compound addition according to the manufacturer's protocol (N2011, Promega). Luciferase activity was measured at 37°C for approximately 60 min using the Victor X4 microplate reader. Background activity of single-transfected LgBiT luciferase activity was subtracted from LgBiT/SmBiT cotransfected cells to obtain PPI-specific luciferase activity.
2.9. Molecular modelling
The structure of pUL97(329-634) was modelled based on the cryo-EM structure of the cyclin H–CDK7 complex (PDB: 7B5O; Greber et al., 2021). The alignment of pUL97 and CDK7 was generated with HHpred (Zimmermann et al., 2018); Modeller (Webb and Sali, 2021) was used for model generation. Prediction of the pUL97(231-280) binding mode was done with ColabFold (Mirdita et al., 2022), which represents a fast implementation of AlphaFold2 (Senior et al., 2020). From a PISA analysis (Krissinel, 2015) of the interfaces, the complex showing the largest cyclin H–pUL97(231-280) interface and the most favorable interaction energy was selected for further characterization. Structural analysis and visualization were done with RasMol (Sayle and Milner-White, 1995).
3. Results
3.1. The physical and functional interaction between vCDK/pUL97, cyclin H and CDK7
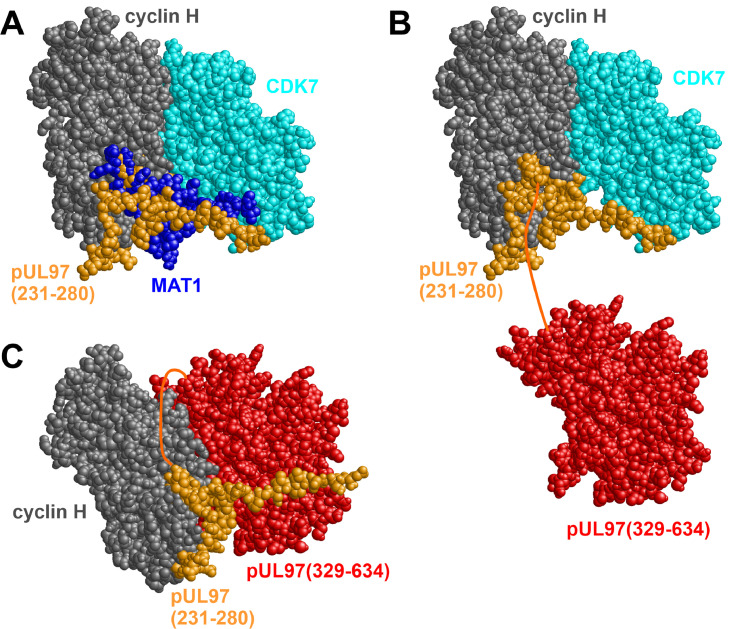
In our previous studies, we and others provided experimental evidence for the relatedness of the HCMV-encoded protein kinase pUL97 with human CDKs, so that the term viral CDK ortholog (vCDK) has been introduced (Couté et al., 2020; Hume et al., 2008; Prichard, 2009; Romaker et al., 2006; Steingruber and Marschall, 2020): Notably, both types of kinases bind to human cyclin H, demonstrating functional relevance compared to other cyclin binding activities (Fisher and Morgan, 1994; Schütz et al., 2022; Schütz et al., 2021; Steingruber et al., 2019). Moreover, the formation of binary, ternary, and even higher-order complexes between vCDK/pUL97, cyclin H, and CDK7 has been suggested based on biochemical and bioinformatic approaches (Schütz et al., 2021; Steingruber et al., 2019). To better understand the underlying structural principles of these interactions, we performed molecular modelling. We first investigated whether cyclin H exhibits a binding site for the 231-280 segment of pUL97, which was previously found important for the interaction between pUL97 and cyclin T1 (Schütz et al., 2021). Of note, the pUL97–cyclin T1 interaction (here becoming clear that this is equally true for cyclin H) has two separable binding interfaces, IF1 (329-634) and IF2 (231-280; Schütz et al., 2021; Steingruber et al., 2016; Steingruber and Marschall, 2020). While IF2 is essential for the interaction, IF1 can be deleted on larger stretches without strict loss of binding and is thus supposed to possess a reinforcing function. Thus, IF2 is considered dominant over IF1 in pUL97–cyclin H/T1 interaction whereas IF1 seems to be auxiliary (Schütz et al., 2021). A refined prediction of this situation of binding sites now suggests that pUL97(231-280) uses a similar surface patch for cyclin H recognition as previously observed for MAT1 (Fig. 1A). MAT1 is a RING finger protein (ménage a trois) that physiologically associates with the CDK7–cyclin H complex and functions as a regulatory cofactor (Patel and Simon, 2010; Rossi et al., 2001). In the context of HCMV-specific complex formation, this short and functionally dominant cyclin binding site (IF2) of pUL97(231-280) is in the immediate vicinity of the cyclin H–CDK7 interface but does not overlap; therefore, simultaneous binding of both kinases, pUL97 and CDK7, to cyclin H should be possible (Fig. 1B). In a further modelling, we investigated whether pUL97 could additionally bind to cyclin H via its additional larger cyclin binding interface (IF1), pUL97(329-634), which includes essential subdomains of the pUL97 kinase domain (Hutterer et al., 2016; Romaker et al., 2006; Steingruber et al., 2016). Principally, such IF1-plus-IF2-based interaction of pUL97 with cyclin H might displace CDK7 from this complex. The corresponding model (Fig. 1C) shows that the distance between the C-terminal residue of IF2 (Ile280) and the N-terminal residue of the kinase domain (Asp329) comprises approximately 40 Å, which can be easily bridged by the connecting 48 residues. In summary, this refined molecular modelling strongly supports our previously suggested mode of interaction between pUL97 and cyclin H involving the two distinct interfaces, IF1 and IF2, which allows the formation of both binary pUL97–cyclin H complexes (Fig. 1C) and ternary pUL97–cyclin H–CDK7 complexes (Fig. 1B).
Fig. 1.
Structural model of the interaction between pUL97 and cyclin H. (A) Predicted binding site of pUL97(231-280) (orange) superimposed with the experimental cyclin H–CDK7–MAT1 complex structure (gray, cyan, and dark blue). This model suggests that pUL97(231-280) uses the same binding pocket as MAT1 for targeting the cyclin H–CDK7 complex. (B) Model of a ternary pUL97–cyclin H–CDK7 complex, in which pUL97 is attached to cyclin H exclusively through IF2 formed by the 231-280 sequence stretch. The pUL97 kinase domain (residues 329-634, marked in red) is connected to the complex by a nonstructured, flexible linker (residues 281-328, indicated as dark orange connecting line). (C) Model of a pUL97–cyclin H complex, in which pUL97 interacts with cyclin H both through IF2, pUL97(231-280) (orange), and the globular kinase domain IF1, pUL97(329-634) (red), thereby displacing CDK7.
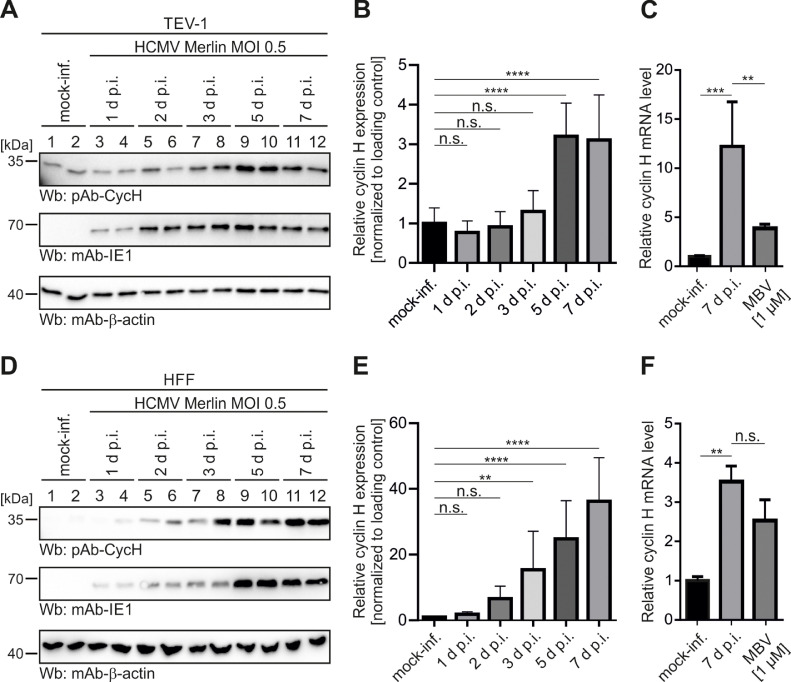
As another striking property in this context of pUL97–cyclin interaction, we identified a consistent upregulation of cyclin H in various human cell types by strains of HCMV. Analyzing protein samples derived from time courses of HCMV infection with several selected strains and cell types, the obtained Wb staining patterns depicted that cyclin H is generally upregulated upon HCMV infection under various conditions, such as viral MOI, time points, virus strain, host environment, and other parameters (Fig. 2; Schütz et al., 2022; Steingruber et al., 2019; Steingruber et al., 2016). It should be emphasized that we focused here on the clinically relevant HCMV strain Merlin, which showed a marked upregulation of cyclin H both in the placental cell type TEV-1 (Fig. 2A) and in primary human fibroblasts HFF (Fig. 2D). Densitometric quantitation of the Wb data (with a second series of Wb stainings, used for the quantitative determination in densitometric quadruplicates, is depicted in Fig. S1) revealed a statistically significant level of upregulation that increased over the time course of a multi-round period of infection (approximately 3 d for 1 round of HCMV replication), with particularly high levels at late time points of 3–7 d p.i. (Fig. 2B and E). This upregulation could also be observed on the RNA level using, for quantitation, a one-step RT-qPCR. As an important result, cyclin H mRNA levels were 12-fold increased in HCMV-infected TEV-1, compared to mock-infected cells (Fig. 2C), and 3.5-fold increased in HCMV-infected HFFs at the time point of 7 d p.i. (Fig. 2F). This upregulation could be substantially reduced by the addition of 1 µM of MBV resulting in a 68% or 28% reduction of the HCMV-induced increase of cyclin H mRNA levels for TEV-1 cells or HFFs, respectively.
Fig. 2.
Cyclin upregulation upon HCMV infection in TEV-1 cells and HFFs. TEV-1 cells (A) or HFFs (D) were seeded in 12-well plates and infected 1 d later with HCMV Merlin at MOI of 0.5. Cells were harvested at indicated time points in duplicates originating from different wells (see duplicate samples indicated by odd- and even-numbered lanes) before cells were lysed and prepared for standard SDS-PAGE and Wb analysis using the indicated antibodies. Two independent SDS-PAGEs/Wbs were used for densitometry measurements in quadruplicate using ImageJ. Cyclin H signal intensity was normalized to β-actin and set in relation to mock-infected controls for infected TEV-1 cells (B) and infected HFFs (E). TEV-1 cells (C) or HFFs (F) were infected in 12-well plates with HCMV Merlin at MOI of 0.5 to assess cyclin H mRNA levels. Optionally, cells were treated with 1 µM of MBV or equal amounts of DMSO. Cells were harvested at the indicated time point, total RNA was extracted and cyclin H mRNA levels were determined by a one-step RT-qPCR using cyclin H-specific primers and probe. The values obtained were normalized to the mock-infected controls. Statistical analysis was performed using an ordinary one-way ANOVA test and post-hoc Tukey correction: ns, not significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
In this context of upregulated cyclin H during viral replication, the previously characterized complexation of pUL97–cyclin H appeared very relevant. Several investigative approaches, including mass spectrometry-based proteomics, CoIP settings, recombinant cellular expression systems, and others, demonstrated this interaction for HCMV-infected cells (Schütz et al., 2021; Steingruber et al., 2019; Steingruber et al., 2016). Here, we add the information that the viral kinase pUL97 consistently interacts with human cyclin H, in both cases of using material derived from HCMV-infected cells (Steingruber et al., 2016) or plasmid-transfected 293T cells (Fig. S2), provided that the expression levels are sufficiently high to ensure the sensitivity of detection. This finding reemphasizes the importance of the pUL97 interface IF2, which was previously identified as the dominant region for pUL97–cyclin T1 interaction and pUL97–pUL97 self-interaction (Graf et al., 2013; Schütz et al., 2021; Steingruber and Marschall, 2020). In this context, it has to be emphasized that an additional larger interface formed by the pUL97 kinase domain (IF1, amino acids 329-634) may also contribute to cyclin interactions (Schütz et al., 2021; Steingruber et al., 2019). Our combined bioinformatic investigations and CoIP deletion mapping experiments strongly suggest a very similar situation of concerted activity between IF2 and IF1 (providing dominant or accessory binding effects, respectively) being jointly responsible for pUL97 interaction with cyclins T1 and H (Schütz et al., 2021; Steingruber et al., 2015; reviewed in Steingruber and Marschall 2020).
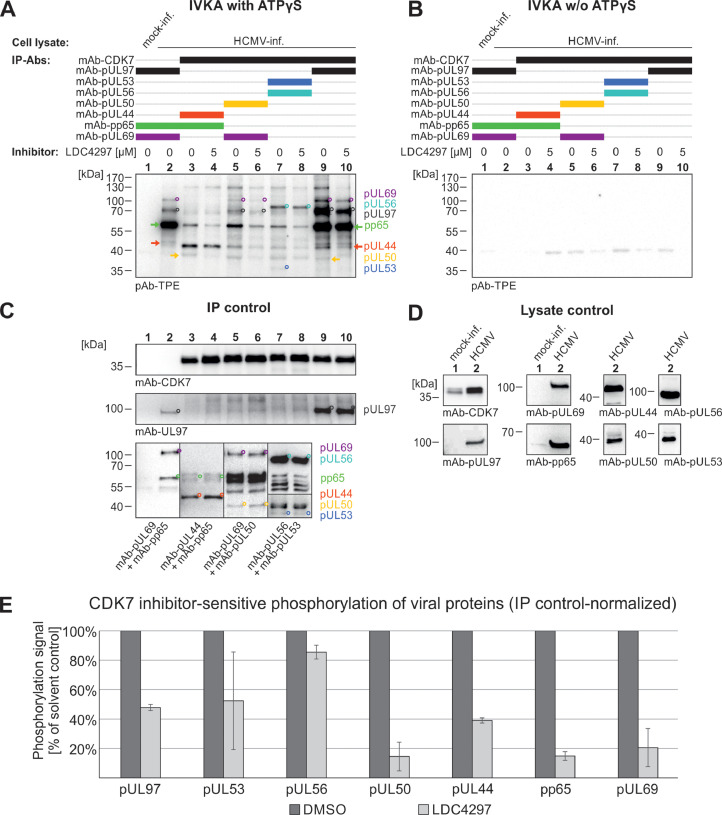
Next, we addressed the question whether cyclin H-associated kinases in HCMV-infected cells (i.e. CDK7 and vCDK/pUL97) may share similar properties of substrate phosphorylation. To this end, we focused on known viral substrate proteins of pUL97, and performed CDK7-specific non-radioactive in vitro kinase assays (IVKAs) using immunoprecipitation samples of this selection of viral proteins to investigate their property as putative CDK7 substrates (Fig. 3). ATPγS was added to the IVKA reaction, which is used by kinases to thiophosphorylate substrates. This thiophosphorylation was then detected by Western blot analysis using a thiophosphate ester (TPE)-specific antibody. The functionality of IVKA reactions was confirmed by the autophosphorylation of pUL97 (Fig. 3A, lane 2). Identical IVKA reactions were used as negative controls in the absence of ATPγS (Fig. 3B). A key finding was that all analyzed viral proteins, i.e. pUL44, pp65, pUL50, pUL53, pUL69, pUL56, and pUL97, showed phosphorylation signals when immunoprecipitated together with CDK7, as depicted in Fig. 3A, lanes 3, 5, 7 and 9. Interestingly, phosphorylation signals for pUL44, pp65, pUL50, pUL69, and pUL97 were also detected in samples lacking specific antibodies for the IP of these proteins, suggesting that they might form relatively stable complexes with each other. However, this implied that the detected phosphorylation of viral proteins was not necessarily solely performed by CDK7 but could additionally be based on the vCDK/pUL97 activity. To specify this question, the selective CDK7-inhibitor LDC4297 (Hutterer et al., 2015) was added to duplicates of the immunoprecipitates shortly before the IVKA reaction. It has to be emphasized that parallel control testings made sure the applied concentration of LDC4297 did not affect pUL97 kinase activity but completely blocked CDK7 activity (Fig. S3). In this SDS-PAGE/Wb-based IVKA determination of substrate specificity, the inhibition of CDK7 resulted in reduced phosphorylation signals for all viral proteins analyzed (Fig. 3A, lanes 4, 6, 8, 10) compared to untreated samples (Fig. 3A, lanes 3, 5, 7, 9). The control Wb detection of all IP samples and total lysate input samples verified the reliability of the measurement (Fig. 3C, D). Densitometric quantitation of the phosphorylation signals confirmed that, upon CDK7 inhibition, all viral proteins analyzed were lower in phosphorylation levels compared to untreated samples (Fig. 3E). Thus, the phosphorylation of the viral proteins pUL97, pUL53, pUL56, pUL50, pUL44, pp65, and pUL69 was, at least in part, sensitive to CDK7 inhibition, indicating that these proteins may indeed be phosphorylation substrates of CDK7. To confirm this point, an additional experiment was performed utilizing material from HFFs infected with a recombinant HCMV that does not express pUL97 (HCMV ΔUL97) in comparison to HCMV WT (Fig. S4). Consistent with the results shown in Fig. 3, phosphorylation was detectable for pUL53, pUL56, pUL50, pUL44, and pp65, when considering this evaluation on a primarily qualitative basis, and thus suggested that no less than these five viral proteins serve as CDK7 phosphorylation substrates in vitro. In the context of previously published data demonstrating their role as viral kinase pUL97 substrates (except for pUL56, detected here for the first time), these five proteins may be dually phosphorylated by both vCDK/pUL97 and CDK7, although quantitative statements or predictions about the preference of phosphorylation by either of the two kinases cannot be made at this stage. Combined, these novel data substantiate the interaction, on both physical and functional levels, between vCDK/pUL97, cyclin H and CDK7. This deepened understanding then raised more specified questions about the molecular, regulatory basis of this interaction, which we subsequently investigated.
Fig. 3.
Viral protein substrate phosphorylation by CDK7. HFFs were infected with HCMV AD169-GFP at MOI of 1 and lysed 3 d.p.i. (A) Viral proteins were coimmunoprecipitated with cellular kinase CDK7 (lanes 3-10) and exposed to the non-radioactive IVKA reaction. Specific phosphorylation signals are marked to indicate single (°) or continuous (arrows in respective colors) bands. Autophosphorylation signals of viral kinase pUL97 served as a positive control (lane 2). Duplicates of each immunoprecipitate were treated with 5 µM of CDK7-inhibitor LDC4297 shortly before the IVKA reaction (lanes 4, 6, 8, 10). (B) An IVKA reaction conducted in parallel without ATPγS served as a negative control. Successful IP (C) and reliable expression levels (D) of all proteins of interest were demonstrated by Wb analysis. (E) Phosphorylation signals from IVKA reactions shown in (A) were quantitated three times with varying background parameters, resulting in triplicate determinations of each sample, and mean values ± SD were then normalized to IP control.
3.2. Increased quantities of cyclin H exert a significant impact on the vCDK/pUL97 kinase activity in vitro
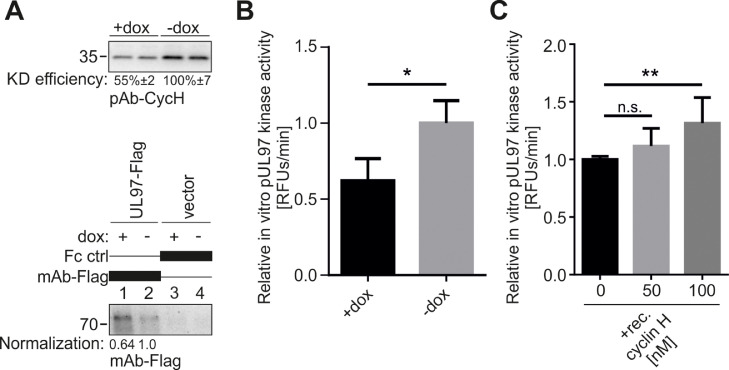
As a next step, we investigated whether cyclin H has a direct effect on vCDK/pUL97 kinase activity in vitro. Therefore, we generated 293T cells with a stable and dox-inducible anti-cyclin H shRNA (sp3) KD. An initial Wb revealed a KD efficiency of approximately 50% compared to non-induced cells (Fig. 4A). Lysate material from cells transfected with either pUL97-Flag or an empty vector was then used for a qSox-IVKA to quantitatively measure pUL97 kinase activity (Schütz et al., 2022). For this purpose, pUL97-Flag was immunoprecipitated from dox-induced or non-induced material using Dynabeads–anti-Flag antibody IP complexes. The quantities of immunoprecipitated proteins were determined and normalized to non-induced samples by Wb analysis (Fig. 4A). Dynabead-bound pUL97, or empty beads derived from mock-transfected material, were used to measure pUL97 kinase activity, in dependence of cyclin H. Strikingly, pUL97 kinase activity was significantly reduced to 62.1%, when immunoprecipitated under conditions of cyclin H KD (+dox), compared to the non-KD (-dox) control (Fig. 4B). To confirm this finding in a reciprocal setting, we immunoprecipitated pUL97-Flag from transfected-cell material and exogenously added recombinant cyclin H to the qSox-IVKA reaction. The exogenous addition of 100 nM recombinant cyclin H increased the in vitro kinase activity of pUL97 by 31.5% (Fig. 4C). These results are consistent with our previously published results and provide further evidence that cyclin H binding has a positive effect on pUL97 kinase activity.
Fig. 4.
The exogenous addition of recombinant cyclin H increases pUL97 in vitro kinase activity. (A) Cyclin H KD in 293T cells was induced by the addition of 500 ng/ml doxycycline (dox) for 5 d. KD efficiency was determined by Wb using the indicated antibodies. Dox-induced and non-induced 293T cells were transiently transfected with plasmids encoding pUL97-Flag or empty vector control. Cells were lysed 2 d post-transfection, and pUL97-Flag was immunoprecipitated using the indicated antibodies; a mouse Fc fragment served as a negative control. Dynabeads-bound proteins were eluted in 100 µl enzyme buffer. Volumes of 25 µl of eluted Dynabeads were denatured in 25 µl 2x loading buffer and subjected to SDS-PAGE and Wb analysis. The quantity of immunoprecipitated pUL97-Flag was determined densitometrically, measuring the mean expression levels derived from two different Wbs, and by subsequent normalization of these mean values towards non-induced reference samples (lane 2). (B) Immunoprecipitated pUL97-Flag and Fc control samples from dox-induced or non-induced 293T cells were subjected to a qSox-IVKA (using the pUL97-specific sensor peptide AQT0258). The background signal of the Fc control was subtracted from the IP-specific pUL97 kinase signal. The resulting pUL97 kinase signal was first multiplied by the normalization factor from (A), to compensate for different quantities in the IVKA reactions, and then normalized to the non-induced reference samples. Mean values ± SD are given, as derived from three independent biological replicates, with measurements in triplicates each. (C) pUL97-Flag derived from lysed WT 293T cells was immunoprecipitated, eluted in 120 µl enzyme buffer, before a qSox-IVKA was performed. A mouse Fc fragment, used instead of an IP antibody, served as a negative control. Additionally, three different concentrations of recombinant cyclin H (100, 50, 0 nM) were applied to the reactions. The background signal of the Fc control was subtracted from the pUL97 kinase signal. The resulting pUL97 signal was normalized to the sample without cyclin H addition. Mean values ± SD are given, as derived from three independent biological replicates, with measurements in triplicates each. Statistical analysis was performed using an ordinary two-way ANOVA: *, p < 0.05; **, p < 0.01; n.s., not significant.
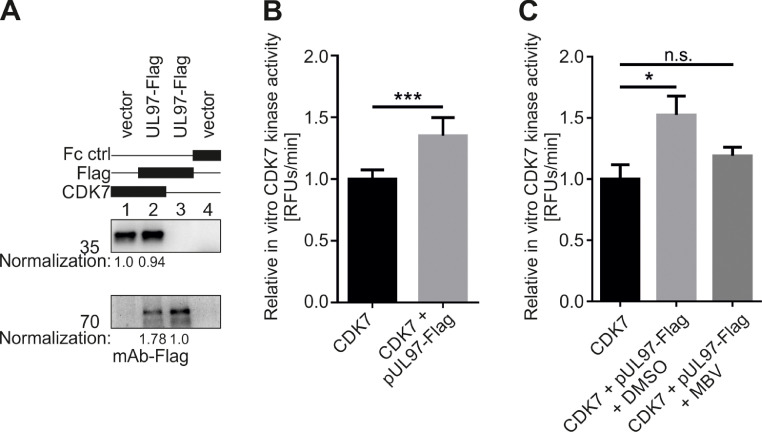
3.3. CDK7 in vitro activity is trans-stimulated by the presence of active vCDK/pUL97
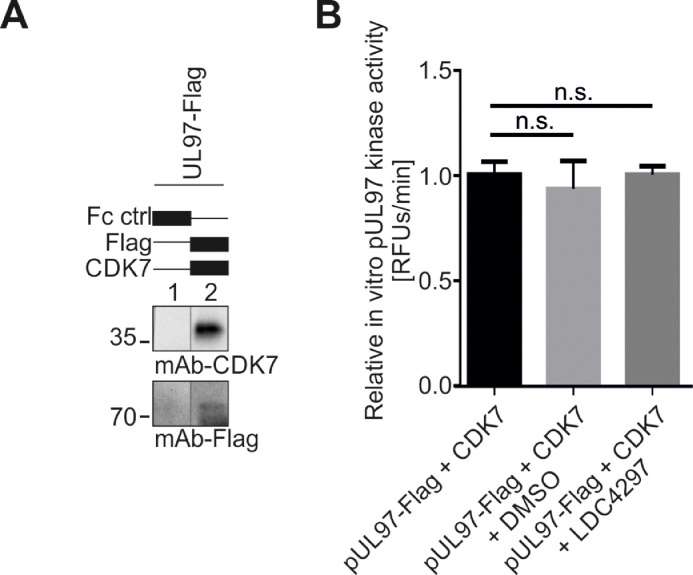
Following these findings, we attempted to investigate whether pUL97, in a complex with cyclin H, which may inherently include the cyclin H-associated CDK7 (see Fig. 1), might have a stimulatory effect on CDK7 kinase activity. As before, we transfected 293T cells with plasmids coding for pUL97-Flag or empty vector control and used the cell material for the subsequent immunoprecipitation of pUL97-Flag, and/or CDK7, or used nonreactive Fc fragment as a negative control. In this setting, the two kinases were either immunoprecipitated separately or simultaneously. The quantity of immunoprecipitated protein was determined and normalized according to Wb analysis (Fig. 5A). For the subsequent qSox-IVKA, a CDK7-specific Sox-peptide (which is strongly recognized by CDK7, but only marginally recognized by pUL97) was used as a kinase sensor. Firstly, the kinase activity of single immunoprecipitates of CDK7 and pUL97 was measured. Secondly, the kinase activity of CDK7/pUL97-Flag coimmunoprecipitation was measured and pUL97 single kinase activity was subtracted from the coimmunoprecipitated activity values, thus revealing the specific kinase activity of CDK7. The activity values of jointly coimmunoprecipitated CDK7 plus pUL97 were normalized to the values derived from CDK7 single immunoprecipitation. Strikingly, the CDK7 activity derived from pUL97-transfected samples was significantly increased by 35.1% compared to the non-transfected control (Fig. 5B). This increased CDK7 kinase activity could be abolished by the addition of 0.35 µM MBV (concentration corresponding to its mean EC50 value in cultured cell-based antiviral assays), thus confirming the activating potential of pUL97 on CDK7 (Fig. 5C). Of note, CDK7 was not inhibited by the concentration of 0.35 µM MBV (corresponding to the pUL97-specific EC50 value), and pUL97 kinase activity was reduced to 50% by this concentration of 0.35 µM, thereby confirming both the pronounced specificity and sensitivity of MBV (Fig. S3). In a reciprocal setting, we also investigated whether CDK7 is likewise capable of activating pUL97. The kinase activity of coimmunoprecipitated pUL97-Flag/CDK7 was measured using a pUL97-specific sensor qSox-peptide. Optionally, 0.01 µM CDK7-specific inhibitor LDC4297 or DMSO as solvent control was added to the reaction (Fig. 6). Interestingly, no change in pUL97 activity was detected in the presence or inhibition of CDK7. Therefore, our data suggest that this mode of kinase trans-stimulation is unidirectional from vCDK/pUL97 to CDK7, but not vice versa.
Fig. 5.
Mode of in vitro trans-stimulation: vCDK/pUL97 can increase the activity of human CDK7 significantly. (A) 293T cells were transiently transfected with plasmids encoding pUL97-Flag or with an empty vector control. Cells were lysed 2 d post-transfection, and pUL97-Flag and/or CDK7 were immunoprecipitated using the indicated antibodies; a mouse Fc fragment served as a negative control. Dynabeads-bound proteins were eluted in 150 µl enzyme buffer. 25 µl of eluted Dynabeads were denatured in 25 µl 2x loading buffer and subjected to SDS-PAGE and Wb analysis. The quantity of immunoprecipitated pUL97-Flag and CDK7 was determined densitometrically, measuring the mean expression levels derived from two different Wbs. Values of CDK7 and pUL97-Flag obtained from the double-immunoprecipitated sample (lane 2) were normalized to those obtained from the single-immunoprecipitated sample of CDK7 (lane 1) and pUL97-Flag (lane 3). (B) Samples based on the immunoprecipitation of CDK7, pUL97-Flag, CDK7 plus pUL97-Flag, as well as Fc control samples, were subjected to a qSox-IVKA (using the CDK7-specific sensor peptide AQT0459). The background signal of the Fc control was subtracted from kinase signals. The resulting kinase-specific signals were multiplied by the normalization factors from (A), to compensate for different quantities of the kinases in the IVKA reactions. Additionally, the kinase signal of single-immunoprecipitated pUL97-Flag was subtracted from double-immunoprecipitated CDK7 plus pUL97-Flag, to obtain a corrected CDK7-specific kinase signal. All kinase signals were then normalized to the kinase signal derived from CDK7 single immunoprecipitation. Mean values ± SD from three independent biological replicates are given, as derived from measurements in triplicates. (C) The concentration of 0.35 µM of pUL97-specific inhibitor maribavir (MBV; corresponds to mean antiviral EC50 value) or DMSO as solvent control was added to CDK7 + pUL97-Flag samples and CDK7-specific kinase activity was measured using a qSox-IVKA (using the CDK7-specific sensor peptide AQT0459). Mean values ± SD are given, as derived from one representative biological replicate, with measurements in triplicates. Statistical analysis was performed using an ordinary two-way ANOVA: ***, p < 0.001; *, p < 0.05; n.s., not significant.
Fig. 6.
Human CDK7 does not increase pUL97 in vitro kinase activity. (A) 293T cells were transiently transfected with plasmids encoding pUL97-Flag. Cells were lysed 2 d post-transfection, and pUL97-Flag and CDK7 were immunoprecipitated using the indicated antibodies; a mouse Fc fragment served as a negative control. Dynabeads-bound proteins were eluted in 150 µl enzyme buffer. 25 µl of eluted Dynabeads were denatured in 25 µl 2x loading buffer and subjected to SDS-PAGE and Wb to confirm successful precipitation of pUL97-Flag and CDK7. (B) Samples derived from immunoprecipitation of CDK7 plus pUL97-Flag, as well as Fc control samples, were subjected to a qSox-IVKA (using the pUL97-specific sensor peptide AQT0258). Optionally, 0.01 µM of CDK7-specific inhibitor LDC4297 (corresponds to the mean antiviral EC50 value) or equal amounts of DMSO were added to the reactions. Background activity determined with the Fc control was subtracted from values of combined pUL97-Flag plus CDK7 kinase activity. Mean values ± SD derived from one representative experiment are given, as derived from measurements in triplicates. Statistical analysis was performed using an ordinary two-way ANOVA: n.s., not significant.
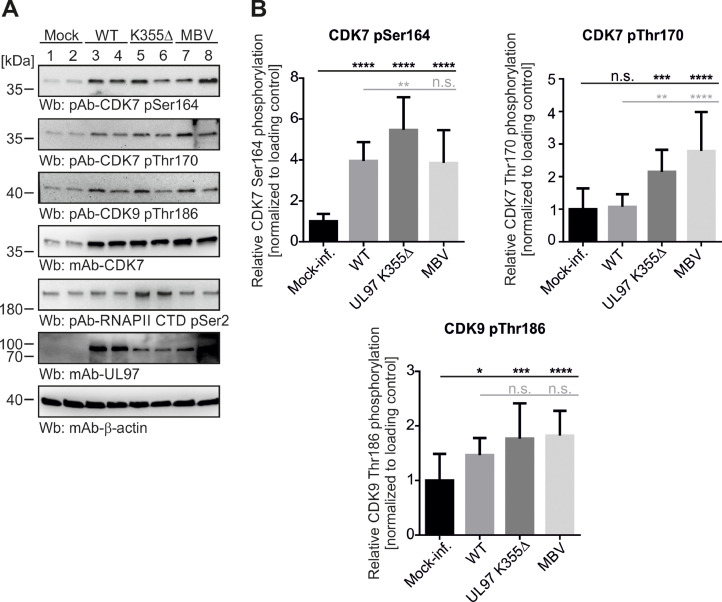
Based on HCMV-infected primary human fibroblasts (HFFs), we identified another remarkable correlation between the phosphorylation patterns of CDK7 (an indicative marker of catalytic activity), on the one hand, and the influences of HCMV infection and pUL97 kinase activity, on the other hand. Wb analyses applying commercially available, phosphosite-specific antibodies against CDK7 T-loop phosphorylation sites pSer164 and pThr170 (as well as antibodies against CDK9 pThr186 or CTD-RNAP II pSer2) were performed. The results indicated the following characteristics: firstly, an upregulation of expression and phosphorylation levels for CDK7 upon HCMV infection was detected (note a modest increase in CDK7 pThr170); secondly, a dependence of the two CDK7 phosphomarkers pSer164 and pThr170 on the presence of active pUL97 kinase (either K355M mutant or MBV treatment; and thirdly, an unexpected modest increase of these two CDK7 phosphomarkers under conditions of an impaired or inhibited activity of viral pUL97 activity (Figs. 7, S5; note that the phosphorylation specificity of antibodies was verified by controls based on alkaline phosphatase-based dephosphorylation of protein samples, see Fig. S6). Especially the latter finding may be explained by a compensatory effect in HCMV-infected cells. Such compensation might be a result of the lack of pUL97 kinase activity. Such counterregulation, in terms of the observed increase of these two CDK7 phosphomarkers in the absence of pUL97, might concern a balancing of CDK7-mediated transcriptional activity in HCMV-infected cells to maintain an activated state required for efficient HCMV replication. This latter scenario, however, lacks supporting data, i.e. specific experimentation should address this point in a follow-up study.
Fig. 7.
Phosphorylation patterns of CDK7 depend on the state of HCMV infection and pUL97 kinase activity. (A) HFFs were seeded in T75 cell culture flasks and infected 1 d later with HCMV AD169, or pUL97 kinase-deficient mutant ORF-UL97 K355Δ at MOI of 0.5, or remained-mock infected. 24 h after infection HCMV AD169 infected cells were treated with 0.35 µM (i.e. EC50) pUL97-specific inhibitor maribavir (MBV), or with equal amounts of DMSO. Cells were harvested 3 d p.i. and samples were subjected to standard SDS-PAGE and Wb analysis. Proteins of interest and site-specific phosphorylation were stained using the indicated antibodies. (B) Mean values ± SD of phosphorylation intensity of CDK7 at sites Ser164 and Thr170, as well as CDK9 Thr186, was determined by quadruplicate densitometric quantitation of those Wbs depicted in (A) together with a second biological replicate (shown in Fig. S5). Statistical analysis was performed using an ordinary one-way ANOVA and post-hoc Tukey correction: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., not significant.
3.4. An inducible knock-down of cyclin H exerts a substantial impact on the HCMV replication efficiency
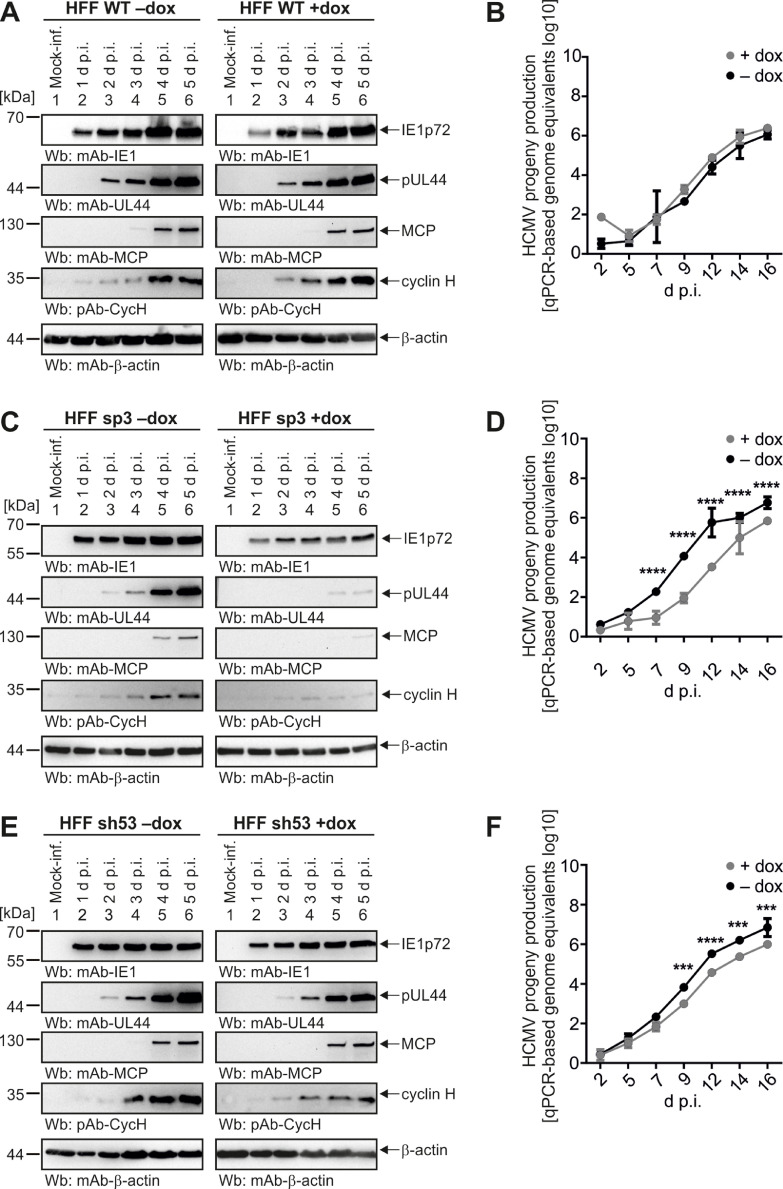
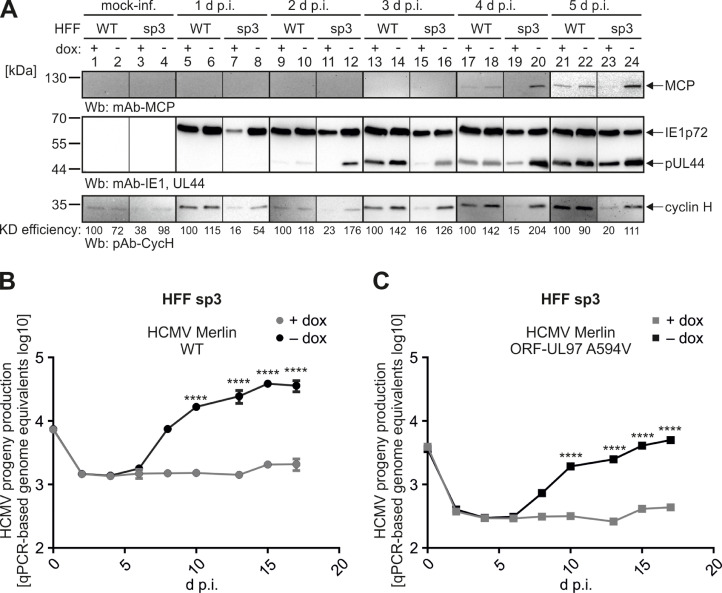
To better understand the functional significance of cyclin H in HCMV infection, we pursued the conditional depletion of cyclin H expression from HCMV-permissive primary HFFs. However, previous attempts to generate a stable and complete CRISPR/Cas9 knock-out of cyclin H proved difficult due to the multiple roles of cyclin H-CDK7 complexes in cell cycle, transcription, and other regulatory functions (Schütz et al., 2022). To overcome this limitation, here we selected five HFF populations with stably transduced doxycycline (dox) inducible anti-cyclin H short hairpin RNAs (shRNAs), as designated sh53, sh107, sp1, sp3, and sp4 (Figs. 8, S7). shRNA was induced for 5 d before infection of HFFs to determine HCMV infection kinetics by Wb analysis and qPCR-based viral genome quantitation. For Wb analysis, dox-induced and non-induced HFF WT, sp3, and sh53 cells were harvested at 1, 2, 3, 4, and 5 d p.i., before immediate early (IE) protein IE1p72, early (E) protein pUL44 and late (L) protein MCP were detected together as viral markers, in addition to cyclin H and β-actin as a loading control. In WT cells, no difference in viral protein expression was observed between dox addition and the control without dox (Fig. 8A). IE protein IE1p72 was strongly detectable from 1 d p.i., E protein pUL44 from 2 d p.i. of infection, and L protein MCP from 4 d p.i.. Similar to Fig. 2, cyclin H was strongly upregulated from 2 d p.i. onwards. The qPCR-based replication kinetics were consistent with the Wb result and revealed no difference in viral genomic copy numbers between +dox and -dox (Fig. 8B). In contrast to WT cells, cyclin H expression in dox-induced sp3 HFFs was close to the detection limit and no upregulation of cyclin H was noted upon infection (Fig. 8C). Consequently, protein levels of IE1p72 were reduced in shRNA-expressing cells, compared to -dox control cells, at all time points analyzed. Strikingly, the expression of E protein pUL44 was strongly delayed and only weakly detectable at 4 d p.i., compared to 2 d p.i. in the control cells. L protein MCP was also delayed and barely apparent at 5 d p.i.. Importantly, also a highly significant, 2-fold log10 reduction in the release of viral genome equivalents was detected by qPCR (Fig. 8D). Concerning infection of HFF sh53 cells, also here viral protein production in dox-induced was reduced compared to non-induced cells, although to a lesser extent than in HFF sp3 cells (Fig. 8E, F). This correlates with the expression of cyclin H, as its HCMV-mediated upregulation upon infection was only slightly affected by the addition of dox, but not completely abolished. The production of viral genome equivalents was also impaired and delayed by about 1-fold log10 in dox-induced HFF sh53 cells compared to non-induced cells.
Fig. 8.
Evaluation of viral replication efficiency in stably transduced HFFs expressing doxycycline-inducible anti-cyclin H shRNAs. HFF WT (A) or stably transduced HFFs, under conditional dox-inducible KD, termed HFF sp3 (C) and HFF sh53 (E) were seeded in 12-well plates before shRNA was induced by the addition of 500 ng/ml dox for 5 d. Next, dox-induced and non-induced HFFs were infected with HCMV AD169 at MOI of 0.25 and were harvested at indicated time points for standard SDS-PAGE and Wb analysis using the indicated antibodies. Additionally, dox-induced and non-induced HFFs were infected with HCMV AD169 at MOI of 0.001 (B, D, F). Viral supernatants were collected at the indicated time points and viral genome equivalents were determined by qPCR. Mean values ± SD of two independent biological replicates, each measured in technical duplicates, are shown. Statistical analysis was performed using an ordinary two-way ANOVA and post-hoc Sidak correction: ***, p < 0.001; ****, p < 0.0001.
Another Wb dataset of HFF sp3 and HFF WT was generated to confirm the cyclin H dependence of HCMV AD169 in terms of viral protein production and viral replication efficiency (Fig. 9A). In addition, the KD efficiency of cyclin H was determined by densitometry and was reduced up to 15% compared to WT cells, while the upregulation of cyclin H in non-induced cells was comparable to WT HFFs. Again, protein production of IE protein IE1p72, E protein pUL44, and L protein MCP was strongly delayed and reduced in HFF sp3 cells compared to non-induced HFF sp3 and WT HFFs. Moreover, we addressed the question of whether cyclin H is also important for the genetically intact, clinically relevant HCMV strain Merlin by applying qPCR-based replication kinetics (Fig. 9B, C). HFF sp3 dox-induced and non-induced cells were infected either with HCMV Merlin WT or the Merlin mutant expressing ganciclovir resistance-conferring ORF-UL97 A594V, before viral genome equivalents released into the supernatant were quantified. Starting at 6 d p.i., a genomic increase could be detected for both HCMV Merlin WT and mutant A594V in the non-induced cells, whereas no increase in viral genome equivalents could be detected in dox-induced cells over the entire time course of the experiment. This complete lack of viral release of genome equivalents was even more pronounced than previously observed for laboratory strain HCMV AD169, suggesting that cyclin H may play an even more crucial role for HCMV Merlin than for HCMV AD169.
Fig. 9.
Viral replication efficiency in transduced HFF cell populations carrying an expression module of inducible anti-cyclin H shRNA sp3. (A) Non-transduced HFFs WT or transduced HFFs carrying the dox-inducible anti-cyclin H shRNA sp3 module were seeded in 12-well plates and shRNA was induced by the addition of 500 ng/ml dox for 5 d. Thereafter, dox-induced and non-induced HFFs were infected with HCMV AD169 at MOI of 0.25 and were harvested at indicated time points for standard SDS-PAGE and Wb analysis using the indicated antibodies. KD efficiency of cyclin H was quantitatively determined by densitometric evaluation. Dox-induced and non-induced HFF sp3 were infected with HCMV Merlin WT (B) or mutant Merlin expressing GCV resistance-conferring A594V variant of pUL97 (Schütz et al., 2022) (C) at MOI of 0.001. Viral supernatants were collected at the indicated time points and viral genome equivalents were determined by qPCR. Mean values ± SD of two independent biological replicates, each measured in technical duplicates, are shown. Statistical analysis was performed using an ordinary two-way ANOVA and post-hoc Sidak correction: ****, p < 0.0001.
3.5. Analysis of covalent inhibitors potentially targeting the pUL97–cyclin interaction
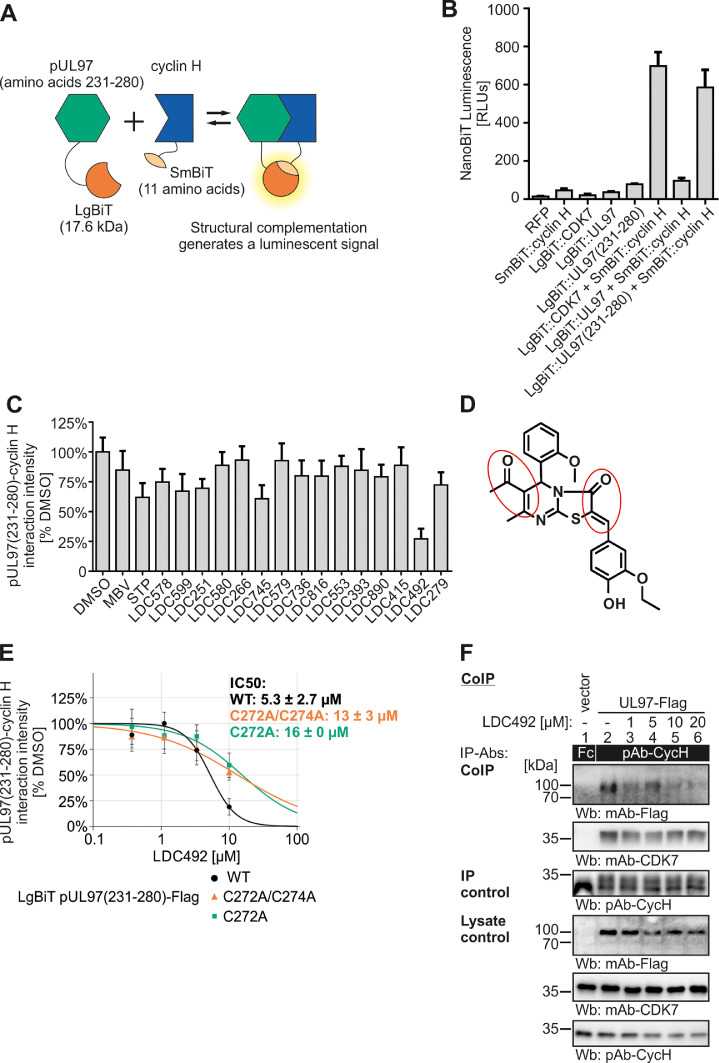
In an approach to validate this important key aspect of virus–host interaction, we analyzed putative PPI inhibitors of the pUL97-cyclin H interaction using the established NanoBiT® system (Fig. 10A). In this assay, proteins of interest are fused to LargeBiT (LgBiT) or SmallBiT (SmBiT) fragments of a recombinant luciferase. Both constructs are transiently coexpressed in 293T cells. The interaction of proteins leads to the structural complementation of LgBiT and SmBiT, generating a functional NanoLuc that converts a chemical substrate and emits a chemoluminescent reporter signal. In a first test, 293T cells were transfected with constructs coding for red fluorescent protein (RFP), SmBiT::cyclin H, LgBiT::CDK7, LgBiT::UL97 or LgBiT::UL97(231-280), either as single transfection controls, or cotransfected with SmBiT::cyclin H together with one of the three LgBiT constructs LgBiT::CDK7, LgBiT::UL97 or LgBiT::UL97(231-280). While single-transfected cells produced only marginal levels of luminescence signal, LgBiT::CDK7 + SmBiT::cyclin H produced a strong signal. Regarding the interaction of vCDK/pUL97 with cyclin H, luminescence levels of full-length pUL97 were very low, whereas the minimally required binding fragment pUL97(231-280) was able to strongly interact with cyclin H and complement to a fully functional NanoLuc luciferase and generate luminescence (Fig. 10B). Apparently, the expression of the minimal interaction fragment in fusion construct LgBiT::UL97(231-280) led to the most prominent PPI signal in this assay system (and a lack of signals with the full-length pUL97 version might refer to a limited stability of overexpressed pUL97, which is consistent with our earlier experience in expressing pUL97 in other systems; Marschall et al., 2002; Schütz et al., 2021; Steingruber et al., 2015; Webel et al., 2014). We then performed a small-scale comparative analysis with a selected series of small molecules, addressing the question of PPI disruption detected by this system (Fig. 10C). For this purpose, we used covalently binding inhibitors, referred to as warhead compounds, which possess a strong inherent reactivity of covalent binding to accessible cysteine residues in target proteins (Petri et al., 2020). Previously, our studies already identified warheads comprising anti-HCMV activity in cultured cell models, in the absence of marked cytotoxicity, in a micromolar range of EC50 values (Tillmanns et al., 2023). Here, we focused on blocking effects on pUL97(231-280)–cyclin H interaction in the NanoBiT by applying inhibitors at a concentration of 10 µM. As a main finding, we identified a strong inhibitory activity of the compound LDC492 (Fig. 10C). The chemical structure and the α, β-unsaturated carbonyl groups responsible for covalent binding to the target are marked by red circles (Fig. 10D). A concentration-dependent evaluation of the mean inhibitory activity revealed an IC50 value of 5.3 µM ± 2.7 µM (Fig. 10E). Of specific relevance is the point that we recognized two possibly structurally exposed cysteine residues in the minimal binding fragment of pUL97–cyclin H (IF2) and generated alanine replacement mutants, by introducing C272A and C272A/C274A into LgBiT::UL97(231-280) using site-directed mutagenesis. Both the single-mutant as well as the double-mutant revealed a marked reduction of the PPI blocking activity of the warhead compound LDC492, as measured with the NanoBiT system under identical conditions (IC50 values of 16 µM ± 0 µM and µM 13 ± 3, respectively, Fig. 10E). This result strongly underlined our concept that this covalent binder, at least in part, is directed to the pUL97–cyclin H interaction. It should be noted that this compound is only considered a preliminary example of an experimental warhead so far, with the potency to block virus-relevant PPI. Nevertheless, a confirmatory experiment, including specificity testing, was undertaken by a CoIP approach, in which we used the identical type of protein samples applied to the NanoBiT experimentation. These samples of transient expression were subjected to a CoIP reaction to confirm the PPI-inhibitory activity of LDC492 towards pUL97-Flag in its interaction with endogenous cyclin H (Fig. 10F). The result shows that the compound is effective in blocking pUL97–cyclin H interaction also in the CoIP approach, especially at the higher concentrations of 10 and 20 µM (Fig. 10F, upper Wb panel), while it is inactive against CDK7–cyclin H interaction (second upper panel). This finding confirms, on the one hand, the blocking activity of the compound against pUL97–cyclin H with an independent, second method (CoIP in addition to NanoBiT), and provides, on the other hand, a binding specificity control, as the compound does not interfere with CDK7–cyclin H. To build on this finding obtained with the compound LDC492, further information on binding characteristics, target specificity, or antiviral developmental potential of this group of small molecules has still to be provided by future analysis. At this stage of investigation, the approach points to the basic option of exploiting this type of PPI, i.e. herpesviral kinase–cyclin interaction, as a novel strategy of antiviral drug targeting.
Fig. 10.
Measurements in the NanoBiT system demonstrate a warhead-mediated inhibition of pUL97(231-280)–cyclin H interaction. (A) Schematic depiction of the NanoBiT system. Proteins of interest are fused to LargeBiT (LgBiT) or SmallBiT (SmBiT) fragments of recombinant luciferase, to be coexpressed in transiently transfected cells. Interaction of proteins leads to the complementation of LgBiT and SmBiT, thereby generating a functional NanoLuc luciferase that converts a chemical substrate and thereby emits a chemoluminescence reporter signal. (B) NanoBiT test measurement of cells expressing red fluorescent protein (RFP), LgBiT::UL97, LgBiT::UL97(231-280), or LgBiT::CDK7 to evaluate the specificity of the system. Additionally, cells expressing LgBiT::CDK7, as well as the complete pUL97 (LgBiT::UL97) or solely the cyclin H interaction interface (LgBiT::UL97(231-280) were tested together with SmBiT::cyclin H to identify the most robust interaction pair. LgBiT::UL97(231-280)–SmBiT::cyclin H was used for further inhibitor analysis. (C) NanoBiT primary small-scale screening of potential inhibitors of the interaction LgBiT::pUL97(231-280) + SmBiT::Cyclin H. LgBiT:pUL97(231-280) and SmBiT::Cyclin H coexpressing cells were treated with a series of experimental compounds (LDC compounds, 10 µM), maribavir (MBV, 10 µM) or staurosporine (STP, 1 µM), the latter two as reference controls, before luciferase signal was measured. Signal intensities were normalized to the DMSO control. (D) The chemical structure of LDC492 is given and the α, β-unsaturated carbonyl groups are marked with red circles (E) 293T cells were transfected with LgBiT::UL97(231-280) WT, point mutants C272A/C274A or C274A, together with SmBiT::cyclin H, and treated with serial concentrations of LDC492 to determine IC50 values of pUL97(231-280)–cyclin H interaction (black curves). Replacement mutants C272A/C274A and C274A of LgBiT::pUL97(231-280) are depicted as orange or green curves, respectively. 293T cells were transiently transfected with plasmids encoding pUL97-Flag or empty vector control (F). Cells were lysed 2 d post-transfection and human cyclin H was immunoprecipitated; a rabbit Fc fragment served as a specificity control. A range of different concentrations of LDC492 or equal amounts of DMSO were added during cell lysis and CoIP. Subsequently, CoIP samples and lysate controls, taken before the addition of IP-Abs were then subjected to standard Wb analysis using the indicated antibodies.
3.6. Conclusion: combined data of the study illustrate the codetermination of efficiency of the HCMV replication by vCDK/pUL97, cyclin H and CDK7
As demonstrated by the findings of this study, the efficiency of HCMV replication is codetermined by viral and cellular CDK-cyclin complexes (summarized in Table 1). Previous reports already illustrated that both, vCDK/pUL97 inhibitors, such as MBV (EC50 0.35 ± 0.42 µM), and CDK7 inhibitors, such as LDC4297 (EC50 0.009 ± 0.002 µM), exert a very pronounced antiviral activity (Biron et al., 2002; Hutterer et al., 2015; Steingruber and Marschall, 2020). Our recent reports also revealed a strong drug synergism between several combination treatments with vCDK/pUL97 and CDK inhibitors (Wild et al., 2022; Wild et al., 2021). Moreover, our current investigations demonstrated a strong functional relevance of cyclin H for HCMV replication in various host cell types (Schütz et al., 2022; Schütz et al., 2021; present study). In particular, the experimental settings of transient cyclin H knock-out (KO) and inducible cyclin H KD showed a significant reduction of HCMV replication compared to control cells (60.9 ± 0.5 / 19.9 ± 5.5 / 6.0 ± 1.2 / 9.4 ± 0.4, respectively, as depending on the experimental conditions). Combined, these results provide evidence for the importance of these regulatory factors as codeterminants of HCMV replication efficiency.
Table 1.
Modes of antiviral interference exemplifying the codetermination of viral replication efficiency by viral and host-specific factors.
| vCDK/pUL97 | CDK7 | Cyclin H | |
|---|---|---|---|
| Impact on HCMV replication |
antiviral drug: EC50 maribavir 0.35 ± 0.42 µM (Wild et al., 2022) |
antiviral drug: EC50 LDC4297 0.009 ± 0.002 µM (Wild et al., 2022) |
transient cyclin H KO: % HCMV vs. control cells AD169: 60.9 ± 0.5 * (Schütz et al., 2022) inducible cyclin H KD: % HCMV vs. control cells AD169: 19.9 ± 5.5* Merlin WT 6.0 ± 1.2* Merlin ORF-UL97 A594V 9.4 ± 0.4* (present study) |
HCMV replication efficiency in HFF KO/KD cells compared to control cells. Primary values of viral genome equivalents obtained from the replication kinetics (i.e. last day transient cyclin H KO AD169 7 d p.i., last day inducible cyclin H KD AD169 16 d pi., last day inducible cyclin H KD Merlin WT/A594V 17 d p.i) were divided by the respective genomic values obtained at 2 d p.i. and finally normalized using their control values (i.e. HFFs WT or non-induced cells).
4. Discussion
4.1. Relevance of CDK–cyclin complexes for host cell regulation
In this research, we substantiated our knowledge about CDK7 and its multiple roles during HCMV infection. Data of this study strongly support the previous statement that cyclin H contributes to the HCMV vCDK/pUL97 activity in concert with host CDK7. As a translational aspect of the current investigations, which is further discussed below, we provide the first experimental evidence that the pUL97–cyclin H interaction, as part of an important regulatory complex constituted by these two components together with CDK7, may be exploited as a drug-accessible target. Regarding the host-specific components of this regulatory complex, it should be mentioned that CDK7 requires two additional proteins to reach its fully activated state, namely cyclin H and the RING-finger protein MAT1 (Peissert et al., 2020; Tassan et al., 1995). This heterotrimeric complex then stimulates the cell cycle CDKs, i.e. primarily CDKs 1, 2, and 4/6, via T-loop phosphorylation and is therefore referred to as the CDK-activating kinase (CAK) complex (Lolli and Johnson, 2005). The CAK also plays an important role in transcriptional regulation by associating with the general transcription factor II H (TFIIH). While phosphorylation is not obligatory for CDK7 activity per se, the T-loop can be phosphorylated at two positions Ser164 and Thr170, thereby enhancing both CDK7 kinase activity and cyclin H binding (Glover-Cutter et al., 2009; Sava et al., 2020). Especially CDK7 phosphorylation at Thr170 appears to enhance CDK7 activity as a part of the transcriptional TFIIH complex towards RNAP II without significantly affecting activity towards CDK2 (Larochelle et al., 2001). Notably, the T-loop phosphorylation-induced increase in activity is solely attributed to an accelerated enzyme turnover (Larochelle et al., 2001). In HCMV infections, the CAK has previously been reported to be more highly expressed and active towards RNAP II CTD than in the uninfected state. Interestingly, CDK7 activity is elevated even before a detectable increase in CDK7 expression levels, suggesting an additional virus-induced mechanism (Tamrakar et al., 2005). In this study, we provide evidence that viral pUL97 activity exerts a stimulatory effect on CDK7 activity, as exemplified by the use of sensitive qSox-IVKA methodology. Based on this finding, it seems plausible that pUL97 might likewise phosphorylate CDK7 in its T-loop, possibly inducing an efficiency increase of RNAP II CTD phosphorylation. In addition, the ternary mode of interaction of pUL97(231-280) close to the cyclin H–CDK7 interface (Fig. 1B) might stabilize this complex, thereby also enhancing CDK7 activity. However, this hypothesis still requires experimental confirmation. In general terms, it appears reasonable that a strong CDK7 phosphorylation, associated with a high level of transcriptional activity, is beneficial for the lytic cycle of HCMV infection. Primarily, CDK7 phosphorylates RNAP II CTD at Ser5 to mediate transcription initiation. In addition, CDK7 activates CDK9, which in turn phosphorylates the CTD of RNAP II at Ser2 to mediate transcription elongation (Sava et al., 2020). Importantly, pUL97 has also been shown to phosphorylate RNAP II at both sites of Ser2 and Ser5 in vitro (Baek et al., 2004). Interestingly, the increased phosphorylation levels of the CDK7 T-loop upon pharmacological inhibition of vCDK/pUL97, observed in the present study, may represent a compensatory mechanism to maintain upregulated CDK7 activity in the absence of pUL97 activity. Another interesting aspect in this context is the fact that pharmacological inhibition of both pUL97 and CDK7, through drug combination treatment, produces strong synergistic effects in antiviral potency, indicating that a lack of both kinase activities cannot be overcome by the virus. Taken together, these findings further underscore the importance of the interplay between CDK7 and vCDK/pUL97 for HCMV replication efficiency.
4.2. Cyclin H is important for the function of both CDK7 and vCDK/pUL97
We reported previously that cyclin H is an important coregulator of HCMV replication (Schütz et al., 2022). However, it remained an open question whether this cyclin H dependence was specifically related to the inactive CAK complex or whether the lack of pUL97–cyclin H interaction may similarly exert a negative effect on HCMV replication. In particular, the efficient inhibition of HCMV replication by CDK7-directed drugs suggested that the inactive CAK might play a major role in the observed cyclin H KO/KD phenotype (Wild et al., 2022). However, here we provide experimental evidence that not exclusively CDK7, but also pUL97 activity, depends on cyclin H in the context of HCMV replication. KD of cyclin H significantly reduced pUL97 kinase activity and the exogenous addition of recombinant cyclin H was able to improve pUL97 kinase activity in vitro. We thus concluded that the KO/KD of cyclin H not only affects the CAK complex but likewise vCDK/pUL97 in its activity. To our knowledge, this is the first evidence that a herpesviral kinase requires human cyclin interaction to achieve full functionality.
4.3. Inhibition of vCDK/pUL97-cyclin H-CDK7 complexes as part of a novel antiviral targeting strategy
Recently, MBV has been clinically approved as the first kinase inhibitor in the entire field of antiviral therapy (Maertens et al., 2019; Marty et al., 2011). This achievement strongly encourages the development of additional kinase inhibitors against HCMV, other herpesviruses, and even unrelated human pathogenic viruses (Steingruber and Marschall, 2020). In particular, the pronounced anti-HCMV efficacy of host-directed CDK inhibitors has been reported and may favor the development of such drugs. It should be noted that the CDK7 inhibitor used in this study, LDC4297, is one of the rare examples of a highly selective kinase inhibitor, as exemplified by the demonstration of a kinome-wide selectivity panel (Hutterer et al., 2015). Based on the herpesviral expression of viral CDK orthologs (e.g. HCMV vCDK/pUL97), approaches for combination treatment with both CDK and vCDK inhibitors may lead to the exploitation of drug synergisms in clinical settings. Beyond this aspect, the findings of the present study illustrated the importance of vCDK-cyclin interaction. This may represent another key point for future drug targeting options. Small molecules with the potential to interfere with cyclin-associated complexes in virus-infected cells, such as pUL97-cyclin H, may prove to be a mechanistically innovative approach to future anti-HCMV drug development. The current report may provide a basis for similar analysis in related herpesviral systems similarly, which should broaden the understanding of virus-host interaction at the molecular and medically oriented translational levels.
Funding
The project was supported by the Interdisciplinary Center of Clinical Research of the Medical Center/Universitätsklinikum Erlangen (IZKF project A88-M.M./H.S.), the Bayerische Forschungsstiftung (grant DeeP CMV/AP-5/M.M.), Wilhelm Sander-Stiftung (grant M.M./H.S. AZ2022.073.1), the Matching Funds Program of Forschungsstiftung Medizin, UKER Medical Center Erlangen & Manfred Roth-Stiftung Fürth (grant M.S./M.M.), and DAAD-Go8 (grants M.M./W.D.R. 2015-16, 2017-18, 2020-21).
CRediT authorship contribution statement
Martin Schütz: Conceptualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing. Christina Wangen: Methodology, Writing – review & editing. Mona Sommerer: Methodology, Writing – review & editing. Melanie Kögler: Methodology, Writing – review & editing. Jan Eickhoff: Project administration, Writing – review & editing. Carsten Degenhart: Project administration, Writing – review & editing. Bert Klebl: Project administration, Funding acquisition, Writing – review & editing. Zin Naing: Project administration, Writing – review & editing. Ece Egilmezer: Project administration, Writing – review & editing. Stuart T. Hamilton: Project administration, Writing – review & editing. William D. Rawlinson: Project administration, Funding acquisition, Writing – review & editing. Heinrich Sticht: Conceptualization, Methodology, Writing – original draft, Funding acquisition, Writing – review & editing. Manfred Marschall: Conceptualization, Writing – original draft, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We like to thank all research group members of the M.M., B.K., H.S., and W.D.R. laboratories for very valuable scientific contributions and continuous competent discussion. We are grateful to Regina Müller and Sabrina Wagner for their excellent technical assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199200.
Contributor Information
Martin Schütz, Email: martin.schuetz@uk-erlangen.de.
Manfred Marschall, Email: manfred.marschall@fau.de.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Baek M.C., Krosky P.M., Pearson A., Coen D.M. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology. 2004;324(1):184–193. doi: 10.1016/j.virol.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Biron K.K., Harvey R.J., Chamberlain S.C., Good S.S., Smith A.A., Davis M.G., Talarico C.L., Miller W.H., Ferris R., Dornsife R.E., Stanat S.C., Drach J.C., Townsend L.B., Koszalka G.W. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob. Agents Chemother. 2002;46(8):2365–2372. doi: 10.1128/AAC.46.8.2365-2372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer P.E., Nimonkar A.V. Herpes virus replication. IUBMB Life. 2003;55(1):13–22. doi: 10.1080/1521654031000070645. [DOI] [PubMed] [Google Scholar]
- Couté Y., Kraut A., Zimmermann C., Büscher N., Hesse A.-M., Bruley C., De Andrea M., Wangen C., Hahn F., Marschall M., Plachter B. Mass spectrometry-based characterization of the virion proteome, phosphoproteome, and associated kinase activity of human cytomegalovirus. Microorganisms. 2020;8(6):820. doi: 10.3390/microorganisms8060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y., Sinha N., Zuber J. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5(6):1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Fellmann C., Zuber J., McJunkin K., Chang K., Malone C.D., Dickins R.A., Xu Q., Hengartner M.O., Elledge S.J., Hannon G.J., Lowe S.W. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 2011;41(6):733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.P., Morgan D.O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78(4):713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R.P., Bentley D.L. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 2009;29(20):5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum F., Britt W., Mocarski E.S. 7th ed. Vol. 2. LWW; Philadelphia, PA, USA: 2021. p. 760. (Fields Virology: DNA Viruses). [Google Scholar]
- Graf L., Feichtinger S., Naing Z., Hutterer C., Milbradt J., Webel R., Wagner S., Scott G.M., Hamilton S.T., Rawlinson W.D., Stamminger T., Thomas M., Marschall M. New insight into the phosphorylation-regulated intranuclear localization of human cytomegalovirus pUL69 mediated by cyclin-dependent kinases (CDKs) and viral CDK orthologue pUL97. J. Gen. Virol. 2016;97(1):144–151. doi: 10.1099/jgv.0.000337. [DOI] [PubMed] [Google Scholar]
- Graf L., Webel R., Wagner S., Hamilton S.T., Rawlinson W.D., Sticht H., Marschall M. The cyclin-dependent kinase ortholog pUL97 of human cytomegalovirus interacts with cyclins. Viruses. 2013;5(12):3213–3230. doi: 10.3390/v5123213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B.J., Remis J., Ali S., Nogales E. 2.5 Å-resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942. Biophys. J. 2021;120(4):677–686. doi: 10.1016/j.bpj.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliesi F., Coscia A., Griffante G., Galitska G., Pasquero S., Albano C., Biolatti M. Where do we stand after decades of studying human cytomegalovirus? Microorganisms. 2020;8(5):685. doi: 10.3390/microorganisms8050685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton S.T., Marschall M., Rawlinson W.D. Investigational antiviral therapy models for the prevention and treatment of congenital cytomegalovirus infection during pregnancy. Antimicrob. Agents Chemother. 2020;65(1) doi: 10.1128/AAC.01627-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume A.J., Finkel J.S., Kamil J.P., Coen D.M., Culbertson M.R., Kalejta R.F. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320(5877):797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- Hutterer C., Eickhoff J., Milbradt J., Korn K., Zeitträger I., Bahsi H., Wagner S., Zischinsky G., Wolf A., Degenhart C., Unger A., Baumann M., Klebl B., Marschall M. A novel CDK7 inhibitor of the Pyrazolotriazine class exerts broad-spectrum antiviral activity at nanomolar concentrations. Antimicrob. Agents Chemother. 2015;59(4):2062–2071. doi: 10.1128/AAC.04534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer C., Hamilton S., Steingruber M., Zeitträger I., Bahsi H., Thuma N., Naing Z., Örfi Z., Örfi L., Socher E., Sticht H., Rawlinson W., Chou S., Haupt V.J., Marschall M. The chemical class of quinazoline compounds provides a core structure for the design of anticytomegaloviral kinase inhibitors. Antivir. Res. 2016;134:130–143. doi: 10.1016/j.antiviral.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Hutterer C., Wandinger S.K., Wagner S., Müller R., Stamminger T., Zeitträger I., Godl K., Baumgartner R., Strobl S., Marschall M. Profiling of the kinome of cytomegalovirus-infected cells reveals the functional importance of host kinases Aurora A, ABL and AMPK. Antivir. Res. 2013;99(2):139–148. doi: 10.1016/j.antiviral.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Iwahori S., Umaña A.C., VanDeusen H.R., Kalejta R.F. Human cytomegalovirus-encoded viral cyclin-dependent kinase (v-CDK) UL97 phosphorylates and inactivates the retinoblastoma protein-related p107 and p130 proteins. J. Biol. Chem. 2017;292(16):6583–6599. doi: 10.1074/jbc.M116.773150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.H. Reversible Michael additions: covalent inhibitors and prodrugs. Mini Rev. Med. Chem. 2012;12(13):1330–1344. doi: 10.2174/13895575112091330. [DOI] [PubMed] [Google Scholar]
- Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43(W1):W314–W319. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S., Chen J., Knights R., Pandur J., Morcillo P., Erdjument-Bromage H., Tempst P., Suter B., Fisher R.P. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 2001;20(14):3749–3759. doi: 10.1093/emboj/20.14.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolli G., Johnson L.N. CAK-cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle. 2005;4(4):572–577. [PubMed] [Google Scholar]
- Lorz K., Hofmann H., Berndt A., Tavalai N., Mueller R., Schlötzer-Schrehardt U., Stamminger T. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 2006;80(11):5423–5434. doi: 10.1128/JVI.02585-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens J., Cordonnier C., Jaksch P., Poiré X., Uknis M., Wu J., Wijatyk A., Saliba F., Witzke O., Villano S. Maribavir for preemptive treatment of cytomegalovirus reactivation. N. Engl. J. Med. 2019;381(12):1136–1147. doi: 10.1056/NEJMoa1714656. [DOI] [PubMed] [Google Scholar]
- Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15(6):122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M., Muller Y.A., Diewald B., Sticht H., Milbradt J. The human cytomegalovirus nuclear egress complex unites multiple functions: recruitment of effectors, nuclear envelope rearrangement, and docking to nuclear capsids. Rev. Med. Virol. 2017;27(4) doi: 10.1002/rmv.1934. 10.1002/rmv.1934. [DOI] [PubMed] [Google Scholar]
- Marschall M., Stein-Gerlach M., Freitag M., Kupfer R., van den Bogaard M., Stamminger T. Direct targeting of human cytomegalovirus protein kinase pUL97 by kinase inhibitors is a novel principle for antiviral therapy. J. Gen. Virol. 2002;83(Pt 5):1013–1023. doi: 10.1099/0022-1317-83-5-1013. [DOI] [PubMed] [Google Scholar]
- Marschall M., Strojan H., Kiener R., Wangen C., Sonntag E., Müller R., Zeitträger I., Wagner S., Stamminger T., Milbradt J., Behrends U., Körber N., Bauer T., Schrödel S., Thirion C., Wagner R., Hutterer C. Differential upregulation of host cell protein kinases by the replication of α-, β-and γ-herpesviruses provides a signature of virus-specific signalling. J. Gen. Virol. 2020;101(3):284–289. doi: 10.1099/jgv.0.001370. [DOI] [PubMed] [Google Scholar]
- Martínez-Alonso D., Malumbres M. Mammalian cell cycle cyclins. Semin. Cell Dev. Biol. 2020;107:28–35. doi: 10.1016/j.semcdb.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Marty F.M., Ljungman P., Papanicolaou G.A., Winston D.J., Chemaly R.F., Strasfeld L., Young J.A., Rodriguez T., Maertens J., Schmitt M., Einsele H., Ferrant A., Lipton J.H., Villano S.A., Chen H., Boeckh M. Maribavir prophylaxis for prevention of cytomegalovirus disease in recipients of allogeneic stem-cell transplants: a phase 3, double-blind, placebo-controlled, randomised trial. Lancet Infect. Dis. 2011;11(4):284–292. doi: 10.1016/S1473-3099(11)70024-X. [DOI] [PubMed] [Google Scholar]
- Milbradt J., Kraut A., Hutterer C., Sonntag E., Schmeiser C., Ferro M., Wagner S., Lenac T., Claus C., Pinkert S. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol. Cell. Proteom. 2014;13(8):2132–2146. doi: 10.1074/mcp.M113.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19(6):679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njue A., Coyne C., Margulis A.V., Wang D., Marks M.A., Russell K., Das R., Sinha A. The role of congenital cytomegalovirus infection in adverse birth outcomes: a review of the potential mechanisms. Viruses. 2020;13(1):20. doi: 10.3390/v13010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.A., Simon M.C. Functional analysis of the Cdk7.cyclin H.Mat1 complex in mouse embryonic stem cells and embryos. J. Biol. Chem. 2010;285(20):15587–15598. doi: 10.1074/jbc.M109.081687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péczka N., Orgován Z., Ábrányi-Balogh P., Keserű G.M. Electrophilic warheads in covalent drug discovery: an overview. Expert Opin. Drug Discov. 2022;17(4):413–422. doi: 10.1080/17460441.2022.2034783. [DOI] [PubMed] [Google Scholar]
- Peissert S., Schlosser A., Kendel R., Kuper J., Kisker C. Structural basis for CDK7 activation by MAT1 and cyclin H. Proc. Natl. Acad. Sci. USA. 2020;117(43):26739–26748. doi: 10.1073/pnas.2010885117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelossof R., Fairchild L., Huang C.H., Widmer C., Sreedharan V.T., Sinha N., Lai D.Y., Guan Y., Premsrirut P.K., Tschaharganeh D.F., Hoffmann T., Thapar V., Xiang Q., Garippa R.J., Rätsch G., Zuber J., Lowe S.W., Leslie C.S., Fellmann C. Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 2017;35(4):350–353. doi: 10.1038/nbt.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri L., Egyed A., Bajusz D., Imre T., Hetényi A., Martinek T., Ábrányi-Balogh P., Keserű G.M. An electrophilic warhead library for mapping the reactivity and accessibility of tractable cysteines in protein kinases. Eur. J. Med. Chem. 2020;207 doi: 10.1016/j.ejmech.2020.112836. [DOI] [PubMed] [Google Scholar]
- Prichard M.N. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 2009;19(4):215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revello M.G., Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 2002;15(4):680–715. doi: 10.1128/CMR.15.4.680-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaker D., Schregel V., Maurer K., Auerochs S., Marzi A., Sticht H., Marschall M. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J. Med. Chem. 2006;49(24):7044–7053. doi: 10.1021/jm060696s. [DOI] [PubMed] [Google Scholar]
- Rossi D.J., Londesborough A., Korsisaari N., Pihlak A., Lehtonen E., Henkemeyer M., Mäkelä T.P. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 2001;20(11):2844–2856. doi: 10.1093/emboj/20.11.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sava G.P., Fan H., Coombes R.C., Buluwela L., Ali S. CDK7 inhibitors as anticancer drugs. Cancer Metastasis Rev. 2020;39(3):805–823. doi: 10.1007/s10555-020-09885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20(9):374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Schregel V., Auerochs S., Jochmann R., Maurer K., Stamminger T., Marschall M. Mapping of a self-interaction domain of the cytomegalovirus protein kinase pUL97. J. Gen. Virol. 2007;88(Pt 2):395–404. doi: 10.1099/vir.0.82393-0. [DOI] [PubMed] [Google Scholar]
- Schütz M., Müller R., Socher E., Wangen C., Full F., Wyler E., Wong D., Scherer M., Stamminger T., Chou S., Rawlinson W.D., Hamilton S.T., Sticht H., Marschall M. Highly conserved interaction profiles between clinically relevant mutants of the cytomegalovirus CDK-like kinase pUL97 and human cyclins: functional significance of cyclin H. Int. J. Mol. Sci. 2022;23(19):11814. doi: 10.3390/ijms231911814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz M., Steingruber M., Socher E., Müller R., Wagner S., Kögel M., Sticht H., Marschall M. Functional relevance of the interaction between human cyclins and the cytomegalovirus-encoded CDK-like protein kinase pUL97. Viruses. 2021;13(7):1248. doi: 10.3390/v13071248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A., Penedones H., Petersen S., Simonyan K., Crossan S., Kohli P., Jones D.T., Silver D., Kavukcuoglu K., Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- Sonntag E., Hamilton S.T., Bahsi H., Wagner S., Jonjic S., Rawlinson W.D., Marschall M., Milbradt J. Cytomegalovirus pUL50 is the multi-interacting determinant of the core nuclear egress complex (NEC) that recruits cellular accessory NEC components. J. Gen. Virol. 2016;97(7):1676–1685. doi: 10.1099/jgv.0.000495. [DOI] [PubMed] [Google Scholar]
- Steingruber M., Keller L., Socher E., Ferre S., Hesse A.-M., Couté Y., Hahn F., Büscher N., Plachter B., Sticht H., Marschall M. Cyclins B1, T1, and H differ in their molecular mode of interaction with cytomegalovirus protein kinase pUL97. J. Biol. Chem. 2019;294(15):6188–6203. doi: 10.1074/jbc.RA118.007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingruber M., Kraut A., Socher E., Sticht H., Reichel A., Stamminger T., Amin B., Couté Y., Hutterer C., Marschall M. Proteomic interaction patterns between human cyclins, the cyclin-dependent kinase ortholog pUL97 and additional cytomegalovirus proteins. Viruses. 2016;8(8):219. doi: 10.3390/v8080219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingruber M., Marschall M. The cytomegalovirus protein kinase pUL97: host interactions. Regul. Mech. Antivir. Drug Target. Microorg. 2020;8(4):515. doi: 10.3390/microorganisms8040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingruber M., Socher E., Hutterer C., Webel R., Bergbrede T., Lenac T., Sticht H., Marschall M. The interaction between cyclin B1 and cytomegalovirus protein kinase pUL97 is determined by an active kinase domain. Viruses. 2015;7(8):4582–4601. doi: 10.3390/v7082834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamrakar S., Kapasi A.J., Spector D.H. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 2005;79(24):15477–15493. doi: 10.1128/JVI.79.24.15477-15493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan J.P., Jaquenoud M., Fry A.M., Frutiger S., Hughes G.J., Nigg E.A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14(22):5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Rechter S., Milbradt J., Auerochs S., Müller R., Stamminger T., Marschall M. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 2009;90(Pt 3):567–578. doi: 10.1099/vir.0.005827-0. [DOI] [PubMed] [Google Scholar]
- Tillmanns J., Häge S., Borst E.M., Wardin J., Eickhoff J., Klebl B., Wagner S., Wangen C., Hahn F., Socher E., Marschall M. Assessment of covalently binding warhead compounds in the validation of the cytomegalovirus nuclear egress complex as an antiviral target. Cells. 2023;12(8):1162. doi: 10.3390/cells12081162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y. Effects of cytomegalovirus infection on embryogenesis and brain development. Congenit. Anom. 2009;49(2):47–55. doi: 10.1111/j.1741-4520.2009.00222.x. [DOI] [PubMed] [Google Scholar]
- Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2021;2199:239–255. doi: 10.1007/978-1-0716-0892-0_14. [DOI] [PubMed] [Google Scholar]
- Webel R., Hakki M., Prichard M.N., Rawlinson W.D., Marschall M., Chou S. Differential properties of cytomegalovirus pUL97 kinase isoforms affect viral replication and maribavir susceptibility. J. Virol. 2014;88(9):4776–4785. doi: 10.1128/JVI.00192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webel R., Milbradt J., Auerochs S., Schregel V., Held C., Nöbauer K., Razzazi-Fazeli E., Jardin C., Wittenberg T., Sticht H., Marschall M. Two isoforms of the protein kinase pUL97 of human cytomegalovirus are differentially regulated in their nuclear translocation. J. Gen. Virol. 2011;92(Pt 3):638–649. doi: 10.1099/vir.0.026799-0. [DOI] [PubMed] [Google Scholar]
- Webel R., Solbak S., Held C., Milbradt J., Groß A., Eichler J., Wittenberg T., Jardin C., Sticht H., Fossen T., Marschall M. Nuclear import of isoforms of the cytomegalovirus kinase pUL97 is mediated by differential activity of NLS1 and NLS2 both acting through classical importin-α binding. J. Gen. Virol. 2012;93(Pt 8):1756–1768. doi: 10.1099/vir.0.040592-0. [DOI] [PubMed] [Google Scholar]
- Wild M., Hahn F., Brückner N., Schütz M., Wangen C., Wagner S., Sommerer M., Strobl S., Marschall M. Cyclin-Dependent Kinases (CDKs) and the human cytomegalovirus-encoded CDK Ortholog pUL97 represent highly attractive targets for synergistic drug combinations. Int. J. Mol. Sci. 2022;23(5):575. doi: 10.3390/ijms23052493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M., Kicuntod J., Seyler L., Wangen C., Bertzbach L.D., Conradie A.M., Kaufer B.B., Wagner S., Michel D., Eickhoff J., Tsogoeva S.B., Bäuerle T., Hahn F., Marschall M. Combinatorial drug treatments reveal promising anticytomegaloviral profiles for clinically relevant pharmaceutical kinase inhibitors (PKIs) Int. J. Mol. Sci. 2021;22(2) doi: 10.3390/ijms22020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430(15):2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.