Abstract
We present evidence, based on extensive mutagenesis, that a hairpin loop at the 5′ end of influenza A virus virion RNA (vRNA) is required for the synthesis of polyadenylated mRNA from model vRNA templates in vitro. The hairpin loop, which we term the vRNA 5′ hook, contains a stem of 2 bp formed by the second and third residues pairing with the ninth and eighth residues, respectively, and a 4-nucleotide loop composed of the intervening residues 4 to 7. Disruption of the base pairs of the vRNA 5′ hook by introducing point mutations prevented polyadenylation, except in two mutants where a G-U base pair reformed. The polyadenylation activity of point mutants could be rescued by constructing double mutants with reformed base pairs in the stem of the vRNA 5′ hook. These results suggest that base pairing rather than a particular nucleotide sequence was critical. We also show that mutation of the analogous region in the 3′ arm of vRNA did not interfere with the synthesis of polyadenylated mRNA, suggesting that a hook structure in the 3′ arm is not required for transcription of polyadenylated mRNA in vitro.
The eight, negative-stranded virion RNA (vRNA) gene segments of influenza A virus are each transcribed to produce polyadenylated mRNA by a virus-encoded RNA polymerase complex composed of three subunits termed PB1, PB2, and PA (11). The termini of each single-stranded RNA segment are conserved in sequence for 13 and 12 nucleotides at the 5′ and 3′ ends, respectively. These ends display partial inverted complementarity (2) and adopt a circular, panhandle conformation in virions and infected cells (9).
The nucleotide sequence requirements controlling influenza virus transcription and replication have been studied extensively (1, 5, 8, 10, 16–18, 20, 24, 25, 29). In vivo studies using influenza virus-like RNAs containing a chloramphenicol acetyltransferase (CAT) reporter gene have demonstrated the importance of both 5′ and 3′ termini of vRNA for overall genome expression and replication (16). In vitro systems initially suggested that the promoter for transcription resided entirely in the 3′ conserved sequence (20, 24, 25). Subsequently, it was shown that the influenza virus polymerase binds strongly to the 5′-terminal sequences and that the segment termini interact during transcription initiation (5, 6, 8, 28).
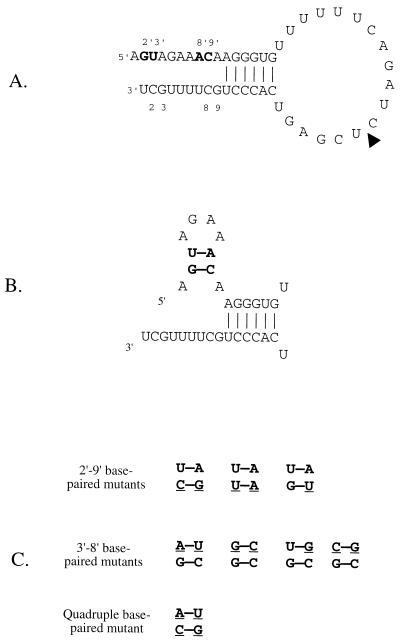
The precise structure of the panhandle formed between the segment termini has been the subject of debate. In vitro transcription studies demonstrated that, although base pairing between residues 10 to 15 of the 3′ arm and residues 11′ to 16′ of the 5′ arm (prime notation is used to distinguish 5′ residues from 3′ residues [5]) was required for promoter activity, base pairing between residues 1 to 9 (3′) and 1′ to 9′ (5′) was not required (5, 6). Indeed, most of the first nine residues of the 3′ arm could be mutated without destroying promoter activity in vitro. Even multiple mutations within the 3′ arm were tolerated (5, 6, 25). These results led to the proposal of the RNA “fork” model, which suggested that the extreme terminal sequences were open, like the prongs of a fork, held together by base pairing between residues further from the ends (5, 6) (Fig. 1A). An in vivo study with CAT reporter constructs suggested an alternative model in which the ends of the vRNA segments each formed a local secondary structure (3, 4). This RNA “corkscrew” model suggested that residues 4 to 7 formed a tetraloop at the end of a 2-bp stem formed by residues 2 and 3 pairing with residues 9 and 8. This stem-loop structure was postulated to be present at both the 3′ and 5′ segment ends.
FIG. 1.
(A) RNA fork representation of vRNA. The sequence is of the 49-mer vRNA-like template used, which is the same as the 717-nt template except for a 668-nt insertion at the position marked by the arrowhead (15, 23). Residues 2′, 3′, 8′, and 9′ are labeled and in boldface. (B) Diagram of the 5′ vRNA hook showing the base pairings of residues 2′ and 3′ with residues 9′ and 8′. (C) Base-paired mutants tested in the present study. The mutated residues are underlined. Only residues 2′, 3′, 8′, and 9′ are shown and are positioned as in panel B.
We have shown recently that polyadenylated mRNA molecules are synthesized from vRNA-like templates in an adenylyl 3′→5′ guanosine (ApG)-primed in vitro transcription system, dependent on the presence of a functional RNA polymerase binding site near the 5′ vRNA terminus of the transcription template (22, 23). Although capped host mRNA is used as a primer by the influenza virus RNA polymerase during viral infection, dinucleotide primers such as ApG have been used extensively in influenza virus in vitro transcription reactions, including for the synthesis of poly(A)-containing transcripts from endogenous vRNA templates (21). ApG-primed synthesis essentially mimics capped primer initiation when transcription is studied in vitro (6, 24, 25).
Here, based on previous evidence from studies on Thogoto virus, a related orthomyxovirus, where a 5′ hook structure was identified (12–14), we investigated whether a hairpin-loop structure near to the 5′ end of influenza virus vRNA is required for the synthesis of polyadenylated mRNA. We found that a 5′ vRNA hook structure, composed of a hairpin loop, is required. Furthermore, we found that an analogous structure in the 3′ end of vRNA suggested by the corkscrew model (3, 4) can be disrupted without loss of mRNA synthesis. This suggests that a 3′ hook is not required for the ApG-primed synthesis of polyadenylated mRNA.
MATERIALS AND METHODS
Preparation of influenza A virus polymerase.
RNA polymerase was isolated from influenza A virus, strain X-31, a reassortant of A/HK/8/68 and A/PR/8/34 as described previously (24). The virus was disrupted with Triton X-100 and lysolecithin; ribonucleoprotein (RNP) was separated by glycerol step gradient centrifugation, and endogenous vRNA was degraded by micrococcal nuclease.
Construction of plasmids.
The 717-nucleotide (nt) wild-type RNA and its mutants were synthesized from pBXPCAT1 (a gift from P. Palese) and its derivatives. Plasmid pBXPCAT1 encodes a 717-nt long RNA (Fig. 1A) with an antisense CAT gene flanked by linker sequences and vRNA terminal sequences derived from segment 8 of influenza virus A/PR/8/34 (15). Mutated plasmids were made as described before (23).
RNA template preparation.
Influenza virus vRNA-like templates were prepared and quantified as described before (23). Briefly, BpuAI-linearized plasmid DNAs were transcribed with 25 U of T7 RNA polymerase at 37°C for 20 min to 2 h. The RNA was extracted with phenol-chloroform and precipitated with ethanol. RNA templates were quantified either by gel electrophoresis, followed by ethidium bromide staining (717-nt RNAs), or by gel electrophoresis after 5′ labeling by [γ-32P]ATP and polynucleotide kinase, followed by PhosphorImager analysis (49-mer RNAs).
In vitro influenza virus transcription.
Transcription was done as described previously (23). Briefly, 5- to 20-μl reaction mixtures contained 0.1 to 1 μg of RNA template (constant within an assay), about 1 μg of nuclease-treated RNP (ca. 5 ng of polymerase protein [26], 500 μM nucleoside triphosphates, 0.5 mM ApG, 50 mM Tris-HCl [pH 7.4], 50 mM KCl, 10 mM NaCl, 5 mM MgCl2, 5 mM dithiothreitol, and 10 U of placental RNase inhibitor). The reactions were incubated at 30°C for 3 h.
[α-32P]ATP incorporation assay.
Reactions were as described previously (23). Briefly, the concentration of the ATP in the in vitro transcription mixtures was reduced to 25 μM, and 2 μCi of [α-32P]ATP (3,000 Ci/mmol) was added in a 5-μl reaction mixture. Reactions were incubated for 3 h at 30°C, after which the products were extracted with phenol-chloroform and precipitated with ethanol. RNA pellets were dissolved in formamide loading dyes, heated at 99°C for 3 min, and analyzed on 16% polyacrylamide–7 M urea gels. To quantify the yield of mRNA products, the gels were dried and the high-molecular-weight smear of mRNA products was examined by PhosphorImager analysis.
RT-PCR assay.
Polyadenylated products from in vitro transcription reactions containing 717-nt templates were assayed as before (23), except that the reverse transcriptase (RT) reactions were assembled at 20°C instead of at 40°C. Briefly, reverse transcription was at 40°C for 20 min in 10-μl reactions containing 50 pmol of 5′ GC-clamped T20 primer (5′-GCCCCGGGATCCT20-3′), 200 μM (each) deoxynucleoside triphosphates, 10 U of placental RNase inhibitor, 100 U of Moloney murine leukemia virus RT in a buffer containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, and 0.1% Triton X-100. Then, 50 pmol of CAT-specific primer (5′-CGGTGAAAACCTGGCCTATTTCCCTAAAGGG-3′) and 1.5 U of Taq polymerase were added to the reverse-transcribed products, which were next amplified by PCR (30 s at 94°C, 30 s at 65°C, and 2 min at 72°C) for 33 cycles. PCR products were analyzed by electrophoresis on 1.2% agarose gels in TAE buffer (40 mM Tris-acetate, 1 mM EDTA) and visualized by ethidium bromide staining.
RESULTS
Mutagenesis of residues 2′ and 9′ of a 717-nt vRNA-like template.
The products transcribed in vitro from 717-nt vRNA-like templates in influenza polymerase reactions were analyzed for the presence of poly(A) tails by using the RT-PCR assay described previously (22, 23). In this assay polyadenylated transcripts are detected as a broad band on ethidium bromide-stained agarose gels (see Materials and Methods). The broad band contains cDNAs of heterogeneous length comprising 320 nt of template (and primer) sequences and poly(A) tails of various lengths derived from randomly primed poly(A) sequences in the mRNA. The broad band has been rigorously characterized before (23).
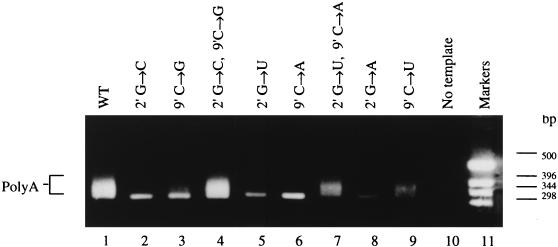
The effects of single and double point mutations at positions 2′ and 9′ of the postulated 5′ hook (Fig. 1B) were investigated (Fig. 2). Lane 1 shows the typical broad band, which is indicative of poly(A) formation, obtained when the transcription products of the wild-type 717-nt vRNA-like template were analyzed. Lanes 2 and 3, resulting from analysis of the products transcribed from templates mutated at positions 2′ (G→C) and 9′ (C→G), respectively, show no evidence of the poly(A)-derived broad band. Instead, a sharp band of 320 bp occurs at a position consistent with mispriming by the T20 primer on the run of six A residues present in cRNA molecules at the polyadenylation junction as seen before (22). In the present study, the RT reactions were assembled at 20°C, rather than at 40°C as before (22, 23) in order to encourage formation of this band, because it serves as a useful control. The yield of the band is variable even for the same template in independent experiments, a result possibly due to slight differences in reaction conditions affecting the efficiency of mispriming.
FIG. 2.
RT-PCR assay of polyadenylated mRNA synthesis from 717-nt vRNA-like templates mutated in the 2′ to 9′ base pair of the 5′ vRNA hook. Lanes: 1, control reaction assaying the products from the wild-type template; 2 to 9, assay of the products synthesized from the mutated templates as shown; 10, no added template control; 11, DNA size markers. Poly(A) marks the broad band indicative of polyadenylated mRNA production.
The double mutation (2′ G→C, 9′ C→G), which can form a “swapped” GC pair between positions 2′ and 9′, rescued activity (Fig. 2, lane 4). Lanes 5 and 6 show the effect of mutating residues 2′ G→U and 9′ C→A, respectively. In both cases, the poly(A)-containing broad band is absent. For the double mutant (2′ G→U, 9′ C→A), a broad band is present (lane 7), indicating that polyadenylated transcripts were synthesized. When position 2′ was mutated (G→A) polyadenylation was also inhibited (lane 8), although when 9′ was mutated (C→U), with position 2′ remaining a G as in wild type, polyadenylated transcripts were again synthesized (lane 9). This last result is explained by the formation of a G-U base pair between the wild-type position 2′ (G) and the mutated position 9′ (C→U) (see Fig. 1C).
Mutagenesis of residues 3′ and 8′ of a 717-nt vRNA-like template.
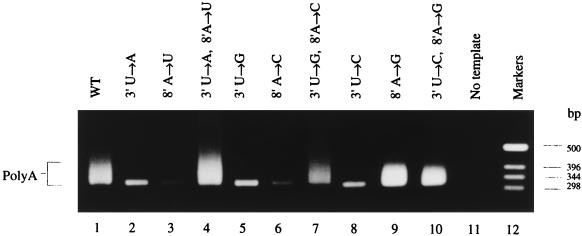
A similar analysis was carried out for residues 3′ and 8′, the residues involved in the second potential base pair of the postulated 5′ hook structure (Fig. 1B). Templates carrying single and double point mutations at positions 3′ and 8′ were transcribed in vitro, and the products were analyzed by RT-PCR assay (see Materials and Methods). Figure 3 (lane 1) shows the broad band containing heterogeneously sized, poly(A)-containing cDNAs present when the wild-type template was transcribed and assayed. All single mutations except 8′ A→G (lane 9, see below) resulted in loss of the broad band (lanes 2, 3, 5, 6, and 8). Instead, these lanes contained the 320-bp band consistent with mispriming on the run of six A residues present in cRNA. Thus, polyadenylated mRNA was not detected in these reactions. In contrast, double mutations which reintroduced base pairs gave rise to polyadenylated mRNA (lanes 4, 7, and 10). Interestingly, the single mutant 8′ A→G (lane 9) also gave rise to polyadenylated products. Again, this single mutation is compatible with the proposed base-paired structure, since a U-G pair would replace the U-A wild-type base pair (see Fig. 1C). The slightly reduced intensity of the broad band in lane 7 was not typical; in replicate experiments the double mutant (3′ U→G, 8′ A→C) gave a broad band similar to the wild type.
FIG. 3.
RT-PCR assay of polyadenylated mRNA synthesis from 717-nt vRNA-like templates mutated in the 3′ to 8′ base pair of the 5′ vRNA hook. Lanes: 1, control reaction containing the wild-type template; 2 to 10, assay of the products synthesized from the mutated templates as shown; 11, no added template control; 12, DNA size markers. Poly(A) marks the broad band indicative of polyadenylated mRNA production.
Representative mutagenesis at positions 2′, 3′, 8′, and 9′ of a 49-mer vRNA-like template.
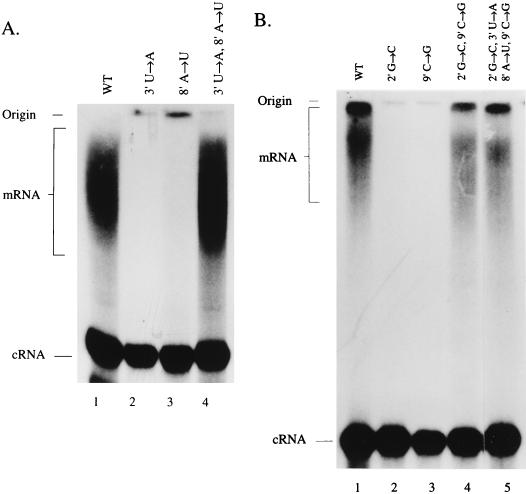
Representative mutations of the above study on the 717-nt template were also introduced into a short, 49-mer vRNA-like template (Fig. 1A) for which a convenient direct [α-32P]ATP incorporation poly(A) assay has been developed (22, 23). In this assay, polyadenylated mRNA molecules appear as a high-molecular-weight smear and are separated from nonpolyadenylated cRNA molecules by electrophoresis through polyacrylamide gels and visualized directly by autoradiography (see Materials and Methods). The mRNA is heterogeneous in size because of the variable lengths of poly(A) tail present (23). The ratio of mRNA to cRNA made in the assay appears to be independent of whether ApG or globin mRNA is used as a primer (data not shown). Figure 4A, lane 1, shows the high-molecular-weight mRNA smear and a discrete cRNA band resulting from transcription of the wild-type 49-mer vRNA-like template. Mutation of either 3′ (U→A) or 8′ (A→U) (lanes 2 and 3, respectively) resulted in loss of the mRNA signal, while cRNA was unaffected. When both positions 3′ and 8′ were mutated to form a double mutant with an A-U instead of a U-A base pair, polyadenylation was restored to wild-type levels (lane 4). Figure 4B (lanes 1 to 4) shows the effects of mutating the 2′-9′ potential base pair of the 5′ hook. Mutating either 2′ G→C or 9′ C→G alone caused the loss of the mRNA signal (lanes 2 and 3, respectively). However, swapping the base pair by introducing the double mutation 2′ G→C 9′ C→G restored polyadenylation (lane 4). Strikingly, simultaneous mutation of all four of the residues in the stem of the 5′ hook, swapping both base pairs (2′ G→C, 9′ C→G and 3′ U→A, 8′ A→U) (see Fig. 1C) also allowed synthesis of polyadenylated mRNA. (Fig. 4B, lane 5). Although the level of polyadenylated product for the base-paired mutants appeared to be reduced compared to wild type (Fig. 4, lanes 4 and 5 compared to lane 1), it should be noted that a duplicate wild-type reaction performed at the same time showed mRNA levels similar to those of the base-paired mutants (results not shown). This suggests that the somewhat reduced mRNA levels observed may not be significant.
FIG. 4.
[α-32P]ATP incorporation assay of polyadenylated mRNA and cRNA synthesized from the 49-mer vRNA-like templates. (A) Effect of mutating the 3′ to 8′ base pair of the 5′ vRNA hook. Lanes: 1, wild-type template products showing the high-molecular-weight polyadenylated mRNA and the cRNA band; 2 and 3, single mutants as shown; lane 4, double mutant as shown. (B) Effect of mutating the base pair 2′ to 9′ of the 5′ vRNA hook. Lanes: 1, wild type; 2 and 3, single mutants as shown; 4, double mutant as shown; 5, quadruple mutant as shown. Only the upper (major) band of the cRNA doublet (22, 23) is shown here.
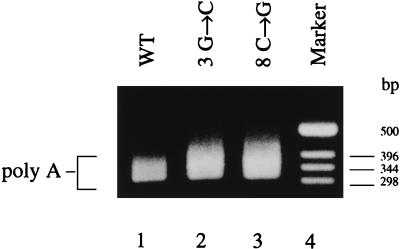
Mutagenesis of positions 3 and 8 of the 3′ arm of vRNA.
We also investigated whether a hairpin loop structure, analogous to that in the 5′ arm, was present in the 3′ arm of vRNA as proposed as part of the corkscrew model (3, 4, 18). Figure 5 shows, by RT-PCR assay (see Materials and Methods), that polyadenylated mRNA synthesis from 717-nt vRNA-like templates mutated at position 3 or 8 from the 3′ end is indistinguishable from that synthesized from the wild-type template. These results contrast clearly with the results of the 5′-arm mutagenesis, where each single point mutation at positions 2′, 3′, 8′, or 9′ (except two examples where a G-U base pair could form) resulted in the loss of polyadenylation function (see Fig. 2, 3, and 4).
FIG. 5.
Effect of mutating positions 3 and 8 in the 3′ arm of the 717-nt vRNA-like template. Lanes: 1, wild-type template; 2 and 3, point mutants as shown; 4, DNA size marker.
DISCUSSION
Previously, we demonstrated that transcription products synthesized in vitro from influenza virus vRNA-like template RNAs by virion-derived RNA polymerase preparations included polyadenylated mRNA in addition to cRNA (23). An extensive mutagenic study identified residues within the conserved 5′ arm of vRNA which were required for mRNA synthesis (22). The roles of the critical residues could be explained by two requirements for polyadenylation. First, residues at the 5′ end of the template, known to be critical for polymerase binding (5), were required for polyadenylation (22, 23). Second, residues involved in base pairing between the 5′ and the 3′ ends of vRNA (6) were required, suggesting that a complex composed of the influenza RNA polymerase and the 5′-terminal vRNA sequences acts in cis on the 3′ end of the same vRNA molecule in order to initiate mRNA synthesis (22).
Those residues essential for polyadenylation because of their involvement in polymerase binding included residues 2′, 3′, 8′, and 9′ (5, 22, 23). These residues had also been shown to be important for CAT expression from influenza-like vRNA templates in reporter assays in vivo (3, 4, 18). These studies had suggested a secondary structure model for the panhandle, known as the corkscrew model, in which two stem-loop structures were present, one in each arm of the panhandle.
Here, we present evidence for a hairpin loop structure at the 5′ end of influenza virus vRNA, which we show is required for the synthesis of polyadenylated mRNA. This vRNA 5′-hook structure has a stem of 2 bp (residues 2′ and 3′ pairing with residues 9′ and 8′, respectively) and a 4-nt loop (residues 4′ to 7′). Disruption of the stem by any single point mutation, except two cases where a G-U pair would form, prevented the synthesis of polyadenylated mRNA—a process known to be dependent on the binding of the polymerase to the 5′ end of vRNA (22, 23). In contrast, introducing alternative base pairs rescued the synthesis of polyadenylated transcripts, thus demonstrating that these templates retained polymerase binding activity. The alternative base-paired structures tested and shown to be active in polyadenylated mRNA synthesis are summarized in Fig. 1C. The results suggest that the base-paired structure, rather than the precise nucleotide sequence of the vRNA 5′ end, is the critical feature allowing the synthesis of polyadenylated mRNA. An alternative explanation for the observed activity of the single mutants at positions 8′ (A→G) and 9′ (C→U), which each allow a G-U pair to form, is that nucleotides 8′ and 9′ are crucial residues per se. However, since RNA G-U base pairs are known to be as stable as A-U base pairs (19) and since all the other data presented point to the importance of base pairing rather than to residue identity, we feel that this alternative explanation is unlikely.
We further show that there is no requirement in ApG-primed synthesis of polyadenylated mRNA in vitro for an analogous hook structure in the 3′ arm of vRNA. Thus, mutagenesis of residues which would form the stem of a 3′ vRNA hook, were it to exist, failed to interfere with the production of polyadenylated transcripts (Fig. 5). This discrepancy with data from the in vivo study (3, 4), which had suggested the presence of hairpin loop structures in both the 5′ and 3′ vRNA ends, may be due to indirect effects in that earlier assay system. Thus, increased CAT activities observed in vivo may have been due not to the presence of a 3′ vRNA hook but to the consequential effects on a 5′ cRNA hook—the sequence of which depends on the vRNA 3′-end sequence. Thus, in the in vivo study, the reference construct which was used had a mutated 3′ vRNA terminus, which would have resulted in a vRNA-like hook being present at the 5′ end of cRNA (3, 4). This might have stimulated CAT production indirectly by increasing replication.
In Thogoto virus, a related orthomyxovirus the promoters of which show striking similarities to those of influenza A virus, the possible secondary structure of the segment termini has been investigated by two different in vitro assays (12–14). Both endonuclease activation and transcription initiation were shown to require base pairing of residues 2′ and 3′ with residues 8′ and 9′ of the 5′ terminus (12, 13), suggesting that a 5′ hook was present in Thogoto virus vRNA. In contrast to vRNA, the Thogoto virus cRNA 5′-arm sequence is not compatible with the formation of a hook (14), although mutagenesis to introduce a 5′ cRNA hook stimulated endonuclease function (14). It is therefore interesting to speculate that the differential activation of endonuclease function by vRNA and cRNA panhandles of influenza A virus (1) may be due to sequence differences within the hook that may control precise interactions with the RNA polymerase complex.
In the present study, it is possible that not all base-paired mutations rescued activity to the same extent. Although the RT-PCR assay is not a quantitative assay, both the 2′ G→U 9′ C→A double mutant and the 9′ C→U (G-U paired) mutant consistently produced weaker broad bands than the wild-type template (Fig. 2, lanes 7 and 9), suggesting that the level of polyadenylated mRNA synthesis from these templates was reduced. Also, although mutation of all four residues of the stem of the 5′ hook in a way which conserved base pairing (Fig. 1C) was compatible with polyadenylated mRNA synthesis in the [α-32P]ATP incorporation assay (Fig. 4B, lane 5), the same mutations inactivated the 717-nt template (data not shown). The reason for this discrepancy between the two assays, the only evident discrepancy between equivalent mutations both here and in previous studies (22, 23), is unknown. This result suggests that features of the template other than the sequence of the ends of the RNA may be important.
It is interesting to speculate what effects swapped base pairs in the 5′ hook might have if live virus could be rescued with such mutations. In a previously identified base-paired region of the vRNA panhandle, specifically residues 11′ to 16′ paired to residues 10 to 15, the in vitro transcription findings (5, 6) were followed up by an in vivo study in which live virus was rescued with swapped base pairs. A transfectant virus with the 11-to-12′ G-C pair mutated to a U-A pair showed reduced levels of mRNA synthesis from the mutated neuraminidase-coding segment 6 (7). The role of this base-paired region and of the hook region in polyadenylation are both consistent with our previous in vitro studies (22, 23). It is not known which, if any, of the 5′ hook base-pair mutants whose activity could be rescued in vitro here (Fig. 1C) could be introduced into viable virus. It is of interest that a swapped mutation at residues 12 to 13′, which was compatible with good levels of transcription in vitro, was not rescued into live virus (4a, 5–7).
The conserved vRNA and cRNA 5′ termini of influenza A, B, and C viruses all have sequences (27) that would allow a 5′ hook to form. Although these viruses also have sequence complementarity between residues 2 and 3 with residues 9 and 8 in the 3′ terminus of each vRNA segment, the present study demonstrates that a 3′ vRNA hook, were it to exist, can be disrupted without affecting the ApG-primed synthesis of polyadenylated mRNA. This implies that the sequence restraints imposed by the 5′ arm of cRNA may be indirectly responsible for the presence of hook-like sequences in the 3′ region of vRNA, rather than there being a direct functional significance to vRNA. We cannot exclude, however, that a 3′ vRNA hook has functional relevance to replication, segment packaging, or some other aspect of the influenza virus life cycle. Neither can we exclude the possibility that the 5′ vRNA hook might also function in cRNA synthesis and replication. It might be possible to investigate these aspects of the hook by characterizing live, transfectant viruses carrying mutations designed to modify the hook.
In summary, we have identified a hairpin loop structure, the 5′ vRNA hook, at the 5′ end of influenza vRNA which is required for the transcription of polyadenylated mRNA. In contrast to a previous in vivo study (4), we find that the 3′ arm of vRNA can be mutated in a way which would disrupt formation of the analogous motif in the 3′ terminus without interfering with the transcription of polyadenylated mRNA. We conclude that a 5′ vRNA hook structure is likely to be required for interaction with the influenza virus RNA polymerase as part of a complex leading to transcription of polyadenylated mRNA. Further work is required to define the precise interactions of the 5′ vRNA hook with the individual components of the RNA polymerase complex.
ACKNOWLEDGMENTS
D.C.P., L.J.D., and M.B.L. were supported by the MRC (programme grant G9523972 to G.G.B.). L.L.M.P. was supported by the Croucher Foundation.
We thank Ervin Fodor for commenting on the manuscript.
REFERENCES
- 1.Cianci C, Tiley L, Krystal M. Differential activation of the influenza virus polymerase via template RNA binding. J Virol. 1995;69:3995–3999. doi: 10.1128/jvi.69.7.3995-3999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′ and 5′ terminal sequences of influenza A, B, and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 3.Flick R, Neumann G, Neumeier E, Hoffmann E, Hobom G. Influenza vRNA interacts with viral polymerase in a “corkscrew” conformation. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 389–409. [Google Scholar]
- 4.Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 4a.Fodor, E. Personal communication.
- 5.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor E, Pritlove D C, Brownlee G G. Characterization of the RNA-fork model of the virion RNA in the initiation of transcription in influenza A virus. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor E, Palese P, Brownlee G G, García-Sastre A. Attenuation of influenza A virus mRNA levels by promoter mutations. J Virol. 1998;72:6283–6290. doi: 10.1128/jvi.72.8.6283-6290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagen M, Chung T D Y, Butcher J A, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu M, Parvin J D, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci USA. 1987;84:8104–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura N, Nishida M, Nagata K, Ishihama A, Oda K, Nakada S. Transcription of a recombinant influenza virus RNA in cells that can express the influenza virus RNA polymerase and nucleoprotein genes. J Gen Virol. 1992;73:1321–1328. doi: 10.1099/0022-1317-73-6-1321. [DOI] [PubMed] [Google Scholar]
- 11.Lamb R F, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1353–1395. [Google Scholar]
- 12.Leahy M B, Dessens J T, Nuttall P A. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J Virol. 1997;71:8352–8356. doi: 10.1128/jvi.71.11.8352-8356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leahy M B, Dessens J T, Pritlove D C, Nuttall P A. An endonuclease switching mechanism in the virion RNA and cRNA promoters of Thogoto orthomyxovirus. J Virol. 1998;72:2305–2309. doi: 10.1128/jvi.72.3.2305-2309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leahy M B, Dessens J T, Pritlove D C, Nuttall P A. The Thogoto orthomyxovirus cRNA promoter functions as a panhandle but does not stimulate cap snatching in vitro. J Gen Virol. 1998;79:457–460. doi: 10.1099/0022-1317-79-3-457. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Palese P. Characterization of the polyadenylation signal of influenza virus RNA. J Virol. 1994;68:1245–1249. doi: 10.1128/jvi.68.2.1245-1249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Amplification, expression, and packaging of a foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 17.Martin J, Albo C, Ortin J, Melero A, Portela A. In vitro reconstitution of active influenza virus ribonucleoprotein complexes using viral proteins purified from infected cells. J Gen Virol. 1992;73:1855–1859. doi: 10.1099/0022-1317-73-7-1855. [DOI] [PubMed] [Google Scholar]
- 18.Neumann G, Hobom G. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol. 1995;76:1709–1717. doi: 10.1099/0022-1317-76-7-1709. [DOI] [PubMed] [Google Scholar]
- 19.Nowakowski S, Tinoco I. RNA structure and stability. Semin Virol. 1997;8:153–165. [Google Scholar]
- 20.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of the influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plotch S J, Krug R M. Influenza virion transcriptase: synthesis in vitro of large, polyadenylic acid-containing complementary RNA. J Virol. 1977;1:24–34. doi: 10.1128/jvi.21.1.24-34.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon L L M, Pritlove D C, Sharps J, Brownlee G G. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J Virol. 1998;72:8214–8219. doi: 10.1128/jvi.72.10.8214-8219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritlove D C, Poon L M, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seong B L, Brownlee G G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992;186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 25.Seong B L, Brownlee G G. Nucleotides 9 to 11 of the influenza A virion promoter are crucial for activity in vitro. J Gen Virol. 1992;73:3115–3124. doi: 10.1099/0022-1317-73-12-3115. [DOI] [PubMed] [Google Scholar]
- 26.Seong B L, Kobayashi M, Nagata K, Brownlee G G, Ishihama A. Comparison of two reconstitution systems for in vitro transcription and replication of influenza virus. J Biochem. 1992;111:496–499. doi: 10.1093/oxfordjournals.jbchem.a123786. [DOI] [PubMed] [Google Scholar]
- 27.Stoeckle M Y, Shaw M W, Choppin P W. Segment-specific and common nucleotides sequences in the non-coding regions of the influenza B virus genome RNAs. Proc Natl Acad Sci USA. 1987;8:2703–2707. doi: 10.1073/pnas.84.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zobel A, Neumann G, Hobom G. RNA polymerase I catalyzed transcription of insert viral cDNA. Nucleic Acids Res. 1993;21:3607–3614. doi: 10.1093/nar/21.16.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]