Abstract
Spiny lizards (genus Sceloporus) have long served as important systems for studies of behavior, thermal physiology, dietary ecology, vector biology, speciation, and biogeography. The western fence lizard, Sceloporus occidentalis, is found across most of the major biogeographical regions in the western United States and northern Baja California, Mexico, inhabiting a wide range of habitats, from grassland to chaparral to open woodlands. As small ectotherms, Sceloporus lizards are particularly vulnerable to climate change, and S. occidentalis has also become an important system for studying the impacts of land use change and urbanization on small vertebrates. Here, we report a new reference genome assembly for S. occidentalis, as part of the California Conservation Genomics Project (CCGP). Consistent with the reference genomics strategy of the CCGP, we used Pacific Biosciences HiFi long reads and Hi-C chromatin-proximity sequencing technology to produce a de novo assembled genome. The assembly comprises a total of 608 scaffolds spanning 2,856 Mb, has a contig N50 of 18.9 Mb, a scaffold N50 of 98.4 Mb, and BUSCO completeness score of 98.1% based on the tetrapod gene set. This reference genome will be valuable for understanding ecological and evolutionary dynamics in S. occidentalis, the species status of the California endemic island fence lizard (S. becki), and the spectacular radiation of Sceloporus lizards.
Keywords: California Conservation Genomics Project, CCGP, de novo genome assembly, Iguania, Phrynosomatidae, reptile, Squamata
Introduction
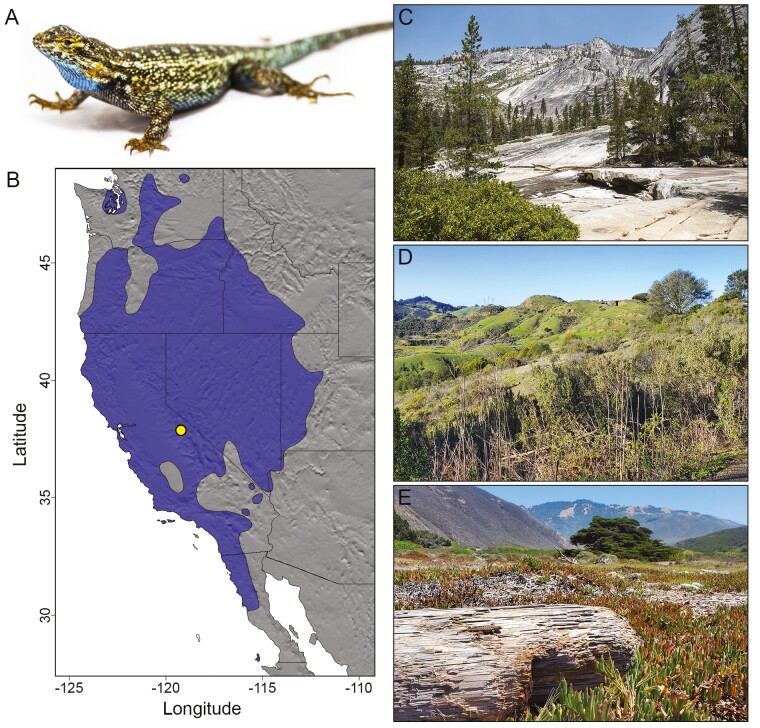
The western fence lizard, Sceloporus occidentalis, is a widespread species found across most of the major biogeographical regions of the western United States (Fig. 1), ranging from northwestern Washington (United States) to northern Baja California (Mexico) and east to central Utah (United States). Western fence lizards occupy a correspondingly broad range of habitats, including grassland, chaparral, sagebrush, woodland, open coniferous forest, urban yards and landscapes, and farmland, from sea level to 3,350 m (Stebbins and McGinnis 2012). Their widespread distribution, often high local abundance, and easily observable nature have made S. occidentalis a model organism for research in areas including behavior, thermal physiology, dietary ecology, and disease vector biology, among others, and it may be the most actively studied North American lizard (Marcellini and Mackey 1970; Goldberg 1974; Rose 1976; Adolph 1990; Sinervo and Losos 1991; Rochester et al. 2010). The species comprises 5 geographically structured subspecies, all of which occur in California, with 1 former subspecies (S. o. becki) recently elevated to species status (S. becki; Salerno et al. 2023). S. occidentalis also exhibits fine-scale genetic structure (Wishingrad and Thompson 2023) and a complex biogeographic history that includes divergence in a ring-like pattern as populations expanded northward across the Sierra Nevada and coast ranges (Bouzid et al. 2022).
Fig. 1.
Photo of the western fence lizard (Sceloporus occidentalis) used to generate the reference genome (A) and a range map (B) for S. occidentalis based on data from the Global Assessment of Reptile Distributions (Roll et al. 2017) with the location for the genome specimen indicated by the circle. Western fence lizards occupy habitat ranging from montane mixed conifer forest (C) to open woodland and grassland (D) to coastal scrub (E). Note the basking S. occidentalis on the log in the foreground of panel E.
The archetypal western fence lizard exhibits light brown to dark gray dorsal coloration with dark brown to black chevrons in two longitudinal lines down the back, yellow to orange coloration on the posterior of all limbs, and bright blue to blue-green ventral patches, particularly in adult males, that make them easily recognizable and have given them the colloquial name “blue-bellies.” However, they display phenotypic variation across their range, including distinct high elevation (Leaché et al. 2010) and urban (Putman et al. 2019) phenotypes, revealing the tight coupling between local environmental conditions and external traits. Despite their large range and ability to persist across many habitat types, as small ectotherms these lizards are sensitive to changes in their thermal environment, and spiny lizards (genus Sceloporus) are projected to experience significant declines due to climate change (Sinervo et al. 2010). Sceloporus occidentalis populations are also frequently affected by urbanization, land use conversion, and habitat fragmentation (Delaney et al. 2010, 2021), making the development of genomic resources in this system valuable for studies of organismal responses to landscape alteration and climate change, for which S. occidentalis could serve as a model system.
Here, we present the first assembled reference genome for S. occidentalis, produced as part of the California Conservation Genomics Project (CCGP; Shaffer et al. 2022; Toffelmier et al. 2022). The S. occidentalis genome assembly joins 42 other lizard reference genomes, including 4 others for Phrynosomatidae, a family with roughly 140 species. This genome assembly will contribute to our understanding of the ecology and evolution of this widespread, ecologically and scientifically important, charismatic species. Because S. occidentalis is an abundant member of most communities in California, this new resource will also contribute to several ongoing and future aspects of California habitat and biodiversity conservation (Amburgey et al. 2021; Fiedler et al. 2022).
Methods
Biological materials
We collected an adult male S. occidentalis (field number: IW3139) from Yosemite National Park, Mariposa County, California (37.72642, −119.38959), in August 2020. The specimen was collected under applicable state (SC-8436) and national park (YOSE-2020-SCI-0058) scientific research permits following a protocol approved by the UC Berkeley Animal Care and Use Committee (AUP-2016-02-8453-2). We removed liver tissue, immediately flash froze it in liquid nitrogen, and stored it at −80 °C until extraction of genomic DNA (gDNA).
DNA extraction
High molecular weight (HMW) gDNA was extracted from 30 mg of liver tissue using the Nanobind Tissue Big DNA kit following the manufacturer’s instructions (Pacific BioSciences—PacBio, Menlo Park, California). DNA purity was estimated using absorbance ratios (260/280 = 1.85 and 260/230 = 2.40) on a NanoDrop ND-1000 spectrophotometer. The final DNA yield (298 ng/µL; 37 µg) was quantified using a Quantus Fluorometer (QuantiFluor ONE dsDNA Dye assay; Promega, Madison, Wisconsin). The size distribution of the HMW DNA was estimated using the Femto Pulse system (Agilent, Santa Clara, California) which indicated that 63% of the fragments were 100 kb or longer.
HiFi library preparation and sequencing
The HiFi SMRTbell library was constructed using the SMRTbell Express Template Prep Kit v2.0 (PacBio, Cat. #100-938-900) according to the manufacturer’s instructions. HMW gDNA was sheared to a target DNA size distribution between 15 and 20 kb. The sheared gDNA was concentrated using 0.45× of AMPure PB beads (PacBio, Cat. #100-265-900) for the removal of single-strand overhangs at 37 °C for 15 min, followed by further enzymatic steps of DNA damage repair at 37 °C for 30 min, end repair and A-tailing at 20 °C for 10 min and 65 °C for 30 min, ligation of overhang adapter v3 at 20 °C for 60 min and 65 °C for 10 min to inactivate the ligase, then nuclease treated at 37 °C for 1 h. The SMRTbell library was purified and concentrated with 0.45× Ampure PB beads (PacBio, Cat. #100-265-900) for size selection using the BluePippin/PippinHT system (Sage Science, Beverly, Massachusetts; Cat. #BLF7510/HPE7510) to collect fragments greater than 7 to 9 kb. The 15 to 20 kb average HiFi SMRTbell library was sequenced at the University of California Davis DNA Technologies Core (Davis, California) using five 8M SMRT cells, Sequel II sequencing chemistry 2.0, and 30-h movies each on a PacBio Sequel II sequencer.
Omni-C library preparation and sequencing
The Omni-C library was prepared using the Dovetail Omni-C Kit (Dovetail Genomics, California) according to the manufacturer’s protocol with slight modifications. First, specimen tissue was thoroughly ground with a mortar and pestle while cooled with liquid nitrogen, after which chromatin was fixed in place in the nucleus. The suspended chromatin solution was then passed through 100 and 40 µm cell strainers to remove large debris. Fixed chromatin was digested under various conditions of DNase I until a suitable fragment length distribution of DNA molecules was obtained. Chromatin ends were repaired and ligated to a biotinylated bridge adapter followed by proximity ligation of adapter-containing ends. After proximity ligation, crosslinks were reversed, and the DNA was purified from proteins. Purified DNA was treated to remove biotin that was not internal to ligated fragments. An NGS library was generated using an NEB Ultra II DNA Library Prep kit (New England Biolabs, Ipswich, Massachusetts) with an Illumina compatible y-adaptor. Biotin-containing fragments were then captured using streptavidin beads. The post-capture product was split into 2 replicates prior to PCR enrichment to preserve library complexity, with each replicate receiving unique dual indices. The library was sequenced at Vincent J. Coates Genomics Sequencing Lab (Berkeley, California) on an Illumina NovaSeq platform (Illumina, San Diego, California) to generate approximately 100 million 2 × 150 bp read pairs per GB of genome size.
Nuclear genome assembly
We assembled the S. occidentalis genome following the CCGP assembly pipeline Version 4, which uses PacBio HiFi reads and Omni-C data to produce high-quality and highly contiguous genome assemblies while minimizing manual curation (outlined in Table 1). We removed remnant adapter sequences from the PacBio HiFi dataset using HiFiAdapterFilt (Sim et al. 2022) and obtained the initial dual or partially phased diploid assembly (http://lh3.github.io/2021/10/10/introducing-dual-assembly) using HiFiasm (Cheng et al. 2021) with the filtered PacBio HiFi reads and the Omni-C dataset. We tagged output haplotype 1 as the primary assembly and output haplotype 2 as the alternate assembly. We identified sequences corresponding to haplotypic duplications, contig overlaps, and repeats on the primary assembly with purge_dups (Guan et al. 2020) and transferred them to the alternate assembly. We scaffolded both assemblies using the Omni-C data with SALSA (Ghurye et al. 2017, 2018).
Table 1.
Assembly pipeline and software used.
| Assembly | Software and options | Version |
|---|---|---|
| Filtering PacBio HiFi adapters | HiFiAdapterFilt | Commit 64d1c7b |
| K-mer counting | Meryl (k = 21) | 1 |
| Estimation of genome size and heterozygosity | GenomeScope | 2 |
| De novo assembly (contiging) | HiFiasm (Hi-C Mode, –primary, output p_ctg.hap1, p_ctg.hap2) | 0.16.1-r375 |
| Remove low-coverage, duplicated contigs | purge_dups | 1.2.6 |
| Scaffolding | ||
| Omni-C scaffolding | SALSA (-DNASE, -i 20, -p yes) | 2 |
| Gap closing | YAGCloser (-mins 2 -f 20 -mcc 2 -prt 0.25 -eft 0.2 -pld 0.2) | Commit 0e34c3b |
| Omni-C contact map generation | ||
| Short-read alignment | BWA-MEM (-5SP) | 0.7.17-r1188 |
| SAM/BAM processing | samtools | 1.11 |
| SAM/BAM filtering | pairtools | 0.3.0 |
| Pairs indexing | pairix | 0.3.7 |
| Matrix generation | cooler | 0.8.10 |
| Matrix balancing | hicExplorer (hicCorrectmatrix correct --filterThreshold -2 4) | 3.6 |
| Contact map visualization | HiGlass | 2.1.11 |
| PretextMap | 0.1.4 | |
| PretextView | 0.1.5 | |
| PretextSnapshot | 0.0.3 | |
| Organelle assembly | ||
| Mitogenome assembly | MitoHiFi (-r, -p 50, -o 1) | 2 Commit c06ed3e |
| Genome quality assessment | ||
| Basic assembly metrics | QUAST (--est-ref-size) | 5.0.2 |
| Assembly completeness | BUSCO (-m geno, -l tetrapoda) | 5.0.0 |
| Merqury | 2020-01-29 | |
| Contamination screening | ||
| Local alignment tool | BLAST+ | 2.1 |
| General contamination screening | BlobToolKit | 2.3.3 |
Software citations are listed in the text.
We generated Omni-C contact maps by aligning the Omni-C data against both assemblies with BWA-MEM (Li 2013) and identified ligation junctions and generated Omni-C pairs using pairtools (Goloborodko et al. 2018). We generated a multiresolution Omni-C matrix with cooler (Abdennur an Mirny 2020) and balanced it with HiCExplorer (Ramírez et al. 2018). We used HiGlass [Version 2.1.11] (Kerpedjiev et al. 2018) and the PretextSuite (https://github.com/wtsi-hpag/PretextView; https://github.com/wtsi-hpag/PretextMap; https://github.com/wtsi-hpag/PretextSnapshot) to visualize the contact maps. We checked the contact maps for major misassemblies, cutting the scaffolds where misassemblies were identified. No further joins were made after this step. Using the PacBio HiFi reads and YAGCloser (https://github.com/merlyescalona/yagcloser) we closed some of the remaining gaps generated during scaffolding. We then checked for contamination using the BlobToolKit framework (Challis et al. 2020). Finally, we trimmed remnants of sequence adaptors and mitochondrial contamination identified during NCBI contamination screening.
Mitochondrial genome assembly
We assembled the mitochondrial genome of S. occidentalis from the PacBio HiFi reads using the reference-guided pipeline MitoHiFi (https://github.com/marcelauliano/MitoHiFi; Allio et al. 2020). An existing mitochondrial sequence for S. occidentalis (NCBI:AB079242.1) was used as the starting sequence. After completion of the nuclear genome, we searched for matches of the resulting mitochondrial assembly sequence in the nuclear genome assembly using BLAST+ (Camacho et al. 2009) and filtered out contigs and scaffolds from the nuclear genome with a percentage of sequence identity >99% and size smaller than the mitochondrial assembly sequence.
Genome size estimation and quality assessment
We generated k-mer counts from the PacBio HiFi reads using meryl (https://github.com/marbl/meryl). The k-mer database was then used in GenomeScope2.0 (Ranallo-Benavidez et al. 2020) to estimate genome features including genome size, heterozygosity, and repeat content. To obtain general contiguity metrics, we ran QUAST (Gurevich et al. 2013). To evaluate genome quality and completeness we used BUSCO (Manni et al. 2021) with the Tetrapoda ortholog database (tetrapoda_odb10), which contains 5,310 genes. Assessments of base-level accuracy (QV) and k-mer completeness were performed using the previously generated meryl database and merqury (Rhie et al. 2020). We further estimated genome assembly accuracy via BUSCO gene set frameshift analysis using the pipeline described in Korlach et al. (2017).
Measurements of the size of the phased blocks are based on the size of the contigs generated by HiFiasm in Hi-C mode. We followed the quality metric nomenclature established by Rhie et al. (2021), with the genome quality code x·y·P·Q·C, where x = log10[contig NG50]; y = log10[scaffold NG50]; P = log10[phased block NG50]; Q = Phred base accuracy QV (quality value); and C = % genome represented by the first “n” scaffolds, following a known karyotype of 2n =2 2 for S. occidentalis (Jackson and Hunsacker 1970). Quality metrics for the notation were calculated on the primary assembly.
Results
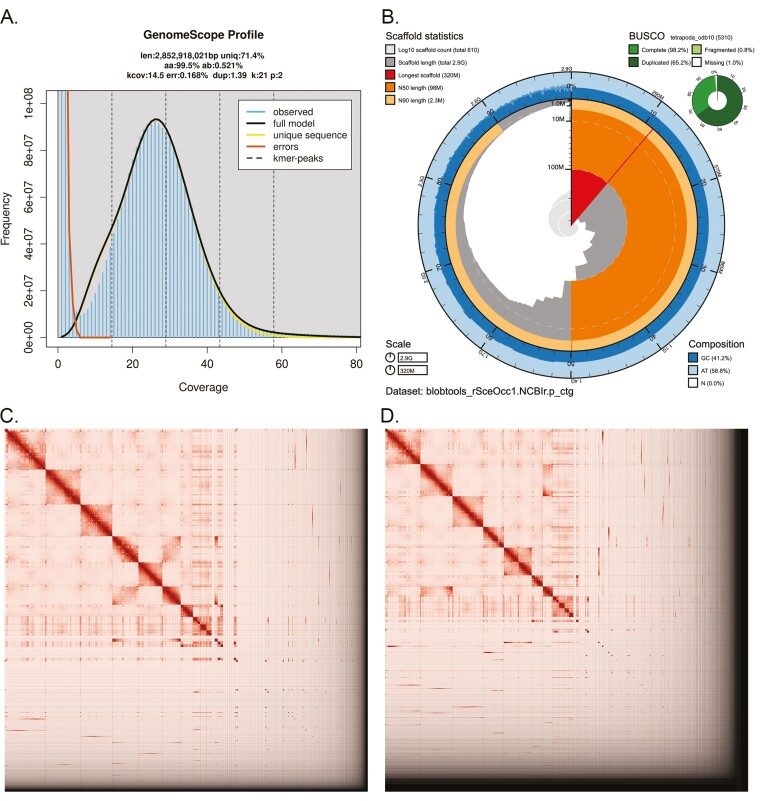
The Omni-C and PacBio HiFi sequencing libraries generated 290 million read pairs and 5.3 million reads, respectively. The latter yielded 29.94-fold coverage (N50 read length 16,535 bp; minimum read length 45 bp; mean read length 16,088 bp; maximum read length of 63,768 bp) based on the GenomeScope2.0 genome size estimation of 2.8 Gb. Based on PacBio HiFi reads, we estimated 0.168% sequencing error rate and 0.521% nucleotide heterozygosity rate. The k-mer spectrum based on PacBio HiFi reads show a bimodal distribution with a major peak at ~29-fold coverage (Fig. 2A), which supports that of a low heterozygosity profile.
Fig. 2.
Visual overview of genome assembly metrics. A) K-mer spectrum output generated from PacBio HiFi data without adapters using GenomeScope2.0. B) BlobToolKit Snail plot showing a graphical representation of the quality metrics presented in Table 2 for the S. occidentalis primary assembly (rSceOcc1). The plot circle represents the full size of the assembly. From the inside out, the central plot covers length-related metrics. The red line represents the size of the longest scaffold; all other scaffolds are arranged in size-order moving clockwise around the plot and drawn in gray, starting from the outside of the central plot. Dark and light orange arcs show the scaffold N50 and scaffold N90 values. The central light gray spiral shows the cumulative scaffold count with a white line at each order of magnitude. White regions in this area reflect the proportion of Ns in the assembly; the dark vs. light blue area around it shows mean, maximum, and minimum GC vs. AT content at 0.1% intervals (Challis et al. 2020). Hi-C contact maps for the primary (C) and alternate (D) genome assembly were generated with PretextSnapshot. Hi-C contact maps translate proximity of genomic regions in 3D space to contiguous linear organization. Each cell in the contact map corresponds to sequencing data supporting the linkage (or join) between 2 such regions. Scaffolds are separated by black lines, and higher density corresponds to higher levels of fragmentation.
The final assembly (rSceOcc1) consists of two pseudo haplotypes, primary and alternate. Genome size for the primary assembly is similar to the estimated value from GenomeScope2.0 (Fig. 2A), but the alternate is ~400 Mb larger. The primary assembly consists of 608 scaffolds spanning 2.8 Gb with contig N50 of 18.9 Mb, scaffold N50 of 98.4 Mb, longest contig of 124.1 Mb and largest scaffold of 320.8 Mb. The alternate assembly consists of 1,822 scaffolds, spanning 3.2 Gb with contig N50 of 17.6 Mb, scaffold N50 of 38.7 Mb, largest contig 115.5 Mb, and largest scaffold of 309.6 Mb. Detailed assembly statistics are reported in Table 2 and graphical representation for the primary assembly in Fig. 2B (see Supplementary Fig. 1 for the alternate assembly). The primary assembly has a BUSCO completeness score of 98.1% using the Tetropoda gene set, a per-base quality (QV) of 61.68, a k-mer completeness of 92.89 and a frameshift indel QV of 48.71, while the alternate assembly has a BUSCO completeness score of 98.3% using the same gene set, a per-base quality (QV) of 61.39, a k-mer completeness of 93.62 and a frameshift indel QV of 49.12.
Table 2.
Sequencing and assembly statistics and accession numbers.
| BioProjects and Vouchers | CCGP NCBI BioProject | PRJNA720569 |
| Genus NCBI BioProject | PRJNA765857 | |
| Species NCBI BioProject | PRJNA777217 | |
| NCBI Bio-sample | SAMN27480378 | |
| Specimen identification number | IW3139 | |
| Genome Sequence | PacBio HiFi long read runs | 1 PacBio SMRT Sequel II run: 5.7 M spots, 85.9 Gbp, 57.2 Gb |
| OmniC Illumina sequencing | 1 Illumina NovaSeq 6000 run: 290 M spots, 87.5 Gbp, 28 Gb | |
| PacBio HiFi NCBI SRA Accession | SRX15651255 | |
| OmniC Illumina NCBI SRA Accession | SRX15651256, SRX15651257 | |
| Genome Assembly Primary (Alternate) |
Assembly identifier | rSceOcc1 |
| HiFi read coverage | 37.14× | |
| Number of contigs (primary/alternate) | 659/1,822 | |
| Contig N50 (bp) | 18,989,278/17,628,953 | |
| Contig NG50b (bp) | 18,989,278/20,278,101 | |
| Longest contigs (primary/alternate) | 124,125,603/115,579,027 | |
| Number of scaffolds (primary/alternate) | 608/1,771 | |
| Scaffold N50 (bp) | 98,418,489/38,771,511 | |
| Scaffold NG50b | 98,418,489/88,220,525 | |
| Size of final assembly (bp) | 2,856,356,971/3,186,658,811 | |
| Gaps per Gbp (# gaps) | 18 (51)/16 (51) | |
| NCBI Genome Assembly Accession | GCA_000XXXXXX.1 | |
| Assembly quality identifiera | 7.7.P7.Q61.C57 | |
| Base pair QV (merqury) | P: Q 61.6835, A: Q 61.4466 | |
| Assembly Qualityc | Indel QV (frameshift analysis) | P: Q 48.7129, A: Q 49.1278 |
| K-mer completeness | P: 92.8931%, A: 93.6217% | |
| BUSCO completeness Primary (C:S:D:F:M) Alternate (C:S:D:F:M) |
98.10%:32.90%:65.20%:0.80%:1.10% 98.30%:31.90%:66.40%:0.70%:1.00% |
|
| Organelles | 1 complete mitochondrial sequence CM041364.1 |
aAssembly quality code x·y·P·Q·C derived notation, from Rhie et al. (2021). x = log10[contig NG50]; y = log10[scaffold NG50]; P = log10[phased block NG50]; Q = Phred base accuracy QV (quality value); C = % genome represented by the first “n” scaffolds, following a known karyotype for SPECIES of 2n = 22. Quality code for all the assembly denoted by primary assembly (rSceOcc1.0.p).
bRead coverage and NGx statistics have been calculated based on the estimated genome size of 2.85 Gb.
c(P)rimary and (A)lternate assembly values.
We identified 2 misassemblies on the alternate assembly and broke the corresponding joins made by SALSA. We were able to close a total of 9 gaps, 7 on the primary assembly and 2 on the alternate. Finally, we filtered out 34 contigs, 2 from the primary and 32 from the alternate assembly, corresponding to mitochondrial contamination. No further contigs were removed. We have deposited both assemblies on NCBI (see Table 2 and Data Availability for details).
We assembled a mitochondrial genome using MitoHiFi. The final mitochondrial genome assembly size is 14,426 bp. The final assembly version has a base composition of A = 39.95%, C = 17.11%, G = 10.38%, T = 32.56% and consists of 20 unique transfer RNAs and 12 protein-coding genes.
Discussion
There are currently 89 other squamate genomes, including 42 lizards and 47 snakes, available on NCBI, ranging in size from 1.13 to 2.74 Gb (mean = 1.64 Gb), making the S. occidentalis genome the largest squamate reference genome assembled so far. At 2.86 Gb, the S. occidentalis genome is 4.2% larger than the next largest, the Japanese gecko, Gekko gecko (Liu et al. 2015). The 89 other squamate reference genomes have an average contig N50 of 14.5 Mb (ranging from 1.1 kb to 127.8 Mb), placing the contig N50 of 19.0 Mb for our S. occidentalis assembly in the upper end of this range, below only 17 other reference genomes.
Three of these existing 89 reference genomes are also phrynosomatid lizards, including two other Sceloporus species (S. tristichus and S. undulatus). The genome assembly sizes of these other species range from 1.82 Gb to 1.91 Gb (mean = 1.87 Gb), so the S. occidentalis genome is considerably larger than those of its congeners. Both of these species and S. occidentalis are members of the same phylogenetic clade within Sceloporus, the undulatus group, which has a crown age of 15 to 20 million years (Leaché et al. 2016). Until recently S. tristichus and S. undulatus were considered conspecific lineages, and so their very similar genome assembly sizes are not surprising. However, the differences in putative genome sizes between these two close relatives and S. occidentalis suggest either substantial genome size evolution within the genus or the under- or overestimation of genome size for one or more of these species. The C value for S. occidentalis is 2.36 Gb, based on Feulgen densitometry (Olmo 1981), suggesting a genome size larger than the 1.82 Gb S. tristichus (Bedoya and Leaché 2021) and 1.91 Gb S. undulatus (Westfall et al. 2021) genome assemblies. The description of the S. undulatus genome indicates that the assembly may be missing some data or has repeat regions condensed (Westfall et al. 2021). It is also possible that our current assembly overestimates the size of the S. occidentalis genome, although this seems unlikely given the high BUSCO completeness (98.4%, see Table 2) and the relatively large estimate based on Feulgen densitometry (Olmo 1981).
The S. occidentalis reference genome provides a valuable resource for studying the conservation, ecology, and evolutionary dynamics of this species and other spiny lizards. The genus Sceloporus constitutes a rapid radiation containing more than 100 species, and the Sceloporus phylogeny as currently understood is characterized by high levels of gene tree discordance (Leaché et al. 2016). This reference genome can help to further resolve the species relationships in this speciose clade of lizards. It will also contribute to understanding patterns of speciation in this group, identifying regions of the genome involved in adaptation to environmental variation and rapid speciation, and examining the evolutionary dynamics in the complex biogeographic history of S. occidentalis.
Supplementary Material
Acknowledgments
PacBio Sequel II library prep and sequencing were carried out at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. Deep sequencing of Omni-C libraries used the Novaseq S4 sequencing platforms at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. We thank the staff at the UC Davis DNA Technologies and Expression Analysis Cores and the UC Santa Cruz Paleogenomics Laboratory for their diligence and dedication to generating high-quality sequence data, and we thank Andrew Crawford for providing helpful comments on the manuscript. We also thank the California Department of Fish and Wildlife (SC-8436) and Yosemite National Park (YOSE-2020-SCI-0058) for granting scientific research permits. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Contributor Information
Anusha P Bishop, Department of Environmental Science, Policy, and Management, University of California, Berkeley, Berkeley, CA, United States; Museum of Vertebrate Zoology, University of California, Berkeley, Berkeley, CA, United States.
Erin P Westeen, Department of Environmental Science, Policy, and Management, University of California, Berkeley, Berkeley, CA, United States; Museum of Vertebrate Zoology, University of California, Berkeley, Berkeley, CA, United States.
Michael L Yuan, Center for Population Biology, University of California, Davis, Davis, CA, United States.
Merly Escalona, Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, CA, United States.
Eric Beraut, Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, CA, United States.
Colin Fairbairn, Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, CA, United States.
Mohan P A Marimuthu, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, Davis, CA, United States.
Oanh Nguyen, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, Davis, CA, United States.
Noravit Chumchim, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, Davis, CA, United States.
Erin Toffelmier, Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States; La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States.
Robert N Fisher, U.S. Geological Survey Western Ecological Research Center, San Diego, CA, United States.
H. Bradley Shaffer, Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States; La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States.
Ian J Wang, Department of Environmental Science, Policy, and Management, University of California, Berkeley, Berkeley, CA, United States; Museum of Vertebrate Zoology, University of California, Berkeley, Berkeley, CA, United States.
Funding
This work was supported by the California Conservation Genomics Project, with funding provided to the University of California by the State of California, State Budget Act of 2019 [UC Award ID RSI-19-690224], with additional support from a National Science Foundation CAREER grant [DEB-1845682] to IJW.
Data availability
Data generated for this study are available under NCBI BioProject PRJNA720569. Raw sequencing data for sample IW3139 (NCBI BioSample SAMN27480378) are deposited in the NCBI Short Read Archive (SRA) under SRX15651255, SRX15651256, and SRX15651257. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: www.github.com/ccgproject/ccgp_assembly.
References
- Abdennur N, Mirny LA.. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36(1):311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph SC. Influence of behavioral thermoregulation on microhabitat use by two Sceloporus lizards. Ecology. 1990;71:315–327. [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F.. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 2020;20:892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amburgey SM, Miller DAW, Rochester CJ, Delaney KS, Riley SPD, Brehme CS, Hathaway SA, Fisher RN.. The influence of species life history and distribution characteristics on species responses to habitat fragmentation in an urban landscape. J Anim Ecol. 2021;90:685–697. [DOI] [PubMed] [Google Scholar]
- Bedoya AM, Leaché AD.. Characterization of a pericentric inversion in plateau fence lizards (Sceloporus tristichus): evidence from chromosome-scale genomes. G3. 2021;11:jkab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid NM, Archie JW, Anderson RA, Grummer JA, Leaché AD.. Evidence for ephemeral ring species formation during the diversification history of western fence lizards (Sceloporus occidentalis). Mol Ecol. 2022;31:620–631. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M.. BlobToolKit—interactive quality assessment of genome assemblies. G3. 2020;10:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Jarvis ED, Fedrigo O, Koepfli K-P, Urban L, Gemmell NJ, Li H.. Robust haplotype-resolved assembly of diploid individuals without parental data, arXiv [q-bio.GN], arXiv:2109.04785, 2021, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Delaney KS, Busteed G, Fisher RN, Riley SPD.. Reptile and amphibian diversity and abundance in an urban landscape: impacts of fragmentation and the conservation value of small patches. Ichthyol Herpetol. 2021;109:424–435. [Google Scholar]
- Delaney KS, Riley SPD, Fisher RN.. A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS One. 2010;5:e12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler PL, Erickson B, Esgro M, Gold M, Hull JM, Norris J, Shapiro B, Westphal M, Toffelmier E, Shaffer HB.. Seizing the moment: the opportunity and relevance of the California Conservation Genomics Project to state and federal conservation policy. J Hered. 2022;113:589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Pop M, Koren S, Bickhart D, Chin C-S.. Scaffolding of long read assemblies using long range contact information. BMC Genomics. 2017;18:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Koren S.. Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Computational Biology. 2018;15(8):e1007273. doi: 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SR. Reproduction in mountain and lowland populations of the lizard Sceloporus occidentalis. Copeia. 1974;1974:176–182. [Google Scholar]
- Goloborodko A, Abdennur N, Venev S, Brandao HB, Fudenberg G. mirnylab/pairtools: v0.2.0. (v0.2.0) Zenodo.2018. doi: 10.5281/zenodo.1490831 [DOI]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R.. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Hunsaker D.. Chromosome morphology of sceloporine lizards (Sceloporus occidentalis and S. graciosus). Experientia. 1970;26:198–199. [DOI] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Gehlenborg N.. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin C-S, Howard JT, Audet J-N, Cantin L, Jarvis ED, Jarvis ED.. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience. 2017;6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Banbury BL, Linkem CW, de Oca AN-M.. Phylogenomics of a rapid radiation: is chromosomal evolution linked to increased diversification in North American spiny lizards (genus Sceloporus)? BMC Evol Biol. 2016;16:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Helmer DS, Moritz C. Phenotypic evolution in high-elevation populations of western fence lizards (Sceloporus occidentalis) in the Sierra Nevada Mountains. Biological Journal of the Linnean Society. 2010;100(3):630–641. [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, arXiv, doi: 10.3389/fnhum.2015.00181, March 16,2013, preprint: not peer reviewed. [DOI] [Google Scholar]
- Liu Y, Zhou Q, Wang Y, Luo L, Yang J, Yang L, Liu M, Li Y, Qian T, Zheng Y, et al. Gekko japonicus genome reveals evolution of adhesive toe pads and tail regeneration. Nat Commun. 2015;6:10033–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM.. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes, Mol Biol Evol. 2021;38(10):4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellini D, Mackey JP.. Habitat preferences of the lizards, Sceloporus occidentalis and S. graciosus (Lacertilia, Iguanidae). Herpetologica. 1970;26:51–56. [Google Scholar]
- Olmo E. Evolution of genome size and DNA base composition in reptiles. Genetica. 1981;57:39–50. [Google Scholar]
- Putman BJ, Gasca M, Blumstein DT, Pauly GB.. Downsizing for downtown: limb lengths, toe lengths, and scale counts decrease with urbanization in western fence lizards (Sceloporus occidentalis). Urban Ecosyst. 2019;22:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Manke T.. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9. doi: 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM.. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester CJ, Brehme CS, Clark DR, Stokes D, Hathaway SA, Fisher RN.. Reptile and amphibian responses to large-scale wildfires in southern California. J Herpetol. 2010;44:333–351. [Google Scholar]
- Roll U, Feldman A, Novosolov M, Allison A, Bauer AM, Bernard R, Böhm M, Castro-Herrera F, Chirio L, Collen B, et al. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat Ecol Evol. 2017;1:1677–1682. [DOI] [PubMed] [Google Scholar]
- Rose BR. Habitat and prey selection of Sceloporus occidentalis and Sceloporus graciosus. Ecology. 1976;57:531–541. [Google Scholar]
- Salerno PE, Chan LM, Pauly GB, Funk WC, Robertson JM.. Near-shore island lizard fauna shaped by a combination of human-mediated and natural dispersal. J Biogeogr. 2023;50:116–129. [Google Scholar]
- Shaffer HB, Toffelmier E, Corbett-Detig RB, Escalona M, Erickson B, Fiedler P, Gold M, Harrigan RJ, Hodges S, Luckau TK, et al. Landscape genomics to enable conservation actions: the California Conservation Genomics Project. J Hered. 2022;113:577–588. [DOI] [PubMed] [Google Scholar]
- Sim SB, Corpuz RL, Simmonds TJ, Geib SM. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genomic. 2022;23(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinervo B, Losos JB.. Walking the tight rope: arboreal sprint performance among Sceloporus occidentalis lizard populations. Ecology. 1991;72:1225–1233. [Google Scholar]
- Sinervo B, Méndez-de-la-Cruz F, Miles DB, Heulin B, Bastiaans E, Cruz MVS, Lara-Resendiz R, Martínez-Méndez N, Calderón-Espinosa ML, Meza-Lázaro RN, et al. Erosion of lizard diversity by climate change and altered thermal niches. Science. 2010;328(5980):894–899. [DOI] [PubMed] [Google Scholar]
- Stebbins RC, McGinnis SM.. Field guide to amphibians and reptiles of California. Berkeley (CA): University of California Press; 2012. [Google Scholar]
- Toffelmier E, Beninde J, Shaffer HB.. The phylogeny of California, and how it informs setting multi-species conservation priorities. J Hered. 2022;113:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall AK, Telemeco RS, Grizante MB, Waits DS, Clark AD, Simpson DY, Klabacka RL, Sullivan AP, Perry GH, Sears MW, et al. A chromosome-level genome assembly for the Eastern Fence Lizard (Sceloporus undulatus), a reptile model for physiological and evolutionary ecology. GigaScience. 2021;10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishingrad V, Thomson RC.. Biogeographic inferences across spatial and evolutionary scales. Mol Ecol. 2023;32:2055–2070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are available under NCBI BioProject PRJNA720569. Raw sequencing data for sample IW3139 (NCBI BioSample SAMN27480378) are deposited in the NCBI Short Read Archive (SRA) under SRX15651255, SRX15651256, and SRX15651257. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: www.github.com/ccgproject/ccgp_assembly.