Abstract
Dittrichia graveolens (L.) Greuter, or stinkwort, is a weedy annual plant within the family Asteraceae. The species is recognized for the rapid expansion of both its native and introduced ranges: in Europe, it has expanded its native distribution northward from the Mediterranean basin by nearly 7 °C latitude since the mid-20th century, while in California and Australia the plant is an invasive weed of concern. Here, we present the first de novo D. graveolens genome assembly (1N = 9 chromosomes), including complete chloroplast (151,013 bp) and partial mitochondrial genomes (22,084 bp), created using Pacific Biosciences HiFi reads and Dovetail Omni-C data. The final primary assembly is 835 Mbp in length, of which 98.1% are represented by 9 scaffolds ranging from 66 to 119 Mbp. The contig N50 is 74.9 Mbp and the scaffold N50 is 96.9 Mbp, which, together with a 98.8% completeness based on the BUSCO embryophyta10 database containing 1,614 orthologs, underscores the high quality of this assembly. This pseudo-molecule-scale genome assembly is a valuable resource for our fundamental understanding of the genomic consequences of range expansion under global change, as well as comparative genomic studies in the Asteraceae.
Keywords: Asteraceae, California, Dovetail Omni-C, organelle genomes, Pacific Biosciences HiFi, range shift
Introduction
Dittrichia graveolens (L.) Greuter (stinkwort) is an annual plant in the Inulae tribe within the Asteraceae family (Fig. 1A–C), growing in open, disturbed habitat including roadsides, agricultural land, industrial lots, and hillsides (Brullo and de Marco 2000). The species’ native distribution lies around the Mediterranean Basin, extending to the Middle East and India and northward to central France (Lustenhouwer and Parker 2022). Germinating in response to heavy rains, D. graveolens grows vegetatively as a rosette until bolting in late spring and summer and flowering in late summer and fall, producing up to several thousand seed heads per plant at the end of the growing season (USDA 2013; Lustenhouwer et al. 2018). Plants have yellow radiate flowers and wind-dispersed fruits (Brownsey et al. 2013b).
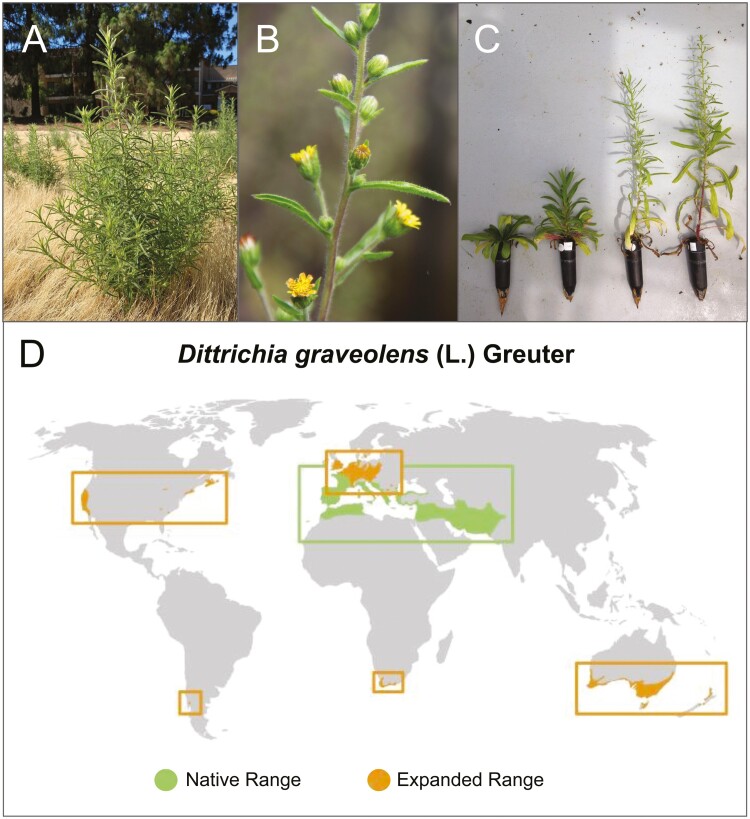
Fig. 1.
Photos of D. graveolens and its global range: A) growing along a footpath in Santa Clara County, California, USA; B) closeup of leaves, buds, and flowers; C) life stages: juvenile rosette through reproductive adult; D) map of global distribution of D. graveolens with both native and expanded ranges. Data sources detailed in Lustenhouwer and Parker (2022), GBIF (2020), and Santilli et al. (2021). Photo sources: A) Andrew Lopez, B) Nicky Lustenhouwer, and C) Miranda K. Melen. Map projection: Robinson.
D. graveolens has expanded its native range northward by nearly 7 °C latitude since the mid-20th century, now occurring in the Netherlands, southern Sweden, and Poland (Kocián 2015; Lustenhouwer et al. 2018; Lustenhouwer and Parker 2022). Although the onset of this native range expansion coincided with climate change in central Europe (Lenoir et al. 2008), population spread extended far beyond expectations based on climate tracking alone, resulting in a niche expansion to more temperate climates (Lustenhouwer and Parker 2022). A common garden study showed that plants at the range edge rapidly evolved earlier flowering time than core populations in response to the shorter growing season in the north, increasing plant fitness and promoting further range expansion (Lustenhouwer et al. 2018). Existing genomic resources for D. graveolens consist of ddRAD sequencing data for 190 individuals originating along a latitudinal gradient from southern France to the Netherlands (European Nucleotide Archive accession PRJEB22978), used in reference-free analyses to characterize population structure in this common garden study. The results showed a decline in genetic diversity toward the northern range limit (Lustenhouwer et al. 2018).
Outside its native range, D. graveolens has been introduced and successfully established elsewhere around the world, including Australia and New Zealand, South Africa, and North and South America (Parsons and Cuthbertson 2001; Brownsey 2012; DiTomaso and Brownsey 2013; Santilli et al. 2021; Fig. 1D). In California and Australia, the species is considered a noxious weed (Parsons and Cuthbertson 2001; California Department of Food and Agriculture 2021); it contains a sesquiterpene lactone that causes contact dermatitis in humans (Thong et al. 2008; Ponticelli et al. 2022), and its seeds have a barbed pappus that can damage the intestines of livestock (Philbey and Morton 2000). The combination of high spread capacity (short generation time, high fecundity, and dispersal ability) and high potential impact on agricultural and restoration land makes D. graveolens an invasive species of particular concern (USDA 2013). No niche shifts beyond the Mediterranean climate of the historic native range have yet been identified in California or Australia (Lustenhouwer and Parker 2022). Nonetheless, given the high evolutionary potential of the species in its native range, it is possible that genomic changes driving other traits play a role in the ongoing invasions.
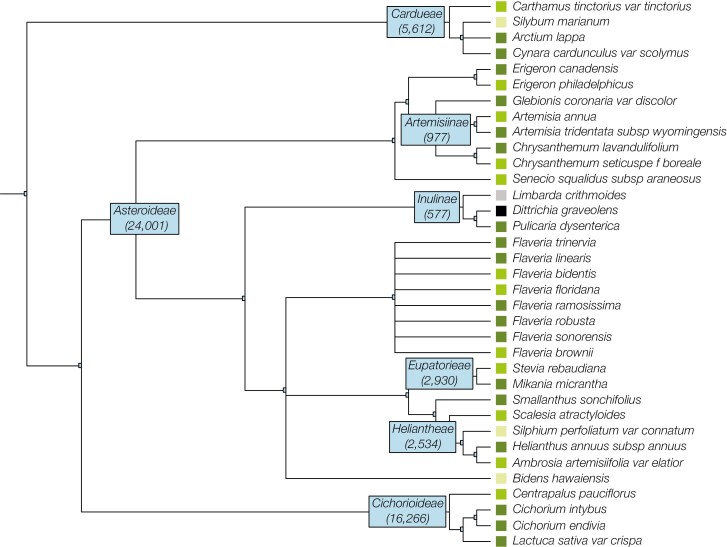
Asteraceae is a superfamily comprising 20,000 to 30,000 species (~5% to 10% of angiosperm species), many of which are economically useful and/or ecologically important. Reference genomes are currently available for 33 taxa in Asteraceae, representing 1% of genera in the family (22/2,128 genera per Open Tree of Life phylogeny ott3.3). Assembly level varies between species, with 18 chromosome-level, 12 scaffold-level, and 3 contig-level genomes (Fig. 2). The available genomes are primarily species of great agricultural importance such as Helianthus annuus (sunflower) and Lactuca sativa (lettuce), several medicinal aromatic species (Artemisia annua, Erigeron breviscapus, Centaurea cyanus, Glebionis coronaria), as well as fellow invasive annuals Ambrosia artemisiifolia and Erigeron canadensis. Within the Inuleae tribe, the only species with a reference genome is Pulicaria dysenterica (Genbank accession GCA_947179395.2). The closest relative to D. graveolens with a high-quality chloroplast assembly is Limbarda crithmoides (Genbank accessionNC_066031.1; Fig. 2).
Fig. 2.
Taxa in the Asteraceae family for which reference genomes are available. Colors indicate assembly level (light green: contig, medium green: scaffold, dark green: chromosome). D. graveolens is labeled in black and the closest relative with a chloroplast assembly (L. crithmoides) is in gray. Numbers at each node (blue boxes) indicate the total number of taxa descending from that subfamily/tribe/subtribe. Taxonomy from Open Tree of Life ott3.3 (OpenTreeofLife et al. 2019; in agreement with Mandel et al. 2019).
Asteraceae species have high sequence diversity, low synteny, and evidence of ancient whole-genome duplication or triplication at different points in time (Fan et al. 2022). This sequence divergence impedes using confamilial species as reference genomes for mapping. Greater species-level resolution in the Asteraceae is necessary to support quality conservation and population genomics and the discovery of genetic mechanisms of secondary metabolite diversity. Asteraceae are well known for including many of the world’s worst invasive species (Baker 1974; Hodgins et al. 2016). The reasons for D. graveolens’ rapid range expansion and invasion success are as yet undetermined, and the genetic consequences of that range expansion remain to be studied. Here, we present the first high-quality de novo genome assembly of D. graveolens based on Pacific Biosciences (PacBio) HiFi long reads and scaffolded with Dovetail Omni-C data. This genome will help fill gaps in comparative genomic resources for the Inulae tribe and the Asteraceae, and it will be used in planned molecular ecological and functional analysis along range expansion gradients in D. graveolens’ Mediterranean home range and along its invasion path in California.
Methods
Biological materials
The plant used for this reference genome originates from a population located in the native range of D. graveolens in southern France. Seeds were sampled from a highway service station, Aire du Merle Sud, along the A54 near Salon-de-Provence (43.63790 N, 4.98474 E) on 1 November 2018. The Nagoya Protocol was followed to collect genetic resources in France (certificate of compliance TREL2302365S/653). The collected seeds were used in a greenhouse common garden experiment at the University of California, Santa Cruz (UCSC), in February to December 2019. Plants were bagged with a light-permeable mesh prior to budding to ensure self-fertilization. Seeds produced by these plants were then used to grow a second generation of this experiment in March to December 2020, which provided the biological material for this study (plant identifier in experiment: id 3264, maternal family 2 from population FS-MER). The reference genome specimen is thus a direct descendant of field-collected seeds after 1 generation of selfing. A tissue voucher specimen of this plant is stored in a frozen collection at the UCSC Department of Ecology and Evolutionary Biology, and is cross-listed at the Norris Center (UCSC_NL_FS-MER_Block4_2020_SAMPLE3264).
Additional short-read sequencing from a different sample was used for the mitochondrial genome assembly (sample reference CEJES6B739). This plant was grown from seed collected at a roadside population along California State Route 26 near the community of Mokelumne Hill (38.3176 N, 120.6525 W) at the invasion front in California on 2 December 2018. D. graveolens is classed as a noxious weed by the California Department of Food and Agriculture (California Code of Regulations 4500) and such plant material may be collected from state highways for research without additional permissions.
Short-read mitogenome sequencing
To prepare Illumina short-read libraries for sample CEJES6B739, DNA was extracted from fresh leaf tissue using the Qiagen DNeasy Plant DNA Kit (Qiagen Inc, Valencia, CA), mixing the leaf in the lysis buffer in a microcentrifuge tube using a scored micropestle. A Tn5-based tagmentation and dual-indexed DNA library were made following the method described in Russell et al. (2020), and this was sequenced with 2 × 150 bp reads on an Illumina NextSeq 550 (Illumina, San Diego, CA) at the UCSC Paleogenomics Laboratory.
Omni-C library preparation and sequencing
We used the Dovetail Omni-C Kit (Dovetail Genomics, Scotts Valley, CA) following the manufacturer’s protocol with slight modifications to prepare the Omni-C library. Young leaf tissue was ground with a mortar and pestle in liquid nitrogen. Nuclear chromatin was fixed in place and then digested under different conditions of DNAse and a suitable fragment length distribution of DNA molecules was chosen. Chromatin end-repair was followed by biotinylated bridge adapter ligation and then proximity ligation of adapter-containing ends. Crosslinks were reversed, DNA purified from proteins, and biotin that was not internal to the ligated fragments was removed. The library was enriched using the NEB Ultra II DNA Library Prep Kit (New England Biolabs, Ipswich, MA) with Illumina-compatible y-adaptor, then split and separately indexed to increase complexity. Biotin-containing fragments were captured using streptavidin beads and size selection was performed to remove fragments >1,000 bp. Both size-selected libraries were sequenced at the UCLA Technology Center for Genomics and Bioinformatics (Los Angeles, CA) on an Illumina NovaSeq S4 aiming to generate 100 million 2 × 150 bp reads.
HMW DNA extraction for PacBio
High molecular weight (HMW) genomic DNA (gDNA) was extracted from a single individual (190 mg; sample 3,264) using the method described in Inglis et al. (2018), with the following modifications. Sodium metabisulfite (1%, w/v) was used instead of 2-mercaptoethanol (1%, v/v) in the sorbitol wash buffer and the lysis buffer. Using a mortar and pestle, frozen tissue was pulverized in liquid nitrogen for 15 min and gently resuspended in 10 mL of sorbitol wash buffer. The suspension was centrifuged at 3,500 × g for 5 min at room temperature, and the supernatant was discarded. The ground tissue pellet was gently resuspended using a paintbrush in 10 mL of sorbitol wash buffer. The wash step was repeated 5 times to remove potential contaminants that may coprecipitate with DNA. Instead of the standard 65 °C, the lysis step was performed at 45 °C for 1 h with gentle inversion every 15 min. The DNA purity was estimated using absorbance ratios (260/280 = 1.80 and 260/230 = 2.45) on a NanoDrop ND-1000 spectrophotometer. The final DNA yield (26 µg) was quantified using a Quantus Fluorometer (QuantiFluor ONE dsDNA Dye assay; Promega, Madison, WI). The size distribution of the HMW DNA was estimated using the Femto Pulse system (Agilent, Santa Clara, CA), where 38% of the DNA fragments were found to be 50 kb or longer.
The PacBio HiFi data generation was performed at UC Davis DNA Technologies Core (Davis, CA). The HiFi SMRTbell library was constructed using the SMRTbell Express Template Prep Kit v2.0 (Pacific Biosciences, Menlo Park, CA; Cat. #100-938-900) according to the manufacturer’s instructions (PN 101-853-100 Version 05, August 2021). HMW gDNA was sheared using Diagenode’s Megaruptor 3 system (Diagenode, Belgium; Cat. B06010003) to the size distribution mode between 15 and 20 kb. The sheared gDNA was then put through the following enzymatic steps: single-strand overhangs removal, DNA damage repair, end-repair and A-tailing, overhang adapter v3 ligation, and nuclease treatment. AMPure PB Beads (Pacific Biosciences, Menlo Park, CA; Cat. #100-265-900) were used for all purification steps for sheared gDNA and SMRTbell library. Size selection for the SMRTbell library was done using the PippinHT system (Sage Science, Beverly, MA; Cat. #HPE7510) to collect fragments greater than 7 to 9 kb. Finally, the HiFi SMRTbell library was sequenced using one 8M SMRT cell, Sequel II sequencing chemistry 2.0, and 30-h movies on a PacBio Sequel II sequencer. Quality control of reads was conducted with FastQC (Andrews 2010) and LongQC (Fukasawa et al. 2020).
Genome size and ploidy estimation
Raw PacBio HiFi reads were provided as input to KMC to count k-mers (k = 21) and create a histogram (Kokot et al. 2017). The histogram was used for estimation of genome size, heterozygosity, and repeat content in GenomeScope 2.0, and Smudgeplot v0.2.2 was used for ploidy estimation with L = 16 and U = 1,800 (Ranallo-Benavidez et al. 2020).
Nuclear genome assembly
HiFi reads were filtered for HiFi-specific adapters using HiFiAdapterFilt (Sim et al. 2022). Filtered reads were checked for contaminants using Centrifuge (Kim et al. 2016) with an index containing archaea, bacteria, fungi, and virus according to NCBI taxonomy (Schoch et al. 2020). No contaminants were filtered out as those identified were minimal and primarily bacterial taxa with sequence similarity to chloroplast. Adapter-filtered HiFi reads were used to create a draft assembly in hifiasm using the -l0 parameter due to the estimated low heterozygosity (Cheng et al. 2021). Raw Omni-C reads (--h1 and --h2) were also included to aid in primary and alternate haplotype organization. The resulting draft assemblies, primary (hap1) and alternate (hap2), were filtered for chloroplast sequence with BLAST (Camacho et al. 2009) using the publicly available L. crithmoides (Sayers et al. 2021), the closest related chloroplast genome assembly available. Scaffolding followed previously described methods (https://aidenlab.org/assembly/manual_180322.pdf). Omni-C reads were aligned to the primary draft assembly with BWA (Li and Durbin 2009) and used to create a pairs file filtered for duplicates with Juicer (Durand et al. 2016). 3D-DNA (Dudchenko et al. 2017) default parameters were used for scaffolding, as it leverages the long-distance read pair alignments to order and orient contigs. Results were visualized in JuiceBox (Dudchenko et al. 2018) followed by YAGCloser to close gaps with support of HiFi reads (https://github.com/merlyescalona/yagcloser). The resulting gap-filled scaffolds were sorted in order of decreasing size and named/numbered accordingly. Scaffolding of the alternate draft assembly followed identical methods. The partial mitochondrial genome was identified within a scaffold in both nuclear assemblies and removed by breaking the scaffolds at those coordinates (Table 1).
Table 1.
Assembly pipeline and software used.
| Nuclear assembly | Software | Version |
|---|---|---|
| Read Quality control | FastQC | 0.11.8 |
| Read Quality control | LongQC | 1.2.0c |
| Omni-C adapter trimming | Cutadapt | 1.18 |
| K-mer counting | KMC | 3.1.1 |
| Estimation of genome size and heterozygosity | GenomeScope | 2 |
| Filtering PacBio HiFi adapters Identifying contaminants |
HiFiAdapterFilt | 2.0.1 |
| Contaminant identification | Centrifuge | 1.0.4 |
| De novo assembly (contigging) | Hifiasm | 0.16.1-r375 |
| Contig detection of organelle sequence | BLAST | 2.13.0 |
| Omni-C mapping and pairing | BWA-MEM | 0.7.17-r1188 |
| Samtools | 1.9,1.16.1 | |
| Juicer | 1.6 | |
| Juicer Tools | 2.20.00 | |
| Scaffolding | 3D-DNA | 180922 |
| Assembly visualization | JuiceBox | 1.11.08 |
| Gap closing | YAGCloser https://github.com/merlyescalona/yagcloser |
Commit 0e34c3b |
| Benchmarking | ||
| Basic assembly stats | QUAST | 5.2.0 |
| Assembly completeness | BUSCO | 5.4.3 |
| Minimap2 | 2.24-r1122 | |
| Meryl | 1.3 | |
| Merqury | 1.3 | |
| Organelle assemblies | ||
| Chloroplast | Seqtk | 1.3-r106 |
| Flye | 2.9 | |
| GetOrganelle | 1.7.4 | |
| Mitochondria | NOVOPlasty | 4.3.1 |
| BLAST | 2.13.0 | |
Software citations are listed in the text.
Assembly quality assessment
At each step in the assembly and scaffolding process, assembly statistics and completeness were tracked with QUAST (Gurevich et al. 2013) and BUSCO (Manni et al. 2021). Final genomes were further evaluated by aligning the HiFi reads used to generate the assembly to the resulting assembly with Minimap2 (Li 2018). The Omni-C reads filtered for adapters with Cutadapt v1.18 (Martin 2011) were used to obtain the quality value (QV) with Merqury (Rhie et al. 2020).
Organelle genome assemblies
Adapter-filtered HiFi reads were aligned to the L. crithmoides chloroplast with minimap2 (-ax map-hifi) and filtered for Q20 (samtools view -F4 -q20). These reads were subset with seqtk subseq and contigged with Flye (Kolmogorov et al. 2019). A circular assembly was achieved using the assembly graph from Flye as input for GetOrganelle (Jin et al. 2020) using the L. crithmoides chloroplast as a reference (--genes).
We used short-read sequencing of sample CEJES6B739 to construct a partial mitochondrial genome in NOVOPlasty (Dierckxsens et al. 2017) with the Cox1 gene from GetOrganelleDB as a seed. We used this mitochondrial genome as a BLAST query to identify the corresponding mitochondrial genome in a contig of the nuclear genome of the reference individual.
Results
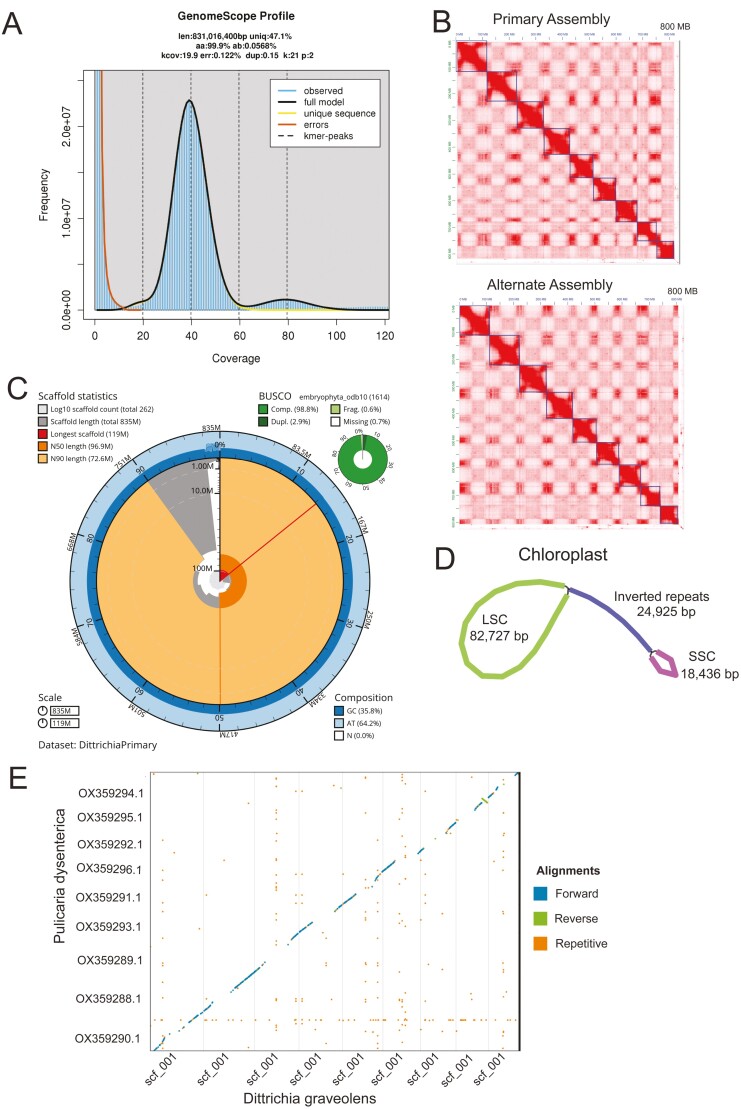
HiFi sequencing produced 2.4 M reads with an N50 of 14 kb and approximately 41× coverage. From this sequencing, the genome length estimate was 831 Mbp, 47.1% of which is unique, non-repetitive sequence, with a heterozygosity of 0.0568% (Fig. 3A). Omni-C sequencing produced 84 M 150 bp paired-end reads totaling 15× in coverage. HiFi sequencing with Omni-C for phasing generated primary and alternate draft assemblies ~835 Mbp in length with contig N50s of ~75 Mbp. After scaffolding with Omni-C and gap closing with HiFi, the final primary genome was 835 Mbp long, with an N50 of 96.9 Mb and 98.1% of its sequence organized into 9 pseudo-molecules, and with an average of 8.69 Ns per 100 kb (Fig. 3B and C). The BUSCO score was 98.8% complete (2.9% duplicates; Fig. 3C) and the QV score was 34.2913. The alternate genome was 824 Mbp long, with an N50 of 96.4 Mb 97.8% organized into 9 pseudo-molecules, and 6.49 Ns per 100 kb. The alternate BUSCO was 98.6% (2.9% duplicate), with 2 extra genes missing that were complete in the primary genome.
Fig. 3.
A) K-mer spectra created with HiFi reads, k = 21, to estimate genome size. B) Contact map of primary and alternate assemblies showing 9 pseudo-molecules ordered largest to smallest. C) Snail plot visualizing the primary assembly contiguity and completeness metrics. D) Assembly graph of the complete chloroplast genome. E) Whole-genome alignment of P. dysentarica to D. graveolens.
The chloroplast assembly produced a circular assembly 151,013 bp in length (Fig. 3D). Illumina short-read paired-end sequencing used for the mitochondrial assembly consisted of 14.2 M reads and the resulting contig was 22,084 bp. Only 1 contig was produced by NOVOPlasty, but this likely does not represent the complete mitogenome.
Detailed statistics for each assembly are summarized in Table 2.
Table 2.
Sequencing and assembly statistics, and accession numbers.
| BioProjects & Vouchers | |
| CCGP NCBI BioProject | PRJNA720569 |
| Species NCBI BioProject | PRJNA919063 |
| NCBI BioSample | SAMN32619994 |
| Specimen identification number | UCSC_NL_FS-MER_Block4_2020_SAMPLE3264 |
| Genome sequence | |
| PacBio HiFi reads | 1 PACBIO_SMRT (Sequel II) run: 2433439 spots, 34.0 G bases |
| Omni-C Illumina sequencing | 6 Illumina NextSeq 550 runs: 11.9 M spots, 3.6 G bases 11.5 M spots, 3.4 G bases 12.2 M spots, 3.7 G bases 3.5 M spots, 1.0 G bases 2.7 M spots, 818 M bases |
| Illumina PE 150 bp | 1 Illumina NextSeq 550 run: 7.1 M spots, 2.1 M bases |
| PacBio HiFi NCBI SRA | SRR23002284 |
| Omni-C Illumina NCBI SRA | SRR23002279-83 |
| Illumina PE SRA | SRR23255570 |
| Genome assembly—primary (alternate) | |
| Assembly identifier | Dgra_v1_pri_scf_ (Dgra_v1_alt_scf_) |
| HiFi read coverage | 41× |
| Number of contigs | 235 (158) |
| Contig N50 (bp) | 74,911,303 (74,485,925) |
| Longest contigs | 111,114,853 (110,676,008) |
| Number of scaffolds | 262 (292)a |
| Scaffold N50 (bp) | 96,898,007 (96,379,130) |
| Size of final assembly (bp) | 834,904,512 (824,346,690) |
| N’s per 100 kbp | 8.68 (6.49) |
| Base pair QV (Merqury) | P: 34.2913, A: 34.2439 |
| k-mer completeness | P: 93.41%, A: 93.34% |
| BUSCO completeness | Embryophyta_obd10 (n: 1,614) |
| Primary (C:S:D:F:M) | 98.8%:95.9%:2.9%:0.6%:0.6% |
| Alternate (C:S:D:F:M) | 98.6%:95.7%:2.9%:0.6%:0.6% |
| Assigned chromosomes % | 98.1% (97.8%) |
| Organelles | 1 complete chloroplast (in primary assembly) 2 partial mitochondria (primary and alternate) |
aThe number of scaffolds is higher than contigs due to small fragments created during misassembly correction in 3D-DNA.
Discussion
Prior to our sequencing effort, the genome size of D. graveolens was estimated to be 2C = 1.99 pg, consisting of 18 chromosomes (Zonneveld 2019). Our results are in concordance with these estimates as 835 Mbp and 9 base chromosomes (2n = 18). This is a relatively small and simple genome compared with others in the Asteraceae (Garcia et al. 2013) but similar in size and synteny to the closest relative with a reference genome, P. dysenterica (Fig. 3E). Our heterozygosity is 0.0568%, lower than that of sequenced medicinal plants A. annua at 1% to 1.5% (Shen et al. 2018), and E. breviscapus (2.04%; He et al. 2021), but the individual we selected was grown from seed produced after 1 generation of selfing. Re-analysis of published ddRAD-seq data for D. graveolens from the same source region in southern France (Lustenhouwer et al. 2018), corrected for fragment length, shows an average observed heterozygosity of 0.12% (Appendix 1). Given that 1 generation of selfing would result in halved heterozygosity, this value closely corresponds to the reference genome presented here.
The rapid population spread of D. graveolens in new areas is aided by its annual lifecycle, self-compatibility, high fecundity, wind dispersal, and high germination rate (Rameau et al. 2008; Brownsey et al. 2013a; Lustenhouwer et al. 2018). The species thrives in human-disturbed habitat such as roadsides and industrial sites which is facilitated by tolerance to environmental stressors such as high salinity (Grašič et al. 2016; Šajna et al. 2017), heavy metals (Shallari et al. 1998), and possibly to arsenic as found in D. viscosa (De Paolis et al. 2022). It has a high secondary metabolite diversity and toxicology with 262 compounds discovered to date (Ponticelli et al. 2022). Along with its small diploid genome, these traits make D. graveolens a candidate model in the Inulae for studying the genetic basis of invasiveness in the Asteraceae.
D. graveolens is a particularly successful example of apparently climate-driven poleward range expansion, which will be key for many native species to persist under climate change (Urban et al. 2016). D. graveolens’ rapid native range expansion is associated with the evolution of earlier phenology at the range limit and a niche expansion toward more temperate climates, enabling spread well beyond the expectation under climate tracking alone (Lustenhouwer et al. 2018; Lustenhouwer and Parker 2022). The reference genome presented here provides an opportunity to study the genomic basis of these impactful eco-evolutionary processes and will advance the emerging field of climate change genomics in plants (Lancaster et al. 2022).
In less than 40 years, D. graveolens has spread rapidly across California and is a potential threat to rangelands and wildlands undergoing fire recovery and restoration. There is now a strong urgency to understand and control the rapid spread of D. graveolens in its invaded range (Brownsey et al. 2013b). Comparative, population and landscape genomic analyses will indicate patterns of selection and evolution in these invading populations, and will help determine how evolution may contribute to invasion risk and impact on both wild and managed ecosystems. We are actively researching these topics and encourage community development and collaboration across the field of invasive species genomics.
Supplementary Material
Acknowledgments
We thank the UCSC Greenhouse staff, especially Sylvie Childress and Jim Velzy, for facilities and plant care. We are grateful to the staff at the UCSC Paleogenomics Laboratory for the Omni-C library preparation and for providing servers for assembly and assistance, particularly Beth Shapiro, Ed Green, and Merly Escalona. PacBio HiFi sequencing was carried out by the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. We are grateful for the help of Ruta Sahasrabudhe who coordinated the work at UC Davis. We thank Natalie Gonzalez for extracting the DNA of the California plant. We thank Matthew Hartfield for assisting with the fieldwork in France. Finally, we thank Calflora for the excellent database curation that helped us locate California candidate collection sites.
Contributor Information
Susan L McEvoy, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States; Department of Conservation and Research, Santa Barbara Botanic Garden, Santa Barbara, CA, United States.
Nicky Lustenhouwer, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States; School of Biological Sciences, University of Aberdeen, Aberdeen, United Kingdom.
Miranda K Melen, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Oanh Nguyen, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, CA, United States.
Mohan P A Marimuthu, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, CA, United States.
Noravit Chumchim, DNA Technologies and Expression Analysis Core Laboratory, Genome Center, University of California, Davis, CA, United States.
Eric Beraut, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Ingrid M Parker, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Rachel S Meyer, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, United States.
Funding
This work was funded by the United States Department of Agriculture, National Institute of Food and Agriculture “Agriculture and Food Research Initiative Grant” [2020-67013-31856]. NL acknowledges support from the Swiss National Science Foundation [P2EZP3_178481] and Natural Environment Research Council [NE/W006553/1].
Conflict of interest
The authors declare that they have no conflicts of interest.
Data availability
Data generated for this study are available under NCBI BioProject PRJNA919063. Raw sequencing data for sample DitGraRef (NCBI BioSample SAMN32619994) are deposited in the NCBI Short Read Archive (SRA) under SRR23002279-84. Primary and alternate genome assemblies are available in PRJNA919087-8. Raw sequencing data for sample DitGraMitoRef used in the mitochondrial genome assembly is under SRR23255570. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: https://doi.org/10.5281/zenodo.7508737.
References
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- Baker HG. The evolution of weeds. Annu Rev Ecol Syst. 1974;5:1–24. [Google Scholar]
- Brownsey RN. Biology of Dittrichia graveolens (stinkwort): implications for management. Davis: University of California; 2012. [Google Scholar]
- Brownsey RN, Kyser GB, DiTomaso JM.. Seed and germination biology of Dittrichia graveolens (stinkwort). Invasive Plant Sci Manage. 2013a;6:371–380. [Google Scholar]
- Brownsey RN, Kyser G, DiTomaso J.. Stinkwort is rapidly expanding its range in California. Calif Agric. 2013b;67:110–115. [Google Scholar]
- Brullo S, de Marco G.. Taxonomical revision of the genus Dittrichia (Asteraceae). Port Acta Biol. 2000;19:341–354. [Google Scholar]
- California Department of Food and Agriculture. CDFA—Plant Health—Encycloweedia: data sheets for California noxious weeds.2021. [accessed 2021 Jun 22]. https://www.cdfa.ca.gov/plant/ipc/encycloweedia/weedinfo/winfo_table-sciname.html
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H.. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paolis A, De Caroli M, Rojas M, Curci LM, Piro G, Di Sansebastiano G-P.. Evaluation of Dittrichia viscosa Aquaporin Nip1.1 gene as marker for arsenic-tolerant plant selection. Plants. 2022;11:1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTomaso JM, Brownsey R.. Invasive stinkwort (Dittrichia graveolens) is quickly spreading in California (No. 9). Sacramento: California Weed Science Society Research Update and News; 2013. p. 1–2. [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aideen AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Shamim MS, Batra SS, Durand NC, Musial NT, Mostofa R, Pham M, Hilaire BGS, Yao W, Stamenova E, et al. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000, bioRxiv, doi: 10.1101/254797, 2018, 254797, preprint: not peer reviewed. [DOI]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL.. Juicer provides a One-Click System for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Wang S, Wang H, Wang A, Jiang F, Liu H, Zhao H, Xu D, Zhang Y.. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae palaeo-polyploidization history and plant inulin production. Mol Ecol Resour. 2022;22:3124–3140. [DOI] [PubMed] [Google Scholar]
- Fukasawa Y, Ermini L, Wang H, Carty K, Cheung M-S.. LongQC: a quality control tool for third generation sequencing long read data. G3. 2020;10:1193–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S, Vigo J, Garnatje T, Vallès J.. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 40 genera, 3 tribes and 2 subfamilies. Plant Biosyst. 2013;147:1219–1227. [Google Scholar]
- GBIF.org. GBIF occurrence download for Dittrichia graveolens (L.) Greuter. 2020; 10.15468/dl.s0qcs0. [DOI] [Google Scholar]
- Grašič M, Anžlovar S, Strgulc Krajšek S.. Germination rate of stinkwort (Dittrichia graveolens) and false yellowhead (D. viscosa) in relation to salinity. Acta Biol Slov. 2016;59:5–11. [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Dong X, Zhang G, Fan W, Duan S, Shi H, Li D, Li R, Chen G, Long G, et al. High quality genome of Erigeron breviscapus provides a reference for herbal plants in Asteraceae. Mol Ecol Resour. 2021;21:153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins KA, Bock DG, Hahn MA, Heredia SM, Turner KG, Rieseberg LH.. Comparative genomics in the Asteraceae reveals little evidence for parallel evolutionary change in invasive taxa. In: Barrett SC, Colautti RI, Dlugosch KM. Rieseberg LH, editors. Invasion genetics. West Sussex, UK: John Wiley & Sons, Ltd; 2016. p. 283–299. [Google Scholar]
- Inglis, PW, Pappas MDCR, Resende, LV, Grattapaglia D. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PloS ONE. 2018;13:e0206085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z.. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song L, Breitwieser FP, Salzberg SL.. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocián P. (L.) Greuter—a new alien species in Poland. Acta Mus Siles Sci Natur. 2015;64:193–197. [Google Scholar]
- Kokot M, Długosz M, Deorowicz S.. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017;33:2759–2761. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA.. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. [DOI] [PubMed] [Google Scholar]
- Lancaster LT, Fuller ZL, Berger D, Barbour MA, Jentoft S, Wellenreuther M.. Understanding climate change response in the age of genomics. J Anim Ecol. 2022;91:1056–1063. [DOI] [PubMed] [Google Scholar]
- Lenoir J, Gégout JC, Marquet PA, de Ruffray P, Brisse H.. A significant upward shift in plant species optimum elevation during the 20th century. Science. 2008;320:1768–1771. [DOI] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustenhouwer N, Parker IM.. Beyond tracking climate: niche shifts during native range expansion and their implications for novel invasions. J Biogeogr. 2022;49:1481–1493. [Google Scholar]
- Lustenhouwer N, Wilschut RA, Williams JL, van der Putten WH, Levine JM.. Rapid evolution of phenology during range expansion with recent climate change. Glob Change Biol. 2018;24:e534–e544. [DOI] [PubMed] [Google Scholar]
- Mandel JR, Dikow RB, Siniscalchi CM, Thapa R, Watson LE, Funk VA.. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc Natl Acad Sci USA. 2019;116:14083–14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM.. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021;38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):Article 1. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- OpenTreeofLife, Cranston KA, Redelings B, Reyes LLS, Allman J, McTavish EJ, Holder MT.. Open Tree of Life Taxonomy (3.2). Zenodo. 2019. doi: 10.5281/zenodo.3937751 [DOI] [Google Scholar]
- Parsons WT, Cuthbertson EG.. Noxious weeds of Australia. 2nd ed. Collingwood, Australia: CSIRO Publishing; 2001. p. 281–283. [Google Scholar]
- Philbey A, Morton A.. Pyogranulomatous enteritis in sheep due to penetrating seed heads of Dittrichia graveolens. Aust Vet J. 2000;78:858–860. [DOI] [PubMed] [Google Scholar]
- Ponticelli M, Lela L, Russo D, Faraone I, Sinisgalli C, Mustapha MB, Esposito G, Jannet HB, Costantino V, Milella L.. Dittrichia graveolens (L.) Greuter, a rapidly spreading invasive plant: chemistry and bioactivity. Molecules. 2022;27:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau J-C, Mansion D, Dumé G, Gauberville C.. Flore forestière française tome 3, région méditerranéenne: Guide écologique illustré. Paris, France: CNPF-IDF; 2008. [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11:Article 1. doi: 10.1038/s41467-020-14998-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM.. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Pepper-Tunick E, Svedberg J, Byrne A, Castillo JR, Vollmers C, Beinart RA, Corbett-Detig R.. Horizontal transmission and recombination maintain forever young bacterial symbiont genomes. PLoS Genet. 2020;16:e1008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šajna N, Adamlje K, Kaligarič M.. Dittrichia graveolens—how does soil salinity determine distribution, morphology, and reproductive potential? Ann Ser Hist Nat. 2017;27:7–12. [Google Scholar]
- Santilli L, Lavandero N, Montes O, Lustenhouwer N.. First record of Dittrichia graveolens (Asteraceae, Inuleae) in Chile. Darwiniana. 2021;9:Article 1. doi: 10.14522/darwiniana.2021.91.938 [DOI] [Google Scholar]
- Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;50:D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020;2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallari S, Schwartz C, Hasko A, Morel JL.. Heavy metals in soils and plants of serpentine and industrial sites of Albania. Sci Total Environ. 1998;209:133–142. [PubMed] [Google Scholar]
- Shen Q, Zhang L, Liao Z, Wang S, Yan T, Shi P, Liu M, Fu X, Pan Q, Wang Y, et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol Plant. 2018;11:776–788. [DOI] [PubMed] [Google Scholar]
- Sim SB, Corpuz RL, Simmonds TJ, Geib SM.. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genomics. 2022;23:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong H-Y, Yokota M, Kardassakis D, Maibach HI.. Allergic contact dermatitis from Dittrichia graveolens (L.) Greuter (stinkwort). Contact Dermatitis. 2008;58:51–53. [DOI] [PubMed] [Google Scholar]
- Urban MC, Bocedi G, Hendry AP, Mihoub J-B, Pe’er G, Singer A, Bridle JR, Crozier LG, De Meester L, Godsoe W, et al. Improving the forecast for biodiversity under climate change. Science. 2016;353:aad8466. [DOI] [PubMed] [Google Scholar]
- USDA. Weed risk assessment for Dittrichia graveolens (L.) Greuter (Asteraceae)—stinkwort. Raleigh, North Carolina: Animal and Plant Health Inspection Service, Plant Protection and Quarantine; 2013. [Google Scholar]
- Zonneveld BJM. The DNA weights per nucleus (genome size) of more than 2350 species of the Flora of The Netherlands, of which 1370 are new to science, including the pattern of their DNA peaks. Forum Geobotanicum. 2019;8:24–78. doi: 10.3264/FG.2019.1022 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are available under NCBI BioProject PRJNA919063. Raw sequencing data for sample DitGraRef (NCBI BioSample SAMN32619994) are deposited in the NCBI Short Read Archive (SRA) under SRR23002279-84. Primary and alternate genome assemblies are available in PRJNA919087-8. Raw sequencing data for sample DitGraMitoRef used in the mitochondrial genome assembly is under SRR23255570. Assembly scripts and other data for the analyses presented can be found at the following GitHub repository: https://doi.org/10.5281/zenodo.7508737.