Abstract
Background:
Improved understanding of the asymptomatic malaria parasite reservoir is a prerequisite to pursue malaria elimination efforts. We therefore characterised temporal trends and transporter polymorphisms in asymptomatic Plasmodium infections during the transition from high to low transmission in Zanzibar.
Methods:
Healthy individuals participating in cross-sectional surveys conducted 2005–2013 were screened for asymptomatic malaria by PCR. Complexity/diversity of infection and transporter polymorphisms were assessed in Plasmodium falciparum positive samples. Symptomatic samples were included for comparison of polymorphisms in 2013.
Results:
PCR-determined parasite prevalence declined from 21.1% (CI95% 17.4–24.9) to 2.3% (CI95% 1.7–2.9) from 2005 to 2013. P. falciparum remained the predominant species; prevalence was highest in children and young adults aged 5–25 years. Parasite densities and complexity of infection, but not population genetic diversity of P. falciparum, decreased from 2005–2009. pfcrt 76T (99.2–64.7%, p < 0.001) and pfmdr1 86Y frequencies (89.4–66.7%, p = 0.03) decreased over time. Pfmdr1 (a.a.86,184,1246) YYY and YYD haplotypes were more frequent in asymptomatic than symptomatic infections in 2013 (p < 0.001).
Conclusions:
There is a declining, albeit persistent, reservoir of parasites present at low-densities in asymptomatic individuals in Zanzibar. This study revealed important characteristics of the remaining parasite population, including intriguing temporal trends in molecular markers associated with antimalarial resistance, which need to be further investigated.
Keywords: Asymptomatic, Plasmodium, Low transmission, Molecular surveillance, Antimalarial drug resistance markers
1. Introduction
Substantial reductions in malaria transmission, temporally associated with deployment of effective vector control and improved case management, have been observed in Africa during 2000–2010 (Noor et al., 2014; WHO, 2010). The considerable shift in epidemiology occurring in areas undergoing transition from malaria control to pre-elimination requires new tools and strategies for detection of malaria infections (Alonso and Tanner, 2013; Cotter et al., 2013; Tietje et al., 2014). The relative proportion of low-density infections that fall beneath the detection level of conventional microscopy and malaria rapid diagnostic tests (mRDTs) increases as transmission declines (Okell et al., 2009). These subpatent infections are likely to be asymptomatic and therefore missed by passive surveillance, but are estimated to potentially fuel 20–50% of human-to-mosquito transmission in pre-elimination settings (Okell et al., 2012). Low-density asymptomatic infections may therefore constitute an important reservoir for continued malaria transmission that needs to be targeted to achieve malaria elimination (Bousema et al., 2014).
Zanzibar was one of the first regions in sub-Saharan Africa to deploy artemisinin-based combination therapies (ACT) free of charge to all age groups through public health facilities. Artesunate–amodiaquine has been the first-line treatment for uncomplicated malaria in Zanzibar since September 2003. Deployment of artesunate–amodiaquine, together with mass distribution of long lasting insecticide-treated nets to high-risk groups, resulted in a substantial decline in Plasmodium falciparum malaria among febrile children, from approximately 30% to 1–2%, and a reduction of crude child mortality of approximately 50% between 2003 and 2006 (Bhattarai et al., 2007). LLINs were mass distributed in 2006 to high-risk groups, after which 90% of children under the age of five were reported to sleep under a LLIN (Bhattarai et al., 2007). Vector control was further strengthened by mass-distribution of two LLINs per household in 2008–2009, and annual rounds of IRS targeting all households in Zanzibar (excluding Stone Town) between 2006 and 2009 (Björkman et al. submitted). Coverage of vector control interventions remained high in 2009 despite reduced perceived threat of malaria among caretakers, with 70% of under-five children reported to sleep under a bed net and 94% living in a house targeted with IRS. Combined, 98% of children were covered by at least one of the vector control methods (Beer et al., 2013). Furthermore, mRDTs were introduced in 2006 for improved case detection, and targeting of treatment to patients with confirmed malaria infections. Malaria RDTs were shown to improve management of fever patients, providing adequate treatment and health outcomes without increased cost per patient (Msellem et al., 2009). Zanzibar is presently in a state of pre-elimination (Björkman et al. submitted), and the Zanzibar Ministry of Health has officially declared the aim of malaria elimination.
Deployment of ACT in Zanzibar has been a corner stone in the recent success in malaria control and monitoring continued efficacy is paramount 10 years after the introduction of artesunate–amodiaquine as first-line treatment. As resistance to ACT partner drugs has historically manifested before that of artemisinins (Venkatesan et al., 2014), molecular markers associated with amodiaquine resistance may serve as an important tool for surveillance of antimalarial drug resistance in Zanzibar (Froberg et al., 2012). Single nucleotide polymorphisms in the P. falciparum chloroquine resistance transporter (pfcrt) and P. falciparum multidrug resistance 1 (pfmdr1) transporter genes have been associated, both in vitro and in vivo, with resistance to amodiaquine (Echeverry et al., 2007; Folarin et al., 2011; Picot et al., 2009). Selection of pfcrt 76T and pfmdr1 86Y alleles, as well as pfmdr1 1246Y and the pfmdr1 (a.a.86,184,1246) YYY haplotype has been shown in recurrent infections after treatment with artesunate–amodiaquine or amodiaquine alone (Djimde et al., 2008; Duraisingh et al., 1997; Holmgren et al., 2006, 2007; Humphreys et al., 2007; Nsobya et al., 2007; Venkatesan et al., 2014).
The aim of this study was to describe temporal trends in the asymptomatic Plasmodium reservoir during the transition from high to low transmission in Zanzibar. Infections were characterised with regards to Plasmodium species, geographic- and age distribution, qPCR-determined parasite densities, complexity and diversity of infection, and temporal trends in P. falciparum SNPs associated with artesunate–amodiaquine resistance.
2. Materials and methods
2.1. Study sites and collection of samples
Finger prick blood samples for malaria screening by microscopy or mRDT were collected from healthy individuals during cross-sectional household surveys conducted in May–July 2005, 2009, 2011 and 2013, in North A (Unguja island) and Micheweni (Pemba island), two sentinel districts in Zanzibar (Bhattarai et al., 2007), (Björkman et al., submitted). In addition, blood spots for molecular analysis were collected (from the same finger prick) on Whatman 3 mm filter paper and stored at room temperature. Epidemiological data in forms of demographic and clinical information were also collected at each survey.
2.2. Malaria diagnosis in cross-sectional surveys
For malaria diagnosis in 2005 and 2009 examination of thick blood smears was conducted by experienced microscopists in Zanzibar according to standard WHO procedures (Bhattarai et al., 2007), (Björkman et al., submitted). In 2011 Paracheck-Pf (Orchid Biomedical Systems, India), a P. falciparum specific histidine-rich protein 2 based mRDT, replaced microscopy. In 2013, Paracheck-Pf was replaced by SD-Bioline Malaria Ag P.f/Pan® (Standard Diagnostic, Republic of Korea), a combo mRDT detecting both P. falciparum HRP2 and pan-Plasmodium lactate dehydrogenase.
2.3. Sample selection for molecular analysis
In 2009, 2011 and 2013 all filter paper samples collected (2423, 2977 and 3038, respectively) were screened by real-time PCR for Plasmodium infections (Table 1). For the 2005 survey a selection of 534 samples were screened including all 72 available blood slide positive samples and 462 blood slide negative samples randomly selected from 2141 filter papers. All molecular analyses were conducted at Karolinska Institutet, Stockholm, Sweden.
Table 1.
Baseline demographic characteristics of study participants, and community prevalence of malaria by microscopy/mRDT and PCR in Zanzibar, 2005–2013.
| Asymptomatic subjects |
Symptomatic subjects | ||||
|---|---|---|---|---|---|
| Year | 2005 | 2009 | 2011a | 2013 | 2013 |
|
| |||||
| Sample material | Filter paper | Filter paper | Filter paper | Filter paper | mRDT |
| Number screened by PCR | 534 | 2423 | 2977 | 3038 | 102 |
| North A (%) | 327 (61.2) | 1013 (41.8) | 1599 (53.7) | 1463 (48.2) | 36 (35.3) |
| Micheweni (%) | 207 (38.8) | 1410 (58.2) | 1378 (46.3) | 1575 (51.8) | 66 (64.7) |
| Age group (years) | |||||
| <5 (%) | 129 (24.2) | 567 (23.4) | 456 (15.3) | 646 (21.3) | 2 (2.0) |
| 5–15 (%) | 152 (28.5) | 546 (22.5) | 583 (19.6) | 872 (28.7) | 8 (7.8) |
| 15–25 (%) | 85 (15.9) | 369 (15.2) | 370 (12.4) | 459 (15.1) | 11 (10.8) |
| >25 (%) | 168 (31.5) | 842 (34.8) | 696 (23.4) | 1046 (34.4) | 15 (14.7) |
| Unknown (%) | 0 | 99 (4.1) | 872 (29.3) | 15 (0.5) | 66 (64.7) |
| Sex | |||||
| Male (%) | 222 (41.6) | 928 (38.3) | 891 (29.9) | 1257 (41.4) | – |
| Female (%) | 312 (58.4) | 1422 (58.7) | 1257 (42.2) | 1766 (58.1) | – |
| Unknown (%) | 0 | 73 (3.0) | 829 (27.9) | 15 (0.5) | 102 (100) |
| Parasite prevalence | |||||
| By microscopy/mRDT % (95% CI) | 7.5 (6.4–8.6) | 0.0 (0–0.2) | 0.4 (0.2–0.7) | 0.3 (0.1–0.7) | – |
| By PCR % (95% CI) | 21.1b (17.4–24.9) | 3.3 (2.6–4.1) | 2.2 (1.6–2.8) | 2.3 (1.7–2.9) | – |
A labelling issue occurred in the field in 2011, resulting in a large number of samples with unknown age and sex data.
Prevalence adjusted to account for the sample selection.
2.4. DNA extraction from filter paper
DNA from a Ø 3 mm filter paper disc (≈3–5 μL blood) was extracted with Chelex® 100 resin (Bio-Rad Laboratory, USA) (Wooden et al., 1993) (Xu et al., 2015). The supernatant containing genomic DNA was stored at −20 °C until use; PCR screening was conducted within 3 months of DNA extraction. Older samples collected in 2005, which were positive in the initial real-time PCR screening, were re-extracted with the QIAmp DNA mini kit (Qiagen Gmbh, Germany) in order to improve the quality of the DNA (Hwang et al., 2012). DNA was extracted from three 3 mm filter paper discs according to the protocol “DNA purification from dried blood spots” (Qiagen, 2012).
2.5. Plasmodium detection by qPCR
Parasite detection and species determination was conducted by a SYBR Green real-time PCR restriction fragment length polymorphism assay (cytb-qPCR) targeting the cytochrome b gene of the four major human Plasmodium species (P. falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale) (Xu et al., 2015). Positivity was confirmed by gel-electrophoresis on a 1.5% agarose gel stained with GelRed (Biotium Inc., USA). Positive and negative controls were included in each run. Plasmodium species was determined by RFLP on positive cytb-qPCR products and gel-electrophoresis (Xu et al., 2015); results were documented with a GelDoc™ system (Bio-Rad Laboratory, USA). The PCR detection limit for the cytb-qPCR assay has previously been determined in blood samples collected on filter paper as 1 p/μL (p/μL) for P. falciparum and 2 p/μL for P. malariae (Xu et al., 2015).
2.6. Parasite densities estimated by quantitative PCR (qPCR)
Parasite densities were estimated by a modified version of a probe-based qPCR method targeting Plasmodium 18S rRNA genes. In brief, all cytb-qPCR positive samples were subjected to the 18S rRNA qPCR using published protocol, primers and probe (Kamau et al., 2011). The reaction was carried out in a final volume of 20 μL, containing 5 μL of extracted DNA, 0.25 μM of each primer, 0.125 μM probe, and 1× SsoAdvanced Universal Probes Supermix (Bio-Rad Laboratory, USA). Parasite densities were determined against a standard curve generated by a Chelex-extracted, 10-fold serial dilutions (10,000–1 p/μL) of P. falciparum and P. malaria spotted on filter paper. The PCR detection limit for the 18S rRNA qPCR has previously been determined in blood samples collected on filter paper as 2 p/μL for P. falciparum and 5 p/μL for P. malariae (Xu et al., 2015).
2.7. Estimating the complexity and diversity of P. falciparum infections
The complexity or ‘multiplicity’ of infection (MOI) was determined by analysis of seven previously published microsatellites with modified conditions (Supplementary Table 1). Samples were amplified using 0.4 U of Phusion Hot Start II (Thermo Fisher Scientific, USA) with 1× HF buffer, 0.2 mM dNTP’s, 0.5 mM of each primer, and 1 μL of DNA in a final reaction volume of 5 μL. Amplicon size was determined by capillary electrophoresis and analysed using Genemapper v5 software (Life technologies, USA). Reproducible artefacts were automatically filtered based on relative peak height and allele size using an in-house software trained on lab strain controls. MOI was defined as the second highest allele frequency for each sample, to minimise influence of outliers. A subset of P. falciparum positive samples from individuals <15 years of age, from Micheweni district, from the 2005, 2009 and 2011 surveys were analysed; samples from 2013 were not included as they were collected after analysis of microsatellites had been completed.
2.8. Molecular genotyping of single nucleotide polymorphisms
Samples containing P. falciparum were genotyped for SNPs at positions pfcrt K76T, pfmdr1 N86Y, Y184F and D1246Y. Genotyping was conducted by nested PCR-RFLP methods (Supplementary Table 2) as previously described (Froberg et al., 2012). PCR negative samples were repeated twice with increased amounts of DNA template. A no template control was included in each PCR. Seven microlitres of each PCR product were digested overnight with ApoI, MluCI or EcoRV restriction enzymes, following manufacturers’ instruction (New England Biolabs, UK). RFLP products were run on 2–2.5% agarose gel stained with GelRed and documented with a GelDoc™ system. Lab clones 3D7, Dd2 and 7G8 were used as positive and negative restriction controls.
2.9. Collection, DNA extraction and genotyping of symptomatic samples
The prevalence of P. falciparum specific SNPs in the 2013 cross-sectional survey was compared with the prevalence in mRDT positive fever patients (hereafter referred to as symptomatic samples) presenting at primary health care facilities January–July 2013 in the two study districts. Positive mRDTs (SD-Bioline Malaria Ag P.f/Pan®) were passively collected at the health facilities and stored at room temperature. Chelex-100 extraction was used to extract DNA from the mRDTs as previously described (Ishengoma et al., 2011; Morris et al., 2013). Samples were genotyped as described above.
2.10. Ethical considerations
All participants of cross-sectional surveys provided written informed consent prior to blood sampling; in case of children a proxy-consent was provided from caretakers. Ethical approvals for this study were obtained from the ethical committees in Zanzibar (ZHRC/RAPD/03/2004, ZAMEC/0005/09, ZAMREC/0001/JUNE/011, ZAMREC/0001/APRIL/013) and the regional ethics committee in Stockholm (2009/387-31).
2.11. Statistical analyses
For prevalence calculations, the number of blood slide positive samples screened by PCR in 2005 was adjusted by 462/2141, stratifying by district and age group, to account for the sample selection (see 2.3). Prevalence in the two sentinel districts was compared by Fisher’s exact test. Odds ratios (OR) were calculated for the different age groups by univariate logistic regression using children aged <5 years as the reference group for each year. Quantitative PCR-determined parasite densities and MOI were compared between the years by Wilcoxon rank-sum test. Expected heterozygosity was calculated, as an indicator of genetic diversity, for each locus as , where of the ith allele. was compared between the years by two-sided student’s -test. The parasite populations were compared by pairwise fixation index based genetic distances. SNP frequencies were defined as the proportion of isolates containing alleles associated with antimalarial drug resistance (including mixed infections). Isolates containing mixed SNP results at more than one position were excluded from the pfmdr1 haplotype analysis; frequencies were calculated as for the SNPs. Longitudinal SNP and haplotype frequencies, and the SNP frequencies in the 2013 asymptomatic (cross-sectional) and symptomatic (fever patient) samples were compared by Fishers’ exact test. Due to small sample sizes the 2009–2013 asymptomatic samples were pooled for comparison of haplotype frequency with symptomatic samples by Fisher’s exact test. Analyses were repeated after excluding mixed infections to ensure these were not biasing the results. was calculated with Arlequin 3.5 software (Swiss Institute of Bioinformatics, Switzerland); all other statistical analyses were conducted using Stata/SE 12.1 (StataCorp LP, USA). Statistical significance was determined as .
3. Results
3.1. Parasite prevalence by microscopy/mRDT and PCR
The baseline demographics of all individuals included in the molecular screening of malaria are shown in Table 1. The prevalence of PCR-detectable malaria was 9.3-fold lower in 2013 as compared to 2005. However, as the prevalence of malaria declined, the relative proportion of subpatent infections only detected by PCR increased. In 2005 microscopy detected 35.7% (50/140) of the PCR-positive samples, whereas in 2013 only 8.8% (6/68) were also detected by mRDT.
3.2. Species, geographic- and age distribution of malaria
P. falciparum remained the predominant species present in 78.5–100% of the infections (including mixed infections). P. malariae was present in 12.2–43.2% of the infections (Fig. 1A). No cases of P. ovale or P. vivax were identified.
Fig. 1.
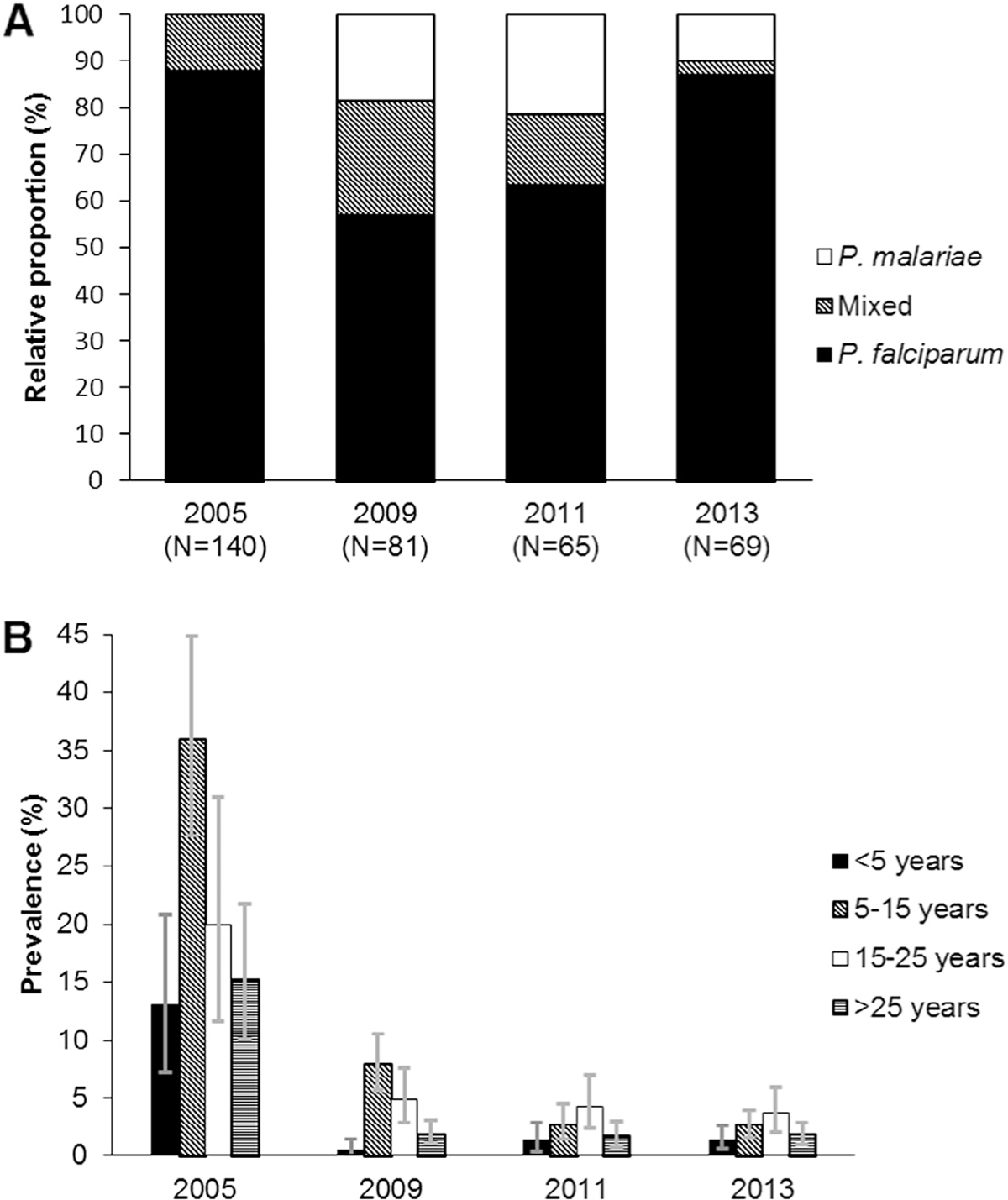
(A) Relative proportion of Plasmodium species in asymptomatic infections determined by cytb-qPCR RFLP assay. (B) PCR-determined prevalence of asymptomatic malaria by age group. Error bars represent 95% confidence intervals.
Between 2005 and 2013 there was an 8.8-fold decrease in North A and 10.7-fold decrease in Micheweni in PCR-determined malaria prevalence (Table 2); the prevalence was higher in Micheweni than in North A for all years, although the difference was not significant in 2013. Children aged 5–15 years and young adults 15–25 years generally appeared to have higher prevalence of malaria as compared to children <5 years and adults (Fig. 1B). In 2005 children aged 5–15 were most likely to have malaria (OR 3.8; CI95% 1.9–7.4), whereas in 2011 and 2013 the burden was highest in young adults aged 15–25 (OR 3.4; CI95% 1.3–8.8 and OR 3.7; CI95% 1.8–7.5, respectively) (Supplementary Table 3).
Table 2.
PCR-determined prevalence of asymptomatic malaria by district.
| Year | PCR positive/samples tested |
Prevalence % (CI 95%) |
p a | ||
|---|---|---|---|---|---|
| North A | Micheweni | North A | Micheweni | ||
|
| |||||
| 2005b | 47/289 | 54/189 | 16.3 (12.2–21.1) | 28.6 (22.2–35.6) | 0.002 |
| 2009 | 10/1013 | 71/1410 | 1.0 (0.4–1.9) | 5.0 (3.9–6.4) | <0.001 |
| 2011 | 20/1599 | 45/1378 | 1.3 (0.7–2.0) | 3.3 (2.3–4.4) | <0.001 |
| 2013 | 27/1463 | 42/1575 | 1.9 (1.2–2.7) | 2.7 (1.9–3.6) | 0.14 |
p calculated by Fisher’s exact test.
Prevalence adjusted to account for the sample selection.
3.3. Parasite densities estimated by qPCR
The majority of asymptomatic infections had parasite densities lower than 10 p/μL (Fig. 2). Densities ranged from <1 to 28,918 p/μL in P. falciparum monoinfections and 1–8 p/μL in P. malariae monoinfections (Table 3). The median parasite density was significantly higher in 2005 than in the subsequent years (p < 0.01). In 2005 there was a significant difference (p < 0.001) in the densities between the microscopy positive (geometric mean: 222, range: 1–247,241) and microscopy negative samples (geometric mean: 9, range: <1–777). Parasites densities could only be determined by 18S rRNA qPCR in 44.9% of the samples in 2013 compared to 88.6% in 2005, suggesting there has been an increase over time in the proportion of samples with parasite densities lower than the 18S rRNA qPCR detection limit.
Fig. 2.
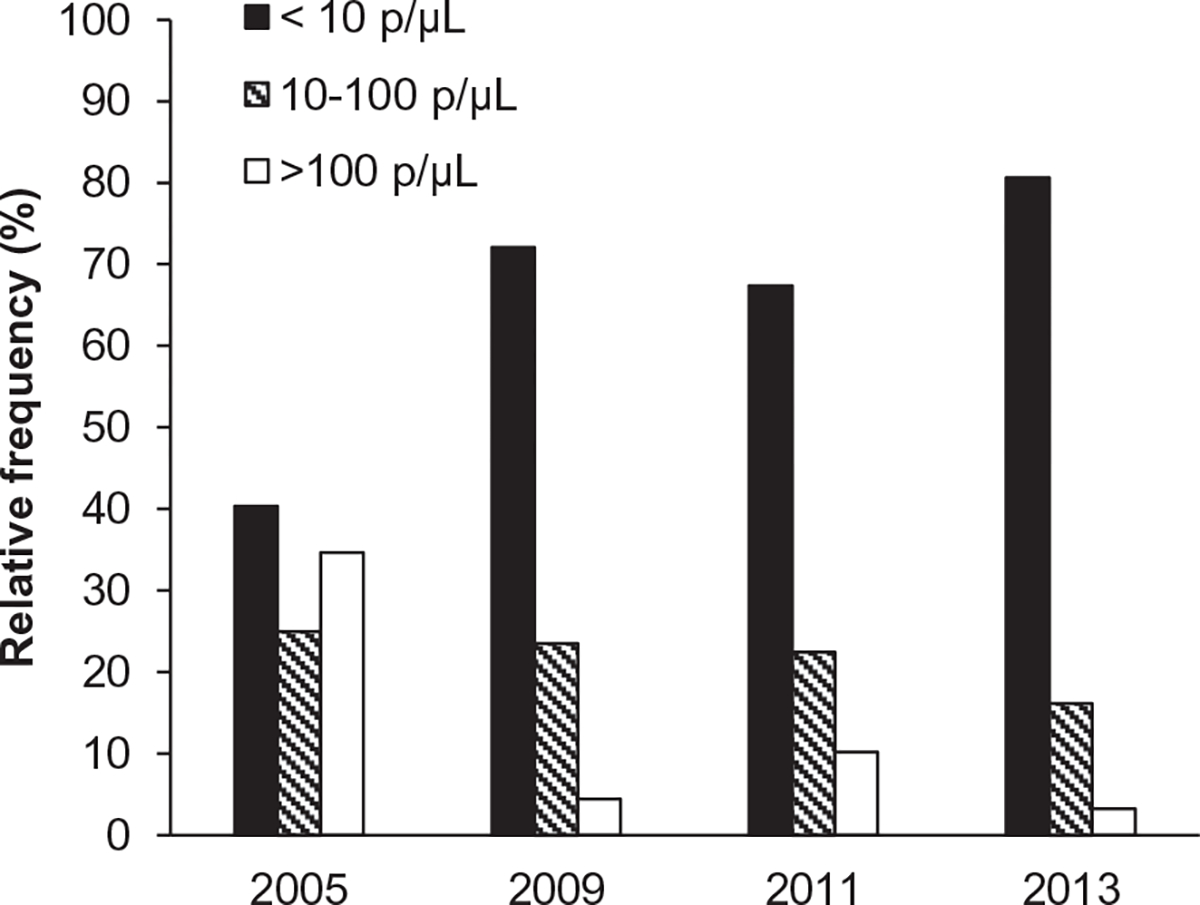
Quantitative PCR-determined parasite densities among asymptomatic persons. A Frequency histogram showing the relative proportion of samples with parasite densities of <10 p/μL, 10–100 p/μL and >100 p/μL by year.
Table 3.
Quantitative PCR-determined parasite densities among asymptomatic persons.
| Parasite densities | 2005 | 2009 | 2011 | 2013 |
|---|---|---|---|---|
|
| ||||
| Geometric mean (range) p/μLa | 31 (<1–247,241) | 6(1–232) | 9(1–4973) | 4 (<1–190) |
| Pfb geometric mean (range) p/μL | 28 (<1–28,918) | 7(1–232) | 13 (1–4973) | 4 (<1–190) |
| Pmc geometric mean (range) p/μL | - | 2 (1–4) | 2(1–3) | 3 (2–8) |
| 18s rRNA qPCR positive/cytb-qPCR positive (%) | 88.6 (124/140) | 84.0(68/81) | 75.4 (49/65) | 44.9 (31/69) |
p/μL = parasites per microlitre.
Pf = P. falciparum monoinfections.
Pm = P. malariae monoinfections.
3.4. Complexity and diversity of P. falciparum infections
The median MOI decreased significantly from 2005 to 2009 (p = 0.01), but no difference was observed between 2009 and 2011 (p = 0.73) (Table 4). There was no significant difference in MOI in children <5 years and those 5–15 years (p = 0.75) in all years. Heterozygosity in the analysed loci was high, with no significant difference between any two years (p > 0.05). analysis did not detect any changes in the population of parasites from one year to another.
Table 4.
Complexity and diversity of asymptomatic P. falciparum infection.
| 2005 (N = 33) | 2009 (N =19) | 2011 (N =23) | |
|---|---|---|---|
|
| |||
| Multiplicity of infection | |||
| MOI = 1 (%) | 21.2 | 52.6 | 60.9 |
| MOI = 2 (%) | 30.3 | 26.3 | 17.4 |
| MOI ≥ 3 (%) | 48.5 | 21.1 | 21.7 |
| Mean MOI; range | 2.8; 1–7 | 1.7; 1–4 | 1.7; 1–4 |
| Heterozygosity | |||
| Ara2New | 0.75 | 0.84 | 0.80 |
| B7M19 | 0.53 | 0.62 | 0.22 |
| PFG377 | 0.61 | 0.79 | 0.46 |
| TA1 | 0.86 | 0.92 | 0.90 |
| TA109 | 0.80 | 0.75 | 0.85 |
| TA81 | 0.82 | 0.82 | 0.76 |
| TA87 | 0.83 | 0.86 | 0.79 |
| Mean heterozygosity | 0.74 | 0.80 | 0.68 |
3.5. Molecular genotyping of single nucleotide polymorphisms
The molecular genotyping data for pfcrt K76T, pfmdr1 N86Y, Y184F and D1246Y are shown in Fig. 3. A significant decrease occurred in the asymptomatic samples in the pfcrt 76T allele frequency between 2005 and 2013 (99.2–64.7%, p < 0.001) and in the pfmdr1 86Y allele frequency between 2009 and 2013 (89.4–66.7%, p = 0.03). There was no significant difference in the pfmdr1 haplotype frequencies over time. The pfmdr1 YYY and YYD haplotypes were most common, followed by pfmdr1 NFD and NYD (Table 5).
Fig. 3.
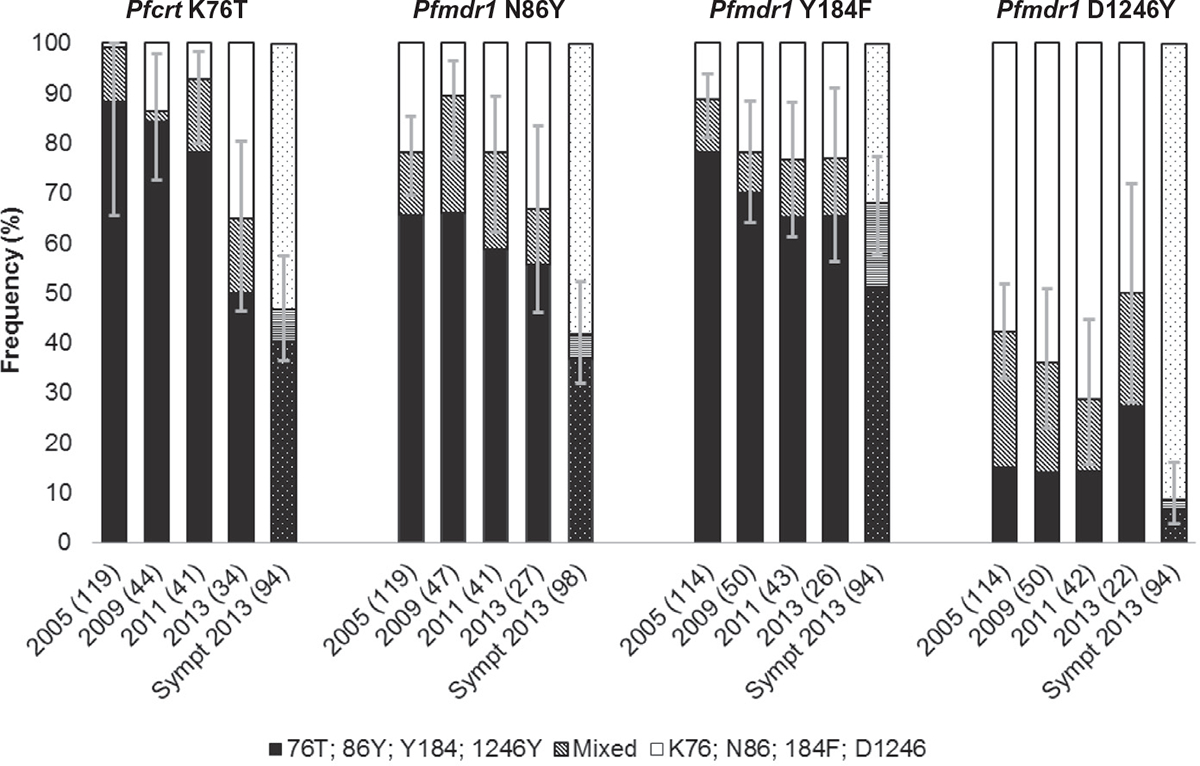
Molecular genotyping of SNPs in P. falciparum infections. Clustered, stacked bar chart showing the frequency of polymorphisms associated with amodiaquine resistance. Error bars represent 95% confidence intervals of the proportion of isolates containing resistance alleles (either alone or in mixed infections). The numbers of individual infections genotyped for each locus are shown in brackets for each year. Solid: asymptomatic samples; patterned symptomatic samples.
Table 5.
Pfmdr1 haplotype frequencies.
|
Pfmdr1 haplotype |
Total (%) N = 171 |
2005 (%) N = 64 |
2009 (%) N = 35 |
2011 (%) N = 31 |
2013 (%) N = 18 |
p a | 2009–2013b (%) N = 84 |
2013 symptomatic samples (%) N = 87 |
p c |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| YFY | 0.8 | 0.0 | 0.0 | 3.2 | 5.6 | - | 2.4 | 0.0 | - |
| NFY | 1.1 | 2.1 | 0.0 | 0.0 | 0.0 | - | 0.0 | 1.1 | - |
| NYY | 1.1 | 2.1 | 0.0 | 0.0 | 5.6 | - | 1.2 | 0.0 | - |
| YFD | 3.4 | 3.2 | 5.7 | 9.7 | 5.6 | - | 7.1 | 0.0 | - |
| NYD | 21.1 | 18.1 | 20.0 | 16.1 | 11.1 | 0.91 | 16.7 | 28.7 | 0.07 |
| NFD | 23.0 | 11.7 | 8.6 | 16.1 | 16.7 | 0.68 | 13.1 | 44.8 | <0.001 |
| YYY | 23.8 | 33.0 | 34.3 | 22.6 | 33.3 | 0.72 | 29.8 | 8.1 | <0.001 |
| YYD | 53.6 | 66.0 | 77.1 | 48.4 | 50.0 | 0.06 | 60.7 | 33.3 | <0.001 |
p calculated by Fishers exact test, comparing frequencies between all year.
Merged haplotype data from 2009–2013 to increase sample size.
p calculated by Fisher’s exact test, comparing merged 2009–2013 asymptomatic frequencies with 2013 symptomatic samples.
The frequencies of pfmdr1 86Y and 1246Y were significantly higher in the 2013 asymptomatic samples when compared with the symptomatic samples (p = 0.03 and p < 0.001, respectively). There was no significant difference in the frequency of pfcrt 76T (p = 0.11). The pfmdr1 YYY and YYD haplotypes were significantly more frequent in the asymptomatic samples (p < 0.001), whereas the pfmdr1 NFD was more frequent in the symptomatic samples (p < 0.001) (Table 5).
The statistical analyses were repeated after excluding mixed infections to ensure these were not biasing the results; removing mixed infections did not significantly alter the results.
4. Discussion
There is a declining, albeit persistent, reservoir of parasites present at low-densities in asymptomatic individuals in Zanzibar. The ability to accurately detect and subsequently treat all parasite carriers is fundamental for malaria elimination efforts.
As the prevalence of PCR-determined malaria declined in Zanzibar so did the relative proportion of infections that were detectable by microscopy or mRDT. High prevalence of sub-microscopic malaria has been reported in many other endemic areas (Ganguly et al., 2013; Golassa et al., 2013; Manjurano et al., 2011; Roper et al., 1996; Stresman et al., 2014). Okell et al. estimated that in low-transmission settings, where PCR-determined prevalence is less than 10%, microscopy would only detect on average 12% of all PCR-detectable infections (Okell et al., 2009, 2012). Our data confirm this and, importantly, our qPCR data showed that the majority of infections had densities much lower than the detection limits of both microscopy (50–100 p/μL) and mRDT (100 p/μL).
Several studies have demonstrated the transmission potential of low-density malaria infections (Bousema et al., 2014; Mawili-Mboumba et al., 2013), although their contribution to overall transmission has been debated (Lin et al., 2014). In Zanzibar, where it appears that subpatent infections constitute a large proportion of the parasite reservoir, it is important that infections are monitored using appropriate molecular tools (Cook et al., 2014). Despite the high sensitivity of PCR, infections with densities below or close to the PCR detection level are still missed. This limitation is also apparent in the low reproducibility of PCR-detectable low-density infections (Costa et al., 2014), and is important in discussions on the optimal strategy to clear remaining parasite reservoirs in malaria pre-elimination settings, by active infection detection or mass drug administration.
P. falciparum remained the predominant species throughout the study period. The prevalence was highest in older children and young adults (aged 5–25 years); with differential distribution between the two study districts. As malaria prevalence declines the epidemiology of malaria shifts, resulting in temporal heterogeneity and concentration of malaria in particular localities and demographic groups (Cotter et al., 2013). Characterisation of submicroscopic parasite populations requires highly sensitive molecular methods, but may provide important information for elimination programmes for scale-up and targeting of elimination strategies in remaining pockets of malaria infection (Tietje et al., 2014).
A significant reduction in the complexity of infection was observed between 2005 and 2009. MOI provides an alternative measure of transmission intensity (Stresman et al., 2014; Tusting et al., 2014), and our results correlate well with the reduction of transmission in Zanzibar. The heterozygosity, on the other hand, remained high over the study period with no significant difference between the years. The results are comparable to other geographical areas of sub-Saharan Africa (Anderson et al., 2000; Mwingira et al., 2011; Zhong et al., 2007), and suggest that the remaining parasite population is of high diversity. analysis showed no detectable reduction in the effective population size or bottlenecks during the transition from high to low transmission in Zanzibar. Similar findings have been reported from the highlands in western Kenya and Malawi (Sisya et al., 2015; Zhong et al., 2007), in contrast to the low genetic diversity and high genetic differentiation that has been observed in low transmission settings in parts of Asia and South America (Anderson et al., 2000). Changes in HE occur slowly over time, and recent changes in malaria transmission combined with small sample sizes may therefore not provide the means to detect subtle changes in as a measure of genetic diversity (Sisya et al., 2015).
Artesunate–amodiaquine was deployed as a first-line treatment for uncomplicated malaria in all public health facilities in 2003. Intriguingly, a substantial decline in the pfcrt 76T and pfmdr1 86Y frequency occurred during the study period among PCR-determined parasite carriers, even though these SNPs have been associated, both in vitro and in vivo, with resistance to amodiaquine (Echeverry et al., 2007; Folarin et al., 2011; Picot et al., 2009; Venkatesan et al., 2014). A similar reduction was observed in febrile cases during the period 2002–2010 in Zanzibar (Froberg et al., 2012). These apparently controversial results may be attributed to the fitness costs associated with these SNPs (Babiker et al., 2009; Rosenthal, 2013). Resistance-mediating polymorphisms, that are associated with substantial fitness costs, result in the selection of wild-type alleles both in vitro and in areas of diminishing drug pressure (Babiker et al., 2009; Froberg et al., 2013; Rosenthal, 2013). A typical example is the return of the chloroquine sensitive form (pfcrt K76) in areas where chloroquine use has been discontinued due to high levels of resistance (Gharbi et al., 2013; Kublin et al., 2003; Ndiaye et al., 2012; Nkhoma et al., 2007; Wurtz et al., 2012). Genetic dilution by imported parasites from the African main-land could be another contributing factor to the decline in frequency since Artemether–lumefantrine, which selects in the opposite direction of artesunate–amodiaquine (pfcrt K76, pfmdr1 N86, 184F, D1246) (Venkatesan et al., 2014), is the first-line treatment in main-land Tanzania and Kenya. Importation of parasites may also explain the sustained high heterozygosity in Zanzibar (Zhong et al., 2007).
Interestingly, in the to-date most extensive report (a pooled analysis of individual patient data), none of the analysed pfcrt or pfmdr1 parasite genotypes were significant risk factors for recrudescent infections (treatment failures), but pfmdr1 86Y, 1246Y were selected in re-infections after treatment with artesunate–amodiaquine (Venkatesan et al., 2014). Furthermore, in patients treated with artesunate–amodiaquine, parasites carrying pfmdr1 86Y, 1246Y, or pfcrt 76T appeared earlier during follow-up than those carrying pfmdr1 N86, D1246, or pfcrt K76, indicating an advantage of harbouring these mutations when under direct drug pressure (Venkatesan et al., 2014). However, prolonged use of artesunate–amodiaquine does not seem to select for molecular markers that have been associated with amodiaquine resistance (Froberg et al., 2012), which is important for the continued efficacy of this ACT since treatment outcome is largely dependent on the long half-life partner drug (Venkatesan et al., 2014).
Frequencies of the pfmdr1 86Y, 1246Y SNPs and the pfmdr1 YYY and YYD haplotypes were significantly higher in asymptomatic infections than in symptomatic infections in 2013. Similar findings have recently been reported in Uganda and Benin, where the prevalence of wild-type genotypes were significantly higher in febrile children compared to asymptomatic children, suggesting greater virulence for wild type parasites (Ogouyemi-Hounto et al., 2013; Tukwasibwe et al., 2014). It could be that asymptomatic infections represent an important reservoir for resistance genes that confer a fitness disadvantage relative to wild-type alleles and may therefore contribute to the epidemiology of drug resistant malaria (Brown et al., 2012). However, additional studies are required to determine if fitness is associated with virulence, and how it may influence onward transmission and spread of antimalarial drug resistance. Most likely these interactions are dependent on a complex interplay between, for example, parasite fitness, host immunity, transmission intensity, and drug pressure (Rosenthal, 2013).
There are several limitations with this study. Asymptomatic malaria was identified by PCR in healthy individuals participating in cross-sectional surveys. This study design gives a “snap-shot in time” and limits the understanding of the history, progression and dynamics of these infections. Importantly, reliable information on fever and treatments histories was not available for the study subjects, and the lack of gametocyte carriage data gives little insight regarding the contribution of these infections to malaria transmission. There are also several inherent limitations with PCR, such as the risk of false positivity and the limited sensitivity of RFLP assays to detect mixed infections. Finally, genotyping of molecular markers may be a useful tool for early detection of selection/development of resistance but should ideally be followed up by clinical trials to assess the true efficacy of artesunate–amodiaquine in Zanzibar.
5. Conclusions
There is a declining, albeit persistent, reservoir of parasites present at low-densities in asymptomatic individuals in Zanzibar. This study revealed important characteristics of the remaining parasite population, including intriguing temporal trends in molecular markers associated with antimalarial resistance, which need to be further investigated.
Supplementary Material
Acknowledgements
We would like to thank all participants, staff members, and ZAMEP employees involved in the cross-sectional surveys for their dedicated participation. This work was supported by the Swedish Civil Contingencies Agency (MSB) [Grant number 2010-7991], the Swedish Medical Research Council (VR) [Grant numbers 2009-3785 and 2013-6594], and Goljes Foundation. In memoriam of Ali K. Abass, a much missed friend and colleague.
Footnotes
Conflict of interest
All authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2015.04.018.
References
- Alonso PL, Tanner M, 2013. Public health challenges and prospects for malaria control and elimination. Nat. Med. 19, 150–155. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP, 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17, 1467–1482. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Hastings IM, Swedberg G, 2009. Impaired fitness of drug-resistant malaria parasites: evidence and implication on drug-deployment policies. Expert Rev. Anti Infect. Ther. 7, 581–593. [DOI] [PubMed] [Google Scholar]
- Beer N, Ali AS, Shakely D, Elfving K, Al-Mafazy AW, Msellem M, Petzold M, Bjorkman A, Kallander K, 2013. High effective coverage of vector control interventions in children after achieving low malaria transmission in Zanzibar Tanzania. Malar. J. 12, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai A, Ali AS, Kachur SP, Martensson A, Abbas AK, Khatib R, Al-Mafazy AW, Ramsan M, Rotllant G, Gerstenmaier JF, Molteni F, Abdulla S, Montgomery SM, Kaneko A, Bjorkman A, 2007. Impact of artemisinin-based combination therapy and insecticide-treated nets on malaria burden in Zanzibar. PLoS Med. 4, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousema T, Okell L, Felger I, Drakeley C, 2014. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat. Rev. Microbiol. [DOI] [PubMed] [Google Scholar]
- Brown T, Smith LS, Oo EK, Shawng K, Lee TJ, Sullivan D, Beyrer C, Richards AK, 2012. Molecular surveillance for drug-resistant Plasmodium falciparum in clinical and subclinical populations from three border regions of Burma/Myanmar: cross-sectional data and a systematic review of resistance studies. Malar. J. 11, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J, Xu W, Msellem M, Vonk M, Bergstrom B, Gosling R, Al-Mafazy AW, McElroy P, Molteni F, Abass AK, Garimo I, Ramsan M, Ali A, Martensson A, Bjorkman A, 2014. Mass screening and treatment using a falciparum-specific rapid diagnostic test did not reduce malaria incidence in Zanzibar. J. Infect. Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa DC, Madureira AP, Amaral LC, Sanchez BA, Gomes LT, Fontes CJ, Limongi JE, de Brito CF, Carvalho LH, 2014. Submicroscopic malaria parasite carriage: how reproducible are polymerase chain reaction-based methods? Mem. Inst. Oswaldo Cruz 109, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter C, Sturrock HJ, Hsiang MS, Liu J, Phillips AA, Hwang J, Gueye CS, Fullman N, Gosling RD, Feachem RG, 2013. The changing epidemiology of malaria elimination: new strategies for new challenges. Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde AA, Fofana B, Sagara I, Sidibe B, Toure S, Dembele D, Dama S, Ouologuem D, Dicko A, Doumbo OK, 2008. Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am. J. Trop. Med. Hyg. 78, 455–461. [PubMed] [Google Scholar]
- Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Targett GA, Greenwood BM, Warhurst DC, 1997. Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology 114 (Pt 3), 205–211. [DOI] [PubMed] [Google Scholar]
- Echeverry DF, Holmgren G, Murillo C, Higuita JC, Bjorkman A, Gil JP, Osorio L, 2007. Short report: polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am. J. Trop. Med. Hyg. 77, 1034–1038. [PubMed] [Google Scholar]
- Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AM, Happi CT, 2011. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 120, 224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froberg G, Jornhagen L, Morris U, Shakely D, Msellem MI, Gil JP, Bjorkman A, Martensson A, 2012. Decreased prevalence of Plasmodium falciparum resistance markers to amodiaquine despite its wide scale use as ACT partner drug in Zanzibar. Malar. J. 11, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froberg G, Ferreira PE, Martensson A, Ali A, Bjorkman A, Gil JP, 2013. Assessing the cost–benefit effect of a Plasmodium falciparum drug resistance mutation on parasite growth in vitro. Antimicrob. Agents Chemother. 57, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Saha P, Guha SK, Biswas A, Das S, Kundu PK, Maji AK, 2013. High prevalence of asymptomatic malaria in a tribal population in eastern India. J. Clin. Microbiol. 51, 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi M, Flegg JA, Hubert V, Kendjo E, Metcalf JE, Bertaux L, Guerin PJ, Le Bras J, Members of the French National Reference Centre for Imported Malaria, S., Aboubaca A, Agnamey P, Angoulvant A, Barbut P, Basset D, Belkadi G, Bellanger AP, Bemba D, Benoit-Vica F, Berry A, Bigel ML, Bonhomme J, Botterel F, Bouchaud O, Bougnoux ME, Bouree P, Bourgeois N, Branger C, Bret L, Buret B, Casalino E, Chevrier S, Conquere de Monbrison F, Cuisenier B, Danis M, Darde ML, De Gentile L, Delarbre JM, Delaunay P, Delaval A, Desoubeaux G, Develoux M, Dunand J, Durand R, Eloy O, Fauchet N, Faugere B, Faye A, Fenneteau O, Flori P, Fontrouge M, Garabedian C, Gayandrieu F, Godineau N, Houze P, Houze S, Hurst JP, Ichou H, Lachaud L, Lebuisson A, Lefevre M, LeGuern AS, Le Moal G, Lusina D, Machouart MC, Malvy D, Matheron S, Maubon D, Mechali D, Megarbane B, Menard G, Millon L, Aiach MM, Minodier P, Morelle C, Nevez G, Parola P, Parzy D, Patey O, Patoz P, Penn P, Perignon A, Picot S, Pilo JE, Poilane I, Pons D, Poupart M, Pradines B, Raffenot D, Rapp C, Receveur MC, Sarfati C, Senghor Y, Simon F, Siriez JY, Taudon N, Thellier M, Thouvenin M, Toubas D,. Longitudinal study assessing the return of chloroquine susceptibility of Plasmodium falciparum in isolates from travellers returning from West and Central Africa, 2000–2011. Malar. J. 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golassa L, Enweji N, Erko B, Aseffa A, Swedberg G, 2013. Detection of a substantial number of sub-microscopic Plasmodium falciparum infections by polymerase chain reaction: a potential threat to malaria control and diagnosis in Ethiopia. Malar. J. 12, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Bjorkman A, 2006. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect. Genet. Evol. 6, 309–314. [DOI] [PubMed] [Google Scholar]
- Holmgren G, Hamrin J, Svard J, Martensson A, Gil JP, Bjorkman A, 2007. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect. Genet. Evol. 7, 562–569. [DOI] [PubMed] [Google Scholar]
- Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL, 2007. Amodiaquine and artemether–lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51, 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Jaroensuk J, Leimanis ML, Russell B, McGready R, Day N, Snounou G, Nosten F, Imwong M, 2012. Long-term storage limits PCR-based analyses of malaria parasites in archival dried blood spots. Malar. J. 11, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishengoma DS, Lwitiho S, Madebe RA, Nyagonde N, Persson O, Vestergaard LS, Bygbjerg IC, Lemnge MM, Alifrangis M, 2011. Using rapid diagnostic tests as source of malaria parasite DNA for molecular analyses in the era of declining malaria prevalence. Malar. J. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF, 2011. Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J. Clin. Microbiol. 49, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV, 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187, 1870–1875. [DOI] [PubMed] [Google Scholar]
- Lin JT, Saunders DL, Meshnick SR, 2014. The role of submicroscopic parasitemia in malaria transmission: what is the evidence? Trends Parasitol. 30, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjurano A, Okell L, Lukindo T, Reyburn H, Olomi R, Roper C, Clarke TG, Joseph S, Riley EM, Drakeley C, 2011. Association of sub-microscopic malaria parasite carriage with transmission intensity in north-eastern Tanzania. Malar. J. 10, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawili-Mboumba DP, Nikiema R, Bouyou-Akotet MK, Bahamontes-Rosa N, Traore A, Kombila M, 2013. Sub-microscopic gametocyte carriage in febrile children living in different areas of Gabon. Malar. J. 12, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris U, Aydin-Schmidt B, Shakely D, Martensson A, Jornhagen L, Ali AS, Msellem MI, Petzold M, Gil JP, Ferreira P, Bjorkman A, 2013. Rapid diagnostic tests for molecular surveillance of Plasmodium falciparum malaria - assessment of DNA extraction methods and field applicability. Malar. J. 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msellem MI, Martensson A, Rotllant G, Bhattarai A, Stromberg J, Kahigwa E, Garcia M, Petzold M, Olumese P, Ali A, Bjorkman A, 2009. Influence of rapid malaria diagnostic tests on treatment and health outcome in fever patients, Zanzibar: a crossover validation study. PLoS Med. 6, e1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck HP, Snounou G, Felger I, Olliaro P, Mugittu K, 2011. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar. J. 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye M, Faye B, Tine R, Ndiaye JL, Lo A, Abiola A, Dieng Y, Ndiaye D, Hallett R, Alifrangis M, Gaye O, 2012. Assessment of the molecular marker of Plasmodium falciparum chloroquine resistance (Pfcrt) in Senegal after several years of chloroquine withdrawal. Am. J. Trop. Med. Hyg. 87, 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkhoma S, Molyneux M, Ward S, 2007. Molecular surveillance for drug-resistant Plasmodium falciparum malaria in Malawi. Acta Trop. 102, 138–142. [DOI] [PubMed] [Google Scholar]
- Noor AM, Kinyoki DK, Mundia CW, Kabaria CW, Mutua JW, Alegana VA, Fall IS, Snow RW, 2014. The changing risk of Plasmodium falciparum malaria infection in Africa: 2000–10: a spatial and temporal analysis of transmission intensity. Lancet 383, 1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ, 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate–amodiaquine in Uganda. Antimicrob. Agents Chemother. 51, 3023–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogouyemi-Hounto A, Ndam NT, Kinde Gazard D, d’Almeida S, Koussihoude L, Ollo E, Azagnandji C, Bello M, Chippaux JP, Massougbodji A, 2013. Prevalence of the molecular marker of Plasmodium falciparum resistance to chloroquine and sulphadoxine/pyrimethamine in Benin seven years after the change of malaria treatment policy. Malar. J. 12, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okell LC, Ghani AC, Lyons E, Drakeley CJ, 2009. Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J. Infect. Dis. 200, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Okell LC, Bousema T, Griffin JT, Ouedraogo AL, Ghani AC, Drakeley CJ, 2012. Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat. Commun. 3, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P, 2009. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar. J. 8, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiagen, 2012. QIAamp DNA mini and Blood Mini Handbook. Third ed., p. 42. [Google Scholar]
- Roper C, Elhassan IM, Hviid L, Giha H, Richardson W, Babiker H, Satti GM, Theander TG, Arnot DE, 1996. Detection of very low level Plasmodium falciparum infections using the nested polymerase chain reaction and a reassessment of the epidemiology of unstable malaria in Sudan. Am. J. Trop. Med. Hyg. 54, 325–331. [DOI] [PubMed] [Google Scholar]
- Rosenthal PJ, 2013. The interplay between drug resistance and fitness in malaria parasites. Mol. Microbiol. 89, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisya TJ, Kamn’gona RM, Vareta JA, Fulakeza JM, Mukaka MF, Seydel KB, Laufer MK, Taylor TE, Nkhoma SC, 2015. Subtle changes in Plasmodium falciparum infection complexity following enhanced intervention in Malawi. Acta Trop. 142, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresman GH, Stevenson JC, Ngwu N, Marube E, Owaga C, Drakeley C, Bousema T, Cox J, 2014. High levels of asymptomatic and subpatent Plasmodium falciparum parasite carriage at health facilities in an area of heterogeneous Malaria transmission intensity in the Kenyan highlands. Am. J. Trop. Med. Hyg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietje K, Hawkins K, Clerk C, Ebels K, McGray S, Crudder C, Okell L, LaBarre P, 2014. The essential role of infection-detection technologies for malaria elimination and eradication. Trends Parasitol. 30, 259–266. [DOI] [PubMed] [Google Scholar]
- Tukwasibwe S, Mugenyi L, Mbogo GW, Nankoberanyi S, Maiteki-Sebuguzi C, Joloba ML, Nsobya SL, Staedke SG, Rosenthal PJ, 2014. Differential prevalence of transporter polymorphisms in symptomatic and asymptomatic falciparum malaria infections in Uganda. J. Infect. Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusting LS, Bousema T, Smith DL, Drakeley C, 2014. Measuring changes in Plasmodium falciparum transmission: precision, accuracy and costs of metrics. Adv. Parasitol. 84, 151–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, Price RN, Martensson A, Rosenthal PJ, Dorsey G, Sutherland CJ, Guerin P, Davis TM, Menard D, Adam I, Ademowo G, Arze C, Baliraine FN, Berens-Riha N, Bjorkman A, Borrmann S, Checchi F, Desai M, Dhorda M, Djimde AA, El-Sayed BB, Eshetu T, Eyase F, Falade C, Faucher JF, Froberg G, Grivoyannis A, Hamour S, Houze S, Johnson J, Kamugisha E, Kariuki S, Kiechel JR, Kironde F, Kofoed PE, LeBras J, Malmberg M, Mwai L, Ngasala B, Nosten F, Nsobya SL, Nzila A, Oguike M, Otienoburu SD, Ogutu B, Ouedraogo JB, Piola P, Rombo L, Schramm B, Some AF, Thwing J, Ursing J, Wong RP, Zeynudin A, Zongo I, Plowe CV, Sibley CH, Group AMMS, Wwarn AL, 2014. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum Malaria after artemether–lumefantrine and artesunate–amodiaquine. Am. J. Trop. Med. Hyg. 91, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2010. World malaria report. Summary. [Google Scholar]
- Wooden J, Kyes S, Sibley CH, 1993. PCR and strain identification in Plasmodium falciparum. Parasitol. Today 9, 303–305. [DOI] [PubMed] [Google Scholar]
- Wurtz N, Fall B, Pascual A, Diawara S, Sow K, Baret E, Diatta B, Fall KB, Mbaye PS, Fall F, Dieme Y, Rogier C, Bercion R, Briolant S, Wade B, Pradines B, 2012. Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal. Malar. J. 11, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Morris U, Aydin-Schmidt B, Msellem MI, Shakely D, Petzold M, Bjorkman A, Martensson A, 2015. SYBR green real-time PCR-RFLP assay targeting the Plasmodium cytochrome b gene - a highly sensitive molecular tool for malaria parasite detection and species determination. PloS one 10, e0120210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, Temu EA, Yan G, 2007. Plasmodium falciparum genetic diversity in western Kenya highlands. Am. J. Trop. Med. Hyg. 77, 1043–1050. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


