Abstract
The human immunodeficiency virus type 1 (HIV-1) integrase protein (IN) is essential for integration of the viral DNA into host cell chromosomes. Since IN is expressed and assembled into virions as part of the 160-kDa Gag-Pol precursor polyprotein and catalyzes integration of the provirus in infected cells as a mature 32-kDa protein, mutations in IN are pleiotropic and may affect virus replication at different stages of the virus life cycle in addition to integration. Several different phenotypes have been observed for IN mutant viruses, including defects in virion morphology, protein composition, reverse transcription, nuclear import, and integration. Because the effects of mutations in the IN domain of Gag-Pol can not always be distinguished from those of mutations in the mature IN protein, there remains a significant gap in our understanding of IN function in vivo. To directly analyze the function of the mature IN protein itself, in the context of a replicating virus but independently from that of Gag-Pol, we used an approach developed in our laboratory for incorporating proteins into HIV virions by their expression in trans as fusion partners of either Vpr or Vpx. By providing IN in trans as a Vpr-IN fusion protein, our analysis revealed, for the first time, that the mature IN protein is essential for the efficient initiation of reverse transcription in infected cells and that this function does not require the IN protein to be enzymatically (integration) active. Our findings of a direct physical interaction between IN and reverse transcriptase and the failure of heterologous HIV-2 IN protein to efficiently support reverse transcription indicate that this novel function occurs through specific interactions with other viral components of the reverse transcription initiation complex. Studies involving complementation between integration- and DNA synthesis-defective IN mutants further support this conclusion and reveal that the highly conserved HHCC motif of IN is important for both activities. These findings provide important new insights into IN function and reverse transcription in the context of the nucleoprotein reverse transcription complex within the infected cell. Moreover, they validate a novel approach that obviates the need to mutate Gag-Pol in order to study the role of its individual mature components at the virus replication level.
The retroviral integrase (IN) protein catalyzes integration of the provirus and is essential for persistence of the infected state in vivo. Significant progress has been made in our understanding of this critical enzyme, especially its protein structure and the biochemical mechanism of the catalytic integration reaction (5, 14, 30). Human immunodeficiency virus type 1 (HIV-1) IN is expressed and assembled into the virus particle as a part of a larger, 160-kDa Gag-Pol precursor polyprotein (Pr160Gag-Pol) that contains other Gag (matrix, capsid, nucleocapsid, and p6) and Pol (protease, reverse transcriptase [RT], and IN) components. After assembly, Pr160Gag-Pol is proteolytically processed by the viral protease to liberate the individual Gag and Pol components, including the 32-kDa IN protein (for a review, see reference 48). Recent studies on IN function using replicating virus (in vivo) have suggested that in addition to catalyzing integration of the viral cDNA, IN may have other effects on virus replication (23, 35, 41). In studies with proviral clones, it is obvious that IN gene mutations can affect virus replication at multiple levels. Mutations in the IN gene can affect the Gag-Pol precursor protein and alter assembly, maturation, and other subsequent viral events. IN gene mutations can also affect the mature IN protein and its organization within the virus particle and the nucleoprotein preintegration complex. Therefore, such mutations are pleiotropic and may alter virus replication through various mechanisms and at different stages in the virus life cycle. At least in part, this likely explains the diverse phenotypes that have been reported for IN mutant viruses. These have included viruses with defects in assembly, virion morphology, reverse transcription, nuclear import, and integration of the provirus (3, 7, 16, 44, 46). While it is obvious that a full understanding of IN function requires analysis in higher-ordered systems that accurately reproduce both the viral and host cell environments, the pleiotropic nature of IN mutations has complicated such studies, and thus there remains a significant gap in our understanding of IN function in vivo.
Numerous in vitro studies have examined the biochemical and genetic properties of retroviral IN proteins and have provided most of the information for the currently accepted mechanism of the integration reaction. Using purified IN and oligonucleotides that represent the viral DNA ends, the in vitro integration reaction proceeds in two steps: IN removes two nucleotides from the 3′ terminus of the viral DNA (terminal cleavage), which is then joined to a break in the cellular DNA (strand transfer) (6, 22, 43). Through amino acid sequence alignment and in vitro activity studies of wild-type and mutant IN proteins, distinct functional domains that are conserved among retroviruses have been identified (12, 15, 33, 53). In the case of HIV-1, the N-terminal domain (residues 1 to 50) contains a highly conserved HHCC motif. Mutation of this motif has variable effects on 3′ processing and strand transfer, and its function remains poorly understood (15, 36, 51, 52). The central region (residues 51 to 212) contains the invariant acidic residues D64, D116, and E152. Mutation of any of these residues causes a loss of all IN activity in vitro, suggesting that this region is the catalytic center of the enzyme (15, 36, 51). The carboxyl-terminal region (residues 213 to 288) is least conserved and possesses nonspecific DNA binding properties. Certain mutations within the C-terminal region may not significantly affect the activity of IN in vitro, while causing a dramatic loss of virus infectivity (10, 16, 56).
Reverse transcription is catalyzed by RT, and although reverse transcription can occur in vitro with recombinant RT, template, and primer, the process is more complex in vivo. In the context of a replicating virus, complete synthesis of the viral cDNA is not as simple as putting together different proteins and nucleic acids; rather, it is a complex, multistep process involving a number of transitional structures. Within the infected cell, reverse transcription takes place in the context of a nucleic acid-protein (nucleoprotein) complex that includes other viral and cellular factors (7, 18, 19, 29, 42). Moreover, synthesis of the viral cDNA is greatly dependent on the proper execution of numerous molecular events that precede reverse transcription. In the case of HIV-1, several viral regulatory proteins are known to affect reverse transcription. Nef mutant viruses exhibit a 5- to 50-fold reduction in DNA synthesis (2, 45). Pseudotyping with vesicular stomatitis virus glycoprotein (VSV-G) complements this defect, indicating that Nef affects uncoating and, in turn, reverse transcription (1). Vif mutant viruses produced by primary cells (nonpermissive) are defective in viral DNA synthesis (47, 54). It remains unknown whether this is due to a direct or indirect effect of Vif on reverse transcription (9, 38). Recently, Tat was shown to be required for efficient reverse transcription in infected cells (25). The nucleocapsid protein, which facilitates strand transfer, may also increase the efficiency of reverse transcription (24, 37). Mutations of the critical proline residues within the capsid domain of p55Gag, which are important for binding cyclophilin A, result in virions that enter cells normally but fail to initiate viral DNA synthesis (4, 21, 40, 49). Taken together, these results have shown that reverse transcription can be affected by multiple factors and at various levels of the virus life cycle. In the absence of a detailed understanding of the molecular mechanisms involved in the formation of infectious virus and the structure and composition of the reverse transcription complex, it is difficult to differentiate between factors that are specific and directly affect reverse transcription and those that involve other steps in virus replication, such as assembly, maturation, and uncoating.
Most in vivo studies of IN function have utilized virus derived from IN mutant proviral DNA, where detection of an integrated provirus was the primary marker for IN activity. Since mutations in the HIV-1 IN gene can cause defects in virus replication prior to integration, assays that rely on integration are not always useful for dissecting the function of IN at the virus replication level. By monitoring for products of reverse transcription in infected cells, certain HIV-1 IN mutants have been found to be defective in steps at or prior to the viral DNA synthesis stage. IN deletion mutant virus or those with mutations in the HHCC motif of IN have been shown to produce 10- to 20-fold less viral DNA following infection (16, 17, 35, 41). Although these viruses are normal in proteolytic processing, virion protein composition, encapsidation of the genomic RNA, and virion-associated RT activity, it has remained unknown at what level(s) such IN mutations affect the virus life cycle (35, 41). The production of infectious retroviral particles occurs through a highly coordinated sequence of events, and numerous examples have illustrated that even subtle changes in this process can have dramatic affects on events early in the virus life cycle, such as reverse transcription. The sensitivity of viral DNA synthesis to events that occur earlier is reflected by the high proportion of virions that are unable to initiate reverse transcription. Thus, it is apparent that an impairment in viral DNA synthesis may be a consequence of other defects and may not necessarily represent an intrinsic defect in reverse transcription itself.
To directly analyze the function of the mature IN protein itself, we used an approach developed in our laboratory that utilizes HIV accessory proteins (Vpr and Vpx) as vehicles to incorporate other proteins into HIV virions by their expression in trans as fusion proteins (39, 57–59). By expression as fusion partners of Vpr, we recently demonstrated that fully functional RT and IN could be efficiently incorporated into HIV-1 particles independently of the Gag-Pol precursor protein (39, 57). Moreover, we have demonstrated that virions derived from an RT- and IN-minus proviral clone were infectious and replicated through a complete cycle of infection when complemented in trans with Vpr-RT and IN fusion proteins. These findings have enabled us to unlink the function of the mature IN protein from that of the Gag-Pol precursor protein and suggested that it was possible to directly analyze IN protein function by introducing mutations into IN without interfering with Gag-Pol function and other late-stage events that could affect reverse transcription. More recent studies from our laboratory and others have shown that the defect in the replication of various IN mutant viruses could be complemented by the Vpr-IN fusion protein (20, 39). However, these studies did not address the specific nature of the defect or the mechanism by which the trans-IN protein was able to complement the impaired phenotype. In this study, we expressed and incorporated functional IN protein into virions in trans to analyze at what stage in the virus life cycle different IN mutations affect HIV-1 reverse transcription. Our analysis has revealed, for the first time, that the mature IN protein itself is required for viral DNA synthesis in vivo. Our analysis also demonstrated that this function of the IN protein is mediated through specific interactions with other components of the nucleoprotein reverse transcription complex after virus entry and uncoating but prior to or at the initiation stage of viral DNA synthesis. These findings provide important new insights into IN function and the reverse transcription process as it occurs in vivo.
MATERIALS AND METHODS
Cells, HIV-1 clones, and expression plasmids.
The 293T, HeLa-CD4, and HeLa CD4-LTR/β-gal indicator cell lines (32) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U of penicillin and 0.1 mg of streptomycin per ml. The wild-type pSG3wt, RT-IN-minus pSG3S-RT, IN-minus pSG3S-IN, and IN mutant pSG3D116A proviral clones have been described previously (38, 57). The RNase H mutant pSG3D443N proviral clone was constructed by replacing the aspartic acid residue with an asparagine residue at position 443 of RT, using PCR methods. This mutation inactivated the RNase H activity of RT. The pHy-SG3 INAA35A clone was constructed by inserting alanine residues into the hygromycin-resistant pHy-SG3 clone at each of the three amino acid positions that comprise the catalytic center of the IN protein (D64A, D116A, and E152A). The Vpr-IN, Vpr-RT, and Vpr-RT-IN expression vectors were described earlier (38, 57). Vpr-INH12A, Vpr-INH16A, Vpr-INΔ22, and Vpr-ΔPCIN were constructed by PCR-based mutagenesis of pLR2P-vprIN. The Vpr-ΔPCIN plasmid was constructed to disrupt normal proteolytic cleavage and liberation of IN protein. The leucine residue at position P1′ of the RT-IN cleavage site was replaced with an isoleucine residue. The Vpr-INF185A expression vector was constructed by inserting the F185A mutant IN (provided by Alan Engelman) into pLR2P-vprIN. The Vpr-HIV-2 IN expression plasmid (pLR2P-vprIN2) was constructed by inserting a BglII/XhoI IN-containing DNA fragment of HIV-2ST into pLR2P-vprIN. The PCR-amplified IN fragment included 30 bp of RT sequence, which was included to preserve the natural protease cleavage site at the N terminus of IN.
Transfections and virus purification.
DNA transfections were performed with monolayer cultures of 293T cells by the calcium phosphate DNA precipitation method according to the recommendations of the manufacturer (Stratagene). Unless otherwise noted, all transfections were performed with 4 μg of each plasmid. Supernatants from the transfected cultures were collected after 48 h, clarified by low-speed centrifugation (1,000 × g, 10 min), and analyzed for RT activity as described previously (13) and for HIV-1 capsid protein concentration by p24 antigen enzyme-linked immunosorbent assay (ELISA) (Coulter Inc.). Virions were pelleted by ultracentrifugation through cushions of 20% sucrose with a Beckman SW41 rotor (125,000 × g, 2 h).
Semiquantitative detection of viral DNA.
The PCR technique used to monitor the synthesis of viral DNA in infected cells was similar to those described earlier (4, 41, 54). Briefly, 500-ng equivalents (p24 antigen) of transfection-derived virus were used to infect one million HeLa-CD4 cells. To control for variation in virus entry by the different mutant viruses, the intracellular p24 antigen concentration of each virus was determined 4 h after infection as described earlier (39). At 4 and 18 h after infection, cells were lysed and total DNA was extracted by organic methods. The DNA extracts were resuspended in 200 μl of distilled water and treated with the DpnI restriction endonuclease to digest bacterially derived plasmid DNA (from transfection). The viral cDNA synthesized de novo following infection is resistant to cleavage by DpnI. To eliminate any effect of differential virus entry on the detection of viral DNA products in infected cells, the DNA extracts were normalized to 250 pg of p24 antigen for PCR amplification. The wild-type DNA extract was adjusted to 250 pg (100%), 100 pg (40%), 40 pg (16%), 16 pg (6.4%), and 6.4 pg (2.5%). The DNA extracts were then subjected to 30 rounds of PCR amplification with primers designed to detect early (R-U5 [sense nucleotides 1 to 22, 5′-GGTCTCTCTGGTTAGACCAGA-3′; antisense nucleotides 181 to 157, 5′-CTGCTAGAGATTTTCCACACTGAC-3′]), intermediate (U3-U5 [sense nucleotides 8687 to 8709, 5′-ACACACAAGGCTACTTCCGTGA-3′; antisense nucleotides 181 to 157, 5′-CTGCTAGAGATTTTCCACACTGAC-3′]), and late (R-gag [sense nucleotides 1 to 22, 5′-GGTCTCTCTGGTTAGACCAGA-3′; antisense nucleotides 355 to 334, 5′-ATACTGACGCTCTCGCACCCAT-3′]) products of reverse transcription. The PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining.
Analysis of RT-IN interaction.
Expression of the recombinant glutathione S-transferase (GST) and GST-IN proteins was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the bacterial pellet was lysed in buffer Y–0.2 M NaCl (50 mM HEPES [pH 7.0], 1 mM EDTA, 0.5% IGEPAL CA-630, 200 mM NaCl, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 2 μg of pepstatin A per ml, 18 μg of phenylmethylsulfonyl fluoride per ml) via six freeze-thaw cycles, followed by lysozyme (0.2 g/liter) treatment for 30 min. The lysates, recovered via centrifugation at 101,000 × g for 30 min, were bound to a 0.5-ml suspension of freshly prepared glutathione beads (G-beads). The beads bound to GST and GST-IN were washed three times with 50 ml of buffer Y–0.2 M NaCl, followed by quantitation of the protein along with standards by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The expression of recombinant HIV-1 RT heterodimers was induced and lysis was performed as described previously (31), except that the lysis buffer contained 300 mM NaCl and HEPES (pH 7.2). After extraction, the samples were centrifuged at 101,000 × g for 30 min, and the lysates were recovered as supernatants. Fifty microliters of beads bound with equimolar quantities of GST or GST-IN proteins was incubated with 100 μl of crude bacterial lysates containing RT heterodimer in HND buffer (20 mM HEPES [pH 7.0], 120 mM NaCl, 4 mM MgCl2, 5 mM dithiothreitol, 0.1% IGEPAL, 100 mg of bovine serum albumin per ml, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 2 μg of pepstatin A per ml, 18 μg of phenylmethylsulfonyl fluoride per ml). The reaction was allowed to proceed with slow mixing for 1 h at 4°C. The beads were washed five times with 1 ml of buffer Y–50 mM NaCl. The washed pellets were resuspended in SDS-PAGE loading buffer, boiled for 10 min, cleared by centrifugation, and applied to SDS-polyacrylamide gels.
RESULTS
Certain IN mutant viruses are impaired in reverse transcription.
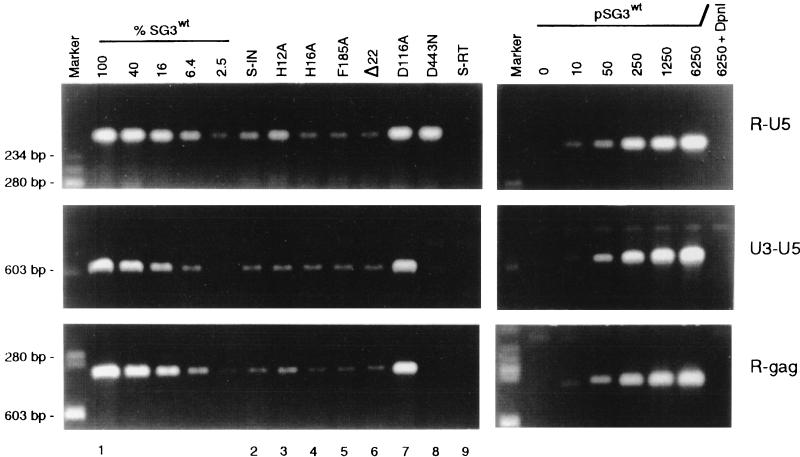
To study the function of the mature IN protein, we first generated and characterized different IN mutant viruses for their ability to synthesize viral DNA. These included S-IN (IN minus), H12A and H16A (the mutations disturb the conserved HHCC motif located in the N terminus), F185A (the mutation is structurally positioned near the catalytic center), Δ22 (the mutation deletes 22 amino acids from the C terminus), and D116A (the mutation destroys enzymatic activity). HeLa-CD4 cells were infected with 500 ng of each virus and analyzed 18 h later for the presence of early (R-U5), intermediate (U3-U5), and late (R-gag) DNA products of reverse transcription. Ten- to 20-fold less early (R-U5) DNA was detected in cells infected with all of the IN mutant viruses, with the exception of D116A (Fig. 1, lanes 2 to 7). Similar changes were detected for the intermediate and late DNA products. No viral DNA was detected in cells that were infected with the control S-RT virus (lacking both RT and IN [57]). The RNase H-defective RT mutant virus (D443N) produced the early R-U5 DNA product in amounts similar to those produced by wild-type virus, but the levels of the intermediate and late DNA products were dramatically reduced, indicating that the strong-stop DNA product is relatively stable for at least 18 h in infected cells. At 4 h after infection, the relative proportions of wild-type and IN mutant DNA products were similar to those measured at 18 h (data not shown). The reverse transcription products (detected by PCR) were confirmed to have been synthesized within the infected cells, since zidovudine completely inhibited the detection of mutant and wild-type viral DNAs (data not shown). Similar concentrations of intracellular CA protein were detected for both the wild-type and mutant viruses, indicating that the impaired DNA synthesis of the IN mutants was not due to a block at the level of virus entry (Table 1). Also, all of the other mutant viruses (except the S-IN mutant) exhibited normal levels of virion-associated RT activity (Table 1). The analysis for two-long-terminal-repeat circular viral DNA (data not shown) confirmed that the nuclear import of nascent viral cDNA of each IN mutant was not impaired in HeLa cells (dividing cells).
FIG. 1.
Mutations in IN can impair reverse transcription. Wild-type (pSG3wt) and mutant (S-IN, H12A, H16A, F185A, Δ22, D116A, S-RT, and D443N) proviral clones were introduced into 293T cells by calcium phosphate DNA transfection methods. Forty-eight hours later, culture supernatants were filtered through 0.45-μm-pore-size filters and analyzed by HIV-1 p24 antigen ELISA (Coulter Inc.). The virus-containing culture supernatants were normalized to 500 ng of p24 antigen (CA), treated with RNase-free DNase H (20 U/ml for 2 h) (Promega Corp.), and placed on cultures of HeLa-CD4 cells at 37°C. After 4 h, the cell monolayers were washed, trypsinized, resuspended in fetal bovine serum, and divided into two aliquots. One aliquot set (which contained 1/10 of the total number of cells) was lysed in phosphate-buffered saline containing 1% Triton X-100 and analyzed by p24 antigen ELISA to quantify intracellular CA protein (Table 1). The other aliquot set was placed back in culture medium at 37°C for an additional 14 h. The cells were then washed, and total DNA was extracted by organic methods. For each DNA extract, 250-pg equivalents (p24 antigen) were analyzed by PCR methods for early (R-U5), intermediate (U3-U5), and late (R-gag) viral DNA products of reverse transcription. The amplified products were resolved on 1.5% agarose gels and stained with ethidium bromide. To assess the relative amount of each of the amplified DNA products, four serial 2.5-fold dilutions of the wild-type (SG3wt) DNA were analyzed in parallel. The undiluted 250-pg sample was arbitrarily set to 100. As standards, 10 to 6,250 copies of the pSG3wt clone were also analyzed by PCR under identical conditions. As a control for the efficiency of DpnI cleavage of potential carryover plasmid DNA, 6,250 copies of pSG3wt DNA were analyzed after digestion with DpnI as described previously (26). The virus origin of the ethidium bromide-stained DNA products was confirmed by Southern blot analysis with a homologous nick-translated probe (data not shown). The ethidium bromide staining intensity of each amplified DNA produce was measured with a Lynx 5000 molecular biology workstation (Applied Imaging, Santa Clara, Calif.). The data shown are from a representative experiment that was repeated three times, each time with independent transfection-derived virus preparations.
TABLE 1.
Analysis of Vpr-IN-complemented and noncomplemented HIV-1 IN mutant viruses
| IN mutant virus | Vpr-INa | RT activitybc | Virion produc-tioncf | RT/CA ratio | Entrydf | Infectivityef |
|---|---|---|---|---|---|---|
| S-IN | − | 103 | 334 | 0.31 | 11.8 | 13 (0.1) |
| + | 107 | 357 | 0.30 | 11.6 | 1,896 (15) | |
| H12A | − | 457 | 1,016 | 0.45 | 10.3 | 76 (0.6) |
| + | 368 | 837 | 0.44 | 9.7 | 3,076 (24) | |
| H16A | − | 393 | 929 | 0.42 | 10.2 | 18 (0.1) |
| + | 378 | 829 | 0.45 | 9.9 | 2,760 (22) | |
| F185A | − | 377 | 868 | 0.43 | 46 | 19 (0.4) |
| + | 321 | 784 | 0.41 | 10.1 | 7,436 (58) | |
| Δ22 | − | 415 | 941 | 0.44 | 11.1 | 62 (0.5) |
| + | 341 | 812 | 0.42 | 10.6 | 3,974 (31) | |
| D116A | − | 401 | 886 | 0.45 | 10.3 | 1,538 (12) |
| SG3 | − | 410 | 934 | 0.44 | 10.4 | 12,820 (100) |
Four micrograms of DNA of each of the viral clones was transfected into 293T cells, either alone (−) or together with 2 μg of the pLR2P-vprIN expression plasmid (+), by calcium phosphate DNA precipitation methods. Forty-eight hours later, the culture supernatants were harvested, clarified by low-speed centrifugation, filtered through 0.45 μm-pore-size filters, and saved as stocks.
RT activity (counts per minute; 103/25 μl) of culture supernatant virus stocks.
HIV-1 p24 antigen (CA protein) concentration (nanograms per milliliter) in culture supernatants. The supernatant stocks were analyzed by HIV-1 antigen ELISA as described by the manufacturer (Coulter Inc.).
Virus entry was quantified by measuring the intracellular HIV-1 CA protein concentration 4 h after infection of the HeLa-CD4 cells with 500-ng equivalents (p24 antigen) of each virus stock. The results represent nanograms of p24 antigen per 106 cells.
Virus infectivity was measured by the MAGI assay as described earlier (32). The infectivity of mutant virus relative to that of wild-type virus is indicated in parentheses. The infectivity of wild-type SG3 virus was arbitrarily set to 100.
Values represent means from at least two independent assays.
Since circular forms of viral DNA in the nuclei of infected cells can express Tat protein, we exploited the HeLa-CD4-LTR/β-gal cell line (32) as a biological indicator for a defect in viral DNA synthesis. Table 1 shows that the infectivity of the IN mutant viruses was decreased 20- to 100-fold compared to that of the integration-defective D116A virus (which supports wild-type levels of viral DNA synthesis). These results indicated that the loss of infectivity could not be explained solely by a defect in integration, but rather, they suggested a defect at the level of viral DNA synthesis. These results are consistent with our analysis for viral DNA. Taken together, these results show that mutations in certain regions of IN can impair DNA synthesis in infected cells.
trans-IN protein restores viral DNA synthesis to IN mutant viruses.
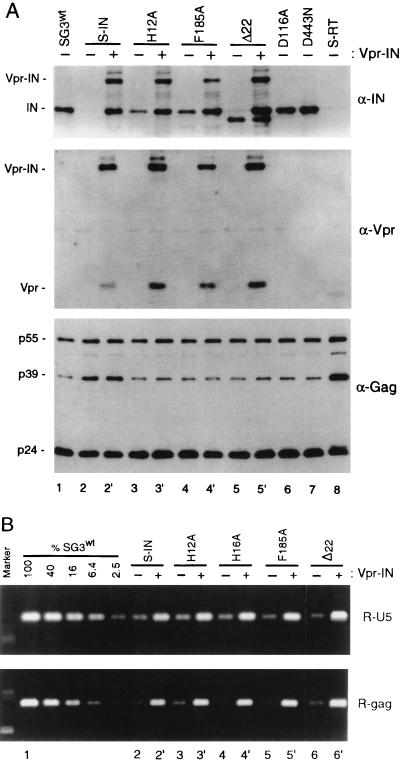
Mutations in the IN gene may affect virus replication at multiple levels. To examine whether changes in the Gag-Pol precursor protein or the IN protein were responsible for the defect in reverse transcription, the Vpr-IN fusion protein was expression in trans with each of the different IN mutant viruses. Figure 2A confirms that the Vpr-IN fusion protein was efficiently packaged and processed by the viral protease to liberate the mature 32-kDa IN protein. Mutant viruses that contained the Vpr-IN fusion protein (trans-IN) exhibited a 5- to 10-fold increase in the synthesis of early, intermediate, and late viral DNA products (Fig. 2B). These results demonstrate that the Vpr-IN fusion protein, which was assembled into virions together with mutant Gag-Pol precursor protein (S-IN, H12A, H16A, F185A, or Δ22, respectively), restored viral DNA synthesis. Using the MAGI assay, we confirmed that Vpr-IN also restored virus infectivity, to between 15 and 58% of that of wild-type virus (Table 1). It is important to note that while the trans-IN protein complemented viral DNA synthesis and infectivity, it did not correct the defect in Gag processing (excess p39) or virion-associated RT activity (Fig. 2A and Table 1).
FIG. 2.
Analysis of Vpr-IN-complemented virions. Four micrograms of the wild-type and mutant proviral DNA clones was individually transfected (−) into 293T cells and cotransfected (+) with the pLR2P-vprIN expression plasmid. Forty-eight hours later, the culture supernatants were collected, passed through 0.45-μm-pore-size filters, and analyzed for HIV-1 p24 antigen concentration by ELISA. (A) Immunoblot analysis. One-half of the filtered supernatant was centrifuged (125,000 × g for 2 h) over cushions of 20% sucrose. The pellets were lysed and examined by immunoblot analysis with anti-IN (α-IN) (top), anti-Vpr (middle), and anti-Gag (bottom) antibodies as described earlier (57). Vpr-IN-containing H16A virions were identical to the H12A virions (data not shown). (B) The trans-IN protein rescues viral DNA synthesis. Five hundred nanograms of wild-type virus and each of the mutant viruses was used to infect cultures of HeLa-CD4 cells. After 4 h, the cell monolayers were washed, trypsinized, resuspended in fetal bovine serum, and divided into two aliquots. One aliquot set was analyzed by p24 antigen ELISA as described for Fig. 1. The other aliquot set was placed back in culture medium at 37°C. At 18 h postinfection, the cells were washed and total DNA was extracted by organic methods. The extracts were normalized for intracellular CA protein concentration and analyzed by PCR for viral DNA products of reverse transcription as described for Fig. 1. The data are from a representative experiment that was repeated three times, each time with independent virus preparations.
trans-IN protein acts after virus assembly to promote viral DNA synthesis.
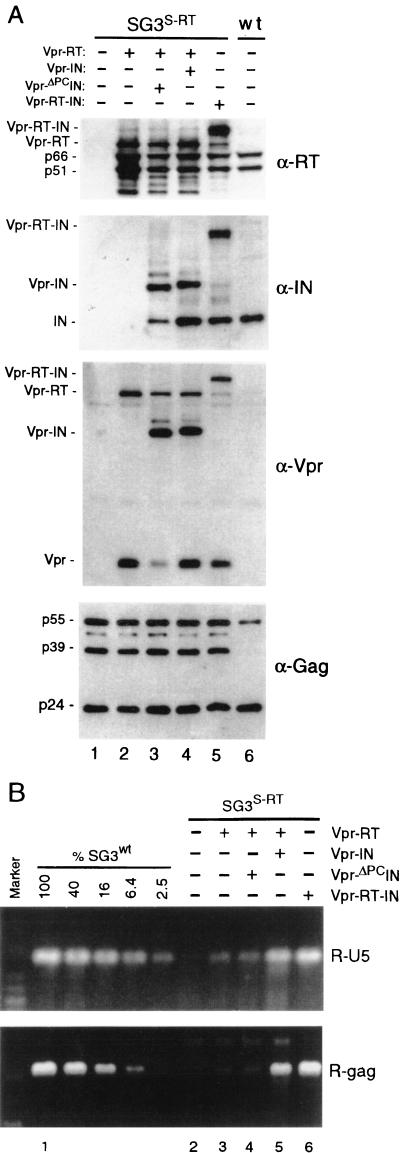
To further analyze the effect of the IN protein on reverse transcription, the RT-IN-minus provirus (S-RT) (57) was complemented with the Vpr-RT fusion protein (Fig. 3A). While high RT activity levels were associated with the progeny virions, they remained severely defective in DNA synthesis. However, when S-RT virus was complemented with both Vpr-RT and Vpr-IN together or with Vpr-RT-IN, viral DNA synthesis was increased 40- to 80-fold compared with that of Vpr-RT-complemented virions (Fig. 3B). By performing complementation experiments with the same virus background (S-RT), we were able to directly examine the effect of the IN protein on reverse transcription. Taken together, these results strongly suggest that the failure of the IN mutant viruses to efficiently support reverse transcription was not due to a defect at the level of Pr160Gag-Pol but, rather, that the mature IN protein is important for viral DNA synthesis in vivo.
FIG. 3.
The trans-IN protein functions after virus assembly and proteolytic processing. Four micrograms of pSG3S-RT DNA was transfected into 293T cells (−) or cotransfected (+) with the Vpr-RT, Vpr-ΔPCIN, and Vpr-RT-IN expression vectors, respectively. (A) Immunoblot analysis. Transfection-derived virions were concentrated from the culture supernatants by ultracentrifugation (125,000 × g for 2 h) through cushions of 20% sucrose. The pellets were lysed and examined by immunoblot analysis with anti-RT (α-RT), anti-IN, anti-Vpr, and anti-Gag antibodies as indicated. (B) The trans-IN protein is required for viral DNA synthesis. Five hundred nanograms of the transfection-derived viruses was used to infect cultures of HeLa-CD4 cells. DNA products of reverse transcription were prepared and analyzed exactly as described above. The data are from a representative experiment that was repeated three times.
Additional evidence arguing against the notion that the trans IN protein complements a defect during assembly comes from our analysis of the cleavage-deficient Vpr-ΔPCIN fusion protein. In parallel with the experiment described above, S-RT virions were complemented with both Vpr-RT and Vpr-ΔPCIN. Figure 3A (lane 3) shows that these virions contained processed RT and minimal amounts of processed Vpr-ΔPCIN (liberated IN protein was reduced by approximately 10-fold [compare lanes 3 and 4]). For Vpr-ΔPCIN-containing virions, no significant increase in the synthesis of viral DNA compared with that of S-RT virions that were complemented with only Vpr-RT was detected (Fig. 3B, compare lanes 3 and 4). Since the Vpr-IN and Vpr-ΔPCIN fusion proteins are isogeneic (except for the amino acid substitution at position P1′ of the cleavage site) and both assemble into virions as an uncleaved 47-kDa fusion protein, this result indicates that free IN protein is necessary for efficient reverse transcription.
Complementation between IN mutants.
By incorporating IN mutant proteins that exhibited different phenotypes into virions, their interdependence in supporting integration and reverse transcription activities was analyzed. Figure 4A shows that the integration-defective trans-IND116A protein restored DNA synthesis to each of the DNA synthesis-defective (H12A, H16A, F185A, and Δ22) mutant viruses. To examine whether the S-IN, H12A, H16A, F185A, and Δ22 IN mutants could support integration of the provirus, each mutant was incorporated as a Vpr-IN mutant fusion protein into the D116A mutant virus, which is DNA synthesis positive and integration defective. Figure 4B shows that some but not all of the Vpr-IN mutants were able to rescue viral DNA integration. The trans-INF185A and trans-INΔ22 mutants markedly increased the integration frequency. This result demonstrated that the F185A and Δ22 mutants still possessed the integration activity necessary to catalyze provirus formation in vivo and that the integration and reverse transcription functions of IN can occur independently. In contrast, the trans-INH12A and trans-INH16A mutants did not efficiently support integration, indicating that mutations in the highly conserved HHCC motif disturb both the reverse transcription and integration functions.
FIG. 4.
Complementation between different IN mutants. (A) Enzymatically defective trans-IN protein supports reverse transcription. Four micrograms of the S-IN, H12A, H16A, F185A, and Δ22 IN mutant proviral clones was transfected alone and separately cotransfected into 293T cells with 2 μg of the Vpr-IND116A or Vpr-IN expression vector. Forty-eight hours later, supernatant virions were prepared and used to infect HeLa-CD4 cells exactly as described in the legend to Fig. 1. The infected cells were washed 18 h later, and total DNA was extracted and treated with DpnI endonuclease. The late R-gag DNA product of reverse transcription was PCR amplified and analyzed as described above. The data are from a representative experiment that was repeated two times. (B) Complementation of proviral DNA integration. The D116A IN mutant was inserted into the SG3 hygromycin-resistant clone, generating Hy-SG3D116A. The Hy-SG3D116A mutant virus produces wild-type levels of viral DNA yet is integration defective. Four micrograms of Hy-SG3D116A was transfected with 2 μg of the control vector (pLR2P) and individually cotransfected with 2 μg of the Vpr-IN, Vpr-INH12A, Vpr-INH16A, Vpr-INF185A, and Vpr-INΔ22 IN mutant expression vectors, respectively. Since the env region of Hy-SG3D116A contains the hygromycin resistance marker, the virions were pseudotyped by including the pCMV-VSV-G env vector in the transfection reactions. Forty-eight hours after transfection, the culture supernatants were filtered through 0.45-μm-pore-size filters and analyzed for HIV-1 p24 antigen concentration by ELISA. Twenty-five nanograms (p24 antigen) of each pseudotyped virus stock was used to infect cultures of HeLa cells. The infected cells were maintained in hygromycin selection medium for 12 days and then stained to identify resistant colonies. These results were highly reproducible in three independent experiments. The data shown are from a single representative experiment.
Virus type-specific IN is required for efficient viral DNA synthesis.
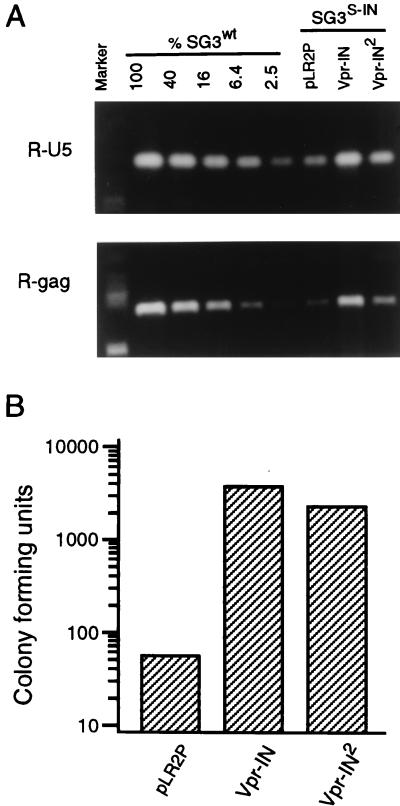
To examine the specificity of these IN functions, the HIV-2 IN protein (IN2) was incorporated into IN-minus (S-IN) virions by expression as a Vpr-IN2 fusion protein. Despite efficient virion incorporation and proteolytic processing of Vpr-IN2 (data not shown), only a modest (2- to 3-fold) increase in HIV-1 DNA synthesis was observed, compared with a 10- to 20-fold increase induced by the homologous IN (Fig. 5A). However, when integration-defective mutant virus was complemented with Vpr-IN2, the integration frequency was increased nearly 100-fold (Fig. 5B). This result indicated that the HIV-2 IN protein was able to associate with the HIV-1 reverse transcription complex but that this alone was not sufficient to support DNA synthesis. This strongly suggests that specific interactions between the homologous IN and other viral components of the reverse transcription complex are required to promote viral cDNA synthesis in vivo.
FIG. 5.
Analysis of heterologous IN. (A) HIV-2 IN protein (IN2) does not efficiently support HIV-1 reverse transcription. Four micrograms of pSG3S-IN was cotransfected into 293T cells with 2 μg of the Vpr-IN, Vpr-IN2, and pLR2P (vector only) expression vectors, respectively. Four micrograms of pSG3wt was also transfected as a control. Forty-eight hours later, supernatant virions were prepared and used to infect HeLa-CD4 cells exactly as described in the legend to Fig. 1. The infected cells were washed 18 h later, and total DNA was extracted and treated with DpnI endonuclease. Early (R-U5) and late (R-gag) viral DNA products of reverse transcription were amplified by PCR and analyzed as described above. The data are from a representative experiment that was repeated three times, each time with independent virus preparations. (B) Complementation of proviral DNA integration. To directly compare the ability of the heterologous trans-IN2 protein to support integration of the provirus with that of the homologous IN protein, the hygromycin-resistant, integration-defective Hy-SG3 INAA35A clone was used for analysis. Hy-SG3 INAA35A contains a mutation in each of the three residues that comprise the catalytic center of the IN protein (D64A, D116A, and E152A) and efficiently synthesizes viral DNA after entry. Four micrograms of Hy-SG3AA35A was cotransfected with 2 μg of the Vpr-IN, and Vpr-IN2 expression plasmids, respectively. The virions were pseudotyped by including the pCMV-VSV-G env vector in the transfection reactions. Forty-eight hours after transfection, the culture supernatants were filtered through 0.45-μm-pore-size filters and analyzed for HIV-1 p24 antigen concentration by ELISA. Twenty-five nanograms (p24 antigen) of each of the pseudotyped virus stocks was used to infect cultures of HeLa cells. The infected cells were maintained in hygromycin selection medium for 12 days and then stained to identify resistant colonies as described earlier (35). These results were highly reproducible in three independent experiments. The data shown are from a single representative experiment.
Direct physical interaction between the HIV-1 RT and IN proteins.
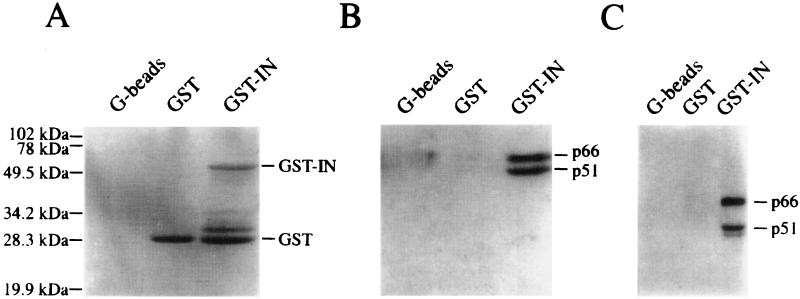
One explanation for the effect of IN on reverse transcription is that IN affects RT via a direct physical interaction. Several observations suggest that HIV RT and IN may form a heterodimeric complex: (i) the two proteins are known to coexist as a complex in some retroviruses (30, 50), (ii) the carboxy-terminal domain of RT (RNase H) and the central core domain of IN are structurally similar (11, 14), and (iii) in murine leukemia virus, IN and RT proteins can be coimmunoprecipitated with antibodies to either protein (27). Therefore, we examined whether a GST-HIV IN fusion protein would interact with an HIV-1 RT heterodimer by using an in vitro binding assay. Figure 6B shows that the GST-IN protein efficiently pulls down recombinant RT heterodimer protein from crude bacterial lysates. The specificity of RT-IN interaction was indicated by the inability of empty G-beads or GST protein-bound G-beads to pull down RT. The possibility that nucleic acids facilitated the association of RT and IN proteins was ruled out by first pretreating the RT-IN reaction mixture with micrococcal nuclease, which did not decrease the amount of RT pulled down (Fig. 6C). The demonstration of a physical interaction between RT and IN suggests that the two proteins exist as a complex within the nucleoprotein reverse transcription complex. While HIV-1 nuclear preintegration complexes have been shown to contain RT (8, 42), no specific role for IN in reverse transcription has been previously demonstrated.
FIG. 6.
Interaction between recombinant HIV-1 RT and IN proteins. (A) Coomassie blue-stained gel showing the G-bead-bound GST and GST-IN proteins used to pull-down the RT heterodimer. Equal quantities of G-beads bound to no protein (lane 1), GST (lane 2), or GST-IN (lane 3) were incubated with crude bacterial lysates in HND buffer containing RT heterodimer. Following extensive washing, the bound proteins were analyzed by SDS-PAGE. (B) Immunoblot analysis showing RT-IN interaction. A duplicate gel run in parallel to that shown in panel A was transferred to nitrocellulose and probed with the 5B2B2 anti-RT monoclonal antibody. The positions of p66 and p51 polypeptides are indicated. (C) The RT-IN interaction is resistant to micrococcal nuclease digestion. The experiment was similar to that described above except that the HND buffer contained 50 mM Tris-HCl (pH 8.0) and 1 mM CaCl2, and prior to addition of the bacterial lysates, the samples were preincubated with 100 U of micrococcal nuclease at 37°C for 10 min, and the nuclease was inactivated with EGTA.
DISCUSSION
From assembly to integration of the provirus, the infectious HIV structure progresses through a succession of precisely coordinated events involving many intra- and intermolecular interactions and rearrangements. A detailed understanding at the virus replication level of the molecular mechanisms that are involved in assembly, maturation, uncoating, and the early stages of reverse transcription remains obscure. In part, this is due to the nature of the process by which the virion proteins are assembled. In the later stages of the virus life cycle, the structural proteins of the virion are synthesized and assembled as precursor polyproteins. Each precursor plays a specific role in the assembly process. After assembly, the structures of the Gag and Gag-Pol precursor polyproteins change due to proteolytic processing. Processing of the Gag and Gag-Pol precursors drives the metamorphosis of the immature (noninfectious) virion into one with a condensed, mature core structure containing the diploid single-stranded viral RNA genome, nucleocapsid, RT, integrase, and primer tRNA (for a review, see reference 48). In the early stage of the virus life cycle, after entry into the host cell, the virus core structure undergoes additional rearrangements (uncoating) to form a nucleoprotein complex structure that supports reverse transcription. After reverse transcription is completed, IN catalyzes integration of the nascent viral cDNA into the host cell chromosomes. It is obvious that mutations in IN (and other domains within Gag and Pol) can affect both late- and early-stage events. As a result, it is inherently difficult to specifically define the effect of such mutations on the virus life cycle. In this study we used trans-complementation methods to distinguish between the effects of mutations in the mature IN protein and its function during the early events of the virus life cycle versus the effects of mutations in the Gag-Pol precursor protein and late stage events. For the first time, our results show that the mature IN protein itself is required for efficient reverse transcription, independent of its enzymatic function. Moreover, our data indicate that the IN protein promotes the initiation step of reverse transcription through virus type-specific interactions with other components that comprise the reverse transcription initiation complex.
Our analysis indicated that a change in the structure and function of the Gag-Pol precursor protein was not responsible for the impairment of viral DNA synthesis. Strong evidence for this comes from experiments showing that trans-IN protein restores infectivity (Table 1) and viral DNA synthesis (Fig. 2 and 3) to viruses that contain a mutated Gag-Pol precursor protein. By analyzing IN mutant virions that were complemented in trans with the cleavage-deficient Vpr-ΔPCIN fusion protein, we further ruled out an assembly defect that could have been complemented by the Vpr-IN fusion protein. The Vpr-IN and Vpr-ΔPCIN fusion proteins are identical, except for the single amino acid change at the P′ position (in the RT-IN cleavage site of Vpr-IN). Therefore, their expression, transport to the surface of the infected cell, and assembly into virions would likely be the same. However, the Vpr-ΔPCIN fusion protein is unable to restore infectivity and viral DNA synthesis above the levels of those for IN-minus virus, indicating that the IN protein supports viral DNA synthesis after virus assembly and proteolytic processing. It is noteworthy that some mature IN protein is detected in virions complemented with the Vpr-ΔPCIN fusion protein (Fig. 3A), yet the virions remain noninfectious. It is likely that the mutation at the P′ residue reduces not only the total extent of cleavage but also the rate at which IN is liberated, which in turn could alter the proper association of IN with the reverse transcription complex during condensation of the virus core. Additional evidence that changes in the structure of the Gag-Pol precursor protein were not responsible for the defect in viral DNA synthesis comes in part from our earlier results, which demonstrated that the Vpr-RT-IN fusion protein, but not the Vpr-RT fusion protein, could efficiently complement (>80% of wild-type levels) the defect in infectivity of RT-IN-minus virus (S-RT) (57). We have now extended those findings by analyzing whether the S-RT mutant virions synthesized viral DNA when complemented with only the Vpr-RT fusion protein. Our data clearly show that in the absence of IN, RT is not sufficient to overcome the defect, even though proteolytic processing of the Gag precursor protein, maturation of the virus particle, and RT/p24 ratios were similar to those for RT-IN-minus virions that were complemented with either Vpr-RT and Vpr-IN together or with Vpr-RT-IN. These results are consistent with the failure of IN-minus virus (S-IN) to synthesize viral DNA. Taken together, these results indicate that the defect in reverse transcription was principally due to changes in the mature IN protein.
Our data suggest that IN is an integral component of the nucleoprotein reverse transcription complex and that through interactions with other components (such as RT, genomic RNA, or tRNA), it is necessary for efficient initiation of viral DNA synthesis. Mutations in the IN protein could change the structure of the reverse transcription complex, which could in turn have a negative effect on reverse transcription. On the other hand, it is also possible that the IN protein directly promotes reverse transcription through specific interactions with other crucial components of the reverse transcription complex. The fact that the wild-type trans-IN protein (Vpr-IN) restores DNA synthesis to IN mutant viruses (H12A, H16A, F185A, and Δ22) indicates that mutant IN protein does not irreversibly disrupt the nucleoprotein complex. However, this result does not exclude the possibility either that the IN mutants fail to associate with the nucleoprotein complex or that they are displaced by the wild-type trans-IN protein. On the contrary, our results indicate that IN mutant proteins do associate with the nucleoprotein preintegration complex. Figure 4 shows that the trans-F185A IN mutant (which did not support reverse transcription) restores integration to D116A mutant virions. Moreover, complementation of the IN mutant viruses (F185A and Δ22) with the trans-D116A IN mutant (Vpr-IND116A) restored DNA synthesis and integration (Fig. 4). Similarly, the heterologous HIV-2 trans-IN protein also efficiently supported integration of HIV-1 DNA but did not support reverse transcription, indicating that the HIV-2 IN protein does associate with the nucleoprotein preintegration complex (Fig. 5). The results of our trans-complementation experiments and our data showing a direct physical interaction between IN and RT clearly show that the mere association of the IN protein with the nucleoprotein complex is not sufficient for DNA synthesis but rather that the IN protein promotes reverse transcription through virus-specific (not cellular) interactions with other viral components in the reverse transcription complex.
Recent studies have suggested that reverse transcription may be regulated at three defined stages: initiation, transition (the point between initiation and elongation), and elongation (34). The efficiency of initiation requires specific and multiple interactions between viral and cellular components, including the viral RNA genome, RT, nucleocapsid, and primer tRNA. Also included are interactions of the primer tRNA with the primer binding site and an A-rich loop located 12 to 17 nucleotides upstream of the primer binding site (28, 55). Disturbances of any of these interactions may cause defects in the initiation of reverse transcription in vivo. Initiation is a slow process and proceeds at a highly reduced processivity compared with elongation (34). This functional distinction suggests that the structures and compositions of the initiation and elongation complexes are different. Our analysis of viral DNA elongation shows that for the H12A, H16A, F185A, and Δ22 IN mutant viruses, the minus-strand strong-stop DNA product was produced in amounts similar to those of the intermediate and late DNA products. Also, it was shown that the trans-IN protein supported the synthesis of minus-strand strong-stop DNA to an extent similar to that for later DNA products. These results indicate that the IN protein is required either prior to or at the initiation stage of reverse transcription. Our data clearly exclude a defect at the level of virus entry (Table 1). Moreover, it seems unlikely that IN protein supports uncoating after virus entry. Recent studies with nef and certain gag mutant viruses have shown that defects in uncoating, which impair virus DNA synthesis, can be overcome if the normal virus entry pathway is bypassed via pseudotyping with the VSV-G envelope (1). Our data show that VSV-G pseudotyping of IN mutant viruses did not overcome the defect in infectivity. Our findings that show a direct physical interaction between the IN and RT proteins strongly suggest that the IN protein is directly involved in reverse transcription in vivo. In the case of the avian retroviruses, the IN protein comprises an integral component of the RT heterodimer; an RT-IN polypeptide makes up the beta subunit (30, 50). Taken together, these results suggest that the IN protein forms an integral part of the reverse transcription initiation complex and specifically promotes interactions between RT, the genomic RNA, and primer tRNA3Lys that facilitate initiation.
The apparent fragility of the reverse transcription initiation complex (34) and the sensitivity of reverse transcription to mutations in any of the three IN subdomains may suggest that the DNA synthesis function of IN could be particularly vulnerable to anti-IN compounds. Of particular interest is the fact that disturbances in the highly conserved HHCC motif caused defects in virus replication at two levels, in both reverse transcription and integration (Fig. 4). Therefore, it is possible that drugs which target this motif could inhibit virus replication at both levels. Finally, our findings validate a novel and powerful approach to dissect important molecular processes of HIV-1 at the virus replication level.
ACKNOWLEDGMENTS
We thank Alan Engelman (Division of Human Retroviruses, Dana-Farber Cancer Institute) for providing the HIV-1 proviral clone mutated at position 185 (F185A) of IN and Stephen Hughes (ABL-Basic Research Program, NCI, Frederick Cancer Research and Development Center) for review of the manuscript and helpful discussions.
This research was supported by National Institutes of Health grants CA73470 and AI39951 (to G.V.K.), and facilities of the Central AIDS Virus and Protein Expression Cores of the Birmingham Center for AIDS Research (grant P30-AI-27767). This research was also supported by a Merit Review Award funded by the Office of Research and Development, Medical Research Services, Department of Veterans Affairs.
REFERENCES
- 1.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari-Lari M A, Gibbs R A. Expression of human immunodeficiency virus type 1 reverse transcriptase in trans during virion release and after infection. J Virol. 1996;70:3870–3875. doi: 10.1128/jvi.70.6.3870-3875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the virus life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukovsky A, Göttlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camuar D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon P M, Byles E D, Kingsman S M, Kingsman A J. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J Virol. 1996;70:651–657. doi: 10.1128/jvi.70.1.651-657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J F, Hostomska Z, Hostomsky Z, Jordan S R, Matthews D A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 12.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 13.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 15.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman A, Liu Y, Chen H, Farzan M, Dyda F. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J Virol. 1997;71:3507–3514. doi: 10.1128/jvi.71.5.3507-3514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMGI(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 19.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher T M, III, Soares M A, McPhearson S, Hui H, Wiskerchen M, Muesing M A, Shaw G M, Leavitt A D, Boeke J D, Hahn B H. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16:5123–5138. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franke E K, Yuan H E, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 23.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 24.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrich D, Ulich C, Gracia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu S C, Court D L, Zweig M, Levin J G. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J Virol. 1986;60:267–274. doi: 10.1128/jvi.60.1.267-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNALys,3. J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 29.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 30.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 31.Kew Y, Qingbin S, Prasad V R. Subunit-selective mutagenesis of Glu-89 residue in human immunodeficiency virus reverse transcriptase. J Biol Chem. 1994;269:15331–15336. [PubMed] [Google Scholar]
- 32.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanchy J-M, Ehresmann C, Le Grice S F J, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcription markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 35.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 37.Li X, Quan Y, Arts E J, Li Z, Preston B D, DeRocquigny H, Roques B P, Darliz J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Wu X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Wu X, Xiao H, Conway J A, Kappes J C. Incorporation of functional human immunodeficiency virus type 1 integrase into virions independent of the Gag/Pol precursor protein. J Virol. 1997;71:7701–7710. doi: 10.1128/jvi.71.10.7704-7710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luban J, Bossolt K, Franke E, Kaplan G V, Goff S P. Human immunodeficiency virus type 1 gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1068. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 41.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pauza C D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 44.Quillent C, Borman A M, Paulous S, Dauguet C, Clavel F. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology. 1996;219:29–36. doi: 10.1006/viro.1996.0219. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz O, Marechal V, Danos O, Heard J-M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin C-G, Taddeo B, Haseltine W A, Farnet C M. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with Vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 49.Thali M, Bukovsky M A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 50.Trentin B, Rebeyrotte N, Mamoun R Z. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-integrase proteins for its activity. J Virol. 1998;72:6504–6510. doi: 10.1128/jvi.72.8.6504-6510.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9601. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vink C, Plasterk R H A. The immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–437. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 54.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakefield J K, Kang S-M, Morrow C D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiskerchen M, Muesing M A. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, Liu H, Xiao H, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Liu H, Xiao H, Conway J A, Kappes J C. Inhibition of human and simian immunodeficiency virus protease function by targeting Vpx-protease-mutant fusion protein into viral particles. J Virol. 1996;70:3378–3384. doi: 10.1128/jvi.70.6.3378-3384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, Liu H O, Xiao H, Kim J, Seshaiah P, Natsoulis G, Boeke J D, Hahn B H, Kappes J C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]