Abstract
The infectious salmon anemia virus (ISAV) is an orthomyxovirus-like virus infecting teleosts. The disease caused by this virus has had major economic consequences for the Atlantic salmon farming industry in Norway, Canada, and Scotland. In this work, we report the cloning and sequencing of an ISAV-specific cDNA comprising 2,245 bp with an open reading frame coding for a predicted protein with a calculated molecular weight of 80.5 kDa. The putative protein sequence shows the core polymerase motifs characteristic of all viral RNA-dependent RNA polymerases. Comparison of the conserved motifs with the corresponding regions of other segmented negative-stranded RNA viruses shows a closer relationship with members of the Orthomyxoviridae than with viruses in other families. The putative ISAV polymerase protein (PB1) has a length of 708 amino acids, a charge of +22 at neutral pH, and a pI of 9.9, which are consistent with the properties of the PB1 proteins of other members of the family. Calculations of the distances between the different PB1 proteins indicate that the ISAV is distantly related to the other members of the family but more closely related to the influenza viruses than to the Thogoto viruses. Based on these and previously published results, we propose that the ISAV comprises a new, fifth genus in the Orthomyxoviridae.
Infectious salmon anemia (ISA) is a severe disease affecting farmed Atlantic salmon with typical pathological changes characterized by severe anemia, leukopenia, ascitic fluids, hemorrhagic liver necrosis, and petecchiae of the viscera (8, 36, 37). Until recently, the disease was not diagnosed outside Norway, but now the ISA virus (ISAV) has been detected in samples from farmed Atlantic salmon on the Canadian east coast and in Scotland (22, 32). The disease is caused by an enveloped virus, with a diameter of approximately 100 nm, budding from endothelial cells lining the heart and blood vessels, as well as from polymorphonuclear leukocytes (13, 27). Electron micrographs of negatively stained virus particles grown in the SHK-1 cell line show round or oval particles ranging from 45 to 140 nm in diameter, with 10-nm-long surface projections (6).
It has recently been reported that the virus has a negative-stranded RNA genome consisting of eight segments ranging from 1 to 2.3 kb in size, which has led to a description of the ISAV as orthomyxovirus-like (20). The relationship to the members of the Orthomyxoviridae is also supported by a recent work characterizing the biochemical, physiochemical, and morphological properties of the ISAV compared to the influenza viruses (9). One ISAV-specific sequence, which is shown to hybridize with genome segment 8, has been cloned (20). However, database searches with the nucleotide and translated putative amino acid sequences did not reveal any significant similarity to any other sequences (20).
The most conserved orthomyxovirid protein has been shown to be the PB1 protein (14, 17, 18, 40), which makes it a good candidate to evaluate the evolutionary relationship between the ISAV and members of the Orthomyxoviridae. The occurrence of consensus regions in the RNA-dependent DNA polymerases has led to the assumption that the sequence similarities may be linked to the existence of a common ancestral genetic element bearing a polymerase function, which emerged only once during the evolution (31). This assumption has been questioned by Zanotto et al. (41). The creation of higher supergroups is especially controversial, whereas the assignment of new virus isolates into genera and families is not ambiguous (41).
In this study, the complete cDNA sequence of the ISAV PB1 is presented. The predicted properties of this protein were compared with those of the polymerases of the recognized members of the Orthomyxoviridae. The core polymerase module of ISAV was aligned with those of other viruses, including members of the segmented negative-stranded RNA virus families Bunyaviridae, Arenaviridae, Paramyxoviridae, and Orthomyxoviridae. From this alignment, phylogenetic trees showing taxonomic allocations of the ISAV were constructed.
MATERIALS AND METHODS
Virus isolation.
Blood collected from ISA-affected Atlantic salmon (Salmo salar L.) in two fish farms (Vinneskarven [1992] and Turøy [1993]) was used as the inoculum in an experimental challenge. Blood and ascites fluid were collected from Atlantic salmon showing typical signs of ISA and were filter sterilized, diluted 1:30 in L-15 medium (BioWhittaker), and used as an inoculum in SHK-1 cells (6). In the course of the present experiments, the virus was passed six times in the SHK-1 cells. In addition, the virus was successfully grown in a head kidney primary cell culture (Atlantic salmon kidney cells [ASK]) from our laboratory.
Virion purification.
The ISA virion was purified in a continuous sucrose gradient and pelleted as previously described (20).
Extraction of RNA.
Virion RNA was extracted by use of TRIzol reagent according to the manufacturer’s recommendations (Gibco-BRL). The RNA was resuspended in diethyl pyrocarbonate (Sigma)-treated water and precipitated for a second time by using 0.5 volumes of 7 M ammonium acetate and 2.5 volumes of 100% ethanol. Total RNA from cell cultures was extracted from flasks of both infected (2 days postinfection) and uninfected cells by the addition of TRIzol reagent and precipitated as described above. The concentration of RNA was estimated by measuring the optical density at 260 nm.
Construction of cDNA libraries.
A total of 2 μg of virion RNA was reverse transcribed into double-stranded cDNA (Universal RiboClone cDNA Synthesis System; Promega). The cDNA fragments were blunt-end ligated into the SmaI site of the pBC KS− phagemid (Stratagene). Two cDNA libraries were also constructed from infected and uninfected cells, using the CapFinder PCR cDNA Synthesis Kit (Clontech). First-strand cDNA was synthesized from 1 μg of total RNA using Moloney murine leukemia virus (MMLV) reverse transcriptase primed by a poly(dT) primer with an anchor sequence. The terminal transferase activity of the MMLV enzyme adds a few deoxycytidine nucleotides to the 3′ end of the cDNA. A primer containing an oligo(G) sequence at the 3′ end anneals to this overhang, and the MMLV reverse transcriptase switches template and continues elongation to the end of the oligonucleotide, which means that primer sites are introduced in both ends of the fragment. Subsequently, the single-stranded cDNA served as template in PCR (15 s at 95°C and 5 min at 68°C for 15 cycles). The pools of PCR products representing the mRNA of ISAV-infected cells and uninfected cells were separately ligated into pGEM-T vectors (Promega).
Assembly of the ISAV polymerase gene.
The cDNA library of infected cells was used as a template for PCR amplifications by using one vector primer (T7 or SP6) in combination with one of two ISAV polymerase-specific primers. The two ISAV-specific primers used were 5′-CAG GTC TAC TGT TGT AGT GAA GGG G (sense) and 5′-CGA ACA TAG AGT TGA ACT CGA AGC TC (antisense). Standard PCR conditions recommended by the supplier of Taq polymerase (Pharmacia) were used. The PCR products were cloned into TA vectors (Invitrogen). The two resulting overlapping fragments were combined to give the complete cDNA sequence.
Northern blot analysis.
Northern blotting was performed according to the instructions in the protocol of the ECL direct nucleic acid labelling and detection system (Amersham). Briefly, approximately 10 μg of total RNA from ISAV-infected (2 days postinfection) or control cells was separated by formaldehyde-agarose gel electrophoresis and blotted onto a Hybond N+ nylon membrane. Approximately 300 ng of cDNA was labelled with horseradish peroxidase and hybridized with the immobilized RNA samples, and signals were detected on Hyperfilm-ECL.
DNA purification and sequencing.
Plasmids were purified with the Wizard Plus SV Miniprep DNA Purification System (Promega). Sequencing reactions were performed on double-stranded plasmid templates with the Thermo Sequenase kit and/or the BigDye Sequencing kit designed for automatic sequencing (Amersham).
Sequence analysis.
GenBank searches were done with BLASTX (3, 11). Protein sequences were analyzed with programs of the University of Wisconsin sequence analysis package (Genetics Computer Group, Madison, Wis.). Multiple alignments were performed with CLUSTAL X (35). Phylogenetic analyses were performed using PAUP 3.1 (34) and programs in PHYLIP 3.5 (10). For each data set, 100 resampled data sets were generated with SEQBOOT. Distance matrices were generated with the PROTDIST program, using the “categories” distance model. Phylogenetic trees were constructed from the distance matrices by the Fitch-Margoliash, neighbor-joining, and UPGMA clustering methods. Additionally, bootstrap analyses were conducted by the parsimony methods with PROTPARS and PAUP 3.1.
Nucleotide sequence accession number.
The nucleotide sequence of the ISAV PB1 gene has been deposited in the EMBL, GenBank, and DDBJ databases under accession no. AJ002475.
RESULTS
Cloning and sequence determination of the ISAV polymerase gene.
From the ISAV genome library, a positive clone with an insert 434 bp long was sequenced and found to be similar to the PB1 protein of orthomyxoviruses (Fig. 1). The similarity was found to be in the central core region motifs previously described for many viral polymerases (31). The full-length sequence of the putative polymerase was obtained by using two partly overlapping clones spanning 2,245 nucleotides [not including the poly(A) tail]. DNA probes from these two clones were used in hybridization reactions with total RNA extracts from ISAV-infected and control cells. The hybridization reactions resulted in one distinct band of about 2.3 kb in size (Fig. 2), which is in accordance with the length of the cDNA sequence. The sequence contains an open reading frame coding for a protein of 708 amino acids with a calculated molecular weight of 80.5 kDa, which is about the same size as for the orthomyxoviruses (Table 1). The charge at neutral pH and the pI were calculated to be +22 and 9.9, respectively, which are very close to the values for the influenza A virus used in this study (Table 1). A pairwise comparison of the orthomyxovirid PB1-like proteins revealed that the ISAV polymerase sequence has 21 to 25% amino acid identity and 44 to 48% amino acid similarity to the other orthomyxoviruses (Table 2).
FIG. 1.
Amino acid sequence alignment of orthomyxovirid PB1 proteins. Motifs preA, A, B, C, D, and E of Poch et al. (31) and Müller et al. (21) are indicated above the sequences. Gaps introduced to optimize the alignment are represented by dashes (–). Conserved residues among the orthomyxoviruses are indicated by asterisks (∗) below the sequences. Residues that are invariant for all RNA polymerases are shown in bold. Residues strictly conserved among negative-stranded RNA viruses are shown in bold and are underlined. Those that are specifically conserved for segmented negative-stranded RNA virus polymerases are underlined. Abbreviations and accession numbers for viruses other than those defined in the text are as follows: InfA, influenza A/PR/8/34 (J02151); InfB, influenza B/AnnArbor/1/66 (M20170); InfC, influenza C/JJ/50 (M28060); THO, Thogoto (AF004985); DHO, Dhori (M65866).
FIG. 2.

Northern blot with two overlapping clones, representing the complete PB1 gene, as probes. Lanes: 1, total RNA from noninfected ASK cells; 2, total RNA from ISAV-infected ASK cells. Molecular size standards (in kilobases) are indicated on the left.
TABLE 1.
Comparison of properties of the orthomyxovirid PB1 proteins
TABLE 2.
Pairwise comparisons of orthomyxovirid PB1 proteinsa
| Virus | ISAV | FluA | FluB | FluC | DHO | THO |
|---|---|---|---|---|---|---|
| ISAV | 44.9 | 46.0 | 47.6 | 46.2 | 45.4 | |
| FluA | 20.8 | 76.2b | 57.8b | 52.8b | 52.2b | |
| FluB | 23.3 | 60.9b | 62.7b | 50.6b | 50.1b | |
| FluC | 24.1 | 39.1b | 42.0b | 53.7b | 53.4b | |
| DHO | 21.8 | 30.7b | 26.6b | 29.8b | 76.6b | |
| THO | 23.2 | 29.1b | 27.6b | 29.9b | 62.3b |
Values in the upper right portion of the table are percentages of amino acid similarity, and values in the lower left portion are percentages of amino acid identity. Virus abbreviations are as defined in the legend for Fig. 1. Percentages of similarity and identity were determined with the University of Wisconsin Genetics Computer Group program GAP.
Data taken from Leahy et al. (17).
Taxonomic allocation of the ISAV.
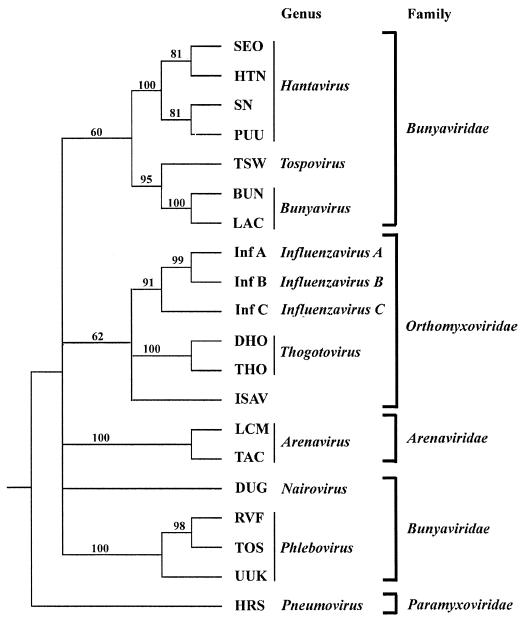
To determine allocation at the family level, a previously published alignment spanning the central conserved core region of viral polymerases from members of the Arenaviridae, Bunyaviridae, Orthomyxoviridae, and Paramyxoviridae (outgroup) was used (19). To this alignment the polymerase sequences of the Thogoto virus (17) and ISAV were added, and phylogenetic trees were constructed. The different methods gave different tree topologies. The two parsimony methods (PROTPARS and PAUP 3.1) and the UPGMA distance method that were used clustered the ISAV with the Orthomyxoviridae. The bootstrap support for this topology was 90% (result not shown), 70% (result not shown), and 62% (Fig. 3) for the PROTPARS, UPGMA, and PAUP methods, respectively. Two of the distance methods, Fitch-Margoliash and neighbor joining, did not cluster the ISAV with any other viruses with more than 50% bootstrap support. However, it is known that different tree building methods have different powers to resolve phylogenies, but the power increases if a large number of characters is used, i.e., more than 1,000 (24). The failure of these two methods to group the ISAV with the orthomyxoviruses, or even the influenza C virus with the influenza A and B viruses (result not shown), is therefore probably due to the relatively few characters used in the alignment (363, including 257 amino acids for the ISAV polymerase).
FIG. 3.
A 50% majority rule consensus tree of 100 bootstrap replicates carried out by maximum parsimony (PAUP 3.1) using a heuristic search option. Numbers on the tree indicate the percentage of bootstrap replicates which contained that topology. The basis for the phylogenetic analysis is an alignment published by Marriott and Nuttall (19) added to the Thogoto and ISAV RdRp sequences. Human respiratory syncytial virus was used as an outgroup. Abbreviations of virus names not defined in the text are as follows: SEO, Seoul; HTN, Hantaan; SN, Sin Nombre; PUU, Puumala; TSW, tomato spotted wilt; BUN, bunyamwera; LAC, La Crosse; LCM, lymphocytic choriomeningitis; TAC, Tacaribe; DUG, Dugbe; RVF, Rift Valley fever; TOS, Toscana; UUK, Uukuniemi; HRS, human respiratory syncytial.
To resolve the intrafamily relationship, an alignment including the complete polymerase sequences of all orthomyxoviruses was used (Fig. 1). From this alignment, the different methods gave the same tree topology, placing the ISAV as a distinct taxon in the Orthomyxoviridae. The results obtained by the Fitch-Margoliash and neighbor-joining methods indicate that the ISAV is more closely related to the influenza viruses than to the Thogoto viruses. The UPGMA method, however, places the ISAV an equal distance from the other two virus groups.
Conserved end sequences.
Common features in the genomes of segmented negative-stranded RNA viruses are conserved sequences at the 5′ and 3′ ends. We have studied the 5′ cDNA ends of the clones from two ISAV segments representing the polymerase sequence (segment 2) and the sequence of segment 8 in the ISAV genome. The ISAV 5′ cDNA ends, which are heterogeneous in both length and sequence, are followed by a stretch of eight conserved nucleotides between the two segments (Table 3). The conserved sequence 5′-AGCAAAGA is shorter than what is found in the influenza and Thogoto viruses, but the sequence is clearly related to both, as indicated by its beginning with AGC (Table 4).
TABLE 3.
Sequences at the 5′ ends of ISAV mRNAs corresponding to virus segments 2 and 8
| Segment | Sequence at 5′ mRNA endsb |
|---|---|
| 2 | g a a u g u a a c u a u A G C A A A G A |
| 8 | g a g a c a g g c a g u g c A G C A A A G A |
| 8a | g a a u u c u u c c – G C A A A G A |
The conserved 5′ terminal sequence of positive-sense mRNAs is shown in capital letters, while the lowercase letters show the heterogeneous sequence, possibly derived through a cap-snatching mechanism. The dash (–) is inserted to align the conserved residues.
TABLE 4.
Comparison of conserved nucleotides at 5′ mRNA ends in orthomyxoviruses
DISCUSSION
The data presented here indicate that the ISAV shares an ancestor with members of Orthomyxoviridae and should probably be included in this family, representing a new genus. This conclusion is based on comparison of the predicted RNA polymerase from ISAV with RNA polymerases from other negative-stranded RNA viruses.
The predicted protein encoded by ISAV segment 2 contains the amino acid sequence motifs present in all polymerases that show RNA template specificity and most likely form the active sites for RNA synthesis (21, 31, 39). Some of these motifs can be used to distinguish between viruses of different genome polarity and organization. The ISAV sequence contains two conserved residues, a glutamic acid (E) and a lysine (K) between premotif A and motif A (Fig. 1), which are specific for negative-stranded RNA viruses with both segmented and nonsegmented genomes (21). In motif C, the ISAV sequence is SDD (Fig. 1), which is a signature for segmented negative-stranded RNA viruses. In contrast, the sequences GDN, GDD, and MDD are typical in the negative-stranded nonsegmented RNA viruses, the positive-stranded RNA viruses, and the retroviruses, respectively (4). The critical function of motif C was demonstrated in a mutational analysis where the SDD sequence of an influenza virus was changed to GDD, as found in positive-strand RNA viruses (4). This mutation drastically reduced the function of the polymerase (4). Furthermore, the ISAV sequence also contains the tetrapeptide EFXS in motif E (Fig. 1), which is conserved in segmented negative-stranded RNA virus polymerases only (21). This tetrapeptide is suggested to be involved in the specific transcriptional initiation with cell-derived capped primers (21). The predicted ISAV protein contains all of the previously described conserved motifs and strictly conserved residues; hence, we suggest that ISAV segment 2 encodes the RNA-dependent RNA polymerase (RdRp).
The size of the ISAV PB1 protein is comparable to the size of the polymerases of the other members of the family (Table 1). The high positive charge that is characteristic of the PB1 proteins of the influenza viruses is also found with the ISAV protein. However, the biological significance of the charge has been questioned due to the variability found between the various PB1-like, as well as PA-like, proteins in the family (17). Most remarkable is that the Dhori virus protein is acidic, and the name Pα has therefore been suggested (18).
In the present study, the conserved motifs of the ISAV RdRp are compared to the homologous regions in other negative-stranded RNA viruses from Arenaviridae, Bunyaviridae, and Orthomyxoviridae, using a member of the Paramyxoviridae as an outgroup. The results of the phylogenetic analysis clearly support the idea that ISAV should be considered for inclusion in the Orthomyxoviridae, as proposed in two recent works characterizing the genome organization and physiochemical properties of the virus (9, 20).
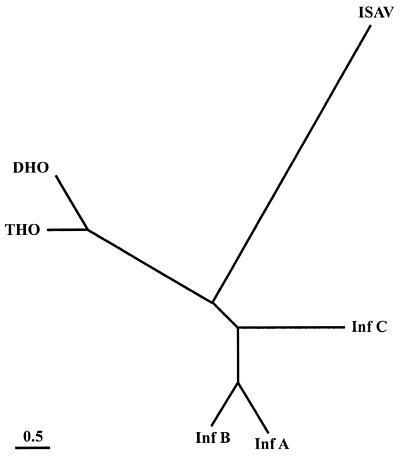
To examine the intrafamily relationship, genetic distances were calculated based on the alignment including the complete PB1 sequences. The neighbor-joining tree that was drawn clearly shows that the ISAV branch is the longest in the tree (Fig. 4), indicating a distant relationship with the other orthomyxoviruses. However, the distance calculations indicate that the ISAV is more closely related to the influenza viruses than to the Thogoto viruses. On this basis, we propose that the ISAV should represent a new genus in the Orthomyxoviridae.
FIG. 4.
Genetic distance tree drawn by the neighbor-joining method. Branch lengths are drawn to scale. Virus abbreviations are as given in the legend for Fig. 1.
A characteristic feature of the influenza viruses is their cap-snatching mechanism, which consists of the initiation of transcription with capped primers generated by cleavage of host-cell RNA polymerase II transcripts (15, 29, 30). Host-cell primers are generated in the nucleus by a cap-dependent endonuclease that cleaves the capped cellular RNAs 10 to 15 nucleotides from their 5′ ends, preferentially at a purine residue (30). The heterogeneous sequence originating from cellular RNAs is then followed by a stretch of 12 to 16 conserved nucleotides in the different genome segments in the influenza viruses (7). The situation for the Thogoto virus is somewhat different: the mRNAs are found to be homogeneous in both length and sequence at their 5′ ends (2, 16, 38). Using an in vitro polymerase assay, Leahy et al. (16) demonstrated that the Thogoto virus preferentially initiates transcription with a cap-A structure, which can base pair with the 3′ ultimate uracil of the vRNA. It has been speculated that the mechanism used by the Thogoto virus for mRNA transcription initiation is an ancient feature which then possibly evolved to the mechanism found in influenza viruses and other segmented negative-sense RNA viruses (2). In our sequencing shown in Table 3, the 5′ cDNA sequences from segments 2 and 8 start with a heterogeneous sequence 10 to 14 nucleotides long, followed by 7 to 8 conserved nucleotides closely related to the conserved nucleotides in the other members of the family (Table 4). Whether the conserved sequence consists of 7 or 8 nucleotides remains to be determined by sequencing several segments. The adenine residue at the start, lacking in segment 8 as determined by Mjaaland et al. (20), could be due to an artifact since that sequence starts with an EcoRI site and may not be full length. Another possibility is that the adenine residue by coincidence ends the heterogeneous sequence in the two segments sequenced in the present study. However, the results presented here indicate that the ISAV uses a transcription mechanism similar to that of the influenza viruses, supporting the distance calculations that indicate a closer relationship to the influenza viruses than to the Thogoto viruses. Whether the 5′ mRNAs are capped has not been addressed in this study. However, Falk et al. (9) demonstrated actinomycin D sensitivity, which, together with the similarities in mRNA 5′-end sequences reported in this study, suggests the existence of a cap-snatching mechanism.
To conclude, the data presented in this study support the findings in other recent work in which the ISAV has been described as orthomyxovirus-like and a candidate for inclusion in the Orthomyxoviridae (9, 20). The polymerase (PB1) sequence reported in this study is the first from the ISAV that shares significant sequence homology with other viruses, and phylogenetic studies using this sequence group the ISAV with members of the Orthomyxoviridae. Distance calculations based on the polymerase sequences clearly indicate that the ISAV is distantly related to the other members of the family. The ISAV has been shown to have a psycrophilic nature, being unable to replicate at temperatures above 25°C and with a optimum replication temperature of 15°C, which probably restricts the host range to cold-blooded animals (9). So far, the ISAV has been observed to replicate in Atlantic salmon (Salmo salar L.) (13), brown trout (Salmo trutta L.) (26), rainbow trout (Onchorhynchus mykiss, Walbaum) (28), and herring (Clupea harengus) (personal observation). In addition, orthomyxovirus-like particles have been observed in eel (Anguilla anguilla) (1, 23, 25) and bluegill (Lepomis macrochirus) (12), although no genetic characterization of these viruses has been done. It is tempting to speculate, on the basis of these observations and the genetic distance calculations, that the ISAV may represent an old group of aquatic orthomyxoviruses. We therefore propose the creation of a new genus in which the ISAV is designated the type species. We also propose a new genus to be named Aquaorthomyxovirus to reflect the host range of the ISAV as well as the proposed family allocation.
ACKNOWLEDGMENTS
We thank Birgit H. Dannevig for providing the SHK-1 cells and Julia Mullins for improving the English of the manuscript.
This work was supported by grant 107131/122 from the Norwegian Research Council.
REFERENCES
- 1.Ahne W, Thomsen I. The existence of three different viral agents in a tumour-bearing European eel (Anguilla anguilla) Zentbl Vet Med B. 1985;32:228–235. doi: 10.1111/j.1439-0450.1985.tb01960.x. [DOI] [PubMed] [Google Scholar]
- 2.Albo C, Martin J, Portela A. The 5′ ends of Thogoto virus (Orthomyxoviridae) mRNAs are homogeneous in both length and sequence. J Virol. 1996;70:9013–9017. doi: 10.1128/jvi.70.12.9013-9017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S K, Nayak D P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clerx J P M, Fuller F, Bishop D H L. Tick-borne viruses structurally similar to Orthomyxoviruses. Virology. 1983;127:205–219. doi: 10.1016/0042-6822(83)90384-7. [DOI] [PubMed] [Google Scholar]
- 6.Dannevig B H, Falk K, Namork E. Isolation of the causal virus of infectious salmon anemia (ISA) in a long-term cell-line from Atlantic salmon head kidney. J Gen Virol. 1995;76:1353–1359. doi: 10.1099/0022-1317-76-6-1353. [DOI] [PubMed] [Google Scholar]
- 7.Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′- and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 8.Evensen Ø, Thorud K E, Olsen Y A. A morphological study of the gross and light microscopic lesions of infectious anaemia in Atlantic salmon (Salmo salar L.) Res Vet Sci. 1991;51:215–222. doi: 10.1016/0034-5288(91)90017-i. [DOI] [PubMed] [Google Scholar]
- 9.Falk K, Namork E, Rimstad E, Mjaaland S, Dannevig B H. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) J Virol. 1997;71:9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 11.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman G L, Dunbar C E, Wolf K, Zwillenberg L O. Epitheliocystis, a new infectious disease of the bluegill (Lepomis macrochirus) Antonie Van Leeuwenhoek J Microbiol Serol. 1969;35:146–158. doi: 10.1007/BF02219125. [DOI] [PubMed] [Google Scholar]
- 13.Hovland T, Nylund A, Watanabe K, Endresen C. Observation of infectious salmon anaemia virus in Atlantic salmon, Salmo salar L. J Fish Dis. 1994;17:291–296. [Google Scholar]
- 14.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krug R M. Priming of influenza viral RNA transcription by capped heterologous RNAs. Curr Top Microbiol Immunol. 1981;93:125–149. doi: 10.1007/978-3-642-68123-3_6. [DOI] [PubMed] [Google Scholar]
- 16.Leahy M B, Dessens J T, Nuttall P A. In vitro polymerase activity of Thogoto virus: evidence for a unique cap-snatching mechanism in a tick-borne orthomyxovirus. J Virol. 1997;71:8347–8351. doi: 10.1128/jvi.71.11.8347-8351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leahy M B, Dessens J T, Weber F, Kochs G, Nuttall P A. The fourth genus in the Orthomyxoviridae: sequence analyses of two Thogoto virus polymerase proteins and comparison with influenza viruses. Virus Res. 1997;50:215–224. doi: 10.1016/s0168-1702(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 18.Lin D A, Roychoudhury S, Palese P, Clay W C, Fuller F J. Evolutionary relatedness of the predicted gene product of RNA segment 2 of the tick-borne Dhori virus and the PB1 polymerase gene of influenza viruses. Virology. 1991;182:1–7. doi: 10.1016/0042-6822(91)90641-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marriott A C, Nuttall P A. Large RNA segment of Dugbe nairovirus encodes the putative RNA polymerase. J Gen Virol. 1996;77:1775–1780. doi: 10.1099/0022-1317-77-8-1775. [DOI] [PubMed] [Google Scholar]
- 20.Mjaaland S, Rimstad E, Falk K, Dannevig B H. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J Virol. 1997;71:7681–7686. doi: 10.1128/jvi.71.10.7681-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller R, Poch O, Delarue M, Bishop D H L, Bouloy M. Rift valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol. 1994;75:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- 22.Mullins J E, Groman D, Wadowska D. Infectious salmon anaemia in salt water Atlantic salmon (Salmo salar L.) in New Brunswick, Canada. Bull Eur Assoc Fish Pathol. 1998;18:110–114. [Google Scholar]
- 23.Nagabayashi T, Wolf K. Characterization of EV-2, a virus isolated from European eels (Anguilla anguilla) with stomatopapilloma. J Virol. 1979;30:358–364. doi: 10.1128/jvi.30.1.358-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei M. Phylogenetic analysis in molecular evolutionary genetics. Annu Rev Genet. 1996;30:371–403. doi: 10.1146/annurev.genet.30.1.371. [DOI] [PubMed] [Google Scholar]
- 25.Neukirch M. Isolation of an orthomyxovirus-like agent from European eel (Anguilla anguilla) Bull Eur Assoc Fish Pathol. 1985;5:12. [Google Scholar]
- 26.Nylund A, Alexandersen S, Rolland J B, Jakobsen P. Infectious salmon anemia virus (ISAV) in brown trout. J Aquat Anim Health. 1995;7:236–240. [Google Scholar]
- 27.Nylund A, Krossøy B, Watanabe K, Holm J A. Target cells for the ISA virus in Atlantic salmon (Salmo salar L.) Bull Eur Assoc Fish Pathol. 1996;16:68–72. [Google Scholar]
- 28.Nylund A, Kvenseth A M, Krossøy B, Hodneland K. Replication of the infectious salmon anaemia virus (ISAV) in rainbow trout (Oncorhynchus mykiss, Walbaum, 1792) J Fish Dis. 1997;20:275–279. [Google Scholar]
- 29.Plotch S J, Bouloy M, Krug R M. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci USA. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 31.Poch O, Sauvagnet I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding element. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodger H D, Turnbull T, Muir F, Millar S, Richards R H. Infectious salmon anaemia (ISA) in the United Kingdom. Bull Eur Assoc Fish Pathol. 1998;18:115–116. [Google Scholar]
- 33.Staunton D, Nuttall P A, Bishop D H L. Sequence analysis of Thogoto viral RNA segment 3: evidence for a distant relationship between an arbovirus and members of the Orthomyxoviridae. J Gen Virol. 1989;70:2811–2817. doi: 10.1099/0022-1317-70-10-2811. [DOI] [PubMed] [Google Scholar]
- 34.Swofford D L. Phylogenetic analysis using parsimony (PAUP), version 3.1.1. Champaign, Ill: University of Illinois; 1993. [Google Scholar]
- 35.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorud K. Infectious salmon anaemia virus. Thesis. Oslo, Norway: Norwegian College of Veterinary Medicine; 1991. [Google Scholar]
- 37.Thorud K, Djupvik H O. Infectious salmon anaemia in Atlantic salmon (Salmo salar L.) Bull Eur Assoc Fish Pathol. 1988;8:109–111. [Google Scholar]
- 38.Weber F, Haller O, Kochs G. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J Virol. 1996;70:8361–8367. doi: 10.1128/jvi.70.12.8361-8367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita M, Krystal M, Palese P. Comparison of the three large polymerase proteins of influenza A, B, and C viruses. Virology. 1989;171:458–466. doi: 10.1016/0042-6822(89)90615-6. [DOI] [PubMed] [Google Scholar]
- 41.Zanotto P M D, Gibbs M J, Gould E A, Holmes E C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J Virol. 1996;70:6083–6096. doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]