Abstract
The transcriptional enhancer of the lymphomagenic mouse retrovirus SL3 contains a binding site for the transcription factor core binding factor (CBF; also called AML1, PEBP2, and SEF1). The SL3 CBF binding site is called the core. It differs from the core of the weakly lymphomagenic mouse retrovirus Akv by one nucleotide (the sequences are TGTGGTTAA and TGTGGTCAA, respectively). A mutant virus called SAA that was identical to SL3 except that its core was mutated to the Akv sequence was only moderately attenuated for lymphomagenicity. In most SAA-infected mice, tumor proviruses contained either reversions of the original mutation or one of two novel core sequences. In 20% of the SAA-infected mice, tumor proviruses retained the original SAA/Akv core mutation but acquired one of two additional mutations (underlined), TGCGGTCAA or TGTGGTCTA, that generated core elements called So and T*, respectively. We tested whether the novel base changes in the So and T* cores were suppressor mutations. SL3 mutants that contained So or T* cores in place of the wild-type sequence were generated. These viruses induced T-cell lymphomas in mice more quickly than SAA. Therefore, the mutations in the So and T* cores are indeed second-site suppressor mutations. The suppressor mutations increased CBF binding in vitro and transcriptional activity of the viral long terminal repeats (LTRs) in T lymphocytes to levels comparable to those of SL3. Thus, CBF binding was increased by any of three different nucleotide changes within the sequence of the SAA core. Increased CBF binding resulted in increased LTR transcriptional activity in T cells and in increased viral lymphomagenicity.
Transcriptional enhancers in murine leukemia viruses (MuLVs) are crucial genetic elements for determining viral pathogenicity. Enhancer sequences located in the unique 3′ region (U3) in the long terminal repeat (LTR) determine the tissue specificity of virally induced disease, the fraction of the mice that develop tumors, and the length of latency prior to the appearance of tumors. MuLV enhancers usually contain tandem repeat units that are located starting approximately 170 bp upstream of the transcription initiation site in the 5′ LTR. Binding sites for various transcription factors are present within the repeats and the sequences flanking them (2, 3, 7, 12, 13, 21, 28, 33, 35–37, 41–45).
One element within the enhancers of MuLVs and related type C mammalian retroviruses including feline leukemia virus and gibbon ape leukemia virus is known as the enhancer core. The core was first identified in the simian virus 40 enhancer (19). Although the sequences of the 9-bp core elements are similar among simian virus 40, polyomavirus, and type C retroviruses, they are not identical. Mutagenesis studies showed that core elements are very important for the pathogenicity of the T-cell lymphomagenic MuLVs, Moloney MuLV (Mo-MuLV), and SL3 (14, 27, 34). In Mo-MuLV, point mutations in the core reduced viral potency and changed the disease specificity from T-cell lymphoma to erythroleukemia (34). The LTR enhancer of the lymphomagenic mouse retrovirus SL3 contains two 72-bp tandem repeats. Each repeat contains two slightly different core elements. Based on its position relative to other transcription factor binding sites, one of these (core I) corresponds to the core element of Mo-MuLV (27). The SL3 and Mo-MuLV cores differ slightly in sequence (TGTGGTTAA and TGTGGTAAG, respectively). In addition, SL3 contains a second core element, termed core II, that has the sequence AGCGGTCTG (14, 27, 38). Mutation of the core I element of SL3 strongly decreased the pathogenicity of the virus (14, 27). Mutation of the core II element by itself had little effect on pathogenicity (14). However, when both core elements were mutated, the virus was only weakly pathogenic and most of the tumors were B-cell lymphomas (10).
Although all type C mammalian retroviruses have an identifiable core element positioned equivalently to the SL3 core I element, the actual sequences of the elements vary somewhat among the different viruses (12). These differences can have large effects on viral pathogenicity. The core I element of SL3 (hereinafter termed the core element of this virus) and the core element of the weakly pathogenic MuLV Akv differ by 1 bp (the sequences are TGTGGTTAA and TGTGGTCAA, respectively). SAA is an engineered mutant of SL3 that is identical to SL3 except for the T to C change within the core elements in both enhancer repeat units in the viral LTR. Although SAA induced T-cell lymphomas in mice, the lymphomagenicity of SAA was reduced compared to that of SL3 as the latency period to disease onset was increased (27). Thus, the 1-bp difference between the SL3 and Akv/SAA cores was important for viral pathogenicity. Nonetheless, it was surprising that SAA was as potent as it was, because the T to C change substantially decreased the transcriptional activity of the SL3 LTR in T lymphocytes (27). Moreover, a 3-bp mutation in the SL3 core substantially inhibited viral lymphomagenicity (14). To account for these discrepancies, it was hypothesized that the 1-bp mutations in the cores of SAA had reverted, thus restoring the original SL3 sequence (27). These reversions were hypothesized to have occurred as the virus replicated during the period before the appearance of lymphomas (27). To test this possibility, a reversion analysis was performed. LTRs of the proviruses in SAA-induced tumors were PCR amplified and sequenced. This analysis revealed that the original mutation in the enhancer core had reverted to the wild-type SL3 sequence in 68% of the mice with tumors (27).
Interestingly, the tumor proviruses in several of the mice retained the original C mutation in the core but also acquired one of two mutations elsewhere within the core element. In 3 of 38 mice with lymphomas, the proviruses contained the core sequence TGCGGTCAA (the novel nucleotide is underlined). This sequence was identical to that of the core element of Soule MuLV (So-MuLV), a T-cell lymphomagenic virus (6). This core element was termed So (27). Six of 38 mice had proviruses with a core sequence TGTGGTCTA (the novel nucleotide is underlined) that generated a core element that was termed T* (27). Since both So and T* retained the original C mutation and occurred independently in multiple mice, we hypothesized that they are core elements with second-site suppressor mutations.
In the study reported here, we tested this hypothesis. The LTR sequences of proviruses in tumors from mice containing So and T* core elements were recovered and used to replace the corresponding sequences of SL3 (27). The lymphomagenic potential of these viruses in mice was analyzed. To elucidate how the mutations generating So and T* might affect viral lymphomagenicity, we also assessed their effects on binding of core binding factor (CBF; also called AML1, Runt, PEBP2, and SEF1) (8, 17, 18, 24, 37, 40) and on the transcriptional activity of the viral LTR in T lymphocytes.
MATERIALS AND METHODS
Generation of viral genomes containing the So and T* cores.
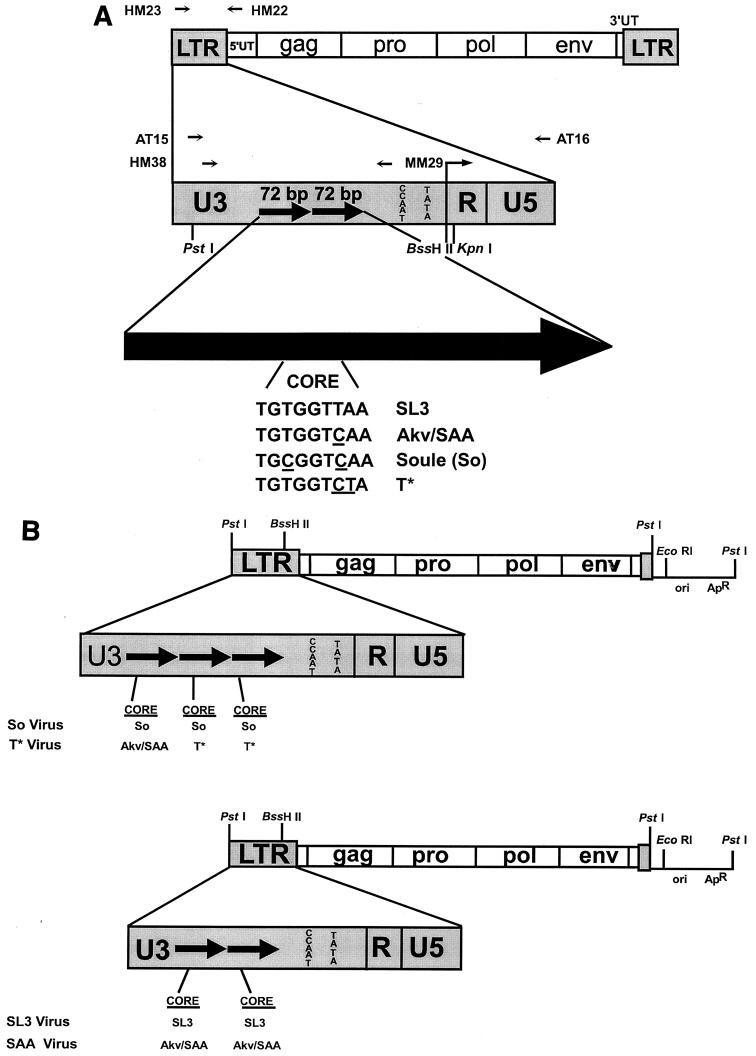
Infectious clones of the viral genome were generated in a manner similar to that previously described (27, 29, 30). The approach is summarized in Fig. 1. LTR sequences containing the So or T* cores were PCR amplified from proviruses present in DNA isolated from lymphomas induced by SAA (27). PCR products were generated with the primers HM23 (5′ TTCATAAGGCTTAGCCAGCTAACTGCAG 3′) and HM22 (5′ GATGCCGGCACACACACACACACTCTCCC 3′) at positions −470 to −443 and +272 to +244, respectively, relative to the transcriptional initiation site (Fig. 1) (27). PCR conditions involved a 30-cycle program of 1 min at 94°C, 1 min at 64°C, and 2 min at 72°C. The PCR products were digested with PstI and KpnI (Fig. 1A) and then subcloned into the corresponding sites of the pGEM 3Z(−) vector (Promega). LTR plasmid subclones were digested with BssHII and EcoRI, and a fragment containing the remainder of the SL3 genome including the gag, pro, pol, and env genes was inserted at those sites (Fig. 1B). This resulted in the formation of a plasmid subclone that contained the complete viral genome with a single LTR (Fig. 1B). These plasmids were cleaved with PstI to separate the viral and plasmid vector sequences. The viral fragments were self-ligated to form concatemers. This resulted in viral genomes that contained two identical LTRs, as previously described (27, 29, 30). Infectious virus was generated by transfection of viral genomes into NIH 3T3 mouse fibroblasts. 106 cells per 60-mm2 plate were seeded 24 h prior to transfection with Lipofectin (GIBCO BRL). Cells were passaged 1:10 every third day. Supernatants collected from transfected cells at each passage were tested for reverse transcriptase activity (11). Approximately 3 weeks after transfection, viral stocks reached maximum reverse transcriptase levels. Aliquots of virus were frozen at this point, and the infectious virus titers were determined by XC plaque assays (31). PCR and sequencing of proviral DNA (described below) from the infected NIH 3T3 cells confirmed the presence of each mutation.
FIG. 1.
Structures of the viruses and viral plasmids used in these studies. (A) Structures of the viral LTRs and sequences of the viral core elements. The top diagram shows the structure of the genome of proviruses of SL3 and the mutants derived from SL3. HM23 and HM22 are the PCR primers that were used to amplify the viral LTRs from tumor cell DNAs. The second diagram shows the structure of the viral LTRs. PCR primers used for sequencing studies are shown above the diagram. Restriction sites used for cloning experiments are shown below the diagram. The large arrow at the bottom represents one 72-bp enhancer repeat. The sequences of the cores present in the SL3, SAA, So, and T* enhancers are shown. Nucleotides that differ relative to the SL3 core are underlined. (B) Structures of the plasmids used to generate infectious virus particles. The top diagram represents the viruses with So and T* core mutations. The bottom diagram represents structures of the SL3 and SAA virus clones. In each diagram, the viral sequences are shown as boxes while the plasmid vector sequences are shown as lines. LTR sequences are enlarged to show the numbers of 72-bp enhancer repeats, which are indicated as black arrows. The sequence of the core in each repeat is shown below the LTR. Akv and SAA have the same core sequence; thus, this core is represented as Akv/SAA.
Tumorigenicity assays.
Newborn NIH/Swiss mice (<1.5 days) were injected intraperitoneally with 0.1 ml of virus (104 PFU, XC plaque assay). The diseased animals were sacrificed and necropsied. Gross pathological examination always revealed enlargement of the thymus, spleen, peripheral lymph nodes, mesenteric lymph nodes, or liver. Enlarged organs were stored frozen at −80°C until DNA was prepared from them. Southern blotting with a T-cell receptor β probe was used to test whether the tumors were of T-cell origin (1, 15).
Analysis of viral enhancer sequences in infected cells.
Proviral LTR DNA was amplified from tumors by using PCR primers HM22 and HM23 (Fig. 1A) as described above. PCR products were electrophoretically resolved on a nondenaturing 5% polyacrylamide gel (27). The individual bands differed by multiples of 72 bp. Each band was excised and the DNA was isolated by using Qiaex II (Qiagen). The bands were reamplified by PCR under the same conditions by using primer pair HM38 (5′ AAGGCTTAGCCAGCTAACTGCAGTAACGCC 3′), at positions −466 to −436, and HM22, at +272 to +244 (Fig. 1A). Products were isolated by using QIAquick (Qiagen). The resulting PCR products were sequenced directly by using primer MM29 (5′ TCATCTGGGGAACCTTGAGAC 3′) at positions −136 to −115 relative to the transcription initiation start site (Fig. 1A).
CAT plasmids and assays.
Proviral core enhancer sequences that were PCR amplified from tumors as described above and subcloned into a pGEM3Z(−) (Promega) plasmid were cleaved with PstI. The 3′ overhang was removed with T4 DNA polymerase, and then the DNA was digested with BssHII. BssHII cleaved SL3 at the U3-R boundary in the LTR. This fragment was used to replace the corresponding sequences in an SL3-chloramphenicol acetyltransferase (SL3-CAT) plasmid clone containing the CAT gene (3, 32, 39). SL3-CAT was digested with NdeI, and the 5′ terminus was filled in by using the Klenow fragment of Escherichia coli DNA polymerase I. SL3-CAT was then cleaved with BssHII to generate a 3.5-kb vector fragment. The vector and LTR insert fragments were ligated together to generate CAT plasmids containing the LTR sequences described in the text. Transfection of cell lines was performed by the DEAE-dextran method as previously described (3, 32). 5.0 × 106 cells per plate were pelleted and resuspended in 1 ml of TD (25 mM Tris-HCl [pH 7.4], 0.7 mM Na2HPO4, 5.1 mM KCl, 137 mM NaCl) containing 250 μg of DEAE-dextran per ml, 5 μg of reporter plasmid DNA, and 1 μg of a Rous sarcoma virus LTR-luciferase plasmid used as an internal control. The reagents were incubated at room temperature for 15 min. Five milliliters of medium supplemented with 10% fetal bovine serum was added, and incubation was continued for 20 min at 37°C. Cells were pelleted and resuspended in 5 ml of medium with serum. Cells were plated in 60-mm2 dishes and harvested at 48 h. Cells were lysed by three cycles of freeze-thawing, and protein concentrations were determined by Bradford assays (Biorad) (4). Aliquots of cell lysates were used for CAT or luciferase assays as previously described (45) except that the protocol included the use of 15 μl of fluorescent BODIPY FL chloramphenicol substrate, FastCAT (Molecular Probes) instead of 14C-labeled chloramphenicol. Silica gel thin-layer chromatography was performed in a sealed, equilibrated chromatography chamber containing chloroform:methanol (9:1, vol/vol). CAT activity was quantified by calculating the percentage of chloramphenicol that was acetylated by using the STORM imager that scans for blue fluorescence. Each sample was normalized with its corresponding luciferase assay. All samplings were performed in duplicate and at multiple times. Means and standard deviations were calculated and plotted.
Cell lines.
SL3H and Jurkat cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 10 mg of streptomycin per ml, and 2 mM glutamine. L691-6 cells were propagated in Dulbecco’s modified Eagle media (DMEM) supplemented as described above. NIH 3T3 cells were grown in DMEM with 10% calf serum and other supplements as previously mentioned. All cells were maintained at 37°C in 100% humidity and 7.5% CO2.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed with a double-stranded, 31-bp radiolabeled probe that contained the SL3 core I sequence as previously described (3). Nuclear protein extract from the mouse T-cell line WEHI 7.1 provided the source of CBF protein (3). Double-stranded, unlabeled oligonucleotides served as competitor DNAs. The sequences of the competitors and probe were as follows (only the sequences of the plus strands are shown): for SL3, 5′ ATCTGTGGTTAAGCACTAGGGCCCCGGCCCA 3′; for Akv, 5′ ATCTGTGGTCAAGCACTAGGGCCCCGGCCCA 3′; for So, 5′ ATCTGCGGTCAAGCACTAGGGCCCCGGCCCA 3′; for T*, 5′ ATCTGTGGTCTAGCACTAGGGCCCCGGCCCA 3′; and for mutated SL3 core (MUT), 5′ ATCTGCCGTTAAGCACTAGGGCCCCGGCCCA 3′. The binding reaction mixtures contained 10,000 dpm of SL3 probe labeled with [γ-32P]ATP and T4 polynucleotide kinase, 5% glycerol, 50 mM NaCl, 10 mM Tris (pH 8.0), 5 mM EDTA (pH 8.0), 1 mM dithiothreitol, 2 μg of poly(dI-dC) · (dI-dC), and 1 μg of sonicated salmon sperm DNA. Competitor DNAs were added as indicated. The binding reaction mixtures were incubated at room temperature for 15 min. The samples were run on a 5% polyacrylamide gel at 110 V for 193 Vh at room temperature. The recirculated electrophoresis buffer contained 6.7 mM Tris hydrochloride (pH 7.5), 3.3 mM sodium acetate, and 1 mM EDTA (pH 8.0) (3, 45). The gels were dried and exposed to a PhosphorImager screen for 24 h. The data were quantified and plotted.
RESULTS
Lymphomagenicity of second-site suppressor mutations generating So and T* cores.
To test the hypothesis that the So and T* cores contain suppressor mutations, we engineered recombinant viruses containing these elements. The So and T* enhancer core sequences were detected initially in proviruses that were present in tumors of SAA-inoculated mice. Previous observations showed that the So and T* cores were present in proviruses where the LTR enhancers contained three 72-bp tandem repeats (27). LTR enhancers of MuLVs usually contain two tandem repeats (12). For example, the infectious molecular clone of SL3 contains two tandem repeats (20). However, viral genomes with more than two repeats or with a single unit have been identified (5, 16, 27). Infectious virus derived by transfection of NIH 3T3 fibroblasts with the molecular clone of SL3 contained a mixture of genomes with a variable number of repeats (27). LTRs with two-repeat units predominated, but LTRs with one and three repeats were also detectable. In lymphomas induced by SL3, proviruses with three enhancer repeats were observed and were often the predominant form (27). Thus, SL3 exists as a quasispecies of isoforms with enhancers that contain variable numbers of tandem repeats within the U3 region of the LTR (27). Three LTR enhancer repeat units were also reported to be present in tumors induced by a mutant of Mo-MuLV and in a tumor induced by feline leukemia virus (5, 25).
When proviruses in lymphomas induced by SAA contained three repeat units, they usually contained reversions or one of the putative suppressor mutations in at least two of the repeats (27). This was interpreted to mean that single base mutations in the core sequence occurred first and that this was followed by changes in the number of enhancer repeat units (27). We reasoned that viruses with the putative suppressor mutations present in more than one repeat unit would function as the most potently lymphomagenic viruses. Therefore, we isolated the LTR sequences that had So or T* cores present in multiple repeat units from proviruses present in SAA-induced lymphomas (27). Consequently, the viruses that were tested were the viruses that we hypothesize actually caused the tumors in SAA-inoculated mice.
LTRs containing the So and T* cores were PCR amplified from DNA isolated from SAA-induced lymphomas (Fig. 1A). For the So core, a viral genome that had the mutation in three tandem repeat units was identified (27). For the T* core, a viral genome that had the mutation in the two promoter-proximal repeats of the three present in the provirus was identified (27). Restriction fragments containing the U3s of the amplified LTRs were used to replace the U3 sequences of an infectious clone of SL3 (Fig. 1B). Each of the plasmid clones contained a single LTR (Fig. 1B). Viral sequences were excised from the plasmids by digestion with PstI, self-ligated to generate concatenates, and used to transfect NIH 3T3 fibroblasts as previously described (27). This resulted in two identical LTRs in the progeny proviruses. Viral RNA transcribed from the transfected DNA generated infectious virus that spread and infected all the cells in the culture. After several passages of the cells over 3 weeks, the reverse transcriptase levels in the culture supernatants reached a maximum level. XC cell assays indicated that both recombinant virus stocks had titers of about 104 infectious virus units per ml. Stocks of SL3 and SAA were generated in parallel. These attained the same titers with the same kinetics as the mutants. Therefore, the differences among the core elements did not affect viral replication in fibroblasts.
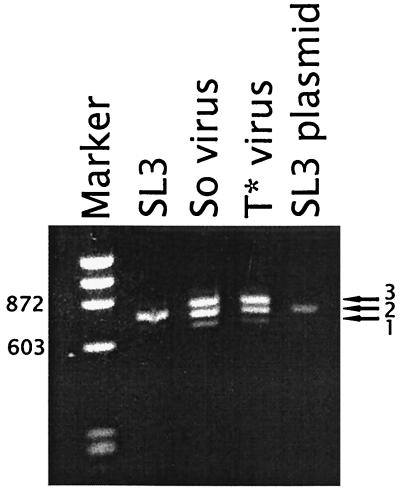
To verify that the viruses that contained the So and T* cores maintained the actual mutations, proviral DNA was PCR amplified from the infected NIH 3T3 cells and sequenced. We previously showed that PCR amplification of viral DNA from infected cells resulted in multiple bands that came from proviruses with variable numbers of repeat units (27). Southern blotting confirmed that the bands were derived from proviruses and were actually present in the genomic DNA rather than resulting from polymerase jumping artifacts during PCR (27). DNA amplified from a control culture transfected with the infectious SL3 clone exhibited bands corresponding to LTRs with one, two, and three repeats (Fig. 2). The two-repeat structure predominated. Proviruses in the cultures infected with the recombinants containing So and T* core elements also showed bands corresponding to one, two, and three repeats (Fig. 2). Although the plasmids used to initiate the infection contained three repeat units, proviruses with two repeats were also abundant. We interpret this result to mean that reductions in the number of enhancer repeats occurred due to polymerase slippage during viral replication. Most likely, viruses with the two LTR repeats have some replicative advantage over those with three repeats. Sequencing of the PCR-amplified bands indicated that the original mutations were indeed present in the viruses that replicated in NIH 3T3 cells. Therefore, the viral stocks used to infect mice consisted of mixtures of viral genomes with varying numbers of tandem repeat units that retained the original core sequences.
FIG. 2.
LTR sequences amplified by PCR from proviral DNA isolated from infected NIH 3T3 cells. LTR sequences from cells infected with SL3, So-core-containing virus (So virus), or T*-core-containing virus (T* virus) are shown. LTR sequences from an SL3 plasmid are shown as a control for the size of a PCR product with two 72-bp repeats. Arrows on the right indicate the positions of fragments with one, two, or three 72-bp repeats. Marker is φX174 DNA digested with HaeIII. Numbers on the left indicate the sizes expressed in numbers of base pairs of two of the fragments in the marker lane.
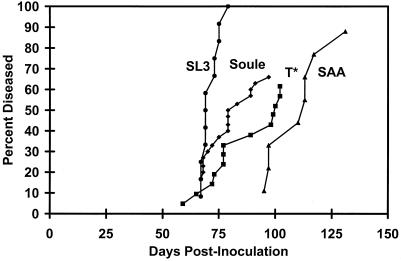
Lymphomagenicity of the viruses containing the So and T* cores was examined by injecting the viruses into newborn NIH/Swiss mice (Fig. 3). Parallel control studies were performed with SL3 and SAA viruses. SL3 virus caused tumors in 100% of infected mice, with a mean latency period of 69 days. SAA virus induced tumors in 88% of inoculated mice, with a mean latency of 112 days. It is unclear why only a fraction of the mice of this strain developed tumors after inoculation with the mutant. However, this effect was observed previously with other SL3 mutants in NIH/Swiss mice (29, 30). Of mice inoculated with the virus containing So core, 66% developed tumors, with a mean latency of 79 days. The decrease in the latency compared to that in mice inoculated with SAA was highly significant in a Student t test (P < 10−6). Of the mice injected with the virus containing T* core, 62% developed tumors. The mean interval until onset of disease in the infected mice was 98 days. This result was also significant (P < 0.001). The increased potency of the So- and T*-core-containing viruses strongly supported the argument that the single base pair substitutions relative to the Akv core in SAA were indeed second-site suppressor mutations.
FIG. 3.
Tumorigenicity of the So- and T*-core-containing viruses in NIH/Swiss mice. Parallel control experiments were performed with SL3 and SAA.
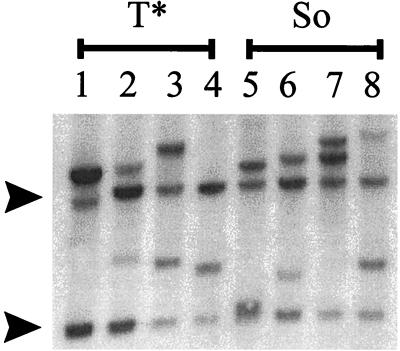
Previous studies showed that mutations within the core elements sometimes altered the type of tumor that the virus caused (34). Therefore, it was important to test whether the tumors induced by the So- and T*-core-containing viruses were T-cell lymphomas. Pathologically, the tumors induced by both these viruses were identical to those induced by SL3 and SAA, and the affected mice showed grossly enlarged thymuses, spleens, lymph nodes, and livers. Southern blotting confirmed that the tumors contained rearrangements in T-cell receptor β chains (Fig. 4). Thus, the tumors induced by the So- and T*-core-containing viruses were T-cell lymphomas.
FIG. 4.
Southern blot analysis of T-cell receptor β rearrangements in tumors induced by the So- and T*-core-containing viruses. Four different lymphomas induced by the T*- (lanes 1 to 4) or the So-core-containing (lanes 5 to 8) virus were analyzed. Arrows to the left of the blot indicate the positions of the germline fragments detected by the probe.
Enhancer sequences of proviruses in So and T* virus-induced tumors.
It was important to test whether the proviruses in the lymphomas induced by the So- and T*-core-containing viruses retained the suppressor mutations within the proviral core sequences. Genomic DNA was prepared from four separate tumors induced by each virus. Viral LTR sequences were PCR amplified and directly sequenced. The results are summarized in Table 1. All four lymphomas induced by the So virus contained proviruses with two and three LTR enhancer repeats. Sequencing analysis showed that all the cores in the amplified bands contained the So core (Table 1). LTRs with two repeats were amplified from all four lymphomas induced by the virus containing the T* core. In each case, the T* core sequence was present (Table 1). Three of the four tumors induced also contained proviruses with three LTRs. In one case, the structure was the same as the LTR of the virus in the original plasmid clone with T* cores in the promoter-proximal two repeats and an Akv/SAA core in the distal repeat (Table 1). In the other two cases, all three repeats had T* cores. In summary, the sequencing analysis showed that the viruses that were present in the tumors retained the original mutations. This observation provided additional strong evidence that the So and T* cores indeed contained suppressor mutations.
TABLE 1.
Sequences of core elements in proviral enhancers from tumor tissue
| Individual mouse no. | Sequence of core elementa
|
|
|---|---|---|
| 2-Repeat | 3-Repeat | |
| So infected | ||
| 2 | So, So | So, So, So |
| 3 | So, So | So, So, So |
| 5 | So, So | So, So, So |
| 16 | So, So | So, So, So |
| T* infected | ||
| 4 | T*, T* | Not present |
| 7 | T*, T* | T*, T*, T* |
| 8 | T*, T* | C, T*, T* |
| 9 | T*, T* | T*, T*, T* |
The sequence of the core element in each of the repeats is shown; the first abbreviation represents the core of the promoter-distal repeat, and the last represents the core in the promoter-proximal repeat. The sequence of So is TGCGGTCAA, that of T* is TGTGGTCTA, and that of C is the sequence found in the Akv/SAA core, TGTGGTCAA.
The enhancer core is one of many elements responsible for tumorigenicity in retrovirus-induced tumors. Mutations or deletions of other factor binding sites can affect disease (9, 29, 34). In the 16 sequences that were obtained (Table 1), a total of two mutations were detected within the enhancer repeats. Both were outside the core element.
Effects of the So and T* enhancer cores on transcriptional activity.
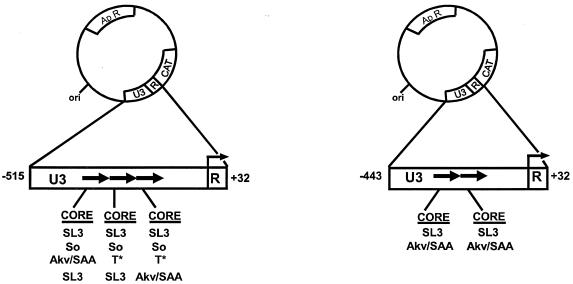
The lymphomagenicity study showed that So and T* enhancer cores rendered the viruses containing them more potent than the virus with the Akv/SAA core. The effects of MuLV LTR enhancers on transcription in the target cells for disease generally reflect the effects on tumorigenicity (32). We therefore expected that the transcriptional activities of the So- and T*-core-containing enhancers would exceed that of SAA but be less than that of SL3 in T cells. To test this, LTR CAT plasmids (Fig. 5) were constructed with the LTRs of SL3, Akv, SAA, and the SAA-derived viruses with the suppressor mutations. U3 and R region sequences were inserted upstream of the CAT reporter gene as previously described (3, 32). We chose to test our set of CAT plasmids in T-cell lines because the SL3 virus caused disease specifically in T lymphocytes. In previous studies in T cells, SL3-CAT exhibited the greatest transcriptional activity, SAA-CAT displayed intermediate activity, and Akv-CAT had relatively low levels (27, 29, 30). In non-T-cell lines, the activities of SL3-CAT, SAA-CAT, and Akv-CAT were approximately equal (27). The same So- and T*-containing LTRs as were used to generate infectious virus (Fig. 1) were used to generate the CAT plasmids (Fig. 5). The abbreviations SoSoSo and CT*T* were used to symbolize these LTRs, where the symbols So, T*, and C represent the So, T*, and Akv/SAA cores, respectively. Each symbol represents the sequence of the core in an individual enhancer repeat. The number of symbols indicates the number of 72-bp enhancer repeats in the LTR, with the first symbol representing the promoter-distal repeat and the last representing the promoter-proximal repeat. In the So-CAT LTR, each of the three repeats had a So core. In the T*-CAT LTR, two of the repeats had T* cores while the promoter-distal repeat retained the Akv core that was in the original mutant in the SAA-infected mouse. Because the So-CAT and T*-CAT constructs each had three 72-bp repeats, we generated for use as controls additional LTR-CAT plasmids that contained SL3 core sequences and three enhancer repeats. TTT-CAT contained three repeats, all with SL3 cores, and TTC-CAT contained three repeats, the two distal ones containing SL3 cores and the promoter-proximal one containing an Akv/SAA core. Viral genomes containing these LTRs were initially identified in proviruses in SAA-induced tumors (27) and were obtained by PCR amplification of the viral genomes from the tumor samples. These constructs allowed us to determine if a three-repeat structure had greater transcriptional activity than a two-repeat structure. Thus, we could distinguish between the effect of repeat number and the actual sequence of the core.
FIG. 5.
Structures of the plasmids used to measure transcriptional activities of the viral LTRs. Positions of the U3 and R sequences from the viral LTRs are shown. The CAT gene, ampicillin resistance gene (Apr), and plasmid origin of replication (ori) are also indicated. The arrow at the U3-R boundary indicates the direction of transcription. Numbers represent distance from transcription initiation site. Black arrows in the boxes below the circles indicate the numbers of 72-bp repeats in the viral LTRs that were tested. The enhancer core sequences within each LTR of the plasmids are depicted below the arrows.
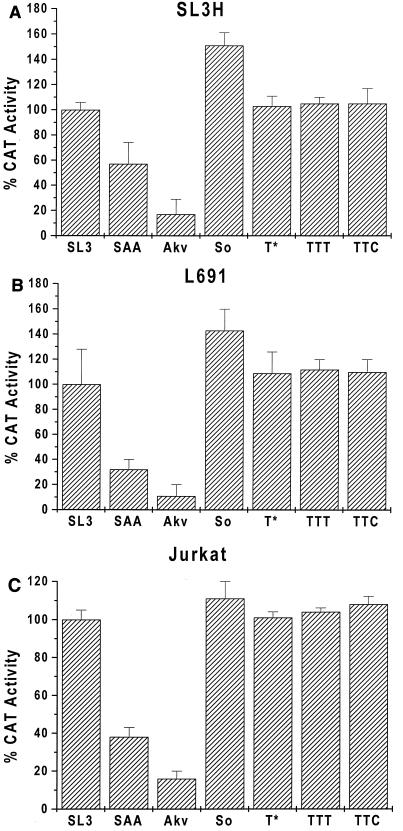
Wild-type SL3-CAT was used as a standard arbitrarily set at 100% activity. In the three T-cell lines tested, Akv-CAT had 10 to 20% as much activity as SL3-CAT (Fig. 6). SAA-CAT exhibited activity intermediate between those of SL3-CAT and Akv-CAT (Fig. 6). TTT-CAT and TTC-CAT had activities similar to that of SL3-CAT (Fig. 6). Therefore, the presence of a third SL3 core- or Akv core-containing repeat did not significantly alter the transcriptional activity of the LTR.
FIG. 6.
Transcription assays in (A) SL3H, (B) Lb91, and (C) Jurkat T-lymphocyte cell lines. Activities of the LTRs of the suppressor mutants are shown together with those of SL3, SAA, Akv, TTT, and TTC controls. The activity of the SL3 LTR in each cell line was set at 100%. Each error bar indicates one standard deviation.
Both So-CAT and T*-CAT had higher transcriptional activity than SAA-CAT. T*-CAT exhibited activity comparable to those of SL3-CAT and the other three-repeat-containing LTRs (Fig. 6). So-CAT also had activity similar to those of SL3-CAT, TTT-CAT, and TTC-CAT, although it was consistently slightly higher than the other activities (Fig. 6). We conclude that changes in the core sequence increase the activity of the viral LTR in T cells to a level similar to that of SL3. Presumably, the second-site suppressor mutations in the core element affect viral lymphomagenicity by increasing the transcriptional activity of the viral LTR in T cells.
Effects of second-site suppressor mutations on transcription factor binding.
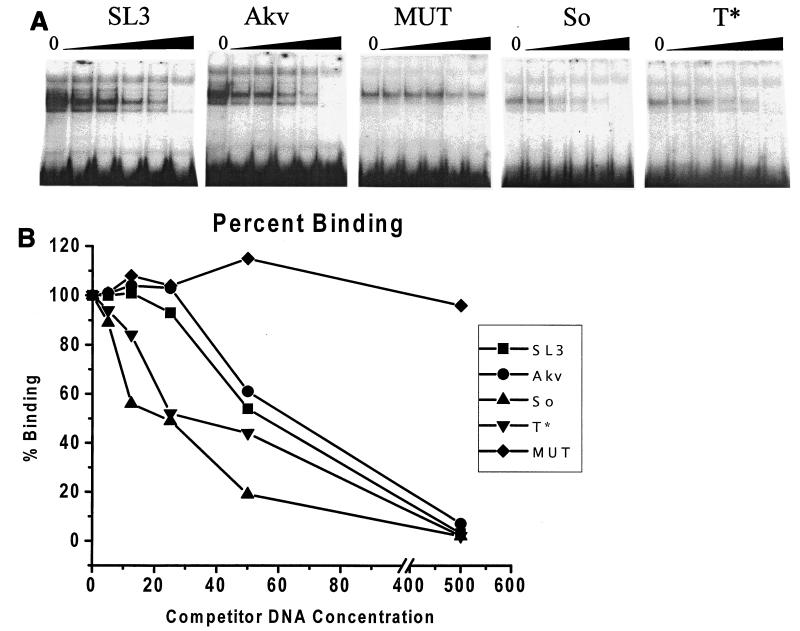
The enhancer core of SL3 binds the transcription factor CBF. Therefore, we tested whether the second-site suppressor mutations might increase transcriptional activity of the LTR in T cells by increasing CBF binding. CBF binding was assessed by using an EMSA. A nuclear extract from WEHI 7.1 T cells was used as the source of CBF protein. A radiolabeled probe containing the core element of SL3 was tested in the presence of increasing amounts of unlabeled competitor DNAs that were identical except for the mutations in the core element. The competitor DNAs contained the SL3, Akv, So, or T* cores. A mutated SL3 core (MUT) that does not bind CBF (43) was also tested as a negative control. Quantitation of the effects of the competitors showed that the SL3 core bound CBF slightly better than the Akv core (Fig. 7), consistent with previous observations (44). Quantitation of the EMSAs indicated that the So and T* cores bound CBF better than the Akv core and even slightly better than the SL3 core (Fig. 7). Therefore, increased binding of CBF to the So and T* cores correlated with the increased transcriptional activity in T cells and increased viral lymphomagenicity. When the oligonucleotides containing the So and T* cores were radiolabeled and used as probes in the EMSAs, no additional factors that bound to these sequences were detected (22). Thus, the So and T* enhancer cores appear to bind only CBF, implying that it is the critical transcription factor for the increased transcriptional and lymphomagenic effects of the suppressor mutations.
FIG. 7.
Binding of CBF to viral enhancer core sequences. (A) A radiolabeled oligonucleotide containing the core element from the SL3 LTR enhancer was used in EMSAs with crude nuclear extract from the WEHI 7.1 mouse T-cell line as a source of CBF. Five different, unlabeled, competitor DNAs were tested in increasing amounts of 0, 5, 12.5, 25, 50, and 500 ng per reaction as indicated. Competitor DNAs were isogenic with the probe except for the sequence of the core element. SL3, Akv, So, and T* indicate competitor DNAs from the corresponding viruses. MUT was a competitor DNA from SL3 with a 2-bp mutation that was previously shown to prevent CBF binding (43). (B) The amount of binding was quantified by PhosphorImager analysis and plotted. The amount of binding detected in each experiment with no competitor DNA was set at 100%.
DISCUSSION
The lymphomagenicity data provided strong evidence that the So- and T*-core-containing viral variants contained second-site suppressor mutations within their core elements. The suppressor mutations were originally selected during the lymphomagenic process in SAA-infected mice. They occurred within the same CBF binding sites as the original mutations and were detected in 20% of the lymphomas that developed in the SAA-infected mice. We previously showed that the SAA/Akv core mutation was reverted in viruses present in 70% of the lymphomas in SAA-inoculated mice (27). Thus, both reversions and suppressor mutations counteracted the detrimental effects on pathogenicity of the original mutation.
Ethelberg et al. (9) identified second-site suppressor mutations that occurred in a mutant of SL3 in which both the core I and core II elements were altered by substitutions at three positions. However, in those studies, suppression was caused by a deletion of a binding site for the transcription factor nuclear factor 1 (NF-1) within the LTR enhancer repeats. In our original analysis of tumor proviruses in 39 SAA-inoculated mice (27), in five different mice we detected proviruses that contained deletions of various sizes in the enhancer region. These deletions encompassed the NF-1 site in the promoter-distal, 72-bp repeat (26). Thus, it is likely that two distinct mechanisms, nucleotide substitutions within the core element and deletion of the NF-1 site, can lead to suppression of the single nucleotide mutation in SAA. However, Ethelberg et al. (9) did not detect any nucleotide substitutions within the core element from mice inoculated with the core I-core II mutant. We hypothesize that this was because the mutant that was tested in those studies had multiple nucleotide substitutions within the core. Presumably, these had such a large effect on CBF binding that the mutations generating So and T* core elements failed to restore sufficient CBF binding and viral lymphomagenic activity for viruses containing the suppressor mutations to be selected.
The original So- and T*-core-containing virus clones that we generated in this study contained three 72-bp repeats within their LTRs (Fig. 1). Upon brief passage in NIH 3T3 fibroblasts, viruses with two repeats were abundant (Fig. 2). When the SL3 clone with two repeats was passaged in the same cells, genomes with three repeats were detectable but not nearly as abundant as genomes with two repeats (27). These observations suggest that the viruses with two repeats had some replicative advantage over those with three. Perhaps the genomes with two repeats are packaged more efficiently.
The presence of genomes with two repeats in the viral stocks used to inoculate mice means that the mice received a mixture of viruses with a variety of enhancer structures. In the case of the So-core-containing virus, the stock included viruses with enhancer repeats with SoSoSo and SoSo structures, where So indicates individual 72-bp units with a So core sequence. Proviruses with both types of enhancers were present in the lymphomas induced by So virus (Table 1). In the case of the T*-core-containing virus, the mixture was more complex. The original clone had CT*T* enhancer structure. The two-repeat-containing viruses that formed by backward slippage during reverse transcription had the T*T* and CT* structures. These in turn could generate viruses with three repeats with the structures T*T*T*, CCT*, and the original CT*T* by forward slippage during reverse transcription in a subsequent replicative cycle. If multiple rounds of reverse and forward slippage occurred, then viruses with repeat structures CC and CCC should also have formed. Thus, mice inoculated with T*-core-containing virus probably received a mixture of isoforms differing in their LTR enhancer structures. However, only T*T*T*, T*T*, and CT*T* were detected in the lymphomas that occurred in these mice (Table 1). This observation emphasizes the strong selective pressure that the lymphomagenic process applies on viral enhancer structure.
The increased lymphomagenicity of the So-core-containing virus was correlated with increased CBF binding by the So core (TGCGGTCAA) compared to the SAA/Akv core (TGTGGTCAA) (Fig. 7). Thornell et al. (38) found that the mutation generating the So core increased CBF binding in the context of the SL3 enhancer (TGCGGTTAA versus TGTGGTTAA). Therefore, this mutation increased CBF binding in the context of either the SL3 or the SAA/Akv core. Using selected and amplified binding analysis, Melnikova et al. (23) also found CBF preferentially bound to DNA molecules containing a C at the position of the mutation generating So. However, a C occurs at this position in only 4 of 35 C-type mammalian retroviruses that were analyzed in an extensive comparison of viruses in this genus (12). The remaining 31 viruses have the T at this position (12), and several of these viruses were potent, T-cell lymphomagenic viruses. This suggests that the effect of a viral core element on pathogenicity depends on additional parameters than just the affinity of the binding site for CBF.
The mutation generating T* (TGTGGTCTA) also increased CBF binding compared to the SAA/Akv core (Fig. 7). However, Thornell et al. (38) found that this mutation in the context of the SL3 core (TGTGGTTTA versus TGTGGTTAA) had no effect on CBF binding. We interpret these observations to mean that the mutation generating T* affected CBF binding only when the preceding nucleotide in the core element was a C. Curiously, the selected and amplified binding analysis of Melnikova et al. (23) revealed that CBF bound to DNA molecules with a T preferentially to molecules with an A at the position where the substitution led to the T* core element, even though almost all C-type retroviruses have the A at this position in their cores (12). One possibility to explain this is that neighboring nucleotides might affect which nucleotides at this position function best for CBF binding. Alternatively, the absence of a T at this position in all 35 C-type viruses analyzed might again reflect the possibility that the pathogenic activity of a viral core element depends on additional parameters than just the affinity of the binding site for CBF.
The increased binding of CBF due to the mutations generating So and T* core elements was correlated with increased transcriptional activity of the viral LTRs in T cells relative to that of the SAA LTR (Fig. 6). The So- and T*-core-containing virus LTRs exhibited transcriptional activities in T cells comparable to that of SL3. The So-containing LTR appeared even slightly more active than the SL3 LTR. These results are consistent with the idea that relatively high levels of LTR transcriptional activity in T cells are necessary for T-cell lymphomagenicity of the virus.
Although the So and T* cores were comparable to the SL3 core in CBF binding and transcriptional activity, the viruses containing them were slightly less lymphomagenic than SL3. Both viruses differed from SL3 in that they induced disease in fewer than 100% of inoculated mice (Fig. 3). T*-core-containing virus induced lymphomas with a statistically significantly longer mean latency period than SL3 (P = 0.02). Although the mean latency periods to disease onset were not significantly different in the SL3 virus and So-core-containing virus (P = 0.16), the last mice infected with the latter virus to develop disease did so more slowly than any of the SL3-infected mice. One possible explanation for the lower lymphomagenicity of the viruses with So and T* cores is that the differences in CBF binding and transcriptional activity among the SL3, So, and T* cores were small and thus may have been affected by experimental variations. However, we were able to detect small differences between the CBF binding by the SL3 core and that by the SAA/Akv core that were consistent with those previously reported (44). Likewise, we also observed differences in transcriptional activity among LTRs containing the SL3, SAA, and Akv cores that were consistent with those previously observed (3, 27, 29, 30). Thus, it is conceivable that the So and T* cores indeed bind CBF and drive LTR transcriptional activity as effectively as the SL3 core. If so, then this raises the question why the viruses with So and T* core elements were less lymphomagenic than SL3. Perhaps a certain level of CBF binding and transcription in T cells is necessary for viral lymphomagenicity, but it is not sufficient for maximum viral lymphomagenicity. If so, then this would suggest that the effect of the core on viral pathogenicity is determined by CBF binding affinity plus some additional process. Another possibility is that cultured lymphoma cell lines that were used to test the transcriptional activity of the viral LTRs did not precisely reflect the normal T lymphocytes in the mice.
In summary, suppressor mutations within the core element restored part or most of the lymphomagenic activity of SAA. Multiple selective pressures may have led to the presence of viruses with the mutations producing So and T* core elements in tumors in SAA-infected mice. The suppressor mutations may have allowed the viruses to replicate better in T cells, leading them to outgrow the original SAA mutant. It is also possible that proviruses that had suppressor mutations were more effective at activating cellular protooncogenes and thus causing proliferation of the tumor cells.
ACKNOWLEDGMENTS
We thank Angel Nieves and Joseph Pantginis for help with these studies.
This work was supported by NIH grants CA44822 and CA57337 to J.L. and by American Cancer Society grant RPG-94-012-VM to L.S.L. M.J.M. was supported by NIH training grant GM07491. Core facilities for oligonucleotide synthesis, PhosphorImager analysis, and DNA sequencing were supported by NIH Cancer Center Grant CA13330 to the Albert Einstein College of Medicine.
REFERENCES
- 1.Athas G, Choi B, Prabhu S, Lobelle-Rich P, Levy L S. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology. 1995;214:431–438. doi: 10.1006/viro.1995.0053. [DOI] [PubMed] [Google Scholar]
- 2.Barat C, Rassart E. Members of the GATA family of transcription factors bind to the U3 region of Cas-Br-E and Graffi retroviruses and transactivate their expression. J Virol. 1998;72:5579–5588. doi: 10.1128/jvi.72.7.5579-5588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boral A L, Okenquist S A, Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989;63:76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brightman B K, Farmer C, Fan H. Escape from in vivo restriction of Moloney mink cell focus-inducing viruses driven by the Mo+PyF101 long terminal repeat (LTR) by LTR alterations. J Virol. 1993;67:7140–7148. doi: 10.1128/jvi.67.12.7140-7148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran L M, Adams J M, Dunn A R, Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984;37:113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 7.Corneliussen B, Thornell A, Hallberg B, Grundström T. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers. J Virol. 1991;65:6084–6093. doi: 10.1128/jvi.65.11.6084-6093.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson P, Gao J, Chang K-S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 9.Ethelberg S, Hallberg B, Lovmand J, Luz A, Grundström T, Pedersen F S. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997;71:1196–1206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ethelberg S, Lovmand J, Schmidt J, Luz A, Pedersen F S. Increased lymphomagenicity and restored disease specificity of AML1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer variant evolved in vivo. J Virol. 1997;71:7273–7280. doi: 10.1128/jvi.71.10.7273-7280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golemis E A, Speck N A, Hopkins N. Alignment of U3 sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunther C V, Graves B J. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994;14:7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallberg B, Schmidt J, Luz A, Pedersen F S, Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedrick S M, Cohen D I, Nielsen E A, Davis M M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 16.Holland C A, Thomas C Y, Chattopadhyay S K, Koehne C, O’Donnell P V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989;63:1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 18.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- 20.Lenz J, Celander D, Crowther R L, Patarca R, Perkins D W, Haseltine W A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- 21.Manley N R, O’Connell M, Sun W, Speck N A, Hopkins N. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J Virol. 1993;67:1967–1975. doi: 10.1128/jvi.67.4.1967-1975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martiney, M. J., and J. Lenz. Unpublished results.
- 23.Melnikova I A, Crute B E, Wang S, Speck N A. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura T, Shibuya M, Tsujimoto H, Fukusawa M, Hayami M. Molecular cloning of a feline leukemia provirus integrated adjacent to the c-myc gene in a feline T-cell leukemia cell line and the unique structure of its terminal repeat. Virology. 1989;169:458–461. doi: 10.1016/0042-6822(89)90172-4. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, H. L., and J. Lenz. 1998. Unpublished results.
- 27.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen A L, Norby P L, Pedersen F S, Jorgensen P. E-box sequence and context-dependent TAL1/SCL modulation of basic helix-loop-helix protein-mediated transcriptional activation. J Biol Chem. 1996;271:31463–31469. doi: 10.1074/jbc.271.49.31463. [DOI] [PubMed] [Google Scholar]
- 29.Nieves A, Levy L S, Lenz J. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J Virol. 1997;71:1213–1219. doi: 10.1128/jvi.71.2.1213-1219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantginis J, Beaty R M, Levy L S, Lenz J. The feline leukemia virus long terminal repeat contains a potent determinant of T-cell lymphomagenicity. J Virol. 1997;71:9786–9791. doi: 10.1128/jvi.71.12.9786-9791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;12:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 32.Short M K, Okenquist S A, Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987;61:1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speck N A, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speck N A, Renjifo B, Golemis E, Fredrickson T, Hartley J, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 35.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor β-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun W, O’Connell M, Speck N A. Characterization of a protein that binds multiple sequences in mammalian type C retrovirus enhancers. J Virol. 1993;67:1976–1986. doi: 10.1128/jvi.67.4.1976-1986.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornell A, Hallberg B, Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988;8:1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornell A, Hallberg B, Grundström T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991;65:42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trubetskoy, A. M., S. A. Okenquist, and J. Lenz. R region sequences in the long terminal repeat of a murine leukemia virus specifically increase expression of unspliced RNAs. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 40.Wang S, Speck N A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992;12:89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimura F K, Diem K. Characterization of nuclear protein binding to a site in the long terminal repeat of a murine leukemia virus: comparison with the NFAT complex. J Virol. 1995;69:994–1000. doi: 10.1128/jvi.69.2.994-1000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimura F K, Cankovic M, Smeltz R, Ibrahim S. Identification of nucleotide sequences that regulate transcription of the MCF13 murine leukemia virus long terminal repeat in activated T cells. J Virol. 1997;71:2572–2576. doi: 10.1128/jvi.71.3.2572-2576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaiman A L, Lenz J. Transcriptional activation of a retrovirus enhancer by CBF (AML1) requires a second factor: evidence for cooperativity with c-Myb. J Virol. 1996;70:5618–5629. doi: 10.1128/jvi.70.8.5618-5629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaiman A L, Lewis A F, Crute B E, Speck N A, Lenz J. Transcriptional activity of core binding factor α (AML1) and β subunits on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaiman A L, Nieves A, Lenz J. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J Virol. 1998;72:3129–3137. doi: 10.1128/jvi.72.4.3129-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]